Abstract
The Ebola virus VP35 protein was previously found to act as an interferon (IFN) antagonist which could complement growth of influenza delNS1 virus, a mutant influenza virus lacking the influenza virus IFN antagonist protein, NS1. The Ebola virus VP35 could also prevent the virus- or double-stranded RNA-mediated transcriptional activation of both the beta IFN (IFN-β) promoter and the IFN-stimulated ISG54 promoter (C. Basler et al., Proc. Natl. Acad. Sci. USA 97:12289-12294, 2000). We now show that VP35 inhibits virus infection-induced transcriptional activation of IFN regulatory factor 3 (IRF-3)-responsive mammalian promoters and that VP35 does not block signaling from the IFN-α/β receptor. The ability of VP35 to inhibit this virus-induced transcription correlates with its ability to block activation of IRF-3, a cellular transcription factor of central importance in initiating the host cell IFN response. We demonstrate that VP35 blocks the Sendai virus-induced activation of two promoters which can be directly activated by IRF-3, namely, the ISG54 promoter and the ISG56 promoter. Further, expression of VP35 prevents the IRF-3-dependent activation of the IFN-α4 promoter in response to viral infection. The inhibition of IRF-3 appears to occur through an inhibition of IRF-3 phosphorylation. VP35 blocks virus-induced IRF-3 phosphorylation and subsequent IRF-3 dimerization and nuclear translocation. Consistent with these observations, Ebola virus infection of Vero cells activated neither transcription from the ISG54 promoter nor nuclear accumulation of IRF-3. These data suggest that in Ebola virus-infected cells, VP35 inhibits the induction of antiviral genes, including the IFN-β gene, by blocking IRF-3 activation.
Ebola viruses cause sporadic outbreaks of viral hemorrhagic fever in humans, such as recent outbreaks in Gabon and the Republic of Congo (89-91). Fatality rates for Ebola hemorrhagic fever have ranged from 50 to 90% in different outbreaks (88). Because it is highly lethal and causes dramatic symptoms, and because effective vaccines and therapies are lacking (14), Ebola virus is classified by the Centers for Disease Control and Prevention and the National Institutes of Health as a category A bioterrorism agent (17, 41).
The molecular mechanisms contributing to the high virulence of Ebola virus are only beginning to be unraveled (21, 78). It is likely that virus-encoded proteins that inhibit the host cell interferon (IFN) response play a significant role in Ebola virus pathogenesis (6, 12). Data from several other viruses have demonstrated an essential role for virus-encoded IFN antagonists in virulence. For example, mutants of influenza A viruses (23, 80), Bunyamwera virus (15, 87), paramyxoviruses (22), vaccinia virus (8, 11), and herpes simplex virus type 1 (19, 42, 49), which lack or which possess altered viral IFN antagonists, are attenuated in mice. Evidence also suggests that full virulence of Rift Valley Fever virus requires the anti-IFN activity of the viral NSs protein (10).
Ebola virus infection has been shown to block host cell responses to IFNs, to inhibit double-stranded RNA (dsRNA)-mediated induction of antiviral gene expression, and to block alpha/beta IFN (IFN-α/β) production in infected cells (7, 13, 27-29), although Ebola virus infection has been reported to induce IFN production under some circumstances (31). We previously demonstrated that the Ebola virus VP35 protein, an essential component of the Ebola virus RNA replication machinery (52, 53), functions as an IFN antagonist (7). VP35 expression rescued growth of a mutant influenza virus, influenza delNS1 virus, which lacks the influenza A virus NS1 protein, a previously described inhibitor of the IFN response (7), demonstrating the ability of VP35 to counteract the antiviral effects of the host IFN system. Furthermore, expression of VP35 was able to inhibit the dsRNA or virus-induced activation of the beta IFN (IFN-β) promoter or the IFN-inducible ISG54 promoter (7). However, these experiments did not identify specific component(s) of the IFN response affected by VP35.
IFN regulatory factor 3 (IRF-3) is a constitutively expressed transcription factor. Inactive IRF-3 shuttles between the nucleus and cytoplasm and is predominantly cytoplasmic. However, following its activation by phosphorylation, IRF-3 accumulates in the nucleus, where it acts as a transcription factor (40, 45, 94). Activation of IRF-3 is associated with serine/threonine phosphorylation near its carboxy terminus, on serines 385 and 386 (94) and on a cluster of five serine and threonine residues (Ser 396, 398, 402, and 405 and Thr 404) (45) in response to virus infection (40, 45, 55, 74, 84, 94). This carboxy-terminal serine/threonine phosphorylation of IRF-3 is thought to promote dimerization, the ability of IRF-3 to bind DNA in a sequence-specific fashion, and IRF-3 interaction with the histone acetyltransferases CBP and p300 (45, 46, 94). It is not completely clear what specific products of virus infection lead to IRF-3 activation, although IRF-3 can be activated by dsRNA (73, 85, 86) and by at least some viral nucleocapsid proteins (34, 82, 84, 86, 94). Two recent publications provide evidence that the virus-activated kinase(s) responsible for activation of IRF-3 are IKKɛ and TBK-1 (21a, 74a). IRF-3 activation has also been reported following activation of Toll-like receptors 3 and 4, leading to IFN production (38, 58, 59, 75). Phosphorylation near the amino terminus of IRF-3 has also been reported following activation of stress-induced signaling pathways (36), although the functional significance of this observation is not fully clear (72).
IRF-3 contributes to the transcriptional activation of a number of genes, including the IFN-β gene and select IFN-α genes (84, 94). The initial production of IFN-α/β, primarily IFN-β in most cell types, induces expression of numerous IFN-responsive genes, including expression of IRF-7. IRF-7 is also dsRNA and virus induced and activates a wider set of IFN-α genes than does IRF-3 (48, 70). As a consequence, the initial activation of IRF-3 can ultimately result in a dramatic up-regulation of IFN and IFN-induced genes, resulting in a potent antiviral state. IRF-3 also induces a subset of genes previously described as IFN-inducible (26, 35, 45, 50, 54, 63, 84). The degree to which this direct action of IRF-3 contributes to the establishment of an antiviral state is not clear, although the expression of the PKR and OAS genes, two important IFN-induced genes with known antiviral activity, is not directly activated by IRF-3 (54). Activated IRF-3 may also promote apoptosis, another potential antiviral mechanism (32, 85).
Because of its central role in the initiation of the antiviral state, several viruses encode inhibitors of IRF-3. These include influenza virus, human papillomavirus 16, human herpesvirus 8 (HHV-8), and vaccinia virus (44, 67, 79, 92). Interestingly, paramyxovirus V proteins previously reported to block IFN-α/β-induced signaling by promoting STAT protein degradation (1, 20, 39, 60, 61, 64, 95) have been found to also inhibit IFN-β gene expression and to block IRF-3 activation (30, 64). In this report, we demonstrate that the Ebola virus protein VP35 specifically targets the function of IRF-3, blocking a pathway that leads to IFN-α/β gene expression, and that VP35 does not block IFN-α/β-induced signaling. We further demonstrate that VP35 prevents activation of IRF-3 and does so by blocking virus-induced phosphorylation of IRF-3. Finally, consistent with these observations, Ebola virus infection is shown to not induce an IRF-3 responsive promoter and to not induce IRF-3 nuclear translocation. The IRF-3 inhibitory activity of VP35 may make an important contribution to Ebola virus pathogenesis.
MATERIALS AND METHODS
Cells and viruses.
293T cells, 293 cells, and Vero cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum. Virus-infected cells were maintained in DMEM supplemented with 0.3% bovine serum albumin. Sendai virus (SeV) strain Cantell was grown in 10-day-old embryonated chicken eggs at 37°C for 48 h. Influenza delNS1 virus was grown in 7-day-old embryonated chicken eggs at 37°C for 48 h. Ebola virus Zaire was grown in VeroE6 cells at either the U.S. Army Medical Research Institute of Infectious Diseases (USAMRIID) or the Philipps-Universität Marburg BSL-4 laboratories.
Plasmids.
The influenza A/PR/8/43 (H1N1) virus NS1 protein expression plasmid has been described previously (79). The Ebola virus Zaire VP35, NP, and VP24 expression plasmids pcDNA3-Ebo VP35, pcDNA3-EboNP and pcDNA3.1-VP24 have also been described (7). The Ebola Reston virus VP35 open reading frame (ORF) was amplified by reverse transcription PCR of RNA extracted from Ebola virus Reston-infected cells and cloned into the mammalian expression plasmid pCAGGS (57). pCAGGS-HA-VP35(R) was constructed by placing a hemagglutinin (HA) tag sequence encoding the amino acids MYPYDVPDYA upstream of the Ebola virus Reston VP35 ORF and inserting this construct into pCAGGS (57). The human IRF-3 cDNA was amplified by PCR from oligo(dT)-primed cDNA reverse transcribed from HeLa cell total RNA. The expression plasmid pCAGGS-hIRF3 was constructed by inserting a human IRF-3 cDNA, reverse transcription PCR amplified from HeLa cells with the primers hIRF-3/EcoRI/5′ CCGGGAATTCATAATGGGAACCCCAAAG and hIRF-3/XhoI/3′ CCGG CTCGAG TCAGCTCTCCCCAGGGCC, into the plasmid pCAGGS (57). The IRF-3 region of this plasmid was mutated to the described sequence to generate a constitutively activated IRF-3 5D mutant in the pCAGGS expression plasmid, pCAGGS-IRF-3 5D. The plasmid pEGFP-C1-hIRF3 expresses green fluorescent protein (GFP) fused to human IRF-3. This plasmid was constructed by PCR amplification of IRF-3 from pCAGGS-IRF-3 and insertion of the IRF-3 ORF in frame with the GFP ORF in pEGFP-C1 (Clontech).
