Abstract
Temperate bacteriophage can transfer toxin-encoding genes between bacteria, often resulting in acquired pathogenicity. However, little is known regarding the effects of the eukaryotic host on the phage-pathogen interaction. Using Streptococcus pyogenes as a model, we demonstrate, both in vitro and in vivo, that the eukaryote mediates the efficient induction of toxin-encoding temperate phage and the resultant conversion of Tox− flora to Tox+. Furthermore, we show that both phage induction and subsequent conversion need not happen in the same mammalian host, as host-to-host phage transmission can result in toxigenic conversion within the secondary host. Ultimately, our findings demonstrate that the eukaryotic host serves as an essential component in the phage-mediated evolution of virulence within the microbial population.
In both gram-positive and gram-negative bacteria, temperate bacteriophage encode toxins and virulence factors in addition to their essential viral proteins (8). Lysogeny provides evolutionary advantages to both species, as phage-endowed virulence allows the pathogen to more successfully proliferate within the eukaryote and the prophage to be maintained via vertical transmission with the host chromosome. Streptococcus pyogenes is a multiply lysogenized organism whose phage constitute ∼10% of the total genome and encode a wide variety of putative and established virulence factors, including a large class of pyrogenic exotoxins (10, 24). Recent comparative genomic studies have demonstrated that streptococcal bacteriophage represent the major variation (up to 71%) between strains of S. pyogenes (3, 21) and potentially account for the distinct disease pathologies associated with otherwise similar strains. In addition to modulating the virulence of organisms found within a common species of pathogenic bacteria, toxin-encoding phage produced by such pathogens have been shown to toxin convert both environmental and commensal bacteria, generating pathogenic Tox+ microbes (4, 9, 19). Thus, bacteriophage represent key vectors for the dissemination of bacterial virulence and the conversion of bacteria from nonpathogenic to pathogenic.
While integrated prophage are well-documented sources of genetic diversity within the bacterial population, the events that ultimately result in this diversity have not been observed. We have previously shown that the human pharyngeal cell releases a soluble factor that stimulates the lytic activation of S. pyogenes prophage (6). During streptococcal disease, it is likely that phage are induced in vivo (1, 12, 13, 23). In the present study, we demonstrate that the mammalian host promotes both efficient bacteriophage induction at the mucosal surface and the subsequent lysogenic conversion of flora occupying the same niche. Thus, we establish that the continuing evolution and phage-mediated diversification of streptococci not only occur at the mucosal surface but are dependent on it.
MATERIALS AND METHODS
Construction of S. pyogenes CS112(ΦCS112 Tox+ Kmr) mutant.
Allelic exchange of the kanamycin resistance-encoding gene aacA (also called aphD) for the phage-encoded nonessential spd1 gene in S. pyogenes strain CS112 (an M− phenotypic variant of CS110 type M76 [22]) was achieved via homologous recombination. A PCR was used to amplify a 1-kb DNA element (with engineered 5′ HindIII and 3′ XbaI sites) from immediately upstream of the structural spd1 gene. The PCR product was directionally ligated into HindIII/XbaI-digested plasmid pBAD18 and then transformed into Escherichia coli XL1-Blue. The resultant plasmid was then digested with KpnI/EcoRI and ligated with a 1-kb PCR product (with engineered 5′ KpnI and 3′ EcoRI sites) that was amplified from immediately downstream of spd1. This pBAD18 recombinant plasmid was then digested with SmaI and ligated with the SmaI/AluI pFW13 fragment containing the aacA kanamycin resistance gene (18). The final plasmid, containing the aacA gene flanked by the upstream and downstream elements of spd1, was linearized by HindIII digestion, and the linear product was separated by agarose gel electrophoresis, followed by DNA gel extraction purification. This purified linear DNA was used to transform S. pyogenes strain CS112 in accordance with the protocol of Simon and Ferretti (20). Potential Spd1− mutants were selected for on Proteose Peptone blood agar plates containing 150 μg of kanamycin per ml. Several Kmr clones were picked, and PCR amplification with oligonucleotides designed to anneal upstream and downstream of the spd1 gene was used to verify the appropriate-size product for a double-recombination event resulting in the complete deletion of spd1.
In vitro lysogenic conversion experiments.
