Abstract
Triple-helix-forming oligonucleotides (TFOs), which can potentially modify target genes irreversibly, represent promising tools for antiviral therapies. However, their effectiveness on endogenous genes has yet to be unambiguously demonstrated. To monitor endogenous gene modification by TFOs in a yeast model, we inactivated an auxotrophic marker gene by inserting target sequences of interest into its coding region. The genetically engineered yeast cells then were treated with psoralen-linked TFOs followed by UV irradiation, thus generating highly mutagenic covalent crosslinks at the target site whose repair could restore gene function; the number of revertants and spectrum of mutations generated were quantified. Results showed that a phosphoramidate TFO indeed reaches its target sequence, forms crosslinks, and generates mutations at the expected site via a triplex-mediated mechanism: (i) under identical conditions, no mutations were generated by the same TFO at two other loci in the target strain, nor in an isogenic control strain carrying a modified target sequence incapable of supporting triple-helix formation; (ii) for a given target sequence, whether the triplex was formed in vivo on an endogenous gene or in vitro on an exogenous plasmid, the nature of the mutations generated was identical, and consistent with the repair of a psoralen crosslink at the target site. Although the mutation efficiency was probably too low for therapeutic applications, our results confirm the validity of the triple-helix approach and provide a means of evaluating the effectiveness of new chemically modified TFOs and analogs.
Keywords: oligonucleotide, psoralen, HIV-1 polypurine tract
Triple-helix-forming oligonucleotides (TFOs), which recognize double-stranded DNA targets in a sequence-specific manner, are promising tools for a variety of applications (1, 2). In particular, TFOs can be used to introduce chemical modifications into selected targets in a sequence-specific manner. For example, a TFO can be covalently linked to a psoralen molecule. Psoralen is a bifunctional photoreagent that, upon irradiation at 365 nm, introduces a covalent crosslink into the target sequence (3, 4). Thus TFOs have the potential to irreversibly modify target genes, such as integrated retroviral genomes, and are envisioned as promising antiviral molecules.
TFOs have been shown to be efficient in modulating gene activity when assessed on exogenous reporter plasmids, when the triple helix is preformed outside the cells. However, formation of triple helices is not favored under physiological conditions, and TFOs must be chemically modified to increase their affinity for the target sequence. Data suggesting that TFOs can modify endogenous gene activity are scarce (5–9), and major improvements must be made for these products to attain satisfactory levels of efficiency. More importantly, a triplex-induced mode of action remains to be demonstrated by using proper negative controls. Indeed, oligonucleotides can have unexpected side effects in live cells. For example, it is well known that they can bind to intracellular proteins in a sequence-specific manner (10, 11) and thus may have biological effects that are related to their sequence but irrelevant to triple-helix formation (12, 13). For negative controls, it is thus critical to change the sequence of the target rather than that of the oligonucleotide.
Here, we have used an assay designed to detect and quantify the functional interaction between a TFO and an endogenous target sequence. Yeast, in which genes can be easily modified, was chosen as a model system. The polypurine tract (PPT), an oligopurine/oligopyrimidine sequence flanked by a 5′-TpA-3′ site (Fig. 1A) from the HIV-1 genome, was used as a target. The PPT was introduced into the yeast URA3 gene, an auxotrophic marker that was rendered inactive by an Ochre mutation in the process. A TFO covalently attached to a psoralen molecule was used to introduce crosslinks into the target sequence, at the site of the Ochre mutation. TFO-targeted crosslinks are highly mutagenic in eukaryotic cells (14, 15), and mutations reverting the negative phenotype of the auxotrophic marker can be easily detected in yeast by selection of the cells on an appropriate medium, as demonstrated by preliminary experiments using exogenous plasmids (16). In those studies, the number and nature of the mutations were found to depend on the orientation of the target sequence with reference to the URA3 gene promoter, due to transcription-coupled repair and the specific orientation of the psoralen crosslink in the triple-helix complex (17).