Reporter gene assays.
Transient transfection of Vero cells was performed by using Lipofectamine 2000 (LF2000) (Invitrogen) according to the manufacturer's instructions. Cells (1 × 106) were transfected with 1 μg of the ISG54 promoter-driven CAT reporter vector pHISG-54-CAT or of the human ISG56 promoter-driven firefly luciferase reporter plasmid ISG56 prom/pGL3b (26), 1 μg of the constitutive expression plasmid Renilla luciferase reporter vector pRL-tk (Promega), and 4 μg of the indicated expression plasmid. At 24 h posttransfection, cells were infected with SeV at a multiplicity of infection (MOI) of 4 or were mock infected. Twenty-four hours postinfection, cells were harvested and resuspended in phosphate-buffered saline (PBS). The resuspended cells were divided into two parts and lysed in reporter lysis buffer (Promega) for chloramphenicol acetyltransferase (CAT) assays and in Renilla luciferase assay lysis buffer (Promega) for Renilla luciferase assays. CAT activity was determined as described previously (68). Luciferase assays were performed by using the Promega Renilla luciferase assay system according to the manufacturer's directions. Relative CAT activities were normalized to relative luciferase activities.
To measure the ability of the constitutively activated mutant IRF-3 5D (45) to activate gene expression, 293T cells were transfected by the calcium phosphate method with 0.5 μg each of the ISG56 prom/pGL3b and pRL-tk, with 0.5 μg of pCAGGS-IRF-3 5D, and with 5 μg of either empty vector (pCDNA3), pcDNA3-EboVP35(Z), or pCAGGS-HA-VP35(R). Twenty-four hours posttransfection, the cells were harvested and assayed for firefly and Renilla luciferase activities.
To measure the ability of Ebola viruses to activate the ISG-54 promoter, 1 × 106 293T cells were transfected in suspension with 0.3 μg of the plasmid pHISG-54-CAT by using the calcium phosphate method. Transfected cells were seeded into 10-cm-diameter dishes and incubated for 24 h at 37°C. Thereafter, cells were washed two times with DMEM and infected with either SeV strain Cantell, Ebola virus Zaire, or Ebola virus Reston, each at an MOI of 3, or mock infected. After an incubation period of 24 h in DMEM with 2.5% fetal calf serum, the cells were harvested and lysed in reporter lysis buffer (Promega), and CAT or luciferase activities were determined.
To measure activation of the mouse IFN-α4 promoter, transient transfection of 293T cells was performed by using the calcium phosphate method (68). Each transfection included 0.5 μg each of the mouse IFN-α4 promoter reporter plasmid pIFNα4-CAT and the constitutively expressed pRL-tk and 5 μg of the indicated protein expression plasmid. Transfections also contained 1 μg of IRF-3 expression plasmid (pCAGGS-hIRF-3) or empty vector (pCAGGS), as indicated. One day posttransfection, the cells were washed once with PBS and infected with SeV or influenza delNS1 virus at an MOI of 1. One day postinfection, CAT and Renilla luciferase assays were performed.
Subcellular localization of IRF-3 and VP35.
To analyze the subcellular localization of IRF-3, Vero cells were transfected using LF2000 with 0.2 μg of pEGFP-C1-hIRF3, which expresses from a human cytomegalovirus promoter a human IRF-3-GFP fusion protein, and 0.4 μg of the indicated expression plasmid. Translocation of IRF-3 was induced by SeV infection at a MOI of 10. The percentage of cells with nuclear GFP-IRF-3 localization was then determined by examining 200 to 300 green fluorescent cells per sample. To analyze the colocalization of IRF-3 and VP35, cells were transfected with 0.8 μg of pEGFP-C1-hIRF3 and 0.4 μg of pCAGGS-VP35(R) HA, which encodes an HA-tagged VP35 derived from Ebola virus Reston. Cells were transfected in suspension and seeded in individual wells of 8-well chamber slides (Lab-Tek) at 1 × 105 cells per well. Twenty-four hours posttransfection, translocation of IRF-3 was induced by SeV infection at an MOI of 10. Cell monolayers were then fixed for 5 min with methanol and permeabilized for 30 s with acetone. Fixed cells were washed with PBS, and where indicated, the HA-tagged Ebola virus VP35(R) protein was detected with a mouse anti-HA monoclonal antibody 12CA5 (Hybridoma Core Facility, Mount Sinai School of Medicine) at a dilution of 1:100. As a secondary antibody, Texas red-conjugated anti-mouse immunoglobulin G (IgG) (Rockland) was used at a dilution of 1:100. All antibodies were diluted in PBS with 3% bovine serum albumin. Cells were affixed to slides using ProLong antifade agent (Molecular Probes). Samples were examined under an Olympus IX-70 fluorescence microscope.
Localization of IRF-3 in Ebola virus-infected cells.
VeroE6 cells were transfected, in suspension, with 5 μg of pEGFP-c1-IRF-3 by using LF2000. Transfected cells were plated on 8-well chamber slides (Lab-Tek) and incubated for 24 h. Chamber slides were transferred into the BSL-4 laboratory at USAMRIID and infected with Ebola virus Zaire at an MOI of 1. Infected cells were washed with PBS, and then fresh viral growth medium was added. Twenty-four and forty-eight hours postinfection, infected cells were fixed for 24 h with 10% formalin before the slides were developed for the Ebola virus GP.
Slides were washed with water for 10 min. Fixed cells were further washed extensively with 1× PBS, treated with proteinase K for 10 min at room temperature, and washed again with 1× PBS. Cells were incubated with a mouse monoclonal antibody directed against Ebola virus Zaire GP (dilution, 1:2,000). Washed cells were then incubated with an anti-mouse IgG (dilution, 1:400) secondary antibody (Alexa Fluor 594, Molecular Probes). All antibodies were diluted with an antibody diluent containing background-reducing components (DAKO). Coverslips were affixed to glass slides using mounting media containing 4′,6′-diamidino-2-phenylindole (DAPI) (Vector). Slides were imaged using a Nikon TE2000 inverted fluorescent microscope. Ebola virus infections were conducted within the Biological Safety Level 4 (BSL-4) Laboratory at USAMRIID. Personnel wore positive-pressure protective suits (ILC, Dover, Frederica, Del.) equipped with high-efficiency particulate air filters and supplied with umbilical-fed air.
Western blot analyses.
293T cells were transfected using LF2000. Each transfection of 1 × 106 cells contained 1 μg of the hIRF-3 expression vector pCAGGS-hIRF3 and 2 μg of the indicated expression plasmid. Twenty-four hours posttransfection, cell monolayers were infected with SeV at a MOI of 10 or mock infected. Four hours postinfection, cell lysates were then made in NP-40 lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% NP-40, 1 mM sodium orthovanadate, 0.1 mg of leupeptin/ml, and 1 mM phenylmethylsulfonyl fluoride [PMSF]), as described previously (34). These lysates were then subjected either to native gel electrophoresis as described previously (34) or to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 7.5% gel. After electrophoresis, the proteins were transferred to a nitrocellulose membrane in a buffer containing 40 mM glycine, 50 mM Tris base, 0.037% SDS and 20% methanol for 2 h. Human IRF-3 was detected by using a mouse anti-IRF-3 monoclonal antibody SL-12 (kindly provided by Peter M. Howley, Harvard Medical School) at a dilution of 1:1,000 in a solution of 5% nonfat dry milk in PBS. The secondary antibody was anti-mouse IgG conjugated to horseradish peroxidase (Boehringer Mannhein Corp.) (1:10,000 dilution in a solution of 5% nonfat dry milk in PBS). The Western blots were developed with Lightning chemiluminescence reagent (PerkinElmer).
Orthophosphate labeling and immunoprecipitation.
293 cells were transfected by using LF2000 transfection reagent and 1 μg of pCAGGS or pCAGGS-hIRF-3 plus either empty vector (pcDNA3), pcDNA3-EboVP35(Z), pCAGGS-HA-VP35(R), or pcDNA3-EboNP. Eighteen hours posttransfection, the cells were mock infected or infected with SeV at an MOI of 4. From 2 to 4 h postinfection, the cells were grown in phosphate-free MEM. From 4 to 6 h postinfection, the cells were labeled with 2 mCi of [32P]orthophosphate/ml (PerkinElmer). At 6 h postinfection, cells were washed with PBS and lysed in NP-40 lysis buffer (see above), and equal portions of each extract were immunoprecipitated with polyclonal mouse antiserum generated against a glutathione S-transferase fusion to amino acids 328 to 427 of human IRF-3.
RESULTS
VP35 blocks IFN-independent virus-induced activation of the ISG54 promoter.
