Abstract
The nonstructural proteins of hepatitis C virus (HCV) have been shown previously to localize to the endoplasmic reticulum (ER) when expressed singly or in the context of other HCV proteins. To determine whether the expression of HCV nonstructural proteins alters ER function, we tested the effect of expression of NS2/3/4A, NS4A, NS4B, NS4A/B, NS4B/5A, NS5A, and NS5B from genotype 1b HCV on anterograde traffic from the ER to the Golgi apparatus. Only the nominal precursor protein NS4A/B affected the rate of ER-to-Golgi traffic, slowing the rate of Golgi-specific modification of the vesicular stomatitis virus G protein expressed by transfection by approximately threefold. This inhibition of ER-to-Golgi traffic was not observed upon expression of the processed proteins NS4A and NS4B, singly or in combination. To determine whether secretion of other cargo proteins was inhibited by NS4A/B expression, we monitored the appearance of newly synthesized proteins on the cell surface in the presence and absence of NS4A/B expression; levels of all were reduced in the presence of NS4A/B. This reduction is also seen in cells that contain genome length HCV replicons: the rate of appearance of major histocompatibility complex class I (MHC-I) on the cell surface was reduced by three- to fivefold compared to that for a cured cell line. The inhibition of protein secretion caused by NS4A/B does not correlate with the ultrastructural changes leading to the formation a “membranous web” (D. Egger et al., J. Virol. 76:5974-5984, 2002), which can be caused by expression of NS4B alone. Inhibition of global ER-to-Golgi traffic could, by reducing cytokine secretion, MHC-I presentation, and transport of labile membrane proteins to the cell surface, have significant effects on the host immune response to HCV infection.
All positive-strand RNA viruses replicate their genomes on intracellular membranes of infected host cells. Formation of the RNA replication complexes of most of these viruses results in dramatic rearrangement and derangement of the secretory pathways of the host cells. For example, mammalian cells infected with Kunjin virus, a flavivirus, display various membrane morphologies that have been termed convoluted membranes and vesicle packets, which contain markers from the intermediate compartment and the trans-Golgi network, respectively (52, 53, 77). Cells infected with poliovirus, a picornavirus, accumulate large numbers of membranous vesicles derived from the endoplasmic reticulum (ER) that range in diameter from 150 to 400 nm (6, 7, 13, 27, 72-74, 76) and that display characteristics similar to those of cellular autophagic vacuoles (18, 68, 72). Finally, cells infected with bovine viral diarrhea virus (BVDV), a pestivirus, contain tubules and spherical vesicles that are 100 to 200 nm in diameter and that are thought to be derived from the rough ER (33).
Hepatitis C virus (HCV) infects approximately 170 to 200 million people worldwide (1, 79) and is the agent formerly responsible for the majority of transfusion-associated non-A, non-B hepatitis cases. Most of these infections develop into chronic hepatitis, which often progresses to liver cirrhosis and hepatocellular carcinoma. Studies on HCV have been hindered by the relatively low virus titer in infectious sera, the low expression of antigens in infected tissues or cells, the lack of a reliable tissue culture system to propagate the virus, and the lack of a good animal model. Most of our knowledge about HCV structure and genome organization and the functions of viral proteins comes from experimental infections of chimpanzees (69), molecular cloning of HCV cDNAs, recombinant expression systems, and sequence analogy to other viruses (reviewed in references 4, 19, and 70). From these studies, HCV has been classified in the genus Hepacivirus of the family Flaviviridae (41, 54), which includes flaviviruses such as Kunjin virus and Yellow fever virus and pestiviruses such as BVDV. The development of systems in which the replication of partial or full-length HCV RNAs can be monitored in tissue culture has provided a major breakthrough to the molecular and cell biology of HCV (10, 11, 34, 39, 51).
HCV is a small, enveloped virus with a positive-strand linear RNA genome of approximately 9,600 nucleotides. The genome contains a single large open reading frame encoding a single polyprotein of about 3,010 amino acids. Proteolytic processing of the polyprotein occurs co- and posttranslationally, creating the structural proteins Core (C), E1, E2, and p7 and the nonstructural (NS) proteins NS2, NS3, NS4A, NS4B, NS5A, and NS5B. These NS proteins are the limit digestion products, but it is likely that larger proteins have functions as well (reviewed in references 4 and 63). Processing of the C protein and the two membrane-associated glycoproteins, E1 and E2, is mediated by host signal peptidases (47), whereas cleavage at the junctions of the NS proteins (NS2, NS3, NS4A, NS4B, NS5A, and NS5B) requires viral proteases NS2-3 and NS3-4A (35, 36, 43). All HCV NS proteins studied thus far have been localized to the ER (37, 38, 42, 57, 78). It is believed, therefore, that HCV RNA replication occurs on membranes derived from the ER or on the ER itself (reviewed in reference 24). NS5A has been shown in isolation to induce certain aspects of the ER stress response (75). There have been descriptions of an altered ultrastructural morphology termed the membranous web in cells that express NS4B alone or in the context of other HCV proteins, as well as in liver biopsy samples (28). In this report, we have examined the effect of the expression of individual NS proteins of HCV genotype 1b on one aspect of ER function, the transport of protein cargo to the Golgi apparatus. We found that nominal precursor protein NS4A/B slows the rate of ER-to-Golgi traffic. A comparable reduction in protein secretion rate is seen for several cargo proteins during NS4A/B expression and for major histocompatibility complex class I (MHC-I) molecules in the presence of a full-length HCV replicon.
MATERIALS AND METHODS
Cells.
The Huh7 C5B 2-3 clonal cell line was derived from human hepatoma (Huh7) cells and contains a replicating, dicistronic selectable RNA (NNeo-C5B) derived from the genotype 1b HCV-N strain of HCV (39). To cure these cell lines of the replicating HCV RNA, we cultured them in the presence of 100 IU of alpha interferon 2b/ml in the absence of G418 selection for 2 weeks. The cured cells (termed 2-3c) lacked demonstrable G418 resistance. A more complete characterization of these paired cell lines will be presented elsewhere (K. Li et al., unpublished data). Human kidney (293T), 2-3, and 2-3c cell lines were grown as monolayers in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 100 U of penicillin/ml, and 100 mg of streptomycin/ml at 37°C in a 5% CO2 incubator. In addition, the media for 2-3c and 2-3 cell lines contained nonessential amino acids (NEAA) or NEAA and 500 μg of G418 (Geneticin)/ml, respectively. All reagents were purchased from Life Technologies/GIBCO-BRL, Rockville, Md.
Construction of plasmids and recombinant vaccinia virus.
To construct dicistronic plasmids that carry the HCV genes of interest in the first cistron and the green fluorescent protein (GFP) gene in the second cistron, the desired HCV gene was amplified by PCR from a full-length cDNA from strain HC-J4 of genotype 1b HCV, in plasmid pCV-J4LS6S (81), kindly provided by Jens Bukh and Robert Purcell (National Institutes of Health, Bethesda, Md.). The GFP gene was included to ensure that transfection efficiencies from different plasmids were comparable (data not shown). For each HCV coding region, deoxyoligonucleotide primers were designed to introduce an EcoRI site, an XhoI site, and an AUG start codon immediately upstream of the coding region and an epitope tag, the stop codons UGA and UAA, and a NotI site immediately downstream of the coding region. The epitope recognized by mouse monoclonal antibody Penta-His (Qiagen, Inc., Valencia, Calif.) added six His residues to the C termini of HCV NS proteins NS4B, NS4A/B, NS4B/5A, and NS5A. The epitope recognized by the FLAG mouse monoclonal antibody (Sigma, St. Louis, Mo.) added eight amino acids (DYKDDDDK) to the C termini of NS2/3/4A, NS4A, and NS5B.
The resulting PCR products were cloned into cloning vector pCR 2.1 TOPO (Invitrogen, Carlsbad, Calif.), and their sequences were confirmed. Recombinant plasmids that harbored the genes for NS4B, NS4A/B, NS2/3/4A, NS4A, and NS5B were digested with EcoRI and NotI, and plasmids that harbored the genes for NS4B/5A and NS5A were digested with XhoI and NotI. The coding region for enhanced GFP (16) under the translational control of the poliovirus internal ribosome entry site (IRES) was obtained from plasmid pTRE-3A/GFP (20) and isolated by cleavage with NotI and XbaI. The purified HCV protein-encoding fragments and the IRES-GFP-encoding fragment were cloned into an EcoRI- and XbaI-cleaved or an XhoI- and XbaI-cleaved pIRES expression vector (Clontech, Palo Alto, Calif.). pEGFP-N3 (kindly provided by Richard Scheller, Genentech, South San Francisco, Calif.) encodes a temperature-sensitive vesicular stomatitis virus G (VSV-Gts045) fused to GFP (12, 66). The temperature-sensitive mutation in VSV-G causes VSV-G to accumulate in the ER when cells are incubated at 40°C. The cells can then be shifted to 33°C to allow the VSV-Gts045/GFP fusion protein to leave the ER (49).
For recombinant vaccinia viruses expressing HCV NS4A/B and GFP from a dicistronic mRNA, the pIRES expression vector (Clontech), encoding NS4A/B-GFP, was cleaved with EcoRI, and XbaI and subcloned into EcoRI- and NheI-cleaved pRB21, a shuttle vector for vaccinia virus recombination (9). 293T cells were transfected with pRB21-NS4A/B-GFP and infected with the plaque-deficient vaccinia virus vRB12 (8). Individual vaccinia virus recombinant plaques (vv-NS4A/B-GFP) were purified as described previously (9), and HCV NS4A/B expression from vv-NS4A/B-GFP was confirmed by immunoblotting, whereas GFP expression was visualized by fluorescence microscopy. Recombinant vaccinia virus including GFP (vv-GFP) (20) was used as a control.