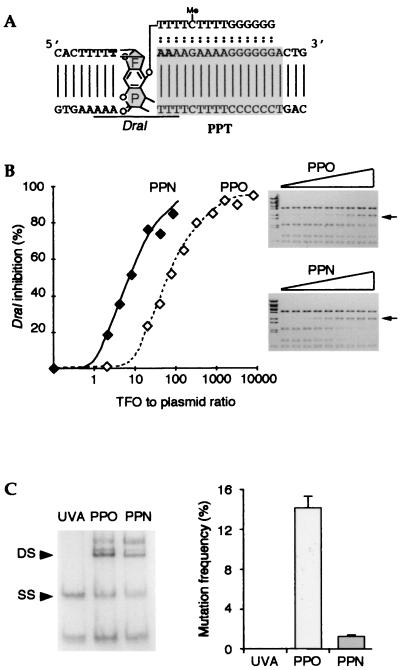
Figure 1.
Analysis of TFOs in vitro and on exogenous plasmids. (A) Sequences of the target gene and the TFO (shown bound on the target gene); two variants of the TFO were used with identical length and sequence but differing in their chemical modification (see text). The ochre codon is indicated in bold. The cytosine in the oligonucleotide is methylated. The furane (F) and pyrone (P) sides of the adduct are indicated, as is the position of the DraI restriction site. (B) The PPN TFO binds to the target sequence with greater affinity than does the PPO TFO. A plasmid including the target sequence (YEplac181_ura3∷hiv1pur) was incubated with increasing doses of TFOs, irradiated with near UV light, and submitted to DraI restriction. Shown are dose curves of DraI inhibition by the PPN or the PPO oligonucleotides, as indicated. (C) Frequency of the mutations generated on an exogenous episomal plasmid (YEplac112_ura3∷hiv1pyr) by the PPO TFO, the PPN TFO, or in the absence of TFO (UVA). Triple helices were preformed on plasmids in vitro. Samples were irradiated, resulting in the introduction of covalent crosslinks into a high proportion of the target plasmids—proportions, which, under the conditions used, were similar for both TFOs as shown by renaturing gel electrophoresis analysis (Left; DS, double-stranded, crosslinked species; SS, single-stranded, noncrosslinked species). The plasmids were used to transform ura3–52 yeast cells. The mutation frequency was defined as the ratio between the number of induced URA3+ revertants and the total number of transformants. Shown is the mean value of three independent experiments.
In the present study, we have used two TFOs, identical in length and sequence, designed to recognize the HIV-1 PPT: a natural phosphodiester (PPO) and a chemically modified, N3′–P5′ phosphoramidate (PPN) (18) oligonucleotide. This last variant displays a better affinity for the target sequence when assayed in vitro and is more resistant to nucleases (19). Both TFOs were able to introduce mutations when used on exogenous targets, but only the higher affinity PPN TFO was active on an endogenous target inserted into the yeast genome. The mutations generated by the PPN TFO were indeed caused by triple-helix formation, because (i) modification of the target sequence abolished their generation, and (ii) mutations generated in an endogenous sequence were identical in nature to those generated on an exogenous plasmid on which the triple helix was preformed in vitro and showed the same dependence on the orientation of the target sequence.
However, the efficiency of gene modification by the PPN TFO was quite low (the mutation frequency was increased only 10- to 100-fold above the background). Furthermore, when assayed on two related endogenous targets that differ by 3 nt without any reversion from pyrimidine to purine, the PPN TFO showed some level of cross-reactivity. Further chemical modifications thus will be needed to improve TFOs and render therapeutic applications possible. The experimental approach described here, which can be adapted to any target of choice, should be useful to monitor those improvements both quantitatively and qualitatively.
Materials and Methods
Oligonucleotides.
The PPO TFO (sequence shown in Fig. 1) coupled to psoralen (16, 17) was obtained from Appligen Oncor (Illkirch, France). The PPN TFO (identical sequence, see Fig. 1) (20) coupled to psoralen was obtained from Lynx Therapeutics (Foster City, CA). Unmodified oligonucleotides that were used for constructions, PCR and sequencing, were purchased from Eurogentec (Seraing, Belgium).
Construction of Modified URA3 Alleles.
