Abstract
The genome of Borrelia burgdorferi, the etiologic agent of Lyme disease, is composed of a linear chromosome and more than 20 linear and circular plasmids. Typically, plasmid content analysis has been carried out by pulsed-field gel electrophoresis and confirmed by Southern hybridization. However, multiple plasmids of virtually identical sizes (e.g., lp28 and cp32) complicate the interpretation of such data. The present study was undertaken to investigate the complete plasmid complements of B. burgdorferi clinical isolates cultivated from patients from a single region where early Lyme disease is endemic. A total of 21 isolates obtained from the skin biopsy or blood samples of Lyme disease patients were examined for their complete plasmid complements by Southern hybridization and plasmid-specific PCR analysis. All clinical isolates harbored at least six of the nine previously characterized cp32s. Fourteen isolates harbored all B31-like linear plasmids, and seven isolates simultaneously lacked lp56, lp38, and some segments of lp28-1. The distinctive plasmid profile observed in these seven isolates was specific to organisms that had ribosomal spacer type 2 and pulsed-field gel type A, which implies a clonal origin for this genotype. The presence of nearly identical complements of multiple linear and circular plasmids in all of the human isolates suggests that these plasmids may be particularly necessary for infection, adaptation, and/or maintenance in the infected host.
Lyme disease is the most prevalent vector-borne infectious disease in the United States (39). The spirochete Borrelia burgdorferi is the etiologic agent of the disease. B. burgdorferi sensu lato consists of 10 distinct species; only B. burgdorferi sensu stricto, Borrelia afzelii, Borrelia garinii, and Borrelia bissettii have been found to be pathogenic to humans (64). B. burgdorferi is the only pathogenic species isolated from humans in North America. The genetic heterogeneity of B. burgdorferi has been established by analysis of varied molecular targets. North American B. burgdorferi isolates from ticks, rodents, and humans are heterogeneous with regard to their macrorestriction chromosomal restriction fragment length polymorphism (RFLP) patterns (32), sequences of various coding and noncoding regions (24, 28, 30, 65), plasmid contents (8, 9, 12, 13, 27, 44, 68, 69), and protein profiles (20, 21, 41). Earlier studies employing three different typing methods with a large number of clinical isolates cultured from erythema migrans lesions or from the blood of Lyme disease patients in Westchester County, N.Y., revealed substantial genetic diversity of B. burgdorferi in this single region of endemicity (23, 28-30).
The genome sequence of B. burgdorferi B31, the B. burgdorferi sensu stricto type strain, has been elucidated (16). The genome is unusual in that it includes a linear chromosome 910 kb in size and numerous linear and circular plasmids totaling an additional 610 kb (12, 16). The linear plasmids are present at a copy number of approximately one per chromosome (16, 19). Linear plasmid content analysis of tick and wildlife isolates showed overall uniformity (11, 40). Long-term culture of B. burgdorferi results in the loss of some plasmids and has been associated with the inability of the spirochete to infect laboratory animals (33, 37, 46, 69), suggesting that the plasmids encode important proteins involved in virulence (13, 48, 50, 53, 71).
Extensive plasmid content analyses have been reported for several North American tick isolates (strains B31 and N40), and the results were compared to those of European isolates (B. afzelii and B. garinii) (12, 40). Studies of the plasmid contents of isolates from Lyme disease patients are limited; only human cerebrospinal fluid (CSF) strain 297 has been well characterized (2, 9). The present study was undertaken to elucidate the complete plasmid complements for clinical isolates of B. burgdorferi sensu stricto from a primary site (erythema migrans lesions) and a disseminated site (blood) of patients with early Lyme disease by using a combination of plasmid-specific PCR and Southern hybridization.
MATERIALS AND METHODS
Strain source and cultivation.
B. burgdorferi isolates were obtained either from skin biopsy samples (diameter, 2 mm) from primary erythema migrans lesions or from the blood of patients presenting at the Lyme Disease Diagnostic Center of the Westchester Medical Center (Valhalla, N.Y.). Both skin biopsy and blood samples were obtained on the same day for cultivation in Barbour-Stoenner-Kelly II medium as described previously (36, 47, 66, 67). Clinical isolates were numbered with a prefix designating tissue source, namely, B for skin biopsy sample and BL for blood. Low-passage-number cultures (those with fewer than five passages) were grown in Barbour-Stoenner-Kelly H medium supplemented with 6% rabbit serum (Sigma Chemical Co., St. Louis, Mo.) for 2 to 4 weeks at 33°C.
DNA isolation and PCR amplification.
DNA was isolated from 10 ml of B. burgdorferi culture (5 × 107 to 1 × 108 cells/ml) by using a commercial nucleic acid extraction kit (IsoQuick; Orca Research, Bothell, Wash.). Isolated DNA was resuspended in 100 μl of nuclease-free water. PCR primers specific for each circular and linear plasmid have been described previously (44, 52) and are listed in Table 1.
TABLE 1.
Oligonucleotides used for plasmid content analysis
| Plasmid | Primer sequences, 5′ to 3′ (position)a | Predicted fragment size (bp) | Strain B31 source, genome designation, or reference |
|---|---|---|---|
| lp56 | TTTCTTAAGCTGAAATCTTAGGGG(3586) | 380 | From cloneb |
| TTACCAAAGAGGAGATATTTGCTC(3109) | |||
| lp56 | ACTATTAAGACGAGCAATAAAAAGTCCA(42929) | 596 | BBQ67 |
| GACGAAGCAAAGAAGGATTTGGATCACCC(43524) | |||
| lp54 | GGGAATAGGTCTAATATTAGCC(9410) | 770 | BBA15 |
| TTTCAACTGCTGACCCCTC(10160) | |||
| lp38 | GGTCTCAATATCTTGTGTTCATG(6134) | 720 | BBJ09 |
