Abstract
Human herpesvirus 7 (HHV-7), which belongs to the betaherpesvirus subfamily, infects mainly CD4+ T cells in vitro and infects children during infancy. After the primary infection, HHV-7 becomes latent. HHV-7 contains two genes (U12 and U51) that encode putative homologs of cellular G-protein-coupled receptors. To analyze the biological function of the U12 gene, we cloned the gene and expressed the U12 protein in cells. The U12 gene encoded a calcium-mobilizing receptor for the EBI1 ligand chemokine-macrophage inflammatory protein 3β (ELC/MIP-3β) but not for other chemokines, suggesting that the chemokine selectivity of the U12 gene product is distinct from that of the known mammalian chemokine receptors. These studies revealed that U12 activates distinct transmembrane signaling pathways that may mediate biological functions by binding with a β-chemokine, ELC/MIP-3β.
Human herpesvirus 7 (HHV-7) was isolated in 1990 from a healthy individual whose cells were stimulated with an antibody against CD3 and then incubated with interleukin-2 (IL-2) (17). HHV-7 is a ubiquitous virus that is similar to HHV-6. The primary infection of HHV-7, like that of HHV-6, causes exanthem subitum or high fever, although the apparent infection rate seems to be very low (47, 51, 53). The median age of children with primary HHV-7 infection is 26 months, significantly older than that of children with primary HHV-6 infection (median, 9 months) (5, 10, 48). HHV-7 can frequently be isolated from the saliva of healthy adults (51, 58).
The complete DNA sequences of HHV-7 have been reported (3, 37). HHV-7 is closely related to the T-lymphotropic betaherpesviruses, which share a common genomic organization and are composed of a single unique component that is bounded by direct repeats. HHV-7 is tropic for CD4+ T lymphocytes, uses the CD4+ molecule as at least part of its receptor mechanism (29), and has an antagonistic effect on human immunodeficiency virus infection of CD4+ cells in vitro (13).
Herpesviruses such as HHV-6, -7, and -8 and human cytomegalovirus (HCMV) encode chemokine receptor homologs (16, 18, 19, 26, 33, 44, 50). HHV-7 encodes two G-protein-coupled receptor (GPCR) homologs (U12 and U51), which are the respective positional and structural homologs of HHV-6 U12 and U51 and of HCMV UL33 and UL78. The HHV-6 and HHV-7 U12 products show the closest sequence similarity to cellular GPCRs (27). In humans, five CXC chemokine receptors, ten CC chemokine receptors, one C chemokine receptor, and one CX3C chemokine receptor have been identified, and their ligand specificities have been defined (1, 6, 11, 12, 21, 28, 32, 34-36, 39-43, 45, 46, 49, 52, 54). We previously showed that regulated upon activation, normal T expressed and secreted (RANTES), monocyte chemoattractant protein 1 (MCP-1), and macrophage inflammatory proteins 1α and 1β (MIP-1α and MIP-1β) induce the HHV-6 U12 product signal transduction responses, as measured by Ca2+ flux (27).
In this study, we focused on an analysis of the U12 gene of HHV-7. This study was undertaken to evaluate whether U12 expressed in HHV-7-infected T cells acts as a chemokine receptor whose signal is induced by binding with EBI1 ligand chemokine (ELC)/MIP-3β.
MATERIALS AND METHODS
Cell lines and cell culture.
SupT1 cells (NIH AIDS Research and Reference Reagent Program, Rockville, Md.), which are derived from a lymphoblastoid CD4+ T-cell line, were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS) (Life Technologies, Rockville, Md.), 200 mM l-glutamine, 100 U of penicillin, and 100 U of streptomycin. The cell density was maintained between 2 × 105 and 1 × 106 cells/ml. SupT1 cells were collected by centrifugation at 400 × g for 5 min and suspended in fresh medium at a density of 2 × 105 or 5 × 105 cells/ml. 293/EBNA-1 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% FCS and 200 mM l-glutamine. Nonadherent K562 human erythroleukemia cells (CCL 243) from the American Type Culture Collection were grown in RPMI 1640 medium supplemented with 10% FCS and 200 mM l-glutamine (complete medium).
Virus and virus preparation.
HHV-7 strain KHR, which was isolated from a patient with exanthem subitum (47), was grown in the immature continuous T-lymphoblastoid cell line SupT1. When more than 80% of the HHV-7-infected cells showed cytopathic effects, the culture was frozen and thawed twice, and after centrifugation at 300 × g for 10 min, the supernatant was stored at −80°C as a cell-free virus stock. For virus adsorption, 1 ml of the virus stock was thawed and mixed with 2 × 106 SupT1 cells, and then the suspension was spun at 300 × g for 1 h at 37°C.
Preparation of RNA.
SupT1 cells (approximately 107) were infected with HHV-7 strain KHR at a multiplicity of infection of 0.1 and spun at 1,500 × g for 1 h at 37°C for virus adsorption. After being washed twice with phosphate-buffered saline, the cells were cultured for 3 days in RPMI 1640 medium supplemented with 10% FCS. Virus-infected and mock-infected cells were pelleted by centrifugation and suspended in Sepasol-RNA I solution (Nacalai Tesque, Kyoto, Japan) for the preparation of RNA.
Cloning of the HHV-7 U12 gene.
A DNA fragment corresponding to U12 was amplified from the HHV-7 cDNA-infected SupT1 lymphoblastoid CD4+ T-cell line by PCR, using primers containing a KpnI site at the 5′ end and an HindIII site at the 3′ end for subsequent cloning. The U12 DNA was amplified by PCR with a 7U12-KpnI primer (5′-GGTACCGACATGGACACTCTAATTGA) and a 7U12-HindIII primer (5′-GCAAGCTTCACTGTTCAATTTTTTATTG). This primer pair was designed based on the predicted genomic structure of HHV-7 U12 (14). PCR was performed with approximately 0.5 ng of cDNA containing 0.2 mM deoxynucleoside triphosphates, 10 pmol of each primer, and 2.5 of U ExTaq in a 50-μl reaction volume (Takara, Kyoto, Japan). The PCR conditions were a 5-min preincubation at 94°C followed by 25 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min. The amplified products were subcloned directly into a pCR2.1 vector (Invitrogen, San Diego, Calif.) and sequenced by using a SequiTherm long-read cycle sequencing kit and a 4000L DNA sequencer (Li-Cor, Inc., Lincoln, Nebr.). After sequencing, the fragments were digested with KpnI and HindIII and subcloned into a pCEP4 vector (Invitrogen).
