Abstract
Resistance to neutralization is an important characteristic of primary isolates of human immunodeficiency virus type 1 (HIV-1) that relates to the potential for successful vaccination to prevent infection and use of immunotherapeutics for treatment of established infection. In order to further elucidate mechanisms responsible for neutralization resistance, we studied the molecular mechanisms that determine the resistance of the primary virus isolate of the strain HIV-1 MN to neutralization by soluble CD4 (sCD4). As is the case for the global neutralization resistance phenotype, sCD4 resistance depended upon sequences in the amino-terminal heptad repeat region of gp41 (HR1), as well as on multiple functional interactions within the envelope complex. The functional interactions that determined the resistance included interactions between the variable loop 1 and 2 (V1/V2) region and sequences in or near the CD4 binding site (CD4bs) and with the V3 loop. Additionally, the V3 loop region was found to interact functionally with sequences in the outer domain of gp120, distant from the CD4bs and coreceptor-binding site, as well as with a residue thought to be located centrally in the coreceptor-binding site. These and previous results provide the basis for a model by which functional signals that determine the neutralization resistance, high-infectivity phenotype depend upon interactions occurring across the surface of the gp120 core structure and involving variable loop structures and gp41. This model should be useful in efforts to define epitopes that may be important for primary virus neutralization.
Resistance to antibody-mediated neutralization of infectivity is characteristic of naturally occurring strains of human immunodeficiency virus type 1 (HIV-1) that have not been manipulated in the laboratory (15, 18). Such strains are commonly referred to as primary isolates. This property is a limiting factor with respect to efforts to develop a vaccine that may protect by induction of potent neutralizing antibodies. Induction of potent neutralizing antibodies is a common property of successful viral vaccines (26). Insights into the mechanisms of neutralization resistance of primary isolates of HIV-1 may indicate possible approaches to neutralization of such viruses and facilitate vaccine development.
Our laboratory has previously reported studies of the MN strain of HIV-1 that have begun to address this issue (14, 21-23). The original isolate of the MN virus was propagated in a continuous T-cell line and is considered T-cell line adapted (TCLA) (12, 30). It is unusual among both primary and TCLA strains of HIV-1 in its high sensitivity to neutralization by antibodies in sera of infected patients and monoclonal antibodies (MAbs) against multiple linear and conformation-sensitive epitopes on the envelope glycoproteins (4, 7, 40, 48). In an early study a neutralization-resistant variant of this strain was studied that was produced by growth of the MN strain virus in the presence of a highly neutralizing human serum (22). Through evaluation of env genes cloned from the parent strain (MN-TCLA) and neutralization-resistant virus (MN-E6), it was determined that the neutralization resistance was due to mutations that altered functional interactions between gp120 and gp41 (21-23). Moreover, the responsible mutations included amino acid substitutions in the CD4 and putative coreceptor binding domains of gp120 and in the first heptad repeat (HR1) of gp41. Based on the requirement for sequences in gp41 for phenotypic effects of mutations in gp120 and the additional observation that neutralization resistance was correlated with high infectivity, it was considered likely that the mechanism responsible for neutralization resistance depended upon enhanced efficiency of envelope mediation of virus-cell membrane interactions.
More recently, studies have been conducted involving env clones derived from primary MN virus (MN-P) (14, 45). The MN-P clone is substantially more neutralization resistant and infectious than MN-E6. Specifically, MN-E6 differs from MN-TCLA in each phenotypic characteristic by approximately 10-fold, while MN-P differs from MN-TCLA by 250- to 1,000-fold in the two characteristics. Both of these characteristics of MN-P again depend upon sequences in HR1 but also require specific sequences from other regions of env. Functional interactions were demonstrated between HR1 and the amino terminus of gp120, the carboxy terminus of gp120, and the carboxy terminus of gp41, each of which contributed to the neutralization-resistant, high-infectivity phenotype (14). Among numerous clones constructed for studying the genetic bases for these phenotypic characteristics, there is a very strong correlation between neutralization resistance and infectivity. The neutralization resistance is global, in the sense that it pertains to neutralization by MAbs directed against conformation-sensitive epitopes in the CD4 binding site (CD4bs), the putative coreceptor-binding site, and variable region 3 (V3). These comparisons of MN-P and MN-TCLA confirmed and extended the previous evidence that the mechanism of neutralization resistance involves functional interactions throughout multiple regions of the envelope glycoprotein complex, with HR1 playing a central role, and that it results from enhanced efficiency of function of the envelope in mediating interaction between the virus and cell membrane.
The manner in which mutations causing neutralization resistance affect interaction of gp120 with CD4 is of interest. Primary isolates are commonly resistant to neutralization by MAbs directed against the CD4bs, with the exception of high concentrations of MAb b12 (8, 17, 20, 35, 36, 38, 39, 49, 50). It has recently been shown that a portion of the antigen-combining site of b12 projects deeply into the pocket of the CD4bs (34). Thus, epitopes within the pocket may be accessible to b12, but not as readily to other MAbs. Steric constraints on MAb binding to the CD4bs pocket may not apply to soluble CD4 (sCD4), since the latter is monomeric and less bulky. At one point, it had been hoped that sCD4 might be useful as a competitive inhibitor of HIV-1 infection in vivo. Unfortunately, that did not turn out to be the case. Many primary isolates of HIV-1 were found to be resistant to neutralization by sCD4, and exposure of the viruses to sCD4 in vitro often resulted in enhanced infectivity (9, 18). With a rare exception, HIV-1 requires CD4 binding in order to infect target cells (45). Therefore, the association of resistance to neutralization by sCD4 with enhanced infectivity would seem paradoxical, if the mechanism of resistance to sCD4 neutralization involved decreased ability of the HIV-1 envelope to interact with CD4. An alternative mechanism is suggested by our previous studies. The MN-P clone is approximately 50-fold more resistant to neutralization by sCD4 than is MN-TCLA (14, 45). The MN-P envelope was found to undergo a substantial gp120-gp41 dissociation response to sCD4 binding, whereas no such response was observed with MN-TCLA. These findings raise the possibility that capacity to undergo conformational change in response to CD4 binding may be a basis for primary virus resistance to neutralization by sCD4.
A large number of distinguishing mutations makes analysis of the genetic bases for differences in phenotypic characteristics between the MN-P and MN-TCLA clones somewhat complex (14, 45). The clones differ from each other at approximately 8% of amino acid residues. However, many of the mutations can be considered in groups, based on their spatial relationships to each other, as surmised by consideration of the published atomic structures of parts of gp120 and gp41. Such a grouping was useful in identification of a number of residues clustered in the inner domain that form a putative gp41-binding site on gp120 (14). The present report describes the use of a similar approach to evaluation of the genetic bases for the resistance of MN-P to neutralization by sCD4. As was the case for the global neutralization resistance phenotype, in general, resistance to sCD4 neutralization was found to depend on interactions between sequences in the amino- and carboxy-terminal halves of gp120 with the gp41 HR1 domain, although HR1 sequences did not in themselves cause the phenotype. Surprisingly, seven mutations in gp120 located in or near the CD4 binding pocket contributed a relatively small effect to resistance to sCD4 neutralization. The contribution of mutations in the amino-terminal half of gp120 to sCD4 resistance appeared to be due to interactions between V1/V2 and mutations near the CD4bs, while the contribution of mutations in the carboxy-terminal half appeared to be due to interactions of V3 with residues in the gp120 outer domain distant from the CD4bs or coreceptor-binding site. The resistance was also modulated by interaction between V1/V2 and V3. Mutations that determined interaction between the proximal limb of V3 and the gp120 core outer domain also determined a global neutralization resistance phenotype. The mechanism of resistance to neutralization by sCD4 appears to be based on similar interactions between regions of the envelope glycoprotein complex that determine global neutralization resistance in general. The interactions demonstrated in these studies between different domains of the envelope suggest the possibility that the neutralization resistance, high-infectivity phenotype may depend upon interactions occurring across the surface of the envelope glycoprotein complex. Such a mechanism may explain, at least in part, how functional interactions within the envelope complex may occur at a distance. Moreover, the studies reveal how targeted mutagenesis may be used to modify the immunological reactivity of conserved neutralization epitopes that may be important for primary virus neutralization and may have implications for elucidation of the mechanisms of HIV entry into cells.