Detroit 562 human pharyngeal cells were grown to confluence in six-well plates and then washed three times with serum-free minimal essential medium (MEM). S. pyogenes strains CS112(ΦCS112 Tox+ Kmr) and CS24 Smr (type M12 [22]) were grown overnight in Todd-Hewitt broth containing 1% yeast extract (THY) at 37°C and suspended in phosphate-buffered saline. After adjustment of the optical density at 650 nm to 1.0, the bacteria were centrifuged and resuspended in serum-free MEM. The pharyngeal cells were then inoculated with ∼108 CFU of each bacterial strain. The mixed bacterial and pharyngeal cell coculture was allowed to incubate for 3 h at 37°C under 5% CO2. Each well was treated with 0.25% trypsin and 0.0125% Triton X-100 to lyse the pharyngeal cells and collect any bacteria that might have adhered to the cell surface. This lysate was pooled with the supernatant from the same well, and dilutions were plated on Proteose Peptone blood agar plates containing either kanamycin (150 μg/ml), streptomycin (200 μg/ml), or both antibiotics (150 and 200 μg/ml, respectively). The number of phage-lysogenized recipient strains [i.e., CS24 Smr(ΦCS112 Tox+ Kmr)] was defined as the total number of CFU demonstrating a double-antibiotic resistance phenotype. Thus, the frequency of lysogenization was determined as the ratio of the number of Smr Kmr CFU to the total number of Smr CFU. Additionally, the phage donor and recipient strains, CS112(ΦCS112 Tox+ Kmr) and CS24 Smr, were mixed in the absence of pharyngeal cells, as well as independently of each other. Lysogenization frequencies were determined for these experiments as well.
To test the effects of pharyngeal cell culture supernatant on streptococcal lysogenic conversion, we performed experiments in which the phage donor and recipient bacteria were incubated in cell-free spent pharyngeal cell supernatant. Detroit 562 pharyngeal cells were grown to confluence and then incubated in serum-free MEM for 3 h at 37°C under 5% CO2. The spent medium was collected and centrifuged (4,000 × g for 30 min). Strain CS112(ΦCS112 Tox+ Kmr) and CS24 Smr cells were incubated in this medium, and newly lysogenized bacteria were enumerated by the acquired antibiotic resistance phenotype.
To determine if the pharyngeal cells promoted lysogenic conversion by sensitizing the phage recipient bacteria to the temperate phage, we performed the following experiment. Confluent Detroit 562 pharyngeal cells were inoculated with ∼108 CFU of CS24 Smr bacteria. Following a 30-min incubation at 37°C under 5% CO2, we inoculated the coculture with ∼105 PFU of cell-free ΦCS112 Tox+ Kmr and then allowed the mixture to further incubate for another 3 h. As a control, the same experiment was performed with no pharyngeal cells present. Following the incubations, frequencies of newly lysogenized bacteria were determined as previously described.
Phage lysogenization in vivo.
Experiments were performed to determine if the murine airway could support the lysogenization events observed in vitro. S. pyogenes strains CS112(ΦCS112 Tox+ Kmr) and CS24 Smr were grown overnight, washed in phosphate-buffered saline, and adjusted to an optical density at 650 nm of 2.0. The strains were mixed 1:1 (vol/vol), and the mixture was used to oronasally challenge (30 μl orally, 30 μl nasally) four 6-week-old female BALB/c mice. After 24 h, the mice were orally swabbed and the swabs were streaked onto Proteose Peptone blood agar plates containing kanamycin (150 μg/ml), streptomycin (200 (μg/ml), or both antibiotics (150 μg/ml and 200 μg/ml, respectively). Additionally, the mice were euthanized and decapitated. Each mouse head was incubated in THY broth containing both kanamycin (150 μg/ml) and streptomycin (200 μg/ml) to enrich for any lysogens that might have been created but were not detectable by oral swabbing. Following overnight incubation, the broth was streaked onto Proteose Peptone blood agar plates containing kanamycin and streptomycin. These plates were then incubated, and beta-hemolytic Smr Kmr colonies were identified.
In vivo toxigenic conversion with isolated phage.
Four 6-week-old female BALB/c mice were challenged with S. pyogenes strain CS24 Smr as described previously. After 6 h, each mouse was challenged with 60 μl of sterile filtered ΦCS112 Tox+ Kmr (107 PFU/ml). After 24 h, we detected Kmr lysogens by oral swabbing or by enrichment from within the mouse head, as described earlier.