Construction of HHV-7 U12-pEGFP and establishment of the stable cell line.
To express the U12-EGFP (enhanced green fluorescent protein) fusion protein, an expression plasmid was constructed by using the GFP expression plasmid pEGFP-N1, purchased from Clontech (Palo Alto, Calif.). The U12 cDNA from the infected SupT1 cells was amplified by PCR with ExTaq (Takara). The primers used were 7U12-KpnI (5′-GGTACCGACATGGACACTCTAATTGA) and 7U12-BamHI (5′-TCGGATCCGGATTTTTTATTGTCAG). The amplification product was digested with KpnI and BamHI and inserted into the pEGFP-N1 plasmid vector.
K562 cells (107) in log phase were electroporated in the presence of 20 μg of HHV-7 U12-pEGFP plasmid DNA with a Gene Pulser (Bio-Rad Laboratories). The electroporation conditions were 300-μl volume, 250 V, and 960 μF, with a 0.4-cm-gap electroporation cuvette. Transfected cells were cultured in complete medium, and 48 h later the cells were seeded at 105/ml in complete medium containing 250 μg of G418 per ml and selected for 5 days. Subsequently, the cells were maintained in complete medium with 200 μg of G418 per ml.
Cloning of the CCR1, CXCR1, CCR5, EBI1/CCR7, and CCR8 genes.
CC chemokine receptor 1 (CCR1) and CXCR1 were constructed previously (27), and the CCR5, EBI1/CCR7, and CCR8 genes were cloned essentially as described previously (27). CCR5 and CCR8 genes were amplified from the genomic DNA of the promyelocytic cell line HL-60, and EBI1/CCR7 was amplified by PCR from the pCAGGS neo-CCR7 plasmid vector, which was kindly provided by T. Imai (KAN Research Institute Inc.). The PCR was performed with the following primer sets: CCR5-Met-HindIII (5′-ATAAGCTTATGGATTATCAAGTGTCAAGTC) and CCR5-Ter-BamHI (5′-TCGGATCCTCACAAGCCCACAGATATTTC); CCR7-Met-XhoI (5′-GCCCTCGAGTGTGGTTTTACCGCCCAGAGA) and CCR7-Ter-BamHI (5′-GGGGGATCCCCCTCTAGTCCAGGCAGAAGA); and CCR8-Met-XbaI (5′-GCTCTAGATCTGTGACCAGGTCCCGCTGCC) and CCR8-Ter-EcoRI (5′-CGGAATTCATATTTAGTCTTCATTGATCCT). The PCR conditions and sequencing analysis were as described above.
Establishment of stable transfected cell lines.
The genes for CCR5, EBI1/CCR7, and CCR8 were recloned from the plasmids described above into the HindIII and BamHI, XhoI and BamHI, and XbaI and EcoRI sites of the hygromycin B-selectable, stable episomal vector pCEP4 (Invitrogen), and the resulting constructs were designated pCECCR5, pCECCR7, and pCECCR8, respectively. K562 cells (107) in log phase were electroporated in the presence of 20 μg of plasmid DNA with a Gene Pulser (Bio-Rad Laboratories). Electroporation conditions were 300-μl volume, 250 V, and 960 μF, with a 0.4-cm-gap electroporation cuvette. Transfected cells were cultured in complete medium, and 48 h later the cells were seeded at 105/ml in complete medium containing 250 μg of hygromycin B per ml and selected for 5 days. Subsequently, the cells were maintained in complete medium with 150 μg of hygromycin B per ml. K562 cells were cultured as described above.
Production of ELC/MIP-3β-SEAP-His6 and SEAP-His6.
To produce the ELC/MIP-3β-SEAP-His6 and SEAP-His6 fusion proteins, 293/EBNA-1 cells were transfected with the expression vectors by using Lipofectamine (Qiagen, Hilden, Germany) as described elsewhere (54). ELC/MIP-3β-SEAP-His6 and SEAP-His6 were expressed as soluble fusion proteins with a modified secreted placental alkaline phosphatase (SEAP) and six histidines at the C terminus. The transfected cells were incubated for 72 h in Dulbecco's modified Eagle's medium supplemented with 10% FCS. The conditioned medium was spun, filtered (0.45 μm), and stored at 4°C with 20 mM HEPES, pH 7.4, and 0.02% sodium azide. The fusion proteins were purified by affinity chromatography on ProBond resin (Invitrogen). The SEAP-His6 protein was also expressed and purified from culture supernatants by affinity chromatography on ProBond resin for use as a negative control in the binding assay. This SEAP-His6 protein contains the complete signal peptide sequence at its N terminus. The concentrations of the ELC/MIP-3β-SEAP-His6 and SEAP-His6 proteins were estimated by using a bicinchoninic acid protein assay reagent kit (Pierce). Five to ten micrograms of ELC/MIP-3β-SEAP-His6 and SEAP-His6 was purified from 50 ml of supernatant in 293/EBNA-1 cells transfected with ELC/MIP-3β-SEAP-His6 or SEAP-His6. The ELC/MIP-3β-SEAP-His6 and pDREF-SEAP-His6 vectors were kindly provided by T. Imai (KAN Research Institute Inc.) (24, 25).
Optical sectioning.
Briefly, K562 cells transfected with HHV-7 U12-pEGFP or pEGFP cells (107 cells) were cultured with RPMI 1640. The cells were collected, washed in PBS (pH 7.4), and spotted on slides in circles with 5-mm inner diameters, 24 circles per slide. The living cells were observed directly. Three-dimensional data sets were collected by using a Deltavision microscope (Applied Precision, Issaquah, Wash.) with a Photometrics PXL cooled charge-coupled device camera. The data sets were “deconvolved” and projected onto a single plane, and the contrast was adjusted before the images were exported as TIFF files to Adobe Photoshop. Deconvolve uses a constrained interative deconvolution algorithm to remove out-of-focus blur in fluorescence optical sections.
Intracellular [Ca2+] measurements.
