Abstract
Actinobacillus pleuropneumoniae is a strict respiratory tract pathogen of swine and is the causative agent of porcine pleuropneumonia. We have used signature-tagged mutagenesis (STM) to identify genes required for survival of the organism within the pig. A total of 2,064 signature-tagged Tn10 transposon mutants were assembled into pools of 48 each, and used to inoculate pigs by the endotracheal route. Out of 105 mutants that were consistently attenuated in vivo, only 11 mutants showed a >2-fold reduction in growth in vitro compared to the wild type, whereas 8 of 14 mutants tested showed significant levels of attenuation in pig as evidenced from competitive index experiments. Inverse PCR was used to generate DNA sequence of the chromosomal domains flanking each transposon insertion. Only one sibling pair of mutants was identified, but three apparent transposon insertion hot spots were found—an anticipated consequence of the use of a Tn10-based system. Transposon insertions were found within 55 different loci, and similarity (BLAST) searching identified possible analogues or homologues for all but four of these. Matches included proteins putatively involved in metabolism and transport of various nutrients or unknown substances, in stress responses, in gene regulation, and in the production of cell surface components. Ten of the sequences have homology with genes involved in lipopolysaccharide and capsule production. The results highlight the importance of genes involved in energy metabolism, nutrient uptake and stress responses for the survival of A. pleuropneumoniae in its natural host: the pig.
Actinobacillus pleuropneumoniae causes porcine pleuropneumonia, a highly contagious, often fatal, respiratory disease. Pleuropneumonia can occur in pigs of all ages, and there are no known associations with predisposing viral or bacterial infections. A. pleuropneumoniae is an obligate parasite of the porcine respiratory tract, and transmission is by aerosol or direct contact with infected pigs. The presentation of disease may be acute, where it is characterized by increasingly severe pulmonary distress that may progress rapidly to death, or chronic infection where it results in a failure to thrive.
Treatment of pigs with acute pleuropneumonia requires parenteral antibiotics (ideally selected after antibiotic resistance testing of the isolated strain), but this is labor intensive, time consuming, and expensive, and can be of limited use due to the rapid progression of the disease. Evidence from field and experimental studies indicates that infection with one serotype of A. pleuropneumoniae provides complete protection against subsequent infection with the homologous serotype and at least partial protection against heterologous infection (19, 37, 68), suggesting that vaccination might be a feasible alternative to antibiotic treatment. However the development of an effective vaccine has been hampered by the antigenic diversity observed between the 15 different serotypes of A. pleuropneumoniae, and existing whole-cell vaccines provide limited protection (17, 31).
To improve our understanding of the pathogenesis of A. pleuropneumoniae infection and to identify genes necessary for virulence, we have performed a large-scale signature-tagged mutagenesis (STM) study. STM is a refinement of classical transposon mutagenesis that allows multiple individually sequence-tagged transposon mutants to be screened in a single animal. Since its first description in 1995 (41), STM has been used to study the in vivo growth and pathogenesis of a variety of microorganisms. These screens have resulted in the identification of large numbers of both classical virulence genes, and what might otherwise be described as housekeeping genes that are essential for growth in vivo. By necessity, STM screens of primary human pathogens have relied on surrogate animal models of infection, typically the mouse. It is clear from looking at these studies that similar genes or biosynthetic pathways are required by different pathogens for survival in the murine host. However, one limitation of this approach is that it may not identify genes involved in specific interactions of a pathogen with its natural host. Here we describe the application of STM to A. pleuropneumoniae to identify genes involved in growth and survival in its natural host, the pig. To our knowledge this is the largest STM screen in a large animal setting with a natural bacterial pathogen.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
A spontaneously nalidixic acid-resistant derivative of the virulent A. pleuropneumoniae serotype 1 strain 4074 was selected using conventional methods and designated 4074 Nalr. The virulence of this strain was verified by passage in pigs. A. pleuropneumoniae strains were routinely propagated at 37°C with 5% CO2 on brain heart infusion plates supplemented with 10% Levinthal's base (BHIL) or at 37°C in Columbia broth with β-NAD (5 μg/ml) and 11 mM CaCl2. The Escherichia coli strains used in this study were S17 λ pir [recA thi pro hsdR−M+ RP4::2-Tc::Mu::Km Tn7 lysogenized with λ pir phage] (62), CC118 λ pir [Δ(are-leu) araD ΔlacX74 galE galK phoA20 thi-1 rpsE rpoB argE recA1 lysogenized with λ pir phage] (22) and XL1-Blue {recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIq ZΔ M15 Tn10 (Tetr)]}. E. coli strains were maintained in Luria-Bertani (LB) medium with appropriate antibiotics. For both E. coli and A. pleuropneumoniae, the following antibiotics were used at the indicated concentrations: ampicillin, 100 μg/ml; kanamycin, 100 μg/ml; and nalidixic acid, 20 μg/ml.
DNA manipulations, amplification, labeling, and hybridization of DNA tags.
Chromosomal DNA was isolated from A. pleuropneumoniae by using a QIAamp mini DNA kit (Qiagen) and plasmid DNA isolated from E. coli by using a Qiagen plasmid midi kit, according to the manufacturer's instructions. DNA manipulation was performed according to standard techniques (87). Probes for Southern blots were labeled with digoxigenin (DIG)-11-dUTP by using a PCR DIG Probe synthesis kit as described by the manufacturer (Roche). Amplification and labeling of DNA signature tags was performed essentially as described by Hensel et al. (41). Tags were initially amplified from genomic DNA by PCR with primers P6 (5′-CGACTACAACCTCAAGCT-3′) and P7 (5′-CGACCATTCTAACCAAGC-3′). The amplified tags were purified on a 2% Nusieve GTG agarose gel and radiolabeled with [32P]dCTP in a second PCR using P6 and P7. The cycling conditions for both PCRs were as follows: denaturation and enzyme activation for 12 min at 95°C, followed by 20 cycles of 95°C for 30 s, 50°C for 45 s, and 72°C for 10 s. Reactions were performed using 1 U of AmpliTaq Gold (Perkin-Elmer) in a Perkin-Elmer 2400 thermocycler. Prior to use, the invariant flanking DNA was separated from the variable signature tag by digestion with HindIII for 18 h at 37°C.
For colony blots, E. coli S17 λ pir strains harboring each of the pLOF/TAG1-48 plasmids were grown to mid-exponential phase and 100 μl of each culture was transferred to the wells of a microtiter dish. Bacteria were transferred from the microtiter dish to a nylon membrane (Hybond N) using a replica plater (Sigma) and the inoculated membrane was placed on the surface of a dried LB agar plate containing ampicillin (100 μg/ml). After overnight growth at 37°C, the membranes were treated with 1.5 M NaCl-0.5 M NaOH for 10 min. DNA was fixed to the membrane by microwaving at maximum power for 30 s, and bacterial debris was removed by washing in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Prehybridization and hybridization were performed at 65°C in Rapid-hyb buffer (Amersham) for 1 h and overnight, respectively. Blots were washed according to standard procedures. To identify potentially attenuated mutants, hybridization signals on a blot probed with labeled tags prepared from the input pool (inoculum) were compared to those on identical blots hybridized with labeled tags prepared from the corresponding output pools from at least two different animals.
Construction of DNA signature tags and pLOF/TAG1-48.
Double-stranded DNA signature tags were obtained by PCR amplification of the variable oligonucleotide pool RT1 (41) using primers P3SAL (5′-CGCCATGTCGACCATTCTAACCAAGCTT-3′) and P5SAL (5′-CGCCTAGTCGACTACAACCTCAAGCTT-3′) that contain recognition sites for the restriction endonuclease SalI at their 5′ termini. To facilitate cloning of the signature tags in plasmid pLOF/Km, the plasmid was modified to incorporate a unique SalI site. Briefly, complementary oligonucleotides SALTOP (5′-GTCGACCCT-3′) and SALBTM (5′-GTCGACAGG-3′) were annealed to give a double-stranded linker containing a SalI site, flanked by SfiI-compatible ends. Plasmid pLOF/Km was digested with SfiI, treated with calf intestinal alkaline phosphatase, and ligated to the phosphorylated double-stranded linker to give plasmid pLOF/Sal. The PCR-amplified double-stranded DNA signature tags were digested with SalI; gel purified; and cloned into SalI-digested, calf intestinal alkaline phosphatase-treated pLOF/Sal to generate a library of uniquely tagged pLOF/TAG plasmids. Individual transformants in E. coli strain CC118 λ pir were screened by colony blot hybridization with their corresponding [32P]dCTP-labeled tags to identify 100 tags that amplified and labeled efficiently. These tags were then tested for absence of cross-hybridization and 48 plasmids (pLOF/TAG1-48) were selected for further use.
Construction of the STM bank.
The plasmid pLOF/Km was modified to accept signature tags by the insertion of a unique SalI site in the mini-Tn10 element. PCR amplified random signature tags were cloned into the SalI site, and a bank of ca. 30,000 tagged plasmids in E. coli CC118 λ pir was obtained. These transformants were screened by colony blotting with their corresponding radiolabeled tags, and signature-tagged plasmids with strong hybridization signals were selected. Plasmids with cross-hybridizing tags were eliminated and a pool of 48 unique signature-tagged plasmids was assembled. These plasmids (pLOF/TAG1-48) were transformed into E. coli S17 λ pir and independently transferred to A. pleuropneumoniae strain 4074 Nalr by conjugation. A. pleuropneumoniae strain 4074 Nalr and E. coli S17 λ pir harboring each of the pLOF/TAG1-48 plasmids were grown overnight at 37°C to the stationary phase of growth. Bacteria were harvested by centrifugation for 15 min at 3,000 × g, washed twice, and resuspended to the original volume in cold 10 mM MgSO4. A. pleuropneumoniae recipient (400 μl) and E. coli donor (100 μl) cells were mixed, concentrated, and spread on to 0.22-μm-pore-size filters. Filters were placed onto BHIL plates containing 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and incubated at 37°C with 5% CO2 for 5 h. Thereafter, the bacteria were removed into 1 ml of sterile phosphate-buffered saline and aliquots were plated on to BHIL supplemented with nalidixic acid (20 μg/ml) and kanamycin (100 μg/ml). Four separate matings were performed for each of the 48 signature-tagged mini-Tn10 transposons in order to reduce the likelihood of selecting siblings. Transconjugant mutants from the four matings were combined for each tag, and all mutants were screened for susceptibility to ampicillin to eliminate strains carrying cointegrants of the suicide vector, inserted into the chromosome. In total, 2,064 transposon mutants from 48 individual matings were isolated and arranged into 43 pools of 48 mutants. Mutants were stored in 1.5-ml cryotubes with 15% glycerol at −80°C.