The URA3 defective alleles containing the HIV-1 PPT, the HIV-2 PPT, or a modified version of the HIV-1 PPT used as a control were constructed by inserting 42-bp fragments immediately downstream from the ATG initiation codon in the two possible orientations, generating, respectively, ura3∷hiv1pur, ura3∷hiv1pyr, ura3∷hiv2pur, ura3∷hiv2pyr, ura3∷TpApur, and ura3∷TpApyr (see Fig. 4A for the sequence of the pyr alleles). YEplac181 and YEplac112 episomal plasmids carrying these alleles were constructed for use in experiments on exogenous DNA (16, 17). Strains containing these alleles were derived from FF18 733 (Mat a, ura3–52, trp1–289, leu2-, his7–2, lys1–1), by homologous recombination using standard procedures. In addition, two strains containing the HIV-1 PPT in direct or inverted orientations were derived from CmY826 (Mat a, ura3–52, trp1Δ163, leu2Δ1, his3-200, lys2–801, ade2–101, bar1∷HIS3). Replacement of the endogenous URA3 allele by the modified versions was verified by Southern analysis and direct nucleotide sequencing.
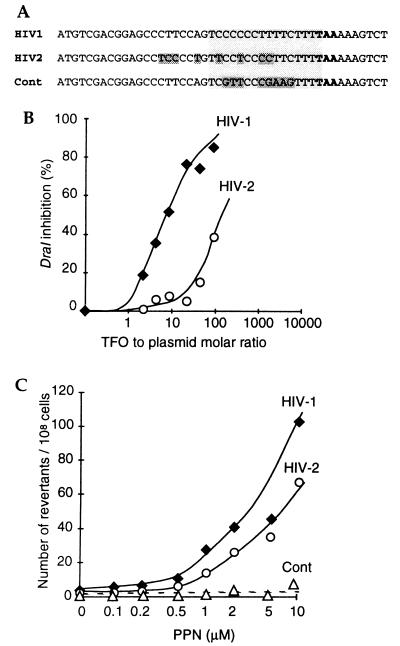
Figure 4.
Cross-reaction between HIV-1 and HIV-2 oligopurine sequences. (A) Sequences of the HIV-1, HIV-2, and control (Cont) targets. The triple-helix target is boxed, and the nucleotides that differ from the HIV-1 sequence are indicated by darker boxes in the HIV-2 and control target sequences. (B) Association of PPN TFO with HIV-1 and HIV-2 in vitro. Dose curves of DraI restriction enzyme inhibition by the PPN oligonucleotide on plasmids including the HIV-1 or the HIV-2 target (YEplac181_ura3∷hiv1pur and _ura3∷hiv2pur), as indicated. (C) Number of revertants generated at the target site by the PPN TFO on HIV-1, HIV-2, or control endogenous targets, as indicated. FF18 733-derived cells harboring the HIV-1, HIV-2, or control sequence shown in Fig. 4A were electroporated, irradiated, and selected as in Fig. 2.
Assays on Exogenous Plasmids.
Triplex formation and irradiation, DraI restriction analysis, renaturing gel electrophoresis, and yeast transformation experiments were performed as described (16, 17).
Assays on Endogenous Genes.
Cells were made competent for electroporation as described (21). For each assay, 108 cells were used in a total volume of 50 μl. A single square pulse of 2 kV/cm and 12 ms was delivered by using a Bio-Rad electroporator with an RF module. After the pulse, cells were allowed to recover for 10 min. They then were resuspended in PBS (108 cells/ml), irradiated for 5 min at room temperature by using a 365-nm monochromatic lamp (Bioblock, Illkirch, France) and plated on medium lacking uracil, adenine, or lysine.
Molecular Analysis of Mutants.
DNA from mutants was isolated by using standard procedures (22). The URA3 allele was amplified by PCR with primers specific for the endogenous allele or primers specific for the plasmid-borne allele. PCR products were purified by using a QiaQuick PCR purification kit (Qiagen, Chatsworth, CA) and sequenced by using nested primers.
Results
The PPN and PPO TFOs Generate Mutations on Exogenous Plasmids.
