Abstract
ClC chloride channels are found in all three kingdoms of life though little is known about their functions in prokaryotes. Here we investigated the role of a Vibrio cholerae ClC channel in acid resistance and intestinal colonization. The putative V. cholerae ClC channel was found to confer mild resistance to acid when pH was adjusted with HCl, but not with other acids. Surprisingly, a ClC channel deletion mutant exhibited enhanced intestinal colonization in infant mice.
In multicellular organisms, at least three structurally distinct classes of Cl− channels have been identified. The ClC family of Cl− channels, which are generally voltage gated, have been found in organisms as diverse as mammals, plants, and yeast (7, 12, 20). In mammals, nine ClC genes have been described, and roles for several of their products have been determined (12). These channels are required for the transepithelial movement of salt and water in the gastrointestinal tract and kidney, for the regulation of electrical excitability in muscle and nerves, and for regulation of cell volume and intracellular pH (12). Mutations in human ClC genes underlie several familial forms of myotonia and nephropathy (7).
The complete genomic sequences of a large number of bacteria led to the surprising realization that ClC type channels are found in prokaryotes as well as eukaryotes (11, 15). Unlike the eukaryotic ClC channels, several microbial ClC proteins could be overexpressed and purified sufficiently to enable the determination of their molecular structures. The X-ray structures of ClC channels from Salmonella enterica serovar Typhimurium and Escherichia coli were reported recently (5). As was predicted from earlier work (10, 18, 19), ClC channels were found to be “double-barreled.” The channels are homodimers whose subunits each contain a conduction pore, an ion-binding site, and a potential gating region (5). Each subunit also contains 18 alpha helices that lie within, but often do not span the entire width of, the membrane. The end of one helix projects into the cytoplasm and may provide a means for cytoplasmic processes to regulate channel function.
The functions of prokaryotic ClC channels have only just begun to be investigated. Maduke et al. found that liposomes reconstituted with an E. coli ClC channel designated EriC exhibited selective anion permeability (15). Recently, it was reported that deletion of eriC and of mriT, which encodes another ClC orthologue in E. coli, rendered the resulting strain sensitive to extreme acid (pH 2.5) challenge (9). These deletions were also found to reduce the ability of membrane-localized antiporters to deliver substrates to and remove products from cytoplasmic decarboxylases. Decarboxylation, which consumes protons, is thought to protect cells from H+ ions that leak into cells in low-pH environments. The reduced activity of the decarboxylase-linked antiporter in ClC-deficient cells led Iyer et al. to propose that E. coli ClC channels function as electrical shunts to prevent the hyperpolarization of the inner membrane, which would otherwise occur as antiporters export positively charged substrates (9).
Vibrio cholerae is a gram-negative rod that causes the severe and sometimes lethal diarrheal disease cholera. V. cholerae is a facultative human pathogen that lives outside of human hosts, often in estuarine environments. Humans become infected with this organism by eating or drinking contaminated food or water. Following passage through the ordinarily acidic environment of the stomach, V. cholerae multiplies within the small bowel. The secretory diarrhea characteristic of cholera is caused by the activity of cholera toxin, a protein exotoxin secreted by V. cholerae during infection (14). This A-B subunit toxin causes secretion of Cl− by intestinal crypt cells and decreased absorption of NaCl by villus cells (22). Water exits from epithelial cells into the bowel lumen across an osmotic gradient, and diarrhea results.
Here we investigated the role of a V. cholerae ClC channel in acid resistance and intestinal colonization. The putative V. cholerae ClC channel was found to confer mild resistance to acid when pH was adjusted with HCl, but not with other acids. Surprisingly, a ClC channel deletion mutant exhibited enhanced intestinal colonization in infant mice.
V. cholerae ClC orthologue.
Investigators who primarily study eukaryotic ClC channels identified a single unannotated open reading frame (ORF) in the V. cholerae genome that encoded a protein, VCA0526, with a mild degree of similarity to the eukaryotic ClC channels (11, 15). The ORF lies on the small V. cholerae chromosome and appears to be monocistronic, as the two ORFs that surround it are divergently transcribed. Pairwise BLAST analysis indicates that the putative amino acid sequence of VCA0526 is highly similar to those of EriC and MriT, the two E. coli ClC channel proteins (205 of 442 identities, E value of e−111 for EriC; 51 of 151 identities, E value of 2e−16 for MriT). Although the degree of similarity of VCA0526 to the human ClC channels is much less (e.g., 41 of 141 identities, E value of 1e−04 for the human ClC-1), there is near identity in four short stretches of the V. cholerae, E. coli, and human proteins in regions that were implicated as ion-binding sites in the crystal structure of EriC (5).
