Abstract
We examined the way the major outer membrane protein OmpA of Salmonella enterica serovar Typhimurium is recognized by the mouse immune system, by raising a panel of 12 monoclonal antibodies (MAbs) against this protein. Interaction between OmpA and these MAbs is competitively inhibited with several-hundredfold dilutions of mouse polyclonal sera obtained by immunization with live or heat-killed whole cells, suggesting that OmpA is one of the immunodominant antigens of serovar Typhimurium. All of the MAbs were specific for an identical epitope(s) located on the C-terminal domain of OmpA, as indicated by the use of OmpA fragments generated by protease or cyanogen bromide treatment and by competitive inhibition enzyme-linked immunosorbent assay. This epitope was highly conserved within (but not outside) the family Enterobacteriaceae. The strong immunogenicity of this epitope was surprising because the C-terminal domain of OmpA, usually thought to be located in the periplasm, is not expected to be exposed on the bacterial cell surface. A MAb, however, reacted in a cytofluorometry assay more strongly with outer-membrane-permeabilized cells than with untreated cells, a result supporting the predominantly periplasmic localization of the epitope. Significant, though low-level, reactivity of intact cells nevertheless suggests that in some cells the C-terminal domain of OmpA is exposed on the surface, a result consistent with the proposal that OmpA can fold into one of the two alternate conformations.
Natural or experimental infections of animals with Salmonella result in stimulation of both humoral and cell-mediated immunity (8, 10). These immune responses primarily occur against the lipopolysaccharide (LPS) and major outer membrane (OM) proteins, including porins and OmpA (1, 7, 21, 28, 40, 42). However, the number and variety of antibodies with distinct specificities and the identity of epitopes that they recognize are not well understood. Previous studies (reviewed in references 40 and 42) established the importance of O-antigen-specific antibodies in immunity to murine salmonellosis. The precise role of porins, however, in humoral immunity is controversial (reviewed in reference 40).
OmpA, like porins and LPS, is also a target of the host immune response (1, 19, 28, 31, 48), but its role in immunoprotection is not clearly understood. Some studies suggest that antibodies specific for OmpA or its homolog do not confer passive protection (13, 20, 49, 51). On the other hand, several investigators have shown that the C-terminal domain of OprF, the OmpA homolog in Pseudomonas aeruginosa, contains an important protective epitope (14, 15, 50).
The OmpA protein, a major Salmonella enterica serovar Typhimurium OM protein that is 94% identical to Escherichia coli OmpA (12), is of particular interest for immune recognition analysis. The majority conformers of OmpA fold into a structure with two large domains, the N-terminal domain (residues 1 to 170 in E. coli) and the C-terminal domain (residues 196 to 325). The N-terminal domain of E. coli OmpA was crystallized as an eight-stranded β-barrel (30), and this domain is believed, like other β-barrel-structured porins, to be inserted into the OM. In contrast, the C-terminal domain of OmpA and homologs contains a peptidoglycan-association motif (17; R. De Mot and J. Vanderleyden, Letter, Mol. Microbiol. 12:333-334, 1994), apparently forms an α-helix-rich structure (47), and is located in the periplasmic space. The N-terminal β-barrel cannot form a large channel (30). However, Sugawara and Nikaido (46) showed that OmpA also contains a minority conformer, estimated to comprise about 2 to 3% of the population, that forms channels allowing the diffusion of solutes up to several hundred daltons in size, explaining the low-efficiency porin activity of OmpA and OprF. More-recent studies showed that these minority conformers are formed only when the C-terminal domains were present (2, 6), suggesting that the C-terminal domains participate in the production of larger β-barrels, thus presumably exposing portions of the C-terminal domains on the cell surface. The presence of these two conformers may be reflected in the way anti-OmpA antibodies react with the surface of intact cells.
In this study we report the isolation and characterization of a panel of monoclonal antibodies (MAbs) against Salmonella OmpA and show that a single, highly conserved, sequential epitope on the C-terminal domain of OmpA was immunodominant in the mouse response to infection by serovar Typhimurium. Furthermore, our data suggest that the C-terminal domain is often hidden in the periplasmic space but may also become exposed, less frequently, on the cell surface.
MATERIALS AND METHODS
Mice.
BALB/c mice were used for preparation of anti-OmpA MAbs, whereas CAF1 (BALB/cJ × A/J) F1 mice (Ityr) were used for preparation of immune polyclonal sera and immunoprotection studies. Both strains of mice were obtained from Jackson Laboratories, Bar Harbor, Maine.
Bacterial strains and growth conditions.
Serovar Typhimurium strains WB600 (wild-type) and SH5014 (rfa mutant) and E. coli strain HN705 (ΔompC ompF::Tn5) have been previously described (37, 40, 42, 45); serovar Typhimurium strains SA1627 (rfb [26]) and SL1917 (galE496 ompA202 [44]) were provided by Ken Sanderson and Bruce Stocker, respectively. Clinical isolates of enteric and nonenteric bacteria, as well as the culture media and growth conditions for enteric and nonenteric bacteria, were previously described (41).
Salmonellae for injection were grown from frozen stocks (40), harvested, washed once, and suspended in sterile Ringer's lactate solution (Abbott Laboratories). The number of CFU per milliliter was determined by viable counts on blood agar and bismuth sulfite agar (Difco).
Isolation and purification of OmpA, porins, OM, and LPS.
Attempts were made to purify native OmpA proteins from cell envelopes of serovar Typhimurium SH5014 and E. coli HN705, following the protocol of Sugawara et al. (47). However, OmpA from Salmonella was contaminated with porins and thus had to be further purified by electroelution from sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) slab gels (29). The protein was dialyzed against water containing 0.1% SDS and then concentrated with solid polyethylene glycol 20000 (Fisher). Salmonella porins, OM, and LPS were isolated and purified from serovar Typhimurium strain SH5014 as previously described (38).
Anti-Salmonella MAbs.
BALB/c mice were immunized intraperitoneally (i.p.) with heat-killed whole cells of strain SH5014 (four times with 108 cells at 2-week intervals), and spleens were removed on the third day after the last injection. Mice were also immunized via the footpads with purified OmpA. Cell fusion was carried out as previously described (37). Hybridomas were selected in hypoxanthine-aminopterin-thymidine medium. Culture fluids from wells with colonies were assayed by enzyme-linked immunosorbent assay (ELISA) against OmpA, porins, OM, LPS, and whole cells of serovar Typhimurium strains SH5014 (rfa), SA1627 (rfb), and SL1917 (galE ompA). Hybridomas of interest were cloned twice by limiting dilution and injected into BALB/c mice for production of ascitic tumors. The class and subclass of MAbs were determined by ELISA with goat antisera against mouse heavy and light chains μ, γ1, γ2a, γ2b, γ3, κ, and λ (Fisher). Antibody concentrations of ascites fluid were calculated from a standard curve using isotype-specific, affinity-purified goat anti-mouse immunoglobulins (Ig) as previously described (40).
MAb purification and biotinylation.
Anti-OmpA MAbs were purified by protein G affinity chromatography and biotinylated with Immunopure NHS-LC-Biotin (Pierce) as previously described (41, 42).
ELISA.
Competitive inhibition ELISA.