In preliminary experiments, the selection system was used to demonstrate, quantify, and analyze the effects of psoralen-linked TFOs after preforming triplexes on exogenous plasmids harboring the target PPT inserted into a URA3 coding sequence in such a way as to inactivate it by an ochre mutation at the psoralen insertion site (Fig. 1A, TAA). Two TFOs, a PPO and a PPN, were used. They were of identical length and sequence and previously have been used to bind to the PPT sequence and target psoralen crosslink formation after UVA irradiation (365 nm) (23, 24). The affinities of the PPO and PPN TFOs for the target were compared in vitro, using an assay in which digestion of the target by DraI, at the site overlapping the psoralen insertion sequence (see Fig. 1A), is inhibited by the crosslinked TFO. Results (Fig. 1B) indicated that the PPN TFO was 10-fold more efficient in inhibiting DraI at the target site, indicating that it displays a better affinity for the sequence than the PPO TFO. Moreover, DraI digestion at other sites carried by the plasmids was not affected, demonstrating the specificity of the crosslink targeting (Fig. 1B Right). To test the ability of the TFOs to modify a gene when linked to their target sequence, the two TFOs were next crosslinked to a target plasmid by irradiation at 365 nm by using saturating concentrations of TFOs, and thus under conditions that resulted in similar efficiencies of crosslinking (Fig. 1C Left). Standardized amounts of crosslinked plasmids were introduced into yeast cells, and URA3 revertants were selected on an appropriate medium. PPO and PPN-treated plasmids had similar transformation efficiencies. Both oligonucleotides generated mutations in the target URA3 gene, although the PPN TFO generated fewer mutations than the PPO TFO (Fig. 1C). These mutations were similar in nature and were generated by similar repair pathways (ref. 16 and data not shown).
The PPN TFO Generates Mutations in an Endogenous Target.
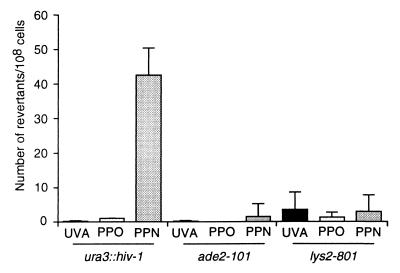
The two TFOs were analyzed next on the PPT target inserted into the endogenous URA3 coding sequence. TFOs were electroporated (21) into the genetically modified yeast. Cells then were irradiated, and URA3 revertants were selected on medium lacking uracil. The PPN TFO generated a significant number of mutations in the target gene (Fig. 2, ura3∷hiv-1). This was not observed at two other loci corresponding to auxotrophic marker genes harboring either the same ochre mutation (Fig. 2, ade2–101) or an amber mutation (Fig. 2, lys2–801). Most importantly (Table 1), no mutations were observed on a sequence inserted into the URA3 gene at the same location as the cognate sequence but which impedes triplex formation in vitro, even though it contains the 5′-TpA-3′ psoralen target site of the PPT (Control, Table 1). This result indicates that the mutations generated in the PPT target sequence (Table 1, Fig. 2) resulted from the interaction with the TFO. Under the same conditions, the PPO oligonucleotide did not give rise to mutagenesis above background levels. UVA irradiation was found to be necessary for PPN-targeted mutagenesis (data not shown), and the mutation spectrum was consistent with psoralen crosslink-induced mutagenesis (Fig. 3C). In all experiments oligonucleotides designed for gene conversion at the URA3 locus (21) were used as positive controls to test for reversion of the ura3-deficient alleles. The oligonucleotide designed for gene conversion of the HIV-1 target generated numbers of transformants that were similar to those obtained through the PPN TFO (data not shown).
Figure 2.
A PPN TFO generates mutations in a cognate endogenous target sequence and not in unrelated genes. Number of revertants generated by a PPO TFO, a PPN TFO, or in the absence of TFO (UVA), as indicated, on the HIV-1 target sequence inserted into the endogenous URA3 gene (ura3∷hiv-1), or on irrelevant genes (ade2–101 and lys2–801) of a CmY826-derived strain. TFOs were electroporated into intact yeast cells, cells were irradiated, and URA3+, ADE2+, or LYS2+ revertants were selected on the appropriate medium. Shown is the mean value of three independent experiments. The HIV-1 target sequence is shown in Fig. 4A.
Table 1.
Generation of mutations is caused by triple-helix formation
| Target
|
||
|---|---|---|
| HIV-1* | Control* | |
| UVA | 4.6 (3-3-6-5-6)† | 2 (0-3-2-4-1)† |
| PPN, 1 μM | 22 (20-28-18)† | 3.6 (5-5-1)† |
| PPN, 10 μM | 73.5 (104-43)† | 5.5 (8-3)† |
Both strains were derived from FF18 733. The sequences of the HIV-1 and control target are shown in Fig. 4A (HIV-1, Cont).
† Number of transformants per 108 cells: mean. Individual results of independent experiments are indicated in parenthesis.
