Abstract
Epstein-Barr virus nuclear antigen 2 (EBNA2) is a transcriptional activator involved in the immortalization of B lymphocytes by the virus. EBNA2 is targeted to the promoters of its responsive genes, via interaction with cellular DNA-binding proteins. Using chromatin immunoprecipitation assays, we show for the first time the conditional recruitment of EBNA2 on two specific viral promoters in vivo and demonstrate a correlation between this recruitment and a local change in the acetylation of histones H3 and H4, which is promoter dependent.
Epstein-Barr virus (EBV) is a human herpesvirus associated with several malignancies, including Burkitt's lymphoma, nasopharyngeal carcinoma, Hodgkin's disease, and various lymphoproliferations in immunosuppressed individuals. In vitro infection of quiescent B lymphocytes by EBV, as well as explant culture of lymphocytes from seropositive individuals, gives rise to continuously proliferating cell lines, lymphoblastoid cell lines (LCL), in which EBV persists as multiple episomal copies. A limited set of viral proteins is expressed in these cells, and six of them (EBNA1, EBNALP, EBNA2, EBNA3A, EBNA3C, and LMP1) are required for the immortalization process (for reviews, see references 15 and 22).
Among these, the nuclear protein EBNA2 transactivates the expression of all the viral genes that are involved in the immortalization process, as well as cellular genes such as CD21, CD23, and c-fgr (for a review, see reference 31). EBNA2 does not bind directly to DNA but is recruited to the promoter of target genes through direct interaction with sequence-specific DNA-binding proteins, such as the ubiquitous cellular factor RBP-Jκ (also called CBF1 or KBF2) (4, 7, 30, 37), a primary nuclear effector of the Notch pathway (for a review, see reference 1). RBP-Jκ is a transcription factor that binds the conserved core sequence GTGGGAA and is involved in both gene activation and repression, depending on the recruitment of distinct corepressor and coactivator complexes (for a review, see reference 16). RBP-Jκ functions as a repressor by interfering with the interaction between TFIIA and TFIID (20) and by recruiting histone deacetylase (HDAC) corepressor complexes to the promoter. These corepressor complexes may include the corepressor proteins SMRT and HDAC1 (12, 29); CIR, SAP30, and HDAC2 (8); and SKIP (36). Binding of EBNA2 to RBP-Jκ is believed to displace the corepressor complex from RBP-Jκ (12). EBNA2 also interacts, via its activation domain, with several components of the RNA polymerase II transcription complex (25, 27); with a coactivator called p100 (26, 35); and with the p300, CBP, and P/CAF histone acetyltransferase coactivators (9, 32). In addition, EBNA2 targets the human SWI-SNF chromatin-remodeling complex to specific promoters by associating with the hSNF5/INI1 subunit of hSWI/SNF (33, 34). Thus, EBNA2 appears to act as an adapter molecule, which targets various multiprotein complexes to specific promoters, each of these complexes contributing to stimulate transcription via a different mechanism. One of these mechanisms could be the control of histone acetylation. The acetylation of histones is believed to increase the accessibility of transcription factors to nucleosomal DNA and correlates with transcriptional activity in vivo (6, 24). EBNA2 may thus affect the local level of histone acetylation, both by competing with the HDAC-containing complex bound on RBP-Jκ and by recruiting proteins with histone acetylase activity, such as CBP/p300.
In order to assess whether the recruitment of EBNA2 onto specific promoters correlates with a local change in histone acetylation in the viral chromatin in vivo, we made chromatin immunoprecipitation (ChIP) assays (21) using antibodies directed against acetylated forms of histones H3 and H4. We focused our analysis on two viral promoter regions from which transcription is regulated by EBNA2: the Cp promoter, at which the transcription of all of the EBNA genes is initiated; and the bidirectional LMPp promoter, at which transcription of both LMP1 and LMP2B genes is initiated. For both Cp and LMPp, we quantified the amount of acetylated histones H3 and H4 along the promoter and flanking sequences, in the absence or presence of functional EBNA2. For these experiments, we used an LCL conditional for EBNA2, ER/EB2-5 (a generous gift from B. Kempkes). In this cell line, the EBNA2 gene is deleted from the resident EBV genome, but EBNA2 is expressed in trans from a mini-EBV plasmid as a chimeric fusion protein with the hormone-binding domain of the estrogen receptor (ER) (14). The functions of the ER-EBNA2 fusion protein (transactivation of reporter gene expression from LMP1, LMP2A, and LMP2B promoter constructs; expression of the cell surface markers CD21 and CD23; and interaction with RBP-Jκ) appear to be strictly dependent on estrogen (β-estradiol) (13). Lymphoblastoid cells expressing ER-EBNA2 proliferate in the presence of estrogen, but stop cycling in estrogen-depleted medium (14).