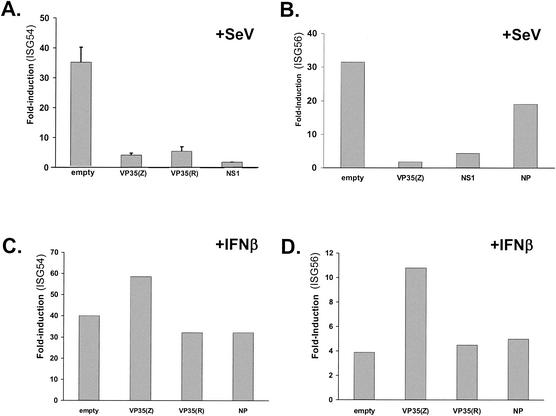
It was previously demonstrated that VP35 could inhibit in 293 cells the activation of the IFN-responsive ISG54 promoter in response to dsRNA treatment or to SeV infection (7). In these previous experiments, activation of the ISG54 reporter could have been mediated by either of two mechanisms. First, IFN produced in response to dsRNA treatment or to viral infection might have activated the ISG54 promoter. Alternatively, because it has been reported that IRF-3 can directly activate the promoters of several IFN-stimulated genes (ISGs), including the ISG54 and ISG56 promoters (26, 63, 70), virus- or dsRNA-mediated activation of the transcription factor IRF-3 could have directly stimulated the ISG54 promoter. We therefore asked whether VP35 can block the virus-induced activation of the ISG54 or ISG56 promoters independently of IFN production. Vero cells, which do not produce IFN-α/β, were transfected with an ISG54-CAT reporter plasmid or a human ISG56-luciferase reporter plasmid plus a constitutively expressed Renilla luciferase plasmid and a mammalian expression plasmid. The expression plasmids were empty vector, a positive control plasmid encoding the influenza A virus NS1 protein (which inhibits IRF-3 activation [80]), an Ebola virus Zaire VP35 expression plasmid, or a plasmid encoding an HA-tagged version of the Ebola virus Reston VP35 protein. Cells were subsequently mock infected or infected with SeV, a potent viral activator of IRF-3 (45), at an MOI of 4. Infection of cells transfected with empty vector resulted in a dramatic up-regulation of expression from either the ISG54 or the ISG56 promoters compared with that seen for uninfected, empty vector-transfected cells (Fig. 1A and B). Expression of the influenza virus NS1 protein or the VP35 protein of Ebola virus Zaire dramatically decreased the activation of either reporter gene (Fig. 1A and B). Interestingly, the HA-tagged VP35 protein of Ebola virus Reston also blocked ISG54 activation in Vero cells (Fig. 1A). Similar results were obtained with an untagged Ebola virus Reston VP35 clone (data not shown). Transfection of a different Ebola virus protein, the nucleoprotein (NP), did not significantly affect virus-induced reporter gene expression (Fig. 1B). That NP is expressed from this plasmid has been previously demonstrated (7). To determine whether VP35 also affects IFN-induced gene activation, Vero cells were again transfected with either the ISG54 or the ISG56 promoter reporter genes in the presence of empty vector, an Ebola virus Zaire VP35 plasmid, an HA-tagged Ebola virus Reston VP35 plasmid, or an Ebola virus NP plasmid. One day posttransfection, the cells were treated with IFN-β and subsequently assayed for reporter gene expression. Neither the Zaire or Reston VP35 proteins were able to inhibit IFN-β-induced activation of the ISG54 or the ISG56 promoters (Fig. 1C and D). Transfection, in parallel, of a plasmid encoding the Nipah virus V or W proteins, previously shown to inhibit IFN-α/β signaling, was able to block IFN-β-mediated ISG54 promoter activation (data not shown) (62). We therefore conclude that the Ebola virus VP35 protein is able to block the virus-mediated, but not the IFN-mediated, activation of the ISG54 and ISG56 promoters.
FIG. 1.
Expression of the Ebola virus VP35 protein inhibits SeV-mediated activation but not the IFN-β-mediated activation of the ISG54 and ISG56 promoters. Shown are the levels of induction of an ISG54 promoter-driven CAT reporter gene (A and C) or of an ISG56 promoter-driven firefly luciferase reporter gene (B and D) in cells transfected with the indicated protein expression plasmids following SeV infection (A and B) or following treatment with IFN-β (C and D). Each transfection included an empty expression plasmid (empty vector) or an expression plasmid producing the viral proteins Ebola virus Zaire VP35 [VP35(Z)] or HA-tagged Ebola virus Reston VP35 [HA-VP35(R)], the influenza virus NS1 protein (NS1), or the Ebola virus Zaire NP. Vero cells were transfected with 4 μg of the indicated expression plasmid, namely, either 1 μg of an ISG-54 promoter-CAT reporter plasmid or 1 μg of an ISG56 promoter plasmid, plus 1 μg of the constitutively expressed Renilla luciferase reporter plasmid pRL-TK (an internal transfection control). At 24 h posttransfection, the cells were mock infected or infected with SeV at a multiplicity of 4 (A and B) or mock treated or treated with 1,000 units of human IFN-β/ml (C and D). Twenty-four hours postinfection, CAT and luciferase activities were determined. Activities of the inducible promoters were normalized to the luciferase activity from the Renilla luciferase plasmid. Fold induction refers to the level of induction relative to an empty plasmid-transfected, mock-infected transfection.
VP35 blocks the IRF-3 dependent, virus-induced activation of the mouse IFN-α4 promoter.
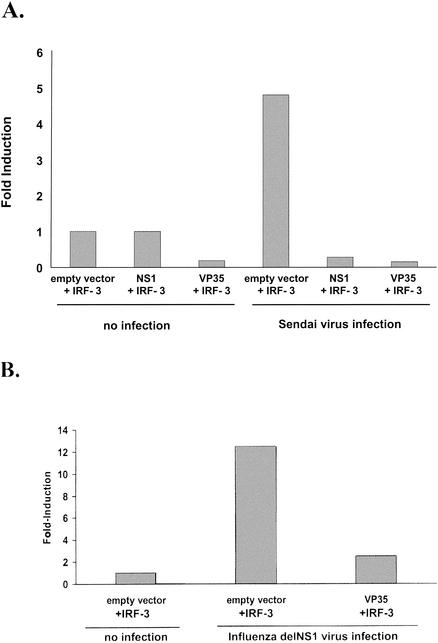
The ISG54 and ISG56 promoters can both be transcriptionally activated directly by IRF-3, without production of IFN, in response to viral infection (26, 54, 63), suggesting that VP35 blocks IRF-3-dependent transcription. We therefore investigated the ability of VP35 to prevent the induction of an additional IRF-3-stimulated promoter, the mouse IFN-α4 promoter. Overexpression of IRF-3 has previously been reported to cooperate with viral infection to activate the IFN-α4 promoter (35). We were therefore able to establish a transfection system where the virus-induced activation of the IFN-α4 promoter was dependent on overexpression of IRF-3, because in the absence of IRF-3 overexpression, SeV infection did not lead to reporter gene induction (data not shown). Using this system, the Ebola virus Zaire VP35 was tested for its ability to inhibit the virus-induced activation of mouse IFN-α4 promoter in the presence of overexpressed IRF-3. Cells were transfected with an IFN-α4-CAT plasmid, an IRF-3 expression plasmid, and either empty vector, NS1 expression plasmid, or VP35 expression plasmid. In the absence of viral infection, little CAT activity was detected, and in the presence of VP35, the basal CAT activity was lower than in empty vector-transfected cells (Fig. 2A). When the cells were infected with SeV, activation of the IFN-α4 promoter increased fivefold. The presence of VP35 completely blocked this virus-induced activation (Fig. 2A). As expected, a previously described IRF-3 inhibitor, the influenza A virus NS1 protein, also blocked promoter activation (Fig. 2A). Similar results were found when influenza delNS1 virus, a mutant influenza virus lacking the IFN antagonist NS1 protein (23), was used instead of SeV to induce IFN-α4 promoter activity (Fig. 2B). As in the SeV experiment, almost no CAT activity could be detected in cells, whether mock infected or delNS1 virus-infected, which were not overexpressing IRF-3 (data not shown). Combined, these data suggest that the Ebola virus VP35 protein can block virus-induced, IRF-3-dependent gene expression.
FIG. 2.
The VP35 protein of Ebola virus inhibits IRF-3-dependent induction of the mouse IFN-α4 promoter. The relative levels of IRF-3-dependent, virus-induced expression from a mouse IFN-α4 promoter reporter plasmid in the presence of empty vector, influenza virus NS1 protein, or Ebola virus VP35 are indicated following infection with SeV (A) or influenza virus delNS1 virus (B). 293T cells were calcium phosphate transfected with a mouse IFN-α4 promoter-driven CAT reporter plasmid and a constitutively expressed Renilla luciferase expression plasmid (pRL-tk), an IRF-3 expression plasmid, and an excess of either empty vector, an influenza virus NS1 expression plasmid, or VP35 expression plasmid. Eighteen hours posttransfection, the cells were infected with SeV (MOI = 1) (A) or influenza delNS1 virus (MOI = 1) (B). All transfections were normalized to the value for uninfected, empty vector-transfected cells that received the IRF-3 expression plasmid (left-most column). No reporter gene expression could be detected in cells in which IRF-3 was not overexpressed, either in mock-infected or in virus-infected cells.
VP35 blocks IRF-3 nuclear translocation.
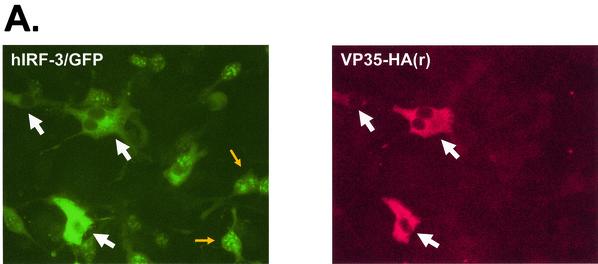
Given the ability of VP35 to inhibit IRF-3-mediated gene expression, it was important to define how IRF-3 function was blocked. Virus infection can induce serine/threonine phosphorylation near the carboxy-terminus of IRF-3 (45, 46, 74, 94). This phosphorylation leads to the dimerization and nuclear accumulation of IRF-3 (40, 46, 74, 94). In order to begin to assess the mechanism by which VP35 inhibits IRF-3-dependent gene activation, we assessed the ability of VP35 to inhibit the nuclear accumulation of IRF-3 by using GFP-IRF-3 fusions, which have been used by a several investigators to monitor the nuclear accumulation of IRF-3 (30, 40, 45, 79). To demonstrate that inhibition of GFP-IRF-3 nuclear accumulation correlated with VP35 expression, Vero cells were transfected with an excess of GFP-IRF-3 and limiting amounts of an HA-tagged Ebola virus Reston VP35 expression plasmid (Fig. 3A). All cells expressing HA-tagged Ebola virus Reston VP35 showed exclusively cytoplasmic GFP-IRF-3 localization following SeV infection. Of those cells not expressing VP35, the large majority showed nuclear accumulation of IRF-3, providing further evidence that VP35 expression blocks virus-induced nuclear accumulation of IRF-3 (Fig. 3A).