DNA transfections.
For protease activity, VSV-G traffic, immunoblotting, and electron microscopy (EM) experiments, plasmid DNA was introduced into 293T cells with Lipofectamine transfection reagents (Life Technologies/GIBCO-BRL). For each experiment, human 293T cells were trypsinized and grown overnight in 100-mm-diameter tissue culture dishes (coated with poly-d-lysine; Sigma) to obtain 50 to 80% confluent monolayer cells. Before transfection, cells were washed with phosphate-buffered saline (PBS) and fed with 3.5 ml of OptiMEM (Life Technologies/GIBCO-BRL) for approximately 2 h. The DNA and Lipofectamine Plus were mixed with 750 μl of OptiMEM, and the mixture was incubated at room temperature for 15 min. Lipofectamine was diluted in 750 μl of OptiMEM and added to the DNA-Lipofectamine Plus mixture, which was incubated for an additional 15 min. The mixture was then added to each dish, and the cells were incubated at 37°C. After 4 h of incubation, 5 ml of OptiMEM containing 20% fetal bovine serum was added to each dish and the cells were incubated for 48 h.
Labeling of cells and preparation of lysates.
Transfected 293T cells were washed with PBS and incubated in 2 ml of DMEM lacking methionine (Life Technologies/GIBCO-BRL) for 30 min at 40°C. [35S]methionine-[35S]cysteine (Expre35S35S label; New England Nuclear, Gaithersburg, Md.) was then added, and the cells were incubated for 15 min at 40°C, washed once with PBS, and placed in a complete medium containing 0.23 mM unlabeled methionine. After the periods of chase at 33°C indicated in Fig. 3, the cells were washed and harvested in ice-cold PBS. The cells were collected by centrifugation at 210 × g for 10 min, the supernatants were removed, and the pellets were lysed by vortexing in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 50 mM Tris [pH 8.0], 1 mM EDTA, 1% NP-40, 0.1% sodium dodecyl sulfate [SDS]) containing 1 mM phenylmethylsulfonyl fluoride (PMSF). Nuclei and insoluble debris were pelleted by centrifugation at 500 × g for 10 min, and the supernatants were collected for further analysis. For NS2/3/4A protease assays, preincubation, labeling with [35S]methionine, and chase with cold methionine were all done at 37°C.
FIG. 3.
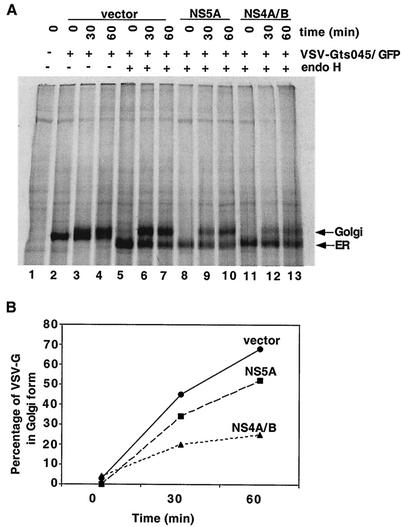
Effect of HCV proteins NS4A/B and NS5A on acquisition of Golgi-specific modifications by VSV-G protein in the Golgi apparatus. (A) 293T cells were cotransfected with plasmids that expressed VSV-Gts045/GFP and the indicated HCV proteins. At 48 h after transfection at 37°C, the cells were labeled with [35S]methionine for 15 min at 40°C. After a 30- or 60-min chase in the presence of unlabeled methionine at 33°C, the permissive temperature for VSV-Gts045 transport, cell lysates were prepared. Immunoprecipitated VSV-G was digested with no enzyme or with endo H overnight, and the resulting products were separated by SDS-7.5% PAGE and analyzed with a PhosphorImager. Arrows, ER and Golgi forms of the VSV-G protein. (B) Quantitation of the acquisition of a Golgi-specific modification by VSV-G protein coexpressed with the indicated HCV proteins at various times of incubation at 33°C.
Northern blotting for HCV RNA.
Total cellular RNA was isolated with the RNAqueous RNA extraction kit (Ambion, Austin, Tex.) and quantified by spectrophotometry at 260 nm. Ten micrograms of total RNA was separated by denaturing gel electrophoresis, transferred to a Hybond-N+ nylon membrane (Amersham-Pharmacia Biotech, Mississauga, Ontario, Canada), and hybridized with a digoxigenin-labeled NS5B antisense riboprobe. The hybridized targets were detected by an anti-digoxigenin-alkaline phosphatase, followed by visualization by chemiluminescence (CSPD; Roche).
Western blotting.
Cells were lysed in a buffer containing 150 mM NaCl, 1% NP-40, 0.1% SDS, 50 mM Tris, 1 mM PMSF, 2 mg of aprotinin/ml, and 1 mg of leupeptin/ml. Protein concentrations were determined with a bicinchoninic acid kit (Sigma). Samples were subjected to SDS-7.5% polyacrylamide gel electrophoresis (PAGE), followed by transfer onto Hybond-ECL nitrocellulose membranes (Amersham Pharmacia Biotech). Membranes were blocked with 5% skim milk in PBS and incubated with pooled mouse monoclonal antibodies against HCV NS3 (Novocastra) and actin (Sigma) diluted in PBS containing 1% skim milk. Following three washes in PBS, membranes were incubated with a secondary horseradish peroxidase-conjugated anti-mouse antibody (Amersham Pharmacia Biotech) diluted in PBS containing 1% skim milk for 1 h at room temperature. Following three additional washes in PBS, proteins bound to the antibody were visualized by enhanced chemiluminescence (ECL; Amersham Pharmacia Biotech).
Immunoprecipitations and protein secretion assays.
For the protease and secretion assays, total cell lysates were used for immunoprecipitation. Prior to immunoprecipitation, the extracts were precleared by incubation for 1 h at 4°C on a tube rotator with 50 μl of protein A-agarose beads (Life Technologies/GIBCO-BRL) that had been washed in RIPA buffer and resuspended to a concentration of 20% (w/vol) in RIPA buffer. The protein A-agarose beads were collected by centrifugation at 500 × g for 10 min at 4°C. The supernatants were recovered, mixed with the primary antibody (5 μg of the anti-Penta-His antibody/ml or 10 to 20 μg of the anti-VSV-G antibody (Sigma)/ml), incubated for 2 to 12 h at 4°C, mixed with 50 μl of protein A-agarose, and incubated for 2 h at 4°C. Protein A-agarose beads with bound immune complexes were collected by centrifugation at 14,000 × g for 10 min and washed once with RIPA buffer, once with RIPA buffer containing 500 mM NaCl, and once more in RIPA buffer.
To examine the immunoprecipitates by SDS-PAGE, the pelleted protein A-agarose beads bound to the immune complexes were resuspended in 30 μl of loading buffer (45), heated at 95°C for 6 min, and centrifuged at 14,000 × g for 2 min and the supernatants were analyzed by SDS-PAGE. For experiments involving endoglycosidase H (endo H) digestion, bound immune complexes were resuspended in 10 μl of resuspension buffer (0.1 M sodium acetate [NaOAc] [pH 5.6], 0.3% SDS, 0.3 M β-mercaptoethanol), heated at 95°C for 6 min, and centrifuged at 14,000 × g for 2 min. The supernatants were collected, 20 μl of a solution containing 0.1 M NaOAc (pH 5.6), with or without endo H (20 U/μl; New England Biolabs, Inc., Beverly, Mass.), was added, and the reaction mixtures were incubated overnight at 37°C. Samples were then heated at 95°C for 6 min and centrifuged at 14,000 × g for 2 min, and the supernatant was analyzed by SDS-PAGE. The gels were dried, and labeled proteins were visualized and quantitated on a PhosphorImager (Storm 860; Amersham Pharmacia Biotech. Inc. and Molecular Dynamics, Piscataway, N.J.).
Assay for cell surface expression of newly synthesized proteins.
To monitor the rate of transport of populations of cell surface proteins in cells that did and did not express HCV NS4A/B, 1.5 × 106 cured Huh7 cells were seeded overnight and infected with recombinant vv-NS4A/B or with control vv-GFP at a multiplicity of infection of 3 PFU/cell for 3 h. For each sample, cells from two 100-mm plates were pooled. The cells were washed with PBS and starved for 30 min in serum-free DMEM lacking methionine. The cells were then washed with PBS, labeled with 800 μCi of [35S]methionine-[35S]cysteine (Expre35S35S label; New England Nuclear) for 15 min, washed once with PBS, and fed with complete medium containing 0.23 mM cold methionine. After periods of chase, the cells were washed three times with cold PBS, pH 8, scraped, and spun down at 210 × g for 10 min at 4°C. Except for negative control or total lysate, the cells were resuspended in 200 μl of ice-cold PBS (pH 8) containing 0.5 mg of EZ-Link sulfo-NHS-SS-biotin [sulfosuccinimidyl 2-(biotinamido)-ethyl-1,3-dithiopropionate; Pierce Biotechnology, Inc., Rockford, Ill.]/ml, incubated on ice for 30 min, and washed three times with ice-cold PBS, pH 8. Cells were lysed with 0.5 ml of lysis buffer (150 mM NaCl, 50 mM Tris [pH 8.0], 1 mM EDTA, 1% NP-40, 1 mM PMSF) and centrifuged at 834 × g for 10 min at 4°C, and the supernatant was transferred to a fresh tube. The lysate was precleared with 50 μl of protein G-agarose (Life Technologies/GIBCO-BRL) at 4°C for 1 h and spun down at 500 × g for 10 min at 4°C, and the recovered supernatant was incubated overnight at 4°C with 30 μl of 50% streptavidin-Sepharose 4B (Zymed Laboratories, Inc., South San Francisco, Calif.). Streptavidin-Sepharose beads with bound biotinylated proteins were collected by centrifugation at 14,000 × g for 10 min and washed once with lysis buffer, once with lysis buffer containing 500 mM NaCl, and once more in lysis buffer. Elution of the pellet with 30 μl of PBS (pH 8)-50 mM dithiothreitol for 10 min at room temperature was followed by centrifugation at 14,000 × g for 1 min. The eluate was added to 30 μl of sample buffer (20 μl of 8 M urea-1% SDS, 10 μl of 4× loading buffer) (45), and one-half of each sample was separated by SDS-10% PAGE.