Approximately 108 ER/EB2-5 cells grown continuously in the presence of 1 μM β-estradiol per ml or 108 ER/EB2-5 cells maintained for 3 days in hormone-depleted medium were fixed with 0.1 volume of 11% formaldehyde solution. Soluble chromatin (sheared by sonication to an average size of 500 bp) was then prepared as described by Orlando et al. (21). The chromatin fragment preparation (1/20 of the initial preparation per assay) was then mock immunoprecipitated or immunoprecipitated with either 5 μl each of anti-acetylated histone H3 (Ac9/14; reference 06-599; Upstate Biotechnology) or anti-acetylated histone H4 antibody (tetra Ac; reference 06-866; Upstate Biotechnology) rabbit polyclonal antibody or 15 μl of anti-EBNA2 monoclonal antibody (NCL-EBV-PE2; Novocastra) in ChIP dilution buffer (0.1% sodium dodecyl sulfate [SDS], 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1], 150 mM NaCl) at 4°C overnight. Immune complexes were recovered by incubation at 4°C for 2 h with 40 μl of 50% (vol/vol) protein G Sepharose, which had been preincubated with 100 μg of sonicated herring sperm DNA and 1 mg of bovine serum albumin per ml overnight. Precipitates were then successively washed with the ChIP dilution buffer, a high-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1], 500 mM NaCl), an LiCl wash buffer (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl [pH 8.1]), and (twice) Tris-EDTA (TE). Immune complexes were eluted from the protein G Sepharose by incubation for 30 min with 1% SDS-0.1 M NaHCO3 elution buffer. The formaldehyde cross-links were then reversed by incubating the samples at 65°C for 4 h, and the DNA was recovered after several phenol-chloroform extractions and ethanol precipitated in the presence of glycogen carrier. Aliquots of the input chromatin were similarly treated. Pellets were resuspended in 100 μl of TE, and the immunoprecipitated DNA was analyzed by quantitative PCR with a set of primer pairs that hybridize either along the Cp promoter region or along the LMPp promoter region (Table 1). For comparison, we also used a primer pair that hybridizes in the Zp promoter of the viral gene BZLF1, which is only expressed after reactivation of the viral productive cycle. PCRs were performed with aliquots of each immunoprecipitated sample, as well as various dilutions of the DNA purified from the input chromatin, in order to generate a standard curve in a linear range. [α-32P]dCTP (0.1 μCi) was added to each PCR mixture, and the PCR products were fractionated on 5% polyacrylamide gels. An example of the data obtained is shown in Fig. 1, which summarizes the PCR amplifications of DNA fragments from the LMP promoter, following chromatin immunoprecipitation with the anti-EBNA2 antibody. For each PCR, bands were subsequently quantified with a Storm PhosphorImager as detailed in the legend to Fig. 1. The totality of the quantified results obtained for the LMPp and Cp promoters is shown as graphs in Fig. 2 and 3, respectively.
TABLE 1.