A competitive inhibition ELISA was performed to identify the epitopes that are recognized by both the anti-OmpA MAbs and the polyclonal immune sera (41, 42). Antigen-coated 96-well plates were blocked and washed as described above. Serial twofold dilutions of polyclonal serum (100 μl) were added to each well (rows 1 to 11) as competitor of the binding by the biotinylated MAb, added subsequently to the antigen. A preselected quantity of biotinylated MAb (100 μl) that produced an ELISA absorbance of 0.5 to 1.0 optical density units compared to the homologous antigen was added to each well (rows 1 to 12), mixed, and incubated overnight at 4°C. The plates were washed and developed with streptavidin-alkaline phosphatase and p-nitrophenylphosphate as described above. Unlabeled homologous antibody was used as a positive control for competitive inhibition. As negative controls, an unrelated mouse antibody (of the same isotype) or preimmune polyclonal serum was incubated with the biotinylated antibody. Plates containing antigen, conjugate and substrate, but without serum or biotinylated antibody, were also included throughout the experiment. The percent binding, calculated by dividing the absorbance obtained in rows 1 to 11 by the absorbance in row 12 (control), was plotted against the dilution of “cold” antiserum.
Proteolytic digestion of OmpA.
The Triton-insoluble envelope fraction (1 mg of protein/ml) (9), which contains porins and OmpA, was incubated with 500 μg of trypsin/ml in 10 mM Tris-HCl, pH 8.0, for 1 h at 37°C. The reaction was terminated with 2 mM phenylmethylsulfonyl fluoride, and the envelope was recovered by centrifugation at 13,000 rpm for 30 min in a microcentrifuge at 4°C. The pellet, shown to contain the N-terminal domain of OmpA by SDS-PAGE, was washed once in 10 mM Tris-HCl, pH 8.0, and was resuspended in the same buffer for ELISA.
Cyanogen bromide (CNBr) digestion of OmpA.
Five milligrams of OmpA monomer was precipitated, washed with acetone, and digested with 250 mg of CNBr in 500 μl of 70% (vol/vol) trifluoroacetic acid as previously described (38, 39). The digested sample was lyophilized twice and then suspended in SDS sample buffer (1 mg/ml) and stored at −80°C for later use in gel electrophoresis.
Immune polyclonal sera.
Immune polyclonal sera were prepared in CAF1 mice against live and heat-killed cells of serovar Typhimurium wild-type strain WB600 and rfa mutant strain SH5014, as previously described (42).
Passive immunization of mice.
Generally, groups of 5 to 10 CAF1 mice received i.p. injections with 0.2 ml of a 1/10 dilution of ascites fluid containing selected anti-OmpA MAbs. Control mice received either 0.2 ml of sterile Ringer's lactate, 1/10 dilution of P3 × 63-Ag8.653 (the non-Ig secreting fusion partner) ascites, CM 12.1 (an unrelated mouse MAb of the IgG2a isotype [38, 40]), normal mouse serum, or immune serum (as a positive control). The mice were subsequently challenged (i.p.) 60 min later with 0.2 ml of WB600 suspension containing 10 or 100 50% lethal doses (12,500 and 125,000 CFU, respectively). The protective efficacies of anti-OmpA MAbs were determined from the 21-day survival data. The median length of survival of each group of mice in which more than 50% of the animals died was estimated, and statistical analyses were performed as previously described (40).
Flow cytometry.
Intact and OM-permeabilized bacteria were stained with anti-OmpA MAbs and fluorescein-conjugated goat anti-mouse Ig (Fisher) and analyzed by cytofluorometry as previously described (41). The OM permeabilization was carried out by an extensive modification (52) of the Ca2+ treatment protocol (5) that was developed for the purpose of introducing proteins into the periplasm of living bacterial cells. It should be noted that the harsh treatment of cells in 300 mM Ca2+ in the original protocol is replaced here by a mild treatment of cells with 10 mM CaCl2, 10 mM MgCl2, and 16% sucrose for 30 min on ice, followed by washing with 0.9% NaCl containing 0.001 M EGTA.
Other methods.
Protein concentrations were measured with the Micro BCA protein assay reagent (Pierce), with bovine serum albumin as the standard. N-terminal sequencing of polypeptides was performed as previously described (38, 39), after first transferring the SDS-PAGE-separated peptides onto Immobilon P polyvinylidene difluoride membranes (Millipore). SDS-PAGE was performed on either 16.5% Tris-Tricine (Bio-Rad) or 10 to 20% linear gradient polyacrylamide gels. Western blotting was performed as described previously (37), after the electrophoretic transfer of the proteins onto nitrocellulose membrane.
RESULTS
MAb specificity to OmpA.
We tested a panel of 12 anti-Salmonella OmpA MAbs (Table 1). Most of these MAbs were derived from fusions against heat-killed SH5014 whole cells (SH), but the panel also included one that was generated by the use of purified Salmonella OmpA (PA). Among the 12, we found 6 IgG2a and 6 IgG2b isotypes (Table 1); all contained κ light chains. In the ascites fluid Ig concentrations were 4 to 7 mg/ml, and ELISA titers to OmpA were 104 to 105 (data not shown).
TABLE 1.
Reactivities of anti-OmpA MAbs with Salmonella OmpA, OM, and whole cells in ELISA
| MAba | Isotype | MAb reactivityb
|
|||
|---|---|---|---|---|---|
| OmpAc | OMc | SH5014 WC | SA1627 WC | ||
| PA 24.1 | IgG2b | +++ | +++ | ++ | ++ |
| SH 16.2 | IgG2a | +++ | +++ | +++ | ++ |
| SH 68.8 | IgG2a | +++ | +++ | +++ | +++ |
| SH 267.15 | IgG2a | +++ | +++ | +++ | +++ |
| SH 269.1 | IgG2b | +++ | +++ | +++ | +++ |
| SH 333.12 | IgG2a | +++ | +++ | +++ | ++ |
| SH 377.16 | IgG2b | +++ | +++ | +++ | + |
| SH 471.7 | IgG2b | +++ | +++ | +++ | +++ |
| SH 591.1 | IgG2a | +++ | +++ | +++ | +++ |
| SH 621.1 | IgG2b | +++ | +++ | +++ | +++ |
| SH 720.1 | IgG2a | +++ | +++ | +++ | +++ |
| SH 785.1 | IgG2b | +++ | +++ | +++ | +++ |
MAbs with the prefixes PA and SH were produced from mice immunized with purified OmpA and heat-killed SH 5014 whole cells, respectively.
Antibodies for ELISA were used at a dilution which resulted in an optical density at 405 nm within the range of 1.0 to 1.5 when tested with homologous antigen (immunogen) as previously described (37). Absorbance values for ascites from the cell fusion partner P3 × 63-Ag8.653, which is a nonsecretor of Igs (control), were always <0.10 at the antibody dilutions tested. Similarly, the absorbance values for the anti-OmpA MAbs against Salmonella porins, LPS, and whole cells of strain SL 1917 (OmpA−) were also <0.1 at the antibody dilutions tested. Abbreviations: OM, outer membrane; WC, whole cells. Symbols for ELISA reactions: +, weakly positive (absorbance, 0.25 to 0.5); ++, moderately positive (absorbance, 0.5 to 1.0); +++, strongly positive (absorbance, >1.0).
OmpA and OM were isolated and purified from S. enterica serovar Typhimurium strain SH5014.