FIG. 3.
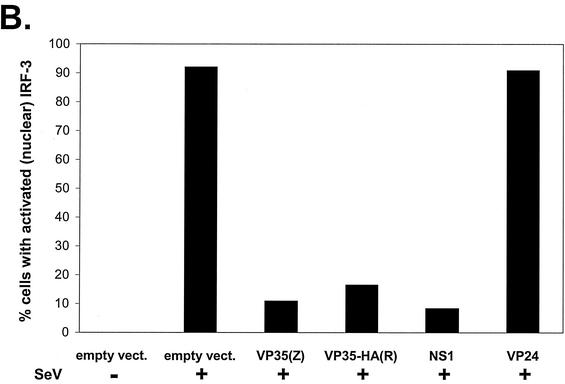
The Ebola virus VP35 protein prevents the nuclear translocation of hIRF-3 after SeV infection. (A) Fluorescence images showing expression of HA-tagged VP35(R) protein (red) and the corresponding distribution of GFP-hIRF-3 (green). Cells expressing both HA-tagged VP35 and GFP-IRF-3 are indicated by the large white arrows. Examples of cells with nuclear GFP-IRF-3 are indicated by the small yellow arrows. Vero cells were transfected with 0.4 μg of VP35(R) expression plasmid plus 0.8 μg of pEGFP-C1-hIRF3 and infected 24 h later with SeV. Eight hours postinfection, cells were fixed and stained with anti-HA monoclonal antibody (red). (B) The percentage of GFP-IRF-3-expressing cells with nuclear GFP-IRF-3 in cells transfected with the indicated plasmids and either mock infected or infected with SeV is shown. Vero cells were transfected with 0.4 μg of empty vector or expression plasmids for Ebola virus Zaire VP35 [VP35(Z)], HA-tagged Ebola virus Reston VP35 [HAVP35(R)], influenza virus NS1 protein (NS1), or Ebola virus Zaire virus VP24 protein (VP24), plus 0.2 μg of pEGFP-C1-hIRF3. At 24 h posttransfection, the cells were mock infected or infected with SeV at an MOI of 10. Eight hours postinfection, the cells were examined for GFP localization. The results are the average of two independent experiments where 200 to 300 cells were counted per transfection.
Vero cells were also transfected with an excess of either protein expression plasmid or empty vector and a limiting amount of GFP-IRF-3 plasmid. In the absence of additional treatment, the vast majority of GFP-expressing cells displayed exclusively cytoplasmic IRF-3 localization (Fig. 3B). When cultures transfected with GFP-IRF-3 and empty vector were subsequently infected with SeV, approximately 90% of GFP-positive cells showed nuclear accumulation of IRF-3 (Fig. 3B). In contrast, in cells cotransfected with Ebola virus Zaire VP35 or HA-tagged Ebola virus Reston VP35, nuclear accumulation of IRF-3 was drastically reduced (Fig. 3B). Similarly, expression of the influenza virus NS1 protein, previously reported to inhibit nuclear accumulation of IRF-3 in response to viral infection, also reduced GFP-IRF-3 nuclear accumulation (Fig. 3B). In contrast, a different Ebola virus protein, VP24, did not block virus-induced nuclear accumulation of GFP-IRF-3 (Fig. 3B). Our ability to differentiate nuclear from cytoplasmic IRF-3 is illustrated in Fig. 3A.
VP35 blocks virus-induced IRF-3 dimerization and phosphorylation.
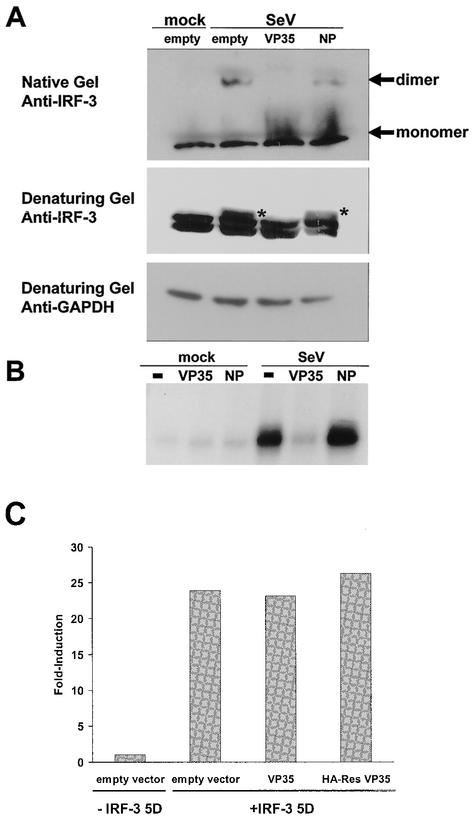
Experiments were then performed to determine whether the VP35-mediated inhibition of IRF-3 nuclear localization might be due to an inhibition of IRF-3 phosphorylation and/or dimerization. 293T cells were transfected with a human IRF-3 expression plasmid and either empty vector or plasmids expressing Ebola virus Zaire VP35, HA-tagged Ebola virus Reston VP35, or Ebola virus Zaire NP proteins and subsequently mock infected or infected with SeV. We employed a previously described native PAGE method which separates IRF-3 monomers and dimers and detects IRF-3 by immunoblotting (34). This is a sensitive method to distinguish inactive (monomeric) from active (dimeric) IRF-3 (34). By this method, SeV infection was found to induce formation of IRF-3 dimers by 4 h postinfection. Expression of VP35 blocked IRF-3 dimerization, whereas that of Ebola virus NP did not (Fig. 4A, upper panel). The same cell extracts used for native gel electrophoresis were also analyzed by standard SDS-PAGE and Western blotting. Infection of empty vector-transfected cells with SeV induced a slower migrating form of IRF-3 (Fig. 4A, middle panel). Similar virus infection-induced shifts in IRF-3 have previously been ascribed to phosphorylation of serine and threonine residues near its carboxy terminus (72). The expression of VP35 blocked the formation of the slower migrating form of IRF-3, whereas that of Ebola virus NP proteins did not (Fig. 4A, middle panel), suggesting that VP35 inhibits IRF-3 phosphorylation. Further confirmation that VP35 blocks IRF-3 phosphorylation was shown by immunoprecipitating IRF-3 from SeV-infected cells radiolabeled with [32P]orthophosphate. Whereas IRF-3 phosphorylation was greatly enhanced by SeV infection in empty vector-transfected cells or NP-expressing cells, IRF-3 phosphorylation was completely blocked in VP35-expressing cells (Fig. 4B).
FIG. 4.
VP35 blocks dimerization and phosphorylation of IRF-3 in response to SeV infection. (A) Immunoblot analysis of IRF-3 in mock-infected (mock) and SeV-infected (SeV) 293 cells transfected with empty vector, VP35 expression plasmid, or NP expression plasmid. IRF-3 monomers and dimers were visualized following native gel electrophoresis, and different IRF-3 forms, including virus-induced, phosphorylated forms (indicated by asterisks), were visualized following SDS-PAGE of the same extracts used on the native gel. An anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) blot is shown as a loading control. (B) Phosphorylated IRF-3 detected by [32P]orthophosphate labeling and immunoprecipitation of IRF-3 from mock- or SeV-infected 293 cells previously transfected with 1 μg of IRF-3 expression plasmid and 2 μg of empty vector, VP35 expression plasmid, or NP expression plasmid. (C) The relative ability of IRF-3 5D to activate an ISG56 promoter-driven reporter gene in Vero cells cotransfected with empty vector, VP35 expression plasmid, or NP expression plasmid. The values are reported as fold induction of the reporter gene relative to induction in cells receiving empty vector and no IRF-3 5D plasmid. All values were normalized to expression from a cotransfected, constitutively expressed Renilla luciferase reporter gene.
In order to determine whether the ability of VP35 to inhibit IRF-3 transcriptional activation is dependent upon its ability to block IRF-3 phosphorylation, we asked whether VP35 could block transcriptional activation mediated by a constitutively activated IRF-3 mutant, IRF-3 5D. In this mutant, phosphomimetic aspartic acid residues are substituted for five serine and threonine residues (Ser 396, 398, 402, and 405 and Thr 404) which become phosphorylated in response to virus infection (45). IRF-3 5D activates transcription in 293 cells independent of virus infection (but reportedly did not constitutively promote gene expression in L929 cells [77]). A tenfold excess of either VP35 or HA-tagged Reston VP35 expression plasmid was unable to inhibit the IRF-3 5D-mediated activation of the human ISG56 promoter (Fig. 4C). This strongly suggests that VP35 blocks IRF-3 activation by preventing IRF-3 phosphorylation.
IRF-3 is not activated in Ebola virus-infected Vero cells.