Genetic analysis of the Smr Kmr phenotype.
To discount the possibility that random mutation gave rise to the observed phenotypes of the lysogenized species, genetic analysis of each strain was performed. Genomic DNA was extracted from S. pyogenes CS112(ΦCS112 Tox+ Kmr), CS24 Smr, and the strains designated CS112(ΦCS112 Tox+ Kmr) that were selected as converted lysogens following incubations in a pharyngeal cell coculture, pharyngeal cell supernatant, and the mouse nasopharynx. Genomic DNA samples served as the templates for PCR amplification of the phage-encoded speC and int1 genes, which served as markers for the integrated ΦCS112 prophage. Additionally, oligonucleotide primers (17) were used to PCR amplify the variable-length emm gene from each S. pyogenes strain. The genetic traits of the Smr Kmr strains, thought to be CS24 Smr(ΦCS112 Tox+ Kmr), were compared to the traits of the two parental strains.
RESULTS
In vitro lysogenic conversion.
We tested whether bacteriophage induced from S. pyogenes in the presence of pharyngeal cells could mediate toxigenic conversion of a phage-naive Tox− recipient strain. Lysogenic S. pyogenes strain CS112(ΦCS112 Tox+), containing the streptococcal pyrogenic exotoxin C gene within an inducible prophage, served as a phage donor. A gene encoding Kmr was inserted into the ΦCS112 Tox+ genome [CS112(ΦCS112 Tox+ Kmr)] for use as a selectable marker for detection of toxin-converted species by an acquired Kmr phenotype.
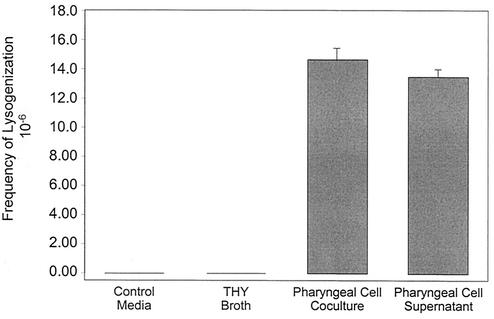
Cultured pharyngeal cells were then inoculated with both phage donor strain CS112(ΦCS112 Tox+ Kmr) and nonlysogenized, streptomycin-resistant S. pyogenes strain CS24 Smr, which served as the phage recipient. During a 3-h coculture, we detected the induction of the donor phage ΦCS112 Tox+ Kmr (∼105 phage/ml) and subsequent lysogenization of the phage-naive recipient strain, CS24 Smr, creating the double-antibiotic-resistant toxigenic organism CS24 Smr(ΦCS112 Tox+ Kmr). Lysogenic conversion of CS24 Smr occurred at a frequency of 1.46 × 10−5. Additionally, incubation of the phage donor and recipient strains in cell-free pharyngeal supernatant (i.e., conditions that we have previously shown to promote phage induction [6]) yielded a similar lysogenization frequency of 1.33 × 10−5 (Fig. 1). This horizontal transfer could not be detected after incubation of both CS112(ΦCS112 Tox+ Kmr) and CS24 Smr for 3 h in minimal medium without serum or THY broth (Fig. 1) and was therefore dependent on the pharyngeal cells. Similar results were obtained in minimal medium containing serum (data not shown). Although these phage transfer events were rare, they were significant (P < 0.001) compared to the conversion frequency in culture medium alone. Additionally, the induced phage titer is ∼1,000 times lower than the streptococcal CFU concentration, meaning that, of the ∼108 phage recipient streptococci present, a maximum of ∼105 could be infected by the temperate phage. Using these considerations, we calculated the frequency of lysogenization per singly infected bacterium to be ∼10−3, meaning that 1 in every 1,000 streptococci that are infected with ΦCS112 Tox+ Kmr becomes lysogenized.
FIG. 1.
Pharyngeal-cell-mediated horizontal gene transfer. The frequency of lysogenized recipient bacteria is plotted for phage transfer events occurring in minimal medium, THY broth, pharyngeal cell coculture, or pharyngeal cell supernatant. The values plotted are mean concentrations of duplicate experiments ± the standard deviations.
