Abstract
Group B streptococci (GBS) remain the most significant bacterial pathogen causing neonatal sepsis, pneumonia, and meningitis in the United States despite the chemoprophylaxis strategies for preventing infection recommended by the Centers for Disease Control and Prevention. Using signature-tagged transposon mutagenesis to screen for novel virulence factors, we identified the rpoE gene as essential for development of sepsis in a neonatal rat model of GBS infection. An rpoE allelic replacement mutant displayed attenuated virulence in the sepsis model of infection identical to that of the transposon mutant, confirming linkage of the phenotype to the mutation in rpoE. The rpoE mutants also displayed increased sensitivity to killing in whole-blood bactericidal assays, which may explain the attenuated virulence. The mutants were otherwise phenotypically identical to the wild-type strain, including growth rate in plasma, indicating that a growth defect is not responsible for the attenuated virulence. rpoE is found only in gram-positive bacterial species and encodes the delta peptide, a subunit of RNA polymerase. Previous in vitro studies in other bacteria suggest that the delta peptide plays a role in maintaining transcriptional fidelity by blocking RNA polymerase binding at all but the strongest promoters, thereby inhibiting initiation of transcription. Despite the availability of rpoE mutants for several gram-positive bacterial species, a role for the peptide in vivo has not been defined, though it has been postulated that the delta peptide may be important for long-term survival in vitro or during growth phase transitions. Our data represent the first report of a phenotype relevant to virulence for rpoE mutants.
Streptococcus agalactiae (group B streptococci [GBS]) is the leading cause of neonatal sepsis, pneumonia, and meningitis in the United States and Western Europe and is an emerging pathogen in immunocompromised adults (31, 32). GBS associated with human disease are almost invariably encapsulated, belonging to one of the nine recognized capsular serotypes: Ia, Ib, or II to VIII. The type-specific capsule of GBS has been extensively studied and is an important determinant for both virulence and immunity (30, 36). Other studies of GBS virulence have examined adherence to and invasion of epithelial and endothelial barriers and interactions with host immune pathways and have focused on a few specific factors, such as hemolysin, alpha and beta proteins, and C5a peptidase (for a review, see reference 24). Recognition of the prevalence and severity of human neonatal and adult disease emphasizes the significance of investigating the mechanisms important to the pathogenesis of GBS infections.
We previously reported the use of signature-tagged transposon mutagenesis (STM) as a screening tool in a neonatal rat sepsis infection model to identify new virulence factors for GBS (13). The most attenuated mutant identified in the STM screen had a transposon insertion in the rpoE gene, encoding the delta subunit of RNA polymerase (RNAP). On the basis of this phenotype, this mutant was subjected to further characterization.
Bacterial RNAP consists of a multisubunit enzyme complex that is the main target for the regulation of gene expression (19). RNAPs from diverse bacterial species display a highly conserved heteromeric structure consisting of a catalytic core enzyme complexed to one of a family of sigma factors (σ). Initiation of transcription requires core RNAP and several different protein factors (18). The minimal core RNAP consists of four subunits (ββ′α2) and is sufficient to catalyze the polymerization of nucleoside triphosphates into RNA (16). Ancillary proteins modify RNAP during promoter binding, initiation, elongation, and termination steps to effect changes in gene expression (19). Core enzyme interacts with sigma factors that provide specificity, to form ββ′α2 σ, which has the capacity to initiate transcription at promoters. Other proteins, including the delta protein encoded by rpoE, are frequently isolated as components of the purified RNAP, but their roles in transcription are less clear (2, 16).
On the basis of genome database sequence information, rpoE appears to be ubiquitous among gram-positive bacterial species. Despite the availability of rpoE mutants in other well-studied gram-positive bacteria such as Bacillus subtilis and Staphylococcus aureus, the function of the delta protein in vivo has not been determined. Phenotypes for rpoE mutants range from extended lag phase growth for B. subtilis mutants (19) to defects in starvation-induced stationary-phase survival or recovery for S. aureus mutants (34). The relevance of these observations from other bacterial species to GBS virulence is unclear at present.
Here we report the construction and characterization of a GBS rpoE allelic exchange mutant. These studies show that the activity of the delta subunit of RNAP appears to be required for resistance of GBS to phagocytic killing and for survival in the host.
MATERIALS AND METHODS
Bacterial strains and growth media.
The bacterial strains and plasmids used in this study are described in Table 1. GBS strains were grown in Todd-Hewitt broth (THB; Difco Laboratories) or in chemically defined medium (CDM) (37) in 5% CO2 at 37°C unless otherwise specified. For antibiotic selection, 1 μg of erythromycin (ERY) per ml or 800 μg of kanamycin (KAN) per ml was added to the medium. GBS strain A909 is a type Ia capsular polysaccharide clinical isolate (21). Strain A909ΔcpsE was generously provided by Larry Madoff. The cpsE gene (glucosyl transferase) was deleted, rendering the strain capsule deficient (L. Madoff, personal communication). Escherichia coli strains were grown in Luria-Bertani broth (LB; Difco Laboratories) with aeration. For antibiotic selection, 300 μg of ERY per ml, 10 μg of chloramphenicol per ml, or 40 μg of KAN per ml was added to the medium.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| S. agalactiae | ||
| A909 | Wild-type, serotype Ia | 21 |
| AJ8D3 | A909 Tn917::rpoE | 13 |
| AJ200 | A909ΔrpoE, allelic exchange mutant, Kmr | This study |
| A909ΔcpsE | A909ΔcpsE, acapsular mutant | L. Madoff |
| E. coli MC1061 | F− araD139 Δ(ara-leu)7696 Δ(lac)X74 galU galK hsdR2 (rK− mK+) mcrA mcrB1 rpsL(Strr) | 35 |
| Plasmids | ||
| pDC123 | Cmr, phoZ, cloning vector | 5 |
| pCIV2 | Kmr/ΩKm-2 | 25 |
| pHY304 | Emr temperature-sensitive pVE6007Δ derivative, lacZα/multiple cloning site of pBluescript | H. H. Yim |
| pAJ020 | pDC123 with 2.5-kb insert containing rpoE gene and flanking DNA sequence | This study |
| pAJ024 | Kmr pAJ020 with rpoE deleted and replaced with ΩKm-2 from pCIV2 | This study |
| pAJ042 | Emr Kmr pHY304 with insert containing ΩKm-2 and flanking DNA sequence | This study |
DNA methods and sequencing analysis.