Sequences of primer sets used to amplify specific sequences along the LMPp and Cp promoter regions or in the Zp promoter
| PCR fragment | Oligonucleotide sequences | Positions relative to +1 of promotera |
|---|---|---|
| LMPp promoter | ||
| L1 | 5′-TCATCAGTGTTGTCAGGGTCCTG-3′ | + 978, + 956 |
| 5′-TAACTCCAACGAGGGCAGACAC-3′ | + 857, + 868 | |
| L2 | 5′-CGTGGTGGTGTTCATCACTGTG-3′ | + 788, + 767 |
| 5′-TTGGAGATGCTCTGGCGACTTG-3′ | + 575, + 596 | |
| L3 | 5′-TACCAAGTAAGCACCCGAAGATG-3′ | + 478, + 456 |
| 5′-GAGTAAGTATTACACCCTTTGCCCC-3′ | + 309, + 333 | |
| L4 | 5′-CCAGTCCAGTCACTCATAACGATG-3′ | + 188, + 165 |
| 5′-CCTACATAAGCCTCTCACACTGCTC-3′ | −31, −8 | |
| L5 | 5′-AGCGGCGGTGTGTGTGTGC-3′ | −98, −121 |
| 5′-TCCAGCCTTGCCTCACCTGAAC-3′ | −346, −330 | |
| L6 | 5′-CAATCAGAAGGGGGAGTGCG-3′ | −265, −284 |
| 5′-ACAGCCTTGCCTCACCTGAAC-3′ | −405, −385 | |
| L7 | 5′-AGCAGCAGACGGCGGATATG-3′ | −467, −486 |
| 5′-CCTCCACTTTTTCCAGGAATGC-3′ | −617, −596 | |
| L8 | 5′-GCAATGGAGCGTGACGAAGG-3′ | −960, −979 |
| 5′-CTTTGTCAGGGTTGCCTGTGTC-3′ | −1079, −1058 | |
| Cp promoter | ||
| C1 | 5′-GGTATGGAGCGAAGGTTAGTGGTC-3′ | −1604, −1581 |
| 5′-CATCCAAGGTAGCCCTTAAAGTCC-3′ | −1384, −1407 | |
| C2 | 5′-GGTGAAAATCTAAAGACCCTACGGC-3′ | −1286, −1261 |
| 5′-AATGCACCCATCTCCTGCTTG-3′ | −1074, −1053 | |
| C3 | 5′-CCCCCCTTTCGACTGTCATTTAC-3′ | −1041, −1020 |
| 5′-CCACTTCTCTTCCCGTTAAGCTG-3′ | −797, −819 | |
| C4 | 5′-TAGGCTGACAAGGGGACAAGTG-3′ | −664, −643 |
| 5′-AGAGTGGAATATGTGAGTGGACACG-3′ | −471, −449 | |
| C5 | 5′-GTGTCCCAATTAGAAACCCAAGC-3′ | −433, −411 |
| 5′-CCGCCAACAAGGTTCAATTTTC-3′ | −266, −245 | |
| C6 | 5′-AACCTTGTTGGCGGGAGAAG-3′ | −258, −239 |
| 5′-GGCGAATTAACTGAGCTTGCG-3′ | −56, −32 | |
| C7 | 5′-CGTGTTCGATTTCGGGGTCAC-3′ | + 491, + 511 |
| 5′-AGGCATCTGAAGCCCAGTTTGG-3′ | + 700, + 679 | |
| Zp promoter | ||
| Z | 5′-CTTCAGCAAAGATAGCAAAGGTGG-3′ | −38, −15 |
| 5′-TGGGCTGTCTATTTTTGACACCAG-3′ | −182, −205 |
The position of the primers is indicated relative to the +1 position of transcription of either the LMP1, EBNA, or BZLF1 gene, respectively.
FIG. 1.
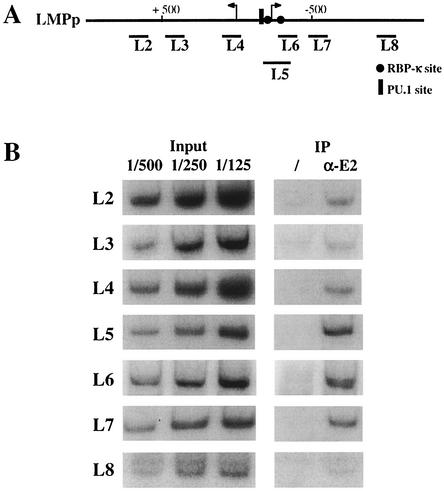
Specific recruitment of EBNA2 onto its responsive element in the LMP promoter. (A) Schematic representation of the EBV genomic region carrying the LMPp bidirectional promoter. Arrows indicate the starts of transcription. Binding sites for RBP-Jκ and PU.1 are also indicated. The positions of the PCR fragments (L2 to L8) used for the ChIP analysis of the LMPp promoter are shown underneath. (B) Primary PCR data obtained from an anti-EBNA2 ChIP experiment. Chromatin fragments (cross-linked with formaldehyde) were prepared from ER/EB2-5 cells cultivated in the presence of β-estradiol and then either immunoprecipitated with an anti-EBNA2 antibody (α-E2) or mock-immunoprecipitated (/). The coimmunoprecipitated DNA was PCR amplified with the various primer pairs (L2 to L8) listed in Table 1, in the presence of 0.1 μCi of [α-32P]dCTP. For quantification, various dilutions of DNA purified from the original chromatin preparation (input) were concomitantly PCR amplified with the same primer pairs. The PCR-amplified fragments were then analyzed on 5% polyacrylamide gels and autoradiographed. The bands were subsequently quantified with a Storm PhosphorImager. The amount of DNA amplified from the mock-immunoprecipitated chromatin was systematically subtracted from the amount of DNA amplified from the specific immunoprecipitated chromatin, and the results are expressed as a percentage of immunoprecipitated DNA relative to the quantity of DNA present before immunoprecipitation (input). The results of such a quantification for the EBNA2 coimmunoprecipitated LMPp DNA fragments are presented in Fig. 2B.