MATERIALS AND METHODS
MAbs.
The human MAbs specific for the CD4bs, CD4-induced epitopes, and the V3 loop (15e, 48d, and 19b, respectively) were gifts from James Robinson (2, 5, 19, 37, 44). The following antibodies were obtained through the National Institutes of Health AIDS Research and Reference Reagent Program: anti-CD4bs human MAbs b12 (provided by D. Burton and C. Barbas [3, 6, 32]) and F105 (provided by M. Posner [24, 25]).
Plasmid constructs and chimeric env plasmid construction.
Envelope genes from the primary MN strain (MN-P) and the TCLA MN strain (MN-TCLA) have been described previously (12, 14, 22, 45). Additional mutant envelope genes were constructed to contain specific segments or sequences of each of the parental genes. The construction of chimera C has been described previously (14). Chimera P was constructed by replacing the V1/V2 region sequence of chimera C with the corresponding sequence from MN-P. To prepare for construction of chimera P, an EcoRV site was introduced by mutagenesis into the MN-P sequence at nucleotide 636. The DraIII-EcoRV sequence from the modified MN-P sequence was transferred into the corresponding site of chimera C. Chimera P/7CD4bs (defined below) was constructed by transferring the EcoRV-SacI fragment from chimera C/7CD4bs (defined below) into the corresponding site of chimera P.
Site-directed mutagenesis.
Mutagenesis procedures were carried out by using primers designed with single or double nucleotide changes and Pfu polymerase (Quick Change mutagenesis kit; Stratagene) according to the instructions of the manufacturer, as described previously (11). The reactions were performed in an automated thermal cycler (Perkin-Elmer model 2400; Perkin-Elmer Corp., Norwalk, Conn.). Nucleotide sequences of each mutation introduced were confirmed by sequencing by the ABI PRISM dye terminator method (Perkin-Elmer).
Pseudovirus construction and assays for infectivity and neutralization.
Pseudoviruses expressing envelope glycoproteins derived from various env plasmids were constructed by using pSV7d-env and pNL4-3.Luc.E-R- plasmids, as described previously (14, 21-23, 27, 28, 45, 46). Briefly, the env plasmid and pNL4-3.Luc.E-R- DNAs were cotransfected into 30% confluent 293T cell cultures by the calcium phosphate method (Promega, Madison, Wis.). The culture medium was replaced with fresh medium containing 1 μM sodium butyrate at 18 to 24 h posttransfection. At 44 to 48 h after transfection, the pseudovirus-containing supernatants were harvested, filtered through a 45-μm-pore-size sterile filter (Millipore Corp., Bedford, Mass.), supplemented with additional fetal bovine serum to a final concentration of 20%, and stored at −80°C if not used immediately. To measure the infectivity of pseudoviruses, a luminescence assay with HOS-CD4-CXCR4 cells was used as previously described. HOS-CD4-CXCR4 cells (104 cells/ml) were inoculated with a serially diluted pseudovirus in 96-well plates with flat-bottomed wells. The cultures were incubated for 3 days at 37°C with 5% CO2, after which the cells were washed with 150 μl of phosphate-buffered saline (pH 7.4) and lysed with 15 μl of cell lysis buffer (Promega) for 30 min. The amount of luciferase activity in each well was determined with 50 μl of substrate (Promega) in an EG & G Berthold MicroLumat Plus luminometer (Wallac, Inc., Gaithersburg, Md.).
The neutralization phenotype of each pseudovirus was tested in a manner similar to that of the infectivity assay, except that aliquots of serially diluted serum or antibody were mixed with appropriately diluted pseudovirus and incubated for 1 h at 4°C, after which HOS-CD4-CXCR4 cell suspensions were added and incubated for 3 days at 37°C with 5% CO2.
Locations of the gp120 core mutations in the gp120 atomic structure.
The portable document format file used for the gp120 atomic structure is from the Protein Data Bank, 1GC1 (13), and was drawn with PC Molecule 2 (version 2.0.0; Molecular Ventures, Inc.). Figures representing the locations of the mutations were drawn with PC Molecule 2 (version 2.0.0) and with Corel Draw (version 8.369).
RESULTS
Regions of the MN-P gene contributing to resistance to neutralization by sCD4.
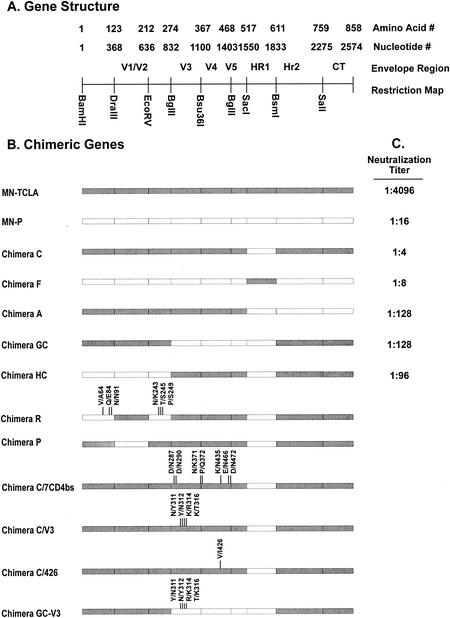
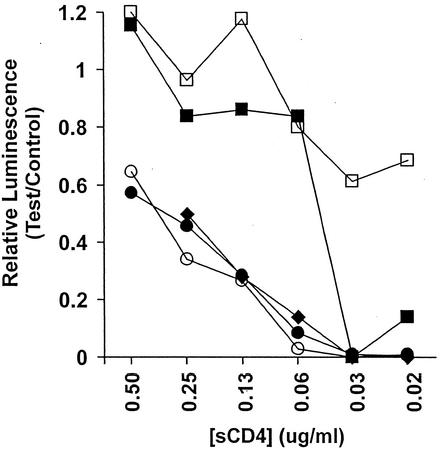
Studies previously reported from this laboratory demonstrated that the neutralization resistance, high-infectivity phenotype of the MN-P clone is dependent upon functional interactions between sequences from the region of the MN-P gene encoding the gp41 HR1 with sequences in other regions of gp120 and gp41. Figure 1 illustrates the structure of chimeric genes C, F, A, GC, and HC, which we used in that previous study, as well as the neutralization titers reported therein with an HIV-1 immune human serum and viruses pseudotyped with the respective envelopes. Figure 1 also illustrates the structures of additional chimeric genes which were used in the present study to analyze intramolecular interactions that determine resistance to neutralization by sCD4. Chimera C has MN-P HR1 region sequences in the MN-TCLA backbone. Chimera F is the reciprocal construct, with the MN-TCLA HR1 region in the MN-P backbone. The amino terminus of gp120, the carboxy terminus of gp120, and the carboxy terminus of gp41 from MN-P were each introduced into chimera C to form chimeras HC, GC, and A, respectively. These three chimeras were used so that we could evaluate whether functional interactions between different regions of gp120 or gp41 and the HR1 region contributed to resistance to neutralization by sCD4. The reasons for construction of chimeras P and R, shown at the bottom of Fig. 1, will be discussed below. As shown in Fig. 2, chimera GC was significantly more resistant to neutralization by sCD4 than was chimera C but less resistant than was MN-P. Chimeras A and HC were similar to chimera C. These results suggested that functional interactions between the carboxy-terminal half of gp120 and HR1 contributed to resistance to neutralization by sCD4 but that interactions not reflected in the chimeric genes included in these comparisons also contributed to the resistance.