Acid sensitivity of a V. cholerae ClC mutant.
YD1, a derivative of El Tor strain N16961 (the V. cholerae genome strain), was constructed by standard means (3, 4) to study whether VCA0526 played a role in V. cholerae acid resistance. In YD1, an in-frame deletion removes all but 10 codons of vca0526. This mutation, whose presence was confirmed by Southern analysis, does not impair growth in vitro in Luria broth (LB).
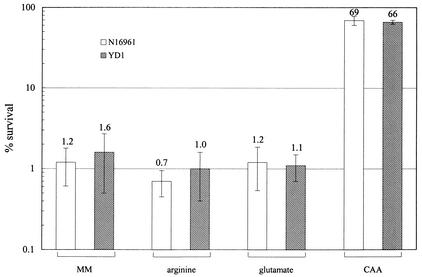
Several experimental methods for measuring acid resistance in bacteria have been reported (6). The extreme acid resistance test protocol is thought to mimic conditions present in the highly acidic environment of the human stomach. With this method, stationary-phase organisms are challenged in low-pH minimal media containing arginine or glutamate. While the viability of wild-type E. coli is not significantly reduced after a pH 2.5 challenge (9), we found that V. cholerae does not survive this acid challenge when the media contained either glutamate or arginine. Wild-type El Tor N16961 cells were not detectable after 1 h of incubation in pH 2.5 media. In fact, even in pH 5 minimal media supplemented with arginine or glutamate, approximately 1% of wild-type cells survived for 30 min (Fig. 1). These observations are consistent with clinical studies that suggest that the infectious dose of V. cholerae is much larger than that of pathogenic E. coli (21). We found that V. cholerae can survive a pH 5 acid challenge when minimal media are supplemented with 1% Casamino Acids. However, N16961 and YD1 did not differ in their sensitivity to challenge in pH 5 minimal media supplemented with 1% Casamino Acids (Fig. 1). This finding suggests that the putative V. cholerae ClC channel encoded by vca0526 may not function in stationary-phase cells in the same manner as the E. coli ClC channels.
FIG. 1.
Survival of stationary-phase N16961 and YD1 in pH 5.0 minimal medium (40 mM KCl, 80 mM KH2PO4, 1.7 mM sodium citrate, 20 mM glucose, using H3PO4 to adjust to pH 5.0) alone (MM), or minimal medium supplemented with 1 mM arginine, 1 mM glutamate, or 1% Casamino Acids (CAA) after 30 min. N16961 and YD1 were grown overnight in LB at 30°C and then diluted 1:100 in low-pH challenge media. After 30 min of incubation at room temperature, cell numbers were enumerated by plating. The percent survival was calculated by comparing the numbers of cells present at time zero and after 30 min in the challenge media. Approximately 5 × 107 cells were used in each assay. Each bar represents the mean with standard error for three to five independent experiments.
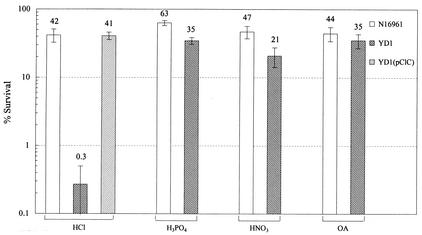
We developed an assay using exponential-phase cells to further investigate YD1 acid sensitivity. In this assay system, cells were grown in LB to an optical density at 600 nm of ∼0.5 and then were placed in lower-pH LB challenge media. Cell viability, determined by plating, was evaluated after 60 min of incubation in the low-pH challenge media. There was minimal reduction (approximately twofold) in N16961 viability after incubation at pH 4.4 when the challenge medium pH was adjusted with HCl (Fig. 2). In contrast, there was an ∼330-fold reduction in the viability of YD1 at this time. Acid sensitivity of YD1 was attributable to the deletion of vca0526, since the expression of this gene from a plasmid (pCLC) in YD1 restored the acid resistance of this strain (Fig. 2).
FIG. 2.