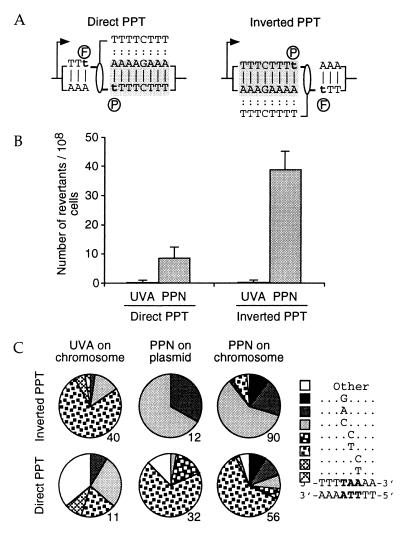
Figure 3.
The frequency and types of mutations generated by the PPN TFO depend on the orientation of the target sequence with respect to the gene promoter. (A) Sequence of the target in the direct and inverted orientations showing the preferential orientation of the psoralen crosslink positioned by the TFO. (B) Number of revertants generated at the target site in the direct and inverted orientations. CmY826-derived cells carrying a direct or inverted HIV-1 PPT were electroporated and selected as in Fig. 2. (C) Mutations generated on exogenous or endogenous targets are similar in nature. Graphs indicate the proportion of each type of mutation, determined by sequencing the mutants obtained through UVA irradiation (UVA on chromosome), through action of the PPN TFO on endogenous HIV-1 PPT targets (PPN on chromosome), or in experiments in which the triplex was preformed on plasmids (YEplac112_ura3∷hiv1pur and _ura3∷hiv1pyr) in vitro (PPN on plasmid). The number of mutants analyzed is indicated below each graph. A similar effect of the orientation of the target was observed in the FF18 733 background, so that sequencing results obtained in both the CmY826 and the FF18 733 backgrounds were pooled.
Comparison Between Mutations Generated in Exogenous and Endogenous Targets.
Further demonstration of a triple-helix-induced effect comes from a comparison of the mutations observed on the endogenous gene on the one hand, and the mutations generated on an exogenous plasmid, on which the triple helix was preformed before introduction into the cells, on the other hand. In the experiments on exogenous plasmids, the number and type of lesions generated by irradiation of the triplex can be precisely assessed in vitro, so that induced mutagenesis can be unambiguously attributed to triple-helix-directed psoralen crosslinks (16, 17).
Using the PPO oligonucleotide, we previously had demonstrated (17) that the frequency of the mutations generated on an exogenous plasmid depended on the orientation of the target sequence with respect to the gene promoter (Fig. 3A): about three times more mutations were generated in the “inverted” orientation (by reference to the orientation of the sequence in the HIV-1 genome) than in the direct orientation. In addition, the orientation of the sequence also influenced the nature of the mutations generated by crosslinks introduced through a TFO (17). This asymmetry was shown to be the result of (i) the preferential repair of the transcribed strand, (ii) the differential repair of the furane and the pyrone adducts of the psoralen, and (iii) the preferential orientation of the psoralen molecule in the triple-helix-targeted crosslink, with the furane side of the psoralen linked to the purine strand of the target sequence (Fig. 1A). The same asymmetry was observed with the PPN TFO assayed on exogenous plasmids (data not shown). Strikingly similar results were obtained with the target sequence inserted into the endogenous URA3 gene (Fig. 3B). The inverted orientation resulted in four times more mutations than the direct orientation. Furthermore, the spectrum of mutations generated in a given orientation on the exogenous plasmid matched that generated on the endogenous gene in the same orientation (Fig. 3C). In addition, these mutations were distinct from those observed in control cells treated with UV only (UVA). Taken together, these results indicate that the mutations in the endogenous gene result from the formation of a triple helix at the target site.
Specificity of Gene Modification.
The specificity of the interaction in live cells is a very important issue for oligonucleotide-based therapeutic applications. To address this issue, we have compared two homologous target sequences inserted at the same locus in isogenic yeast strains: the HIV-1 and HIV-2 PPTs differ by a few nucleotides without any interruption in the oligopurine tract (Fig. 4A). The PPN TFO discriminates between these two sequences in vitro, and, in the DraI inhibition assay, the ID50 (dose at which a 50% inhibition is observed) on HIV-2 is about 20-fold higher than that on HIV-1 (Fig. 4B). In vivo, however, the difference between the two sequences was less pronounced, and the dose curves of generated mutants were hardly distinguishable (Fig. 4C). Note that the PPN TFO did not modify a control sequence with pyrimidine to purine reversion (Fig. 4C, Cont). This result indicates that the mutations in HIV-1 and HIV-2 derived from triple-helix formation, which in vivo occurs on the two HIV PPTs.