| CAAGGCCACAACTTCTTGTAC(6831) | |||
| lp36 | TTCTTATCCCTGACTTTCACTTTCACTTTTGAGG(7247) | 381 | BBK11-K12 |
| TCCTTTACTTCTATGTTTTTACTTTCCTTGGT(7627) | |||
| lp28-1 | CGGGGATCCAGCCAAGTTGCTGATAAGGACGACCC | 263 | 44 |
| ACGGCAGTTCCAACAGAACCTGTACTATCT | |||
| AGTAGTACGACGGGGAAACCA(370) | 670 | 70 | |
| ACTTTGCGAACTGCAGAC(1040) | |||
| lp28-1 | TTCTGATGGCACTGAGCAAACCA(1309) | 150 | BBF01 |
| AACCCTTTACACTTTCTTCGATTGCGCT(1457) | |||
| lp28-2 | CCCTCATCAAGTTTTTCCATGTGTTTTT(13824) | 453 | BBG17 |
| AGGTGGCCTTTCCGAGCTTGTACCTTAC(14277) | |||
| lp28-3 | TAAACAAAAAAGGCCAGCGAAGCATCAA(11432) | 395 | BBH16-H17 |
| GCCCCCATGCAGAAAGAAACCCTATAAC(11826) | |||
| lp28-4 | TCACCTCAGCTAATCTATTTATCGACAC(9005) | 215 | BBI17-I18 |
| AAGCGCGGAGTTTTCGGCTG(9219) | |||
| lp25 | AGAATTATGTCGGTGGCGTTGT(14663) | 260 | BBE21-E22 |
| ATTAAAGCCGCCTTTTCCTTGGT(14899) | |||
| lp17 | ACTGCAATCTGCCCAAGCTACATAATCT(7657) | 283 | BBD12 |
| AAGGTAAGGACGGTTGTCTACATGGATT(7939) | |||
| cp26 | GGGAAAGATGGGAATACATCTGC(16971) | 524 | BBB19 |
| GCTTACAAGCCCTGTTGTGGCAG(17495) | |||
| cp32-1 | ACGATAGGGTAATATCAAAAAAGG(21583) | 471 | BBP33 |
| AGTTCATCTAATAAAAATCCCGTG(22054) | |||
| cp32-2/7 | GGAATGTATTAATTGATAATTCAG(20986) | 370 | BBO32 |
| GCGAAATAAATAGTGCCTTATGGG(21356) | |||
| cp32-3 | TTACGAAAAAATAGAAAAACTAGG(21412) | 328 | BBS35 |
| TTTCCACTGCCCACTTTTCAGCCG(21740) | |||
| cp32-4 | AGATCCTCAAAATAGTTTAACCAG(20737) | 373 | BBR33 |
| TTAATATTGGCAGAGAGTCTACAG(21110) | |||
| cp32-5 | ACTGATAATGATGTTATGGTTAGG(3221) | 365 | From clonec |
| TTTCTTAAGCTGAAATCTTAGGGG(3586) | |||
| cp32-6 | GACTTTACATAGTATAAATGCTTTTGG(21008) | 273 | BBM32 |
| TCTCGTTATTATAAAATAAGTAGG(21281) | |||
| cp32-8 | GAAGATTTAAACAAAAAAATTGCG(21376) | 235 | BBL32-L33 |
| GTAATCACTTCTTTTTTACCATCG(21610) | |||
| cp32-9 | TATCAAAAAAGTGCTGTTTTATAG(21473) | 248 | BBN32-N33 |
| TAATCTCAAATATTCTTCTTTATG(21721) | |||
| cp32-11 | ATTTAAGCTTACATATGCTTAACG(286) | 595 | From cloned |
| CGTTGTCCTTTTCTTCCAAATTTC(881) |
For each primer pair, the top sequence is the forward primer and the bottom sequence is the reverse primer. Numbers in parentheses indicate the position of the 5′ nucleotide of each primer in the relevant plasmid sequence.
GenBank accession number X87201.
GenBank accession number X87202.
GenBank accession number AY090888.
Amplification of nine linear plasmids and cp26 was accomplished by multiplex PCRs in three separate reaction mixtures. Each PCR mixture contained 10 ng of DNA, 250 μM deoxynucleoside triphosphate, 10 ng of each primer, and 1.25 U of Taq DNA polymerase (Roche Molecular Biochemicals, Indianapolis, Ind.) in 10 mM Tris-HCl-1.5 mM MgCl2-50 mM KCl, pH 8.3. For primer set A, three amplification cycles of 94°C for 30 s, 48°C for 30 s, and 72°C for 30 s were carried out with primers specific for lp38, cp26, and lp28-3. Primers for lp17 and lp28-4 were then added, and amplification was continued with the same cycling parameters for an additional 35 cycles. Amplification for lp54, lp28-2, and lp25 (primer set B) and for lp36 and lp28-1 (primer set C) involved 35 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 30 s.
For cp32 content analysis, the orfC-orf3 region (PF-32-PF-49, plasmid maintenance locus) of each cp32 and lp56 was amplified separately by using 10 ng of DNA and 10 ng of each primer (Table 1), as described above. The PCR conditions were 95°C for 2 min followed by 35 cycles of 94°C for 30 s, 48°C for 30 s, 72°C for 30 s, and a final extension at 72°C for 2 min.
Restriction enzyme digestion and Southern hybridization.
One microgram of total genomic DNA obtained from B. burgdorferi clinical isolates was digested for 2 h at 37°C with 10 U of EcoRI in 50 mM Tris-HCl-100 mM NaCl-10 mM MgCl2-1 mM dithioerythritol, pH 7.5. Digested genomic DNA was electrophoresed on a 0.8% agarose gel in 1× Tris-borate-EDTA buffer, stained with ethidium bromide, denatured in 0.4 N NaOH-1.5 M NaCl for 30 min, and neutralized with 1.0 M Tris-HCl-1.5 M NaCl (pH 8.0) for 30 min. The DNA was transferred to positively charged nylon membranes (Roche Molecular Biochemicals) by using 10× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.4]) as a transfer buffer. After transfer, DNA was cross-linked to the membrane by UV irradiation for 20 s and dried at room temperature. Membranes were hybridized overnight at 55°C with digoxigenin-labeled lp56, lp28-1, or cp32 PCR products. Membranes were washed with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate (SDS) followed by 0.5× SSC-0.1% SDS at 55°C and treated with blocking reagent containing 2% milk in 0.1 M Tris-HCl-0.15 NaCl, pH 7.5, for 1 h at room temperature. The membranes were incubated in a 1:10,000 dilution of digoxigenin-labeled anti-alkaline phosphatase antibody and washed twice in 0.1 M Tris-HCl-0.15 NaCl, pH 7.5. The blots were then washed in 100 mM Tris-HCl-100 mM NaCl-50 mM MgCl2, pH 9.5, for 15 min. The chemiluminescent substrate CDP-Star (1:100 dilution) was added, and the membranes were exposed to X-ray film for 2 to 5 min to visualize the hybridization pattern. One microgram of purified PCR product for the BBQ67 (lp56), BBF01 (lp28-1), BBF32 (lp28-1), or cp32 gene was labeled with digoxigenin-11-dUTP using a random prime labeling method as described by the manufacturer (Roche Molecular Biochemicals). Twenty microliters of digoxigenin-labeled DNA diluted in 10 ml of 5× SSC-0.1% lauryl sarcosine-0.02% SDS-1% milk powder was used as a probe for Southern analysis.
Nucleotide sequencing.
PCR products for lp28-1 (vlsE region, BBF32) and lp56 (BBQ67 region) from representative isolates were sequenced commercially (Davis Sequencing, Davis, Calif.). Sequences obtained in this manner were compared to those in GenBank by means of the BLAST homology search algorithm from the National Center for Biotechnology Information (available at http://www.ncbi.nlm.nih.gov).
RESULTS
Plasmid content of B. burgdorferi sensu stricto clinical isolates.