Survival of mid-log-phase N16961, YD1, and YD1(pClC) in LB adjusted to pH 4.4 with either HCl, H3PO4, or HNO3, or to pH 4.9 with an organic acid cocktail (OA) after 60 min of incubation. Cells were grown in LB to an optical density at 600 nm of around 0.5 and then spun down and resuspended in the challenge media. After 1 h of incubation at room temperature, cell numbers were enumerated by plating. The percent survival was calculated by comparing the numbers of cells present at time zero and after 60 min in the challenge media. OA challenge medium was LB supplemented with a 0.1× organic acid cocktail. The 1× cocktail was 87 mM acetic acid, 25 mM butyric acid, and 37 mM propionic acid. Approximately 8 × 107 cells were used in each assay. Each bar represents the mean with standard error for three to five independent experiments.
The acid sensitivity of YD1 was influenced by the acid used to adjust the pH of the challenge media. For example, in contrast to the marked sensitivity of YD1 to HCl-adjusted media, pH 4.4, the viability of this strain was hardly reduced after incubation in media adjusted to this pH with either H3PO4 or HNO3 (Fig. 2). YD1 was somewhat more susceptible to acid than N16961 when the pH of the challenge media was adjusted to 4.2 with either H3PO4 or HNO3 (data not shown); however the significance of this observation is difficult to interpret given the fact that the viability of N16961 is greatly reduced (<0.1% survival) in media adjusted to pH 4.2 with either of these acids. N16961 was more sensitive to acid conditions in the presence of organic acids (a cocktail of butyric acid, acetic acid, and propionic acid at concentrations that approximate those in the human intestine; see the Fig. 2 legend) (2, 17). For example, less than 0.1% of these cells survived challenge in pH 4.7 LB supplemented with organic acids. Approximately 50% of these cells survived after 60 min in media adjusted to pH 4.9 with organic acids. There was no difference in the susceptibilities of YD1 and N16961 to this acid challenge (Fig. 2).
Our findings reveal that the V. cholerae ClC channel encoded by vca0526, like the E. coli channels EriC and MriT, plays some role in acid resistance. However, while the mechanistic basis for the acid sensitivity of the E. coli mriT eriC deletion mutant is known, for YD1 it is not. Surprisingly, in our assay, the anion component of the acid used to adjust the pH of the challenge media influenced the acid sensitivity of YD1. Similar observations have not been reported for the E. coli ClC deletion mutant. Since we used exponentially growing cells and rich media to study acid sensitivity of the V. cholerae ClC channel deletion mutant, we cannot directly compare our findings with those obtained from studies of the acid sensitivity of the E. coli eriC deletion mutant. However, if the V. cholerae ClC channel acts as an electrical shunt, as was proposed for the E. coli ClC channel, it is difficult to explain the anion specificity that we observed. The amount of Cl− added to LB challenge media to adjust the pH to 4.4 does not significantly alter the Cl− concentration of the media.
Intestinal colonization of suckling mice by YD1.
We speculated that vca0526 might play some role in V. cholerae pathogenicity for two reasons. First, acid resistance may facilitate intestinal colonization. Second, V. cholerae might utilize a Cl− channel to detect and/or respond to the Cl−-rich fluid in the intestinal lumen that is induced by cholera toxin. A competition assay was used to assess the intestinal colonization properties of YD1 in the suckling mouse model of cholera. In this assay, approximately equal numbers of YD1 (which is lacZ+) and NLAC, a lacZ derivative of N16961, were intragastrically inoculated into 4- to 5-day-old CD-1 mice. After ∼22 h of intraintestinal growth, the ratio of YD1 to NLAC in small bowel homogenates was determined. Surprisingly, the ratio of YD1 to NLAC in intestinal homogenates was significantly greater than 1; in two independent experiments, the ratio of YD1 to NLAC in the homogenates varied from ∼9 to 16 (Table 1). This was not the case with in vitro competition assays. When the identical inocula used for the intestinal colonization assays were inoculated into LB, YD1 did not outcompete NLAC; instead, the ratio of YD1 to NLAC cells after these in vitro competition assays was close to one (Table 1).
TABLE 1.
Intestinal colonization of YD1 and N16961 in suckling micea
| Exptb | No. of YD1 cells/no. of NLAC cells
|
CId
|
|||
|---|---|---|---|---|---|
| Input | In vivo (SEM)c | In vitro | In vivo | In vitro | |
| 1 | 0.94 | 8.9 (1.9) | 0.7 | 9.4 | 0.8 |
| 2 | 1.24 | 16.5 (6.0) | 0.6 | 12.5 | 0.5 |
Competition assays were conducted as previously described (1). Briefly, NLAC, a lacZ derivative of N16961, and YD1 cells were grown overnight in LB at 30°C. After 1:1,000 dilution, the two strains were mixed 1:1 and then intragastrically inoculated into 4- to 5-day-old CD-1 mice (inoculum, 105 to 106 CFU of each strain) and into 3.0 ml of LB. After 20 to 24 h of intraintestinal or in vitro growth at 37°C, the numbers of YD1 and NLAC cells in intestinal homogenates and in LB cultures were determined by plating on LB plates containing 200 μg of streptomycin and 40 μg of 5-bromo-4-indolyl-β-d-galactoside (X-Gal)/ml.