Cell surface expression of newly synthesized MHC-I molecules.
To monitor the rate of transport of newly synthesized MHC-I molecules to the surfaces of cells that did and did not contain the genome length HCV replicon, 5 × 105 replicon-containing Huh7 cells (39) and 5 × 105 cured cells were seeded for 46 h, pulse-chased with [35S]methionine, and subjected to surface biotinylation as described above. The MHC-I protein was collected by incubating the eluate from the streptavidin-Sepharose beads with 470 μl of lysis buffer and 2 μl of anti-MHC-I primary antibody (0.8 μg of W6/32/ml; Sigma-Aldrich, St. Louis, Mo.) at 4°C for 2 h. Subsequently, 50 μl of protein G-agarose (Life Technologies/GIBCO-BRL) was added, followed by overnight incubation at 4°C. Protein G-agarose beads with bound immune complexes were collected by centrifugation at 14,000 × g for 10 min and washed once with lysis buffer, once with lysis buffer containing 500 mM NaCl, and once more with lysis buffer. The pellet was resuspended in a mixture containing 20 μl of 8 M urea-1% SDS and 10 μl of 4× loading buffer (45). The samples were boiled for 5 min and separated by SDS-10% PAGE. The gel was dried and exposed to a PhosphorImager plate, and the MHC-I protein was visualized with a Storm 860 (Amersham Biosciences Corp., Piscataway, N.J.).
SDS-glycine or Tricine gels and immunoblot analysis.
Lysates of transfected 293T cells were separated by SDS-PAGE on 13% polyacrylamide gels (45, 67) and transferred to Immobilon-P membranes (Millipore, Bedford, Mass.) for 1 h. After drying, the membranes were treated for 1 h with 3% nonfat dry milk (NFDM)-0.25% Tween 20 in PBS (PBST), washed three times with PBST, and incubated with a mouse monoclonal antibody to the His6 tag (1:1,000 dilution) or FLAG tag (1:1,000 dilution) overnight in PBST with 1% NFDM. The membranes were washed three times with PBST and incubated for 1 h in PBST with 1% NFDM and a secondary antibody (1:10,000 dilution; alkaline phosphatase-conjugated rabbit anti-mouse immunoglobulin G; Zymed Laboratories Inc.). After three washes, the immune complexes were detected by enhanced chemifluorescence (Amersham Pharmacia Biotech) and visualized with a PhosphorImager.
High-pressure freezing, freeze substitution, and EM.
For cryofixation and EM, 293T cells from a 100-mm tissue culture dish were incubated for 2 days after transfection, washed three times with PBS, trypsinized, and collected by centrifugation. The cell pellet was resuspended in 0.15 M mannitol in PBS and centrifuged. Aliquots of the resulting pellet were frozen in a Balzers HPM 10 high-pressure freezing apparatus as described previously (68) and stored in liquid nitrogen. To observe the cellular morphology, samples were freeze substituted in 0.1% tannic acid in acetone at −80°C, rinsed in acetone, and then warmed at −20°C in the presence of 2% osmium tetroxide in acetone for 16 h and incubated at 4°C for 4 h. After being rinsed in acetone at 4°C, samples were embedded in Epon-Araldite resin. Thin sections were stained with 2% uranyl acetate and lead citrate and then imaged at 80 kV in a JEOL 100C or Philips CM10 electron microscope.
Immunogold labeling of high-pressure-frozen cells.
For immunostaining, high-pressure-frozen cell samples were freeze substituted in 0.1% glutaraldehyde-0.05% uranyl acetate in acetone and then embedded in Lowicryl HM20. Sections were mounted on Formvar-coated nickel grids and immunolabeled with a mouse monoclonal antibody against the C-terminal His6 tag. Grids were floated on a drop of blocking solution containing 1% NFDM in PBST and the anti-His6 monoclonal antibody (1:25 dilution) as described previously (22, 68). Mouse monoclonal primary antibodies were detected with goat anti-mouse secondary antibodies coupled to gold particles 15 nm in diameter (Ted Pella, Inc., Redding, Calif.).
RESULTS
Cloning, expression, and processing of HCV NS proteins and precursors.
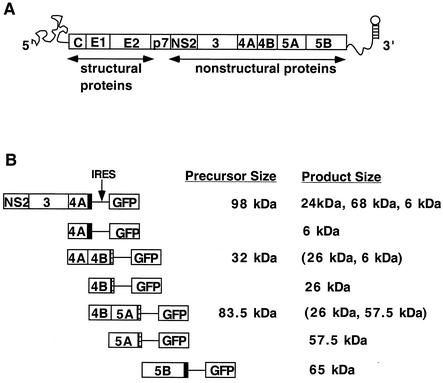
To determine whether any of the HCV NS proteins or precursors modify the function of the ER in anterograde protein traffic, we cloned several coding regions so that they would be expressed in mammalian cells as dicistronic transcripts (Fig. 1B). Specifically, the coding regions for NS2/3/4A, NS4A, NS4A/B, NS4B, NS4B/5A, NS5A, and NS5B of HCV genotype 1b (81) were cloned under the transcriptional control of a human cytomegalovirus immediate-early promoter such that, for each construct, the HCV sequence of interest was expressed in the first cistron and enhanced GFP was expressed in the second cistron under the translational control of the poliovirus IRES. An epitope tag was added to the C terminus of each HCV coding region: a FLAG tag for NS2/3/4A, NS4A, and NS5B and a His6 tag for NS4A/B, NS4B, NS4B/5A, and NS5A.
FIG. 1.
Features of the HCV genome and bicistronic expression constructs. (A) Schematic representation of the HCV genome, which is translated into a single polyprotein that is processed by host signal peptidases (29, 35, 47, 65) and viral proteases (3, 31, 80) to create structural proteins C, E1, E2, and p7 and NS proteins NS2, NS3, NS4A, NS4B, NS5A, and NS5B. (B) Organization of the bicistronic constructs used to express the various HCV NS proteins. HCV NS proteins were expressed in the first cistron, and GFP was expressed in the second cistron, downstream of the IRES from poliovirus. Each bicistronic transcript is under the transcriptional control of a human cytomegalovirus immediate-early promoter. Solid bars, C-terminal FLAG tags; hatched bars, C-terminal His6 tags. The sizes of the expected protein products are indicated. The sizes of both the precursors and the products are also indicated. Products that should not accumulate unless the HCV NS3/4A protease is coexpressed are shown in parentheses.
To monitor the synthesis of proteins of interest, immunoprecipitates of 35S-labeled proteins were examined. Human embryonal kidney 293T cells were transfected with HCV protein-expressing plasmids and metabolically labeled with [35S]methionine for 15 min. Figure 2 shows an example of such an analysis, in which His6-tagged proteins were collected by immunoprecipitation. Proteins NS4A/B, NS4B/5A, and NS5A, each with a C-terminal His6 tag, were found to accumulate during the 15-min labeling period (Fig. 2, lanes 3 to 5). The accumulation of NS2/3/4A could not be observed in this experiment, because the protein contained a C-terminal FLAG tag.
FIG. 2.
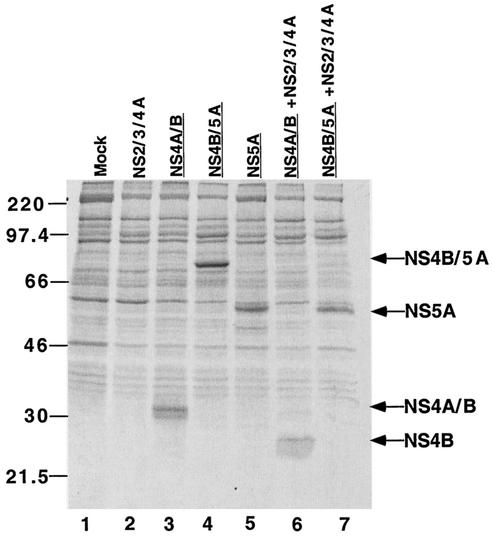
Expression of HCV NS proteins and activity of NS2/3/4A protease in transfected human kidney 293T cells. 293T cells were transfected with plasmids that encode the indicated HCV NS proteins. At 48 h posttransfection, cells were labeled with [35S]methionine for 15 min and then incubated in the presence of cold methionine for an additional 30 min. Immunoprecipitated, labeled His6-tagged proteins were collected from cell lysates, displayed by SDS-10% PAGE, and visualized with a PhosphorImager. Molecular weight standards are indicated at the left. Lanes display proteins from cells that were mock-transfected (lane 1) or transfected with NS2/3/4A-FLAG (lane 2), NS4A/B-His6 (lane 3), NS4B/5A-His6 (lane 4), NS5A-His6 (lane 5), NS4A/B-His6 and NS2/3/4A-FLAG protease (lane 6), and NS4B/5A-His6 and NS2/3/4A-FLAG protease (lane 7). In lanes 6 and 7, where the NS3/4A protease complex is active, only NS4B-His6 and NS5A-His6 products can be detected. His6-tagged proteins are underlined.