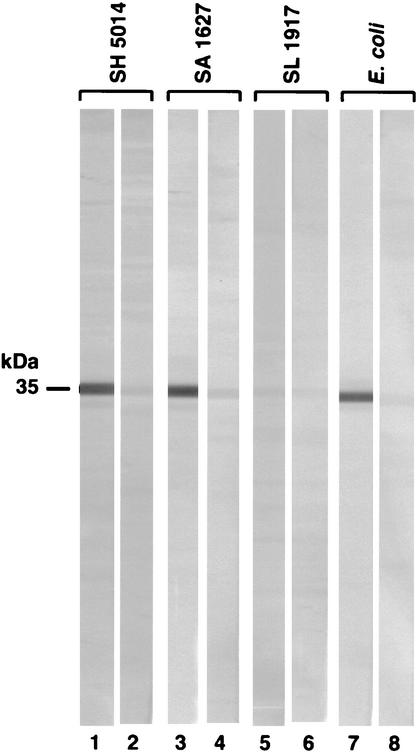
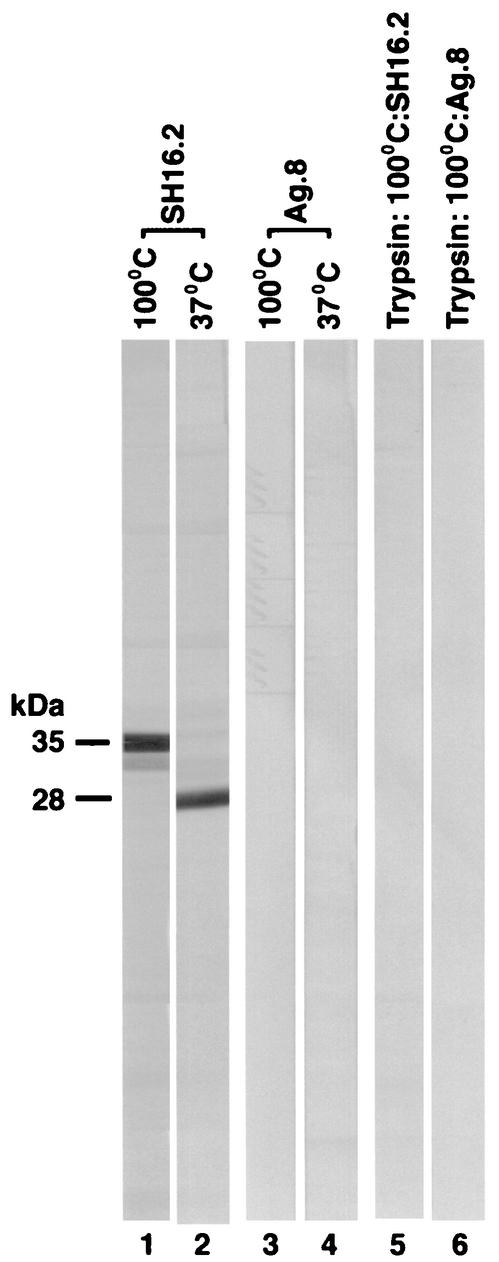
The MAbs were initially examined for their reactivity with Salmonella OmpA, porins, OM, and LPS, as well as whole cells of strains WB600 (wild-type), SH5014 (rfa), SA1627 (rfb), and SL1917 (galE ompA), in ELISA. Positive data are shown in Table 1. All the MAbs in the panel gave strong positive reactions with OmpA, OM, and whole cells of the rough strains (see SH5014 and SA1627 in Table 1), but none bound to Salmonella porins, LPS, or whole cells of wild-type or OmpA− strain (as explained in footnote b of Table 1). Although positive reactions seen in whole-cell ELISA may be due to inadvertent cell lysis or exposure of the antigenic sites on dead cells or OM vesicles shed by the bacteria (15, 49), the ELISA results are consistent with our Western blot and cytofluorometric data (see below). We then examined the MAb specificity by immunoblotting with heated (100°C for 5 min in SDS) and unheated (37°C for 1 h in SDS) cell envelope extracts. The heat treatment was used to identify OmpA because its mobility on SDS-PAGE becomes lower in the completely heat-denatured or heat-modified form than in the partially denatured form present in unheated preparations (27). We found that these antibodies recognized a protein with mobilities corresponding to 35,000 and 28,000 Da in heated and unheated samples, respectively (Fig. 1; Fig. 2, lanes 1 and 2). These data suggested that our panel of MAbs recognized an epitope(s) on the heat-modifiable protein OmpA that was probably sequential in nature since its recognition was not affected by the denaturation of the protein.
FIG. 1.
Western immunoblot reactions of anti-OmpA MAbs. Cell envelopes (or whole cells; see below) from bacterial strains were lysed with SDS by boiling at 100°C for 5 min, subjected to SDS-PAGE, and transferred to nitrocellulose. The paper was cut into strips and incubated with a 1:50 dilution of MAb SH16.2 (lanes 1, 3, 5, and 7). Control strips (lanes 2, 4, 6, and 8) were probed with ascites fluid from the cell fusion partner P3-63-Ag.8.653. The reaction was developed with alkaline phosphatase-labeled goat anti-mouse Ig-nitroblue tetrazolium plus bromochloroindolyl phosphate. Other bacterial strains (whole cells) showing positive reaction patterns included serovar Typhimurium WB600, C. amalonaticus, S. boydii, Klebsiella pneumoniae, Enterobacter cloacae, Serratia marcescens, Edwardsiella tarda, Hafnia alvei, and Yersinia enterocolitica. On the other hand, Proteus vulgaris, Morganella morganii, and Providencia stuartii gave negative reactions.
FIG. 2.
Effect of heat and trypsin treatment on the Western immunoblot reactivity of anti-OmpA MAbs. Triton-insoluble cell envelope of serovar Typhimurium strain SH5014 before (lanes 1 to 4) and after (lanes 5 and 6) treatment with trypsin was suspended in SDS sample buffer and then either boiled at 100°C for 5 min (lanes 1, 3, 5, and 6) or solubilized at 37°C for 1 h (lanes 2 and 4). Proteins were separated by SDS-PAGE and immunoblotted (Fig. 1). Nitrocellulose strips were probed with either MAb SH16.2 (lanes 1, 2, and 5) or Ag.8 ascites (lanes 3, 4, and 6).
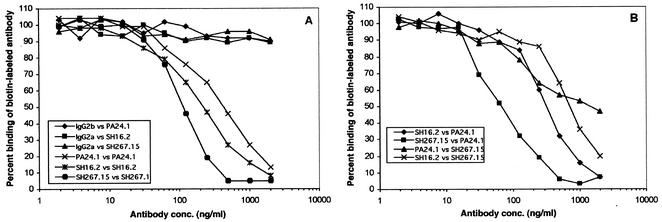
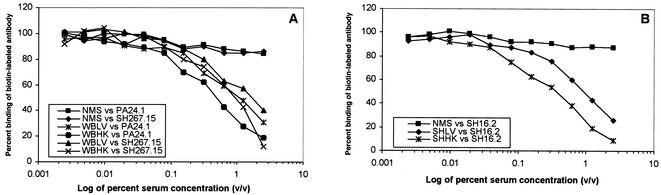
We performed competitive binding assays (see Materials and Methods) with anti-OmpA MAbs to determine whether these antibodies recognized the same or different epitopes on OmpA. We found the binding of each biotinylated MAb was decreased by the presence of the other, nonbiotinylated MAbs (Fig. 3B; controls shown in Fig. 3A), suggesting that all antibodies tested recognized the same, common epitope. On the other hand, we did not observe any competition between anti-OmpA MAbs and the previously reported (41, 42) antiporin (CM12.1, CT21.1, and DT20.2) or anti-LPS (SH6.11 and WB60.4) MAbs (data not shown). Unless stated otherwise, all subsequent experiments were carried out with MAbs PA24.1, SH16.2, SH267.15, and/or SH591.1.