Chromosomal DNA from GBS was isolated by the method of Framson et al. (9). Digestion and modification of plasmid and chromosomal DNA were performed using standard protocols (4). Southern hybridization analysis of GBS chromosomal DNA was performed as described previously (4). Plasmid DNA from GBS strains was isolated using a Hi Speed Plasmid Midi or Miniprep kit (Qiagen) according to the manufacturer's directions, except that GBS were first converted to spheroplasts by digestion with endo-N-acetylmuramidase (mutanolysin; 0.5 U/μl) (Sigma) at 37°C for 40 min (9).
DNA sequencing was performed using a Perkin-Elmer Ready Reaction sequencing kit (Perkin-Elmer Cetus) according to the manufacturer's directions. Sequences were analyzed using BLAST analysis (3) and aligned using Sequencher version 3.1 (Gene Codes Corporation, Framingham, Mass.).
Construction of an rpoE allelic exchange mutant.
The GBS rpoE gene and approximately 2 kb of flanking DNA were amplified by high-fidelity PCR (Failsafe; Epicentre Technologies) using A909 chromosomal DNA as a template and primers RopA3 (5′-GAATTTAGTGTTGCCAAAGG-3′) and RopA3R (5′-CACTATAAATCTTACCCGTTG-3′) using the protocol supplied by the manufacturer. The resulting PCR product was ligated to EcoRV-digested pDC123, a streptococcal/E. coli shuttle vector (6) to which T-overhangs had been added using Taq polymerase and dTTP as described previously (4). E. coli MC1061 clones containing the resulting recombinant plasmid pAJ020 were identified on agar containing 10 μg of chloramphenicol per ml and 50 μg of 5-bromo-4-chloro-3-indolylphosphate per ml. The rpoE gene in pAJ020 was replaced with a SmaI restriction enzyme site by inside-out PCR using primers AJRDFS (5′-GCCCCGGGGACTATCATTTTGTTTAGC-3′) and AJRDRS (5′-GCCCCGGGGGAAGAAGAGGAAGAAGAGG-3′), which read outward from the 5′ and 3′ ends of rpoE. A 2.2-kb BamHI restriction fragment of pCIV2 containing the omega (Ω) Km gene was ligated to the SmaI-cut PCR product, generating pAJ024.
pAJ024 was digested with XbaI and XhoI restriction enzymes to release a 2.5-kb insert containing the ΩKm cassette and adjacent flanking DNA. The fragment was cloned into pHY304 (generously provided by H. H. Yim), a temperature-sensitive shuttle vector suitable for obtaining double-crossover mutants in GBS (38), generating pAJ042. pAJ042 was electroporated into strain A909, and transformants were selected at 30°C on Todd-Hewitt agar (THA) containing KAN. Allelic exchange of the rpoE gene on the wild-type chromosomal copy with the ΩKm cassette was achieved in a single-step process. A909 containing pAJ042 was grown at 30°C for 6 h in THB containing KAN. The culture was pelleted, washed in THB prewarmed to 37°C, diluted 1/20 into fresh THB prewarmed to 37°C, and then grown to saturation. This step was repeated a second time, and the culture was plated on THA containing KAN. Individual Kmr colonies were replica plated onto THA containing ERY to allow differentiation between single- and double-crossover events.
Kmr Ems clones were screened by PCR to confirm allelic replacement of the rpoE gene with the ΩKm cassette. Southern blotting analysis of chromosomal DNA was used to further confirm the allelic exchange. Chromosomal DNA isolated from potential allelic exchange mutants was digested with EcoRI, resolved on a 0.8% agarose gel, and transferred to a MagnaGraph membrane (Osmonics). A probe consisting of the insert from pAJ020 (3′ end of ropA, rpoE, and 5′ end of pyrG) was generated by PCR and labeled with the digoxigenin (DIG) Chem-Link Labeling and Detection set (Roche). Hybridization, washing, and detection were performed according to the manufacturer's directions.
Transcriptional analysis.
RNA was isolated from GBS strains grown to logarithmic phase in THB as previously described (39) using a FastPrep FP101 bead beater (Bio 101). RNA dot blots were generated by applying serial dilutions of total cellular RNA to a MagnaGraph membrane using a Minifold I dot blot vacuum apparatus (Schleicher & Schuell) according to the manufacturer's recommendations. RNA probes were generated using PCR products to which the T7 promoter sequence (5′-TAATACGACTCACTATA-3′) had been added as templates, and the DIG RNA Labeling kit (SP6/T7) (Roche) was used according to the manufacturer's instructions.