We also examined for the possibility that the pharyngeal cell may sensitize the phage recipient strain to phage conversion, thus contributing to the observed increase in the lysogenization frequency. In the presence or absence of pharyngeal cells, we incubated CS24 Smr bacteria with known titers of cell-free ΦCS112 Tox+ Kmr bacteriophage and then determined respective frequencies of lysogenization. No significant differences were found (data not shown). These results indicate that the increase in lysogenization frequency observed during pharyngeal cell coculture (Fig. 1) resulted entirely from the induction of bacteriophage that subsequently lysogenized naive bacteria.
Lysogenic conversion in vivo.
Horizontal transfer of ΦCS112 Tox+ Kmr was tested in vivo with a murine model of streptococcal colonization. Essentially, BALB/c mice were challenged oronasally with the phage donor [CS112(ΦCS112 Tox+ Kmr)] and phage recipient (CS24 Smr) strains used in the experiments described above. We failed to detect horizontal phage transfer (S. pyogenes with the Smr Kmr phenotype) by oral swabbing at 24 h (Table 1), which was likely attributable to the limitations of the swab recovery technique and its inability to efficiently capture rare or hidden organisms. The coinfected mice were euthanized at 24 h, and each separated mouse head was then incubated in THY medium to which kanamycin and streptomycin were added. After overnight incubation, we successfully enriched and detected the lysogenized CS24 Smr(ΦCS112 Tox+ Kmr) strain in two of four mice. As a control, the CS24 Smr and CS112(ΦCS112 Tox+ Kmr) strains were mixed together in kanamycin-streptomycin broth. Following an 18-h incubation, no growth was detected, indicating that spontaneous lysogenic conversion did not occur during the overnight lysogen enrichment step. The same results were obtained when this control experiment was performed in the presence of a mouse head, verifying that the lysogenized organisms were created within the viable mouse naso- and oropharynx during the initial challenge.
TABLE 1.
In vivo lysogenic conversion assay results
| Strain or phage and bacterial environment | Primary swaba | After enrichmentb |
|---|---|---|
| CS112(ΦCS112 Tox+ Kmr) | ||
| Mouse 1 | −c | ++++d |
| Mouse 2 | − | − |
| Mouse 3 | − | ++++ |
| Mouse 4 | − | − |
| Cell-free ΦCS112 Tox+ Kmr | ||
| Mouse 1 | − | ++++ |
| Mouse 2 | − | ++++ |
| Mouse 3 | − | ++++ |
| Mouse 4 | − | − |
Lysogens detected by throat swabs after 24 h.
Following the throat swab, mice were euthanized and the heads were incubated overnight to enrich for possible lysogens.
−, No lysogens detected.
++++, Lysogens detected.
In vivo toxigenic conversion with isolated phage.
Just as pathogenic microbes are transmitted through contaminated fluids (e.g., saliva or blood), we reasoned that bacteriophage themselves could be transmitted in the same fashion. Once a transmitted phage enters the new host environment, it may lysogenize pre-existing bacteria, continuing its own life cycle while providing potential virulence factors to its host bacterium. To test this hypothesis, we colonized BALB/c mice with nonlysogenized S. pyogenes CS24 Smr and challenged them with cell-free phage (ΦCS112 Tox+ Kmr). At 24 h after the phage challenge, we attempted to identify lysogenized organisms (marked by Smr Kmr) either by throat swabbing or by further enrichment of Smr Kmr organisms from within the mouse head. While we again were unable to detect any newly lysogenized species via throat swabbing, we did detect such lysogens following enrichment in three of four mice (Table 1).
Genetic characterization of lysogenized bacteria.