K562 cells transfected with CCR1, CCR2B, CCR5, and CCR8 cells were used for intracellular Ca2+ measurements. The cells were washed twice in HEPES-buffered Krebs solution (HBKS) as described previously (27). Briefly, 107 cells were incubated for 30 min at 37°C in the dark in 1 ml of HBKS, which consisted of 124 mM NaCl, 5 mM KCl, 1.24 mM KH2PO4, 1.3 mM MgSO4, 2.4 mM CaCl2, 10 mM glucose, and 25 mM HEPES (pH 7.4) containing 5 mM indo-1 AM {1-[2-amino-5-(6-carboxy-2-indolyl)phenoxy]-2-(2-amino-5-methylphenoxy)ethane-N,N,N′,N′-tetraacetic acid, pentaacetoxymethyl ester; Dojin Chemical Co.}. The cells were subsequently washed twice with HBKS and resuspended at 2.5 to 3 × 106 cells/ml. One milliliter of the cell suspension was placed in a continuously stirred cuvette at 37°C in a CAF-110 fluorometer (Jasco, Tokyo, Japan). Fluorescence was monitored at an excitation wavelength of 355 nm and emission wavelengths of 405 and 485 nm; the data are presented as the ratio of fluorescence detected at 405 and 485 nm. Data were collected every 10 ms. ELC/MIP-3β, RANTES, I-309, MIP-1β, and IL-8 were purchased from PeproTech EC (London, England).
Homology and ORF analysis.
The DNA sequences were analyzed for the presence of open reading frames (ORFs) and for their translation products with MacDNASIS (Hitachi Software Engineering Co., Yokohama, Japan). The FASTA program was used to search the HHV-7 U12 sequences for homologous protein sequences. The protein sequence databases searched with this program were the National Biomedical Research Foundation PIR and SWISS-PROT. The sequences were aligned to homologous genes with the Higgins et al. program in MacDNASIS (Hitachi).
Biomolecular interaction analysis using SPR.
A simple and rapid analytical method for detecting interactions between ELC/MIP-3β and U12 was studied by surface plasmon resonance (SPR) using a biosensor (Biacore, Uppsala, Sweden). ELC/MIP-3β-SEAP-His6 as the ligand was immobilized on a nitrilotriacetic acid (NTA) sensor chip (Biacore). SPR was used to measure changes in the refractive index caused by the capability of the U12 expressed by the K562 cells to bind ELC/MIP-3β. The sensor chips were activated with 500 μM NiCl2 and ELC/MIP-3β-SEAP-His6 was immobilized according to the manufacturer's instructions. ELC/MIP-3β-SEAP-His6 was immobilized at a concentration of 125 nM for 2 min at 25°C. Cells (106) were suspended in 1 ml of running buffer (10 mM HEPES [pH 7.4], 0.15 M NaCl, 3 mM EDTA, 0.005% surfactant P20 [pH 7.4]). K562 cells transfected with the parental pCEP4 vector alone, CCR7, and U12 were used as analytes, and binding to the ELC/MIP-3β immobilized on the chip was monitored in real time. The analysis of the interactions was carried out at a flow rate of 20 μl/min at 25°C by using a Biacore 2000 system. Each sensorgram consists of an association phase (first 180 s), reflecting binding of the injected cells to the ELC/MIP-3β, followed by a dissociation phase (260 s), during which the running buffer is passed over the chip and the bound cells are washed off the ELC/MIP-3β surface. The SPR signal was expressed in terms of resonance units. Data were globally analyzed with BIAevaluation 3.0 software (Biacore).
RESULTS
cDNA analysis and molecular cloning of the U12 gene.
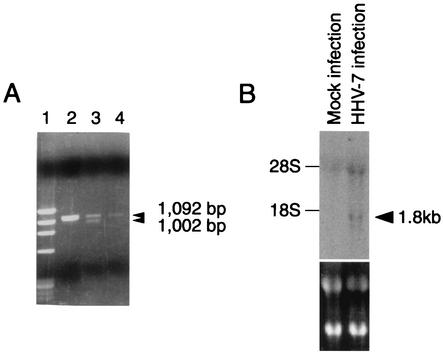
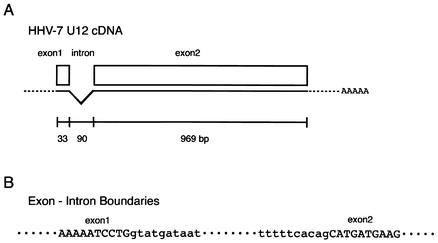
We were interested in the 1,002-bp ORF U12, because the deduced amino acid sequence of the original predicted ORF U12 product had a high degree of similarity to the sequences of the chemokine receptors, including GPCRs. The N-terminal amino acid sequence of the original predicted ORF U12 was shorter than that of HHV-6 U12. The primers 7U12-KpnI and 7U12-BamHI were designed to include a methionine site in a small ORF upstream of the original predicted U12 and the terminal site in the U12 gene, respectively. With these primers, the U12 coding region was amplified from cDNA that was synthesized with an oligo(dT) primer from total RNA purified from HHV-7-infected cells. Two bands of 1,092 and 1,002 bp and of approximately equal intensity were obtained (Fig. 1A), and these DNAs were inserted into pCR2.1. The 1,002-bp band was not observed in the gel containing total RNA purified from HHV-7-infected cells without reverse transcriptase (RT) treatment (Fig. 1A, lane 4) or in the gel containing DNA purified from infected cells with RT treatment (Fig. 1A, lane 2). These results suggest that the virus-infected cells contained at least two species of mRNAs for U12 in approximately equal amounts. The nucleotide sequencing data from five clones for each band showed that 1,092- and 1,002-bp bands were amplified from unspliced RNA or genomic DNA and spliced mRNA, respectively. From the positions of consensus splice donor and acceptor sites, it appears that the shorter mRNA was generated from two exons after a 90-nucleotide intron was spliced out (Fig. 1A and 2).
FIG. 1.
(A) Agarose gel (ethidium bromide stained) showing the results of PCR and RT-PCR amplification of HHV-7 DNA and mRNA, obtained from HHV-7-infected cell lines at day 7 postinfection. A 30-cycle PCR amplification was performed on a 1,102-bp fragment from the U12 gene of HHV-7. Lane 1, DNA size markers (φX174/HaeIII digest); lane 2, DNA from infected SupT1 cells; lane 3, cDNA from infected SupT1 cells treated with RT; lane 4, same as lane 3 but without RT. Arrowheads indicate the position of U12 and the splice variant. (B) Northern blot analysis of U12 expression in SupT1 cells infected with HHV-7. Total RNA was isolated from mock-infected and HHV-7-infected SupT1 cells on day 3 after infection. The lower panel shows ethidium bromide staining. The arrowhead indicates the U12 transcript.
FIG. 2.
Genomic structure of the U12 gene. (A) Schematic description of the U12 cDNA structure. (B) Nucleotide sequences of the exon-intron boundaries. Uppercase indicates exons; lowercase represents introns.
We then analyzed U12 expression by Northern blot analysis using total RNA prepared from HHV-7-infected or uninfected SupT1 cells. As shown in Fig. 1B, U12 was highly expressed in the infected cells.