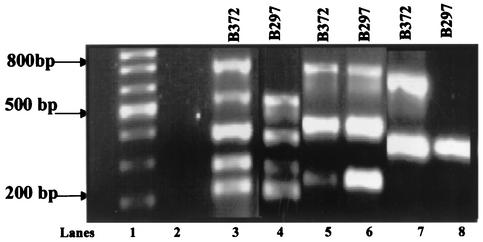
We have previously differentiated B. burgdorferi clinical isolates into three genotypes based on ribosomal DNA spacer sequence type (RST; RST1 through RST3) and demonstrated a significant correlation between RST and both the MluI restriction site polymorphism of chromosomal DNA (pulsed-field gel [PFG] types A through F) and the presence of the linear plasmid lp38 (23, 28). Isolates for the present study were chosen to represent all three RSTs: eight isolates were RST1, seven isolates were RST2, and six isolates were RST3. The total plasmid content for these 21 clinical isolates (16 from the skin and 5 from the blood) was determined by a combination of linear-plasmid-specific multiplex PCR, cp32 plasmid-specific PCR, and Southern hybridization. For multiplex PCR, 10 plasmid-specific primers were combined into one of three different sets. Primer set A contained primers for specific amplification of lp38, lp28-3, lp28-4, lp17, and cp26; primer set B contained primers for lp54, lp28-2, and lp25; and primer set C contained primers for lp36 and lp28-1. Results for the simultaneous detection of these 10 plasmids for two representative isolates by using the above-named primer sets are presented in Fig. 1. Isolate B372 (RST1) yielded PCR products for all 10 plasmids (Fig. 1, lanes 3, 5, and 7), whereas isolate B297 (RST2) lacked an lp38-derived PCR product (Fig. 1, lane 4), as well as a PCR product for lp28-1 (Fig. 1, lane 8). Based on these assays, the presence or absence of all known B31-MI linear plasmids (except lp21 and lp5) and cp26 was determined for these 21 clinical isolates (Table 2).
FIG. 1.
Simultaneous detection of 10 plasmids of B. burgdorferi by multiplex PCR. PCR amplification by using genomic DNA from strain B372 or B297 as a template was performed as detailed in Materials and Methods with the primers listed in Table 1. Products obtained using primer sets A, B, and C are shown in lanes 3 and 4, 5 and 6, and 7 and 8, respectively. Lanes 1 and 2 contain DNA molecular size markers and a control lacking DNA, respectively.
TABLE 2.
Plasmid characteristics and contents of B. burgdorferi clinical isolates
| Isolate(s)a | RFLP RSTb | MluI restriction type (PFG type)b | Plasmid(s) absentc |
|---|---|---|---|
| B349, BL203 | 1 | B | None |
| B352 | 1 | B | None |
| BL206 | 1 | B | cp32-8 |
| B363 | 1 | B | None |
| B372, BL219 | 1 | D | None |
| B373 | 1 | D | cp32-5 |
| B149 | 2 | A | lp56, lp38, segment of lp28-1 |
| B265, BL162 | 2 | A | lp56, lp38, segment of lp28-1 |
| B297 | 2 | A | lp56, lp38, segment of lp28-1 |
| B379, BL224 | 2 | A | lp56, lp38, segment of lp28-1 |
| B376 | 2 | F | lp56 |
| B161 | 3 | A | lp38 |
| B327 | 3 | F | cp32-3 |
| B331 | 3 | C | lp56 |
| B333 | 3 | C | None |
| B348 | 3 | E | Segment of lp28-1 |
| B356 | 3 | E | cp32-1, cp32-8 |
Isolates listed together on a row and isolates B352 and BL206 were obtained from the skin biopsy (B prefix) and blood (BL) samples of the same patient on the same day.
RFLP RSTs and PFG types are as defined in Iyer et al. (23).
“None” indicates that these isolates harbored all 20 plasmids listed in Table 1.
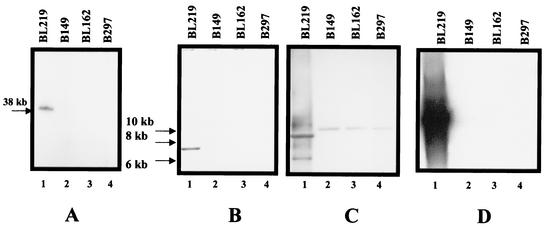
Six of the seven RST2 isolates lacked lp38 and some portion of lp28-1 (Table 2). A negative result on plasmid content analysis by PCR does not rule out the possibility that the plasmid is present, because PCR failure may be due to sequence heterogeneity at the priming sites. To confirm the PCR-based results, representative RST2 isolates were also analyzed by Southern hybridization of total genomic DNA separated by PFG electrophoresis (Fig. 2A). The results confirm that RST2 isolates lack lp38, which is in agreement with previous reports (23).
FIG. 2.
Plasmid analysis for four representative isolates by Southern hybridization. (A) PFG electrophoresis blot hybridized with an lp38-specific probe (ospD); (B) blot hybridized with an lp28-1-specific probe (bbf01); (C) blot in panel B stripped and hybridized with a second lp28-1-specific probe (vlsE); (D) blot hybridized with an lp56-specific probe (bbq67). Lanes 1 through 4 contain undigested genomic DNA (A) or EcoRI-digested genomic DNA (B through D) from isolates BL219, B149, BL162, and B297, respectively.
Plasmid lp28-1.
Two primer sets for detection of lp28-1 were employed, one targeted at vlsE and the other at a conserved portion of the vls cassette region (Table 1) (44, 70). All RST1 isolates (eight of eight) and five of six RST3 isolates yielded PCR products of the predicted size (660 or 260 bp). By contrast, PCR amplification of six of seven RST2 isolates and a single RST3 isolate (with primer set AGTAGTACGACGGGGAAACCA and CGGGGATCCAGCCAAGTTGCTGATAAGGACGACCC) produced a 400-bp product (data not shown). DNA sequencing of the latter amplicon revealed 47% identity (176 and 376 nucleotides) to a region in lp28 that corresponds to the vls cassette region (nucleotides 25490 to 25871 in the strain B31-MI lp28-1 sequence). Southern hybridization of EcoRI-digested genomic DNA using a vlsE probe detected vls-like sequences in all isolates examined (Fig. 2C), which is consistent with an earlier report for a different set of clinical isolates (22).
BBF01 was chosen as a third target for PCR analysis of lp28-1. The same isolates which yielded the 400-bp PCR product on vlsE amplification (six of seven RST2 isolates and one RST3 isolate) produced no product with the BBF01 primers. Southern hybridization confirmed that this BBF01 region is absent in these isolates (Fig. 2B). The remaining 14 isolates yielded the expected PCR product and positive hybridization with the BBF01 probe (e.g., isolate BL219) (Fig. 2B).
cp32 plasmid content.
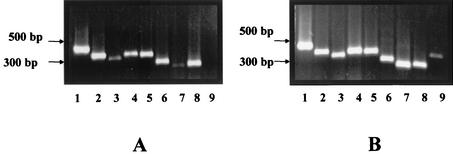
The plasmid maintenance and partition protein region (orfC-orf3) was selected to discriminate among the different cp32s since this region is one of the three hypervariable regions on these plasmids. Primers specific for each cp32 (and for lp56, which contains a complete cp32 sequence) were employed for PCR. Figure 3 displays representative results. Isolate B297 (RST2) lacked only the lp56 PCR product, whereas isolate B372 (RST1) yielded products corresponding to all cp32s and to lp56. A single PCR product of a predicted size (based on the B31-MI genomic sequence) was obtained for all isolates analyzed. The results for all 21 clinical isolates (Table 2) indicated that all cp32s were present in 81% of the isolates studied and that cp32-2/7, -4, -6, -9, and -11 were universally present in the B. burgdorferi sensu stricto isolates examined. One isolate lacked both cp32-1 and cp32-8, one lacked cp32-8, one lacked cp32-3, and another lacked cp32-5.