If VP35 expression blocks activation of IRF-3 in response to infection with heterologous viruses, such as SeV and influenza delNS1 virus, it would be expected that Ebola virus infection would not activate the ISG54 promoter and would not promote nuclear accumulation of IRF-3. To test whether Ebola virus infection activates the ISG54 promoter, 293 cells were transfected with the ISG54-CAT reporter plasmid. One day posttransfection, the cells were infected with either SeV, Ebola virus Zaire, or Ebola virus Reston, each at an MOI of 3, or mock infected. After an incubation period of 24 h, CAT activity was determined. In dishes infected in parallel, the Ebola virus Reston-infected and mock-infected cells were stained for Ebola virus antigen. Despite the efficient infection of the cells by Ebola viruses, no activation of the ISG54 promoter was detected (Fig. 5A). In contrast, SeV infection strongly activated the ISG54 promoter (Fig. 5A). To test whether Ebola virus infection promotes IRF-3 nuclear translocation, Vero cells were transfected with the GFP-IRF-3 expression plasmid and 1 day later were mock infected or infected with Ebola virus Zaire. The cells were then fixed at 24 or 48 h postinfection and stained with antibodies against the Ebola virus surface glycoprotein GP. Infection did not induce significant GFP-IRF-3 nuclear accumulation at either 24 (data not shown) or 48 h postinfection despite the presence of significant amounts of viral antigen (Fig. 5B). These observations are consistent with the presence of an Ebola virus-encoded inhibitor of IRF-3 activation.
FIG. 5.
Ebola virus infection does not induce the activation of the ISG54 promoter or IRF-3 nuclear accumulation. (A) Ebola virus infection does not activate the ISG54 promoter. 293 cells were transfected with the IFN and IRF-3-responsive reporter plasmid pHIG54-CAT and, 24 h posttransfection, infected with the indicated viruses at an MOI of 3. Twenty-four hours posttransfection, the cells were either fixed and stained for the viral GP surface antigen (left panels) or lysed for CAT assays. The mock-infected immunofluorescence shows a background of red staining, but this is clearly distinguishable from the viral antigen staining in the infected cells. (B) Ebola virus infection did not induce GFP-IRF-3 nuclear accumulation. Cells were mock infected or infected with Ebola virus Zaire. Vero cells were transfected with the GFP-IRF-3 expression plasmid. Twenty-four hours posttransfection, the cells were mock infected or infected with Ebola virus Zaire at an MOI of 1. At 48 h postinfection, the cells were fixed and stained with an anti-GP antibody (red) and with DAPI stain (blue) to identify the nucleus. The top panels show GFP-IRF-3 and viral antigen staining; the bottom panels show the same fields as the top panels but with DAPI staining included to identify the nuclei. One field of uninfected cells is shown on the left. Two fields of virus-infected cells are shown, one in the center and one on the right.
DISCUSSION
This study demonstrates that the Ebola virus VP35 protein inhibits the activation of IRF-3 in response to viral infection. IRF-3 is a cellular transcription factor that plays a central role in the initiating the host cell IFN response to viral infection (for recent reviews, see references 3, 5, 43, 66, 72, 81, and 93). Activation of IRF-3 contributes to the immediate activation of IFN-β gene expression and of select IFN-α genes as well as the immediate expression of several other genes with potential antiviral activity (26). Previously, we had found that expression of VP35 could complement the growth of a mutant influenza virus, influenza delNS1 virus, which lacks the influenza virus IFN antagonist NS1 protein (7). VP35 had also been found to block activation of the IFN-β promoter by dsRNA or by virus infection, to inhibit SeV- or influenza delNS1 virus-induced transcription of the endogenous IFN-β promoter, and to inhibit the virus-induced activation of the ISG54 promoter (7). In the present study, we have demonstrated that VP35 expression inhibits the virus-induced activation of both the ISG54 promoter and the ISG56 promoter in Vero cells. In contrast, VP35 was not able to block the activation of either promoter following treatment of cells with IFN-β. Because Vero cells do not produce IFN-α/β, which might otherwise activate the ISG54 and ISG56 promoters (51), and because both the ISG54 and ISG56 promoters can be activated directly by IRF-3 (26, 54), these results suggested that VP35 could block IRF-3 mediated activation of these genes. Furthermore, VP35 expression inhibited virus-induced, IRF-3-dependent activation of the mouse IFN-α4 promoter (Fig. 2). These results are consistent with earlier studies demonstrating that Ebola virus infection blocks host cell responses to both dsRNA, which is often used as an experimental inducer of IFN responses, and IFN (28, 29). They are also consistent with studies showing that Ebola virus infection does not induce production of IFN in some cell types and can block the dsRNA-induced production of IFN-α/β in primary human peripheral blood mononuclear cells and monocytes (27).
Our results also address the manner in which IRF-3 is inhibited by VP35. Our data using a GFP-IRF-3 fusion protein as a marker for cellular IRF-3 localization indicate that the presence of VP35 prevents the virus-induced nuclear accumulation of IRF-3 (Fig. 3). In order to define how VP35 affects the IRF-3 activation process, we examined the dimerization and phosphorylation of IRF-3. Expression of VP35 was able to prevent IRF-3 dimerization and virus-induced phosphorylation of IRF-3 (Fig. 4). That expression of VP35 was unable to inhibit the function of IRF-3 5D, which does not need to be phosphorylated to stimulate gene expression, argues strongly for a model whereby VP35 functions specifically by blocking IRF-3 phosphorylation.
Several viruses, including human cytomegalovirus, are known to activate IRF-3 through interactions with the host cell prior to viral entry (55, 65). Several other viruses encode proteins which are known to inhibit the function of IRF-3. The influenza A virus NS1 protein prevents IRF-3 activation and blocks production of IFN-α/β in influenza virus-infected cells (23, 79). This activity appears to require the dsRNA-binding activity of NS1, suggesting a model where NS1 binds to dsRNA produced during viral infection and prevents the activation of dsRNA-downstream pathways leading to IRF-3 phosphorylation (79). Similarly, the vaccinia virus E3L protein, also a dsRNA-binding protein, inhibits IRF-3 activation (76); whereas a vaccinia virus lacking E3L activates IRF-3 upon infection, wild-type vaccinia virus does not (92). The human papillomavirus 16 E6 protein has been shown to bind IRF-3 and to inhibit IRF-3 transcriptional activity (44, 67). HHV-8 vIRF-1 and vIRF-3 have also been shown to bind to IRF-3 (3, 44). The reported ability of vIRF-1 to interact directly with IRF-3, to associate with CBP/p300, to inhibit CBP/p300 transcriptional activation, and to compete with IRF-3 for binding to CBP/p300, may play a role in the ability of vIRF-1 to affect IRF-3-related transcriptional activity (16, 44, 71). The ability of vIRF-3 to influence virus-induced IFN-α gene expression may be related to its ability to interact with IRF-3 and/or CBP/p300 (3). Recently, the rotavirus NSP1 protein was shown to interact with IRF-3, although it has not yet been reported to inhibit IRF-3 function (25). Also, noncytopathogenic bovine viral diarrhea virus blocks IFN-α/β production by preventing formation of IRF-3-DNA complexes, although the mechanism by which this occurs is, as yet, undefined (2). Finally, the V proteins of paramyxoviruses, which were previously shown to block signaling from the IFN-α/β receptor by promoting degradation of STAT proteins (1, 20, 39, 60, 61), have been reported to inhibit IFN-β gene expression and to block dsRNA-induced nuclear translocation of an IRF-3-GFP fusion protein (30, 64).
Our data suggest that VP35 inhibits IRF-3 activation primarily by inhibiting phosphorylation. The identification of host cell factors that interact with VP35 would shed light on its mechanism of action, and experiments to identify such proteins are under way. Experiments to determine whether VP35 interacts directly with IRF-3 have thus far yielded only negative results. The ability of VP35 to prevent IRF-3 phosphorylation suggests the possibility that VP35 may interact with some component of the virus-activated signaling pathway which leads to IRF-3 activation. It will also be of interest to examine the effect of VP35 on other IRF family members. The influenza virus NS1 and vaccinia virus E3L proteins reportedly also prevent IRF-7 activation (48, 76), and other viral IRF-7 inhibitors have been described (96). IRF-7 and IRF-5 are of particular interest because they are involved in direct virus-induced activation of IFN-α genes (4, 48, 50, 70).
We have found that the Ebola virus Reston VP35 behaves similarly to the Ebola Zaire virus VP35 with regard to IFN antagonist functions, at least in the cell types tested. The Reston VP35 inhibits activation of the ISG54 promoter. Like the Zaire VP35, Reston VP35 blocks GFP-IRF-3 nuclear accumulation and appears to inhibit IRF-3 phosphorylation in response to SeV infection. In addition, infection of human-derived 293 cells with Ebola virus Reston did not activate the ISG54 promoter. Ebola virus Reston causes lethal disease in nonhuman primates but did not cause disease in the few documented cases of human exposure. Animal handlers caring for Ebola Reston virus-infected cynomolgus macaques did not become ill but did seroconvert, suggesting that they experienced subclinical infections (12, 18). In contrast, the Zaire subtype of Ebola virus appears to be the most lethal filovirus in humans, and outbreaks of Ebola virus Zaire have had reported fatality rates approaching 90 percent (69). Based on these observations, some have suggested that the Reston strain is avirulent in humans; however, the possibility exists that Ebola virus Reston may cause human disease under some circumstances (12). Given the possible differences between Ebola Zaire and Ebola Reston viruses in terms of human virulence, comparisons of these viruses are therefore of great interest. At this point our data do not suggest differences in IFN antagonist function for Zaire versus Reston virus VP35s. However, we cannot presently rule out the possibility that there exist species-specific or cell type-specific differences between the VP35s of different filoviruses.