Infection studies.
Pools of 48 A. pleuropneumoniae signature-tagged mutants were grown individually until the mid-exponential phase of growth. At an optical density at 600 nm of 0.3, the individual mutant strains were combined and the pooled culture was diluted in 10 mM HEPES (pH 7.4) to 3 × 106 CFU/ml. Large White Cross piglets, 6 to 10 weeks old and determined to be A. pleuropneumoniae free by bacteriological and serological analyses, were inoculated intratracheally with 3 ml of the diluted culture. The number of CFU in the inoculum was verified by viable counts after plating serial dilutions of the inoculum on selective BHIL. Each mutant pool was used to infect at least two animals. At approximately 24 h postinfection, surviving animals were humanely killed and the lungs were removed. In initial experiments, surviving bacteria were recovered from infected lungs by extensive lavage with Hanks buffered saline solution (HBSS). However, in later experiments lungs were homogenized in 500 ml of HBSS. Aliquots of the bacterial suspension were plated on selective media and following growth overnight at 37°C with 5% CO2, approximately 10,000 colonies were pooled (the “output pool”) and total chromosomal DNA was prepared.
Competitive indices and virulence studies.
To determine the in vivo competitive index (CI) of 14 selected STM mutants, mutant and wild-type bacteria were grown in Columbia broth with NAD (5 μg/ml) and 11 mM CaCl2 to an optical density at 600 nm of 0.3. The cultures were then combined to give approximately equal numbers of mutant and wild-type organisms, diluted to approximately 3 × 106 CFU/ml, and inoculated intratracheally into at least two animals. The ratio of mutant to wild-type bacteria in the inoculum was verified by viable counts after plating serial dilutions of the inoculum in parallel on BHIL plus nalidixic acid (20 μg/ml) (mutant and wild type) and BHIL plus nalidixic acid (20 μg/ml) plus kanamycin (100 μg/ml) (mutant only). Approximately 24 h postinfection, surviving animals were killed and the lungs were removed. The lungs were homogenized in 500 ml of HBSS and aliquots were plated on selective media (see above) to recover surviving bacteria. The in vivo CI was calculated as the output ratio of mutant to wild type divided by the input ratio of mutant to wild-type organisms.
In vitro CIs were determined for all STM mutants identified as consistently attenuated in 4 pigs. Essentially, mutant and wild-type organisms were grown to mid-exponential phase as described above and mixed in equal numbers and the combined culture was incubated, with agitation, at 37°C for a further 3 h. Samples taken at 0 and 3 h were diluted and plated on selective BHIL, and the in vitro CI was calculated as described above.
Virulence gene identification and DNA sequencing.
To identify the genes in which insertions attenuate bacterial survival in vivo, DNA flanking the site of transposon insertion was sequenced after cloning or amplification by inverse PCR. For cloning, chromosomal DNA was digested with a panel of restriction enzymes, separated by agarose gel electrophoresis, transferred to a nylon membrane, and probed in a Southern blot with a DIG-labeled probe specific for the Tn10 kanamycin gene. Suitably sized fragments hybridizing to the probe were excised from a gel, purified, and cloned into appropriately digested plasmid vector pBR322. Transformants in E. coli strain XL1-Blue were selected on LB agar containing 100 μg of kanamycin/ml. To facilitate sequencing with the Tn10- specific oligonucleotide primer BS401 (5′-GACAAGATGTGTATCCACC-3′), the cloned insert was further subcloned into pBCKS (Stratagene). Inverse PCR amplification of DNA flanking the site of transposon insertion was performed essentially as described previously (28). Briefly, chromosomal DNA was digested with SspI (which cuts once in the transposon), and 150 ng was self-ligated for 15 min using the Rapid DNA ligation kit (Roche). The ligated DNA was then amplified using the primer pairs BS401-TEF48 and BS401-TEF62. Reaction conditions for inverse PCR were as described previously. The resultant amplicons were purified following agarose gel electrophoresis using a QIAQUICK gel extraction kit (Qiagen). DNA sequencing reactions were performed using the BigDye terminator cycle sequencing kit (Perkin-Elmer). Sequences from each of the attenuated mutants were used to query the nonredundant public DNA and protein databases using the BLAST set of similarity search programs (2).
RESULTS AND DISCUSSION
Generation of signature-tagged mutants of A. pleuropneumoniae.
A total of 2,064 transposon mutants from 48 individual matings were isolated and arranged into 43 pools of 48 mutants. Southern blot analysis of ten randomly chosen exconjugants with a mini-Tn10-specific probe confirmed single, random insertion of the tagged transposon (data not shown). However, subsequent sequence analysis of attenuated clones revealed that mini-Tn10 insertion in A. pleuropneumoniae was not entirely random (see below).
STM screen of A. pleuropneumoniae in pigs.
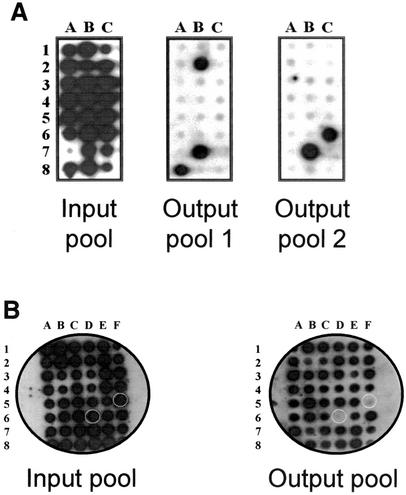
Signature-tagged A. pleuropneumoniae mutants were screened using a porcine intratracheal infection model to identify genes required for virulence. Bacteria were instilled directly into the trachea as previous studies have shown that aerosol particles produced by sneezing are small enough to penetrate into the lower respiratory tract, obviating the need for colonization of the upper respiratory tract (46). A range of experimental factors including the infecting dose and the method of recovery were investigated before we could reproducibly recover the majority of individual mutants following in vivo selection in the porcine host. Using challenge doses of 103, 104, or 105 CFU, there was a poor correlation between the infecting dose and the appearance of clinical signs of disease. Moreover, when different pigs were infected with the same pool, the hybridization patterns of the postpassage pool suggested that different clones were missing after passage of a given pool through different animals (data not shown). This inconsistency was overcome by increasing the challenge dose to 106 to 107 CFU/pig. In an earlier STM study of A. pleuropneumoniae infection in pigs (29), bacteria were recovered by lung lavage following experimental infection. However, we found this method to be unreliable, yielding essentially random recovery of individual signature tags. In contrast, we observed consistent recovery of viable bacteria following homogenization of the entire lung in HBSS. Optimization of these two crucial parameters resulted in the recovery of the majority of individual clones (>90%, 20 to 48 h postinfection), with excellent reproducibility between animals (Fig. 1, compare panels A and B). The initial screen using two pigs per pool identified 550 mutants present in the input pool but missing or significantly reduced in the output pools. Potentially attenuated mutants were reassembled into new pools and each new pool was used to infect a further two animals. Following this second screen, 105 mutants were found to be consistently absent from the output pools and retained for further analysis.
FIG. 1.
Representative hybridizations of paired input and output pools (recovered from pig lungs) of A. pleuropneumoniae STM mutants. (A) Typical results from an initial suboptimal experiment using an input pool of 24 mutants (total inoculum, 105 CFU) instilled intratracheally into two animals with bacteria being recovered by lung lavage 48 h postinfection. Three and two mutant strains were recovered from animal 1 (output pool 1) and animal 2 (output pool 2), respectively. The results are suggestive of clonal expansion. (B) Optimized STM showing recovery of input and output pools from a single animal infected with 107 CFU A. pleuropneumoniae with bacteria being recovered from infected homogenized lungs 24 h postinfection. Hybridization signals for mutants D6 and F5 are not present in the output pool.
Identification and analysis of virulence genes of A. pleuropneumoniae.
Southern blot analysis of all attenuated mutants confirmed the presence of single insertions. The nucleotide sequence of the DNA flanking the site of transposon insertion obtained for each of the 105 attenuated mutants was used to search the GenBank databases for homologous genes. The results of this analysis are shown in Table 1, with the genes assigned to one of five functional categories: cell surface, metabolism, regulation, transport, and stress responses. Insertions in genes encoding proteins with no significant homology to proteins in the databases or homologous to proteins or predicted proteins of unknown function are categorized as unknown. The 105 attenuated mutants contained transposon insertions in 55 individual genes, and a number of hot spots for Tn10 insertion were identified. In particular, seven identical insertions were independently isolated in the A. pleuropneumoniae rfbP gene (Tn insertion at codon 444), five insertions were independently isolated in the apvD (macA) gene homologue (codon 66), and three of four insertions in the putative fur gene were at the same site (codon 128). Interestingly, the DNA sequences flanking the three hot spots identified in this study are similar neither to each other nor to a consensus sequence for Tn10 insertion identified previously in A. pleuropneumoniae (12, 13). However, the hot spot in the rfbP gene was previously identified by Labrie et al. (55), who found 2 of 3 Tn10 insertions in this gene mapped to codon 444.
TABLE 1.