FIG. 3.
Detection of multiple cp32 plasmids by PCR amplification of the orfC-orf3 locus. PCR analysis was performed for each cp32 as described in Materials and Methods by using the primers shown in Table 1. (A) Isolate B297; (B) isolate B372. In each panel, the lanes contain products obtained with primers specific for cp32-1 (lane 1), cp32-2/7 (lane 2), cp32-3 (lane 3), cp32-4 (lane 4), cp32-5 (lane 5), cp32-6 (lane 6), cp32-8 (lane 7), cp32-9 (lane 8), and lp56 (lane 9). Migration positions for DNA molecular size markers are indicated to the left of each panel.
To ensure the accuracy of PCR analysis, Southern hybridization of EcoRI-digested genomic DNA for a set of eight isolates was performed by using orfC-orf3 gene probes specific for each of the nine cp32s (data not shown). In all cases, the data obtained by PCR and Southern hybridization were compatible, indicating that the cp32 content as determined by PCR is accurate. Some differences in the restriction patterns were noted among the isolates for a given cp32 probe, suggesting sequence heterogeneity among these plasmids. Although the probes used in hybridization experiments were obtained from the B31-MI DNA template, the positive hybridization results for all isolates indicated that all B. burgdorferi isolates contained multiple cp32s and that these cp32s are related to those of strain B31 at their partition loci.
Plasmid lp56.
Linear plasmid lp56 contains within it an essentially complete copy of cp32 (12). Two regions were chosen for PCR detection of lp56, one in the putative cp32 region (BBQ40-Q41) and one in the non-cp32 portion of the plasmid (BBQ67). Seven isolates (six of RST2 and one of RST3) yielded no PCR product for the BBQ40-Q41 region. For these seven isolates, amplification with BBQ67 primers resulted in a product of approximately 200 bp rather than the expected 596 bp. This 200-bp product was sequenced and found to be homologous (83% identity) to members of paralogous gene family 1 that are present in lp36, lp25, and lp28-3, but it had no homology to any sequences in lp56. Southern blotting of genomic DNA from these isolates with a BBQ67 probe (generated from B31-MI genomic DNA) did not result in any hybridization (Fig. 2D). Thus, these isolates lacked the evidence of either the linear plasmid portion of lp56 or the integrated cp32. In contrast, representative RST1 and RST3 isolates yielded positive hybridization signals. Taken together, these results indicate that lp56 is missing from these isolates (all RST2 isolates except B376 and RST3 isolate B331).
For one RST2 isolate, B376, a positive PCR product was obtained with both the BBQ40-Q41 and BBQ67 primer sets. However, sequencing of the BBQ67-derived PCR product revealed it to be closely related to paralogous family 156 sequences rather than to the lp56-specific BBQ67 sequence. With a BBQ67 probe generated from B31-MI genomic DNA, no hybridization was detected for B376 genomic DNA, although positive hybridization was observed for representative RST1 isolates. Based on these findings, we conclude that isolate B376 either lacks lp56 or contains an lp56 whose sequence differs substantially from that of B31 MI.
Comparison of skin and blood isolates from the same patient.
Of the 21 isolates examined for plasmid content, 10 were paired isolates (from skin and blood samples) obtained from the same patients at the same time. These are indicated in Table 2. Identical plasmid profiles were obtained for four sets of paired isolates (two RST1 and two RST2). For the remaining isolate pair, the blood isolate (BL206) appears to have lost cp32-8. This isolate is infectious and spreads among mice (61, 62), so cp32-8 is apparently dispensable for infectivity and hematogenous dissemination.
DISCUSSION
Elucidation of the genome sequence of B. burgdorferi strain B31-MI revealed the presence of 12 linear and 9 circular plasmids (12, 16). A number of previous studies have examined plasmid contents in various isolates of B. burgdorferi (27, 33, 40, 44, 49, 69). However, no comprehensive analysis of the complete plasmid complements of clinical isolates from Lyme disease patients has been reported. In the present investigation, the available sequence information for strain B31-MI (RST1, PFG type B) (30, 32) was employed to design probes and primers for analysis of the plasmid complements of 21 isolates from human patients. The analysis involved all the known B31 plasmids except for lp5, lp21, and cp9. All 21 isolates in the present study were examined for the presence of cp9 by amplification of the rev gene (BBC10); no amplification was obtained with any of the RST3 isolates. Stewart et al. (57) reported that the sequence of cp9 of strain N40 (also RST3) varies from that of strain B31 and lacks some of the genes present in the corresponding B31 plasmid. Indeed, strain B31 can carry at least two different 9-kb circular plasmids, with the plasmid designated as cp9 in the B31-MI genome sequence now being designated cp9-1 and a second cp9 being designated cp9-2 (34). The sequences of B31 cp9-1, B31 cp9-2, and N40 cp9 are divergent from each other. For these reasons, cp9 was excluded from this study.
The data presented in Table 2 demonstrate that numerous circular and linear plasmids are nearly ubiquitous among B. burgdorferi clinical isolates from Lyme disease patients. Twelve of the 19 plasmids examined were uniformly present in all isolates. In addition, 14 of 21 isolates either harbored all the plasmids or lacked only a single plasmid. The nearly universal presence of most plasmids in all clinical isolates suggests that these plasmids may be necessary for infection and/or maintenance of B. burgdorferi in humans or adaptation to diverse environments during the infectious cycle. Only three plasmids—lp38, lp56, and segments of lp28-1—were absent from multiple isolates. The absence of all three linear plasmids simultaneously was specific to isolates that shared an identical ribosomal spacer type (RST2) and chromosome macrorestriction type (PFG type A), strongly suggesting a clonal origin for this group of isolates.
Palmer et al. reported the distribution of 12 linear plasmids in 15 B. burgdorferi isolates from geographically diverse sites in the United States (40). Only one of these isolates was of human origin, the remainder having been isolated from either ticks or wildlife hosts. They found that most plasmids were present in all isolates tested, the notable exceptions being lp28-1, lp38, and lp56, which were present in 10, 10, and 3 isolates of 15, respectively. Thus, despite the differences in isolate source between the two studies, the overall results are comparable. Some differences between the findings of the two studies are also noteworthy. lp17 was absent in 5 of 15 isolates analyzed by Palmer et al. but was present in all isolates in the present study. In addition, lp5 and lp21 were present in only three and two isolates, respectively, in the earlier study (40). These plasmids were not tested in the present investigation; however, the absence of these plasmids from most isolates in the previous analysis suggests that they are dispensable for maintenance in either ticks or humans.