The IFN antagonist activity of the VP35 protein is likely to play a significant role in Ebola virus pathogenesis. Data from several viruses have demonstrated that functional IFN antagonists are required for virulence in animal models. For example, an influenza virus lacking the IFN-antagonist NS1 protein, influenza delNS1 virus, is avirulent in BALB/c mice, whereas the isogenic wild-type influenza A/PR/8/34 (H1N1) virus (PR8 virus) readily kills mice (23). However, the influenza delNS1 virus kills STAT1-null mice, which lack a critical component of the IFN-α/β system (23). The ability of influenza delNS1 virus to kill mice with defective IFN-signaling pathways demonstrates that its attenuation in wild-type mice is related to the IFN system. Similarly, we would predict that an Ebola virus lacking an effective IFN antagonist activity would be attenuated, and the inability of wild-type Ebola viruses to cause disease in mice may reflect an inability of Ebola viruses to counteract the mouse IFN system (13). Our present results would predict that in the absence of VP35 IFN antagonist function, large amounts of IFN-α/β would be produced, inducing in cells an antiviral state and leading to inhibition of viral spread. In addition, VP35 may assist Ebola virus in evading other aspects of the host antiviral response, as IFN-α/β appear to influence adaptive immune responses (6). In particular, IFN-α/β production has been shown to influence the production of other immunoregulatory cytokines, including interleukin-15 and IFN-γ, and to influence NK cell and dendritic cell function (reviewed in references 9 and 59). Interestingly, Ebola virus infection has recently been reported to impair human dendritic cell function (47). It is possible that the ability of VP35 to inhibit IFN production may contribute to the ability of Ebola virus to inhibit dendritic cell functions.
While our data provide insights into the mechanism by which VP35 blocks IFN production, the overall impact of the IFN antagonist function of VP35 on Ebola virus replication remains to be determined. Several recently developed technologies are likely to prove useful in elucidating the functional significance of the VP35 IFN antagonist activity. Reverse genetics methods have been established for Ebola virus, permitting the introduction of specific alterations into the viral genome (56, 83). These techniques hold the promise of creating viruses devoid of VP35 anti-IFN activity. However, the prospect of creating such a mutant VP35 virus is complicated by the other functions of VP35, including its essential role in viral RNA synthesis and its role in viral assembly (33, 52, 53). We are seeking to identify mutations that eliminate VP35 anti-IFN function without significantly impairing its other functions. The developing genomics and proteomics technologies also hold great promise in helping to define the impact of viral infection and viral gene expression on the host cell (37). Recently, microarray technology has been employed to study the impact of both wild-type and mutant NS1 IFN-antagonist proteins of influenza A viruses (24). Such techniques could be employed to analyze the IFN response to Ebola virus infection in different cell types and to compare the IFN responses in cells infected with different subtypes of Ebola virus. These techniques could also be used to examine the specific influence of VP35s of different filovirus strains in different cell types. Ultimately, it is hoped that our current and future studies on VP35 will enhance our understanding of the pathogenesis of filoviruses and identify new targets for antiviral therapies.
Acknowledgments
C.F.B. and A.M. contributed equally to this work.
This work was supported by NIH grants to C.F.B., A.G.-S., and P.P. C.F.B. is an Ellison Medical Foundation New Scholar in Global Infectious Diseases. J.P. is a recipient of a National Research Council Fellowship.
We thank Neva Morales and Estanislao Nistal-Villan for expert technical assistance. We thank John Hiscott (McGill University) for helpful discussions and providing the ISG56 promoter reporter plasmid. We thank Peter Howley (Harvard Medical School) for providing the SL-12 antibody.
REFERENCES
- 1.Andrejeva, J., D. F. Young, S. Goodbourn, and R. E. Randall. 2002. Degradation of STAT1 and STAT2 by the V proteins of simian virus 5 and human parainfluenza virus type 2, respectively: consequences for virus replication in the presence of alpha/beta and gamma interferons. J. Virol. 76:2159-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baigent, S. J., G. Zhang, M. D. Fray, H. Flick-Smith, S. Goodbourn, and J. W. McCauley. 2002. Inhibition of beta interferon transcription by noncytopathogenic bovine viral diarrhea virus is through an interferon regulatory factor 3-dependent mechanism. J. Virol. 76:8979-8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes, B., B. Lubyova, and P. M. Pitha. 2002. On the role of IRF in host defense. J. Interferon Cytokine Res. 22:59-71. [DOI] [PubMed] [Google Scholar]
- 4.Barnes, B. J., P. A. Moore, and P. M. Pitha. 2001. Virus-specific activation of a novel interferon regulatory factor, IRF-5, results in the induction of distinct interferon alpha genes. J. Biol. Chem. 276:23382-23390. [DOI] [PubMed] [Google Scholar]
- 5.Basler, C. F., and A. Garcia-Sastre. 2002. Viruses and the type I interferon antiviral system: induction and evasion. Int. Rev. Immunol., in press. [DOI] [PubMed]
- 6.Basler, C. F., and P. Palese. Modulation of innate immunity by filoviruses. In H.-D. Klenk and H. Feldmann (ed.), Molecular and Cellular Biology of Marburg and Ebola Viruses, in press. Horizon Scientific Press, Norfolk, United Kingdom.
- 7.Basler, C. F., X. Wang, E. Muhlberger, V. Volchkov, J. Paragas, H. D. Klenk, A. Garcia-Sastre, and P. Palese. 2000. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc. Natl. Acad. Sci. USA 97:12289-12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beattie, E., K. L. Denzler, J. Tartaglia, M. E. Perkus, E. Paoletti, and B. L. Jacobs. 1995. Reversal of the interferon-sensitive phenotype of a vaccinia virus lacking E3L by expression of the reovirus S4 gene. J. Virol. 69:499-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biron, C. A. 2001. Interferons alpha and beta as immune regulators—a new look. Immunity 14:661-664. [DOI] [PubMed] [Google Scholar]
- 10.Bouloy, M., C. Janzen, P. Vialat, H. Khun, J. Pavlovic, M. Huerre, and O. Haller. 2001. Genetic evidence for an interferon-antagonistic function of rift valley fever virus nonstructural protein NSs. J. Virol. 75:1371-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brandt, T. A., and B. L. Jacobs. 2001. Both carboxy- and amino-terminal domains of the vaccinia virus interferon resistance gene, E3L, are required for pathogenesis in a mouse model. J. Virol. 75:850-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bray, M. 2002. Filoviridae. In D. D. Richman, R. J. Whitely, and F. G. Hayden (ed.), Clinical virology, 2nd ed. ASM Press, Washington, D.C.
- 13.Bray, M. 2001. The role of the type I interferon response in the resistance of mice to filovirus infection. J. Gen. Virol. 82:1365-1373. [DOI] [PubMed] [Google Scholar]
- 14.Bray, M., and J. Paragas. 2002. Experimental therapy of filovirus infections. Antiviral Res. 54:1-17. [DOI] [PubMed] [Google Scholar]
- 15.Bridgen, A., F. Weber, J. K. Fazakerley, and R. M. Elliott. 2001. Bunyamwera bunyavirus nonstructural protein NSs is a nonessential gene product that contributes to viral pathogenesis. Proc. Natl. Acad. Sci. USA 98:664-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burysek, L., W. S. Yeow, B. Lubyova, M. Kellum, S. L. Schafer, Y. Q. Huang, and P. M. Pitha. 1999. Functional analysis of human herpesvirus 8-encoded viral interferon regulatory factor 1 and its association with cellular interferon regulatory factors and p300. J. Virol. 73:7334-7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Centers for Disease Control and Prevention. 2000. Biological and chemical terrorism: strategic plan for preparedness and response. Recommendations of the CDC Strategic Planning Workgroup. Morb. Mortal. Wkly. Rep. 49:1-14. [PubMed] [Google Scholar]
- 18.Centers for Disease Control and Prevention. 1990. Update: filovirus infection in animal handlers. Morb. Mortal. Wkly. Rep. 39:221. [PubMed] [Google Scholar]
- 19.Chou, J., E. R. Kern, R. J. Whitley, and B. Roizman. 1990. Mapping of herpes simplex virus-1 neurovirulence to gamma 134.5, a gene nonessential for growth in culture. Science 250:1262-1266. [DOI] [PubMed] [Google Scholar]
- 20.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of simian virus 5 inhibits interferon signaling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 73:9928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feldmann, H., V. E. Volchkov, V. A. Volchkova, U. Stroher, and H. D. Klenk. 2001. Biosynthesis and role of filoviral glycoproteins. J. Gen. Virol. 82:2839-2848. [DOI] [PubMed] [Google Scholar]
- 21a.Fitzgerald, K. A., S. M. McWhirter, K. L. Faia, D. C. Rowe, E. Latz, D. T. Golenbock, A. J. Coyle, S. M. Liao, and T. Maniatis. 2003. IKKɛ and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491-496. [DOI] [PubMed] [Google Scholar]