FIG. 3.
Recognition of similar epitopes by anti-OmpA MAbs. Studies of MAb competition for binding to OmpA were carried out by competitive inhibition ELISA. Plates were coated with the homologous antigen (purified Salmonella OmpA or heat-killed whole cells of strain SH5014). Unbiotinylated MAbs were added at various concentrations (2.0 to 0.016 μg/ml) followed by the biotinylated MAbs at a preselected concentration that yielded an absorbance of 0.5 to 1.0 at 420 nm versus the homologous antigen. ELISA was then carried out as described in Materials and Methods. Unrelated IgG2a and IgG2b mouse antibodies were used as negative controls (A), whereas unlabeled homologous MAbs were used as positive controls (A) in the competition experiments (B).
MAbs were specific to the C-terminal domain of OmpA.
The majority conformer of OmpA is separated into two large domains, the N-terminal domain and the C-terminal domain. Trypsin treatment of cell envelope is known to degrade the C-terminal domain but leaves the N-terminal domain of approximately 24,000 Da intact (18). In our studies, the anti-OmpA MAbs in the panel did not react with the trypsin-treated crude OM (Triton-insoluble envelope) (Fig. 2; lane 5). Thus, these antibodies recognized an epitope(s) that was probably localized to the C-terminal domain of OmpA.
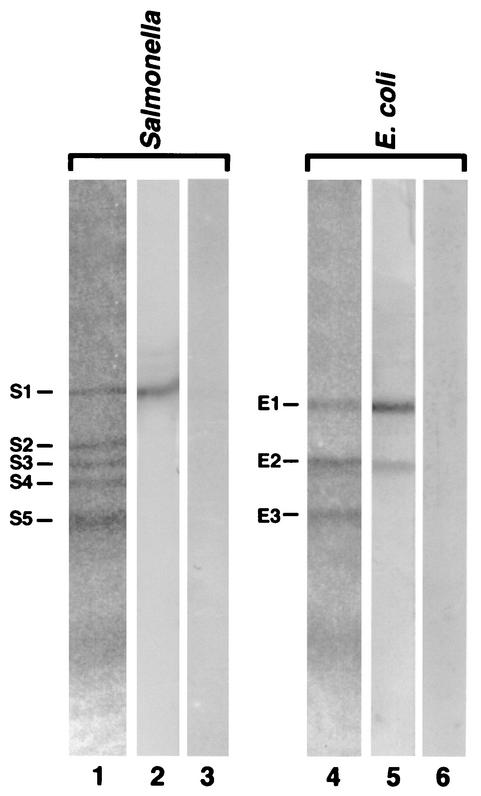
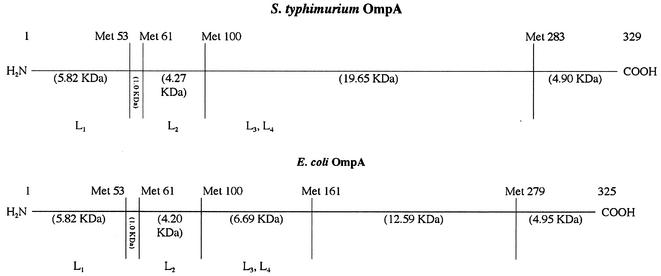
OmpA from serovar Typhimurium and E. coli were split into fragments with CNBr. We detected at least three to five bands (fragments, in E. coli and Salmonella, respectively) with estimated molecular masses ranging from 24,200 to 7,400 Da (Fig. 4; lanes 1 and 4). Analysis of these fragments for their immunoreactivity with anti-OmpA MAbs revealed a strong binding of these antibodies to a band with an approximate molecular mass of 24,200 Da in Salmonella (Fig. 4, band S1, lane 2). In E. coli, the antibodies bound to two bands with an approximate molecular mass of 22,800 Da (band E1) and 16,900 Da (Fig. 4, band E2, lane 5). These protein bands were transferred to polyvinylidene difluoride membranes, excised, and sequenced. The Salmonella fragment S1 (N-terminal sequence: VWRAD…) contained residues 101 to 329 (Fig. 5). The E. coli fragment E2 (N-terminal sequence: LSLGV…) contained residues 162 to 325. Band E1, however, contained two peptides with N-terminal sequences LSLGV… (major component) and GESNP… (minor component). These correspond to residues 162 to 325 and residues 280 to 325, respectively (Fig. 5). The observed molecular weights of S1, E1, and E2 fragments were comparable to the molecular weights of 24,553, 22,494, and 17,545, respectively, expected from the sequences. CNBr thus cut OmpA after Met 100 in Salmonella and Met 161 and Met 279 in E. coli, but apparently the cleavage at Met 283 (and at Met 279) was incomplete (Fig. 5). The association of a small peptide (residues 280 to 325) with a much larger peptide (residues 162 to 325) in fragment E1 of E. coli was unexpected; however, such anomalous electrophoretic behavior has been reported for other OM protein fragments (see reference 39). The E. coli fragment E2 (residues 162 to 325) in any case contains only the C-terminal domain (22, 35), and its reactivity with MAb SH16.2 confirms the location of epitope in the C-terminal domain.
FIG. 4.
CNBr cleavage of OmpA and reaction of the fragments with MAb SH16.2. The CNBr-generated peptides from Salmonella and E. coli were separated by SDS-PAGE and immunoblotted (Fig. 1). Lanes 1 and 4 contain the CNBr peptides as separated on the SDS gel, lanes 2 and 5 contain the immunoblots of MAb SH16.2 with these peptides, and lanes 3 and 6 contain the control strips probed with ascites fluid from Ag.8.
FIG. 5.
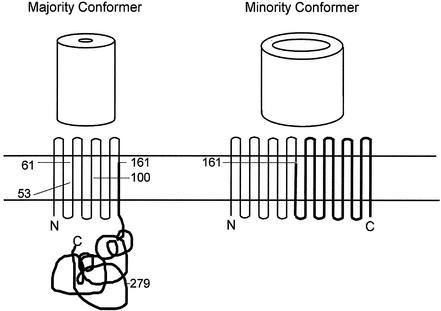
Identification of OmpA domain bound by anti-OmpA MAbs. The Met positions and the corresponding molecular weights of the expected CNBr-generated fragments are shown for mature OmpA of serovar Typhimurium and E. coli in the top panel. Surface-exposed loops L1 (amino acids 17 to 33), L2 (amino acids 61 to 71), L3 (amino acids 107 to 114), and L4 (amino acids 144 to 160) that are localized in the N-terminal domain are also shown. Transmembrane topology for the majority and minority conformers of OmpA is shown at the bottom, with the location of the methionine residues (in the E. coli protein) indicated for the majority conformer. The C-terminal fragment, bearing the major epitope, is shown as thick lines.
The C terminus epitope is conserved.
Western blotting showed that anti-OmpA MAb SH16.2 cross-reacted with at least 9 of the 12 other enterobacterial species tested (see Fig. 1). However, this and other MAbs did not bind to proteins in the nonenterobacterial species such as Aeromonas hydrophila, Bordetella pertussis, Brucella suis, Haemophilus influenzae, Neisseria gonorrhoeae, and P. aeruginosa (data not shown). These results indicated that the OmpA epitope recognized by our panel of MAbs is widely conserved within (but not outside) the family Enterobacteriaceae.