Reverse transcription-PCR (RT-PCR) was used to confirm that transcription of adjacent genes was not disrupted in the allelic exchange mutant. RNA was reverse transcribed using Omniscript reverse transcriptase (Qiagen) and random hexamers (Amersham Pharmacia Biotech) according to the manufacturer's directions and then amplified by PCR using primers RopA3 (5′-GAATTTAGTGTTGCCAAAGG-3′) and RopA4 (5′-TGCTTCAAAGCCTTCTGCAGC-3′) to amplify a portion of ropA, primers RPOEF (5′-GATGATTGAAGTTGCTCGCGC-3′) and RPOER (5′-CCTCTTCTTCCTCTTCTTCC-3′) to amplify a portion of rpoE, and primers CTPF2 (5′-GGTGGTGTGGTCTCTTCT-3′) and CTPR2 (5′-CATCTGAATCTGTAGTTG-3′) to amplify a portion of pyrG.
Growth and phenotypic plate assays.
Growth of isogenic strains was assessed in THB and CDM by monitoring the optical density at 600 nm of cultures grown statically at 37°C in 5% CO2. The ability of the isogenic strains to grow in human plasma was also assessed. Whole-blood samples were collected from laboratory volunteers in accordance with the guidelines of the Children's Hospital and Regional Medical Center Institutional Review Board. Heparin was added to 10 U/ml of blood and plasma obtained by centrifugation. As GBS do not achieve an optical density great enough to monitor by spectrophotometric methods, growth over a 4-h period was determined by plating serial dilutions of the cultures grown in plasma at 37°C for viable colony counts.
Production of hemolysin, CAMP factor, hyaluronidase, and DNase activity by isogenic strains was assessed by standard plate assays (23, 33).
Quantitation of capsular sialic acid.
Capsular sialic acid recovered after acid hydrolysis of equivalent numbers of whole cells grown to mid-logarithmic phase in THB was quantitated by high-performance liquid chromatography (HPLC) as described previously (4) as a measure of the amount of capsule produced by isogenic GBS strains. Sialic acid recovered after hydrolysis for 1 h with 4 N acetic acid was derivatized with fluorescent 1,2-diamino-4,5-methylene-dioxybenzene (DMB) and quantified compared to a standard curve using pure sialic acid (Sigma) similarly derivatized with DMB. Samples were injected into a Bio-logic Duoflow HPLC (Bio-Rad Laboratories) equipped with a fluorescence detector (model RF-530; Shimadzu Scientific Instruments, Columbia, Md.) and an ASI 5-μm-particle-size C18 column (0.4 by 30 cm; Analytical Sciences, Incorporated, Santa Clara, Calif.). The samples were eluted isocratically at 0.9 ml/min, and the derivatized sialic acid was detected using an excitation wavelength of 373 nm, emission wavelength of 448 nm, and a range setting of 16.
Bactericidal assays.
Sensitivity to phagocytosis was assessed using standard Lancefield whole-blood bactericidal assays (17, 27). Mid-logarithmic phase cultures grown in THB were washed in phosphate-buffered saline and incubated with a sample of whole blood or plasma from a nonimmune donor. The amount of killing in whole blood was calculated for a 3-h incubation period at 37°C relative to the growth of each strain in plasma obtained from the same donor.
Animal infection studies.
Time-mated, barrier-sustained, female Sprague-Dawley rats delivered by cesarian section were obtained from Charles River Laboratories. Lethal dose assays using the neonatal rat sepsis model of GBS infection were performed as previously described (13). All procedures were performed in accordance with the Children's Hospital and Regional Medical Center Institutional Animal Care and Use Committee Guidelines.
Nucleotide sequence accession number.
The sequence of the GBS rpoE gene was deposited in GenBank under accession number AY216696.
RESULTS
Genotypic analysis of the rpoE region.
To initiate characterization of GBS AJ8D3, the rpoE STM mutant, chromosomal DNA flanking the left terminal repeat of Tn917stm was cloned to identify the gene disrupted by the transposon insertion as previously described (13). Initial sequence analysis indicated that the transposon was located immediately 5′ to the rpoE gene predicted to encode the delta subunit of RNAP. The transposon insertion mapped within the predicted promoter region of rpoE identified by sequence analysis and a World Wide Web-based Neural Network Promoter Prediction program (http://www.fruitfly.org/seq_tools/promoter.html). By sequence analysis, a ribosome-binding site was identified 5′ to rpoE, and a potential transcriptional terminator was identified 3′ to rpoE, suggesting that the gene was transcribed independently.
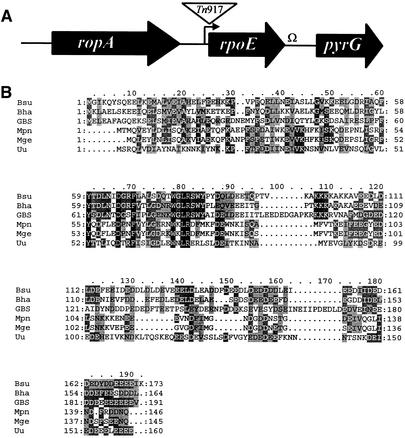
Genomic DNA and arbitrary-primed PCR products (26) were sequenced to further characterize the GBS chromosomal DNA adjacent to the transposon insertion site and to identify other open reading frames in this region. The rpoE chromosomal region is depicted in Fig. 1A. A ropA homolog predicted to encode trigger factor, a putative chaperone found in many gram-positive bacterial species, is located 5′ of rpoE. The pyrG gene, encoding CTP synthase, is located downstream of rpoE. The arrangement of these three genes in GBS is identical to that described for group A streptococci (20).