To monitor the folding of proteolytic precursors and to test the activity of NS3/4A protease, protein products were examined by immunoprecipitation in the presence and absence of NS2/3/4A. NS2 and the N-terminal domain of NS3 are responsible for the proteolytic cleavage of the NS2/3 junction (32, 36), whereas a complex between NS3 and NS4A cleaves the NS3/4A, NS4A/B, NS4B/5A, and NS5A/5B junctions (2, 31, 48, 78). When the NS4A/B- and NS2/3/4A-expressing plasmids were cotransfected, no full-length NS4A/B could be observed (Fig. 2, compare lanes 3 and 6). Instead, a band corresponding to the cleaved NS4B product, the product bearing the His6 tag, was observed. Similarly, when the NS4B/5A- and NS2/3/4A-expressing plasmids were cotransfected, only the cleaved NS5A protein could be observed (Fig. 2, compare lanes 4 and 7). These data indicate that the expression of NS2/3/4A provides a functional NS3/4A protease complex, active within the 15-min labeling period. Furthermore, the NS4A/B and NS4B/5A precursors were folded in a sufficiently native conformation to serve as substrates for their natural protease.
HCV precursor protein NS4A/B reduces the rate of ER-to-Golgi protein traffic.
Possible effects of the HCV nonstructural proteins on anterograde transport from the ER were examined by monitoring the glycosylation state of coexpressed VSV-G. VSV-G acquires N-linked glycosylation at two asparagine residues when it is translocated across the ER membrane (64). When VSV-G reaches the medial Golgi, the high-mannose oligosaccharides of the ER form are modified by Golgi enzymes N-acetylglucosaminyltransferase and mannosidase II (44). The ER and the Golgi forms of VSV-G can be distinguished by treatment with endo H, which specifically cleaves the high-mannose oligosaccharides characteristic of the ER form of VSV-G but not the modified versions found in the Golgi. For these experiments, VSV-G sequences contained mutation ts045, which causes the retention of VSV-G in the ER at temperatures above 39°C (49). The ts045 mutant VSV-G (VSV-Gts045) sequences were expressed as N-terminal extensions of GFP (12). The processing of this VSV-Gts045/GFP fusion protein in the absence of coexpressed HCV proteins can be seen in Fig. 3A (lanes 2 to 7). The acquisition of Golgi-specific modifications can be seen both as a slight decrease in mobility of the VSV-Gts045/GFP band with time in the absence of endo H treatment (Fig. 3A, lanes 2 to 4) and as the loss of endo H sensitivity upon incubation (lanes 5 to 7). As shown quantitatively in Fig. 3B, more than 65% of the pulse-labeled VSV-Gts045/GFP acquired Golgi-specific modifications during a 60-min incubation at the permissive temperature for VSV-Gts045 transport.
Upon coexpression of the VSV-Gts045/GFP protein with HCV protein NS5A, the rate of acquisition of Golgi-specific modifications was only slightly reduced. In contrast, coexpression of VSV-Gts045/GFP with HCV protein precursor NS4A/B resulted in a large (2.7-fold) decrease in the rate of ER-to-Golgi traffic, with only 25% of the labeled VSV-Gts045/GFP protein acquiring Golgi-specific modifications in 60 min (Fig. 3). This finding led to the hypothesis that HCV protein NS4A/B specifically inhibits ER-to-Golgi traffic.
Other HCV NS proteins do not affect the rate of ER-to-Golgi protein traffic.
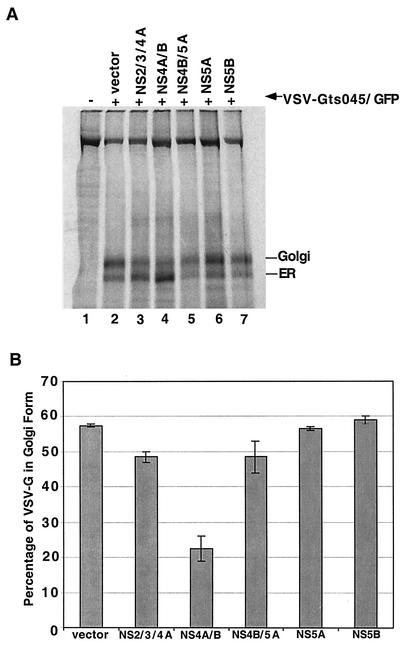
To test whether NS4A/B is unique among HCV proteins in its effect on the ER, we tested the effects of other HCV NS proteins on ER-to-Golgi traffic. VSV-Gts045/GFP was coexpressed with NS2/3/4A, NS4A/B, NS4B/5A, and NS5B and the vector that expressed only GFP in the second cistron. Duplicate transfection products were harvested following labeling with [35S]methionine and incubation with cold methionine for 60 min, after which the VSV-Gts045/GFP immunoprecipitates were subjected to endo H digestion (Fig. 4A). Again, HCV NS4A/B reduced the rate of acquisition of Golgi-specific modifications by two- to threefold in comparison to cotransfections with the vector alone, whereas neither the protease complex NS2/3/4A nor the precursor protein NS4B/5A nor NS5B, the viral RNA-dependent RNA polymerase, substantially affected the apparent rate of ER-to-Golgi traffic (Fig. 4). Expression of all proteins was confirmed by immunoblot analysis (data not shown).
FIG. 4.
Modification of pulse-labeled VSV-G following expression of various HCV NS proteins. (A) 293T cells were cotransfected with plasmids that express VSV-Gts045/GFP and the indicated HCV proteins. At 48 h after transfection at 37°C, the cells were labeled with [35S]methionine for 15 min at 40°C. After a 60-min chase at 33°C, the cell lysates were prepared. Immunoprecipitated proteins were digested with endo H overnight, separated by SDS-7.5% PAGE, and analyzed with a PhosphorImager. Arrows, ER and Golgi forms of the VSV-G marker. (B) Quantitation of the percentages of immunoprecipitated VSV-G in Golgi form after a 60-min incubation at 33°C; the standard errors from duplicate transfections are shown.
Inhibition of ER-to-Golgi traffic by NS4A/B is not reproduced by NS4A or NS4B, singly or coexpressed.
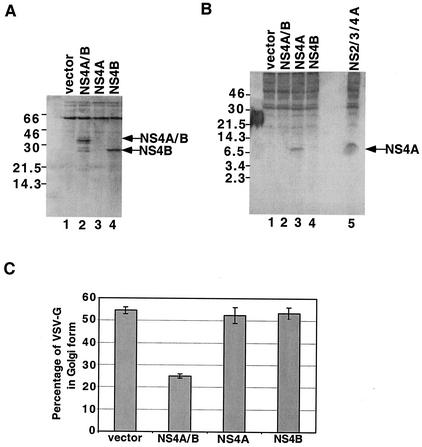
To determine whether the NS4A or NS4B limit digestion products of NS4A/B could inhibit ER-to-Golgi traffic when expressed in isolation, NS4A and NS4B were expressed singly. Successful expression of each of these proteins can be seen in Fig. 5A and B. However, their expression had little effect on the rate of acquisition of Golgi-specific modifications by coexpressed VSV-Gts045/GFP (Fig. 5C). Therefore, the inhibition of ER-to-Golgi traffic by NS4A/B is a specific characteristic of this nominal “precursor” protein.
FIG. 5.
Comparison of effects of the NS4A/B precursor and its products on acquisition of Golgi-specific modifications by VSV-G. 293T cells were cotransfected with plasmids that expressed VSV-Gts045 and either a control plasmid or a plasmid that expressed NS4A/B-His6, NS4A-FLAG, or NS4B-His6. Immunoblot analyses were performed using anti-His6 (A) or anti-FLAG (B) antibodies to confirm expression of the indicated HCV proteins. In panel A, NS2/3/4A cannot be detected because it contains a C-terminal FLAG tag. (C) The percentages of coexpressed, 35S-labeled VSV-G that acquired Golgi-specific modifications after 60 min of transport under permissive conditions were determined as described for Fig. 4. Molecular weight standards (A and B) are shown at the left.
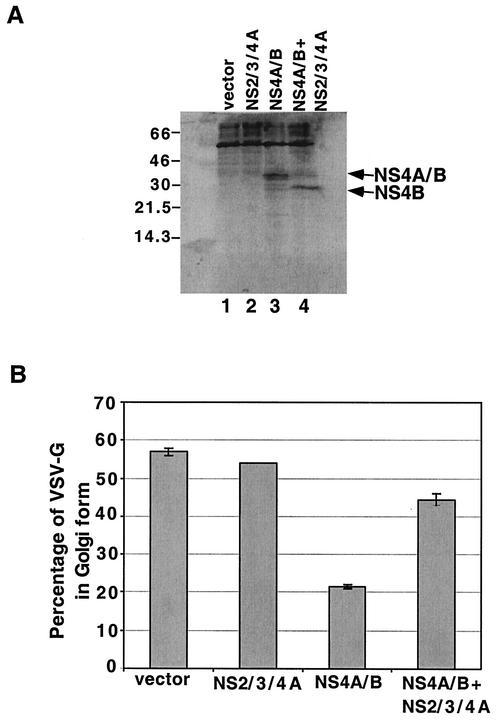
To examine the effect of the processing of NS4A/B within the cell on its ability to inhibit ER-to-Golgi traffic, we monitored the glycosylation state of VSV-Gts045/GFP during coexpression with NS4A/B and the HCV protease complex NS2/3/4A. Immunoblot analysis showed that, under the conditions of this assay, NS4A/B was efficiently processed during coexpression with the NS2/3/4A protease complex (Fig. 6A, lane 4). Furthermore, coexpression of NS4A/B with the protease-expressing vector did not alter the amount of His6-tagged NS4B protein that accumulated. However, when VSV-Gts045/GFP-expressing plasmids were coexpressed with NS4A/B and NS2/3/4A, little inhibition of ER-to-Golgi traffic was observed (Fig. 6B). Therefore, the presence of the intact NS4A/B protein is required for inhibition of ER-to-Golgi traffic.