FIG. 1.
Schematic representations of recombinant env genes used for analysis of the genetic basis for resistance to sCD4 neutralization. (A) Gene organization. (B) Structure of chimeric genes used in the study. Segments of each clone derived from MN-TCLA are shown as shaded bars, and segments derived from MN-P are shown as white bars. The nature of mutations introduced by site-directed mutagenesis is indicated, with locations noted by vertical lines. Construction of the clones and mutagenesis procedures are described in Materials and Methods.
FIG. 2.
Neutralization by sCD4 of viruses pseudotyped with the envelopes of MN-P (□) or chimera A (○), GC (▪), HC (⧫), or C (•). Neutralization sensitivities of MN-TCLA (data not shown) and chimera C were similar. Assays were performed in triplicate, and results shown are means obtained at each sCD4 concentration. Control luminescence was determined based on infections performed in the absence of sCD4. Essentially identical results were obtained in two similar experiments.
Amino acid substitutions in the segment of gp120 contributed by MN-P to chimera GC.
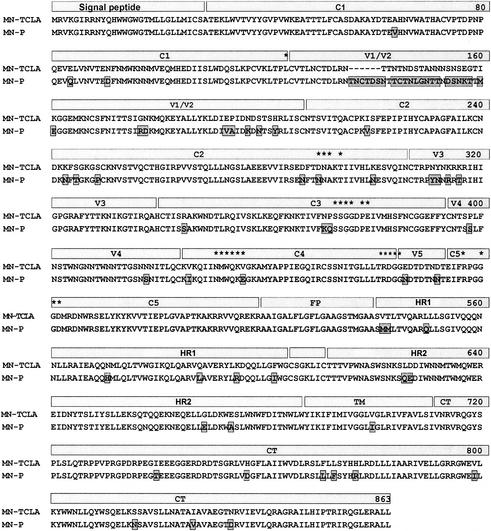
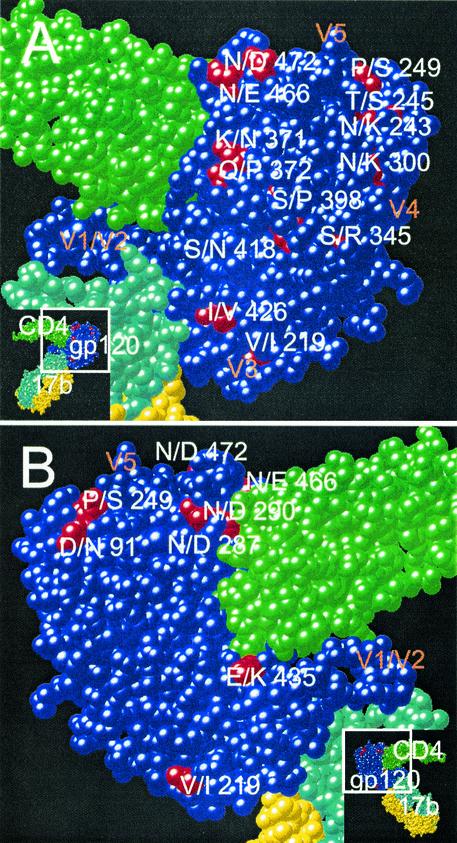
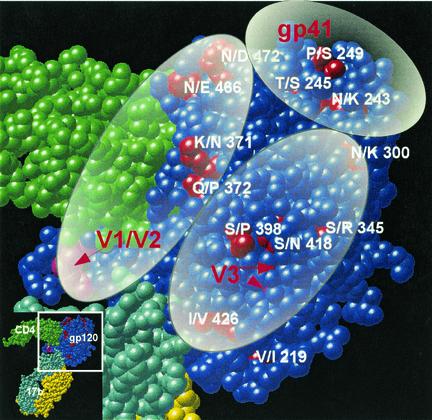
The distribution of mutations distinguishing MN-P from MN-TCLA throughout the linear sequences of the genes and on the core structure of gp120, as described by Kwong et al. (13), is illustrated in Fig. 3 and 4. There were four distinguishing amino acid substitutions in V3 and 12 additional substitutions in the segment of MN-P gp120 included in chimera GC (amino acids 284 to 474 of MN-P). Seven of the 12 non-V3 substitutions were seen on the model of the atomic structure (Fig. 4) to be in or near the CD4 binding pocket of gp120. Two of these, N/D290 and E/K435, involved amino acids that aligned with residues shown by Kwong et al. to form direct contacts with CD4 (13). Two other mutations, KQ/NP371-2 and N/E466, affected residues immediately adjacent to residues that form direct contacts with CD4. In addition, six of the seven substitutions involved charge alterations (D/N287, N/D290, K/N371, E/K435, N/E466, and N/D472), and three involved gain or loss of potential N-linked glycosylation sites (D/N288, K/N371, and N/E466). Of the remaining five gp120 core substitutions in the segment from amino acids 284 to 474, but not within or in high proximity to the CD4bs, one is believed to be located in the center of the coreceptor-binding site (I/V426) (31). This mutation was seen previously in the MN-E6 clone and did not, by itself, confer resistance to neutralization by sCD4 in that context. The remaining four mutations in this segment are located on the surface of the gp120 outer domain, distant from the CD4 and coreceptor-binding sites. Based on these considerations we hypothesized that mutations in or near the CD4 binding pocket altered sensitivity to neutralization by sCD4 through substitutions at residues that directly bond CD4 or at adjacent residues that modify their interactions with CD4, such as through charge alterations, or by alteration of glycosylation sites that may have steric effects on CD4 binding.
FIG. 3.
Alignment of deduced amino acid sequences of MN-TCLA and MN-P envelopes. Regions of the envelope proteins that are indicated schematically above the sequences include conserved regions 1, 2, 3, 4, and 5 (C1, C2, C3, C4, and C5), variable regions 1/2, 3, 4, and 5 (V1/V2, V3, V4, and V5), fusion peptide (FP), heptad repeats 1 (HR1) and 2 (HR2), transmembrane segment (TM), and cytoplasmic tail (CT). Residues identified by Kwong et al. as forming direct bonds with CD4 are marked by asterisks above the sequences (13). Alignment was produced with DNAStar.
FIG. 4.
Localization of mutations distinguishing MN-P and MN-TCLA in the atomic structure of the gp120 core. The space-filling model is based on the report of Kwong et al. (13). CD4 residues are shown in green, and 17b MAb residues are shown in yellow and aqua. Most of the gp120 residues are shown in blue, except for the residues at which mutations distinguish MN-P and MN-TCLA, which are shown in red. The approximate locations of the variable loops are shown as V1 to V5. Insets indicate the portions of the trimolecular complexes that are shown in the larger views. (A) “Front” view; (B) “back” view. Mutated residues considered to be in or close to the CD4bs were 287, 290, 371, 372, 435, 466, and 472. Inner domain residues implicated in the gp41-binding site are 91, 243, 245, and 249. Mutated residues in or near the coreceptor-binding site are 219 and 426. Mutated outer domain residues distant from binding sites are 300, 345, 398, and 418.