The epitope in the C-terminal domain of OmpA is an immunodominant antigen.
The significance of the OmpA epitope in the context of natural or artificial infection was examined first by following the course of humoral immune response in mice with live or heat-killed cells of either S-form WB600 or an Ra mutant SH5014. We found that the highest titer to OmpA was reached on day 21 and the titer remained high up to day 42 (data not shown). The ELISA titer of sera to OmpA varied from approximately 1.0 × 104 to 4.1 × 104 and, in general, was higher when mice were given heat-killed cells than when they were given the live organisms (data not shown). The primary antibody against OmpA belonged to the IgG2a subclass in mice given the live as well as heat-killed cells (data not shown). These data on the humoral immune response are consistent with those reported earlier by Singh et al. for the Salmonella porins (42).
We then examined the content of anti-OmpA antibodies in these polyclonal sera by competition binding assays with biotin-labeled MAbs. The immune sera used were previously shown to also contain antibodies to other OM proteins and LPS (42). Similar competition data were obtained with each of the three polyclonal sera (days 14, 21, and 28) we tested; data obtained with sera from day 21 are shown in Fig. 6. Positive and negative control results from this assay are presented in Fig. 3A; an additional negative control, NMS at a 1:40 dilution, inhibited binding only by approximately 15% (Fig. 6). With all the polyclonal sera (raised to live or heat-killed, WB600 or SH5014 cells), 50% inhibition of the binding of MAbs occurred with serum concentrations between 0.3 and 1% (Fig. 6). In earlier studies using anti-O4 and antiporin MAbs, 50% inhibition was obtained over a similar range of concentrations of the same polyclonal sera, except for one anti-O4 MAb (42). Thus, the epitope(s) located on the C-terminal domain of OmpA is indeed a major immunodominant antigen, comparable to OmpC porin (recognized by MAb CT21.1 [42]) and O4 antigen (recognized by MAb SH6.11 [41]), in the humoral immune response against whole live or killed cells of Salmonella. Unlike SH6.11, however, CT21.1 and anti-OmpA MAbs (PA 24.1, SH16.2, SH267.15, and SH591.1) did not protect or prolong the survival of mice against as little as 10 50% lethal doses of virulent strain WB600 (40) (Table 2).
FIG. 6.
Immune recognition of the OmpA epitope in mice. Competition for binding to the epitope on the C-terminal domain by anti-OmpA MAbs and the immune mouse sera raised against serovar Typhimurium strains WB600 (wild type) (A) and SH5014 (rfa) (B) was measured by competitive ELISA. Plates were coated with the homologous antigen (see Fig. 3), blocked, and filled with serial twofold dilutions of preimmune normal mouse serum (NMS) or immune mouse serum raised against live (LV) or heat-killed (HK) serovar Typhimurium strain WB600 (WB) or SH5014 (SH). A preselected concentration of biotinylated MAb that yielded an absorbance of 0.5 to 1.0 at 420 nm versus the homologous antigen was added to each well, the well contents were mixed, and the plate was incubated overnight at 4°C. The plates were washed and developed with streptavidin-alkaline phosphatase as described in Materials and Methods.
TABLE 2.
Passive immunization with anti-Salmonella OmpA MAbsa
| MAbb | Dilutionc | Median length (days) of survivald (no. of mice alive/total no. receiving injection after challenge dose (LD50)e of:
|
|
|---|---|---|---|
| 10 | 100 | ||
| PA 24.1 | 1:10 | 5 (0/10) | 4 (0/9) |
| SH 16.2 | 1:10 | 6 (0/10) | 4 (0/9) |
| SH 267.15 | 1:10 | 6 (0/10) | 4 (0/7) |
| SH 591.1 | 1:10 | 6 (0/10) | 4 (0/8) |
| Immune mouse serum | 1:10 | >21 (7/7)f | >21 (5/5)f |
| Normal mouse serum | 1:10 | 5 (0/8) | 3 (0/5) |
| Ringer's lactate | 6 (0/7) | 3 (0/3) | |
| Ag. 8 | 1:10 | 6 (0/9) | 4 (0/5) |
| CM 12.1 | 1:10 | 6 (0/7) | 4 (0/5) |
CAF1 female mice (7 to 9 weeks old, weighing 20 to 22 g) received injections (i.p.) with 0.2 ml of individual ascites. Control mice received either 0.2 ml of immune mouse serum (positive control), normal mouse serum, sterile Ringer's lactate, Ag.8 ascites, or an unrelated mouse MAb (CM12.1 [38, 40]) of the IgG2a isotype. The mice were subsequently challenged (i.p.) 60 min later with 0.2 ml containing either 10 (12,500 CFU) or 100 (125,000 CFU) 50% lethal doses (LD50s) of S. enterica serovar Typhimurium strain WB600. The protective efficacies of anti-Salmonella MAbs and the immune polyclonal serum were determined from the 21-day survival data as previously described (40).
See Table 1 for the isotype of anti-OmpA MAbs. Immune polyclonal mouse serum was prepared by infecting mice with sublethal doses of S. enterica serovar Typhimurium WB600 as previously described (40).
All dilutions were made in sterile Ringer's lactate.
The median length of survival of each group of mice in which more than 50% of the animals died was estimated by calculating the reciprocal mean length of survival in days postchallenge of each animal in the group as previously described (40).
S. enterica serovar Typhimurium WB600 cells were grown, harvested, and suspended in sterile Ringer's lactate solution. The number of CFU per milliliter was estimated by measurement of optical density at 420 nm (optical density of 1 = 1.5 × 108 CFU/ml). The actual number of CFU was determined by viable count on blood agar and bismuth sulfite agar.
Statistical analyses were carried out as previously described (40). P ≤ 0.005.
Accessibility of the OmpA epitope on the cell surface.
The C-terminal domain of the majority conformer of OmpA is buried in the periplasm (see the Introduction), and it was unexpected to see MAbs directed to this domain reacting with intact cells of even strains with R-form LPS (SH5014 or SH1627) (Table 1). We therefore studied the issue of surface accessibility of this epitope in depth.
Before we investigated on which side of the membrane this epitope was located, we had to examine whether O chains of LPS hindered access of MAbs to the antigen, as has been the case with the surface epitopes of porin (4). Indeed, by ELISA, the MAbs showed strong binding to whole cells of Salmonella R mutants (SH5014 and SH1627) but did not react with those of the smooth strains of Salmonella (WB600) (Table 1). Thus, the epitope on the cell surface was not readily accessible for MAbs if the O antigen of LPS was present. This is consistent with earlier reports of the physical shielding of surface epitopes of porin proteins by the O chains in wild-type Salmonella (reviewed in reference 40). However, MAb SH16.2 bound to whole cells of E. coli (clinical isolate), Citrobacter amalonaticus, and Shigella boydii by ELISA, suggesting that O chains of these bacteria did not strongly hinder the access of MAb to OmpA (data not shown).