Identification of A. pleuropneumoniae STM mutants
| Class | Strain | Genea | Similarity
|
Known or putative function | CIf
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Geneb | % Identity (% similarity) | Spand | Accession no.e | In vitro | In vivo | ||||
| Cell surface | 9C2 | cpxC | AAD30161 | Capsular polysaccharide export | 1.02 | 3.88 × 10−3 | |||
| 36B1 | cpxB | AAD30162 | Capsular polysaccharide export | 0.77 | |||||
| 26A12 | cpxD | AAD30160 | Capsular polysaccharide export | 0.41 | |||||
| 26A9 | lcbB (Neisseria meningitidis) | 37 (50) | 82 | AAF21951 | LPS or capsule biosynthesis | 1.07 | |||
| 9B7 | galU | AAC28326 | LPS core biosynthesis | 1.75 | |||||
| 25B7 | rfbU | AAG45944 | LPS biosynthesis | 1.40 × 10−3 | |||||
| 10B11 | rmlC | rfbC (Actinobacillus actinomycetemcomitans) | 100 | 15 | AAG49406 | LPS biosynthesis | 0.23 | ||
| 12D5 | rfbN | AAG45942 | LPS biosynthesis | 0.02 | |||||
| 21B8 | rfbP | AAG45943 | LPS biosynthesis | 0.03 | |||||
| 15A9 | PF0798 (Pyrococcus furiosus) | 37 (55) | 270 | NP_578527 | LPS biosynthesis | 0.74 | |||
| 4D4 | lppB (Pasteurella multocida) | 46 (63) | 83 | AAK03698 | Lipoprotein B precursor | 1.87 | |||
| 17A4 | ompP2 (Haemophilus influenzae) | 40 (55) | 237 | Q48221 | OMPP2 precursor | 1.15 | |||
| Metabolism | 10B12 | adpP (Haemophilus influenzae) | 61 (76) | 62 | P44684 | ADP-ribose pyrophosphatase | 1.03 | ||
| 10A11 | argG (Haemophilus influenzae) | 65 (88) | 34 | AAC23373 | Argininosuccinate synthase | 0.80 | |||
| 33C7 | atpA (Haemophilus influenzae) | 89 (95) | 134 | P43714 | ATP synthase | 0.31 | |||
| 17B8 | ccmH (Haemophilus influenzae) | 53 (77) | 166 | P46458 | Cytochrome c biogenesis | 1.10 | |||
| 35D11 | dsbA (Haemophilus influenzae) | 63 (77) | 160 | P31810 | Thiol:disulfide interchange | 1.04 | 3.4 × 10−4 | ||
| 26C3 | hemA (Pasteurella multocida) | 61 (74) | 394 | CAA71452 | Glutamyl-tRNA reductase | 0.05 | |||
| 4B9 | moaA (Haemophilus influenzae) | 67 (74) | 114 | P45311 | Molybdenum cofactor biosynthesis | 1.15 | |||
| 19D5 | mrp | AAD01696 | ATPase | 0.20 | |||||
| 29B11 | napB (Pasteurella multocida) | 55 (70) | 77 | AAK03681 | Periplasmic nitrate reductase | 1.05 | 1.09 | ||
| 0A7 | pntB (Haemophilus influenzae) | 68 (77) | 458 | P43010 | NAD(P) transhydrogenase | 0.79 | |||
| 23C9 | aopA/nqrA | AAC43631 | Na-translocating NADH-ubiquinone oxidoreductase | 0.72 | |||||
| 29A10 | prfC (Haemophilus influenzae) | 73 (81) | 342 | P43928 | Peptide chain release factor | 4.67 | |||
| 29B12 | recR (Haemophilus influenzae) | 81 (87) | 71 | P44712 | Recombination | 3.60 | |||
| 26A10 | thrC (Haemophilus influenzae) | 88 (93) | 190 | P44503 | Threonine synthase | 0.87 | 1.60 | ||
| 0F6 | tonB (Haemophilus ducreyi) | 64 (74) | 288 | O51810 | Energy transducer | 2.49 | 2.2 × 10−2 | ||
| 27A12 | tonB | Y17916 | Energy transducer | 0.90 | 1.54 | ||||
| 26D5 | uroD/hemE (Pasteurella multocida) | 88 (93) | 230 | AAK03818 | Uroporphyrinogen decarboxylase | 0.05 | |||
| 15A11 | visC (Pasteurella multocida) | 55 (82) | 43 | AAK03810 | Monooxygenase | 0.68 | |||
| 26A6 | yibK (Haemophilus influenzae) | 70 (84) | 127 | P44868 | Probable tRNA/rRNA methyltransferase | 0.60 | |||
| 33B7 | yjfH (Haemophilus influenzae) | 77 (85) | 157 | P44906 | Probable rRNA methylase | 0.74 | |||
| 26D12 | guaA (Haemophilus influenzae) | 83 (85) | 43 | P44335 | GMP synthase | 1.59 | |||
| Regulatory | 6C12 | fur (Haemophilus ducreyi) | 79 (83) | 139 | P71333 | Ferric uptake regulator | 0.63 | 2.8 × 10−2 | |
| 26D3 | luxS (Haemophilus influenzae) | 69 (84) | 118 | P44007 | Autoinducer-2 production | 2.06 | |||
| 19B10 | mlcA (Pasteurella multocida) | 45 (69) | 61 | AAK03872 | Negative regulator of rpoE | 1.73 | 6.5 × 10−2 | ||
| Stress | 21D3 | rpoE (Haemophilus influenzae) | 81 (90) | 189 | P44790 | Sigma factor E | 0.79 | 0.88 | |
| 13A3 | dnaJ (Haemophilus ducreyi) | 85 (99) | 14 | P48208 | Chaperone protein | 0.16 | |||
| 26B6 | htpG (Escherichia coli) | 69 (82) | 368 | P10413 | Heat shock protein | 1.25 | |||
| 13D8 | lon (Pasteurella multocida) | 74 (86) | 292 | AAK04062 | ATP-dependent protease | 0.67 | |||
| 13C1 | prc (Pasteurella multocida) | 65 (79) | 290 | AAK02353 | Tail-specific protease | 1.32 | |||
| 22A10 | tig (Pasteurella multocida) | 50 (61) | 152 | AAK04059 | Trigger factor | 0.85 | |||
| 9B5 | pnp (Pasteurella multocida) | 75 (79) | 125 | AAF68421 | Polynucleotide phosphorylase | 1.18 | |||
| Transport | 9D10 | corC (Haemophilus influenzae) | 66 (78) | 248 | Q57368 | Magnesium/cobalt efflux protein | 3.38 | ||
| 19D1 | PM1728 (Pasteurella multocida) | 79 (89) | 110 | AAK03812 | Probable ABC transporter | 1.34 | |||
| 0C5 | mglA (Pasteurella multocida) | 81 (86) | 242 | P44884 | Galactoside ATP transporter | 0.65 | 1.8 × 10−2 | ||
| 0D6 | PM0997 (Pasteurella multocida) | 56 (76) | 210 | AAK03081 | Probable membrane protein | 1.27 | |||
| 32A7 | PM0996 (Pasteurella multocida) | 60 (73) | 370 | AAK03080 | Probable ABC transporter | 0.96 | |||
| 13B12 | yfeB (Pasteurella multocida) | 75 (86) | 185 | AAK02483 | Iron(III) ATP-binding protein | 0.92 | |||
| 35D1 | znuA (Haemophilus ducreyi) | 71 (78) | 176 | AAF00116 | Zinc ABC transporter | 0.45 | |||
| Unknown | 14D5 | No homology A | Unknown | 0.95 | 5.3 × 10−3 | ||||
| 2D5 | No homology B | Unknown | 1.25 | 1.0 × 10−2 | |||||
| 32A11 | PM1184 (Pasteurella multocida) | 31 (52) | 124 | AAK03268 | Unknown | 2.95 | 0.99 | ||
| 4C1 | PM0270 (Pasteurella multocida) | 42 (59) | 327 | AAK02354 | Unknown | 1.11 | 0.94 | ||
| 9A4 | HI0636 (Haemophilus influenzae) | 53 (74) | 91 | P44027 | C4-decarboxylate transport protein homologc | 1.02 | |||
Known A. pleuropneumoniae genes.
Genes with sequences in public databases with highest similarity to STM sequences. Names of source bacteria are shown in parentheses. In some instances, gene designations are from whole genome sequences, giving a two-letter code for the bacterium followed by the sequence number (e.g., PF0798).
The transposon insertion in 9A4 maps to the last codon of a gene encoding a C4-decarboxylate transport protein homolog. However, the downstream sequence (likely affected by the Tn insertion) shows no homology to any known genes.
Span = number of nucleotides over which similarity was detected.
Genbank accession numbers listed are for the sequences similar to the STM sequences, not for the STM sequences themselves.
The CI was calculated as the ratio of (mutant CFU/wild-type CFU) input/(mutant CFU/wild-type CFU) recovered (41).
Of the 55 mutants found to be consistently attenuated in four pigs, only 11 mutants showed a >2-fold reduction in growth in vitro compared to the wild type (Table 1). Mutations located in three genes involved in lipopolysaccharide (LPS) biosynthesis (rfbU, rfbN, and rfbP) and two genes involved in heme biosynthesis (hemA and uroD) resulted in the lowest in vitro CIs, with a >10-fold reduction in growth. The attenuation of these mutants is thus at least in part due to a general growth defect, although it is possible that they are even more attenuated in vivo.
Due to the large number of attenuated mutants identified in the screening process, it was not possible to determine an in vivo CI for each one. Therefore we selected mutants representing each group of affected genes. In calculating the level of attenuation in vivo, we included mutant 9C2 (cpxC::km) as a control, since capsular mutants are known to be highly attenuated in vivo (81, 100). This mutant gave an in vivo CI of 3.88 × 10−3. A further 7 of 14 mutants tested were confirmed as highly attenuated in vivo, with CIs ranging from 6.50 × 10−2 to 3.43 × 10−4 (Table 1). However, the remaining six mutants did not appear attenuated when administered in equal amounts with the wild type despite consistent lack of recovery from the mixed pools. Similar results have been found in other STM studies (4, 60), and may indicate that the affected genes have more subtle influences on virulence that are not seen at higher doses, or in less diverse populations. Alternatively these genes may be false positives from the STM screen. Further in vivo analysis of these and the remaining 41 mutants will help to determine the relative importance of the affected genes in pathogenesis of A. pleuropneumoniae infection.