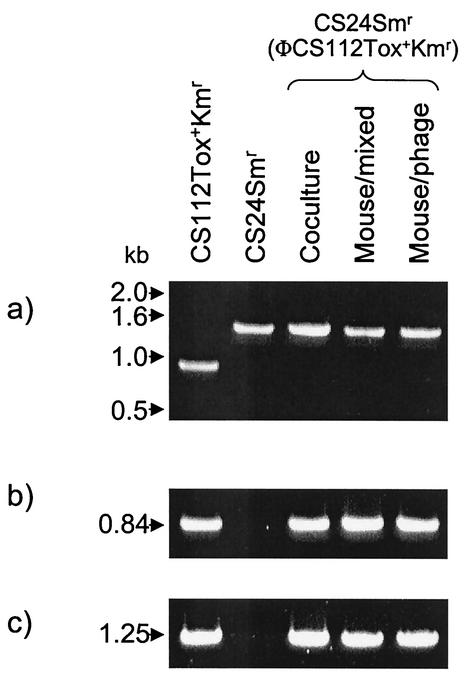
By using antibiotic resistance as an acquired and selective phenotype, we were able to demonstrate horizontal transfer of a toxin-encoding phage within S. pyogenes during incubation in pharyngeal cell cocultures, as well as in the murine nasopharynx itself. Additionally, we show that the same lysogenic conversion occurred within the mouse when bacteria were challenged with isolated phage alone. To discount the possibility that random mutation gave rise to the acquired antibiotic resistance phenotypes, we performed genetic analyses of each of these newly formed lysogens. Because the S. pyogenes emm gene varies in size from strain to strain (15), we amplified this gene by PCR to distinguish the phage donor strain from the phage recipient and lysogenized derivatives thereof. PCR performed on the emm gene from phage donor strain CS112(ΦCS112 Tox+ Kmr) generated a 950-bp fragment, whereas amplification of the emm gene from recipient strain CS24 Smr and each of the suspected lysogenized strains, CS24 Smr(ΦCS112 Tox+ Kmr), generated 1.5-kb products (Fig. 2). Furthermore, PCR amplification of the speC gene phage marker verified the presence of the integrated prophage in the donor strain and in each of the lysogenized strains but not in the parental recipient strain (Fig. 2). PCR amplification of the int1 gene, a gene located on the opposite end of the integrated prophage, substantiated the presence of the fully integrated phage. These results, taken together, verify that the bacteria with the acquired double antibiotic resistance phenotype were the anticipated Tox+ lysogenized organisms and not the result of spontaneous mutation.
FIG. 2.
Analysis of S. pyogenes lysogens. CS112(ΦCS112 Tox+ Kmr) is the parent phage donor strain. CS24 Smr is the parent recipient strain. CS24 Smr(ΦCS112 Tox+ Kmr) is the designation of the newly formed lysogens generated during pharyngeal cell coculture (Coculture), during mixed bacterial culture within the mouse nasopharynx (Mouse/mixed), or following isolated phage challenge of a colonized mouse (Mouse/phage). (a) PCR amplification of the variable-length emm gene from each strain. (b) PCR amplification of the speC gene to detect the integrated prophage. (c) PCR amplification of the int1 gene to further detect the integrated prophage. Molecular sizes are shown on the left.
DISCUSSION
The omnipresence of bacteriophage throughout nature (estimated to be 1030 on earth) empowers these bacterial viruses to be a significant force shaping microbial evolution. Perhaps more than other systems, the evolution of S. pyogenes has been guided by bacteriophage. If we carefully examined them, however, we likely would find that many bacteria have bacteriophage systems. It is clear that phage mediate the exchange of toxin-encoding genes between bacteria, and this toxigenic conversion can have a serious impact on the pathogenic fitness of the organism. Until now, however, our understanding of the phage-microbe relationship has lacked a critical component. We demonstrate that the partnership between a phage and a microbe can be mediated by a third party, i.e., a mammalian eukaryotic host. Both in vitro and vivo, we have found that the mammalian host promotes bacteriophage induction and lysogenic conversion. The impact of this finding is important, as it is evidence that the eukaryotic host actively participates in the diversification and evolution of a bacterial population. Our inability to detect lysogenic conversion when phage donor and recipient organisms are incubated together in medium alone, as well as our previous identification of a eukaryotic phage-inducing molecule (6), demonstrates the essential role of the mammalian host in promoting the transmission of bacteriophage between bacteria. Moreover, the mammalian host coordinates the events resulting in lysogenic conversion by bringing phage donor and recipient strains into proximity at a common biological niche. By demonstrating that phage induction and subsequent lysogenic conversion occur within that niche, our findings suggest that phage have evolved to specifically disseminate at the mucosal surface.