Effects of mutations in and near the CD4bs on resistance to neutralization by sCD4.
To test our hypothesis regarding the significance of mutations in or near the CD4bs, we pursued a strategy using chimera C as a platform for testing the effect of other mutations on resistance to neutralization by sCD4. The comparative sensitivities to neutralization by sCD4 of MN-TCLA, MN-P, and chimera C are shown near the top of Fig. 5. We have previously described the MN-P clone as being more than 50-fold more resistant to sCD4 neutralization than the MN-TCLA clone (14, 45). In most of the assays represented in Fig. 5, the maximum concentration of sCD4 used was 1.0 μg/ml, so that neutralization of MN-P was not achieved. Based on the assays reported here we calculated that the MN-P clone was at least 20-fold more resistant than MN-TCLA. Chimera C was not significantly more resistant to neutralization by sCD4 than was MN-TCLA. The seven mutations located in and near the CD4bs that distinguished MN-P from MN-TCLA were introduced into chimera C, singly and in various combinations, as shown in Fig. 5. Only small differences between chimera C and any of these mutants were noted, including the clone of chimera C containing all seven of the mutations (this clone is referred to subsequently as chimera C/7CD4bs). These results indicated that the resistance of MN-P to neutralization by sCD4 was not due primarily to changes in the CD4bs, negating our hypothesis.
FIG. 5.
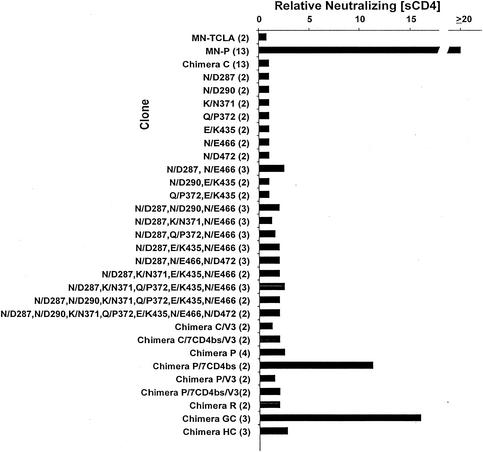
Mutations in and around the CD4bs contribute to sCD4 resistance in the context of MN-P V1/V2 sequences. The relative neutralizing [sCD4] was determined for each clone as follows. The 50% inhibitory concentration of sCD4 was determined by linear regression analysis in Microsoft Excel, as described in Materials and Methods. The relative neutralizing [CD4] was then calculated as test clone 50% inhibitory concentration/chimera C 50% inhibitory concentration. The 50% inhibitory concentrations for MN-P and for chimera C were determined in each experiment. The number of assays for each clone is indicated in parentheses after the clone designation. The structures of chimeras C, P, GC, and HC are illustrated in Fig. 1. Individual clones were constructed that contained one or combinations of the seven CD4bs mutations. Chimeras C/7CD4bs/V3, P/7CD4bs, and P/7CD4bs/V3 each contain the seven CD4bs mutations from MN-P (shown in Fig. 3 and 4) in the respective chimeras. Chimeras C/V3, C/7CD4bs/V3, P/V3, and P/7CD4bs/V3 each contain the four MN-P V3 region mutations, shown in Fig. 3.
Effects of non-CD4bs mutations on resistance to neutralization by sCD4.
We next considered the possible importance of mutations represented in chimera R with respect to resistance to neutralization by sCD4. Chimera R was of interest because of previous studies that demonstrated that mutations in the amino terminus of gp120 determine a functional interaction with HR1 sequences that confers an sCD4 response phenotype (14). MN-TCLA has a high spontaneous gp120-gp41 dissociation phenotype but does not display enhanced dissociation consequent to sCD4 binding. In contrast, MN-P has a low rate of spontaneous gp120-gp41 dissociation, which is increased substantially by sCD4 binding. Chimera R has a gp120-gp41 dissociation phenotype like MN-P. The mutations that distinguish chimera R from chimera C include the four mutations that can be seen to cluster in the inner domain in Fig. 3 (D/N91, N/K243, T/S245, and P/S249), as well as two that are in the extreme amino terminus of gp120 and not visualized in the crystallographically determined gp120 core structure (V/A64 and Q/E84). We have previously suggested, based on their effects on CD4-induced dissociation of gp120 from gp41, that these six residues contribute to a potential gp41-binding site on gp120 (14). Remarkably, even though these mutations determine an sCD4 response phenotype, they did not significantly affect sensitivity to neutralization by sCD4 (Fig. 1 and 5).
We next studied whether sequences in the V1/V2 or V3 regions contributed to sCD4 resistance. Previous reports have demonstrated that variable regions 1 and 2 may partially mask access to the CD4bs (31, 44). The MN-P clone has an unusual duplication in V1, as well as a number of other mutations that distinguish it from the V1/V2 region of MN-TCLA (Fig. 3). To permit testing of the role of the V1/V2 region in sCD4 resistance, we constructed chimera P, which consisted of MN-TCLA sequences throughout most of the gene, except for the V1/V2 and HR1 regions that were derived from MN-P, as illustrated in Fig. 1. Additional genes were constructed using the chimera P backbone, by introduction of the segment containing the seven mutations in or near the CD4bs (chimera P/7CD4bs), four mutations (YN/NY311/12, R/K313, and T/K315 [Fig. 3]) that distinguish the proximal limb of the MN-P V3 region from that of MN-TCLA (chimera P/V3), or both of these sets of mutations (chimera P/7CD4bs/V3). Chimera P was only 1.9-fold more resistant to neutralization by sCD4 than was chimera C (Fig. 5), whereas chimera P/7CD4bs was 11.3-fold more resistant. The mean 50% neutralizing concentrations of sCD4 for chimera C, chimera P, and chimera P/7CD4bs were 16, 31, and 176 ng/ml, respectively. Unexpectedly, addition of V3 region mutations to chimera P/7CD4bs, forming chimera P/7CD4bs/V3, eliminated the neutralization resistance. These results suggested a functional relationship between residues near the CD4bs and the V1/V2 region that contributed to the sCD4 resistance of MN-P, as well as a further functional relationship between these regions and V3.