FIG. 1.
(A) Schematic representation of the GBS rpoE region. The relative positions of the GBS rpoE homolog and flanking genes are represented by arrows. The positions of the Tn917 insertion, putative promoters (arrow), and transcriptional terminators (Ω) are also indicated. (B) Amino acid sequence alignment of delta proteins using CLUSTAL W (11) and BoxShade. Residues conserved in all five proteins are shown on a black background, and residues identical in three or more of the proteins are shown on a dark gray background. Similar residues are shown on a light gray background. Gaps introduced to maximize alignment are indicated by periods in the Bsu to Uu sequences. Species abbreviations: Bsu, B. subtilis; Bha, Bacillus halodurans; Mpn, M. pneumoniae; Mge, M. genitalium; Uu, Ureaplasma urealyticum.
Predicted protein structure.
The GBS delta subunit is predicted to be a 191-amino-acid protein. The tool used to predict pI and molecular mass in the Expert Protein Analysis System (http://us.expasy.org/tools/pi_tool.html) predicts a molecular mass of 22 kDa and a pI of 3.7 for the GBS delta protein. The GBS delta subunit was aligned with those of other organisms using CLUSTAL W (11). As shown in Fig. 1B, the GBS peptide, including an acidic carboxy terminus preceded by a basic region, exhibits considerable homology with those of other organisms. The highest homology in the proteins is seen between residues 60 to 90, which is thought to be the core RNAP binding region. The acidic C-terminal domains of the delta protein are predicted to function as an unstructured polyanionic polymer that mimics single-stranded RNA (18). As expected, the carboxy-terminal amino acids of the GBS delta subunit are highly enriched for aspartate residues (n = 19) and glutamate residues (n = 26).
Construction of an allelic exchange mutant and expression analysis.
To confirm that the transposon insertion in the putative promoter region of rpoE was responsible for the attenuated virulence of the mutant, the rpoE gene in the wild-type strain A909 was deleted by allelic exchange as described in Materials and Methods. Southern blot analysis and PCR were performed to confirm that the rpoE gene had been replaced with the ΩKm cassette (data not shown).
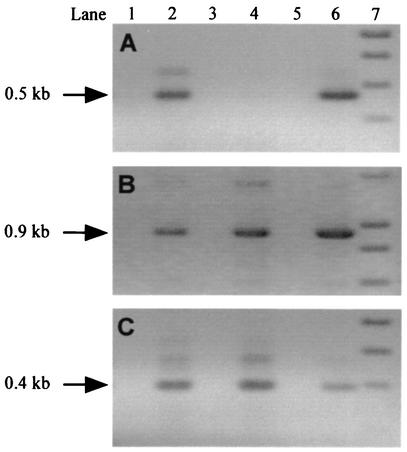
RT-PCR analysis of isogenic strains was performed to assess expression of genes in the rpoE region. As seen in Fig. 2A, rpoE was not transcribed in the allelic exchange mutant AJ200 but was transcribed in wild-type strain A909, as expected. Transcription of both ropA and pyrG in AJ200 was unaffected and was similar to that observed in A909 (Fig. 2B and C). These data confirm that allelic replacement of rpoE does not result in polar effects on transcription of adjacent genes. These observations were confirmed by RNA dot blot analysis (data not shown).
FIG. 2.
RT-PCR analysis of GBS strains A909 and AJ200. cDNA was PCR amplified with primers specific for rpoE (A), ropA (B), and pyrG (C). The position of the expected product is indicated with an arrow. Lanes 1 and 2, A909; lanes 3 and 4, AJ200; lane 5, no-template control; lane 6, DNA control; lane 7, DNA standards. Reverse transcriptase was omitted from samples in lanes 1 and 3, which serve as a control for DNA contamination in the RNA samples.
Virulence of allelic exchange mutants.
The virulence of strain AJ200 was compared to the virulence of strain A909 and the original transposon mutant, AJ8D3, in our neonatal rat sepsis model of GBS infection. Two separate lethal dose experiments were performed. As seen in Table 2, AJ200 displayed attenuated virulence similar to that of AJ8D3. Both mutants had 50% lethal doses (LD50s) 1.5 to 2 log units greater than the LD50 of A909. These data confirmed that the attenuated virulence observed for AJ8D3 and AJ200 was due to disruption of rpoE expression.
TABLE 2.
Lethal dose values for isogenic strains in a neonatal rat sepsis model of infection
| Strain | LD50a
|
|
|---|---|---|
| Expt 1 | Expt 2 | |
| A909 (wild type) | 2.1 × 105 | 3.9 × 105 |
| AJ200 (A909ΔrpoE) | 2.1 × 107 | 1.6 × 107 |
| AJ8D3 (A909 Tn917::rpoE) | 1.3 × 107 | 1.7 × 106 |
LD50 were calculated by the method of Reed and Muench (29). Each strain was tested on five pups per group.
Phenotypic characterization of allelic exchange mutants.