Amino acid sequence homology with human CCR7.
The N-terminal extracellular domain of the U12 product contained a potential N-glycosylation site, as do most known GPCRs. All the extracellular domains of the U12 product contained four highly conserved cysteine residues that are essential to the structure of GPCRs (15). The cDNA contained introns, a feature of most chemokine receptor genes (30), and its sequence was identical to the genomic sequence. The resultant 1,002 nucleotides of coding region could be translated into a 334-amino-acid polypeptide encoding a GPCR homolog (Fig. 3). Since, as described above, there are counterparts of this gene in the HHV-7 and human genomes, we compared the amino acid sequence with human chemokine receptors. U12 also exhibited homology to multiple mammalian GPCRs; the highest was to EBI1/CCR7. The deduced amino acid sequence showed 22% homology to human CCR7 (Fig. 3). Sequence alignment of the U12 product with the EBI1/CCR7 product suggested that HHV-7 U12 contained seven transmembrane regions, four extracellular domains, and four cytoplasmic domains (Fig. 3).
FIG. 3.
Sequence alignment of the U12 cDNA product and human EBI1/CCR7. Dashes indicate gaps that were inserted to optimize the alignment. Overlines mark the amino acid sequences of the transmembrane regions (I to VII). Dark and light gray boxes, identical and homologous amino acids, respectively.
Expression of U12-GFP.
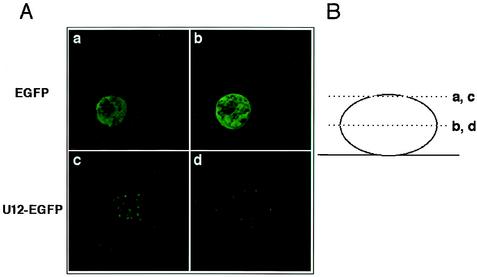
We used fluorescence microscopy to confirm the expression of U12-GFP in each experiment (Fig. 4A). A schematic drawing of the section planes is given in Fig. 4B. A U12-GFP-tagged expression vector was utilized in this experiment because the U12 polyclonal antiserum was not very reactive in Western blot analyses or immunofluorescence assays. The GFP vector, pEGFP-N1, alone was a control in this experiment. GFP expression was localized to the cytoplasm (Fig. 4A, panels a and b) in the control, and the U12-GFP product seemed to be detected on the membrane, given that fluorescence was detected as dots throughout the cells (Fig. 4B, panels c and d).
FIG. 4.
Optical sectioning. (A) Confocal laser microscopy analysis and deconvolution software confirmed that K562 cells were transfected with either the pEGFP-N1 vector alone (a and b) or the U12-pEGFP fusion vector (c and d). EGFP products resulted in a predominantly cytoplasmic distribution with a strong signal, but not all cells were labeled. In contrast, the U12-GFP fusion protein accumulated in large granules that were clearly distinct from the more numerous speckles seen on the cell surface. (B) Cells were observed 24 h after transfection in two planes.
Signaling through the U12 GPCR in response to ELC/MIP-3β.
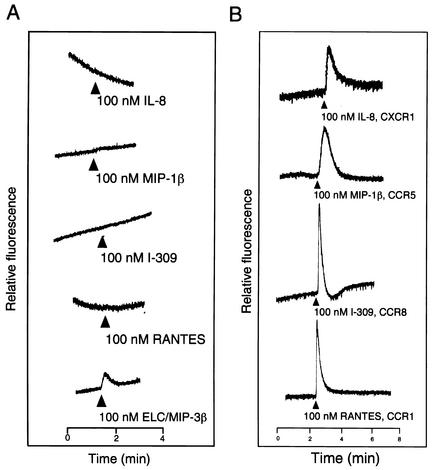
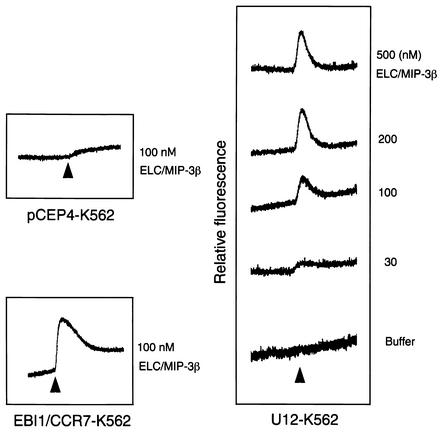
To test whether the U12 product is capable of signal transduction, intracellular Ca2+ levels were monitored by measuring the relative fluorescence of indo-1-loaded cells stimulated with ELC/MIP-3β, RANTES, I-309, MIP-1β, and IL-8, as described previously (27) (Fig. 5). To confirm the positive signaling by a specific ligand and receptor, we used as positive control cells expressing CXCR1, CCR5, CCR8, and CCR1, which responded to IL-8, MIP1-β, I-309, and RANTES, respectively (Fig. 5B). Untransfected and pCEP4-transfected K562 cells did not respond to any of the chemokines tested (data not shown). K562 cells transfected with U12 responded to ELC/MIP-3β but not to the other cytokines (Fig. 5A). Treatment with buffer alone did not induce Ca2+ flux in parental K562 cells or K562 cells transfected with CCR1, CCR8, CCR5, or CXCR1 (data not shown). Treatment of U12 gene transfectants with ELC/MIP-3β induced transient elevations of the intracellular Ca2+ level, with a 50% effective concentration of about 30 nM (Fig. 6).
FIG. 5.
Ca2+ mobilization analysis in K562 cells transfected with U12 and treated with ELC/MIP-3β, RANTES, I-309, MIP-1β, or IL-8 (A) and K562 cells transfected with CCR1, CCR8, CCR5, or CXCR1 and treated with RANTES, I-309, MIP-1β, or IL-8 (B). Indo-1 AM-loaded cells were suspended at 2.5 × 106 to 3 × 106/ml and continuously monitored for fluorescence changes. Peptide agonists were added at the indicated times at a dilution of 1:25 (vol/vol), resulting in final concentrations of 100 nM. The tracings are from a single experiment that was representative of two separate experiments.
FIG. 6.
Dose-response experiments. K562 cells expressing U12 were loaded with indo-1 AM. (Right) Magnitudes of the peak transient calcium increase elicited by the indicated concentrations of MIP-3β. (Left) Parental K562 cells transfected with vector alone did not respond to 100 nM ELC/MIP-3β, while K562 cells transfected with EBI1/CCR7 did respond to 100 nM ELC/MIP-3β.
Binding of ELC/MIP-3β to U12.