FIG. 2.
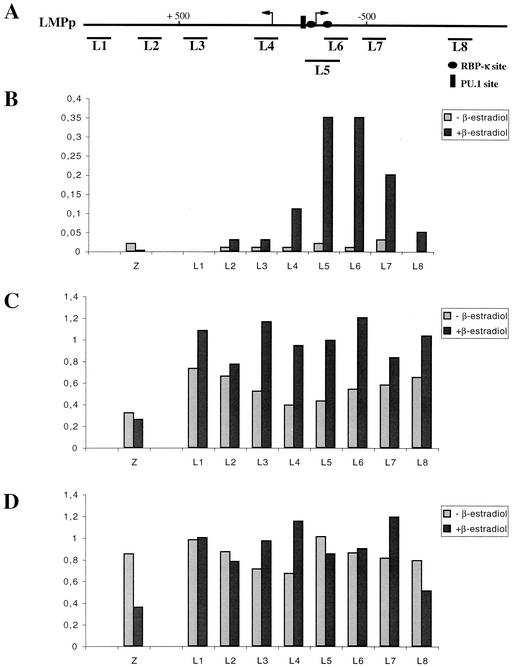
Recruitment of EBNA2 onto the LMPp promoter in vivo correlates with a local increase in the acetylation of histone H3 but not histone H4. (A) Schematic representation of the EBV genomic region carrying the LMPp bidirectional promoter. Arrows indicate the starts of transcription. Binding sites for RBP-Jκ and PU.1 are also indicated. The positions of the PCR fragments (L1 to L8) used for the ChIP analysis of the LMPp promoter are shown underneath. (B) Chromatin from ER/EB2-5 cells cultivated in the presence of β-estradiol (dark gray) or left 3 days in the absence of estrogen (light gray) was fixed by formaldehyde treatment. Chromatin fragments (L1 to L8 for the LMPp region and Z for the control Zp promoter) were precipitated with an antibody specific for EBNA2. The amounts of coimmunoprecipitated DNA determined by quantitative PCR are shown as a percentage of the respective input DNA. (C) Same as panel B except for the use of an antibody specific for diacetylated histone H3 (Ac9/14). (D) Same as panel B except for the use of an antibody specific for tetraacetylated histone H4 (tetra Ac).
FIG. 3.