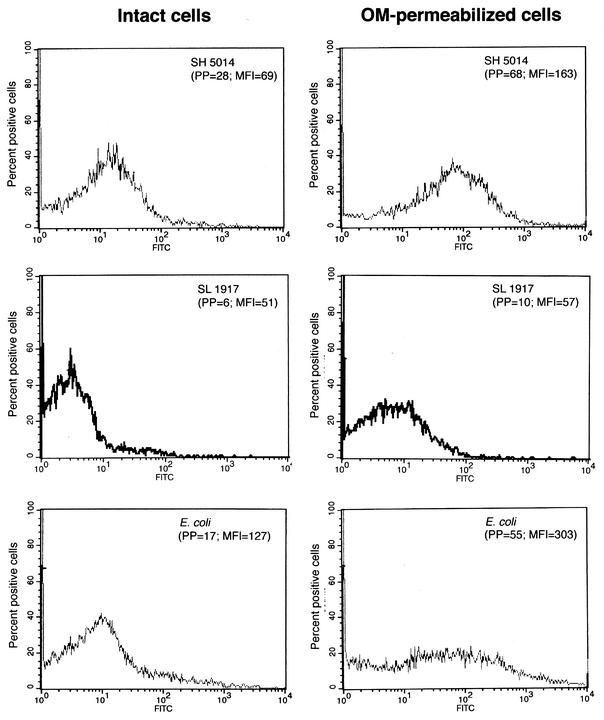
To obtain more-detailed information on the localization of the OmpA epitope, we investigated the binding of SH16.2 to the enteric bacteria at the individual-cell level with cytofluorometry. This method quantitates MAb binding to surface epitopes on intact live bacteria, while excluding cell ghosts and debris from analysis by forward scatter-side scatter gating (16). We found that the mean fluorescence values of serovar Typhimurium rough strain SH5014 (rfa), and E. coli (clinical isolate) were approximately two to two and a half times the signal obtained with serovar Typhimurium strain SL1917 (OmpA−) (Fig. 7). Serovar Typhimurium strains SA1627 (rfb) and WB600 (wild-type) gave mean fluorescence values that were similar to those obtained with SH5014 and SL1917, respectively (data not shown). We further examined the binding of SH16.2 to Salmonella SH5014 and E. coli cells whose OM had been permeabilized for the entry of proteins, as described in Materials and Methods. The data indicate that the OM permeabilization resulted in an ∼2.5- to 3-fold increase in both the number and the mean fluorescence intensity of the cells stained (Fig. 7). These results suggest that a major portion of the SH16.2 epitope is located in the periplasm but a minor portion of it is indeed exposed on the cell surface.
FIG. 7.
Flow cytometry analysis of intact and OM-permeabilized cells with anti-OmpA MAb SH16.2. Histograms of the occurrence of cells with various levels of fluorescence intensity are shown. Intact or OM-permeabilized cells were incubated with a 1/100 dilution of SH16.2, stained with fluorescein isothiocyanate-conjugated goat anti-mouse IgG, and analyzed for intensity of green fluorescence (MAb binding). OM permeabilization of cells was carried out by incubation of cells in 10 mM CaCl2-10 mM MgCl2 and 16% sucrose for 30 min on ice. Permeabilized cells were washed with a buffer containing 0.9% NaCl and 0.001 M EGTA. The total population analyzed was 10,000 bacteria for each strain. Abbreviations: PP, percent of cells staining positive; MFI, mean fluorescence intensity.
DISCUSSION
OmpA is a major OM protein in E. coli and other enteric bacteria (3, 18, 27). In this study, we showed that the major epitope of the Salmonella OmpA is located in its C-terminal half. These data confirm the strong immunogenicity of the C-terminal domain (31) and are also consistent with the observation that some of the major epitopes of OprF, a homolog of OmpA in P. aeruginosa, are located in its C-terminal domain (14, 23, 33). Furthermore, examination of mouse polyclonal sera after immunization with live or heat-killed whole cells of Salmonella showed that the OmpA epitope bound by our panel of MAbs was an immunodominant antigen. This epitope of OmpA appears similar in its importance to other immunodominant epitopes of Salmonella that are present on the surface of OmpC porin and in LPS O4 factor (42). However, anti-OmpA MAbs, like antiporin antibodies, could not protect mice against Salmonella infection, possibly because the access of the antibody to these proteins on the surface of OM was limited by the O chains of the LPS. Vuopio-Varkila et al. (51) also reported that anti-OmpA polyclonal sera failed to protect against challenge with the encapsulated E. coli O18:K1.
Since our panel of anti-OmpA MAbs binds purified OmpA, OM, and intact rough strains of Salmonella and several other enteric bacteria in ELISA and shows some binding to intact cells by flow cytometry, we believe that this epitope is at least to some extent exposed on the cell surface. This is at first sight surprising, (i) because the majority of the OmpA population is known to fold into a two-domain structure, with the N-terminal half forming an eight-stranded β-barrel traversing the OM and the C-terminal half forming a periplasmic, globular domain (17, 22, 30, 47) and (ii) because this model predicts that the C-terminal half, buried in the periplasm, should be inaccessible to MAbs in intact cells. However, Sugawara and Nikaido (46) showed that both OmpA and OprF fold into two alternative conformations, a majority one with essentially closed pores and a minority one with open channels. β-Barrels each containing at least a dozen or more β-strands would be needed to produce open channels that allow diffusion of solutes of several hundred daltons (45), and indeed, Arora et al. (2) showed that the entire OmpA sequence is needed to produce large, open channels. It is therefore likely that the minority, open conformers of OmpA fold like a classical porin, with many β-strands, as was predicted (incorrectly for the majority population) previously (43). In this conformer, the loops in the C-terminal half will be exposed on the external surface of the OM and should be accessible for MAbs. Indeed, site-specific labeling showed that in the open conformers of OprF, an OmpA homolog, parts of the C-terminal half were exposed on the cell surface (Sugawara and Nikaido, unpublished data).
Since 2 to 3% of the OmpA population is in the open conformation (46), and since there are about 50,000 molecules of OmpA per E. coli cell (25), it is understandable that MAb specific for the C-terminal epitope reacted to OmpA in intact cells, presumably to these open conformers (Table 1). However, MAb would be able to react much more strongly if it could gain access to the cognate epitope present in the C-terminal, periplasmic domain of the majority conformers of OmpA. This is presumably reflected by the two- to threefold increase in MAb reactivity in OM-permeabilized intact cells (Fig. 7). We emphasize that the permeabilization of the OM was carried out under very mild conditions, using only 10 mM CaCl2 in the presence of 10 mM MgCl2 and 16% sucrose, conditions that are not expected to remove LPS and thereby increase the accessibility of already surface-exposed OmpA epitopes.
In a homologous system of P. aeruginosa OprF, a folding model composed of porin-like 16 transmembrane β-strands was prevalent, in large part because the MAbs directed to epitopes in the C-terminal half reacted with intact cells of this organism (33). Additionally, peptides from the C-terminal half, corresponding to residues 261 to 274 and 305 to 318 of OprF, elicited whole-cell-reactive antibodies (14, 15, 50), a result not expected if the C-terminal domain is always located in the periplasm. A monoclonal antibody directed against the C terminus also reacted with the intact bacterium (34). However, the present study suggests that qualitative demonstration of antibody binding to intact cells is not sufficient, because the protein may have more than one final conformations; a comparison of untreated versus OM-permeabilized cells would have been instructive. In fact, a circular dichroism study (47) showed clearly that the majority conformer of OprF contains a large α-helical (presumably periplasmic) domain just as OmpA does and that the folding model proposed by Rawling et al. (33) is incorrect. The presence of a minority conformer, which forms large channels by utilizing the entire sequence including the C-terminal half, was recently shown also for OprF (6).