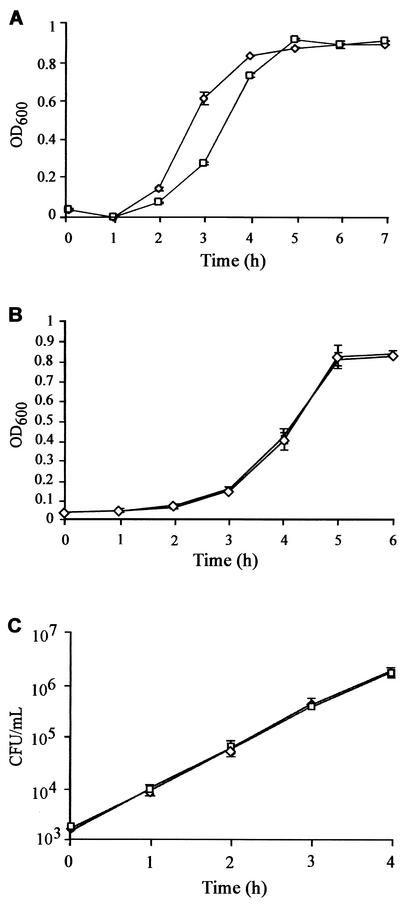
To ensure that the virulence defect observed for strain AJ200 was independent of growth and production of known virulence factors, we compared the phenotypes of strains AJ200 to A909. Significant growth defects causing attenuated virulence were ruled out by comparing the growth curve of AJ200 with that of parent strain A909. In rich medium (THB), AJ200 exhibited a 30- to 60-min delay in entering logarithmic-phase growth compared to A909; this delay was reproduced in experiments (Fig. 3A). Once AJ200 entered logarithmic-phase growth, it displayed a growth rate identical to that of the wild-type strain. Hence, it is unlikely that the extended lag phase in THB is responsible for the virulence defect observed for AJ200, as logarithmic-phase bacteria were used for all virulence assays. Additionally, the final culture density achieved by AJ200 was equivalent to that of A909. In CDM (37), the growth rates of the two strains were essentially identical (Fig. 3B). Each growth experiment was repeated multiple times, and identical results were obtained.
FIG. 3.
(A and B) Growth of isogenic strains in THB (A) and CDM (B). Growth was measured by optical density at 600 nm (OD600). (C) Viable cell counts (CFU ± standard deviation) of isogenic strains grown in nonimmune human plasma. Data were collected from cultures grown in triplicate. □, AJ200; ⋄, A909.
In an attempt to simulate growth in vivo, we compared the growth rates of the isogenic strains in human plasma from a nonimmune donor. As seen in Fig. 3C, the strains grew equally well in plasma, suggesting that a growth defect in plasma was not responsible for the attenuated virulence of AJ200. This experiment was repeated three times with plasma from independent donors.
The type-specific capsule of GBS has been extensively studied and is an important determinant for both virulence and protective immunity (for a review, see reference 24). Previous studies have shown that significant alterations in the amount of capsular polysaccharide affect GBS virulence (30, 36). We compared the amount of capsule on the surfaces of strains AJ200 and A909 by quantitating sialic acid recovered after acid hydrolysis of equivalent numbers of whole cells by HPLC. We were unable to detect significant differences in the amounts of capsular polysaccharide produced by the two strains: A909 produced 240 ± 111 μmol of sialic acid/g (dry wt), and AJ200 produced 173 ± 65 μmol of sialic acid/g (dry wt) (P > 0.05 by Student's t test). In contrast, we recovered 14 ± 6 μmol of sialic acid/g (dry wt) from the acapsular control strain A909ΔcpsE. Thus, we concluded that no changes in expression of capsule had occurred, at least when strains were grown and processed under the conditions that would account for the attenuated virulence of AJ200.
GBS express a number of other potential virulence factors that we examined. Production of hemolysin, CAMP factor, hyaluronidase, and DNase activity by strains AJ200 and A909 were determined by standard plate assays and compared, and no differences were observed.
Resistance to phagocytic killing.
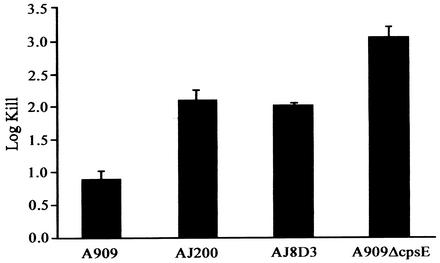
As the rpoE mutant was initially identified on the basis of attenuation in a sepsis model of GBS infection, the ability of the mutants to survive in blood from a nonimmune individual was compared with the wild-type strain A909. In this bactericidal assay, all streptococcal strains survive and grow in plasma, while only strains resistant to phagocytosis survive in whole human blood (17). Strain A909ΔcpsE was used as a positive control, as the absence of capsule renders GBS highly sensitive to killing in whole blood (22). In an immune host, the classical complement pathway promotes or mediates killing of GBS when serotype-specific anticapsule antibody is present (8). By determining survival of GBS in whole blood from a nonimmune individual, the contribution of the classical complement pathway is minimized (22). Hence, these bactericidal assays evaluated the contribution of the rpoE gene to resistance to alternate complement pathway-mediated killing. The log kill in whole blood was calculated relative to the growth of each strain in plasma obtained from the same donor (both log kill and growth were studied over the 3-h incubation period). All strains survived and grew in plasma, as expected. As seen in Fig. 4, AJ200 and AJ8D3 were significantly more sensitive to killing in whole blood than the parent strain (P = 0.007 for A909 versus AJ200 values and P = 0.001 A909 versus AJ8D3 values by Student's t test). The control strain A909ΔcpsE demonstrated a reduction in CFU of more than 3 log units during incubation in whole blood. These results provide evidence that disruption of rpoE expression affects the organism's ability to inhibit the alternate pathway of complement activation, rendering it susceptible to phagocytic killing and therefore less virulent.