Discussion
The ability of TFOs to modify a selected target gene in live cells is an important issue, in view of the potential applications of this strategy, in particular in antiviral therapies. The use of psoralen-modified TFOs to target mutagenesis on an endogenous gene has been described in two other studies, using as a target the HPRT gene (8) or multiple integrated copies of a modified SupF gene (9). These reports can be taken as proofs of concept for the TFO strategy. However, in neither case was demonstrated the absence of effect of the TFO on an isogenic cell line carrying a control sequence in place of the target sequence. In addition, the reports differ by the nature of the observed mutations, and none of these mutations corresponded to those described in the literature after treatment of cells with psoralen. Finally, for Majumbar et al. (8) the mutations generated depended on UVA treatment of the cells, whereas Vasquez et al. (9) found the UVA activation unnecessary. Thus, the mechanism and specificity of action of psoralen-TFOs in those studies remains to be investigated. We here demonstrate without ambiguity that a TFO designed to bind to and to modify a triple-helix target sequence from the HIV retrovirus is indeed able to introduce mutations into the target gene in live cells. These mutations are generated through a triple-helix effect, because the same TFO was unable to modify a control target inserted at the same locus in isogenic strains. Furthermore, mutations were not observed in the absence of UVA, were identical in nature to those obtained after triple-helix formation in vitro, and corresponded to those expected for psoralen crosslinks.
Two oligonucleotides of identical length and sequence were used in this study, a natural PPO and a chemically modified variant (PPN). They differ by their affinity for the target (Fig. 1) and by their resistance to nucleases (19). When the two TFOs were compared for their efficiency in generating mutations on exogenous plasmids, on which the triplex was preformed under optimal conditions in vitro, the natural PPO oligonucleotide was more efficient than the chemically modified PPN. However, opposite results were obtained when the TFOs were assayed on endogenous targets, and the PPN was the only one with which mutations were detected above background levels. This discrepancy might be explained by the difference between the two assays. On exogenous plasmids, on the one hand, the triple helix is preformed in vitro by using saturating concentrations of TFOs, and the TFO is crosslinked to the target before the introduction of the complex into cells. The affinity of the TFO for the sequence thus is not limiting in this assay, and the intracellular stability of the preformed triplex seems to be the limiting parameter. If they are long enough, oligonucleotide tails can affect the repair of a psoralen crosslink in vitro (25) as well as in cell extracts (26). We previously have shown that the 15-mer PPO does not affect the repair process in yeast cells (17). This suggests that the phosphodiester tail at the site of the crosslink is rapidly degraded in these cells (17). The PPN, in contrast, is more resistant to nucleases and might impede the processing of the lesion (repair and replication) and thus the establishment of mutated cell clones, resulting in the observed low number of mutations generated on exogenous plasmids. In the assay on an endogenous target, on the other hand, both the affinity of the TFO for the sequence and the intracellular stability of the TFO are crucial parameters. The PPN, which has a better affinity than the PPO and is more stable, is thus the only one able to form an intracellular covalent triplex and hence to generate mutations.
The PPN TFO generates mutations in vivo in the cognate sequence and not in a modified target, demonstrating that the generation of mutations results from the formation of a triple helix in vivo. However, the frequency of mutations generated (10−6), although it is between 1 and 2 orders of magnitude higher than the background level of mutations, is low. Indeed, in the case of a preformed triple helix using exogenous plasmids with a high proportion of crosslinks (around 90%), the maximal frequency of mutations obtained was on the order of 10−3 (10−2, with about only 10−1 of the plasmids being repaired and replicated). If this value is taken to represent the expected maximal frequency of mutations on an endogenous target, the proportion of targets reached by the TFO thus can be estimated as one in a thousand—a proportion too low to envision any therapeutic applications. This estimation, however, probably represents a minimal figure, because of other influences on triplex formation in live cells. Indeed, the target sequence in the URA3 gene, at least in the native configuration, is located in a nucleosome and not in linker DNA (27). Nucleosome core particles decrease the association of a TFO with its target sequence (28) and thus might reduce the efficiency of the PPN TFO in this study.