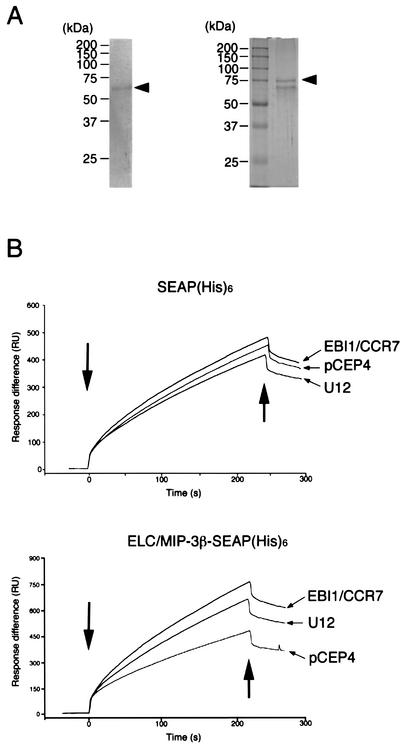
ELC/MIP-3β-SEAP-His6 was secreted as a single major protein with an expected apparent molecular mass of 74 kDa (Fig. 7A). This fusion protein has an alkaline phosphatase activity for quantitative tracing and a His6 tag in its COOH terminus for easy purification and for fixing to the Biacore NTA sensor chip.
FIG. 7.
Expression of ELC/MIP-3β-SEAP-His6 and SEAP-His6 fusion proteins. (A) ELC/MIP-3β-SEAP-His6 and SEAP-His6 were purified from the conditioned medium of transfected cells by metal affinity chromatography, fractionated on 10% polyacrylamide gels, and stained with Coomassie brilliant blue. Arrowheads indicate ELC/MIP-3β-SEAP-His6 (left) and SEAP-His6 (right). The positions of size markers are shown on the left. (B) Real-time analysis of the interaction between ELC/MIP-3β or SEAP-His6 and U12-, EBI1/CCR7-, or pCEP4-expressing K562 cells, as monitored by SPR (Biacore). The SPR signal was expressed as resonance units (RU). ELC/MIP-3β-SEAP-His6 was immobilized on an NTA sensor chip, and K562 cells stably expressing U12, EBI1/CCR7, or pCEP4 were injected. Association and dissociation starts are shown by the left and right arrows, respectively. Representative binding curves for the interaction of ELC/MIP-3β with K562 cells transfected with U12 or EBI1/CCR7 and with parental pCEP4 cells are shown. The tracings are from a single experiment that was representative of three separate experiments.
By using real-time biospecific interaction analysis (BIAcore, Uppsala, Sweden), we studied the binding affinity of the U12 (Fig. 7B). To test whether the U12 product functions as a chemokine receptor, we examined the binding activity of the U12 product with ELC/MIP-3β (Fig. 7B). Stable K562 cell lines expressing pCEP4, EBI1/CCR7, or U12 were used for this assay. The U12-transfected K562 cells expressed specific binding sites for ELC/MIP-3β-SEAP-His6, whereas the pCEP4-transfected K562 cells did not. A typical profile of binding of ELC/MIP-3β-SEAP-His6 to U12-transfected cells is shown in Fig. 7B. The K562 cells transfected with pCEP4, EBI1/CCR7, and U12 bound with relatively equal affinity to SEAP-His6 (Fig. 7B). Under the same experimental conditions, SEAP-His6, used here as a negative control, did not show binding of U12 to ELC/MIP-3β (Fig. 7B).
DISCUSSION
In this paper, we report that the U12 gene of HHV-7 encodes a chemokine receptor that was expressed in HHV-7 infected SupT1 cells, and we characterize the U12 protein. The U12 gene is a 1.8-kb RNA and encodes a protein of 334 amino acid residues that has seven transmembrane domains and a highly conserved GPCR motif at the junction of the third transmembrane domain and second intracellular loop (Fig. 1 and 3) (15). The N-terminal amino acid sequence of HHV-7 U12 was 19 amino acids shorter than that of HHV-6 U12 (27). Homology searches showed that the U12 sequence was 22% identical to that of EBI1/CCR7 (Fig. 3). The U12 receptor was capable of transducing a chemokine signal from ELC/MIP-3β to the cytoplasm that resulted in a transient elevation of the intracellular Ca2+ concentration, with a 50% effective concentration of 30 nM (Fig. 5 and 6). Its signal transduction response was similar to that of EBI1/CCR7 (Fig. 5). U12 is notable in that of the >10 purified chemokines tested, the U12 protein appeared to interact only with ELC/MIP-3β. ELC/MIP-3β shows homologies to other CC chemokines with 20 to 30% identity (54). ELC/MIP-3β is constitutively expressed in various lymphoid tissues, including the thymus, lymph nodes, appendix, and spleen (54, 55). Collectively, these results show that ELC/MIP-3β is a specific high-affinity biological ligand for U12. The present studies strongly suggest that HHV-7-infected cells respond to ELC/MIP-3β via U12.
The EBI1/CCR7 gene was first reported by Birkenbach et al. (4) as a lymphocyte-specific GPCR induced by Epstein-Barr virus (EBV) infection. EBI1/CCR7 is specific for ELC/MIP-3β and secondary lymphoid-tissue chemokine (31, 54) and acts to moderate cell migration and proliferation. Furthermore, Hasegawa et al. showed that the EBI1/CCR7 gene was induced not only by EBV but also by HHV-6 and -7 infection in CD4+ T cells (23).
Because EBI1/CCR7 is constitutively expressed in various lymphoid tissues and on activated T and B lymphocytes (54-57), ELC/MIP-3β and EBI1/CCR7 may play roles not only in inflammatory and immunological responses but also in normal lymphocyte recirculation and homing. Most CC chemokines are potent attractants of monocytes and are known to act via shared receptors (20, 45). Genes for EBI1/CCR7 and EBI2, designated EBV-induced genes 1 and 2, were isolated through their strong up-regulation in EBV-negative Burkitt's lymphoma cells upon infection with EBV (4). Similarly, BLR1 and BLR2, designated as Burkitt's lymphoma virus receptors 1 and 2, were isolated through their induction by EBV infection (4, 46). BLR2, which is identical to EBI1/CCR7, was further shown to be induced by the EBV-encoded transactivator EBNA-2 (7). BLR1 and EBI1/CCR7 are most homologous to the chemokine receptors, whereas EBI2 is most related to the thrombin receptor. Furthermore, herpesviruses such as HCMV (2, 8, 9), herpesvirus saimiri (38), HHV-6 (19, 26), HHV-7 (37), and HHV-8/Kaposi's sarcoma-associated herpesvirus (22) are all known to encode GPCRs that are homologous to chemokine receptors. A chemokine receptor encoded by HHV-8 was found to be constitutively active and to stimulate the proliferation of transfected cells, making it a candidate viral oncogene (2).