FIG. 6.
The effect of proteolytic cleavage of NS4A/B on the acquisition of Golgi-specific modifications by coexpressed VSV-G. (A) 293T cells were cotransfected with plasmids that expressed VSV-Gts045/GFP and either a control plasmid (lane 1), NS2/3/4A (lane 2), NS4A/B (lane 3), or both NS2/3/4A and NS4A/B (lane 4). Immunoblot analysis with an anti-His6 antibody was performed to quantify the expression of NS4A/B and its cleavage product in the presence of NS2/3/4A protease after display by SDS-13% PAGE. (B) Percentages of coexpressed, 35S-labeled VSV-G that acquired Golgi-specific modifications after 60 min of transport under permissive conditions were determined as described for Fig. 4.
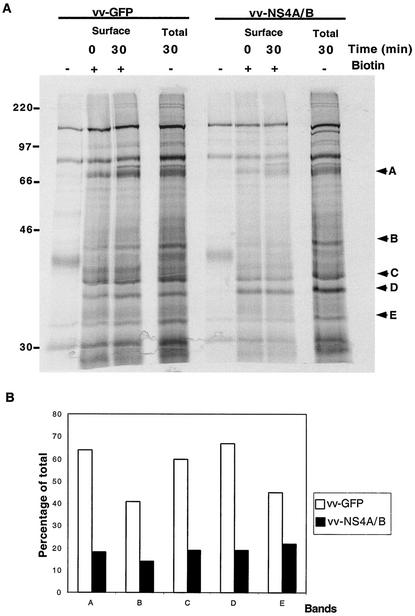
The HCV NS4A/B precursor inhibits traffic of several proteins to the cell surface.
To determine whether traffic of other cargo proteins besides VSV-G was inhibited by NS4A/B expression, we monitored the cell surface expression of proteins in Huh7 cells following infection with recombinant vaccinia virus that expressed either HCV NS4A/B (vv-NS4A/B-GFP) or GFP alone (vv-GFP). Expression of NS4A/B was confirmed by immunoblotting, and expression of GFP was monitored by fluorescence microscopy (data not shown). To examine the appearance of newly synthesized proteins on the cell surface in the presence and absence of NS4A/B, cells were labeled with [35S]methionine and incubated with a cell surface biotinylation reagent after 0 and 30 min of chase in the presence of cold methionine. Biotinylated cell surface proteins were isolated with streptavidin-Sepharose and displayed by SDS-PAGE for comparison with total labeled proteins. As can be seen in Fig. 7A, the amounts of total labeled proteins in the presence and absence of NS4A/B expression were similar. However, the rate of appearance of newly synthesized proteins on the cell surface was reduced by expression of HCV NS4A/B. Indeed, after a 30-min chase, expression of recombinant vv-NS4A/B resulted in a two- to threefold reduction in the rate of traffic of all detectable proteins to the cell surface, as exemplified by representative bands A to E, whose cell surface appearance is quantified in Fig. 7B. Therefore, the inhibition of protein traffic by HCV NS4A/B applies to several cargo proteins and thus is likely to be a general effect on ER function and not cargo specific.
FIG. 7.
Effect of HCV NS4A/B on traffic of proteins to the cell surface. (A) Cured Huh7 cells were infected with recombinant vaccinia virus that expressed GFP (vv-GFP) or both HCV NS4A/B and GFP (vv-NS4A/B). At 3 h after infection at 37°C, the cells were labeled with [35S]methionine for 15 min at 37°C; after a 0- or 30-min chase in the presence of unlabeled methionine, cells were washed with cold PBS, pH 8, and either subjected to surface biotinylation or left untreated. The untreated samples were displayed both as total labeled protein and as a control for nonspecific background. Cell lysates were prepared, and cell surface proteins were pulled down with streptavidin-Sepharose. Eluted proteins were resolved by SDS-10% PAGE and analyzed with a PhosphorImager. Arrows, representative cell surface proteins. (B) Quantitation of the amount of representative newly synthesized proteins present on the cell surface after a 30-min chase in the presence or absence of HCV NS4A/B.
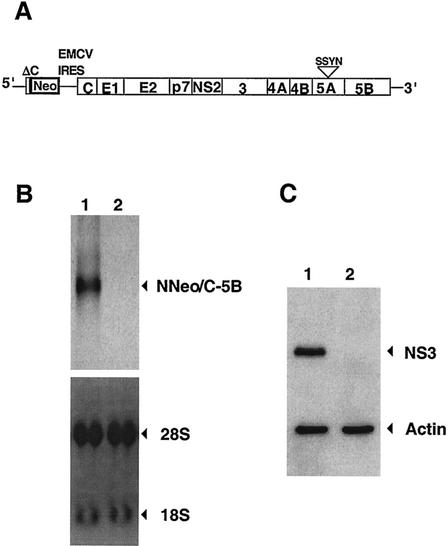
The HCV polyprotein also blocks protein secretion.
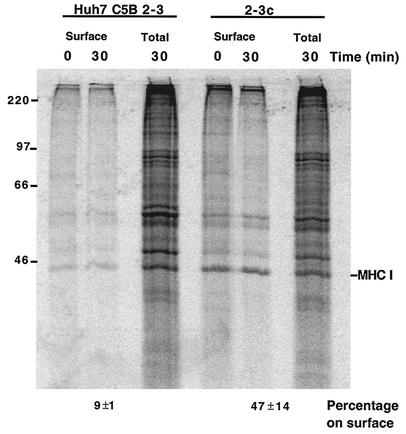
To determine whether host protein traffic is inhibited in the presence of the entire HCV genome, the rate of cell surface expression of MHC-I molecules in Huh7 cells that harbored a replication-competent dicistronic RNA (NNeo-C5B), derived from HCV genotype 1b (Fig. 8A), was monitored and compared to that in control cells that had been cured of the replication-competent RNA by alpha interferon treatment. The elimination of the viral RNA and related proteins was confirmed by Northern blotting (Fig. 8B) and immunoblot detection of HCV NS3 (Fig. 8C). Both cell lines were grown at low density for approximately 46 h to allow maximum expression of the HCV polyprotein (data not shown), labeled with [35S]methionine for 15 min, and incubated with cold methionine for 0 or 30 min. MHC-I α chain molecules were immunoprecipitated after cell surface biotinylation and elution of all cell surface proteins. As can be seen in Fig. 9, the total amounts of MHC-I protein synthesized in the cells that contained replication-competent RNA and the cured Huh7 cells were comparable. However, much less labeled MHC-I protein was expressed on the surfaces of the replication-competent RNA-containing cells after a 30-min chase than was expressed in the cured cells. When amounts of cell surface MHC-I were expressed as percentages of total newly synthesized MHC-I, it was found that cells that contained replicating HCV-derived RNA showed three- to fivefold inhibition of MHC-I cell surface expression relative to the cured control cells. Therefore, like the NS4A/B protein in isolation, the HCV polyprotein inhibits host protein traffic.
FIG. 8.
(A) Organization of the replication-competent dicistronic RNA present in Huh7 C5B 2-3 cell line (39). This replication-competent RNA was derived from an infectious clone of the strain HCV-N (5); like strain HC-J4, the source of the constructs used in Fig. 2 to 6 (81), strain HCV-N belongs to HCV genotype 1b. The two HCV strains display 95% homology throughout their polyprotein sequences and show only four amino acid differences in the sequences of NS4A/B. ΔC, N-terminal segment of the C protein fused in frame to the neomycin phosphotransferase gene. The neomycin gene is under the translational control of the HCV IRES, whereas the HCV polyprotein is regulated by the encephalomyocarditis virus (EMCV) IRES. (B) Northern analysis of HCV-derived RNA in extracts from cells (C5B clone 2-3) that contain the replication-competent dicistronic RNA (NNeo/C5B) (lane 1) and the interferon-cured counterpart cells, clone 2-3c (lane 2). Loading controls are shown at the bottom. (C) Immunoblot detection of NS3 protein in extracts from the 2-3 (lane 1) and 2-3c (lane 2) cell lines.
FIG. 9.
Effect of expression of the replication-competent dicistronic RNA on traffic of intracellular MHC-I to the cell surface. C5B 2-3 and 2-3c cells were labeled with [35S]methionine for 15 min at 37°C. After a 0- or 30-min chase in the presence of unlabeled methionine, cells were subjected to surface biotinylation and biotinylated proteins were collected. Samples were immunoprecipitated with the W6/32 monoclonal antibody against MHC-I, and collected proteins were separated by SDS-10% PAGE and analyzed with a PhosphorImager.
Expression of HCV proteins NS4A/B or NS4B causes the accumulation of “membranous webs.”