The upper respiratory mucosal site is at least one location where lysogenic exchange occurs. Such surfaces often constitute the sole environmental reservoir for a number of pathogenic bacteria (e.g., S. pyogenes, S. pneumoniae, and Haemophilus influenzae). Thus, for successful propagation and survival, temperate phage must reach the mucosal site to access and infect the reservoir of potentially phage-sensitive organisms. For example, a carrier survey of Corynebacterium diphtheriae suggests that spread of the tox gene to infected individuals occurred via phage conversion of a pre-existing colonizing organism, rather than by colonization of those individuals with the challenge strain (16). In the case of S. pyogenes, ∼50% of strains do not harbor a SpeC toxin phage (5) and, barring restriction by other, also related temperate phage, these organisms represent potential toxigenic-conversion targets within the human host. Our findings indicate that isolated phage may be transmitted between individuals, converting the environmental reservoir within the secondary host into a toxigenic population. Such bacteria may then proceed with a more pathogenic lifestyle, resulting in the potential for infection absent challenge from any exogenous microbe.
Up to 25% of S. pyogenes pharyngitis infections result in asymptomatic colonization of the human host, with the bacteria entering into a nonpathogenic carrier state (11). Our work indicates that when a colonized host is challenged with a lysogenized toxigenic strain, phage induction and subsequent toxigenic conversion of the carrier state bacteria can occur, resulting in a novel and potentially pathogenic derivative. Thus, previous work suggests and we provide evidence for the lysogenic conversion of virulence factors into pre-existing bacteria and the consequential creation of a unique pathogenic agent within the host itself.
Taken together, our results indicate that within the mammalian host, bacteria alter not only their gene expression profiles (14) but their genomes themselves. The bacterium-bacteriophage system is in a constant state of flux, with phage inducing from one bacterium and integrating into another. This is supported by data showing that the genome of S. pyogenes is saturated with integrated phage (up to eight integrants per chromosome [7]), which constitute a significant portion of the total genome. We have sought to understand the etiological events resulting in both the genetic diversity found within S. pyogenes and the development of streptococcal virulence itself. Our findings are substantiated by recent streptococcal genomic work indicating the differing outcomes of streptococcal infections to be based on the lysogenic status of the organism (3). Similar phage-based genetic diversity is found in a number of other bacteria, including Staphylococcus aureus (2), and it is likely that the mammalian host plays a similar role in the evolution of those systems as well. Thus, despite the absence of a direct interaction between phage and eukaryote, our findings suggest that in vivo phage conversion represents one method by which bacteriophage can have a direct impact on a mammalian host.
Acknowledgments
We thank Joshua Lederberg and Alexander Tomasz for insightful discussion of the subject matter. Additionally, we thank Patricia Ryan, Raymond Schuch, Daniel Nelson, Pauline Yoong, and Corrie Broudy for critical comments throughout the execution of this work and the preparation of the manuscript. We are also greatly indebted to Clara Eastby for cell culture expertise.
This work was supported by U.S. Public Health Service grant AI11822 to V.A.F.
Editor: A. D. O'Brien
REFERENCES
- 1.Acheson, D. W., J. Reidl, X. Zhang, G. T. Keusch, J. J. Mekalanos, and M. K. Waldor. 1998. In vivo transduction with Shiga toxin 1-encoding phage. Infect. Immun. 66:4496-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. [DOI] [PubMed] [Google Scholar]
- 3.Beres, S. B., G. L. Sylva, K. D. Barbian, B. Lei, J. S. Hoff, N. D. Mammarella, M. Liu, J. C. Smoot, S. F. Porcella, L. D. Parkins, D. S. Campbell, T. M. Smith, J. K. McCormick, D. Y. Leung, P. M. Schlievert, and J. M. Musser. 2002. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc. Natl. Acad. Sci. USA 99:10078-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beutin, L., E. Strauch, and I. Fischer. 1999. Isolation of Shigella sonnei lysogenic for a bacteriophage encoding gene for production of Shiga toxin. Lancet 353:1498. [DOI] [PubMed] [Google Scholar]