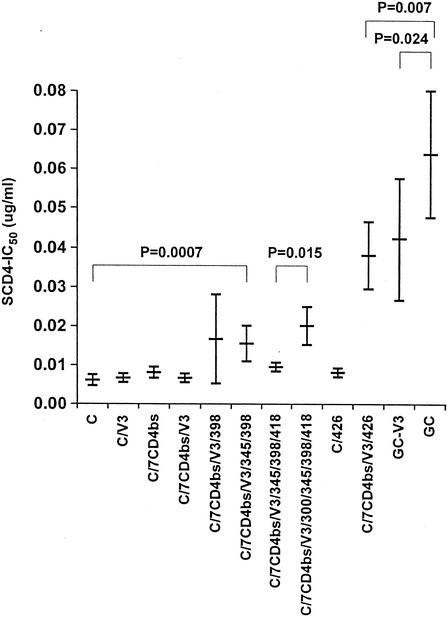
We next considered the structure of chimera GC (Fig. 1), which is relatively resistant to neutralization by sCD4. Chimera GC incorporates carboxy-terminal sequences from MN-P gp120 into chimera C. The gp120 sequences from MN-P in chimera GC include the seven CD4bs mutations discussed above, the V3 region mutations, one mutation thought to be located in the center of the coreceptor-binding site (I/V426), and four mutations localized in a loose cluster in the outer domain of gp120 (N/K300, S/R345, S/P398, and S/N418) (Fig. 4). As shown in Fig. 5, there were only minor differences noted in sCD4 resistance among chimeras C, C/V3, C/7CD4bs/V3, P, P/V3, and P/7CD4bs/V3. We considered it possible, therefore, that interactions between residues in V3 and other residues that are mutated in chimera GC contributed to neutralization resistance. The S/P398, S/R345, S/N418, and N/K300 outer domain mutations were introduced sequentially and studied for effect, as shown in Fig. 6. Among these, both the S/P398 and N/K300 mutations appeared to contribute small but statistically significant effects. The coreceptor-binding site mutation I/V426 had no effect on sCD4 neutralization when introduced into chimera C but had a significant effect when introduced into chimera C/7CD4bs/V3. This mutant, chimera C/7CD4bs/V3/426, was significantly less resistant than chimera GC, indicating that one or more of the four outer domain mutations were required for expression of the full phenotype of chimera GC. Finally, the chimera GC V3 sequences were back-mutated to the MN-TCLA V3 sequence, forming chimera GC-V3. This clone was also less resistant to neutralization than was chimera GC, indicating that V3 sequences were required for the full neutralization resistance phenotype. These results presented in Fig. 6 demonstrate, therefore, the occurrence of functional interactions between the V3 region and mutated residues in the outer domain, as well as with residue 426 in the coreceptor-binding site.
FIG. 6.
Resistance of chimera GC to neutralization by sCD4 depends upon sequences in V3, the mutation at residue 426 in the coreceptor-binding site, and mutations in the outer domain distant from the CD4 and coreceptor-binding sites. The construction of chimeras C, C/V3, C/7CD4bs/V3, and GC is described in Materials and Methods and in the Fig. 5 legend. Mutations at residues 298, 345, 418, and 300, in the outer domain distant from the CD4 and coreceptor-binding sites, were introduced sequentially into chimera C/7CD4bs/V3. The V/I mutation at residue 426 was introduced into both chimera C and chimera C/7CD4bs/V3. Four mutations corresponding to the MN-TCLA V3 region sequence were introduced into chimera GC to form chimera GC-V3. Results shown are means of three assays, each done in triplicate. Statistical comparisons were done using Microsoft Excel by analysis of variance (C versus C/V3 versus C/7CD4bs/V3 versus C/7CD4bs/V3/398 versus C/7CD4bs/V3/345/398) or Student t test (C/7CD4bs/V3/345/398/418 versus C/7CD4bs/V3/300/345/398/418, C/7CD4bs/V3/426 versus GC, and BC-V3 versus GC). Results shown are the means and standard deviations of three experiments, each done in triplicate. IC50, 50% inhibitory concentration.
Comparative sensitivity of clones to neutralization by CD4bs ligands and MAbs to V3 and the coreceptor-binding site.
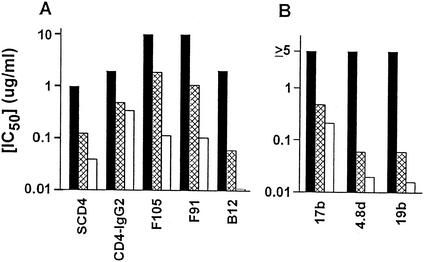
It was of interest to determine if the relative sensitivity of clones to neutralization by various CD4bs ligands was consistent. CD4-immunoglobulin G2 (CD4-IgG2) tends to be more cross-reactive than sCD4 in neutralization of primary envelopes but also has greater bulk, which could limit its access to the CD4bs (47). Among various anti-CD4bs MAbs, b12 tends to be more cross-reactive among primary envelopes. Neutralization of viruses pseudotyped with the MN-P, chimera P/7CD4bs, and chimera P/7CD4bs/V3 clones by various ligands is shown in Fig. 7A. The comparative neutralization of the clones by sCD4 was generally similar to that observed with the MAbs to CD4bs epitopes, including b12. MN-P was more resistant to neutralization by each of the ligands than was either of the other two clones. The greater sensitivity to neutralization of chimera P/7CD4bs/V3 than of chimera P/7CD4bs that was observed with each of the other ligands was not observed with CD4-IgG2. These results indicated that the phenomena that determined resistance to neutralization by sCD4 usually acted similarly in determining resistance to neutralization by various CD4bs ligands and that the mechanism of resistance to neutralization may not have been steric inhibition of ligand binding.
FIG. 7.
Functional interactions between V1/V2 and residues near the CD4bs and V3 affect global neutralization resistance. The clones MN-P (black bars), chimera P/7CD4bs (crosshatched bars), and chimera P/7CD4bs/V3 (white bars) were tested for neutralization. Results shown are averages of two assays, each done in triplicate. (A) Neutralization by the anti-CD4bs ligands sCD4, CD4-IgG2, F105, F91, and b12. (B) Neutralization by antibodies directed against the coreceptor-binding site, 17b and 4.8d, and the V3 region, 19b. In this panel, the maximum concentration of each antibody used was 2.5 μg/ml. There was no neutralization of MN-P at any concentration, so the inhibitory concentration was considered to be ≥5 μg/ml. IC50, 50% inhibitory concentration.
Neutralization by MAbs to the coreceptor-binding site (17b and 4.8d) and V3 (19b) is shown in Fig. 7B. Fifty percent neutralization of the MN-P pseudotyped virus was not achieved at the highest concentration tested of these antibodies, 2.5 μg/ml. Chimera P/7CD4bs/V3 was more sensitive than chimera P/7CD4bs to neutralization by 17b (210 versus 480 ng/ml), 4.8d (20 versus 60 ng/ml), and 19b (16 versus 60 ng/ml). These results indicated that the relative sensitivities of these clones to neutralization by sCD4 and MAbs to non-CD4bs epitopes were similar. Moreover, the functional interactions between V3 and other regions of gp120 that accounted for the comparative sensitivity of chimera P/7CD4bs and chimera P/7CD4bs/V3 also modulated global neutralization sensitivity.
DISCUSSION
We have used genetic analyses to probe the mechanisms that determine the global neutralization resistance phenotype of the HIV-1 MN strain primary virus (MN-P). Previous studies from our laboratory have demonstrated that the resistance of HIV-1 strains to neutralization by human sera can be attributed to multiple functional interactions between various regions of gp120 and gp41 (1, 10, 14, 21-23, 29, 33, 41-43, 45). The studies presented here were initially intended to elucidate the mechanism of resistance of MN-P to neutralization by sCD4. The results that we obtained demonstrated that the resistance of MN-P to neutralization by sCD4 was also determined by multiple functional interactions between regions of gp120 and gp41. Moreover, multiple interactions that determined resistance to neutralization by sCD4 also determined resistance to neutralization by MAbs directed against neutralization epitopes on gp120, demonstrating that the mechanisms responsible for resistance to neutralization by sCD4 pertained more generally to the global neutralization resistance phenotype. As we will discuss below, analyses of the various functional interactions that we identified suggest a model for interpreting how genetic alterations contribute to the neutralization resistance phenotype.