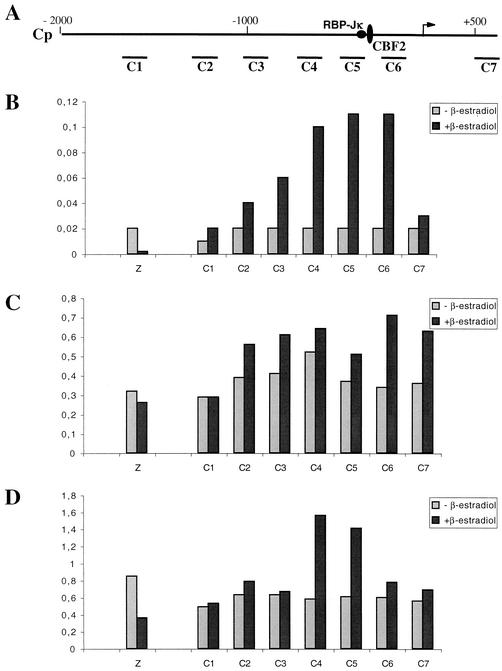
Recruitment of EBNA2 onto the Cp promoter in vivo correlates with a slight increase in the acetylation of histone H3 and a significant and very localized increase in histone H4 acetylation. (A) Schematic representation of the EBV genomic region carrying the Cp promoter. Arrows indicate the start of transcription. Binding sites for RBP-Jκ and CBF-2 are also indicated. The positions of the PCR fragments (C1 to C8) used for the ChIP analysis of the Cp promoter are shown underneath. (B) Chromatin from ER/EB2-5 cells cultivated in the presence of β-estradiol (dark gray) or left for 3 days in the absence of estrogen (light gray) was fixed by formaldehyde treatment. Chromatin fragments (C1 to C7 for the Cp region and Z for the control Zp promoter) were precipitated with an antibody specific for EBNA2. The amounts of coimmunoprecipitated DNA determined by quantitative PCR are shown as a percentage of the respective input DNA. (C) Same as panel B except for the use of an antibody specific for diacetylated histone H3 (Ac9/14). (D) Same as panel B except for the use of an antibody specific for tetraacetylated histone H4 (tetra Ac).
ChIPs using the anti-EBNA2 antibody demonstrated a specific recruitment of EBNA2 on both the LMPp (Fig. 2B) and the Cp (Fig. 3B) promoters but not on the Zp promoter. Moreover, this recruitment was seen only with the chromatin prepared from cells grown in the presence of β-estradiol, although the ER-EBNA2 fusion protein had been shown to be present as well, albeit less abundant, in cells grown without estrogen (14). These results confirm that the ER-EBNA2 protein is functional only in the presence of β-estradiol. Interestingly, the percentages of immunoprecipitated chromatin detected by PCR using primer pairs located along either the LMPp or Cp promoter regions follow a Gaussian curve centered on the cis elements previously described as EBNA2-responsive elements on these two promoters (2, 10, 18, 19, 28). It should be noted that the amount of immunoprecipitated chromatin, which contains LMPp promoter sequences, is approximately three times larger than the amount of immunoprecipitated chromatin, which contains Cp promoter sequences. This could be explained by the different affinities of EBNA2 for the protein complexes bound on the LMPp and Cp promoters, respectively. Indeed, although both promoters contain RBP-Jκ binding sequences and EBNA2 is supposed to be recruited to the DNA by RBP-Jκ, transcriptional activation by EBNA2 of reporter genes placed under the control of either LMPp or Cp promoters has been shown to be largely dependent on other cis-acting elements. In particular, a PU-1 binding site in the LMPp promoter appears to be essential to EBNA2 transcriptional activation (11, 17, 23; unpublished data). Similarly, a sequence adjacent to the RBP-Jκ binding site, which binds a factor initially called “CBF2” (19) but recently shown to be AUF1/hnRNPD (3), is essential to EBNA2 transcriptional activation of the Cp promoter.