FIG. 4.
Whole-blood bactericidal assay. Isogenic GBS strains were incubated in whole blood from a nonimmune donor for 3 h at 37°C. Results are presented as log kill ± standard deviation.
DISCUSSION
In gram-positive bacterial species, RNAP is found in association with an additional protein factor known as the delta subunit, encoded by the rpoE gene. On the basis of the results of in vitro assays, the delta protein is thought to enhance the specificity of transcription by a variety of different RNAP holoenzyme forms (1, 12, 16, 28). Despite the fact that rpoE is ubiquitous in gram-positive bacteria and the availability of rpoE mutants in Bacillus species and S. aureus, a physiological role for the delta protein has not been defined.
We identified the delta subunit as a potential virulence factor during screening for genes required for development of GBS-induced sepsis in neonatal rats. The data presented here confirm that the rpoE gene is required for virulence of GBS. Interestingly, rpoE mutants were also found to be sensitive to phagocytic killing in whole-blood bactericidal assays, a phenotype that may explain the attenuated virulence of rpoE mutants.
The delta subunit of RNAP is thought to play a role in maintaining transcriptional fidelity by enhancing promoter selectivity. This may occur by preferential destabilization of RNAP complexes at weak promoter sites and nonpromoter DNA. In the presence of delta protein, the close association between RNAP and promoter DNA is reduced at weak promoter sites, and RNAP is released (18). This ability may be explained by the unique structure of delta peptides, which has been best characterized in B. subtilis. The tertiary structure of B. subtilis delta protein consists of an amino-terminal half that is folded as an α/β structure and a carboxy-terminal half that has no tertiary structure in solution. The carboxy-terminal region is highly acidic and is essential for displacing RNA bound to RNAP. The results of structural and functional analyses of the delta protein suggest that the carboxy-terminal region is essentially an unstructured polyanion that mimics nucleic acid. This may account for how the delta protein displaces RNAP from nonpromoter and weak promoter sites (18). Lopez de Saro and coworkers (18) reported that B. subtilis delta protein acted like other polyanionic polymers (such as heparin, single-stranded DNA, and amino acid copolymers) and was able to bind to both the template and product-binding sites of RNAP. The delta subunit was able to displace RNA from preformed RNAP-RNA complexes, and efficient displacement required both the amino-terminal (core binding) domain and the polyanionic carboxy-terminal portion. Homologs of the delta protein are present in Mycoplasma genitalium, Mycoplasma pneumoniae, Enterococcus faecalis, and Streptococcus pyogenes. These homologs exhibit statistically significant similarity to the amino-terminal domain of delta protein and are typically associated with an acidic carboxy-terminal domain (18). Our recent database searches also found putative homologs in Streptococcus mutans, Bacillus anthracis, Streptococcus pneumoniae, and Bacillus stearothermophilus. Previous work has revealed functionally equivalent proteins in other Bacillus species (2), and a similarly sized protein was found to copurify with RNAP from S. aureus (7). On the basis of CLUSTAL W alignments (11), the GBS delta protein is predicted to be highly homologous to other described delta proteins.
The delta subunit of RNAP has no known counterpart in gram-negative bacterial species. The primary sigma factor in E. coli, σ70, has two properties: promoter recognition and inhibition of nonspecific complex formation. Since RNAP in gram-positive bacterial species contain a primary σ factor of ∼43 kDa (σA) and a delta subunit of ∼21 kDa, it has been speculated that together these two proteins may contain the activities found in the single 70-kDa σ subunit of E. coli (10, 18). Alignment of the B. subtilis σA and E. coli σ70 proteins reveals extensive homology with the exception of a 245-amino-acid insertion in σ70 (18). This insertion contains an 80-residue acidic region (predicted net charge of −30) with an amino acid composition similar to that of the carboxy terminus of the delta protein. This region may have functional similarity to the delta protein (15, 18).
Our studies revealed that the GBS rpoE deletion mutant displayed an extended lag phase in complex laboratory media, but not in CDM or plasma. These data suggest that a growth defect in plasma is not responsible for the attenuated virulence of the mutant. The significance of this growth defect in complex media is not clear at present, but this finding is in agreement with observations made for B. subtilis rpoE mutants. A B. subtilis rpoE mutant was reported to exhibit a reproducible extended lag phase. The mutant was otherwise phenotypically identical to the wild-type strain (19).
Disruption of rpoE expression rendered GBS more sensitive to phagocytic killing than the wild-type strain as measured by in vitro bactericidal assays. Mutants were otherwise identical to the parent strain, including production of wild-type levels of surface capsular polysaccharide, the major antiopsonin of GBS, which prevents phagocytosis by inhibiting the alternative complement pathway (22). These data suggest that rpoE affects the organism's sensitivity to phagocytic killing independent of the capsular polysaccharide.
While a role for rpoE in virulence has not been demonstrated prior to these studies, some insights into the function of rpoE may be gleaned from phenotypic characterization of rpoE mutants derived from other gram-positive bacterial species. Several groups have suggested that the delta subunit may be important for long-term survival or during growth phase transitions. Watson et al. (34) screened transposon insertion libraries for mutants defective in starvation-induced stationary-phase survival or recovery and identified a S. aureus rpoE mutant. Mutants were identified by using a screen based on their inability to remain culturable on CDM with limiting amounts of amino acids at 25°C for 21 days. This rpoE mutant exhibited a stationary-phase survival defect as well as an increased sensitivity to acid treatment. However, the mutant did not display a survival defect under phosphate or carbon starvation or display an increased sensitivity to oxidative stress (34). Expression analysis of an rpoE::lacZ transcriptional fusion revealed increased expression during mid-exponential phase, suggesting that the rpoE homolog plays a regulatory role in stationary phase (34).