Some new insights into the requirements of this bacterium during acute infection are indicated through identification of highly attenuated mutants, as well as through groups of mutants in which affected genes share related functions. In particular, genes involved in expression of cell surface antigens, energy metabolism, nutrient uptake, and stress response are highlighted below.
Production of a polysaccharide capsule is a common virulence mechanism of bacterial pathogens and previous studies have demonstrated a key role for capsular polysaccharide in the pathogenesis of A. pleuropneumoniae infection (100). Sequence analysis of three attenuated strains (36B1, 9C2, and 26A12) showed that the transposon had inserted in genes involved in the export of capsular polysaccharide (cpxB, cpxC, and cpxD, respectively [99]). In A. pleuropneumoniae, the cpx genes are arranged in an operon in the order cpxD, cpxC, cpxB, cpxA, and it is likely that transposon insertions within the operon would have polar effects on downstream genes.
Alterations in LPS are also known to cause reductions in A. pleuropneumoniae virulence (80), and 5 attenuating lesions were identified in genes implicated in A. pleuropneumoniae O-antigen biosynthesis. Mutant strains 10B11, 25B7, 15A9, 12D5, and 14A1 contain, respectively, insertions in open reading frames (ORFs) 9, 12, 15, 17, and 18 of the A. pleuropneumoniae O-antigen biosynthesis operon (55). These genes encode putative rhamnosyl transferases (rfbC and rfbN), putative glycosyl transferases (rfbU and an unnamed gene), and a putative undecaprenyl-phosphate galactose phosphotransferase gene (rfbP). Interestingly, Labrie et al. (55) found a mutant disrupted in the rfbP gene was not attenuated and had growth in vitro similar to that of the wild-type strain. In contrast, we isolated seven independent mutants defective in this gene that were all identified as attenuated in four pigs each. In addition, we found that this mutant as well as the rfbC, rfbU, and rfbN mutants showed reduced growth in vitro compared to that of the wild type (Table 1). In strain 9B7, the transposon has inserted immediately upstream of galU, a gene known to be involved in synthesis of the LPS core, as well as in virulence, of A. pleuropneumoniae (79). Finally, mutant 26A9 contains an insertion in a gene homologous to lcbB, a putative bifunctional polymerase implicated in LPS or capsule biosynthesis in Neisseria meningitidis serogroup L (GenBank reference no. AF112478 [unpublished data]).
Strains 9A5 and 18A12 contain transposon insertions in genes encoding putative lipoproteins. The disrupted sequence in strain 9A5 encodes a protein with greatest homology to an unknown protein from Pasteurella multocida, but is also similar to lipoproteins and putative lipoproteins of Haemophilus influenzae and Haemophilus somnus (92). H. somnus LppB binds Congo red dye (a structural analogue of heme) and has been suggested to be a virulence factor, but this has not been formally proven (92). In strain 18A12, the transposon has inserted in a gene whose product shares homology with the NlpI lipoprotein gene of E. coli (71). Mutations in NlpI affect cell morphology and render cells osmosensitive. In E. coli and P. multocida the nlpI gene is located immediately downstream from pnp (see mutant 9B5 below), but may be transcribed independently from pnp, at least in E. coli (97). Finally, strain 25B12 contains an insertion in an ORF encoding a protein with homology to the P2 porin of H. influenzae (24). The P2 protein constitutes approximately one-half of the total outer membrane protein of H. influenzae and represents an important target of the immune response to nontypeable H. influenzae infection (103).
Interestingly, in this STM screen, 10 of 21 attenuating mutations identified in genes encoding metabolic functions are located in genes involved in energy metabolism and/or redox balancing. In E. coli, disulfide bond formation in exported proteins is catalyzed by DsbA and DsbB, and is directly coupled to the electron transport chain via reoxidation of DsbB by either ubiquinone or menaquinone, which are in turn reoxidized by cytochrome terminal oxidases (5, 6). Mutant 35D11, with the transposon inserted in dsbA, was 10 times more attenuated than the capsule mutant in vivo, yet showed no difference in growth compared to the wild type in vitro. Mutations in dsbA are pleiotropic, affecting the production of a variety of translocated proteins including bacterial virulence factors, components of type III secretion machinery, and c-type cytochromes such as periplasmic nitrate reductase (NAP) (104). The catalogue of proteins processed by DsbA have not yet been determined in A. pleuropneumoniae, and it may be that the marked reduction in virulence of the DsbA mutant is due to the combined effects of two or more altered proteins. As mentioned previously, ubiquinone, a central component of the electron transport chain under aerobic conditions, functions in the formation of disulfide bonds in periplasmic proteins by facilitating the reoxidation of DsbA by DsbB (5, 51). In strain 15A11, the transposon has inserted in a gene homologous to P. multocida visC, putatively encoding a monooxygenase belonging to the UbiH/CoQ6 family. UbiH is required for the biosynthesis of ubiquinone (66).
One of the proteins likely processed by DsbA is NAP. Strain 29B11 contains a transposon insertion in a napB gene homologue, encoding the smaller, diheme cytochrome c subunit of heterodimeric NAP. As in H. influenzae and Pseudomonas spp, NAP is the sole nitrate reductase in A. pleuropneumoniae and may function to support anaerobic growth in the presence of nitrate (11, 15, 25). Although the in vivo CI of the napB mutant indicates it is not attenuated at a high infectious dose, NAP may play a role in redox balancing, as well as in adaptation to anaerobic metabolism after transition from aerobic conditions, and the utilization of alternate reductants (65). Several of the other inactivated genes identified in this study could also affect the production of NAP, in addition to having other functions. Strain 4B9 contains a transposon insertion in a moaA gene homologue. In E. coli the moaABCDE genes are required for the synthesis of molybdopterin (molybdopterin guanine dinucleotide) from GTP (82). Molybdopterin is the principal cofactor found in prokaryotic molybdoproteins including NAP. Expression of the moa genes is induced by anaerobiosis (3), and Salmonella enterica serovar Typhi moaA mutants are impaired in their ability to replicate within epithelial cells (18). Strain 17B8 contains an insertion in a gene homologous to H. influenzae ccmH, required for the assembly of c-type cytochromes, including NAP, in enteric bacteria (53, 94). CcmH is encoded by the final gene of the ccm operon and with CcmF and CcmG is believed to catalyze the reduction of disulfide bonds in the cytochrome c apoprotein and to facilitate heme attachment (53). Hemes including protoheme are key components of all cytochromes including cytochrome c, and in addition, play important roles as enzyme prosthetic groups in mineral nutrition and oxidative catalysis (9). Two genes involved in heme synthesis, glutamyl-tRNA reductase (hemA) and uroporphyrinogen decarboxylase (uroD/hemE), have been disrupted in strains 26C3 and 26D5, respectively. Glutamyl-tRNA reductase catalyses the reduction of glutamate to glutamate 1-semialdehyde, the first committed step unique to the formation of δ-aminolevulinate (ALA) from glutamate. Downstream from ALA formation, uroporphyrinogen decarboxylase catalyses the decarboxylation of uroporphyrinogen III to yield coproporphyrinogen, the first step in the conversion of uroporphyrinogen III to protoheme via protoporphyrin IX. The critical importance of heme biosynthesis in A. pleuropneumoniae is highlighted by the fact that both the hemA and hemE mutants showed growth defects in vitro.
In strain 0A7, the transposon has inactivated a homologue of the pntB gene that encodes the beta subunit of NAD(P) transhydrogenase. Transhydrogenase couples the redox reaction between NAD(H) and NADP(H) to the translocation of protons across the inner membrane, and plays a key role in energy metabolism, biosynthetic and catabolic activities of the cell (73). The A. pleuropneumoniae gene aopA is inactivated in strain 23C9. The aopA gene has been identified as encoding a 48 kDa immunogenic outer membrane protein common to all serotypes of A. pleuropneumoniae tested (20). Interestingly, AopA is highly homologous (76% identity, 85% similarity over the entire protein sequence) to Nqr1 of H. influenzae (NqrA of Vibrio alginolyticus), encoding the alpha chain of the sodium-translocating NADH-ubiquinone oxidoreductase (NQR) complex (10, 40). The NQR enzyme is a respiration-linked Na+ pump establishing an electrochemical gradient of sodium ions across the membrane. The resultant sodium motive force can be used for solute transport, ATP synthesis, and flagellar rotation. This alternative energy coupling of sodium ions rather than protons enables the bacteria to maintain a cytoplasmic pH near neutrality in an alkaline environment (39). The enzyme ATP synthase is responsible for utilizing the proton electrochemical gradient across the cytoplasmic membrane for ATP synthesis in cells growing aerobically or anaerobically. In strain 33C7, the attenuating lesion was found to occur in a gene homologous to H. influenzae atpA, encoding the F1 alpha subunit of F1F0 ATP synthase. In P. multocida, all eight subunits of the bacterial enzyme are encoded by the atp operon, with atpA flanked by atpH and atpG (61). This gene order is the same in A. pleuropneumoniae, and both atpH and atpG have been previously implicated in the pathogenesis of A. pleuropneumoniae infection (29).
Transduction of energy from the proton motive force (PMF), generated by the electrochemical gradient across the cytoplasmic membrane, to the outer membrane occurs via the cytoplasmic membrane-localized TonB-ExbB-ExbD complex (42, 64). This complex plays a central role uptake of iron and vitamin B12 by coupling energy from the proton motive force to high-affinity outer membrane receptors (42, 64). Some bacterial pathogens, including Vibrio cholerae and Pseudomonas aeruginosa, possess two independent TonB systems that are not entirely redundant in function (70, 105). Our STM screen identified transposon insertions in 2 distinct tonB loci in A. pleuropneumoniae. Mutant 27A12 contains a transposon insertion in a gene previously identified in A. pleuropneumoniae as tonB (96) (now referred to as tonB1). This gene is found immediately upstream of, and is cotranscribed with, exbB, exbD, and the transferrin binding protein (tbp) genes. In contrast, strain 0F6 contains a transposon insertion in an ORF encoding a protein with homology to TonB proteins from P. multocida and Haemophilus sp. (now designated tonB2). Moreover, sequence analysis of the region upstream from the transposon insertion in 0F6 revealed the presence of distinct exbB and exbD gene homologues, suggesting that A. pleuropneumoniae, in common with other bacterial pathogens, including V. cholerae and P. aeruginosa, possesses two independent TonB systems (70, 105). Results of the in vivo CI experiments indicate that mutation of tonB2, but not tonB1, leads to attenuation under conditions of acute infection. This indicates that the two systems may serve different functions in vivo.