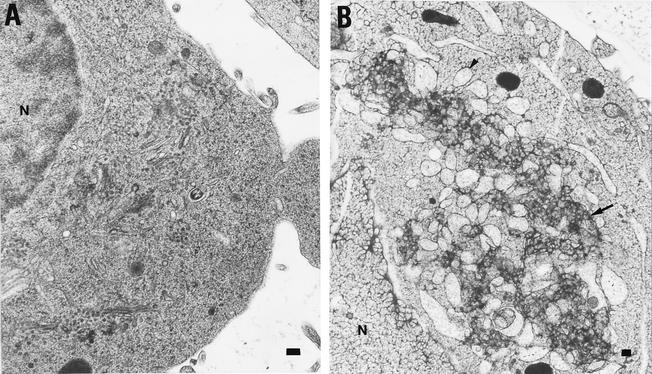
To test whether the HCV NS4A/B protein affected intracellular ultrastructure, human 293T cells that expressed the HCV NS4A/B protein were observed by EM using high-pressure freezing and freeze substitution methods. These methods constitute an improvement over the chemical fixation technique because they allow extremely rapid immobilization and fixation of cellular components as well as improved preservation of labile cellular structures such as membranes (17). Cells were transfected with a control GFP-expressing plasmid and with the plasmid that expressed NS4A/B and GFP on a dicistronic mRNA. As can be seen in Fig. 10B, cells that expressed NS4A/B displayed two types of membrane alteration compared to the control vector-transfected cells: swollen, partially vesiculated membranes and clustered, aggregated membranes.
FIG. 10.
Ultrastructure of 293T cells following transfection with a control plasmid (A) and with a plasmid that expressed NS4A/B (B). Cells were prepared by high-pressure freezing and freeze substitution for electron microscopy. N, nucleus. Bars = 400 nm. Examples of membrane clusters (arrow) and swollen compartments (arrowhead) are indicated.
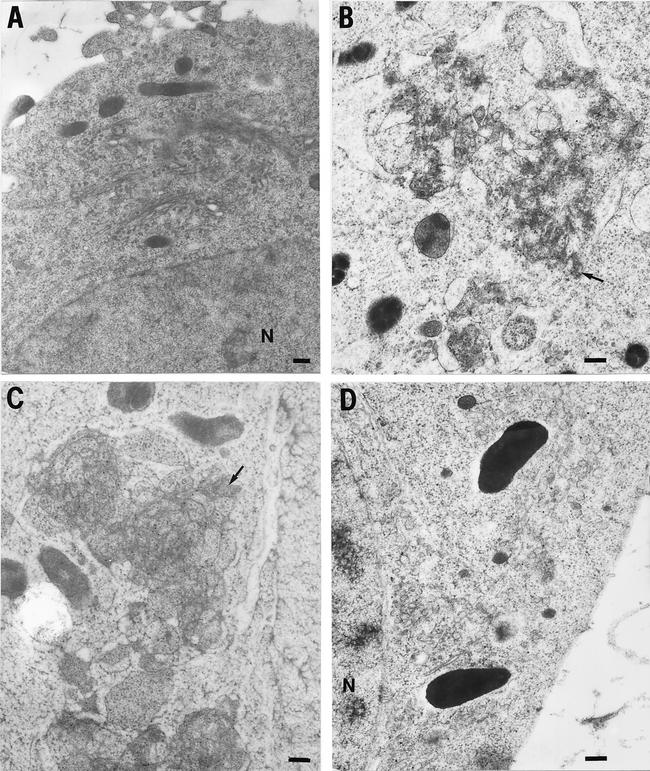
To determine whether the NS4A/B protein was localized to the swollen membranes, the clustered, aggregated membranes, or both, immuno-EM was performed on NS4A/B-transfected cells by using an anti-His6 antibody. No background labeling was observed in cells transfected with the control plasmid (Fig. 11A). In the NS4A/B-expressing cells, the His6-tagged protein was localized exclusively to the clustered, aggregated membranes (Fig. 11B). As shown in Fig. 11C, NS4B-expressing cells displayed similar clustered membranes and the tagged NS4B localized exclusively to these structures. Cells that expressed NS5A (Fig. 11D) displayed some vesiculation compared to control cells but did not display the clustered, aggregated membranes formed by NS4A/B and NS4B.
FIG. 11.
Immunostaining of His6-containing proteins in 293T cells transfected with plasmids that expressed no HCV proteins (A), NS4A/B-His6 (B), NS4B-His6 (C), and HS5A-His6 (D). 293T cells were labeled with 15-nm gold particles conjugated to a secondary antibody against the Penta-His tag monoclonal antibody (Qiagen). Arrows, characteristic locations of NS4B and NS4A/B His-tagged proteins; N, nucleus. Bars = 400 nm.
DISCUSSION
Very little is known about the effect of HCV proteins on the activity of the membranous compartments that form the secretory apparatus. In this study we have expressed several different forms of HCV NS proteins and monitored their effects on one step of intracellular protein traffic: transport from the ER to the Golgi apparatus. There were two reasons to examine this particular step in protein secretion. First, like those of many positive-strand RNA viruses, HCV proteins assemble RNA replication complexes on membranes derived from the ER, making it possible that basic functions of the ER could be deranged. Second, two different proteins encoded by poliovirus, another positive-strand RNA virus, slow the rate of ER-to-Golgi traffic when expressed in isolation, with a protein termed 3A having the stronger effect (23). For poliovirus-infected cells, the reduction in the rate of protein secretion by the wild-type 3A protein is sufficient to reduce the presentation of antigens on the cell surface in the context of MHC-I (20) and the amounts of interleukin-6 (IL-6), IL-8, and beta interferon secreted by three- to fivefold (21).
Here, we showed that HCV NS protein NS4A/B can, in isolation, significantly slow the rate of ER-to-Golgi traffic (Fig. 3 to 6). Only the precursor NS4A/B, not the limit digestion products NS4A or NS4B, singly or in combination, causes this decrease in the rate of protein traffic (Fig. 5 and 6). The effect of NS4A/B is seen for VSV-G, a protein expressed via DNA transfection, and for a variety of other cargo proteins as well (Fig. 7), arguing that the effect of NS4A/B on ER function is not cargo specific.
Sometimes precursor proteins have functions that the limit digestion products do not exhibit. For example, poliovirus proteins 3C and 3D function as a protease and an RNA-dependent RNA polymerase, respectively. However, the nominal precursor protein, 3CD, does not display polymerase activity, binds site specifically to the 5′ noncoding region of the viral RNA, and displays proteolytic substrate specificity distinct from that of the 3C protein (30, 58, 59). For Sindbis virus, the uncleaved precursor nsP1/nsP2/nsP3, in combination with the nsP4 polymerase, supports negative-strand but not positive-strand RNA synthesis, whereas the cleaved forms show increased specificity for positive-strand synthesis (46, 71).
Recent advances in HCV research have led to the construction of self-replicating subgenomic (10, 51) and genomic RNAs (39). To test whether the inhibition of protein secretion observed upon NS4A/B expression could also be seen in cells expressing the HCV polyprotein, we monitored cell surface expression of newly synthesized MHC-I in the presence of a replication-competent, dicistronic HCV-derived RNA. Strain HCV-N (39), from which the replicating RNA was derived, is a genotype 1b strain like HC-J4, the source of the constructs used in Fig. 2 to 6 (81). These two strains display 95% homology throughout their polyprotein sequences and show only four amino acid differences in the sequences of NS4A/B. The rate of MHC-I traffic to the cell surface in cells that harbored the replicating RNA was inhibited by at least threefold in comparison to that in cured derivatives of these cells. This finding is not at variance with the published findings of Moradpour et al. (55), who reported that expressed HCV proteins do not interfere with MHC-I processing and presentation in tissue culture cells, because their study examined the steady-state accumulation of a subset of HLA (A2 and B44) molecules. Due to the long half-lives of some MHC-I molecules on the surfaces of cells (26), steady-state measurements can neglect a feature of MHC-I expression that may be important during active infection, the rate of transfer of newly synthesized MHC-I to the cell surface.
Both NS4A/B and the limit digestion product NS4B cause the accumulation of clustered, aggregated membranes in 293T cells (Fig. 10 and 11) and can be found localized to these membranes (Fig. 11). Only NS4A/B caused the formation of swollen vesicles (Fig. 10), but the protein did not localize to these structures (Fig. 11). Perhaps these membranes are derived from the ER and, as is found when poliovirus 3A is expressed in isolation, became swollen with cargo due to the blockage in ER-to-Golgi traffic (20, 22). The clustered, aggregated membranes common to cells that express NS4A/B and NS4B strongly resemble the membranous web induced in U-2 OS human osteosarcoma cells expressing the polyprotein or NS4B from HCV genotype 1a, strain H77, or in a liver biopsy sample from an HCV-infected chimpanzee (28). Structures such as fragmented rough ERs, ER membranes surrounding and engulfing mitochondria, and convoluted membranes have been described in studies with Huh7 cells harboring a subgenomic replicon (56); these structures were not observed in most of the cells expressing the NS4B or NS4A/B precursor. The roles of the swollen membranes or the membranous web during HCV infection remain to be determined. However, the ultrastructural similarities between the membranes induced by the HCV NS4A/B or NS4B protein and those observed in the liver biopsy sample from the HCV-infected chimpanzee (28, 69) make it likely that the biochemistry and ultrastructure of NS4A/B- and NS4B-expressing cells reported here reflect activities that will be observed during HCV infection in humans.
There have been few reports of putative functions attributed to the NS4A/B and NS4B proteins. The NS4A coding region is a required cofactor for NS3 protease activity (4, 63, 78). Coimmunoprecipitation experiments showed that NS4B and NS4A were found in membrane-associated complexes that contained other HCV NS proteins as well (37). Coimmunofluorescence and fractionation experiments have shown that NS4B colocalizes with other HCV nonstructural proteins on ER membranes (38). Protease susceptibility studies of in vitro-translated NS4B have argued that NS4B is an integral membrane protein, mostly localized on the cytoplasmic side of the ER membrane (38). The nominal precursor NS4A/B has been observed in replicon-expressing cells, although at less abundance than, for example, NS4B/5A or fully processed NS4B (61). In HCV-derived replicons, adaptive mutations that increase the number of replicon-expressing cell colonies obtained after transfection have been identified; one of these adaptive mutations has been identified in the NS4B coding region (50). In BVDV, a mutation in NS4B that converts a cytopathic virus to a noncytopathic phenotype was found (62). These results implicate NS4B or its precursors in cytotoxicity, cell survival, or both, although the mechanisms are not yet known for either viral protein.