GBS encounter dramatic environmental changes during the course of an infection, which we anticipate require changes in growth phase and gene expression in order to adapt and survive. Thus, we hypothesize that the delta peptide is required for GBS to persist in the host and adjust to environmental changes within various host compartments encountered during infection. Further studies are under way to investigate the effects of rpoE on gene expression in GBS.
Regulation of rpoE expression has not been well characterized. In B. subtilis, delta factor was found to be abundant in the cell during all stages of growth and was found at concentrations of approximately 104 copies/cell (19). Recently, Lopez de Saro et al. (19) reported that overexpression of rpoE from a plasmid did not alter patterns of gene expression in B. subtilis cells, but these studies were far from conclusive. The rpoE gene in B. subtilis is constitutively expressed from a single σA-dependent promoter, and no evidence of autoregulation was detected (19). Expression of rpoE in B. subtilis was demonstrated to be maximal at the transition between logarithmic and stationary phase (19).
Collectively, our studies demonstrate that the rpoE gene promotes resistance of GBS to phagocytic killing, a phenotype that may explain the attenuated virulence observed for rpoE mutants in animal models of infection. We anticipate that the effect of rpoE on sensitivity to phagocytosis is likely to be indirect through regulation of expression of other genes required for virulence. Transcriptional fidelity mediated by the delta subunit of RNAP may in fact be important for tight control of virulence gene expression in a host environment, allowing GBS to survive within various host compartments, such as blood.
Our findings are evidence for a novel role for the delta protein, a subunit of RNAP found ubiquitously in gram-positive bacteria. We have previously reported that like other streptococci, GBS use multiple mechanisms to evade phagocytosis and immune clearance (14). The addition of rpoE to the list of genes involved in the pathogenesis of GBS infection, perhaps by contributing to resistance to phagocytosis, emphasizes that evasion of immune clearance by GBS is a multifactorial process worthy of additional study.
Acknowledgments
This work was funded by The Streptococcal Initiative of the National Institutes of Health (grant BWH 811501; contract N01-AI-75326).
We thank Anne Clancy and Brandi Limbago for helpful discussions and critical review of the manuscript and Donald Chaffin for assistance with the HPLC analysis.
Editor: J. N. Weiser
REFERENCES
- 1.Achberger, E. C., M. D. Hilton, and H. R. Whiteley. 1982. The effect of the delta subunit on the interaction of Bacillus subtilis RNA polymerase with bases in a SP82 early gene promoter. Nucleic Acids Res. 10:2893-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Achberger, E. C., M. Tahara, and H. R. Whiteley. 1982. Interchangeability of delta subunits of RNA polymerase from different species of the genus Bacillus. J. Bacteriol. 150:977-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1997. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 5.Chaffin, D. O., S. B. Beres, H. Y. Yim, and C. E. Rubens. 2000. The serotype of type Ia and III group B streptococci is determined by the polymerase gene within the polycistronic capsule operon. J. Bacteriol. 182:4466-4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaffin, D. O., and C. E. Rubens. 1998. Blue/white screening of recombinant plasmids in Gram-positive bacteria by interruption of alkaline phosphatase gene (phoZ) expression. Gene 219:91-99. [DOI] [PubMed] [Google Scholar]
- 7.Deora, R., and R. Misra. 1995. Purification and characterization of DNA dependent RNA polymerase from Staphylococcus aureus. Biochem. Biophys. Res. Commun. 208:610-616. [DOI] [PubMed] [Google Scholar]
- 8.Edwards, M. S., A. Nicholson-Weller, C. J. Baker, and D. L. Kasper. 1980. The role of specific antibody in alternative complement pathway-mediated opsonophagocytosis of type III, group B Streptococcus. J. Exp. Med. 151:1275-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Framson, P., A. Nittayajarn, J. Merry, P. Youngman, and C. Rubens. 1997. New genetic techniques for group B streptococci: high-efficiency transformation, maintenance of temperature-sensitive pWV01 plasmids, and mutagenesis with Tn917. Appl. Environ. Microbiol. 63:3539-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Helmann, J. D., and M. J. Chamberlin. 1988. Structure and function of bacterial sigma factors. Annu. Rev. Biochem. 57:839-872. [DOI] [PubMed] [Google Scholar]
- 11.Higgins, D., J. Thompson, T. Gibson, J. D. Thompson, D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hyde, E. I., M. D. Hilton, and H. R. Whiteley. 1986. Interactions of Bacillus subtilis RNA polymerase with subunits determining the specificity of initiation. Sigma and delta peptides can bind simultaneously to core. J. Biol. Chem. 261:16565-16570. [PubMed] [Google Scholar]
- 13.Jones, A. L., K. M. Knoll, and C. E. Rubens. 2000. Identification of Streptococcus agalactiae virulence genes in the neonatal rat sepsis model using signature-tagged mutagenesis. Mol. Microbiol. 37:1444-1455. [DOI] [PubMed] [Google Scholar]
- 14.Jones, A. L., R. H. V. Needham, K. M. Knoll, and C. E. Rubens. 2003. Penicillin-binding proteins in Streptococcus agalactiae: a novel mechanism for evasion of immune clearance. Mol. Microbiol. 47:247-256. [DOI] [PubMed] [Google Scholar]