The remaining mutants in the metabolic category are affected in various pathways. Strain 10B12 contains a transposon insertion in an ADP-ribose pyrophosphatase (adpP) gene homologue. The E. coli ADP-ribose pyrophosphatase catalyzes the hydrolysis of ADP-ribose to ribose-5-P and AMP, compounds that can be recycled as part of nucleotide metabolism (32). In strain 26D12 the transposon has inactivated a GMP synthase, guaA, gene homologue. GMP synthase catalyzes the synthesis of GMP from XMP as part of the purine biosynthetic pathway. Mutations in the guaBA operon attenuate the virulence of a variety of enteric pathogens, including E. coli, Shigella flexneri, and S. enterica serovar Typhi, and a ΔguaBA S. enterica serovar Typhi derivative shows promise as a live attenuated vector (69, 86, 98). Moreover, previous STM screens in Streptococcus agalactiae and P. multocida have implicated guaA and guaB, respectively, in growth in vivo and pathogenesis (28, 45). In strain 26A10 the transposon has inserted in a gene with extensive homology to thrC of H. influenzae. The thrC gene encodes threonine synthase, the final enzyme in the biosynthetic pathway leading to the production of threonine (72). Mutations in a Streptococcus pneumoniae thrC homologue attenuate the virulence of this organism in a murine model of pneumoniae (56). Strain 10A11 carries an insertion in an argininosuccinate synthase, argG, gene homologue. Argininosuccinate synthase catalyses the conversion of citrulline and aspartate into argininosuccinate, linking the urea cycle and the citric acid cycle via the so-called aspartate-argininosuccinate shunt. The aspartate-argininosuccinate shunt provides a metabolic link between the pathways by which amino groups and the carbon skeletons of amino acids are processed (34).
Mutant 29B12 contains an insertion in the A. pleuropneumoniae recR gene homolog. The RecR protein is involved in the RecF pathway of recombination DNA repair: recR mutants are deficient at filling in daughter strand gaps in newly synthesized DNA (54). Mutations in recR delay induction of the SOS response in E. coli and such mutants are sensitive to UV-irradiation and mitomycin C (48). In strains 26A3 and 33B7 the transposon has inserted in genes with homology to the trmH family of tRNA/rRNA methyltransferases. The interrupted ORF in strain 26A3 shows greatest homology to H. influenzae yibK, while that in strain 33B7 shows most similarity to yjfH of H. influenzae. Strain 29A10 contains a transposon insertion in a gene homologous to prfC of H. influenzae. The prfC gene encodes peptide chain release factor 3 (RF3), a GTPase that, in the presence of GTP, catalyses the removal of RF1 and RF2 from the ribosome after translation termination (27). The efficient recycling of RF1 and RF2 by RF3-GTP, although dispensable for viability in prokaryotes, is required for maximum growth rate (49).
In strain 19D5, the transposon has inserted in the A. pleuropneumoniae mrp gene. The partial A. pleuropneumoniae mrp gene sequence available (GenBank reference no. AAD01696) is highly homologous to those of mrp genes from H. influenzae and E. coli. In H. influenzae, the mrp gene has been implicated in the biosynthesis of the Galα(1-4)βGal component of LPS, while in Salmonella enterica serovar Typhimurium, mutations in mrp affect the synthesis of thiamine (43, 74). In A. pleuropneumoniae, the mrp gene is located approximately 1 kb upstream from the apxIVA gene, encoding the in vivo-induced RTX toxin, ApxIV (88). These genes do not appear to be transcriptionally linked due to the presence of a transcriptional terminator downstream of mrp (88), although like the apxIVA gene, mrp was found to be specifically expressed in vivo in A. pleuropneumoniae (30).
Finally, insertions in the A. pleuropneumoniae luxS homologue (strain 26D3) were observed to attenuate A. pleuropneumoniae virulence. The LuxS protein is required for production of an autoinducer molecule (AI-2) implicated in quorum sensing and gene regulation in a number of gram-negative and gram-positive bacteria (8). Recently, LuxS has been shown to have a central metabolic function in the cleavage of S-ribosylhomocysteine as part of S-adenosylmethionine metabolism (101). It was suggested that AI-2 might not be a signal molecule, but rather a metabolite that is released during early growth and may be taken up at later stages as a nutrient (101). This role in metabolism may be more likely to be responsible for the attenuation detected in the STM screen, as secretion of an autoinducer would be complemented by the other mutants within the pool.
In addition to the TonB systems discussed above, A. pleuropneumoniae expresses a number of factors that are involved in the acquisition and uptake of iron (reviewed in reference 14), attesting to the importance of this metal in cellular metabolism. In this study, we have identified yet another possible iron transport protein. In strain 13B12, the transposon has inactivated a gene with homology to yfeB of P. multocida. The Yfe system has been best characterized in Yersinia pestis where it functions as an ATP-binding protein cassette (ABC) transporter of iron and manganese, and is required for full virulence (35). Uptake of iron needs to be tightly regulated since, although iron is essential for cellular metabolism, excess iron is toxic. The ferric uptake regulation (Fur) protein is a transcriptional repressor of iron-regulated promoters that binds to target DNA when excess iron is available (38). The A. pleuropneumoniae homologue of fur was found to be inactivated in strain 6C12, leading to a high level of attenuation in vivo. Fur or Fur-like proteins have been shown to regulate expression of a large number of proteins, including virulence factors, in a variety of pathogens (16, 75, 89). It is likely that the A. pleuropneumoniae Fur similarly regulates expression of a large number of genes. The mutant generated in this study will be useful in dissecting the Fur regulon of A. pleuropneumoniae.
In addition to the putative iron-manganese ABC transport protein identified in strain 13B12, components of six other putative ABC transport proteins were identified as necessary for virulence of A. pleuropneumoniae. ABC transport systems are involved in the transport (uptake or efflux) of a diverse array of macromolecules across the cytoplasmic membrane of bacteria and eukaryotes. These systems usually consist of three basic parts: one or two ATPases, one or two integral membrane proteins and a substrate-specific binding protein. Strains 13B12, 0C5, 19D1, and 32A7 contain transposon insertions in the ATPase component of ABC transporters. The gene inactivated in strain 0C5 encodes a protein with significant homology to MglA of prokaryotic galactose transporters (78). The in vivo CI of this mutant indicates limited availability of certain sugars within the respiratory tract environment. The interrupted ORFs in strains 19D1 and 32A7 are homologous to hypothetical ATPases from transport systems of unknown specificity in P. multocida (PM1728 and PM0996, respectively). In strain 35D1, the transposon has inactivated a gene with extensive homology to znuA of Haemophilus ducreyi and pzp1 of H. influenzae (57-59). ZnuA (PZP1) is a periplasmic zinc-binding protein that plays a key role in zinc uptake. Inactivation of the znuA gene in H. ducreyi caused a significant decrease in virulence in the rabbit model for experimental chancroid (57). In strain 0D6 the transposon has inserted in a gene encoding a protein with homology to putative integral membrane proteins from P. multocida (PM0997) and N. meningitidis (NMB0549). In P. multocida, PM0997 is located immediately upstream from the ATPase inactivated in strain 32A7, suggesting that they are part of the same transport system (61). This gene was also identified by Fuller et al. (29) as apvD, the predicted product of which shows homology with the macrolide-specific ABC-type efflux protein MacA (50). In mutant 9D10, the disrupted gene sequence encodes a protein with homology to CorC that mediates both influx and efflux of magnesium as well as cobalt efflux (33). Thus, not all of the ABC transport proteins identified are involved in nutrient acquisition.
Members of the third group of genes of related function identified as necessary for full virulence of A. pleuropneumoniae encode proteins involved in stress response. In strain 13A3, the transposon has inserted in a dnaJ gene homologue. DnaJ is an essential component of bacterial Hsp70 chaperone systems where it functions as a cochaperone, providing substrate specificity to its Hsp70 partner, DnaK (85). In addition to their fundamental roles in protein folding and translocation, DnaK and DnaJ constitute the primary stress-sensing and transduction systems of the E. coli heat shock response (95) where they modulate the induction of the heat shock response by altering the stability of the heat shock sigma factor σ32. Significantly, mutations in DnaK have previously been shown to attenuate A. pleuropneumoniae virulence, and to decrease the survival of Brucella suis in human macrophage-like cell lines (29, 52). Strain 26B6 contains an insertion in an htpG gene homologue. HtpG is a heat shock-induced molecular chaperone that, in E. coli, contributes to the correct folding of cytoplasmic proteins in mildly stressed cells (93). It has been suggested that HtpG serves as a holder chaperone, transiently maintaining a subset of de novo synthesized proteins in a conformation that is accessible to DnaK and DnaJ (26). In strain 22A10, the transposon has inserted in a trigger factor (tig) gene homologue that was identified in a previous STM study of A. pleuropneumoniae (29). Trigger factor is a molecular chaperone with peptidyl-prolyl isomerase activity. In E. coli, trigger factor and DnaK cooperate to promote the folding of a variety of proteins (23). A lon gene homologue was also identified as containing the transposon insertion in strain 13D8. Lon (also called protease La) is a heat shock-inducible, ATP-dependent serine protease that plays a major role in the elimination of abnormally folded proteins (36, 91). In addition to its demonstrated role as a stress response protease, Lon also functions as a pleiotropic regulator of gene expression by degrading unstable regulatory proteins, including RcsA, a positive regulator of capsule synthesis (47), and SulA, an inhibitor of cell division and component of the E. coli SOS response (63). Recent studies in Brucella abortus have demonstrated that Lon is required for survival in murine macrophages and wild-type virulence in a mouse model of infection (83). Strain 13C1 carries a transposon insertion in a prc gene homologue. Prc is a periplasmic protease, the natural substrates of which appear to be membrane proteins, including TonB (36). It has been speculated that Prc functions as a chaperone involved in the folding and turnover of proteins in the periplasmic space (7). Strain 9B5 contains an insertion in the pnp gene encoding polynucleotide phosphorylase (PNPase). PNPase is a processive, cold shock-inducible, inorganic phosphate-dependent exoribonuclease, a component involved in mRNA degradation (77). In E. coli and Yersinia enterocolitica, PNPase function is required for the resumption of normal cell growth after cold shock (67, 102). Mutations in pnp affect the virulence of related organisms, including P. multocida and N. meningitidis (28, 90). Finally, strains 21D3 and 19B10 contain transposon insertions in rpoE and downstream mclA (rseA) gene homologues, respectively. The rpoE-encoded sigma factors σE and σ24 are members of the subfamily of sigma factors that function as effector molecules in response to extracytoplasmic stimuli, including heat shock and oxidative stress (1, 21, 76, 84). In S. enterica serovar Typhimurium, σE is an important determinant of virulence (44). MclA (RseA) is an inner membrane protein that acts as a σE-specific anti-σ factor. MclA (RseA) is rapidly degraded in response to extracytoplasmic stress, resulting in increased σE concentration and initiation of the stress response (1). Surprisingly, although the MclA mutant was confirmed as attenuated by in vivo CI, the RpoE mutant was not. Further investigation will be required to determine the relative importance of this system in pathogenesis of A. pleuropneumoniae.