Southern blot analysis of pulsed-field-gel-separated genomic DNA employing probes specific for plasmid lp54 (ospA), lp38 (ospD), and cp26 (ospC) indicated that they hybridize to plasmids of the same size in all isolates tested. This finding demonstrates the constancy of the plasmid structure in these isolates. By contrast, the structure of lp28-1 varied among the isolates. The precise nature of the alteration in lp28-1 is not clear. Although all isolates yielded positive hybridization with a vlsE probe, the sizes of the hybridized bands varied, as reported previously for a different set of clinical isolates (22). vls-like sequences may be carried on other plasmids in these isolates. Alternatively, the structure of lp28-1 may be altered in some of these isolates, e.g., by truncation. This possibility is discussed further below.
Substantial evidence linking plasmid content and pathogenesis of B. burgdorferi has been reported. Repeated culture passage results in loss of plasmids and reduced infectivity (6, 46), and Xu et al. (69) found a correlation between the loss of plasmids of 7.6 kb (probably cp9), 22 kb, and 24.7 kb (matching the restriction pattern of lp25) and a reduced infectivity in mice or hamsters. More recently, plasmids lp25 and lp28-1 were shown to be necessary for a high-infectivity phenotype in clonal isolates of B31-MI (27, 44). However, clonal isolates retaining these two plasmids may also demonstrate reduced infectivity in mice (33), suggesting that factors other than the absence of specific linear plasmids likely contribute to loss of infectivity. The virulence of a number of the clinical isolates analyzed here was tested previously in C3H/HeJ mice. Inoculation with isolate BL203, BL206, or B348 resulted in disseminated infection, high spirochete loads in tissues, and moderate to severe carditis and arthritis. By contrast, inoculation with isolates B331 and B356 did not result in any disseminated infection (62). It is noteworthy that isolate B331 lacks only lp56 and isolate B356 lacks cp32-1 and cp32-8, supporting the view that the presence of lp25 and lp28-1 is not sufficient to confer a highly pathogenic dissemination phenotype. In addition, isolate B348 does not contain a B31-like lp28-1 (Table 2) but does retain pathogenicity in mice. Taken together, these findings underscore the role of factors other than plasmid presence or absence in B. burgdorferi pathogenicity.
The most extensive set of multiple, highly similar sequences in the B. burgdorferi genome are the cp32 plasmids (2, 10, 56). These plasmids possess up to 45 open reading frames, virtually all of which have paralogues in each cp32 (12). Despite this substantial sequence homology, each plasmid contains three relatively variable regions: (i) a plasmid partition and maintenance locus, (ii) the erp locus, and (iii) a bdr/mlp locus (56). In addition, strain B31-MI harbors a linear plasmid, lp56, that contains an intact copy of a cp32-like plasmid (12). cp32-like plasmid content has been elucidated for only a small number of B. burgdorferi sensu stricto isolates (56). The overall cp32 contents and gene organizations are similar in the tick isolates B31 and N40 and in the human CSF isolate 297 (1, 2, 9, 15; B. Stevenson and J. C. Miller, unpublished results). Both isolate N40 and CSF isolate 297 harbor truncated plasmids of approximately 18 kb, referred to as cp18 (9, 53). However, regardless of the isolate, all cp32-like plasmids have a unique plasmid partition and maintenance (orfC-orf3) locus (14, 52, Stevenson and Miller, unpublished). We therefore utilized this region to examine the plasmid contents of clinical isolates.
Most isolates analyzed in this study carried a full complement of B31-like cp32 plasmids (Table 2). Only one isolate (B356) lacked more than one cp32. Thus, the presence of all the cp32 plasmids appears to be a common property of B. burgdorferi sensu stricto rather than a trait peculiar to strain B31. Furthermore, these plasmids are not readily lost upon moving from a natural environment (either reservoir host or tick) to humans. It should be noted that our analysis could reveal only the presence or absence of B31-like cp32s but that the plasmid size was not determined. Therefore, the presence of truncated cp32-like plasmids similar to those observed in strains 297 and N40 (9, 53) cannot be ruled out.
The fact that these highly homologous plasmids are maintained despite extensive sequence identity among many paralogous open reading frames carried on each plasmid suggests that at least some proteins encoded by each plasmid perform functions crucial to spirochete maintenance, growth, transmission, or infectivity. Differential levels of expression for a number of paralogous gene families carried by cp32 have been demonstrated. In strains 297 and B31, some members of the mlp family (paralogous family 113) were not expressed at 23°C, were expressed somewhat at 35°C, and were expressed more substantially at 37°C (2, 42). Studies of multiple borrelial strains have shown that some members of the erp and bdr gene families are regulated by temperature and other mammal-specific factors (1, 3, 7, 17, 45, 51, 52, 55, 58, 60). Interestingly, Miller et al. reported only limited reactivity to B31 Erp proteins in Lyme disease patient sera, although these proteins are highly immunogenic in mice by needle inoculation (35). Several recent studies have demonstrated that Erp proteins of strains B31, 297, and N40 can bind complement inhibitory factor H (4, 5, 18, 25, 26, 54). Expression of multiple Erp proteins in a single spirochete may facilitate protection from complement-mediating killing in a range of wildlife hosts (54). It is tempting to speculate that this may be one reason for maintenance of multiple homologous cp32 plasmids in a single isolate.
In a previous study, isolates from Lyme disease patients were characterized by PCR-RFLP analysis of the 16S-to-23S ribosomal DNA spacer, the macrorestriction pattern of MluI-digested genomic DNA, and the presence or absence of plasmid lp38. There was a significant (P < 0.001) association between the three typing methods, suggesting a clonal origin for each genotype (23). The present data confirm and extend the results of the earlier analysis to include the full plasmid complement. It is striking that six of seven RST2 isolates simultaneously lack lp56, lp38, and a segment of lp28-1 (Table 2). The remaining RST2 isolate (B376) lacks only lp56 but also has a different MluI macrorestriction pattern. Clearly, these three plasmids are not required for human infection and dissemination (two of the studied RST2 isolates were cultured from patient blood). The well-characterized human CSF isolate 297 displays an RST2 pattern (30), is highly pathogenic in mice (1), and was shown to lack lp56 (14). Moreover, primers based on the strain B31 sequences for lp38 and lp28-1 do not amplify the corresponding regions in this human CSF isolate (M. Caimano, C. Eggers, and J. Radolf, personal communication). The absence of lp38 in natural isolates has been reported, and loss of infectivity does not correlate with the absence of this plasmid (31, 37, 38, 43). Very-high-passage-number isolates of strain N40 lacked lp38 and lp28-1 and were not found to be infectious in C3H mice by needle inoculation (59). As described earlier, the loss of infectivity of strain B31 clones in mice correlates with the absence of lp28-1 and lp25 (27, 44). The present studies clearly show that clinical isolates may not harbor lp56, lp38, and segments of lp28-1 yet maintain infectivity in humans.