Data were derived from five to eight suckling mice in each experiment.
Mean of ratios. SEM, standard error of the mean.
CI (competitive index) is calculated by dividing the ratio of YD1 to NLAC cells found in vivo or in vitro by the input ratio.
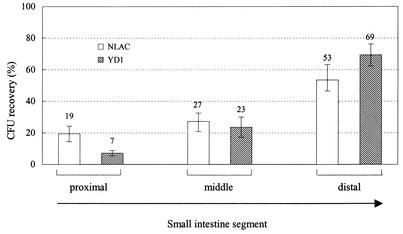
The enhanced growth of YD1 in vivo relative to that of wild-type V. cholerae could reflect either increased growth throughout the mouse intestine or an ability to colonize sites not typically utilized by V. cholerae. For example, V. cholerae chemotaxis mutants, which also have enhanced intestinal colonization, exhibited an aberrant distribution within the small bowel, namely, increased colonization of the proximal small bowel (13). We therefore analyzed the distribution of YD1 within the suckling mouse intestine using the protocol developed by Angelichio et al. (1). Five hours after intraintestinal inoculation, we found that YD1, like N16961, was primarily located within the middle and distal two-thirds of the small intestine (Fig. 3). Therefore, at least at this fairly gross anatomical level of analysis, YD1 localization within the segments of the small bowel is not unusual; aberrant localization of YD1 does not appear to explain why this strain colonizes better than wild-type cells. It is possible that finer dissection of the sites of colonization of YD1 would reveal that these cells can grow in unusual niches. Alternatively, the vca0526 deletion may facilitate the growth of V. cholerae in its usual intestinal niches.
FIG. 3.
Recovery of NLAC and YD1 in segments of the small intestines of suckling mice. Cultures of NLAC and YD1 were grown overnight at 30°C in LB and then diluted 1:1,000. Suckling mice were intragastrically inoculated with 7.0 × 105 to 8.0 × 105 CFU of either NLAC or YD1. After 5 h of intraintestinal growth, the mice were sacrificed. The small intestines were removed and were divided into three segments of equal length. After homogenization in LB, the numbers of CFU in each segment were determined by plating. The mean numbers of CFU recovered from the entire small intestine were 1.8 × 104 for NLAC and 1.1 × 105 for YD1. Each bar represents the mean percentage of CFU found in each segment compared to the total CFU found in the entire small intestine. The error bars represent the standard errors of the means. There were eight mice for each strain in two independent experiments.
The results of the YD1-versus-NLAC competition experiments using suckling mice and LB suggest that vca0526 is deleterious to the growth of V. cholerae in the intestine but not in rich media. The reasons for the enhanced intestinal colonization of YD1 are not known. In suckling mouse small intestine colonization experiments, the numbers of recoverable CFU represent the sums of the cells present in the inoculum and their progeny minus the numbers of cells that were killed or expelled from the small intestine into the large intestine. Therefore, the hypercolonization of YD1 could be accounted for by factors that influence the bacterial growth rate, adhesion, motility of either the bacteria or the gastrointestinal tract, or resistance to host-derived antibacterial factors. In V. cholerae, motility and virulence are linked and both are influenced by sodium flux (8). It is tempting to speculate that Cl− flux mediated by VCA0526 may also regulate these processes. However, since YD1 exhibited normal motility and chemotaxis in vitro (data not shown), Cl− flux-mediated alterations in these phenotypes in vivo would have to be a response to stimuli present only within the intestine. Such stimuli might include the Cl−-rich intestinal secretions induced by cholera toxin. Regardless of the mechanism of the enhanced intestinal colonization of YD1, this phenotype may be useful in the development of live-attenuated vaccine strains of V. cholerae, since prior studies suggest that colonization is a key determinant of immunogenicity (16).
Acknowledgments
We are grateful to Chris Miller for pointing out to us the V. cholerae ClC orthologue and Brigid Davis and Andrew Camilli for critical reading of the manuscript. We thank Anne Kane and the NEMC GRASP Center (P30DK-34928) for preparation of our media.
This work was supported by NIH grant AI-42347 and HHMI.