Taken together, these results suggest that virally encoded putative chemokine receptors play important roles in the infection and life cycle of herpesviruses, especially in vivo. The roles of ELC/MIP-3β and U12 in HHV-7-infected T cells are not known at present, but they may have biological activities in infected cells, such as promoting growth, protecting cells from apoptosis, and/or directing migration into specific anatomical locations in vivo. The identification of ELC/MIP-3β as a specific ligand for U12 now enables us to examine the possible roles of ELC/MIP-3β and U12 in the infection process and life cycle of HHV-7. Such studies may lead to new strategies to prevent herpesvirus infection. Further studies exploring the function of U12 in vivo may provide new insights into the molecular pathogenesis and latency of HHV-7 infection.
In summary, this paper reports the first characterization of HHV-7 U12, as signaling the GPCR. This result was expected, given the properties of HHV-6 U12, but this still represents the first description of this molecule. The most interesting result is the identification of the U12 ligand. The significance of these results stems from the possibility that a signaling receptor may well contribute to pathogenesis.
Acknowledgments
This study was supported in part by a grant-in-aid from the Ministry of Education, Science and Culture of Japan.
We thank T. Imai for his gift of the ΔpDREF-SEAP-His6, ELC/MIP-3β-SEAP-His6, and EBI1/CCR7 expression plasmids and J. Katahira and H. Ozawa for their technical support with the Biacore KK.
REFERENCES
- 1.Baba, M., T. Imai, M. Nishimura, M. Kakizaki, S. Takagi, K. Hieshima, H. Nomiyama, and O. Yoshie. 1997. Identification of CCR6, the specific receptor for a novel lymphocyte-directed CC chemokine LARC. J. Biol. Chem. 272:14893-14898. [DOI] [PubMed] [Google Scholar]
- 2.Bais, C., B. Santomasso, O. Coso, L. Arvanitakis, E. G. Raaka, J. S. Gutkind, A. S. Asch, E. Cesarman, M. C. Gershengorn, E. A. Mesri, and M. C. Gerhengorn. 1998. G-protein-coupled receptor of Kaposi's sarcoma-associated herpesvirus is a viral oncogene and angiogenesis activator. Nature 391:86-89. [DOI] [PubMed] [Google Scholar]
- 3.Berneman, Z. N., D. V. Ablashi, G. Li, M. Eger-Fletcher, M. S. Reitz, Jr., C. L. Hung, I. Brus, A. L. Komaroff, and R. C. Gallo. 1992. Human herpesvirus 7 is a T-lymphotropic virus and is related to, but significantly different from, human herpesvirus 6 and human cytomegalovirus. Proc. Natl. Acad. Sci. USA 89:10552-10556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birkenbach, M., K. Josefsen, R. Yalamanchili, G. Lenoir, and E. Kieff. 1993. Epstein-Barr virus-induced genes: first lymphocyte-specific G protein-coupled peptide receptors. J. Virol. 67:2209-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black, J. B., T. F. Schwarz, J. L. Patton, K. Kite-Powell, P. E. Pellett, S. Wiersbitzky, R. Bruns, C. Muller, G. Jager, and J. A. Stewart. 1996. Evaluation of immunoassays for detection of antibodies to human herpesvirus 7. Clin. Diagn. Lab. Immunol. 3:79-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonini, J. A., S. K. Martin, F. Dralyuk, M. W. Roe, L. H. Philipson, and D. F. Steiner. 1997. Cloning, expression, and chromosomal mapping of a novel human CC-chemokine receptor (CCR10) that displays high-affinity binding for MCP-1 and MCP-3. DNA Cell Biol. 16:1249-1256. [DOI] [PubMed] [Google Scholar]
- 7.Burgstahler, R., B. Kempkes, K. Steube, and M. Lipp. 1995. Expression of the chemokine receptor BLR2/EBI1 is specifically transactivated by Epstein-Barr virus nuclear antigen 2. Biochem. Biophys. Res. Commun. 215:737-743. [DOI] [PubMed] [Google Scholar]
- 8.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison III, T. Kouzarides, J. A. Martignetti, et al. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 9.Chee, M. S., S. C. Satchwell, E. Preddie, K. M. Weston, and B. G. Barrell. 1990. Human cytomegalovirus encodes three G protein-coupled receptor homologues. Nature 344:774-777. [DOI] [PubMed] [Google Scholar]
- 10.Clark, D. A., M. L. Freeland, L. K. Mackie, R. F. Jarrett, and D. E. Onions. 1993. Prevalence of antibody to human herpesvirus 7 by age. J. Infect. Dis. 168:251-252. [DOI] [PubMed] [Google Scholar]
- 11.Combadiere, C., S. K. Ahuja, J. Van Damme, H. L. Tiffany, J. L. Gao, and P. M. Murphy. 1995. Monocyte chemoattractant protein-3 is a functional ligand for CC chemokine receptors 1 and 2B. J. Biol. Chem. 270:29671-29675. [DOI] [PubMed] [Google Scholar]
- 12.Combadiere, C., K. Salzwedel, E. D. Smith, H. L. Tiffany, E. A. Berger, and P. M. Murphy. 1998. Identification of CX3CR1. A chemotactic receptor for the human CX3C chemokine fractalkine and a fusion coreceptor for HIV-1. J. Biol. Chem. 273:23799-23804. [DOI] [PubMed] [Google Scholar]
- 13.Crowley, R. W., P. Secchiero, D. Zella, A. Cara, R. C. Gallo, and O. Lusso. 1996. Interference between human herpesvirus 7 and HIV-1 in mononuclear phagocytes. J. Immunol. 156:2004-2008. [PubMed] [Google Scholar]