As noted earlier, the structure of lp28-1 was unusual in seven isolates. The presence of lp28-1 was scored on the basis of PCR for vlsE, the vls cassette region, or BBF01 and Southern blotting with a vlsE or a BBF01 probe. Both PCR and Southern analysis for BBF01 were negative, suggesting the absence of B31-like lp28-1 in these isolates. By contrast, either PCR or hybridization for vlsE resulted in positive findings. These findings suggest the presence of the vls sequences in these isolates on a plasmid with a structure substantially different than that of B31 lp28-1. The precise nature of this structural alteration is not clear. Since BBF01 is at the left end of lp28-1 and the vls region is at the right end, it is possible that this plasmid is truncated. Alternatively, the vls genes may be located on a different non-lp28-1 plasmid. Although extensive analyses of the vls region of lp28-1 in several isolates have been reported (63), very little is known regarding other regions of lp28-1 in isolates other than B31-MI.
In conclusion, multiple circular and linear plasmids are ubiquitous among the clinical isolates of B. burgdorferi from Lyme disease patients. The absence of three linear plasmids (lp56, lp38, and segments of lp28-1) was specific to isolates that were characterized as RST2 and PFG type A, which suggests a clonal origin for this genotype. The presence of nearly identical complements of plasmids in all human isolates implies that these plasmids may be necessary for infection, adaptation, and/or maintenance of B. burgdorferi in humans during early Lyme disease or during other stages of the spirochete life cycle. Conversely, the absence of plasmids comparable to B31 lp56, lp38, and possibly lp28-1 does not impair either the infectivity or the hematogenous dissemination of B. burgdorferi in humans.
Acknowledgments
We thank Sherwood Casjens, Patti Rosa, Dionysios Liveris, and Guiqing Wang for their valuable insights and Melissa Caimano, Christian Eggers, and Justin Radolf for sharing some of their unpublished observations.
This work was supported by National Institutes of Health grants AI45801 and AR41511 (I.S.), AI44254 (B.S.), and AI37277 (S.N.).
Editor: D. L. Burns
REFERENCES
- 1.Akins, D. R., K. W. Bourell, M. J. Caimano, M. V. Norgard, and J. D. Radolf. 1998. A new animal model for studying Lyme disease spirochetes in a mammalian host-adapted state. J. Clin. Investig. 101:2240-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akins, D. R., M. J. Caimano, X. Yang, F. Cerna, M. V. Norgard, and J. D. Radolf. 1999. Molecular and evolutionary analysis of Borrelia burgdorferi 297 circular plasmid-encoded lipoproteins with OspE- and OspF-like leader peptides. Infect. Immun. 67:1526-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akins, D. R., S. F. Porcella, T. G. Popova, D. Shevchenko, S. I. Baker, M. Li, M. V. Norgard, and J. D. Radolf. 1995. Evidence for in vivo but not in vitro expression of a Borrelia burgdorferi outer surface protein F (OspF) homologue. Mol. Microbiol. 18:507-520. [DOI] [PubMed] [Google Scholar]
- 4.Alitalo, A., T. Meri, H. Lankinen, I. Seppala, P. Lahdenne, P. S. Hefty, D. Akins, and S. Meri. 2002. Complement inhibitor factor H binding to Lyme disease spirochetes is mediated by inducible expression of multiple plasmid-encoded outer surface protein E paralogs. J. Immunol. 169:3847-3853. [DOI] [PubMed] [Google Scholar]
- 5.Alitalo, A., T. Meri, L. Rämö, T. S. Jokiranta, T. Heikkilä, I. J. Seppälä, J. Oksi, M. Viljanen, and S. Meri. 2001. Complement evasion by Borrelia burgdorferi: serum-resistant strains promote C3b inactivation. Infect. Immun. 69:3685-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anguita, J., S. Samanta, B. Revilla, K. Suk, S. Das, S. W. Barthold, and E. Fikrig. 2000. Borrelia burgdorferi gene expression in vivo and spirochete pathogenicity. Infect. Immun. 68:1222-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Babb, K., N. El-Hage, J. C. Miller, J. A. Carroll, and B. Stevenson. 2001. Distinct regulatory pathways control expression of Borrelia burgdorferi infection-associated OspC and Erp surface proteins. Infect. Immun. 69:4146-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbour, A. G. 1988. Plasmid analysis of Borrelia burgdorferi, the Lyme disease agent. J. Clin. Microbiol. 26:475-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caimano, M. J., X. Yang, T. G. Popova, M. L. Clawson, D. R. Akins, M. V. Norgard, and J. D. Radolf. 2000. Molecular and evolutionary characterization of the cp32/18 family of supercoiled plasmids in Borrelia burgdorferi 297. Infect. Immun. 68:1574-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlyon, J. A., and R. T. Marconi. 1998. Cloning and molecular characterization of a multicopy, linear plasmid-carried, repeat motif-containing gene from Borrelia turicatae, a causative agent of relapsing fever. J. Bacteriol. 180:4974-4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casjens, S. 2000. Borrelia genomes in the year 2000. J. Mol. Microbiol. Biotechnol. 2:401-410. [PubMed] [Google Scholar]
- 12.Casjens, S., N. Palmer, R. van Vugt, W. M. Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. M. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490-516. [DOI] [PubMed] [Google Scholar]
- 13.Casjens, S., R. van Vugt, K. Tilly, P. A. Rosa, and B. Stevenson. 1997. Homology throughout the multiple 32-kilobase circular plasmids present in Lyme disease spirochetes. J. Bacteriol. 179:217-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eggers, C. H., M. J. Caimano, M. L. Clawson, W. G. Miller, D. S. Samuels, and J. D. Radolf. 2002. Identification of loci critical for replication and compatibility of a Borrelia burgdorferi cp32 plasmid and use of a cp32-based shuttle vector for the expression of fluorescent reporters in the lyme disease spirochaete. Mol. Microbiol. 43:281-295. [DOI] [PubMed] [Google Scholar]
- 15.El Hage, N., L. D. Lieto, and B. Stevenson. 1999. Stability of erp loci during Borrelia burgdorferi infection: recombination is not required for chronic infection of immunocompetent mice. Infect. Immun. 67:3146-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J. F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. C. Venter, et al. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 17.Hefty, P. S., C. S. Brooks, A. M. Jett, G. L. White, S. K. Wikel, R. C. Kennedy, and D. R. Akins. 2002. OspE-related, OspF-related, and Elp lipoproteins are immunogenic in baboons experimentally infected with Borrelia burgdorferi and in human Lyme disease patients. J. Clin. Microbiol. 40:4256-4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hellwage, J., T. Meri, T. Heikkila, A. Alitalo, J. Panelius, P. Lahdenne, I. J. Seppala, and S. Meri. 2001. The complement regulator factor H binds to the surface protein OspE of Borrelia burgdorferi. J. Biol. Chem. 276:8427-8435. [DOI] [PubMed] [Google Scholar]