Another limiting parameter is the specificity of the interaction. Indeed, the PPN TFO designed to recognize the HIV-1 target was almost as efficient in binding an HIV-2 target, which differs by 3 nt. This finding could have several explanations. First, because of the low efficiency of gene modification, high concentrations of the PPN TFO (resulting in a large excess of TFO in the cells after electroporation) were necessary to observe a significant effect. The tract of 7 nt that the two sequences have in common (see Fig. 4A) might be sufficient to bind the TFO under these conditions. Indeed, a 100-fold excess is sufficient to allow binding of the PPN TFO to the HIV-2 target in vitro (Fig. 4B). Quantitative improvements of the oligonucleotide delivery to the target site are likely to increase the specificity of the interaction in live cells. In addition, the chromatin structure, which could be different in the context of the two sequences, could favor the HIV-2/TFO interaction. Finally, cellular proteins binding to triplexes also could stabilize this interaction (29). In any case, this cross-reaction increases the number of potential target sequences for the HIV-1 TFO and, in vivo, the TFO might reach nondesired target sequences within the host genome.
Taken together, our results demonstrate without ambiguity that TFOs are capable of modifying an endogenous target gene. However, their efficiency is low and their specificity of action not as high as desired. Chemical modifications already have been used to improve TFO efficiency (30). The assay described here should be useful for monitoring the improvements both quantitatively and qualitatively. In addition, a number of parameters of the interaction between the oligonucleotide and the target strongly depend on the sequence; for example, 2′-O-methyl modified oligonucleotides, which have been used against a hypoxanthine phosphoribosyltransferase target sequence (8), cannot form stable triple helices on other targets such as the HIV-1 polypurine track (unpublished observations). The versatility of our assay, which can be adapted to any target sequence of choice, should prove useful in testing triple-helix formation on sequences of particular interest, such as the HIV PPT used here.
Acknowledgments
We thank Serguei Gryaznov for the synthesis of the PPN oligonucleotide and Francis Fabre and Carl Mann for the kind gift of yeast strains. This work was supported by grants from the Agence Nationale pour la Recherche contre le SIDA and Rhône-Poulenc. F.-X.B., a student at the Institut de Formation Supérieure Biomédicale, was a recipient of a fellowship from the Agence Nationale pour la Recherche contre le SIDA. S.A.-S.-A. was awarded a fellowship from the Ligue Nationale contre le Cancer.
Abbreviations
- TFO
triple-helix-forming oligonucleotide
- PPT
polypurine tract
- PPO
phosphodiester
- PPN
phosphoramidate
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.050368997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.050368997
References
- 1.Hélène C, Giovannangeli C, Guieysse-Peugeot A L, Praseuth D. Ciba Found Symp. 1997;209:94–102. doi: 10.1002/9780470515396.ch8. [DOI] [PubMed] [Google Scholar]
- 2.Malvy C, Harel-Bellan A, Pritchard L L, editors. Triple Helix Forming Oligonucleotides. Boston: Kluwer; 1999. [Google Scholar]
- 3.Takasugi M, Guendouz A, Chassignol M, Decout J L, Lhomme J, Thuong N T, Hélène C. Proc Natl Acad Sci USA. 1991;88:5602–5606. doi: 10.1073/pnas.88.13.5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grigoriev M, Praseuth D, Guieysse A L, Robin P, Thuong N T, Hélène C, Harel-Bellan A. Proc Natl Acad Sci USA. 1993;90:3501–3505. doi: 10.1073/pnas.90.8.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Postel E H, Flint S J, Kessler D J, Hogan M E. Proc Natl Acad Sci USA. 1991;88:8227–8231. doi: 10.1073/pnas.88.18.8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orson F M, Thomas D W, McShan W M, Kessler D J, Hogan M E. Nucleic Acids Res. 1991;19:3435–3441. doi: 10.1093/nar/19.12.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McShan W M, Rossen R D, Laughter A H, Trial J, Kessler D J, Zendegui J G, Hogan M E, Orson F M. J Biol Chem. 1992;267:5712–5727. [PubMed] [Google Scholar]