As in most STM studies, we also identified homologues of genes of unknown function in addition to organism-specific sequences with no known homologues. Two mutants (4C1 and 32A11) with transposon insertions within homologues of P. multocida genes encoding proteins of unknown function were not attenuated at a high infectious dose. In contrast, two other mutants (14D5 and 2D5) that contained transposon insertions in genes that shared no homology with any known sequences were highly attenuated. The transposon insertion in 9A4 maps to the last codon of a gene encoding a C4-decarboxylate transport protein homolog. However, the downstream sequence (likely affected by the Tn insertion) shows no homology to any known genes. Determination of the function of these unknown proteins may provide information critical to our understanding of pathogenesis of A. pleuropneumoniae infection.
Conclusions.
This STM study has resulted in the identification of both known virulence genes, such as those involved in biosynthesis of capsule and LPS, and a large number of genes that were not previously implicated in pathogenesis of A. pleuropneumoniae infection. In total we found 105 of 2,064 (5.1%) mutants to be attenuated, with 55 different genes disrupted. In a previous STM screen of A. pleuropneumoniae (29), 110 of 800 (13.8%) potentially attenuated mutants were found, but only 20 different genes were identified, and of these only 3 (tig, pnp, and apvD/macA) are common to that study and ours. In addition, only one of the genes (mrp) identified in an in vivo expression technology study of A. pleuropneumoniae (30) was also found in our STM screen. These results indicate that neither STM screen has been saturating, and there are likely many more factors to be discovered that contribute to the ability of A. pleuropneumoniae to survive in vivo. Interestingly, Fuller et al. (29) found that 28 of 110 inserts were located in 13 different sites within 23S RNA regions, a factor that they considered to be a barrier to saturation mutagenesis. In contrast, we found no inserts in 23S RNA regions, although three other hot spots were identified. It is possible that differences in the methods of bacterial recovery used influenced the number and type of mutants identified in the two studies.
Although many of the genes encoding metabolic and transport proteins are conventionally considered to be housekeeping genes, we have shown that they are specifically necessary for the survival of A. pleuropneumoniae within the lungs of infected pigs. In particular, several of the identified genes may contribute to the organism's ability to grow under anaerobic conditions. Further analysis of the affected metabolic pathways and nutrient acquisition mechanisms may provide a better understanding of the in vivo environment with regard to availability of oxygen and various nutrients during acute pleuropneumonia. To this end, the forthcoming availability of a whole genome sequence should facilitate identification of metabolic operons.
It is well known that iron is an important nutrient that is limited in vivo and that A. pleuropneumoniae, like many pathogenic bacteria, expresses a number of iron acquisition proteins. In this study, we have identified a second TonB/ExbB/ExbD system in A. pleuropneumoniae. Mutation of tonB2 greatly reduced the virulence of A. pleuropneumoniae, indicating that the other TonB/ExbB/ExbD system does not fully complement the function(s) of the secondary system (manuscript in preparation).
Clearly the ability to respond to, and repair damage caused by, environmental stress is an important contributor to the ability of A. pleuropneumoniae to survive and cause disease. Although stress response mechanisms are considered virulence determinants of numerous intracellular pathogens, their role in pathogenesis of A. pleuropneumoniae has not yet been investigated. Studies are now under way in our laboratory to address this important aspect of host-pathogen interaction.
Acknowledgments
This study was supported by grants from The Wellcome Trust and in part by grants from the BBSRC (to P.R.L., J.S.K., and A.N.R.).
B.J.S. and J.T.B. contributed equally to this work.
Editor: J. N. Weiser
REFERENCES
- 1.Ades, S. E., L. E. Connolly, B. M. Alba, and C. A. Gross. 1999. The Escherichia coli sigmaE-dependent extracytoplasmic stress response is controlled by the regulated proteolysis of an anti-sigma factor. Genes Dev. 13:2449-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 15:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, L. A., E. McNairn, T. Leubke, R. N. Pau, and D. H. Boxer. 2000. ModE-dependent molybdate regulation of the molybdenum cofactor operon moa in Escherichia coli. J. Bacteriol. 182:7035-7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Autret, N., I. Dubail, P. Trieu-Cuot, P. Berche, and A. Charbit. 2001. Identification of new genes involved in the virulence of Listeria monocytogenes by signature-tagged transposon mutagenesis. Infect. Immun. 69:2054-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bader, M., W. Muse, D. P. Ballou, C. Gassner, and J. C. Bardwell. 1999. Oxidative protein folding is driven by the electron transport system. Cell 98:217-227. [DOI] [PubMed] [Google Scholar]
- 6.Bader, M. W., T. Xie, C. A. Yu, and J. C. Bardwell. 2000. Disulfide bonds are generated by quinone reduction. J. Biol. Chem. 275:26082-26088. [DOI] [PubMed] [Google Scholar]
- 7.Bass, S., Q. Gu, and A. Christen. 1996. Multicopy suppressors of prc mutant Escherichia coli include two HtrA (DegP) protease homologs (HhoAB), DksA, and a truncated R1pA. J. Bacteriol. 178:1154-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bassler, B. L. 1999. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr. Opin. Microbiol. 2:582-587. [DOI] [PubMed] [Google Scholar]
- 9.Beale, S. I. 1996. Biosynthesis of hemes, p. 731-748. In F. C. Neidhardt, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella, 2nd ed., vol. 1. ASM press, Washington, D.C.
- 10.Beattie, P., K. Tan, R. M. Bourne, D. Leach, P. R. Rich, and F. B. Ward. 1994. Cloning and sequencing of four structural genes for the Na+-translocating NADH-ubiquinone oxidoreductase of Vibrio alginolyticus. FEBS Lett. 356:333-338. [DOI] [PubMed] [Google Scholar]
- 11.Bedzyk, L., T. Wang, and R. W. Ye. 1999. The periplasmic nitrate reductase in Pseudomonas sp. strain G-179 catalyzes the first step of denitrification. J. Bacteriol. 181:2802-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bender, J., and N. Kleckner. 1992. Tn10 insertion specificity is strongly dependent upon sequences immediately adjacent to the target-site consensus sequence. Proc. Natl. Acad. Sci. USA 89:7996-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bossé, J. T., H. D. Gilmour, and J. I. MacInnes. 2001. Novel genes affecting urease activity in Actinobacillus pleuropneumoniae. J. Bacteriol. 183:1242-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bossé, J. T., H. Janson, B. J. Sheehan, A. J. Beddek, A. N. Rycroft, J. S. Kroll, and P. R. Langford. 2002. Actinobacillus pleuropneumoniae: pathobiology and pathogenesis of infection. Microbes Infect. 4:225-235. [DOI] [PubMed] [Google Scholar]
- 15.Brige, A., J. A. Cole, W. R. Hagen, Y. Guisez, and J. J. Van Beeumen. 2001. Overproduction, purification and novel redox properties of the dihaem cytochrome c, NapB, from Haemophilus influenzae. Biochem. J. 356:851-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calderwood, S. B., and J. J. Mekalanos. 1987. Iron regulation of Shiga-like toxin expression in Escherichia coli is mediated by the fur locus. J. Bacteriol. 169:4759-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiers, K., I. van Overbeke, P. De Laender, R. Ducatelle, S. Carel, and F. Haesebrouck. 1998. Effects of endobronchial challenge with Actinobacillus pleuropneumoniae serotype 9 of pigs vaccinated with inactivated vaccines containing the Apx toxins. Vet. Q. 20:65-69. [DOI] [PubMed] [Google Scholar]
- 18.Contreras, I., C. S. Toro, G. Troncoso, and G. C. Mora. 1997. Salmonella typhi mutants defective in anaerobic respiration are impaired in their ability to replicate within epithelial cells. Microbiology 143:2665-2672. [DOI] [PubMed] [Google Scholar]
- 19.Cruijsen, T., L. A. van Leengoed, M. Ham-Hoffies, and J. H. Verheijden. 1995. Convalescent pigs are protected completely against infection with a homologous Actinobacillus pleuropneumoniae strain but incompletely against a heterologous-serotype strain. Infect. Immun. 63:2341-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cruz, W. T., Y. A. Nedialkov, B. J. Thacker, and M. H. Mulks. 1996. Molecular characterization of a common 48-kilodalton outer membrane protein of Actinobacillus pleuropneumoniae. Infect. Immun. 64:83-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dartigalongue, C., D. Missiakas, and S. Raina. 2001. Characterization of the Escherichia coli sigma E regulon. J. Biol. Chem. 276:20866-20875. [DOI] [PubMed] [Google Scholar]