It is surprising that proteins encoded by HCV, an enveloped virus, should slow protein traffic. It is not known how HCV is released from infected cells. E1 and E2, the envelope glycoproteins, contain ER retention signals (14, 15) and localize to the ER when expressed singly or together (25). In fact, in previously reported cell lines that contain a replication-competent full-length HCV-derived RNA, Golgi-specific modification of the E2 glycoprotein was not observed (60). One possibility is that HCV, like rotavirus, exits cells via a nonclassical secretory route that bypasses the Golgi (40); then inhibition of the normal secretory pathway downstream of the ER would not affect viral maturation but would still thwart host defenses. The ability of HCV to escape the host immune response, even temporarily, is one of the mysteries of its replicative cycle and a challenge for treatment. Inhibition of protein traffic by NS4A/B could facilitate this evasion by reducing antigen presentation in the context of MHC-I molecules, reducing the secretion of proinflammatory and antiviral cytokines or reducing the abundance of labile membrane proteins on the surfaces of infected cells.
Acknowledgments
We thank Golnar Vazirabadi for expert technical assistance with electron microscopy and William Jackson, Peter Sarnow, and Elizabeth Stillman for critical reading of the manuscript.
This work was supported by NIH grants AI07328 (to K.K.) and U19-AI40035 (to S.M.L.), the Hutchison Foundation for Translational Research, and the Eli Lilly-Stanford Consortium for Hepatitis C Research. K.V.K. was supported by K01 award CA95086-02 from the National Institutes of Health. K.L. was supported by a J. W. McLaughlin Fellowship.
REFERENCES
- 1.Alter, M. J. 1997. Epidemiology of hepatitis C. Hepatology 26:62S-65S. [DOI] [PubMed]
- 2.Bartenschlager, R., L. Ahlborn-Laake, J. Mous, and H. Jacobsen. 1994. Kinetic and structural analyses of hepatitis C virus polyprotein processing. J. Virol. 68:5045-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartenschlager, R., L. Ahlborn-Laake, J. Mous, and H. Jacobsen. 1993. Nonstructural protein 3 of the hepatitis C virus encodes a serine-type proteinase required for cleavage at the NS3/4 and NS4/5 junctions. J. Virol. 67:3835-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. [DOI] [PubMed] [Google Scholar]
- 5.Beard, M. R., G. Abell, M. Honda, A. Carroll, M. Gartland, B. Clarke, K. Suzuki, R. Lanford, D. V. Sangar, and S. M. Lemon. 1999. An infectious molecular clone of a Japanese genotype 1b hepatitis C virus. Hepatology 30:316-324. [DOI] [PubMed] [Google Scholar]
- 6.Bienz, K., D. Egger, T. Pfister, and M. Troxler. 1992. Structural and functional characterization of the poliovirus replication complex. J. Virol. 66:2740-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bienz, K., D. Egger, Y. Rasser, and W. Bossart. 1983. Intracellular distribution of poliovirus proteins and the induction of virus-specific cytoplasmic structures. Virology 131:39-48. [DOI] [PubMed] [Google Scholar]
- 8.Blasco, R., and B. Moss. 1991. Extracellular vaccinia virus formation and cell-to-cell virus transmission are prevented by deletion of the gene encoding the 37,000-Dalton outer envelope protein. J. Virol. 65:5910-5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blasco, R., and B. Moss. 1995. Selection of recombinant vaccinia viruses on the basis of plaque formation. Gene 158:157-162. [DOI] [PubMed] [Google Scholar]
- 10.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 11.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chao, D. S., J. C. Hay, S. Winnick, R. Prekeris, J. Klumperman, and R. H. Scheller. 1999. SNARE membrane trafficking dynamics in vivo. J. Cell Biol. 144:869-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho, M. W., N. Teterina, D. Egger, K. Bienz, and E. Ehrenfeld. 1994. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology 202:129-145. [DOI] [PubMed] [Google Scholar]
- 14.Cocquerel, L., S. Duvet, J. C. Meunier, A. Pillez, R. Cacan, C. Wychowski, and J. Dubuisson. 1999. The transmembrane domain of hepatitis C virus glycoprotein E1 is a signal for static retention in the endoplasmic reticulum. J. Virol. 73:2641-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cocquerel, L., J. C. Meunier, A. Pillez, C. Wychowski, and J. Dubuisson. 1998. A retention signal necessary and sufficient for endoplasmic reticulum localization maps to the transmembrane domain of hepatitis C virus glycoprotein E2. J. Virol. 72:2183-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cormack, B. P., G. Bertram, M. Egerton, N. A. Gow, S. Falkow, and A. J. Brown. 1997. Yeast-enhanced green fluorescent protein (yEGFP), a reporter of gene expression in Candida albicans. Microbiology 143:303-311. [DOI] [PubMed] [Google Scholar]
- 17.Dahl, R., and L. A. Staehelin. 1989. High-pressure freezing for the preservation of biological structure: theory and practice. J. Electron Microsc. Tech. 13:165-174. [DOI] [PubMed] [Google Scholar]
- 18.Dales, S., H. J. Eggers, I. Tamm, and G. E. Palade. 1965. Electron microscopic study of the formation of poliovirus. Virology 26:379-389. [DOI] [PubMed] [Google Scholar]
- 19.De Francesco, R. 1999. Molecular virology of the hepatitis C virus. J. Hepatol. 31(Suppl. 1):47-53. [DOI] [PubMed] [Google Scholar]
- 20.Deitz, S. B., D. A. Dodd, S. Cooper, P. Parham, and K. Kirkegaard. 2000. MHC I-dependent antigen presentation is inhibited by poliovirus protein 3A. Proc. Natl. Acad. Sci. USA 97:13790-13795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dodd, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2001. Poliovirus 3A protein limits interleukin-6 (IL-6), IL-8, and beta interferon secretion during viral infection. J. Virol. 75:8158-8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doedens, J. R., T. H. Giddings, and K. Kirkegaard. 1997. Inhibition of endoplasmic reticulum-to-Golgi traffic by poliovirus protein 3A: genetic and ultrastructural analysis. J. Virol. 71:9054-9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doedens, J. R., and K. Kirkegaard. 1995. Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J. 14:894-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dubuisson, J., F. Penin, and D. Moradpour. 2002. Interaction of hepatitis C virus proteins with host cell membranes and lipids. Trends Cell Biol. 12:517-523. [DOI] [PubMed] [Google Scholar]
- 25.Duvet, S., L. Cocquerel, A. Pillez, R. Cacan, A. Verbert, D. Moradpour, C. Wychowski, and J. Dubuisson. 1998. Hepatitis C virus glycoprotein complex localization in the endoplasmic reticulum involves a determinant for retention and not retrieval. J. Biol. Chem. 273:32088-32095. [DOI] [PubMed] [Google Scholar]
- 26.Eberl, G., C. Widmann, and G. Corradin. 1996. The functional half-life of H-2Kd-restricted T cell epitopes on living cells. Eur. J. Immunol. 26:1993-1999. [DOI] [PubMed] [Google Scholar]
- 27.Egger, D., N. Teterina, E. Ehrenfeld, and K. Bienz. 2000. Formation of the poliovirus replication complex requires coupled viral translation, vesicle production, and viral RNA synthesis. J. Virol. 74:6570-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Egger, D., B. Wolk, R. Gosert, L. Bianchi, H. E. Blum, D. Moradpour, and K. Bienz. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elbers, K., N. Tautz, P. Becher, D. Stoll, T. Rumenapf, and H. J. Thiel. 1996. Processing in the pestivirus E2-NS2 region: identification of proteins p7 and E2p7. J. Virol. 70:4131-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gamarnik, A. V., and R. Andino. 2000. Interactions of viral protein 3CD and poly(rC) binding protein with the 5′ untranslated region of the poliovirus genome. J. Virol. 74:2219-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grakoui, A., D. W. McCourt, C. Wychowski, S. M. Feinstone, and C. M. Rice. 1993. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J. Virol. 67:2832-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grakoui, A., D. W. McCourt, C. Wychowski, S. M. Feinstone, and C. M. Rice. 1993. A second hepatitis C virus-encoded proteinase. Proc. Natl. Acad. Sci. USA 90:10583-10587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gray, E. W., and P. F. Nettleton. 1987. The ultrastructure of cell cultures infected with border disease and bovine virus diarrhoea viruses. J. Gen. Virol. 68:2339-2346. [DOI] [PubMed] [Google Scholar]
- 34.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hijikata, M., N. Kato, Y. Ootsuyama, M. Nakagawa, and K. Shimotohno. 1991. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc. Natl. Acad. Sci. USA 88:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hijikata, M., H. Mizushima, T. Akagi, S. Mori, N. Kakiuchi, N. Kato, T. Tanaka, K. Kimura, and K. Shimotohno. 1993. Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J. Virol. 67:4665-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang, Y., Y. Uchiyama, T. Fujimura, H. Kanamori, T. Doi, A. Takamizawa, T. Hamakubo, and T. Kodama. 2001. A human hepatoma cell line expressing hepatitis C virus nonstructural proteins tightly regulated by tetracycline. Biochem. Biophys. Res. Commun. 281:732-740. [DOI] [PubMed] [Google Scholar]
- 38.Hugle, T., F. Fehrmann, E. Bieck, M. Kohara, H. G. Krausslich, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. The hepatitis C virus nonstructural protein 4B is an integral endoplasmic reticulum membrane protein. Virology 284:70-81. [DOI] [PubMed] [Google Scholar]