Editor: V. J. DiRita
REFERENCES
- 1.Angelichio, M. J., J. Spector, M. K. Waldor, and A. Camilli. 1999. Vibrio cholerae intestinal population dynamics in the suckling mouse model of infection. Infect. Immun. 67:3733-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baik, H. S., S. Bearson, S. Dunbar, and J. W. Foster. 1996. The acid tolerance response of Salmonella typhimurium provides protection against organic acids. Microbiology 142:3195-3200. [DOI] [PubMed] [Google Scholar]
- 3.Davis, B. M., and M. K. Waldor. 2000. CTXφ contains a hybrid genome derived from tandemly integrated elements. Proc. Natl. Acad. Sci. USA 97:8572-8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dutzler, R., E. B. Campbell, M. Cadene, B. T. Chait, and R. MacKinnon. 2002. X-ray structure of a ClC chloride channel at 3.0 Å reveals the molecular basis of anion selectivity. Nature 415:287-294. [DOI] [PubMed] [Google Scholar]
- 6.Foster, J. W. 2000. Microbial responses to acid stress, p. 99-115. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 7.George, A. L., Jr., L. Bianchi, E. M. Link, and C. G. Vanoye. 2001. From stones to bones: the biology of ClC chloride channels. Curr. Biol. 11:R620-R628. [DOI] [PubMed] [Google Scholar]
- 8.Hase, C. C., and J. J. Mekalanos. 1999. Effects of changes in membrane sodium flux on virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 96:3183-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iyer, R., T. M. Iverson, A. Accardi, and C. Miller. 2002. A biological role for prokaryotic ClC chloride channels. Nature 419:715-718. [DOI] [PubMed] [Google Scholar]
- 10.Jentsch, T. J. 2002. Chloride channels are different. Nature 415:276-277. [DOI] [PubMed] [Google Scholar]
- 11.Jentsch, T. J., T. Friedrich, A. Schriever, and H. Yamada. 1999. The CLC chloride channel family. Pflueg. Arch. Eur. J. Physiol. 437:783-795. [DOI] [PubMed] [Google Scholar]
- 12.Jentsch, T. J., and W. Gunther. 1997. Chloride channels: an emerging molecular picture. Bioessays 19:117-126. [DOI] [PubMed] [Google Scholar]
- 13.Lee, S. H., S. M. Butler, and A. Camilli. 2001. Selection for in vivo regulators of bacterial virulence. Proc. Natl. Acad. Sci. USA 98:6889-6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levine, M. M., J. B. Kaper, R. E. Black, and M. L. Clements. 1983. New knowledge on pathogenesis of bacterial enteric infections as applied to vaccine development. Microbiol. Rev. 47:510-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maduke, M., D. J. Pheasant, and C. Miller. 1999. High-level expression, functional reconstitution, and quaternary structure of a prokaryotic ClC-type chloride channel. J. Gen. Physiol. 114:713-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mekalanos, J. J., and J. C. Sadoff. 1994. Cholera vaccines: fighting an ancient scourge. Science 265:1387-1389. [DOI] [PubMed] [Google Scholar]
- 17.Merrell, D. S., and A. Camilli. 1999. The cadA gene of Vibrio cholerae is induced during infection and plays a role in acid tolerance. Mol. Microbiol. 34:836-849. [DOI] [PubMed] [Google Scholar]
- 18.Miller, C. 1982. Open-state substructure of single chloride channels from Torpedo electroplax. Philos. Trans. R. Soc. Lond. B Biol. Sci. 299:401-411. [DOI] [PubMed] [Google Scholar]
- 19.Miller, C., and M. M. White. 1984. Dimeric structure of single chloride channels from Torpedo electroplax. Proc. Natl. Acad. Sci. USA 81:2772-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20..Mindell, J. A., and M. Maduke. 2001. ClC chloride channels. Genome Biol. 2: 3003.1-3003.6. [DOI] [PMC free article] [PubMed]
- 21.Mintz, E. D., T. Popovic, and P. A. Blake. 1994. Transmission of Vibrio cholerae O1, p. 345-356. In I. K. Wachsmuth, P. A. Blake, and O. Olsvik (ed.), Vibrio cholerae and cholera: molecular to global perspectives. American Society for Microbiology, Washington, D.C.
- 22.Sears, C. L., and J. B. Kaper. 1996. Enteric bacterial toxins: mechanisms of action and linkage to intestinal secretion. Microbiol. Rev. 60:167-215. [DOI] [PMC free article] [PubMed] [Google Scholar]