- 15.Karlin, S. 1993. Unusual charge configurations in transcription factors of the basic RNA polymerase II initiation complex. Proc. Natl. Acad. Sci. USA 90:5593-5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lampe, M., C. Binnie, R. Schmidt, and R. Losick. 1988. Cloned gene encoding the delta subunit of Bacillus subtilis RNA polymerase. Gene 67:13-19. [DOI] [PubMed] [Google Scholar]
- 17.Lancefield, R. C. 1957. Differentiation of group A streptococci with a common R antigen into three serological types with special reference to the bactericidal test. J. Exp. Med. 106:525-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez de Saro, F. J., A. Y. Woody, and J. D. Helmann. 1995. Structural analysis of the Bacillus subtilis delta factor: a protein polyanion which displaces RNA from RNA polymerase. J. Mol. Biol. 252:189-202. [DOI] [PubMed] [Google Scholar]
- 19.Lopez de Saro, F. J., N. Yoshikawa, and J. D. Helmann. 1999. Expression, abundance, and RNA polymerase binding properties of the delta factor of Bacillus subtilis. J. Biol. Chem. 274:15953-15958. [DOI] [PubMed] [Google Scholar]
- 20.Lyon, W. R., C. M. Gibson, and M. G. Caparon. 1998. A role for Trigger Factor and an Rgg-like regulator in the transcription, secretion and procession of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 17:6263-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madoff, L. C., J. L. Michel, and D. L. Kasper. 1991. A monoclonal antibody identifies a protective C-protein alpha-antigen epitope in group B streptococci. Infect. Immun. 59:204-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marques, M. B., D. L. Kasper, M. K. Pangburn, and M. R. Wessels. 1992. Prevention of C3 deposition by capsular polysaccharide is a virulence mechanism of type III group B streptococci. Infect. Immun. 60:3986-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murray, P. R., E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.). 1999. Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 24.Nizet, V., P. Ferrieri, and C. Rubens. 2000. Molecular pathogenesis of group B streptococcal disease in newborns, p. 180-221. In D. L. Stevens and E. L. Kaplan (ed.), Streptococcal infection: clinical aspects, microbiology and molecular pathogenesis. Oxford University Press, New York, N.Y.
- 25.Okada, N., R. T. Geist, and M. G. Caparon. 1993. Positive transcriptional control of mry regulates virulence in the group A streptococcus. Mol. Microbiol. 7:893-903. [DOI] [PubMed] [Google Scholar]
- 26.O'Toole, G., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple convergent signaling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 27.Perez-Casal, J., J. A. Price, E. Maguin, and J. R. Scott. 1993. An M protein with a single C repeat prevents phagocytosis of Streptococcus pyogenes: use of a temperature-sensitive shuttle vector to deliver homologous sequences to the chromosome of S. pyogenes. Mol. Microbiol. 8:809-819. [DOI] [PubMed] [Google Scholar]
- 28.Pero, J., J. Nelson, and T. D. Fox. 1975. Highly asymmetric transcription by RNA polymerase containing phage-SP01-induced polypeptides and a new host protein. Proc. Natl. Acad. Sci. USA 72:1589-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 30.Rubens, C. E., M. R. Wessels, L. M. Heggen, and D. L. Kasper. 1987. Transposon mutagenesis of type III group B streptococcus: correlation of capsule expression with virulence. Proc. Natl. Acad. Sci. USA 84:7208-7212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schuchat, A. 1998. Epidemiology of group B streptococcal disease in the United States: shifting paradigms. Clin. Microbiol. Rev. 11:497-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schuchat, A. 2001. Group B streptococcal disease: from trials and tribulations to triumph and trepidation. Clin. Infect. Dis. 33:751-756. [DOI] [PubMed] [Google Scholar]
- 33.Smith, R. F., and N. P. Willett. 1968. Rapid plate method for screening hyaluronidase and chondroitin sulfatase-producing microorganisms. Appl. Microbiol. 16:1434-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watson, S. P., M. Antonio, and S. J. Foster. 1998. Isolation and characterization of Staphylococcus aureus starvation-induced, stationary-phase mutants defective in survival or recovery. Microbiology 144:3159-3169. [DOI] [PubMed] [Google Scholar]
- 35.Wertman, K. F., A. R. Wyman, and D. Botstein. 1986. Host/vector interactions which affect the viability of recombinant phage lambda clones. Gene 49:253-262. [DOI] [PubMed] [Google Scholar]
- 36.Wessels, M. R., C. E. Rubens, V. J. Benedi, and D. L. Kasper. 1989. Definition of a bacterial virulence factor: sialylation of the group B streptococcal capsule. Proc. Natl. Acad. Sci. USA 86:8983-8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Willet, N. P., and G. E. Morse. 1966. Long-chain fatty acid inhibition of growth of Streptococcus agalactiae in a chemically defined medium. J. Bacteriol. 91:2245-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yim, H. H., and C. E. Rubens. 1998. Site-specific homologous recombination mutagenesis in group B streptococci. Methods Cell Sci. 20:13-20. [Google Scholar]
- 39.Yim, H. H., and C. E. Rubens. 1997. Use of a dental amalgamator to extract RNA from the gram-positive bacterium Streptococcus agalactiae. BioTechniques 23:229-231. [DOI] [PubMed] [Google Scholar]