The molecular mechanism of pore formation by OmpA/OprF family proteins has been controversial (24). However, now all the results, including direct functional fractionation (46), planar bilayer studies (2, 6), X-ray crystallography of the N-terminal half (30), and antibody accessibility studies (reference 33 and the present study), are consistent with a unified model in which the proteins fold into two alternative conformations, a two-domain, closed channel conformation and a one-domain, open channel conformation. (This is likely to be true with any member of this family. Indeed, a recent report suggests that the C-terminal end of MopB, the OmpA homolog in Methylococcus capsulatus, also contains surface-exposed regions [11].) Crystallography obviously favors more abundant conformers, and furthermore the N-terminal half, which has no possibility of forming a channel alone, was used as the material (30). Clearly the successful crystallization of the N-terminal domain is not a proof that the whole protein cannot take another conformation with open channels. The proteins of OmpA/OprF family are known to be important for the stabilization of the envelope structure, presumably because their C-terminal periplasmic domains interact with peptidoglycan (17). This is likely to be the main function of the majority conformer of these proteins.
Finally, it should be added that the OmpA epitope bound by our panel of MAbs, because it is widely conserved, immunodominant, and antibody-accessible at least in partially rough mutants, may provide a site for insertion and expression of foreign genes in Salmonella strains (32, 36).
Acknowledgments
We thank Ken Sanderson and Bruce Stocker for providing bacterial strains; Madhu Singh for compilation of reference material; Carol Williams for assistance with data management and line figures and for typing the manuscript; and John Ward of the NIH Medical Arts and Photography Branch for production of some of the images.
This work was supported by Public Health Service grant GM 08219 to S.P.S.
Editor: A. D. O'Brien
REFERENCES
- 1.Aron, L., G. Faundez, C. Gonzalez, E. Roessler, and F. Cabello. 1993. Lipopolysaccharide-independent radioimmunoprecipitation and identification of structural and in vivo induced immunogenic surface proteins of Salmonella typhi in typhoid fever. Vaccine 11:10-17. [DOI] [PubMed] [Google Scholar]
- 2.Arora, A., D. Rinehart, G. Szabo, and L. K. Tamm. 2000. Refolded outer membrane protein A of Escherichia coli forms ion channels with two conductance states in planar lipid bilayers. J. Biol. Chem. 275:1594-1600. [DOI] [PubMed] [Google Scholar]
- 3.Beher, M. G., C. A. Schnaitman, and A. P. Pugsley. 1980. Major heat-modifiable outer membrane protein in gram-negative bacteria: comparison with the OmpA protein of Escherichia coli. J. Bacteriol. 143:906-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bentley, A. T., and P. E. Klebba. 1988. Effect of lipopolysaccharide structure on reactivity of antiporin monoclonal antibodies with the bacterial cell surface. J. Bacteriol. 170:1063-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brass, J. M. 1986. Calcium-induced permeabilization of the outer membrane: a method for reconstitution of periplasmic binding protein-dependent transport systems in Escherichia coli and Salmonella typhimurium. Methods Enzymol. 125:289-302. [DOI] [PubMed] [Google Scholar]
- 6.Brinkman, F. S. L., M. Bains, and R. E. W. Hancock. 2000. The amino terminus of Pseudomonas aeruginosa outer membrane protein OprF forms channels in lipid bilayer membranes: correlation with a three-dimensional model. J. Bacteriol. 182:5251-5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calderón, I., S. R. Lobos, H. A. Rojas, C. Palomino, L. H. Rodríguez, and G. C. Mora. 1986. Antibodies to porin antigens of Salmonella typhi induced during typhoid infection in humans. Infect. Immun. 52:209-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins, F. M. 1974. Vaccines and cell-mediated immunity. Bacteriol. Rev. 38:371-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diedrich, D. L., A. O. Summers, and C. A. Schnaitman. 1977. Outer membrane proteins of Escherichia coli. V. Evidence that protein 1 and bacteriophage directed protein 2 are different polypeptides. J. Bacteriol. 131:598-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eisenstein, T. K., and B. M. Sultzer. 1983. Immunity to Salmonella infection, p. 261-296. In T. K. Eisenstein, P. Actor, and H. Friedman (ed.), Host defenses to intracellular pathogens. Plenum Press, New York, N.Y.
- 11.Fjellbirkeland, A., V. Bemanian, I. R. McDonald, J. C. Murrell, and H. B. Jensen. 2000. Molecular analysis of an outer membrane protein, MopB, of Methylococcus capsulatus (Bath) and structural comparisons with proteins of the OmpA family. Arch. Microbiol. 173:346-351. [DOI] [PubMed] [Google Scholar]
- 12.Freudl, R., and S. T. Cole. 1983. Cloning and molecular characterization of the ompA gene from Salmonella typhimurium. Eur. J. Biochem. 134:497-502. [DOI] [PubMed] [Google Scholar]
- 13.Gentry-Weeks, C. R., A.-L. Hultsch, S. M. Kelly, J. M. Keith, and R. Curtiss III. 1992. Cloning and sequencing of a gene encoding a 21-kilodalton outer membrane protein from Bordetella avium and expression of the gene in Salmonella typhimurium. J. Bacteriol. 174:7729-7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilleland, H. E., Jr., E. E. Hughes, L. B. Gilleland, J. M. Matthews-Greer, and J. Staczek. 1995. Use of synthetic peptides to identify surface-exposed, linear B-cell epitopes within outer membrane protein F of Pseudomonas aeruginosa. Curr. Microbiol. 31:279-286. [DOI] [PubMed] [Google Scholar]
- 15.Hughes, E. E., L. B. Gilleland, and H. E. Gilleland, Jr. 1992. Synthetic peptides representing epitopes of outer membrane protein F of Pseudomonas aeruginosa that elicit antibodies reactive with whole cells of heterologous immunotype strains of P. aeruginosa. Infect. Immun. 60:3497-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klebba, P. E., S. A. Benson, S. Bala, T. Abdullalh, J. Reid, S. P. Singh, and H. Nikaido. 1990. Determinants of OmpF porin antigenicity and structure. J. Biol. Chem. 265:6800-6810. [PubMed] [Google Scholar]
- 17.Koebnik, R. 1995. Proposal for a peptidoglycan-associating alpha-helical motif in the C-terminal regions of some bacterial cell-surface proteins. Mol. Microbiol. 16:1269-1270. [DOI] [PubMed] [Google Scholar]
- 18.Lugtenberg, B., and L. van Alphen. 1983. Molecular architecture and functioning of the outer membrane of Escherichia coli and other gram-negative bacteria. Biochim. Biophys. Acta 737:51-115. [DOI] [PubMed] [Google Scholar]
- 19.Mahasreshti, P. J., G. L. Murphy, J. H. Wyckoff, I. I. I., S. Farmer, R. E. W. Hancock, and A. W. Confer. 1997. Purification and partial characterization of the OmpA family of proteins of Pasteurella haemolytica. Infect. Immun. 65:211-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marandi, M. V., and K. R. Mittal. 1997. Role of outer membrane protein H (OmpH)- and OmpA-specific monoclonal antibodies from hybridoma tumors in protection of mice against Pasteurella multocida. Infect. Immun. 65:4502-4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Metcalf, E. S., and A. D. O'Brien. 1981. Characterization of murine antibody response to Salmonella typhimurium by a class-specific solid-phase radioimmunoassay. Infect. Immun. 31:33-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morona, R., M. Klose, and U. Henning. 1984. Escherichia coli K-12 outer membrane protein (OmpA) as a bacteriophage receptor: analysis of mutant genes expressing altered proteins. J. Bacteriol. 159:570-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mutharia, L. M., and R. E. W. Hancock. 1985. Characterization of two surface-localized antigenic sites on porin protein F of Pseudomonas aeruginosa. Can. J. Microbiol. 31:381-386. [DOI] [PubMed] [Google Scholar]
- 24.Nikaido, H. 1992. Porins and specific channels of bacterial outer membranes. Mol. Microbiol. 6:435-442. [DOI] [PubMed] [Google Scholar]
- 25.Nikaido, H. 1996. Outer membrane, p. 29-47. In F. C. Neidhardt (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. I. ASM Press, Washington, D.C.