- 22.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deuerling, E., A. Schulze-Specking, T. Tomoyasu, A. Mogk, and B. Bukau. 1999. Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature 400:693-696. [DOI] [PubMed] [Google Scholar]
- 24.Duim, B., J. Dankert, H. M. Jansen, and L. van Alphen. 1993. Genetic analysis of the diversity in outer membrane protein P2 of non-encapsulated Haemophilus influenzae. Microb. Pathog. 14:451-462. [DOI] [PubMed] [Google Scholar]
- 25.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 26.Freeman, B. C., and R. I. Morimoto. 1996. The human cytosolic molecular chaperones hsp90, hsp70 (hsc70) and hdj-1 have distinct roles in recognition of a non-native protein and protein refolding. EMBO J. 15:2969-2979. [PMC free article] [PubMed] [Google Scholar]
- 27.Freistroffer, D. V., M. Y. Pavlov, J. MacDougall, R. H. Buckingham, and M. Ehrenberg. 1997. Release factor RF3 in E. coli accelerates the dissociation of release factors RF1 and RF2 from the ribosome in a GTP-dependent manner. EMBO J. 16:4126-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fuller, T. E., M. J. Kennedy, and D. E. Lowery. 2000. Identification of Pasteurella multocida virulence genes in a septicemic mouse model using signature-tagged mutagenesis. Microb. Pathog. 29:25-38. [DOI] [PubMed] [Google Scholar]
- 29.Fuller, T. E., S. Martin, J. F. Teel, G. R. Alaniz, M. J. Kennedy, and D. E. Lowery. 2000. Identification of Actinobacillus pleuropneumoniae virulence genes using signature-tagged mutagenesis in a swine infection model. Microb. Pathog. 29:39-51. [DOI] [PubMed] [Google Scholar]
- 30.Fuller, T. E., R. J. Shea, B. J. Thacker, and M. H. Mulks. 1999. Identification of in vivo induced genes in Actinobacillus pleuropneumoniae. Microb. Pathog. 27:311-327. [DOI] [PubMed] [Google Scholar]
- 31.Furesz, S. E., B. A. Mallard, J. T. Bosse, S. Rosendal, B. N. Wilkie, and J. I. MacInnes. 1997. Antibody- and cell-mediated immune responses of Actinobacillus pleuropneumoniae-infected and bacterin-vaccinated pigs. Infect. Immun. 65:358-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gabelli, S. B., M. A. Bianchet, M. J. Bessman, and L. M. Amzel. 2001. The structure of ADP-ribose pyrophosphatase reveals the structural basis for the versatility of the Nudix family. Nat. Struct. Biol. 8:467-472. [DOI] [PubMed] [Google Scholar]
- 33.Gibson, M. M., D. A. Bagga, C. G. Miller, and M. E. Maguire. 1991. Magnesium transport in Salmonella typhimurium: the influence of new mutations conferring Co2+ resistance on the CorA Mg2+ transport system. Mol. Microbiol. 5:2753-2762. [DOI] [PubMed] [Google Scholar]
- 34.Glansdorff, N. 1996. Biosynthesis of arginine and polyamines, p. 408-433. In F. C. Neidhardt, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 35.Gong, S., S. W. Bearden, V. A. Geoffroy, J. D. Fetherston, and R. D. Perry. 2001. Characterization of the Yersinia pestis Yfu ABC inorganic iron transport system. Infect. Immun. 69:2829-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gottesman, S. 1996. Proteases and their targets in Escherichia coli. Annu. Rev. Genet. 30:465-506. [DOI] [PubMed] [Google Scholar]
- 37.Haesebrouck, F., A. Van de Kerkhof, P. Dom, K. Chiers, and R. Ducatelle. 1996. Cross-protection between Actinobacillus pleuropneumoniae biotypes-serotypes in pigs. Vet. Microbiol. 52:277-284. [DOI] [PubMed] [Google Scholar]
- 38.Hantke, K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172-177. [DOI] [PubMed] [Google Scholar]
- 39.Hase, C. C., and B. Barquera. 2001. Role of sodium bioenergetics in Vibrio cholerae. Biochim. Biophys. Acta 1505:169-178. [DOI] [PubMed] [Google Scholar]
- 40.Hayashi, M., Y. Nakayama, and T. Unemoto. 1996. Existence of Na+-translocating NADH-quinone reductase in Haemophilus influenzae. FEBS Lett. 381:174-176. [DOI] [PubMed] [Google Scholar]
- 41.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400-403. [DOI] [PubMed] [Google Scholar]
- 42.Higgs, P. I., P. S. Myers, and K. Postle. 1998. Interactions in the TonB-dependent energy transduction complex: ExbB and ExbD form homomultimers. J. Bacteriol. 180:6031-6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.High, N. J., M. E. Deadman, D. W. Hood, and E. R. Moxon. 1996. The identification a novel gene required for lipopolysaccharide biosynthesis by Haemophilus influenzae RM7004, using transposon Tn916 mutagenesis. FEMS Microbiol. Lett. 145:325-331. [DOI] [PubMed] [Google Scholar]
- 44.Humphreys, S., A. Stevenson, A. Bacon, A. B. Weinhardt, and M. Roberts. 1999. The alternative sigma factor, σE, is critically important for the virulence of Salmonella typhimurium. Infect. Immun. 67:1560-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones, A. L., K. M. Knoll, and C. E. Rubens. 2000. Identification of Streptococcus agalactiae virulence genes in the neonatal rat sepsis model using signature-tagged mutagenesis. Mol. Microbiol. 37:1444-1455. [DOI] [PubMed] [Google Scholar]
- 46.Kaltreider, H. B. 1976. Initiation of immune responses in the lower respiratory tract with red cell antigens., p. 73-97. In C. H. Kirkpatrick and H. Y. Reynolds (ed.), Immunologic and infectious reactions in the lung. Marcel Dekker, New York, N.Y.
- 47.Keenleyside, W. J., P. Jayaratne, P. R. MacLachlan, and C. Whitfield. 1992. The rcsA gene of Escherichia coli O9:K30:H12 is involved in the expression of the serotype-specific group I K (capsular) antigen. J. Bacteriol. 174:8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Keller, K. L., T. L. Overbeck-Carrick, and D. J. Beck. 2001. Survival and induction of SOS in Escherichia coli treated with cisplatin, UV-irradiation, or mitomycin C are dependent on the function of the RecBC and RecFOR pathways of homologous recombination. Mutat. Res. 486:21-29. [DOI] [PubMed] [Google Scholar]
- 49.Kisselev, L. L., and R. H. Buckingham. 2000. Translational termination comes of age. Trends Biochem. Sci. 25:561-566. [DOI] [PubMed] [Google Scholar]
- 50.Kobayashi, N., K. Nishino, and A. Yamaguchi. 2001. Novel macrolide-specific ABC-type efflux transporter in Escherichia coli. J. Bacteriol. 183:5639-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kobayashi, T., S. Kishigami, M. Sone, H. Inokuchi, T. Mogi, and K. Ito. 1997. Respiratory chain is required to maintain oxidized states of the DsbA-DsbB disulfide bond formation system in aerobically growing Escherichia coli cells. Proc. Natl. Acad. Sci. USA 94:11857-11862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kohler, S., J. Teyssier, A. Cloeckaert, B. Rouot, and J. P. Liautard. 1996. Participation of the molecular chaperone DnaK in intracellular growth of Brucella suis within U937-derived phagocytes. Mol. Microbiol. 20:701-712. [DOI] [PubMed] [Google Scholar]
- 53.Kranz, R., R. Lill, B. Goldman, G. Bonnard, and S. Merchant. 1998. Molecular mechanisms of cytochrome c biogenesis: three distinct systems. Mol. Microbiol. 29:383-396. [DOI] [PubMed] [Google Scholar]
- 54.Kuzminov, A. 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol. Mol. Biol. Rev. 63:751-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Labrie, J., S. Rioux, M. M. Wade, F. R. Champlin, S. C. Holman, W. W. Wilson, C. Savoye, M. Kobisch, M. Sirois, C. Galarneau, and M. Jacques. 2002. Identification of genes involved in biosynthesis of Actinobacillus pleuropneumoniae serotype 1 O-antigen and biological properties of rough mutants. J. Endotoxin Res. 8:27-38. [PubMed] [Google Scholar]
- 56.Lau, G. W., S. Haataja, M. Lonetto, S. E. Kensit, A. Marra, A. P. Bryant, D. McDevitt, D. A. Morrison, and D. W. Holden. 2001. A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol. Microbiol. 40:555-571. [DOI] [PubMed] [Google Scholar]
- 57.Lewis, D. A., J. Klesney-Tait, S. R. Lumbley, C. K. Ward, J. L. Latimer, C. A. Ison, and E. J. Hansen. 1999. Identification of the znuA-encoded periplasmic zinc transport protein of Haemophilus ducreyi. Infect. Immun. 67:5060-5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lu, D., B. Boyd, and C. A. Lingwood. 1998. The expression and characterization of a putative adhesin B from H. influenzae. FEMS Microbiol. Lett. 165:129-137. [DOI] [PubMed] [Google Scholar]
- 59.Lu, D., B. Boyd, and C. A. Lingwood. 1997. Identification of the key protein for zinc uptake in Haemophilus influenzae. J. Biol. Chem. 272:29033-29038. [DOI] [PubMed] [Google Scholar]
- 60.Maroncle, N., D. Balestrino, C. Rich, and C. Forestier. 2002. Identification of Klebsiella pneumoniae genes involved in intestinal colonization and adhesion using signature-tagged mutagenesis. Infect. Immun. 70:4729-4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.May, B. J., Q. Zhang, L. L. Li, M. L. Paustian, T. S. Whittam, and V. Kapur. 2001. Complete genomic sequence of Pasteurella multocida, Pm70. Proc. Natl. Acad. Sci. USA 98:3460-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mizusawa, S., and S. Gottesman. 1983. Protein degradation in Escherichia coli: the lon gene controls the stability of sulA protein. Proc. Natl. Acad. Sci. USA 80:358-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moeck, G. S., and J. W. Coulton. 1998. TonB-dependent iron acquisition: mechanisms of siderophore-mediated active transport. Mol. Microbiol. 28:675-681. [DOI] [PubMed] [Google Scholar]