- 19.Hinnebusch, J., and A. G. Barbour. 1992. Linear- and circular-plasmid copy numbers in Borrelia burgdorferi. J. Bacteriol. 174:5251-5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hofmeister, E. K., and J. E. Childs. 1995. Analysis of Borrelia burgdorferi sequentially isolated from Peromyscus leucopus captured at a Lyme disease enzootic site. J. Infect. Dis. 172:462-469. [DOI] [PubMed] [Google Scholar]
- 21.Hofmeister, E. K., B. A. Ellis, G. E. Glass, and J. E. Childs. 1999. Longitudinal study of infection with Borrelia burgdorferi in a population of Peromyscus leucopus at a Lyme disease-enzootic site in Maryland. Am. J. Trop. Med. Hyg. 60:598-609. [DOI] [PubMed] [Google Scholar]
- 22.Iyer, R., J. M. Hardham, G. P. Wormser, I. Schwartz, and S. J. Norris. 2000. Conservation and heterogeneity of vlsE among human and tick isolates of Borrelia burgdorferi. Infect. Immun. 68:1714-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iyer, R., D. Liveris, A. Adams, J. Nowakowski, D. McKenna, S. Bittker, D. Cooper, G. P. Wormser, and I. Schwartz. 2001. Characterization of Borrelia burgdorferi isolated from erythema migrans lesions: interrelationship of three molecular typing methods. J. Clin. Microbiol. 39:2954-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jauris-Heipke, S., R. Fuchs, M. Motz, V. Preac-Mursic, E. Schwab, E. Soutschek, G. Will, and B. Wilske. 1993. Genetic heterogenity of the genes coding for the outer surface protein C (OspC) and the flagellin of Borrelia burgdorferi. Med. Microbiol. Immunol. 182:37-50. [DOI] [PubMed] [Google Scholar]
- 25.Kraiczy, P., C. Skerka, V. Brade, and P. F. Zipfel. 2001. Further characterization of complement regulator-acquiring surface proteins of Borrelia burgdorferi. Infect. Immun. 69:7800-7809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kraiczy, P., C. Skerka, P. F. Zipfel, and V. Brade. 2002. Complement regulator-acquiring surface proteins of Borrelia burgdorferi: a new protein family involved in complement resistance. Wien. Klin. Wochenschr. 114:568-573. [PubMed] [Google Scholar]
- 27.Labandeira-Rey, M., and J. T. Skare. 2001. Decreased infectivity in Borrelia burgdorferi strain B31 is associated with loss of linear plasmid 25 or 28-1. Infect. Immun. 69:446-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liveris, D., A. Gazumyan, and I. Schwartz. 1995. Molecular typing of Borrelia burgdorferi sensu lato by PCR-restriction fragment length polymorphism analysis. J. Clin. Microbiol. 33:589-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liveris, D., S. Varde, R. Iyer, S. Koenig, S. Bittker, D. Cooper, D. McKenna, J. Nowakowski, R. B. Nadelman, G. P. Wormser, and I. Schwartz. 1999. Genetic diversity of Borrelia burgdorferi in Lyme disease patients as determined by culture versus direct PCR with clinical specimens. J. Clin. Microbiol. 37:565-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liveris, D., G. P. Wormser, J. Nowakowski, R. Nadelman, S. Bittker, D. Cooper, S. Varde, F. H. Moy, G. Forseter, C. S. Pavia, and I. Schwartz. 1996. Molecular typing of Borrelia burgdorferi from Lyme disease patients by PCR-restriction fragment length polymorphism analysis. J. Clin. Microbiol. 34:1306-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marconi, R. T., D. S. Samuels, R. K. Landry, and C. F. Garon. 1994. Analysis of the distribution and molecular heterogeneity of the ospD gene among the Lyme disease spirochetes: evidence for lateral gene exchange. J. Bacteriol. 176:4572-4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mathiesen, D. A., J. H. Oliver, Jr., C. P. Kolbert, E. D. Tullson, B. J. Johnson, G. L. Campbell, P. D. Mitchell, K. D. Reed, S. R. Telford III, J. F. Anderson, R. S. Lane, and D. H. Persing. 1997. Genetic heterogeneity of Borrelia burgdorferi in the United States. J. Infect. Dis. 175:98-107. [DOI] [PubMed] [Google Scholar]
- 33.McDowell, J. V., S. Y. Sung, M. Labandeira-Rey, J. T. Skare, and R. T. Marconi. 2001. Analysis of mechanisms associated with loss of infectivity of clonal populations of Borrelia burgdorferi B31MI. Infect. Immun. 69:3670-3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller, J. C., J. L. Bono, K. Babb, N. El-Hage, S. Casjens, and B. Stevenson. 2000. A second allele of eppA in Borrelia burgdorferi strain B31 is located on the previously undetected circular plasmid cp9-2. J. Bacteriol. 182:6254-6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller, J. C., N. El-Hage, K. Babb, and B. Stevenson. 2000. Borrelia burgdorferi B31 Erp proteins that are dominant immunoblot antigens of animals infected with isolate B31 are recognized by only a subset of human Lyme disease patient sera. J. Clin. Microbiol. 38:1569-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nadelman, R. B., and G. P. Wormser. 1990. A clinical approach to Lyme disease. Mt. Sinai J. Med. 57:144-156. [PubMed] [Google Scholar]
- 37.Norris, S. J., C. J. Carter, J. K. Howell, and A. G. Barbour. 1992. Low-passage-associated proteins of Borrelia burgdorferi B31: characterization and molecular cloning of OspD, a surface-exposed, plasmid-encoded lipoprotein. Infect. Immun. 60:4662-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Norris, S. J., J. K. Howell, S. A. Garza, M. S. Ferdows, and A. G. Barbour. 1995. High- and low-infectivity phenotypes of clonal populations of in vitro-cultured Borrelia burgdorferi. Infect. Immun. 63:2206-2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orloski, K. A., E. B. Hayes, G. L. Campbell, and D. T. Dennis. 2000. Surveillance for Lyme disease—United States, 1992-1998. Morb. Mortal. Wkly. Rep. CDC Surveill. Summ. 49:1-11. [PubMed] [Google Scholar]
- 40.Palmer, N., C. Fraser, and S. Casjens. 2000. Distribution of twelve linear extrachromosomal DNAs in natural isolates of Lyme disease spirochetes. J. Bacteriol. 182:2476-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Picken, R. N., Y. Cheng, D. Han, J. A. Nelson, A. G. Reddy, M. K. Hayden, M. M. Picken, F. Strle, J. K. Bouseman, and G. M. Trenholme. 1995. Genotypic and phenotypic characterization of Borrelia burgdorferi isolated from ticks and small animals in Illinois. J. Clin. Microbiol. 33:2304-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Porcella, S. F., C. A. Fitzpatrick, and J. L. Bono. 2000. Expression and immunological analysis of the plasmid-borne mlp genes of Borrelia burgdorferi strain B31. Infect. Immun. 68:4992-5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Probert, W. S., and R. B. LeFebvre. 1994. Protection of C3H/HeN mice from challenge with Borrelia burgdorferi through active immunization with OspA, OspB, or OspC, but not with OspD or the 83-kilodalton antigen. Infect. Immun. 62:1920-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Purser, J. E., and S. J. Norris. 2000. Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc. Natl. Acad. Sci. USA 97:13865-13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roberts, D. M., M. Caimano, J. McDowell, M. Theisen, A. Holm, E. Orff, D. Nelson, S. Wikel, J. Radolf, and R. T. Marconi. 