- 39.Ikeda, M., M. Yi, K. Li, and S. M. Lemon. 2002. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J. Virol. 76:2997-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jourdan, N., M. Maurice, D. Delautier, A. M. Quero, A. L. Servin, and G. Trugnan. 1997. Rotavirus is released from the apical surface of cultured human intestinal cells through nonconventional vesicular transport that bypasses the Golgi apparatus. J. Virol. 71:8268-8278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kato, N., M. Hijikata, Y. Ootsuyama, M. Nakagawa, S. Ohkoshi, T. Sugimura, and K. Shimotohno. 1990. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc. Natl. Acad. Sci. USA 87:9524-9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim, J. E., W. K. Song, K. M. Chung, S. H. Back, and S. K. Jang. 1999. Subcellular localization of hepatitis C viral proteins in mammalian cells. Arch. Virol. 144:329-343. [DOI] [PubMed] [Google Scholar]
- 43.Kolykhalov, A. A., E. V. Agapov, and C. M. Rice. 1994. Specificity of the hepatitis C virus NS3 serine protease: effects of substitutions at the 3/4A, 4A/4B, 4B/5A, and 5A/5B cleavage sites on polyprotein processing. J. Virol. 68:7525-7533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kornfeld, R., and S. Kornfeld. 1985. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 54:631-664. [DOI] [PubMed] [Google Scholar]
- 45.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 46.LaStarza, M. W., J. A. Lemm, and C. M. Rice. 1994. Genetic analysis of the nsP3 region of Sindbis virus: evidence for roles in minus-strand and subgenomic RNA synthesis. J. Virol. 68:5781-5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin, C., B. D. Lindenbach, B. M. Pragai, D. W. McCourt, and C. M. Rice. 1994. Processing in the hepatitis C virus E2-NS2 region: identification of p7 and two distinct E2-specific products with different C termini. J. Virol. 68:5063-5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin, C., B. M. Pragai, A. Grakoui, J. Xu, and C. M. Rice. 1994. Hepatitis C virus NS3 serine proteinase: trans-cleavage requirements and processing kinetics. J. Virol. 68:8147-8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lodish, H. F., N. Kong, M. Snider, and G. J. Strous. 1983. Hepatoma secretory proteins migrate from rough endoplasmic reticulum to Golgi at characteristic rates. Nature 304:80-83. [DOI] [PubMed] [Google Scholar]
- 50.Lohmann, V., F. Korner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 52.Mackenzie, J. M., M. K. Jones, and E. G. Westaway. 1999. Markers for trans-Golgi membranes and the intermediate compartment localize to induced membranes with distinct replication functions in flavivirus-infected cells. J. Virol. 73:9555-9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mackenzie, J. M., M. K. Jones, and P. R. Young. 1996. Improved membrane preservation of flavivirus-infected cells with cryosectioning. J. Virol. Methods 56:67-75. [DOI] [PubMed] [Google Scholar]
- 54.Miller, R. H., and R. H. Purcell. 1990. Hepatitis C virus shares amino acid sequence similarity with pestiviruses and flaviviruses as well as members of two plant virus supergroups. Proc. Natl. Acad. Sci. USA 87:2057-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moradpour, D., B. Grabscheid, A. R. Kammer, G. Schmidtke, M. Groettrup, H. E. Blum, and A. Cerny. 2001. Expression of hepatitis C virus proteins does not interfere with major histocompatibility complex class I processing and presentation in vitro. Hepatology 33:1282-1287. [DOI] [PubMed] [Google Scholar]
- 56.Mottola, G., G. Cardinali, A. Ceccacci, C. Trozzi, L. Bartholomew, M. R. Torrisi, E. Pedrazzini, S. Bonatti, and G. Migliaccio. 2002. Hepatitis C virus nonstructural proteins are localized in a modified endoplasmic reticulum of cells expressing viral subgenomic replicons. Virology 293:31-43. [DOI] [PubMed] [Google Scholar]
- 57.Park, J. S., J. M. Yang, and M. K. Min. 2000. Hepatitis C virus nonstructural protein NS4B transforms NIH3T3 cells in cooperation with the Ha-ras oncogene. Biochem. Biophys. Res. Commun. 267:581-587. [DOI] [PubMed] [Google Scholar]
- 58.Parsley, T. B., C. T. Cornell, and B. L. Semler. 1999. Modulation of the RNA binding and protein processing activities of poliovirus polypeptide 3CD by the viral RNA polymerase domain. J. Biol. Chem. 274:12867-12876. [DOI] [PubMed] [Google Scholar]
- 59.Parsley, T. B., J. S. Towner, L. B. Blyn, E. Ehrenfeld, and B. L. Semler. 1997. Poly(rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA 3:1124-1134. [PMC free article] [PubMed] [Google Scholar]
- 60.Pietschmann, T., V. Lohmann, A. Kaul, N. Krieger, G. Rinck, G. Rutter, D. Strand, and R. Bartenschlager. 2002. Persistent and transient replication of full-length hepatitis C virus genomes in cell culture. J. Virol. 76:4008-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pietschmann, T., V. Lohmann, G. Rutter, K. Kurpanek, and R. Bartenschlager. 2001. Characterization of cell lines carrying self-replicating hepatitis C virus RNAs. J. Virol. 75:1252-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qu, L., L. K. McMullan, and C. M. Rice. 2001. Isolation and characterization of noncytopathic pestivirus mutants reveals a role for nonstructural protein NS4B in viral cytopathogenicity. J. Virol. 75:10651-10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reed, K. E., and C. M. Rice. 2000. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr. Top. Microbiol. Immunol. 242:55-84. [DOI] [PubMed] [Google Scholar]
- 64.Rothman, J. E., and H. F. Lodish. 1977. Synchronised transmembrane insertion and glycosylation of a nascent membrane protein. Nature 269:775-780. [DOI] [PubMed] [Google Scholar]
- 65.Rumenapf, T., G. Unger, J. H. Strauss, and H. J. Thiel. 1993. Processing of the envelope glycoproteins of pestiviruses. J. Virol. 67:3288-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Scales, S. J., R. Pepperkok, and T. E. Kreis. 1997. Visualization of ER-to-Golgi transport in living cells reveals a sequential mode of action for COPII and COPI. Cell 90:1137-1148. [DOI] [PubMed] [Google Scholar]
- 67.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 68.Schlegel, A., T. H. Giddings, Jr., M. S. Ladinsky, and K. Kirkegaard. 1996. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J. Virol. 70:6576-6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shimizu, Y. K., S. M. Feinstone, R. H. Purcell, H. J. Alter, and W. T. London. 1979. Non-A, non-B hepatitis: ultrastructural evidence for two agents in experimentally infected chimpanzees. Science 205:197-200. [DOI] [PubMed] [Google Scholar]
- 70.Shimizu, Y. K., A. J. Weiner, J. Rosenblatt, D. C. Wong, M. Shapiro, T. Popkin, M. Houghton, H. J. Alter, and R. H. Purcell. 1990. Early events in hepatitis C virus infection of chimpanzees. Proc. Natl. Acad. Sci. USA 87:6441-6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shirako, Y., and J. H. Strauss. 1994. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J. Virol. 68:1874-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Suhy, D. A., T. H. Giddings, and K. Kirkegaard. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74:8953-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Teterina, N. L., K. Bienz, D. Egger, A. E. Gorbalenya, and E. Ehrenfeld. 1997. Induction of intracellular membrane rearrangements by HAV proteins 2C and 2BC. Virology 237:66-77. [DOI] [PubMed] [Google Scholar]
- 74.Teterina, N. L., D. Egger, K. Bienz, D. M. Brown, B. L. Semler, and E. Ehrenfeld. 2001. Requirements for assembly of poliovirus replication complexes and negative-strand RNA synthesis. J. Virol. 75:3841-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Waris, G., K. D. Tardif, and A. Siddiqui. 2002. Endoplasmic reticulum (ER) stress: hepatitis C virus induces an ER-nucleus signal transduction pathway and activates NF-kappaB and STAT-3. Biochem. Pharmacol. 64:1425-1430. [DOI] [PubMed] [Google Scholar]
- 76.Weed, H. G., G. Krochmalnic, and S. Penman. 1985. Poliovirus metabolism and the cytoskeletal framework: detergent extraction and resinless section electron microscopy. J. Virol. 56:549-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Westaway, E. G., J. M. Mackenzie, M. T. Kenney, M. K. Jones, and A. A. Khromykh. 1997. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J. Virol. 71:6650-6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wölk, B., D. Sansonno, H. G. Kräusslich, F. Dammacco, C. M. Rice, H. E. Blum, and D. Moradpour. 2000. Subcellular localization, stability, and trans-cleavage competence of the hepatitis C virus NS3-NS4A complex expressed in tetracycline-regulated cell lines. J. Virol. 74:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.World Health Organization. 2000. Hepatitis C—global prevalence (update). Wkly. Epidemiol. Rec. 75:18-19. [PubMed] [Google Scholar]
- 80.Xu, J., E. Mendez, P. R. Caron, C. Lin, M. A. Murcko, M. S. Collett, and C. M. Rice. 1997. Bovine viral diarrhea virus NS3 serine proteinase: polyprotein cleavage sites, cofactor requirements, and molecular model of an enzyme essential for pestivirus replication. J. Virol. 71:5312-5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yanagi, M., M. St. Claire, M. Shapiro, S. U. Emerson, R. H. Purcell, and J. Bukh. 1998. Transcripts of a chimeric cDNA clone of hepatitis C virus genotype 1b are infectious in vivo. Virology 244:161-172. [DOI] [PubMed] [Google Scholar]