- 5.Broudy, T. B., V. Pancholi, and V. A. Fischetti. 2002. The in vitro interaction of Streptococcus pyogenes with human pharyngeal cells induces a phage-encoded extracellular DNase. Infect. Immun. 70:2805-2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broudy, T. B., V. Pancholi, and V. A. Fischetti. 2001. Induction of lysogenic bacteriophage and phage-associated toxin from group A streptococci during coculture with human pharyngeal cells. Infect. Immun. 69:1440-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desiere, F., W. M. McShan, D. van Sinderen, J. J. Ferretti, and H. Brussow. 2001. Comparative genomics reveals close genetic relationships between phages from dairy bacteria and pathogenic streptococci: evolutionary implications for prophage-host interactions. Virology 288:325-341. [DOI] [PubMed] [Google Scholar]
- 8.Dobrindt, U., and J. Reidl. 2000. Pathogenicity islands and phage conversion: evolutionary aspects of bacterial pathogenesis. Int. J. Med. Microbiol. 290:519-527. [DOI] [PubMed] [Google Scholar]
- 9.Faruque, S. M., M. M. Rahman, Asadulghani, K. M. Nasirul Islam, and J. J. Mekalanos. 1999. Lysogenic conversion of environmental Vibrio mimicus strains by CTXΦ. Infect. Immun. 67:5723-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gastanaduy, A. S., E. L. Kaplan, B. B. Huwe, C. McKay, and L. W. Wannamaker. 1980. Failure of penicillin to eradicate group A streptococci during an outbreak of pharyngitis. Lancet ii:498-502. [DOI] [PubMed] [Google Scholar]
- 12.Lazar, S., and M. K. Waldor. 1998. ToxR-independent expression of cholera toxin from the replicative form of CTXφ. Infect. Immun. 66:394-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDonnell, M., R. Lain, and A. Tomasz. 1975. “Diplophage”: a bacteriophage of Diplococcus pneumoniae. Virology 63:577-582. [DOI] [PubMed] [Google Scholar]
- 14.Merrell, D. S., and A. Camilli. 2000. Detection and analysis of gene expression during infection by in vivo expression technology. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355:587-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller, L., L. Gray, E. Beachey, and M. Kehoe. 1988. Antigenic variation among group A streptococcal M proteins. Nucleotide sequence of the serotype 5 M protein gene and its relationship with genes encoding types 6 and 24 M proteins. J. Biol. Chem. 263:5668-5673. [PubMed] [Google Scholar]
- 16.Pappenheimer, A. M., Jr., and J. R. Murphy. 1983. Studies on the molecular epidemiology of diphtheria. Lancet ii:923-926. [DOI] [PubMed] [Google Scholar]
- 17.Podbielski, A., B. Melzer, and R. Lutticken. 1991. Application of the polymerase chain reaction to study the M protein(-like) gene family in beta-hemolytic streptococci. Med. Microbiol. Immunol. 180:213-227. [DOI] [PubMed] [Google Scholar]
- 18.Podbielski, A., B. Spellerberg, M. Woischnik, B. Pohl, and R. Lutticken. 1996. Novel series of plasmid vectors for gene inactivation and expression analysis in group A streptococci (GAS). Gene 177:137-147. [DOI] [PubMed] [Google Scholar]
- 19.Schmidt, H., M. Bielaszewska, and H. Karch. 1999. Transduction of enteric Escherichia coli isolates with a derivative of Shiga toxin 2-encoding bacteriophage φ3538 isolated from Escherichia coli O157:H7. Appl. Environ. Microbiol. 65:3855-3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simon, D., and J. J. Ferretti. 1991. Electrotransformation of Streptococcus pyogenes with plasmid and linear DNA. FEMS Microbiol. Lett. 66:219-224. [DOI] [PubMed] [Google Scholar]
- 21.Smoot, J. C., K. D. Barbian, J. J. Van Gompel, L. M. Smoot, M. S. Chaussee, G. L. Sylva, D. E. Sturdevant, S. M. Ricklefs, S. F. Porcella, L. D. Parkins, S. B. Beres, D. S. Campbell, T. M. Smith, Q. Zhang, V. Kapur, J. A. Daly, L. G. Veasy, and J. M. Musser. 2002. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc. Natl. Acad. Sci. USA 99:4668-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spanier, J. G., and P. P. Cleary. 1980. Bacteriophage control of antiphagocytic determinants in group A streptococci. J. Exp. Med. 152:1393-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tiraby, J. G., E. Tiraby, and M. S. Fox. 1975. Pneumococcal bacteriophages. Virology 68:566-569. [DOI] [PubMed] [Google Scholar]
- 24.Weeks, C. R., and J. J. Ferretti. 1984. The gene for type A streptococcal exotoxin (erythrogenic toxin) is located in bacteriophage T12. Infect. Immun. 46:531-536. [DOI] [PMC free article] [PubMed] [Google Scholar]