In order to study the effect of EBNA2 recruitment on the acetylation level of the surrounding chromatin, we used the same set of primer pairs described above to compare the amount of chromatin immunoprecipitated with anti-acetylated histone H3 or H4 antibody under conditions in which cells were grown in either the presence or absence of β-estradiol. Interestingly, recruitment of EBNA2 onto DNA correlates with a modification of the histone acetylation pattern at the targeted promoter, and this modification appears to be dependent on the promoter involved. Recruitment of EBNA2 onto the LMPp promoter correlates with a specific increase in the level of histone H3 acetylation (Fig. 2C). In the absence of β-estradiol, the level of acetylation appears to vary along the promoter, with a net decrease in the amount of acetylated histone H3 detected, centered on the LMP1 TATA box. This result is in agreement with a model in which repressing factors like RBP-Jκ, bound on the LMPp promoter, recruit protein complexes with HDAC activity. In the presence of β-estradiol, EBNA2 is recruited onto the promoter, and we observed the most important increase in the level of histone H3 acetylation (up to 2.4 times) also at the position of the LMP1 TATA box. On the contrary, no significant difference in the level of H4 acetylation at the LMPp promoter between cells cultivated in the presence or absence of β-estradiol was detected (Fig. 2D). When similar experiments were carried out with the Cp promoter, recruitment of EBNA2 correlated with only a very slight increase in the level of histone H3 acetylation (Fig. 3C). However, a significant (up to 2.6 times) and very local increase in histone H4 acetylation centered on the region immediately upstream of the EBNA2-responsive element was observed. In order to ensure the specificity of our observations on the LMPp and Cp promoters, we have also evaluated both the recruitment of EBNA2 and the acetylation of histones H3 and H4 at the Zp promoter. As expected, no recruitment of EBNA2 could be detected on the Zp promoter, and the level of acetylation of histone H3 in the ER/EB2-5 cells grown in either the presence or absence of β-estradiol was not affected. However, the level of acetylation of histone H4 was twice as high in cells grown in the absence of β-estradiol as in cells grown in the presence of β-estradiol. We have previously shown that the BZLF1 gene is probably actively repressed during latency, by a mechanism involving a specific and local deacetylation of histones at the Zp promoter, following the recruitment of class II HDACs by the MEF2D cellular protein (5). It is thus possible that, because of the arrest of cell proliferation when the ER/EB2-5 cells are maintained in the absence of β-estradiol, expression of some of the factors involved in deacetylation at the Zp promoter is affected and the hypoacetylation at Zp is no longer effective.
The ChIP approach described in this paper has allowed us to demonstrate for the first time in vivo the specific recruitment of EBNA2 onto its target promoters in the context of the viral chromatin. Furthermore, the precise localization of the EBNA2 protein in the chromatin can be determined to within a few 100 bp. From the experiments using antibodies to the acetylated H3 or H4 histones, we can conclude that the recruitment of EBNA2 to its target promoters correlates with an increase in the level of histone H3 or H4 acetylation. The type of modified histone involved and the extent of the modification over the promoter region appear to be dependent on the target promoter and are probably a consequence of the diversity of both the protein complexes already present on the promoter in the absence of EBNA2 and the protein complexes recruited by EBNA2, which may also be influenced by the cellular proteins present on the promoter. EBV chromatin appears to be a good model with which to study in vivo the composition of the various protein complexes recruited onto EBV promoters as well as the epigenetic modifications of the viral chromatin following recruitment of specific factors.
Acknowledgments
We thank B. Kempkes for providing us with the ER/EB2-5 cell line and R. Buckland for reading the manuscript.
This work was supported by INSERM and by grants from the Ligue Nationale Contre le Cancer (LNCC), Comité Départemental du Rhône, and the Association pour la Recherche contre le Cancer (ARC). A.S. and E.M. are CNRS scientists, and N.A and E.H. are recipients of fellowships from the Ministère de l'Education Nationale, de la Recherche et des Technologies.
REFERENCES
- 1.Artavanis-Tsakonas, S., M. D. Rand, and R. J. Lake. 1999. Notch signaling: cell fate control and signal integration in development. Science 284:770-776. [DOI] [PubMed] [Google Scholar]
- 2.Fahraeus, R., A. Jansson, A. Sjöblom, T. Nilsson, G. Klein, and L. Rymo. 1993. Cell phenotype-dependent control of Epstein-Barr virus latent membrane protein 1 gene regulatory sequences. Virology 195:71-80. [DOI] [PubMed] [Google Scholar]