- 22.Garcia-Sastre, A. 2001. Inhibition of interferon-mediated antiviral responses by influenza A viruses and other negative-strand RNA viruses. Virology 279:375-384. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Sastre, A., A. Egorov, D. Matassov, S. Brandt, D. E. Levy, J. E. Durbin, P. Palese, and T. Muster. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324-330. [DOI] [PubMed] [Google Scholar]
- 24.Geiss, G. K., M. Salvatore, T. M. Tumpey, V. S. Carter, X. Wang, C. F. Basler, J. K. Taubenberger, R. E. Bumgarner, P. Palese, M. G. Katze, and A. Garcia-Sastre. 2002. Cellular transcriptional profiling in influenza A virus-infected lung epithelial cells: the role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza. Proc. Natl. Acad. Sci. USA 99:10736-10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graff, J. W., D. N. Mitzel, C. M. Weisend, M. L. Flenniken, and M. E. Hardy. 2002. Interferon regulatory factor 3 is a cellular partner of rotavirus NSP1. J. Virol. 76:9545-9550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grandvaux, N., M. J. Servant, B. tenOever, G. C. Sen, S. Balachandran, G. N. Barber, R. Lin, and J. Hiscott. 2002. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J. Virol. 76:5532-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gupta, M., S. Mahanty, R. Ahmed, and P. E. Rollin. 2001. Monocyte-derived human macrophages and peripheral blood mononuclear cells infected with Ebola virus secrete MIP-1alpha and TNF-alpha and inhibit poly-IC-induced IFN-alpha in vitro. Virology 284:20-25. [DOI] [PubMed] [Google Scholar]
- 28.Harcourt, B. H., A. Sanchez, and M. K. Offermann. 1998. Ebola virus inhibits induction of genes by double-stranded RNA in endothelial cells. Virology 252:179-188. [DOI] [PubMed] [Google Scholar]
- 29.Harcourt, B. H., A. Sanchez, and M. K. Offermann. 1999. Ebola virus selectively inhibits responses to interferons, but not to interleukin-1β, in endothelial cells. J. Virol. 73:3491-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He, B., R. G. Paterson, N. Stock, J. E. Durbin, R. K. Durbin, S. Goodbourn, R. E. Randall, and R. A. Lamb. 2002. Recovery of paramyxovirus simian virus 5 with a V protein lacking the conserved cysteine-rich domain: the multifunctional V protein blocks both interferon-beta induction and interferon signaling. Virology 303:15-32. [DOI] [PubMed] [Google Scholar]
- 31.Hensley, L. E., H. A. Young, P. B. Jahrling, and T. W. Geisbert. 2002. Proinflammatory response during Ebola virus infection of primate models: possible involvement of the tumor necrosis factor receptor superfamily. Immunol. Lett. 80:169-179. [DOI] [PubMed] [Google Scholar]
- 32.Heylbroeck, C., S. Balachandran, M. J. Servant, C. DeLuca, G. N. Barber, R. Lin, and J. Hiscott. 2000. The IRF-3 transcription factor mediates Sendai virus-induced apoptosis. J. Virol. 74:3781-3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang, Y., L. Xu, Y. Sun, and G. Nabel. 2002. The assembly of Ebola virus nucleocapsid requires virion-associated proteins 35 and 24 and posttranslational modification of nucleoprotein. Mol. Cell 10:307. [DOI] [PubMed] [Google Scholar]
- 34.Iwamura, T., M. Yoneyama, K. Yamaguchi, W. Suhara, W. Mori, K. Shiota, Y. Okabe, H. Namiki, and T. Fujita. 2001. Induction of IRF-3/-7 kinase and NF-kappaB in response to double-stranded RNA and virus infection: common and unique pathways. Genes Cells 6:375-388. [DOI] [PubMed] [Google Scholar]
- 35.Juang, Y., W. Lowther, M. Kellum, W. C. Au, R. Lin, J. Hiscott, and P. M. Pitha. 1998. Primary activation of interferon A and interferon B gene transcription by interferon regulatory factor 3. Proc. Natl. Acad. Sci. USA 95:9837-9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karpova, A. Y., M. Trost, J. M. Murray, L. C. Cantley, and P. M. Howley. 2002. Interferon regulatory factor-3 is an in vivo target of DNA-PK. Proc. Natl. Acad. Sci. USA 99:2818-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katze, M. G., Y. He, and M. Gale. 2002. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2:675-687. [DOI] [PubMed] [Google Scholar]
- 38.Kawai, T., O. Takeuchi, T. Fujita, J. Inoue, P. F. Muhlradt, S. Sato, K. Hoshino, and S. Akira. 2001. Lipopolysaccharide stimulates the MyD88-independent pathway and results in activation of IFN-regulatory factor 3 and the expression of a subset of lipopolysaccharide-inducible genes. J. Immunol. 167:5887-5894. [DOI] [PubMed] [Google Scholar]
- 39.Kubota, T., N. Yokosawa, S. Yokota, and N. Fujii. 2001. C terminal CYS-RICH region of mumps virus structural V protein correlates with block of interferon alpha and gamma signal transduction pathway through decrease of STAT 1-alpha. Biochem. Biophys. Res. Commun. 283:255-259. [DOI] [PubMed] [Google Scholar]
- 40.Kumar, K. P., K. M. McBride, B. K. Weaver, C. Dingwall, and N. C. Reich. 2000. Regulated nuclear-cytoplasmic localization of interferon regulatory factor 3, a subunit of double-stranded RNA-activated factor 1. Mol. Cell. Biol. 20:4159-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lane, H. C., J. L. Montagne, and A. S. Fauci. 2001. Bioterrorism: a clear and present danger. Nat. Med. 7:1271-1273. [DOI] [PubMed] [Google Scholar]
- 42.Leib, D. A., T. E. Harrison, K. M. Laslo, M. A. Machalek, N. J. Moorman, and H. W. Virgin. 1999. Interferons regulate the phenotype of wild-type and mutant herpes simplex viruses in vivo. J. Exp. Med. 189:663-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levy, D. E., and A. Garcia-Sastre. 2001. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 12:143-156. [DOI] [PubMed] [Google Scholar]
- 44.Lin, R., P. Genin, Y. Mamane, M. Sgarbanti, A. Battistini, W. J. Harrington, Jr., G. N. Barber, and J. Hiscott. 2001. HHV-8 encoded vIRF-1 represses the interferon antiviral response by blocking IRF-3 recruitment of the CBP/p300 coactivators. Oncogene 20:800-811. [DOI] [PubMed] [Google Scholar]
- 45.Lin, R., C. Heylbroeck, P. M. Pitha, and J. Hiscott. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18:2986-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin, R., Y. Mamane, and J. Hiscott. 1999. Structural and functional analysis of interferon regulatory factor 3: localization of the transactivation and autoinhibitory domains. Mol. Cell. Biol. 19:2465-2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mahanty, S., K. Hutchinson, S. Agarwal, M. McRae, P. E. Rollin, and B. Pulendran. 2003. Cutting edge: impairment of dendritic cells and adaptive immunity by Ebola and Lassa viruses. J. Immunol. 170:2797-2801. [DOI] [PubMed] [Google Scholar]
- 48.Marie, I., J. E. Durbin, and D. E. Levy. 1998. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 17:6660-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Markovitz, N. S., D. Baunoch, and B. Roizman. 1997. The range and distribution of murine central nervous system cells infected with the γ134.5− mutant of herpes simplex virus 1. J. Virol 71:5560-5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morin, P., J. Braganca, M. T. Bandu, R. Lin, J. Hiscott, J. Doly, and A. Civas. 2002. Preferential binding sites for interferon regulatory factors 3 and 7 involved in interferon-A gene transcription. J. Mol. Biol. 316:1009-1022. [DOI] [PubMed] [Google Scholar]
- 51.Mosca, J. D., and P. M. Pitha. 1986. Transcriptional and posttranscriptional regulation of exogenous human beta interferon gene in simian cells defective in interferon synthesis. Mol. Cell. Biol. 6:2279-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muhlberger, E., B. Lotfering, H. D. Klenk, and S. Becker. 1998. Three of the four nucleocapsid proteins of Marburg virus, NP, VP35, and L, are sufficient to mediate replication and transcription of Marburg virus-specific monocistronic minigenomes. J. Virol. 72:8756-8764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muhlberger, E., M. Weik, V. E. Volchkov, H. D. Klenk, and S. Becker. 1999. Comparison of the transcription and replication strategies of Marburg virus and Ebola virus by using artificial replication systems. J. Virol. 73:2333-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakaya, T., M. Sato, N. Hata, M. Asagiri, H. Suemori, S. Noguchi, N. Tanaka, and T. Taniguchi. 2001. Gene induction pathways mediated by distinct IRFs during viral infection. Biochem. Biophys. Res. Commun. 283:1150-1156. [DOI] [PubMed] [Google Scholar]
- 55.Navarro, L., K. Mowen, S. Rodems, B. Weaver, N. Reich, D. Spector, and M. David. 1998. Cytomegalovirus activates interferon immediate-early response gene expression and an interferon regulatory factor 3-containing interferon-stimulated response element-binding complex. Mol. Cell. Biol. 18:3796-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neumann, G., H. Feldmann, S. Watanabe, I. Lukashevich, and Y. Kawaoka. 2002. Reverse genetics demonstrates that proteolytic processing of the Ebola virus glycoprotein is not essential for replication in cell culture. J. Virol. 76:406-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 58.Oshiumi, H., M. Matsumoto, K. Funami, T. Akazawa, and T. Seya. 2003. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat. Immunol. 4:161-167. [DOI] [PubMed] [Google Scholar]