2002. Environmental regulation and differential production of members of the Bdr protein family of Borrelia burgdorferi. Infect. Immun. 70:7033-7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwan, T. G., W. Burgdorfer, and C. F. Garon. 1988. Changes in infectivity and plasmid profile of the Lyme disease spirochete, Borrelia burgdorferi, as a result of in vitro cultivation. Infect. Immun. 56:1831-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwartz, I., G. P. Wormser, J. J. Schwartz, D. Cooper, P. Weissensee, A. Gazumyan, E. Zimmermann, N. S. Goldberg, S. Bittker, G. L. Campbell, and C. S. Pavia. 1992. Diagnosis of early Lyme disease by polymerase chain reaction amplification and culture of skin biopsies from erythema migrans lesions. J. Clin. Microbiol. 30:3082-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simpson, W. J., C. F. Garon, and T. G. Schwan. 1990. Analysis of supercoiled circular plasmids in infectious and non-infectious Borrelia burgdorferi. Microb. Pathog. 8:109-118. [DOI] [PubMed] [Google Scholar]
- 49.Simpson, W. J., C. F. Garon, and T. G. Schwan. 1990. Borrelia burgdorferi contains repeated DNA sequences that are species specific and plasmid associated. Infect. Immun. 58:847-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simpson, W. J., M. E. Schrumpf, and T. G. Schwan. 1990. Reactivity of human Lyme borreliosis sera with a 39-kilodalton antigen specific to Borrelia burgdorferi. J. Clin. Microbiol. 28:1329-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stevenson, B., J. L. Bono, T. G. Schwan, and P. Rosa. 1998. Borrelia burgdorferi Erp proteins are immunogenic in mammals infected by tick bite, and their synthesis is inducible in cultured bacteria. Infect. Immun. 66:2648-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stevenson, B., S. Casjens, and P. Rosa. 1998. Evidence of past recombination events among the genes encoding the Erp antigens of Borrelia burgdorferi. Microbiology 144:1869-1879. [DOI] [PubMed] [Google Scholar]
- 53.Stevenson, B., S. Casjens, R. van Vugt, S. F. Porcella, K. Tilly, J. L. Bono, and P. Rosa. 1997. Characterization of cp18, a naturally truncated member of the cp32 family of Borrelia burgdorferi plasmids. J. Bacteriol. 179:4285-4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stevenson, B., N. El-Hage, M. A. Hines, J. C. Miller, and K. Babb. 2002. Differential binding of host complement inhibitor factor H by Borrelia burgdorferi Erp surface proteins: a possible mechanism underlying the expansive host range of Lyme disease spirochetes. Infect. Immun. 70:491-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stevenson, B., T. G. Schwan, and P. A. Rosa. 1995. Temperature-related differential expression of antigens in the Lyme disease spirochete, Borrelia burgdorferi. Infect. Immun. 63:4535-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stevenson, B., W. R. Zuckert, and D. R. Akins. 2000. Repetition, conservation, and variation: the multiple cp32 plasmids of Borrelia species. J. Mol. Microbiol. Biotechnol. 2:411-422. [PubMed] [Google Scholar]
- 57.Stewart, P. E., R. Thalken, J. L. Bono, and P. Rosa. 2001. Isolation of a circular plasmid region sufficient for autonomous replication and transformation of infectious Borrelia burgdorferi. Mol. Microbiol. 39:714-721. [DOI] [PubMed] [Google Scholar]
- 58.Suk, K., S. Das, W. Sun, B. Jwang, S. W. Barthold, R. A. Flavell, and E. Fikrig. 1995. Borrelia burgdorferi genes selectively expressed in the infected host. Proc. Natl. Acad. Sci. USA 92:4269-4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thomas, V., J. Anguita, S. Samanta, P. A. Rosa, P. Stewart, S. W. Barthold, and E. Fikrig. 2001. Dissociation of infectivity and pathogenicity in Borrelia burgdorferi. Infect. Immun. 69:3507-3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wallich, R., C. Brenner, M. D. Kramer, and M. M. Simon. 1995. Molecular cloning and immunological characterization of a novel linear-plasmid-encoded gene, pG, of Borrelia burgdorferi expressed only in vivo. Infect. Immun. 63:3327-3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang, G., C. Ojaimi, R. Iyer, V. Saksenberg, S. A. McClain, G. P. Wormser, and I. Schwartz. 2001. Impact of genotypic variation of Borrelia burgdorferi sensu stricto on kinetics of dissemination and severity of disease in C3H/HeJ mice. Infect. Immun. 69:4303-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, G., C. Ojaimi, H. Wu, V. Saksenberg, R. Iyer, D. Liveris, S. A. McClain, G. P. Wormser, and I. Schwartz. 2002. Disease severity in a murine model of Lyme borreliosis is associated with the genotype of the infecting Borrelia burgdorferi sensu stricto strain. J. Infect. Dis. 186:782-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang, G., A. P. van Dam, and J. Dankert. 2001. Analysis of a VMP-like sequence (vls) locus in Borrelia garinii and Vls homologues among four Borrelia burgdorferi sensu lato species. FEMS Microbiol. Lett. 199:39-45. [DOI] [PubMed] [Google Scholar]
- 64.Wang, G., A. P. van Dam, I. Schwartz, and J. Dankert. 1999. Molecular typing of Borrelia burgdorferi sensu lato: taxonomic, epidemiological, and clinical implications. Clin. Microbiol. Rev. 12:633-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang, I. N., D. E. Dykhuizen, W. Qiu, J. J. Dunn, E. M. Bosler, and B. J. Luft. 1999. Genetic diversity of ospC in a local population of Borrelia burgdorferi sensu stricto. Genetics 151:15-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wormser, G. P., S. Bittker, D. Cooper, J. Nowakowski, R. B. Nadelman, and C. Pavia. 2001. Yield of large-volume blood cultures in patients with early Lyme disease. J. Infect. Dis. 184:1070-1072. [DOI] [PubMed] [Google Scholar]
- 67.Wormser, G. P., J. Nowakowski, R. B. Nadelman, S. Bittker, D. Cooper, and C. Pavia. 1998. Improving the yield of blood cultures for patients with early Lyme disease. J. Clin. Microbiol. 36:296-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu, Y., and R. C. Johnson. 1995. Analysis and comparison of plasmid profiles of Borrelia burgdorferi sensu lato strains. J. Clin. Microbiol. 33:2679-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu, Y., C. Kodner, L. Coleman, and R. C. Johnson. 1996. Correlation of plasmids with infectivity of Borrelia burgdorferi sensu stricto type strain B31. Infect. Immun. 64:3870-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang, J. R., J. M. Hardham, A. G. Barbour, and S. J. Norris. 1997. Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell 89:275-285. [DOI] [PubMed] [Google Scholar]
- 71.Zückert, W. R., and J. Meyer. 1996. Circular and linear plasmids of Lyme disease spirochetes have extensive homology: characterization of a repeated DNA element. J. Bacteriol. 178:2287-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]