- 8.Majumdar A, Khorlin A, Dyatkina N, Lin F L M, Powell J, Liu J, Feiz Z Z, Khripine Y, Watanabe K A, George J, et al. Nat Genet. 1998;20:212–214. doi: 10.1038/2530. [DOI] [PubMed] [Google Scholar]
- 9.Vasquez K M, Wang G, Havre P A, Glazer P M. Nucleic Acids Res. 1999;27:1176–1181. doi: 10.1093/nar/27.4.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feigon J, Dieckmann T, Smith F W. Chem Biol. 1996;3:611–617. doi: 10.1016/s1074-5521(96)90127-1. [DOI] [PubMed] [Google Scholar]
- 11.Famulok M, Jenne A. Curr Opin Chem Biol. 1998;2:320–327. doi: 10.1016/s1367-5931(98)80004-5. [DOI] [PubMed] [Google Scholar]
- 12.Hélène C, Thuong N T, Harel-Bellan A. Ann NY Acad Sci. 1992;660:27–36. doi: 10.1111/j.1749-6632.1992.tb21054.x. [DOI] [PubMed] [Google Scholar]
- 13.Michelotti E F, Tomonaga T, Krutzsch H, Levens D. J Biol Chem. 1995;270:9494–9499. doi: 10.1074/jbc.270.16.9494. [DOI] [PubMed] [Google Scholar]
- 14.Havre P A, Glazer P M. J Virol. 1993;67:7324–7331. doi: 10.1128/jvi.67.12.7324-7331.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang G, Levy D D, Seidman M M, Glazer P M. Mol Cell Biol. 1995;15:1759–1768. doi: 10.1128/mcb.15.3.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barre F X, Giovannangeli C, Hélène C, Harel-Bellan A. Nucleic Acids Res. 1999;27:743–749. doi: 10.1093/nar/27.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barre F X, Asseline U, Harel-Bellan A. J Mol Biol. 1999;286:1379–1387. doi: 10.1006/jmbi.1999.2550. [DOI] [PubMed] [Google Scholar]
- 18.Chen J K, Schultz R G, Lloyd D H, Gryaznov S M. Nucleic Acids Res. 1995;23:2661–2668. doi: 10.1093/nar/23.14.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gryaznov S, Skorski T, Cucco C, Nieborowska-Skorska M, Chiu C Y, Lloyd D, Chen J K, Koziolkiewicz M, Calabretta B. Nucleic Acids Res. 1996;24:1508–1514. doi: 10.1093/nar/24.8.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giovannangeli C, Diviacco S, Labrousse V, Gryaznov S, Charneau P, Hélène C. Proc Natl Acad Sci USA. 1997;94:79–84. doi: 10.1073/pnas.94.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barre F X, Mir L M, Lecluse Y, Harel-Bellan A. BioTechniques. 1998;25:294–296. doi: 10.2144/98252gt04. [DOI] [PubMed] [Google Scholar]
- 22.Johnston J R. Molecular Genetics of Yeast. Oxford: Oxford Univ. Press; 1994. [Google Scholar]
- 23.Giovannangeli C, Thuong N T, Hélène C. Nucleic Acids Res. 1992;20:4275–4281. doi: 10.1093/nar/20.16.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giovannangeli C, Perrouault L, Escudé C, Gryaznov S, Hélène C. J Mol Biol. 1996;261:386–398. doi: 10.1006/jmbi.1996.0471. [DOI] [PubMed] [Google Scholar]
- 25.Duval-Valentin G, Takasugi M, Hélène C, Sage E. J Mol Biol. 1998;278:815–825. doi: 10.1006/jmbi.1998.1728. [DOI] [PubMed] [Google Scholar]
- 26.Wang G, Glazer P M. J Biol Chem. 1995;270:22595–22601. doi: 10.1074/jbc.270.38.22595. [DOI] [PubMed] [Google Scholar]
- 27.Wellinger R E, Thoma F. EMBO J. 1997;16:5046–5056. doi: 10.1093/emboj/16.16.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown P M, Fox K R. Biochem J. 1996;319:607–611. doi: 10.1042/bj3190607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guieysse A L, Praseuth D, Hélène C. J Mol Biol. 1997;267:289–298. doi: 10.1006/jmbi.1997.0884. [DOI] [PubMed] [Google Scholar]
- 30.Giovannangeli C, Hélène C. Antisense Nucleic Acid Drug Dev. 1997;7:413–421. doi: 10.1089/oli.1.1997.7.413. [DOI] [PubMed] [Google Scholar]