- 14.Davis-Poynter, N. J., D. M. Lynch, H. Vally, G. R. Shellam, W. D. Rawlinson, B. G. Barrell, and H. E. Farrell. 1997. Identification and characterization of a G protein-coupled receptor homolog encoded by murine cytomegalovirus. J. Virol. 71:1521-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dixon, R. A., I. S. Sigal, M. R. Candelore, R. B. Register, W. Scattergood, E. Rands, and C. D. Strader. 1987. Structural features required for ligand binding to the beta-adrenergic receptor. EMBO J. 6:3269-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dominguez, G., T. R. Dambaugh, F. R. Stamey, S. Dewhurst, N. Inoue, and P. E. Pellett. 1999. Human herpesvirus 6B genome sequence: coding content and comparison with human herpesvirus 6A. J. Virol. 73:8040-8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frenkel, N., E. C. Schirmer, L. S. Wyatt, G. Katsafanas, E. Roffman, R. M. Danovich, and C. H. June. 1990. Isolation of a new herpesvirus from human CD4+ T cells. Proc. Natl. Acad. Sci. USA 87:748-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao, J. L., and P. M. Murphy. 1994. Human cytomegalovirus open reading frame US28 encodes a functional beta chemokine receptor. J. Biol. Chem. 269:28539-28542. [PubMed] [Google Scholar]
- 19.Gompels, U. A., J. Nicholas, G. Lawrence, M. Jones, B. J. Thomson, M. E. Martin, S. Efstathiou, M. Craxton, and H. A. Macaulay. 1995. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology 209:29-51. [DOI] [PubMed] [Google Scholar]
- 20.Gong, X., W. Gong, D. B. Kuhns, A. Ben-Baruch, O. M. Howard, and J. M. Wang. 1997. Monocyte chemotactic protein-2 (MCP-2) uses CCR1 and CCR2B as its functional receptors. J. Biol. Chem. 272:11682-11685. [DOI] [PubMed] [Google Scholar]
- 21.Grob, P. M., E. David, T. C. Warren, R. P. DeLeon, P. R. Farina, and C. A. Homon. 1990. Characterization of a receptor for human monocyte-derived neutrophil chemotactic factor/interleukin-8. J. Biol. Chem. 265:8311-8316. [PubMed] [Google Scholar]
- 22.Guo, H. G., P. Browning, J. Nicholas, G. S. Hayward, E. Tschachler, Y. W. Jiang, M. Sadowska, M. Raffeld, S. Colombini, R. C. Gallo, and M. S. Reitz, Jr. 1997. Characterization of a chemokine receptor-related gene in human herpesvirus 8 and its expression in Kaposi's sarcoma. Virology 228:371-378. [DOI] [PubMed] [Google Scholar]
- 23.Hasegawa, H., Y. Utsunomiya, M. Yasukawa, K. Yanagisawa, and S. Fujita. 1994. Induction of G protein-coupled peptide receptor EBI 1 by human herpesvirus 6 and 7 infection in CD4+ T cells. J. Virol. 68:5326-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hieshima, K., T. Imai, G. Opdenakker, J. Van Damme, J. Kusuda, H. Tei, Y. Sakaki, K. Takatsuki, R. Miura, O. Yoshie, and H. Nomiyama. 1997. Molecular cloning of a novel human CC chemokine liver and activation-regulated chemokine (LARC) expressed in liver. Chemotactic activity for lymphocytes and gene localization on chromosome 2. J. Biol. Chem. 272:5846-5853. [DOI] [PubMed] [Google Scholar]
- 25.Imai, T., T. Yoshida, M. Baba, M. Nishimura, M. Kakizaki, and O. Yoshie. 1996. Molecular cloning of a novel T cell-directed CC chemokine expressed in thymus by signal sequence trap using Epstein-Barr virus vector. J. Biol. Chem. 271:21514-21521. [DOI] [PubMed] [Google Scholar]
- 26.Isegawa, Y., T. Mukai, K. Nakano, M. Kagawa, J. Chen, Y. Mori, T. Sunagawa, K. Kawanishi, J. Sashihara, A. Hata, P. Zou, H. Kosuge, and K. Yamanishi. 1999. Comparison of the complete DNA sequences of human herpesvirus 6 variants A and B. J. Virol. 73:8053-8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Isegawa, Y., Z. Ping, K. Nakano, N. Sugimoto, and K. Yamanishi. 1998. Human herpesvirus 6 open reading frame U12 encodes a functional beta-chemokine receptor. J. Virol. 72:6104-6112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, Q. J., S. Lu, R. D. Ye, and M. Martins-Green. 2000. Isolation and characterization of a new chemokine receptor gene, the putative chicken CXCR1. Gene 257:307-317. [DOI] [PubMed] [Google Scholar]
- 29.Lusso, P., P. Secchiero, R. W. Crowley, A. Garzino Demo, Z. N. Berneman, and R. C. Gallo. 1994. CD4 is a critical component of the receptor for human herpesvirus 7: interference with human immunodeficiency virus. Proc. Natl. Acad. Sci. USA 91:3872-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy, P. M. 1994. The molecular biology of leukocyte chemoattractant receptors. Annu. Rev. Immunol. 12:593-633. [DOI] [PubMed] [Google Scholar]
- 31.Nagira, M., T. Imai, K. Hieshima, J. Kusuda, M. Ridanpaa, S. Takagi, M. Nishimura, M. Kakizaki, H. Nomiyama, and O. Yoshie. 1997. Molecular cloning of a novel human CC chemokine secondary lymphoid-tissue chemokine that is a potent chemoattractant for lymphocytes and mapped to chromosome 9p13. J. Biol. Chem. 272:19518-19524. [DOI] [PubMed] [Google Scholar]
- 32.Napolitano, M., A. Zingoni, G. Bernardini, G. Spinetti, A. Nista, C. T. Storlazzi, M. Rocchi, and A. Santoni. 1996. Molecular cloning of TER1, a chemokine receptor-like gene expressed by lymphoid tissues. J. Immunol. 157:2759-2763. [PubMed] [Google Scholar]
- 33.Neipel, F., J. C. Albrecht, and B. Fleckenstein. 1997. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J. Virol. 71:4187-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neote, K., D. DiGregorio, J. Y. Mak, R. Horuk, and T. J. Schall. 1993. Molecular cloning, functional expression, and signaling characteristics of a C-C chemokine receptor. Cell 72:415-425. [DOI] [PubMed] [Google Scholar]
- 35.Neote, K., J. Y. Mak, L. F. Kolakowski, Jr., and T. J. Schall. 1994. Functional and biochemical analysis of the cloned Duffy antigen: identity with the red blood cell chemokine receptor. Blood 84:44-52. [PubMed] [Google Scholar]
- 36.Nibbs, R. J., S. M. Wylie, J. Yang, N. R. Landau, and G. J. Graham. 1997. Cloning and characterization of a novel promiscuous human beta-chemokine receptor D6. J. Biol. Chem. 272:32078-32083. [DOI] [PubMed] [Google Scholar]