The strategy that we used for analysis of the mechanism of resistance to neutralization by sCD4 was based on consideration of the localization of residues in the gp120 core structure that are polymorphic in the MN-TCLA and MN-P strains (Fig. 4), as well as other evidence of functional interactions within the HIV-1 envelope protein complex reported previously by our laboratory and others. The existence of important interactions between gp120 and the HR1 region of gp41 has been well documented in our previous studies. The relevance of this interaction to the mechanism of sCD4 resistance was established by the results shown in Fig. 2, which demonstrated that a substantial part, but not all, of the resistance found in MN-P could be attributed to functional interactions between the carboxyl terminus of gp120 and the HR1 sequence, reflected in chimera GC. When we examined the localization of mutations that distinguished the carboxyl termini of MN-P and MN-TCLA, as shown in Fig. 4, we recognized that seven of the relevant mutations were within or very close to the CD4bs on gp120, one was within the presumed coreceptor-binding site, four mutations were in the V3 region, and four were on the gp120 outer domain at sites distant from the two binding sites. In the amino termini, there were two mutations in gp120 that do not appear in the core structure determined by X-ray diffraction, four mutations in the inner domain that we have previously suggested may lie in the gp41-binding site, and multiple mutations in the V1/V2 segment that distinguish the two clones. The relevance of each of these sets of mutations was pursued in tests of a series of hypotheses. Because our previous studies had established the importance of HR1 sequences in determining neutralization resistance, and because the resistance of chimera GC depended on HR1 sequences (Fig. 2), the effects of various gp120 mutations were tested in the context of MN-P HR1 sequences.
The first hypothesis that we pursued was that mutations in or near the CD4bs mediated resistance to sCD4 neutralization. It was surprising that introduction of the seven gp120 mutations located in or near the CD4bs, singly or in various combinations, in the context of the MN-P HR1 sequences had little or no effect on resistance to sCD4 neutralization. Since the V1/V2 region is spatially related to the CD4bs, we also studied its contribution to sCD4 resistance (43). Introduction of V1/V2 sequences from MN-P in the context of MN-P HR1 sequences also had little effect. Simultaneous introduction of the seven CD4bs mutations, V1/V2, and HR1 sequences from MN-P substantially increased resistance to neutralization by sCD4. Conversely, the further introduction of MN-P V3 region sequences into this mutant eliminated the sCD4 resistance conferred by the combined presence of the other mutations. These results demonstrated the existence of an important functional interaction between the V1/V2 region and one or more of the CD4bs mutations studied and a negative effect of V3 region mutations on this interaction. As discussed further below, we consider it likely that these functional interactions reflect structural relationships between the V1/V2 region and residues near the CD4bs and in the V3 region. These results are consistent with a previous report that demonstrated that CD4 binding affects the conformation of the V3 region (16).
Our observations also suggested a role for additional sequences in the carboxy terminus of gp120. A mutant with the full MN-P gp120 carboxy-terminal sequence (chimera GC) was more resistant than was the mutant with the seven CD4bs and four V3 mutations plus the HR1 sequences from MN-P (chimera C/7CD4bs/V3). The studies presented in Fig. 6 demonstrated that the MN-P V3 mutations actually contribute to sCD4 resistance, when present in the context of either four outer domain, non-binding-site mutations or a coreceptor-binding-site mutation of MN-P. Therefore, these studies demonstrated that V3 interacts widely with other structures across the surface of the core structure of gp120.
The observations that the mutations responsible for resistance to sCD4 were disseminated throughout the coding sequences for gp120 and gp41 led us to test whether the same mutations may mediate global neutralization resistance. To test this hypothesis, selected mutants were studied for sensitivity to neutralization by MAbs to conformation-sensitive epitopes in the CD4bs, CD4-induced epitopes in the putative coreceptor-binding site, and an apical V3 conformation-sensitive epitope. Selected mutants were studied, because it was not feasible to test all mutations that contribute to sCD4 resistance individually. Accordingly, we compared MN-P to chimeras P/7CD4bs and P/7CD4bs/V3. These comparisons allowed evaluation of the effects of the functional interactions of the V1/V2 region with the CD4bs region and V3 on resistance to MAb neutralization. We found that resistance to neutralization by these antibodies and sCD4 corresponded well. These findings lead us to believe that the mechanisms responsible for sCD4 resistance and the global neutralization resistance phenotypes are largely the same.
The results of the studies presented here demonstrated that the mechanism of MN-P resistance to sCD4 neutralization, and likely for global neutralization resistance as well, involves multiple functional interactions of the V1/V2 and V3 regions across the surface of the gp120 core structure. The extents of these interactions are illustrated in Fig. 8. Ovals representing the approximate genetic footprints of each of the two loop structures are shown overlaid on the gp120 core. The footprint of the V1/V2 region extends from the stalk of the double loop to the area near the CD4bs and includes interaction with V3. The footprint of V3 extends from the stalk of the loop and includes interaction with V1/V2, with the outer domain mutations distant from the binding sites and with residue 426 in the coreceptor-binding site. Our data do not define structural features of the envelope complex. However, the findings suggest the possibility that the V1/V2 and V3 loop structures may mediate functional interactions at distances among the CD4, coreceptor-, and gp41-binding sites. Binding of CD4 to the CD4bs may induce conformational changes in V1/V2 and gp41 that cause sequential changes in V3 required for coreceptor interaction. The transmission of signals in this manner could well complement changes in the conformation of the gp120 core structure that result from the binding energy changes associated with CD4 interaction. Such an effect might well explain how mutations that are apparently on opposite ends of the complex, such as in the gp41 HR1 and at residue 426 in the coreceptor-binding site, might display very strong functional interactions. The binding of gp120 to each of its ligands results in conformational changes that are transmitted throughout the gp120 core. A reasonable interpretation of our results is that the interactions with the ligands also result in conformational changes, such that functional signals are transmitted across the surface of the complex. We conclude that the primary virus neutralization resistance phenotype evaluated in the present study is the capacity of the HIV-1 envelope complex to undergo various conformational changes, resulting in high-efficiency infectivity. This model may provide a basis for understanding the nature of epitopes that are important for primary virus neutralization and for designing methods for stabilizing particular conformations of the envelope complex.
FIG. 8.
Schematic representation of the functional interactions of the V1/V2 and V3 regions and gp41 over the surface of gp120. The view is the same as the “front” view in Fig. 4A. The cysteine residues at the stalks of the V1/V2 and V3 loops are shown in magenta, and red arrows highlight their locations. See the text for an explanation.
Acknowledgments
This work was supported by National Institutes of Health grants AI37438 and AI48280.