- 3.Fuentes-Pananá, E. M., R. Peng, G. Brewer, J. Tan, and P. D. Ling. 2000. Regulation of the Epstein-Barr virus C promoter by AUF1 and the cyclic AMP/protein kinase A signaling pathway. J. Virol. 74:8166-8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grossman, S. R., E. Johannsen, X. Tong, R. Yalamanchili, and E. Kieff. 1994. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the Jκ recombination signal binding protein. Proc. Natl. Acad. Sci. USA 91:7568-7572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gruffat, H., E. Manet, and A. Sergeant. 2002. MEF2-mediated recruitment of class II HDAC at the EBV immediate early gene BZLF1 links latency and chromatin remodeling. EMBO Rep. 3:141-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grunstein, M. 1997. Histone acetylation in chromatin structure and transcription. Nature 389:349-352. [DOI] [PubMed] [Google Scholar]
- 7.Henkel, T., P. D. Ling, S. D. Hayward, and M. G. Peterson. 1994. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein Jκ. Science 265:92-95. [DOI] [PubMed] [Google Scholar]
- 8.Hsieh, J. J.-D., S. Zhou, L. Chen, D. B. Young, and S. D. Hayward. 1999. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc. Natl. Acad. Sci. USA 96:23-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jayachandra, S., K. G. Low, A.-E. Thlick, J. Yu, P. D. Ling, Y. Chang, and P. S. Moore. 1999. Three unrelated viral transforming proteins (vIRF, EBNA2, and E1A) induce the MYC oncogene through the interferon-responsive PRF element by using different transcription coadaptors. Proc. Natl. Acad. Sci. USA 96:11566-11571. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Jin, X. W., and S. H. Speck. 1992. Identification of critical cis elements involved in mediating Epstein-Barr virus nuclear antigen 2-dependent activity of an enhancer located upstream of the viral BamHI C promoter. J. Virol. 66:2846-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johannsen, E., E. Koh, G. Mosialos, X. Tong, E. Kieff, and S. R. Grossman. 1995. Epstein-Barr virus nuclear protein 2 transactivation of the latent membrane protein 1 promoter is mediated by Jκ and PU.1. J. Virol. 69:253-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kao, H.-Y., P. Ordentlich, N. Koyana-Nakagawa, Z. Tang, M. Downes, C. R. Kintner, R. M. Evans, and T. Kadesh. 1998. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 12:2269-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kempkes, B., M. Pawlita, U. Zimber-Strobl, G. Eissner, G. Laux, and G. W. Bornkamm. 1995. Epstein-Barr virus nuclear antigen 2-estrogen receptor fusion proteins transactivate viral and cellular genes and interact with RBP-Jκ in a conditional fashion. Virology 214:675-679. [DOI] [PubMed] [Google Scholar]
- 14.Kempkes, B., D. Spitkovsky, P. Jansen-Dürr, J. W. Ellwart, E. Kremmer, H.-J. Delecluse, C. Rottenberger, G. W. Bornkamm, and W. Hammerschmidt. 1995. B-cell proliferation and induction of early G1-regulating proteins by Epstein-Barr virus mutants conditional for EBNA2. EMBO J. 14:88-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kieff, E., and A. B. Rickinson. 2001. Epstein-Barr virus and its replication, p. 2511-2573. In D. Knipe and P. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Baltimore, Md.
- 16.Lai, E. C. 2002. Keeping a good pathway down: transcriptional repression of Notch pathway target genes by CSL proteins. EMBO Rep. 3:840-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laux, G., B. Adam, L. J. Strobl, and F. Moreau-Gachelin. 1994. The Spi-1/PU.1 and Spi-B ets family transcription factors and the recombination signal binding protein RBP-Jκ interact with an Epstein-Barr virus nuclear antigen 2 responsive cis-element. EMBO J. 13:5624-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laux, G., F. Dugrillon, C. Eckert, B. Adam, U. Zimber-Strobl, and G. W. Bornkamm. 1994. Identification and characterization of an Epstein-Barr virus nuclear antigen 2-responsive cis element in the bidirectional promoter region of latent membrane protein and terminal protein 2 genes. J. Virol. 68:6947-6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ling, P. D., D. R. Rawlins, and S. D. Hayward. 1993. The Epstein-Barr virus immortalizing protein EBNA-2 is targeted to DNA by a cellular enhancer-binding protein. Proc. Natl. Acad. Sci. USA 90:9237-9241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olave, I., D. Reinberg, and L. D. Vales. 1998. The mammalian transcriptional repressor RBP (CBF1) targets TFIID and TFIIA to prevent activated transcription. Genes Dev. 12:1621-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orlando, V., H. Strutt, and R. Paro. 1997. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods 11:205-214. [DOI] [PubMed] [Google Scholar]
- 22.Rickinson, A. B., and E. Kieff. 2001. Epstein-Barr virus, p. 2575-2627. In D. Knipe and P. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Baltimore, Md.