- 59.Ozato, K., H. Tsujimura, and T. Tamura. 2002. Toll-like receptor signaling and regulation of cytokine gene expression in the immune system. BioTechniques (Suppl.) 33:66-68, 70, 72. [PubMed]
- 60.Parisien, J. P., J. F. Lau, and C. M. Horvath. 2002. STAT2 acts as a host range determinant for species-specific paramyxovirus interferon antagonism and simian virus 5 replication. J. Virol. 76:6435-6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parisien, J. P., J. F. Lau, J. J. Rodriguez, B. M. Sullivan, A. Moscona, G. D. Parks, R. A. Lamb, and C. M. Horvath. 2001. The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2. Virology 283:230-239. [DOI] [PubMed] [Google Scholar]
- 62.Park, M. S., M. L. Shaw, J. Munoz-Jordan, J. F. Cros, T. Nakaya, N. Bouvier, P. Palese, A. Garcia-Sastre, and C. F. Basler. 2003. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J. Virol. 77:1501-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peters, K. L., H. L. Smith, G. R. Stark, and G. C. Sen. 2002. IRF-3-dependent, NFkappa B- and JNK-independent activation of the 561 and IFN-beta genes in response to double-stranded RNA. Proc. Natl. Acad. Sci. USA 99:6322-6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Poole, E., B. He, R. A. Lamb, R. E. Randall, and S. Goodbourn. 2002. The V proteins of simian virus 5 and other paramyxoviruses inhibit induction of interferon-beta. Virology 303:33-46. [DOI] [PubMed] [Google Scholar]
- 65.Preston, C., A. Harman, and M. Nicholl. 2001. Activation of interferon response factor-3 in human cells infected with herpes simplex virus type 1 or human cytomegalovirus. J. Virol. 75:8909-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reich, N. C. 2002. Nuclear/cytoplasmic localization of IRFs in response to viral infection or interferon stimulation. J. Interferon Cytokine Res. 22:103-109. [DOI] [PubMed] [Google Scholar]
- 67.Ronco, L. V., A. Y. Karpova, M. Vidal, and P. M. Howley. 1998. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 12:2061-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sambrook, J., E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., vol. 3. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 69.Sanchez, A., A. S. Khan, S. R. Zaki, G. J. Nabel, T. G. Ksiazek, and C. J. Peters. 2001. Filoviridae: Marburg and Ebola viruses, p. 1279-1304. In D. M. Knipe, P. M. Howley, et al. (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 70.Sato, M., H. Suemori, N. Hata, M. Asagiri, K. Ogasawara, K. Nakao, T. Nakaya, M. Katsuki, S. Noguchi, N. Tanaka, and T. Taniguchi. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13:539-548. [DOI] [PubMed] [Google Scholar]
- 71.Seo, T., D. Lee, B. Lee, J. H. Chung, and J. Choe. 2000. Viral interferon regulatory factor 1 of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) binds to, and inhibits transactivation of, CREB-binding protein. Biochem. Biophys. Res. Commun. 270:23-27. [DOI] [PubMed] [Google Scholar]
- 72.Servant, M. J., N. Grandvaux, and J. Hiscott. 2002. Multiple signaling pathways leading to the activation of interferon regulatory factor 3. Biochem. Pharmacol. 64:985-992. [DOI] [PubMed] [Google Scholar]
- 73.Servant, M. J., N. Grandvaux, B. R. TenOever, D. Duguay, R. Lin, and J. Hiscott. 2003. Identification of the minimal phosphoacceptor site required for in vivo activation of interferon regulatory factor 3 in response to virus and double-stranded RNA. J. Biol. Chem. 278:9441-9447. [DOI] [PubMed] [Google Scholar]
- 74.Servant, M. J., B. ten Oever, C. LePage, L. Conti, S. Gessani, I. Julkunen, R. Lin, and J. Hiscott. 2001. Identification of distinct signaling pathways leading to the phosphorylation of interferon regulatory factor 3. J. Biol. Chem. 276:355-363. [DOI] [PubMed] [Google Scholar]
- 74a.Sharma, S., B. R. tenOever, N. Grandvaux, G. P. Zhou, R. Lin, and J. Hiscott. 2003. Triggering the interferon response through an IKK-related pathway. Science 300:1148-1151. [DOI] [PubMed] [Google Scholar]
- 75.Shinobu, N., T. Iwamura, M. Yoneyama, K. Yamaguchi, W. Suhara, Y. Fukuhara, F. Amano, and T. Fujita. 2002. Involvement of TIRAP/MAL in signaling for the activation of interferon regulatory factor 3 by lipopolysaccharide. FEBS Lett. 517:251-256. [DOI] [PubMed] [Google Scholar]
- 76.Smith, E. J., I. Marie, A. Prakash, A. Garcia-Sastre, and D. E. Levy. 2001. IRF3 and IRF7 phosphorylation in virus-infected cells does not require double-stranded RNA-dependent protein kinase R or Ikappa B kinase but is blocked by vaccinia virus E3L protein. J. Biol. Chem. 276:8951-8957. [DOI] [PubMed] [Google Scholar]
- 77.Suhara, W., M. Yoneyama, T. Iwamura, S. Yoshimura, K. Tamura, H. Namiki, S. Aimoto, and T. Fujita. 2000. Analyses of virus-induced homomeric and heteromeric protein associations between IRF-3 and coactivator CBP/p300. J. Biochem. (Tokyo) 128:301-307. [DOI] [PubMed] [Google Scholar]
- 78.Takada, A., and Y. Kawaoka. 2001. The pathogenesis of Ebola hemorrhagic fever. Trends Microbiol. 9:506-511. [DOI] [PubMed] [Google Scholar]
- 79.Talon, J., C. M. Horvath, R. Polley, C. F. Basler, T. Muster, P. Palese, and A. Garcia-Sastre. 2000. Activation of interferon regulatory factor 3 (IRF-3) is inhibited by the influenza A viral NS1 protein. J. Virol. 74:7989-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Talon, J., M. Salvatore, R. E. O'Neill, Y. Nakaya, H. Zheng, T. Muster, A. Garcia-Sastre, and P. Palese. 2000. Influenza A and B viruses expressing altered NS1 proteins: a vaccine approach. Proc. Natl. Acad. Sci. USA 97:4309-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Taniguchi, T., K. Ogasawara, A. Takaoka, and N. Tanaka. 2001. Irf family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19:623-655. [DOI] [PubMed] [Google Scholar]
- 82.tenOever, B. R., M. J. Servant, N. Grandvaux, R. Lin, and J. Hiscott. 2002. Recognition of the measles virus nucleocapsid as a mechanism of IRF-3 activation. J. Virol. 76:3659-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Volchkov, V. E., V. A. Volchkova, E. Muhlberger, L. V. Kolesnikova, M. Weik, O. Dolnik, and H. D. Klenk. 2001. Recovery of infectious Ebola virus from complementary DNA: RNA editing of the GP gene and viral cytotoxicity. Science 291:1965-1969. [DOI] [PubMed] [Google Scholar]
- 84.Wathelet, M. G., C. H. Lin, B. S. Parekh, L. V. Ronco, P. M. Howley, and T. Maniatis. 1998. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol. Cell 1:507-518. [DOI] [PubMed] [Google Scholar]
- 85.Weaver, B. K., O. Ando, K. P. Kumar, and N. C. Reich. 2001. Apoptosis is promoted by the dsRNA-activated factor (DRAF1) during viral infection independent of the action of interferon or p53. FASEB J. 15:501-515. [DOI] [PubMed] [Google Scholar]
- 86.Weaver, B. K., K. P. Kumar, and N. C. Reich. 1998. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol. Cell. Biol. 18:1359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Weber, F., A. Bridgen, J. Fazakerley, H. Streitenfeld, N. Kessler, R. Randall, and R. Elliott. 2002. Bunyamwera bunyavirus nonstructural protein NSs counteracts the induction of alpha/beta interferon. J. Virol. 76:7949-7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wilson, J. A., C. M. Bosio, and M. K. Hart. 2001. Ebola virus: the search for vaccines and treatments. Cell. Mol. Life Sci. 58:1826-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.World Health Organization. 2002. Ebola haemorrhagic fever in Gabon/The Republic of the Congo. W. H. O. Communicable Disease and Surveillance and Response—Outbreak News. World Health Organization, Geneva, Switzerland.
- 90.World Health Organization. 2003. Ebola haemorrhagic fever in the Republic of the Congo—Update 8. W. H. O. Communicable Disease and Surveillance and Response—Outbreak News, 14 March 2003. World Health Organization, Geneva, Switzerland.
- 91.World Health Organization. 2001. Suspected viral haemorrhagic fever in Gabon. W. H. O. Communicable Disease and Surveillance and Response—Outbreak News. World Health Organization, Geneva, Switzerland.
- 92.Xiang, Y., R. C. Condit, S. Vijaysri, B. Jacobs, B. R. Williams, and R. H. Silverman. 2002. Blockade of interferon induction and action by the E3L double-stranded RNA binding proteins of vaccinia virus. J. Virol. 76:5251-5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yoneyama, M., W. Suhara, and T. Fujita. 2002. Control of IRF-3 activation by phosphorylation. J. Interferon Cytokine Res. 22:73-76. [DOI] [PubMed] [Google Scholar]
- 94.Yoneyama, M., W. Suhara, Y. Fukuhara, M. Fukuda, E. Nishida, and T. Fujita. 1998. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 17:1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Young, D. F., N. Chatziandreou, B. He, S. Goodbourn, R. A. Lamb, and R. E. Randall. 2001. Single amino acid substitution in the V protein of simian virus 5 differentiates its ability to block interferon signaling in human and murine cells. J. Virol. 75:3363-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhu, F. X., S. M. King, E. J. Smith, D. E. Levy, and Y. Yuan. 2002. A Kaposi's sarcoma-associated herpesviral protein inhibits virus-mediated induction of type I interferon by blocking IRF-7 phosphorylation and nuclear accumulation. Proc. Natl. Acad. Sci. USA 99:5573-5578. [DOI] [PMC free article] [PubMed] [Google Scholar]