REFERENCES
- 1.Back, N. K. T., L. Smit, M. Schutten, P. L. Nara, M. Tersmette, and J. Goudsmit. 1993. Mutations in human immunodeficiency virus type 1 gp41 affect sensitivity to neutralization by gp120 antibodies. J. Virol. 67:6897-6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagley, J., P. J. Dillon, C. Rosen, J. Robinson, J. Sodroski, and W. A. Marasco. 1994. Structural characterization of broadly neutralizing human monoclonal antibodies against the CD4 binding site of HIV-1 gp120. Mol. Immunol. 31:1149-1160. [DOI] [PubMed] [Google Scholar]
- 3.Barbas, C. F., J. E. Crowe, Jr., D. Cababa, T. M. Jones, S. L. Zebedee, B. R. Murphy, R. M. Chanock, and D. R. Burton. 1992. Human monoclonal Fab fragments derived from a combinatorial library bind to respiratory syncytial virus F glycoprotein and neutralize infectivity. Proc. Natl. Acad. Sci. USA 89:10164-10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berman, P. W., T. J. Matthews, L. Riddle, M. Champe, M. R. Hobbs, G. R. Nakamura, J. Mercer, D. J. Eastman, C. Lucas, A. J. Langlois, F. M. Wurm, and T. J. Gregory. 1992. Neutralization of multiple laboratory and clinical isolates of human immunodeficiency virus type 1 (HIV-1) by antisera raised against gp120 from the MN isolate of HIV-1. J. Virol. 66:4464-4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boots, L. J., P. M. McKenna, B. A. Arnold, P. M. Keller, M. K. Gorny, S. Zolla-Pazner, J. E. Robinson, and A. J. Conley. 1997. Anti-human immunodeficiency virus type 1 human monoclonal antibodies that bind discontinuous epitopes in the viral glycoproteins can identify mimotopes from recombinant phage peptide display libraries. AIDS Res. Hum. Retrovir. 13:1549-1559. [DOI] [PubMed] [Google Scholar]
- 6.Burton, D. R., C. F. Barbas, M. A. Persson, S. Koenig, R. M. Chanock, and R. A. Lerner. 1991. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc. Natl. Acad. Sci. USA 88:10134-10137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrow, E. W., L. K. Vujcic, W. L. Glass, K. B. Seamon, S. C. Rastogi, R. M. Hendry, R. Boulos, N. Nzila, and G. V. Quinnan. 1991. High prevalence of antibodies to the gp120 V3 region principal neutralizing determinant of HIV-1MN in sera from Africa and the Americas. AIDS Res. Hum. Retrovir. 7:831-838. [DOI] [PubMed] [Google Scholar]
- 8.Cavacini, L. A., M. H. Samore, J. Gambertoglio, B. Jackson, M. Duval, A. Wisnewski, S. Hammer, C. Koziel, C. Trapnell, and M. R. Posner. 1998. Phase I study of a human monoclonal antibody directed against the CD4-binding site of HIV type 1 glycoprotein 120. AIDS Res. Hum. Retrovir. 14:545-550. [DOI] [PubMed] [Google Scholar]
- 9.Daar, E. S., X. L. Li, T. Moudgil, and D. D. Ho. 1990. High concentrations of recombinant soluble CD4 are required to neutralize primary human immunodeficiency virus type 1 isolates. Proc. Natl. Acad. Sci. USA 87:6574-6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.di Marzo Veronese, F., M. S. J. Reitz, G. Gupta, M. Robert-Guroff, C. Boyer-Thompson, A. Louie, R. C. Gallo, and P. Lusso. 1993. Loss of a neutralizing epitope by a spontaneous point mutation in the V3 loop of HIV-1 isolated from an infected laboratory worker. J. Biol. Chem. 268:25894-25901. [PubMed] [Google Scholar]
- 11.Dong, M., P. F. Zhang, F. Grieder, J. Lee, G. Krishnamurthy, T. VanCott, C. Broder, V. R. Polonis, X.-F. Yu, Y. Shao, D. Faix, P. Valente, and G. V. Quinnan, Jr. 2003. Induction of primary virus-cross-reactive human immunodeficiency virus type 1-neutralizing antibodies in small animals by using an alphavirus-derived in vivo expression system. J. Virol. 77:3119-3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gurgo, C., H. G. Guo, G. Franchini, A. Aldovini, E. Collalti, K. Farrell, F. Wong-Staal, R. C. Gallo, and M. S. J. Reitz. 1988. Envelope sequences of two new United States HIV-1 isolates. Virology 164:531-536. [DOI] [PubMed] [Google Scholar]
- 13.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leavitt, M., E. J. Park, I. A. Sidorov, D. S. Dimitrov, and G. V. Quinnan, Jr. 2003. Concordant modulation of neutralization resistance and high infectivity of the primary human immunodeficiency virus type 1 MN strain and definition of a potential gp41 binding site in gp120. J. Virol. 77:560-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levy, J. A. 1993. Pathogenesis of human immunodeficiency virus infection. Microbiol. Rev. 57:183-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mbah, H. A., S. Burda, M. K. Gorny, C. Williams, K. Revesz, S. Zolla-Pazner, and P. N. Nyambi. 2001. Effect of soluble CD4 on exposure of epitopes on primary, intact, native human immunodeficiency virus type 1 virions of different genetic clades. J. Virol. 75:7785-7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McInerney, T. L., and N. J. Dimmock. 2001. Postattachment neutralization of a primary strain of HIV type 1 in peripheral blood mononuclear cells is mediated by CD4-specific antibodies but not by a glycoprotein 120-specific antibody that gives potent standard neutralization. AIDS Res. Hum. Retrovir. 17:1645-1654. [DOI] [PubMed] [Google Scholar]
- 18.Moore, J. P., Y. Cao, L. Qing, Q. J. Sattentau, J. Pyati, R. Koduri, J. Robinson, C. F. Barbas, D. R. Burton, and D. D. Ho. 1995. Primary isolates of human immunodeficiency virus type 1 are relatively resistant to neutralization by monoclonal antibodies to gp120, and their neutralization is not predicted by studies with monomeric gp120. J. Virol. 69:101-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore, J. P., A. Trkola, B. Korber, L. J. Boots, J. A. Kessler, F. E. McCutchan, J. Mascola, D. D. Ho, J. Robinson, and A. J. Conley. 1995. A human monoclonal antibody to a complex epitope in the V3 region of gp120 of human immunodeficiency virus type 1 has broad reactivity within and outside clade B. J. Virol. 69:122-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moulard, M., S. K. Phogat, Y. Shu, A. F. Labrijn, X. Xiao, J. M. Binley, M. Y. Zhang, I. A. Sidorov, C. C. Broder, J. Robinson, P. W. Parren, D. R. Burton, and D. S. Dimitrov. 2002. Broadly cross-reactive HIV-1-neutralizing human monoclonal Fab selected for binding to gp120-CD4-CCR5 complexes. Proc. Natl. Acad. Sci. USA 99:6913-6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park, E. J., and G. V. Quinnan, Jr. 1999. Both neutralization resistance and high infectivity phenotypes are caused by mutations of interacting residues in the human immunodeficiency virus type 1 gp41 leucine zipper and the gp120 receptor- and coreceptor-binding domains. J. Virol. 73:5707-5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park, E. J., L. K. Vujcic, R. Anand, T. S. Theodore, and G. V. Quinnan, Jr. 1998. Mutations in both gp120 and gp41 are responsible for the broad neutralization resistance of variant human immunodeficiency virus type 1 MN to antibodies directed at V3 and non-V3 epitopes. J. Virol. 72:7099-7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park, E. J., M. K. Gorny, S. Zolla-Pazner, and G. V. Quinnan, Jr. 2000. A global neutralization resistance phenotype of human immunodeficiency virus type 1 is determined by distinct mechanisms mediating enhanced infectivity and conformational change of the envelope complex. J. Virol. 74:4183-4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Posner, M. R., L. A. Cavacini, C. L. Emes, J. Power, and R. Byrn. 1993. Neutralization of HIV-1 by F105, a human monoclonal antibody to the CD4 binding site of gp120. J. Acquir. Immune Defic. Syndr. 6:7-14. [PubMed] [Google Scholar]
- 25.Posner, M. R., T. Hideshima, T. Cannon, M. Mukherjee, K. H. Mayer, and R. Byrn. 1991. An IgG human monoclonal antibody that reacts with HIV-1/GP120, inhibits virus binding to cells, and neutralizes infection. Rev. Infect. Dis. 146:4325-4332. [PubMed] [Google Scholar]
- 26.Quinnan, G. V. 1997. Immunization against viral diseases, p. 791-834. In G. Galasso, R. Whitley, and T. C. Merigan (ed.), Antiviral agents and human viral diseases. Raven Press, New York, N.Y.