- 37.Nicholas, J. 1996. Determination and analysis of the complete nucleotide sequence of human herpesvirus. J. Virol. 70:5975-5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicholas, J., K. R. Cameron, and R. W. Honess. 1992. Herpesvirus saimiri encodes homologues of G protein-coupled receptors and cyclins. Nature 355:362-365. [DOI] [PubMed] [Google Scholar]
- 39.Oberlin, E., A. Amara, F. Bachelerie, C. Bessia, J. L. Virelizier, F. Arenzana-Seisdedos, O. Schwartz, J. M. Heard, I. Clark-Lewis, D. F. Legler, M. Loetscher, M. Baggiolini, and B. Moser. 1996. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature 382:833-835. [DOI] [PubMed] [Google Scholar]
- 40.Pardigol, A., U. Forssmann, H. D. Zucht, P. Loetscher, P. Schulz-Knappe, M. Baggiolini, W. G. Forssmann, and H. J. Magert. 1998. HCC-2, a human chemokine: gene structure, expression pattern, and biological activity. Proc. Natl. Acad. Sci. USA 95:6308-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Power, C. A., D. J. Church, A. Meyer, S. Alouani, A. E. Proudfoot, I. Clark-Lewis, S. Sozzani, A. Mantovani, and T. N. Wells. 1997. Cloning and characterization of a specific receptor for the novel CC chemokine MIP-3α from lung dendritic cells. J. Exp. Med. 186:825-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raport, C. J., J. Gosling, V. L. Schweickart, P. W. Gray, and I. F. Charo. 1996. Molecular cloning and functional characterization of a novel human CC chemokine receptor (CCR5) for RANTES, MIP-1β, and MIP-1α. J. Biol. Chem. 271:17161-17166. [DOI] [PubMed] [Google Scholar]
- 43.Roos, R. S., M. Loetscher, D. F. Legler, I. Clark-Lewis, M. Baggiolini, and B. Moser. 1997. Identification of CCR8, the receptor for the human CC chemokine I-309. J. Biol. Chem. 272:17251-17254. [DOI] [PubMed] [Google Scholar]
- 44.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samson, M., O. Labbe, C. Mollereau, G. Vassart, and M. Parmentier. 1996. Molecular cloning and functional expression of a new human CC-chemokine receptor gene. Biochemistry 35:3362-3367. [DOI] [PubMed] [Google Scholar]
- 46.Schweickart, V. L., C. J. Raport, R. Godiska, M. G. Byers, R. L. Eddy, Jr., T. B. Shows, and P. W. Gray. 1994. Cloning of human and mouse EBI1, a lymphoid-specific G-protein-coupled receptor encoded on human chromosome 17q12-q21.2. Genomics 23:643-650. [DOI] [PubMed] [Google Scholar]
- 47.Tanaka, K., T. Kondo, S. Torigoe, S. Okada, T. Mukai, and K. Yamanishi. 1994. Human herpesvirus 7: another causal agent for roseola (exanthem subitum). J. Pediatr. 125:1-5. [DOI] [PubMed] [Google Scholar]
- 48.Tanaka-Taya, K., T. Kondo, T. Mukai, H. Miyoshi, Y. Yamamoto, S. Okada, and K. Yamanishi. 1996. Seroepidemiological study of human herpesvirus-6 and -7 in children of different ages and detection of these two viruses in throat swabs by polymerase chain reaction. J. Med. Virol. 48:88-94. [DOI] [PubMed] [Google Scholar]
- 49.Wang, J. M., A. Hishinuma, J. J. Oppenheim, and K. Matsushima. 1993. Studies of binding and internalization of human recombinant monocyte chemotactic and activating factor (MCAF) by monocytic cells. Cytokine 5:264-275. [DOI] [PubMed] [Google Scholar]
- 50.Welch, A. R., L. M. McGregor, and W. Gibson. 1991. Cytomegalovirus homologs of cellular G protein-coupled receptor genes are transcribed. J. Virol. 65:3915-3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wyatt, L. S., W. J. Rodriguez, N. Balachandran, and N. Frenkel. 1991. Human herpesvirus 7: antigenic properties and prevalence in children and adults. J. Virol. 65:6260-6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamagami, S., Y. Tokuda, K. Ishii, H. Tanaka, and N. Endo. 1994. cDNA cloning and functional expression of a human monocyte chemoattractant protein 1 receptor. Biochem. Biophys. Res. Commun. 202:1156-1162. [DOI] [PubMed] [Google Scholar]
- 53.Yamanishi, K., T. Okuno, K. Shiraki, M. Takahashi, T. Kondo, Y. Asano, and T. Kurata. 1988. Identification of human herpesvirus-6 as a causal agent for exanthem subitum. Lancet i:1065-1067. [DOI] [PubMed] [Google Scholar]
- 54.Yoshida, R., T. Imai, K. Hieshima, J. Kusuda, M. Baba, M. Kitaura, M. Nishimura, M. Kakizaki, H. Nomiyama, and O. Yoshie. 1997. Molecular cloning of a novel human CC chemokine EBI1-ligand chemokine that is a specific functional ligand for EBI1, CCR7. J. Biol. Chem. 272:13803-13809. [DOI] [PubMed] [Google Scholar]
- 55.Yoshida, R., M. Nagira, T. Imai, M. Baba, S. Takagi, Y. Tabira, J. Akagi, H. Nomiyama, and O. Yoshie. 1998. EBI1-ligand chemokine (ELC) attracts a broad spectrum of lymphocytes: activated T cells strongly up-regulate CCR7 and efficiently migrate toward ELC. Int. Immunol. 10:901-910. [DOI] [PubMed] [Google Scholar]
- 56.Yoshida, R., M. Nagira, M. Kitaura, N. Imagawa, T. Imai, and O. Yoshie. 1998. Secondary lymphoid-tissue chemokine is a functional ligand for the CC chemokine receptor CCR7. J. Biol. Chem. 273:7118-7122. [DOI] [PubMed] [Google Scholar]
- 57.Yoshie, O., T. Imai, and H. Nomiyama. 1997. Novel lymphocyte-specific CC chemokines and their receptors. J. Leukoc. Biol. 62:634-644. [DOI] [PubMed] [Google Scholar]
- 58.Yoshikawa, T., Y. Asano, I. Kobayashi, T. Nakashima, T. Yazaki, S. Suga, T. Ozaki, L. S. Wyatt, and N. Frenkel. 1993. Seroepidemiology of human herpesvirus 7 in healthy children and adults in Japan. J. Med. Virol. 41:319-323. [DOI] [PubMed] [Google Scholar]