- 23.Sjöblom, A., A. Jansson, W. Yang, S. Lain, T. Nilsson, and L. Rymo. 1995. PU box-binding transcription factors and a pou domain protein cooperate in the Epstein-Barr virus (EBV) nuclear antigen 2-induced transactivation of the EBV latent membrane protein 1 promoter. J. Gen. Virol. 76:2679-2692. [DOI] [PubMed] [Google Scholar]
- 24.Struhl, K. 1998. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12:599-606. [DOI] [PubMed] [Google Scholar]
- 25.Tong, X., R. Drapkin, D. Reinberg, and E. Kieff. 1995. The 62- and 80-kDa subunits of transcription factor IIH mediate the interaction with Epstein-Barr virus nuclear protein 2. Proc. Natl. Acad. Sci. USA 92:3259-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tong, X., R. Drapkin, R. Yalamanchili, G. Mosialos, and E. Kieff. 1995. The Epstein-Barr virus nuclear protein 2 acidic domain forms a complex with a novel cellular coactivator that can interact with TFIIE. Mol. Cell. Biol. 15:4735-4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tong, X., F. Wang, C. J. Thut, and E. Kieff. 1995. The Epstein-Barr virus nuclear protein 2 acidic domain can interact with TFIIB, TAF40, and RPA70 but not with TATA-binding protein. J. Virol. 69:585-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsang, S.-F., F. Wang, K. M. Izumi, and E. Kieff. 1991. Delineation of the cis-acting element mediating EBNA-2 transactivation of latent infection membrane protein expression. J. Virol. 65:6765-6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waltzer, L., P. Y. Bourillot, A. Sergeant, and E. Manet. 1995. RBP-Jκ repression activity is mediated by a co-repressor and antagonized by the Epstein-Barr virus transcription factor EBNA2. Nucleic Acids Res. 23:4939-4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waltzer, L., F. Logeat, C. Brou, A. Israel, A. Sergeant, and E. Manet. 1994. The human Jκ recombination signal sequence binding protein (RBP-Jκ) targets the Epstein-Barr virus EBNA2 protein to its DNA responsive elements. EMBO J. 13:5633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waltzer, L., F. Meggetto, A. Sergeant, and E. Manet. 1997. EBNA2: a viral transcription factor essential for the immortalization of human B lymphocytes by the Epstein-Barr virus (EBV), p. 133-161. In M. Yaniv and J. Ghysdeal (ed.), Oncogenes as transcriptional regulators: cell cycle regulators and chromosomal translocation, vol. 2. Birkhäuser Verlag, Basel, Switzerland.
- 32.Wang, L., S. R. Grossman, and E. Kieff. 2000. Epstein-Barr virus nuclear protein 2 interacts with p300, CBP, and PCAF histone acetyltransferases in activation of the LMP1 promoter. Proc. Natl. Acad. Sci. USA 97:430-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu, D. Y., G. V. Kalpana, S. P. Goff, and W. H. Schubach. 1996. Epstein-Barr virus nuclear protein 2 (EBNA2) binds to a component of the human SNF-SWI complex, hSNF5/Ini1. J. Virol. 70:6020-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu, D. Y., A. Krumm, and W. H. Schubach. 2000. Promoter-specific targeting of human SWI-SNF complex by Epstein-Barr virus nuclear protein 2. J. Virol. 74:8893-8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang, J., S. Aittomaki, M. Pesu, K. Carter, J. Saarinen, N. Kalkkinen, E. Kieff, and O. Silvennoinen. 2002. Identification of p100 as a coactivator for STAT6 that bridges STAT6 with RNA polymerase II. EMBO J. 21:4950-4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou, S., M. Fujimuro, J. J.-D. Hsieh, L. Chen, A. Miyamoto, G. Weinmaster, and S. D. Hayward. 2000. SKIP, a CBF1-associated protein, interacts with the ankyrin repeat domain of NotchIC to facilitate NotchIC function. Mol. Cell. Biol. 20:2400-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zimber-Strobl, U., L. J. Strobl, C. Meitinger, R. Hinrichs, T. Sakai, T. Furukawa, T. Honjo, and G. Bornkamm. 1994. Epstein-Barr virus nuclear antigen 2 exerts its transactivating function through interaction with recombination signal binding protein RBP-Jκ, the homologue of Drosophila Suppressor of Hairless. EMBO J. 13:4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]