- 26.Nikaido, H., M. Levinthal, K. Nikaido, and K. Nakane. 1967. Extended deletions in the histidine-rough-B region of the Salmonella chromosome. Proc. Natl. Acad. Sci. USA 57:1825-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikaido, H., and M. Vaara. 1985. Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 49:1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ortiz, V., A. Isibasi, E. Garcia-Ortigoza, and J. Kumate. 1989. Immunoblot detection of class-specific humoral immune response to outer membrane proteins isolated from Salmonella typhi in humans with typhoid fever. J. Clin. Microbiol. 27:1640-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parr, T. R., K. Poole, G. W. K. Crockford, and R. E. W. Hancock. 1986. Lipopolysaccharide-free Escherichia coli OmpF and Pseudomonas aeruginosa protein P porins are functionally active in lipid bilayer membranes. J. Bacteriol. 165:523-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pautsch, A., and G. E. Schulz. 1998. Structure of the outer membrane protein A transmembrane domain. Nat. Struct. Biol. 5:1013-1017. [DOI] [PubMed] [Google Scholar]
- 31.Puohiniemi, R., M. Karvonen, J. Vuopio-Varkila, A. Muotiala, I. M. Helander, and M. Sarvas. 1990. A strong antibody response to the periplasmic C-terminal domain of the OmpA protein of Escherichia coli is produced by immunization with purified OmpA or with whole E. coli or Salmonella typhimurium bacteria. Infect. Immun. 58:1691-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rauly, I., L. Goetsch, J.-F. Haeuw, C. Tardieux, T. Baussant, J.-Y. Bonnefoy, and N. Corvaia. 1999. Carrier properties of a protein derived from outer membrane protein A of Klebsiella pneumoniae. Infect. Immun. 67:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rawling, E. G., N. L. Martin, and R. E. W. Hancock. 1995. Epitope mapping of the Pseudomonas aeruginosa major outer membrane porin protein OprF. Infect. Immun. 63:38-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rebecca, S. Y., H. J. Wong, and R. E. W. Hancock. 1993. Linker-insertion mutagenesis of Pseudomonas aeruginosa outer membrane protein OprF. Mol. Microbiol. 10:283-292. [PubMed] [Google Scholar]
- 35.Ried, G., R. Koebnik, I. Hindennach, B. Mutschler, and U. Henning. 1994. Membrane topology and assembly of the outer membrane protein OmpA of Escherichia coli K12. Mol. Gen. Genet. 243:127-135. [DOI] [PubMed] [Google Scholar]
- 36.Roberts, M., S. N. Chatfield, and G. Dougan. 1994. Salmonella as carrier of heterologous antigens, p. 27-58. In D. T. O'Hagen (ed.), Novel delivery systems for oral vaccines. CRC Press, Boca Raton, Fla.
- 37.Singh, S. P., Y. Upshaw, T. Abdullah, S. R. Singh, and P. E. Klebba. 1992. Structural relatedness of enteric bacterial porins assessed with monoclonal antibodies to Salmonella typhimurium OmpD and OmpC. J. Bacteriol. 174:1965-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh, S. P., S. R. Singh, Y. U. Williams, L. Jones, and T. Abdullah. 1995. Antigenic determinants of the OmpC porin from Salmonella typhimurium. Infect. Immun. 63:4600-4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh, S. P., S. Miller, Y. U. Williams, K. E. Rudd, and H. Nikaido. 1996. Immunochemical structure of the OmpD porin from Salmonella typhimurium. Microbiology 142:3201-3210. [DOI] [PubMed] [Google Scholar]
- 40.Singh, S. P., Y. U. Williams, W. H. Benjamin, P. E. Klebba, and D. Boyd. 1996. Immunoprotection by monoclonal antibodies to the porins and lipopolysaccharide of Salmonella typhimurium. Microb. Pathog. 21:249-263. [DOI] [PubMed] [Google Scholar]
- 41.Singh, S. P., S. Miller, Y. U. Williams, P. E. Klebba, P. Macchia, and N. Marshall. 1999. Recognition specificity of monoclonal antibodies that protect mice against Salmonella typhimurium infection. Res. Microbiol. 150:385-394. [DOI] [PubMed] [Google Scholar]
- 42.Singh, S. P., Y. U. Williams, P. E. Klebba, P. Macchia, and S. Miller. 2000. Immune recognition of porin and lipopolysaccharide epitopes of Salmonella typhimurium in mice. Microb. Pathog. 28:157-167. [DOI] [PubMed] [Google Scholar]
- 43.Stathopoulos, C. 1996. An alternative topological model for Escherichia coli OmpA. Protein Sci. 5:170-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stocker, B. A. D., M. Nurminen, and P. H. Mäkelä. 1979. Mutants defective in the 33K outer membrane protein of Salmonella typhimurium. J. Bacteriol. 139:376-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sugawara, E., and H. Nikaido. 1992. Pore-forming activity of OmpA protein of Escherichia coli. J. Biol. Chem. 267:2507-2511. [PubMed] [Google Scholar]
- 46.Sugawara, E., and H. Nikaido. 1994. OmpA protein of Escherichia coli outer membrane occurs in open and closed channel forms. J. Biol. Chem. 269:17981-17987. [PubMed] [Google Scholar]
- 47.Sugawara, E., M. Steiert, S. Rouhani, and H. Nikaido. 1996. Secondary structure of the outer membrane proteins OmpA of Escherichia coli and OprF of Pseudomonas aeruginosa. J. Bacteriol. 178:6067-6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tagawa, Y., M. Haritani, H. Ishikawa, and N. Yuasa. 1993. Characterization of a heat-modifiable outer membrane protein of Haemophilus somnus. Infect. Immun. 61:1750-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tarkka, E., A. Muotiala, M. Karvonen, K. Saukkonen-Laitinen, and M. Sarvas. 1989. Antibody production to a meningococcal outer membrane protein cloned into live Salmonella typhimurium aroA vaccine strain. Microb. Pathog. 6:327-335. [DOI] [PubMed] [Google Scholar]
- 50.von Specht, B., B.-U. Knapp, G. Muth, M. Broker, K.-D. Hungerer, K.-D. Diehl, K. Massarrat, A. Seemann, and H. Domdey. 1995. Protection of immunocompromised mice against lethal infection with Pseudomonas aeruginosa by active or passive immunization with recombinant P. aeruginosa outer membrane protein F and outer membrane protein I fusion proteins. Infect. Immun. 63:1855-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vuopio-Varkila, J., M. Karvonen, and H. Saxen. 1988. Protective capacity of antibodies to outer-membrane components of Escherichia coli in a systemic mouse peritonitis model. J. Med. Microbiol. 25:77-84. [DOI] [PubMed] [Google Scholar]
- 52.Zgurskaya, H. I., and H. Nikaido. 1999. AcrA is highly asymmetric protein capable of spanning the periplasm. J. Mol. Biol. 285:409-420. [DOI] [PubMed] [Google Scholar]