- 27.Quinnan, G. V., Jr., P. F. Zhang, D. W. Fu, M. Dong, and H. J. Alter. 1999. Expression and characterization of HIV type 1 envelope protein associated with a broadly reactive neutralizing antibody response. AIDS Res. Hum. Retrovir. 15:561-570. [DOI] [PubMed] [Google Scholar]
- 28.Quinnan, G. V., P. F. Zhang, D. W. Fu, M. Dong, and J. B. Margolick. 1998. Evolution of neutralizing antibody response against HIV type 1 virions and pseudovirions in multicenter AIDS cohort study participants. AIDS Res. Hum. Retrovir. 14:939-949. [DOI] [PubMed] [Google Scholar]
- 29.Reitz, M. S., C. Wilson, C. Naugle, R. C. Gallo, and M. Robert-Guroff. 1988. Generation of a neutralization resistant variant of HIV-1 is due to selection for a point mutation in the envelope gene. Cell 54:57-63. [DOI] [PubMed] [Google Scholar]
- 30.Reitz, M. S., Jr., H. G. Guo, J. Oleske, J. Hoxie, M. Popovic, E. Read-Connole, P. Markham, H. Streicher, and R. C. Gallo. 1992. On the historical origins of HIV-1 (MN) and (RF). AIDS Res. Hum. Retrovir. 8:1950. [PubMed] [Google Scholar]
- 31.Rizzuto, C. D., R. Wyatt, N. Hernandez-Ramos, Y. Sun, P. D. Kwong, W. A. Hendrickson, and J. Sodroski. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280:1949-1953. [DOI] [PubMed] [Google Scholar]
- 32.Roben, P., J. P. Moore, M. Thali, J. Sodroski, C. F. Barbas, and D. R. Burton. 1994. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J. Virol. 68:4821-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robert-Guroff, M., M. S. J. Reitz, W. G. Robey, and R. C. Gallo. 1986. In vitro generation of an HTLV-III variant by neutralizing antibody. Rev. Infect. Dis. 137:3306-3309. [PubMed] [Google Scholar]
- 34.Saphire, E. O., P. W. Parren, R. Pantophlet, M. B. Zwick, G. M. Morris, P. M. Rudd, R. A. Dwek, R. L. Stanfield, D. R. Burton, and I. A. Wilson. 2001. Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science 293:1155-1159. [DOI] [PubMed] [Google Scholar]
- 35.Shearer, W. T., R. J. Israel, S. Starr, C. V. Fletcher, D. Wara, M. Rathore, J. Church, J. DeVille, T. Fenton, B. Graham, P. Samson, S. Staprans, J. McNamara, J. Moye, P. J. Maddon, W. C. Olson, et al. 2000. Recombinant CD4-IgG2 in human immunodeficiency virus type 1-infected children: phase 1/2 study. J. Infect. Dis. 182:1774-1779. [DOI] [PubMed] [Google Scholar]
- 36.Thali, M., C. Furman, D. D. Ho, J. Robinson, S. Tilley, A. Pinter, and J. Sodroski. 1992. Discontinuous, conserved neutralization epitopes overlapping the CD4-binding region of human immunodeficiency virus type 1 gp120 envelope glycoprotein. J. Virol. 66:5635-5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thali, M., J. P. Moore, C. Furman, M. Charles, D. D. Ho, J. Robinson, and J. Sodroski. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67:3978-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thali, M., U. Olshevsky, C. Furman, D. Gabuzda, J. Li, and J. Sodroski. 1991. Effects of changes in gp120-CD4 binding affinity on human immunodeficiency virus type 1 envelope glycoprotein function and soluble CD4 sensitivity. J. Virol. 65:5007-5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turner, S., R. Tizard, J. DeMarinis, R. B. Pepinsky, J. Zullo, R. Schooley, and R. Fisher. 1992. Resistance of primary isolates of human immunodeficiency virus type 1 to neutralization by soluble CD4 is not due to lower affinity with the viral envelope glycoprotein gp120. Proc. Natl. Acad. Sci. USA 89:1335-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vancott, T. C., V. R. Polonis, L. D. Loomis, N. L. Michael, P. L. Nara, and D. L. Birx. 1995. Differential role of V3-specific antibodies in neutralization assays involving primary and laboratory-adapted isolates of HIV type 1. AIDS Res. Hum. Retrovir. 11:1379-1391. [DOI] [PubMed] [Google Scholar]
- 41.Watkins, B. A., S. Buge, K. Aldrich, A. E. Davis, J. Robinson, M. S. Reitz, and M. Robert-Guroff. 1996. Resistance of human immunodeficiency virus type 1 to neutralization by natural antisera occurs through single amino acid substitutions that cause changes in antibody binding at multiple sites. J. Virol. 70:8431-8437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watkins, B. A., M. S. Reitz, C. Wilson, K. Aldrich, A. E. Davis, and M. Robert-Guroff. 1993. Immune escape by human immunodeficiency virus type 1 from neutralizing antibodies: evidence for multiple pathways. J. Virol. 67:7493-7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson, C., M. S. Reitz, K. Aldrich, P. J. Klasse, J. Blomberg, R. C. Gallo, and M. Robert-Guroff. 1990. The site of an immune-selected point mutation in the transmembrane protein of human immunodeficiency virus type 1 does not constitute the neutralization epitope. J. Virol. 64:3240-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wyatt, R., J. P. Moore, M. Accola, E. Desjardin, J. Robinson, and J. Sodroski. 1995. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 69:5723-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, P. F., P. Bouma, E. J. Park, J. B. Margolick, J. E. Robinson, S. Zolla-Pazner, M. N. Flora, and G. V. Quinnan, Jr. 2002. A variable region 3 (V3) mutation determines a global neutralization phenotype and CD4-independent infectivity of a human immunodeficiency virus type 1 envelope associated with a broadly cross-reactive, primary virus-neutralizing antibody response. J. Virol. 76:644-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang, P. F., X. Chen, D. W. Fu, J. B. Margolick, and G. V. Quinnan, Jr. 1999. Primary virus envelope cross-reactivity of the broadening neutralizing antibody response during early chronic human immunodeficiency virus type 1 infection. J. Virol. 73:5225-5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu, P., W. C. Olson, and K. H. Roux. 2001. Structural flexibility and functional valence of CD4-IgG2 (PRO 542): potential for cross-linking human immunodeficiency virus type 1 envelope spikes. J. Virol. 75:6682-6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zwart, G., N. K. Back, C. Ramautarsing, M. Valk, L. van der Hoek, and J. Goudsmit. 1994. Frequent and early HIV-1MN neutralizing capacity in sera from Dutch HIV-1 seroconverters is related to antibody reactivity to peptides from the gp120 V3 domain. AIDS Res. Hum. Retrovir. 10:245-251. [DOI] [PubMed] [Google Scholar]
- 49.Zwick, M. B., L. L. C. Bonnycastle, A. Menendez, M. B. Irving, C. F. Barbas III, P. W. H. I. Parren, D. R. Burton, and J. K. Scott. 2001. Identification and characterization of a peptide that specifically binds the human, broadly neutralizing anti-human immunodeficiency virus type 1 antibody b12. J. Virol. 75:6692-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zwick, M. B., M. Wang, P. Poignard, G. Stiegler, H. Katinger, D. R. Burton, and P. W. H. I. Parren. 2001. Neutralization synergy of human immunodeficiency virus type 1 primary isolates by cocktails of broadly neutralizing antibodies. J. Virol. 75:12198-12208. [DOI] [PMC free article] [PubMed] [Google Scholar]