- 65.Moreno-Vivian, C., P. Cabello, M. Martinez-Luque, R. Blasco, and F. Castillo. 1999. Prokaryotic nitrate reduction: molecular properties and functional distinction among bacterial nitrate reductases. J. Bacteriol. 181:6573-6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nakahigashi, K., K. Miyamoto, K. Nishimura, and H. Inokuchi. 1992. Isolation and characterization of a light-sensitive mutant of Escherichia coli K-12 with a mutation in a gene that is required for the biosynthesis of ubiquinone. J. Bacteriol. 174:7352-7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Neuhaus, K., S. Rapposch, K. P. Francis, and S. Scherer. 2000. Restart of exponential growth of cold-shocked Yersinia enterocolitica occurs after down-regulation of cspA1/A2 mRNA. J. Bacteriol. 182:3285-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nielsen, R. 1979. Haemophilus parahaemolyticus serotypes. Pathogenicity and cross immunity. Nord. Vet. Med. 31:407-413. [PubMed] [Google Scholar]
- 69.Noriega, F. R., G. Losonsky, C. Lauderbaugh, F. M. Liao, J. Y. Wang, and M. M. Levine. 1996. Engineered ΔguaB-A ΔvirG Shigella flexneri 2a strain CVD 1205: construction, safety, immunogenicity, and potential efficacy as a mucosal vaccine. Infect. Immun. 64:3055-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Occhino, D. A., E. E. Wyckoff, D. P. Henderson, T. J. Wrona, and S. M. Payne. 1998. Vibrio cholerae iron transport: haem transport genes are linked to one of two sets of tonB, exbB, exbD genes. Mol. Microbiol. 29:1493-1507. [DOI] [PubMed] [Google Scholar]
- 71.Ohara, M., H. C. Wu, K. Sankaran, and P. D. Rick. 1999. Identification and characterization of a new lipoprotein, NlpI, in Escherichia coli K-12. J. Bacteriol. 181:4318-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Patte, J.-C. 1996. Biosynthesis of threonine and lysine, p. 528-541. In F. C. Neidhardt, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 73.Penfound, T., and J. W. Foster. 1996. Biosynthesis and recycling of NAD, p. 721-729. In F. C. Neidhardt, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 74.Petersen, L., and D. M. Downs. 1996. Mutations in apbC (mrp) prevent function of the alternative pyrimidine biosynthetic pathway in Salmonella typhimurium. J. Bacteriol. 178:5676-5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Prince, R. W., D. G. Storey, A. I. Vasil, and M. L. Vasil. 1991. Regulation of toxA and regA by the Escherichia coli fur gene and identification of a Fur homologue in Pseudomonas aeruginosa PA103 and PA01. Mol. Microbiol. 5:2823-2831. [DOI] [PubMed] [Google Scholar]
- 76.Raina, S., D. Missiakas, and C. Georgopoulos. 1995. The rpoE gene encoding the sigma E (sigma 24) heat shock sigma factor of Escherichia coli. EMBO J. 14:1043-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rauhut, R., and G. Klug. 1999. mRNA degradation in bacteria. FEMS Microbiol. Rev. 23:353-370. [DOI] [PubMed] [Google Scholar]
- 78.Richarme, G., A. el Yaagoubi, and M. Kohiyama. 1993. The MglA component of the binding protein-dependent galactose transport system of Salmonella typhimurium is a galactose-stimulated ATPase. J. Biol. Chem. 268:9473-9437. [PubMed] [Google Scholar]
- 79.Rioux, S., C. Begin, J. D. Dubreuil, and M. Jacques. 1997. Isolation and characterization of LPS mutants of Actinobacillus pleuropneumoniae serotype 1. Curr. Microbiol. 35:139-144. [DOI] [PubMed] [Google Scholar]
- 80.Rioux, S., C. Galarneau, J. Harel, J. Frey, J. Nicolet, M. Kobisch, J. D. Dubreuil, and J. Jasques. 1999. Isolation and characterization of mini-Tn10 lipopolysaccharide mutants of Actinobacillus pleuropneumoniae serotype 1. Can J. Microbiol. 45:1017-1026. [DOI] [PubMed] [Google Scholar]
- 81.Rioux, S., C. Galarneau, J. Harel, M. Kobisch, J. Frey, M. Gottschalk, and M. Jacques. 2000. Isolation and characterization of a capsule-deficient mutant of Actinobacillus pleuropneumoniae serotype 1. Microb. Pathog. 28:279-289. [DOI] [PubMed] [Google Scholar]
- 82.Rivers, S. L., E. McNairn, F. Blasco, G. Giordano, and D. H. Boxer. 1993. Molecular genetic analysis of the moa operon of Escherichia coli K-12 required for molybdenum cofactor biosynthesis. Mol. Microbiol. 8:1071-1081. [DOI] [PubMed] [Google Scholar]
- 83.Robertson, G. T., M. E. Kovach, C. A. Allen, T. A. Ficht, and R. M. Roop, 2nd. 2000. The Brucella abortus Lon functions as a generalized stress response protease and is required for wild-type virulence in BALB/c mice. Mol. Microbiol. 35:577-588. [DOI] [PubMed] [Google Scholar]
- 84.Rouviere, P. E., A. De Las Penas, J. Mecsas, C. Z. Lu, K. E. Rudd, and C. A. Gross. 1995. rpoE, the gene encoding the second heat-shock sigma factor, sigma E, in Escherichia coli. EMBO J. 14:1032-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rudiger, S., J. Schneider-Mergener, and B. Bukau. 2001. Its substrate specificity characterizes the DnaJ co-chaperone as a scanning factor for the DnaK chaperone. EMBO J. 20:1042-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Russo, T. A., S. T. Jodush, J. J. Brown, and J. R. Johnson. 1996. Identification of two previously unrecognized genes (guaA and argC) important for uropathogenesis. Mol. Microbiol. 22:217-229. [DOI] [PubMed] [Google Scholar]
- 87.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd. ed. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 88.Schaller, A., R. Kuhn, P. Kuhnert, J. Nicolet, T. J. Anderson, J. I. MacInnes, R. P. Segers, and J. Frey. 1999. Characterization of apxIVA, a new RTX determinant of Actinobacillus pleuropneumoniae. Microbiology 145:2105-2116. [DOI] [PubMed] [Google Scholar]
- 89.Schmitt, M. P., and R. K. Holmes. 1991. Iron-dependent regulation of diphtheria toxin and siderophore expression by the cloned Corynebacterium diphtheriae repressor gene dtxR in C. diphtheriae C7 strains. Infect. Immun. 59:1899-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sun, Y. H., S. Bakshi, R. Chalmers, and C. M. Tang. 2000. Functional genomics of Neisseria meningitidis pathogenesis. Nat. Med. 6:1269-1273. [DOI] [PubMed] [Google Scholar]
- 91.Suzuki, C. K., M. Rep, J. M. van Dijl, K. Suda, L. A. Grivell, and G. Schatz. 1997. ATP-dependent proteases that also chaperone protein biogenesis. Trends Biochem. Sci. 22:118-123. [DOI] [PubMed] [Google Scholar]
- 92.Theisen, M., C. R. Rioux, and A. A. Potter. 1993. Molecular cloning, nucleotide sequence, and characterization of lppB, encoding an antigenic 40-kilodalton lipoprotein of Haemophilus somnus. Infect. Immun. 61:1793-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Thomas, J. G., and F. Baneyx. 2000. ClpB and HtpG facilitate de novo protein folding in stressed Escherichia coli cells. Mol. Microbiol. 36:1360-1370. [DOI] [PubMed] [Google Scholar]
- 94.Thony-Meyer, L., F. Fischer, P. Kunzler, D. Ritz, and H. Hennecke. 1995. Escherichia coli genes required for cytochrome c maturation. J. Bacteriol. 177:4321-4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tomoyasu, T., T. Ogura, T. Tatsuta, and B. Bukau. 1998. Levels of DnaK and DnaJ provide tight control of heat shock gene expression and protein repair in Escherichia coli. Mol. Microbiol. 30:567-581. [DOI] [PubMed] [Google Scholar]
- 96.Tonpitak, W., S. Thiede, W. Oswald, N. Baltes, and G. F. Gerlach. 2000. Actinobacillus pleuropneumoniae iron transport: a set of exbBD genes is transcriptionally linked to the tbpB gene and required for utilization of transferrin-bound iron. Infect. Immun. 68:1164-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Toone, W. M., K. E. Rudd, and J. D. Friesen. 1991. deaD, a new Escherichia coli gene encoding a presumed ATP-dependent RNA helicase, can suppress a mutation in rpsB, the gene encoding ribosomal protein S2. J. Bacteriol. 173:3291-3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang, J. Y., M. F. Pasetti, F. R. Noriega, R. J. Anderson, S. S. Wasserman, J. E. Galen, M. B. Sztein, and M. M. Levine. 2001. Construction, genotypic and phenotypic characterization, and immunogenicity of attenuated ΔguaBA Salmonella enterica serovar Typhi strain CVD 915. Infect. Immun. 69:4734-4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ward, C. K., and T. J. Inzana. 1997. Identification and characterization of a DNA region involved in the export of capsular polysaccharide by Actinobacillus pleuropneumoniae serotype 5a. Infect. Immun. 65:2491-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ward, C. K., M. L. Lawrence, H. P. Veit, and T. J. Inzana. 1998. Cloning and mutagenesis of a serotype-specific DNA region involved in encapsulation and virulence of Actinobacillus pleuropneumoniae serotype 5a: concomitant expression of serotype 5a and 1 capsular polysaccharides in recombinant A. pleuropneumoniae serotype 1. Infect. Immun. 66:3326-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Winzer, K., K. R. Hardie, N. Burgess, N. Doherty, D. Kirke, M. T. Holden, R. Linforth, K. A. Cornell, A. J. Taylor, P. J. Hill, and P. Williams. 2002. LuxS: its role in central metabolism and the in vitro synthesis of 4-hydroxy-5-methyl-3(2H)-furanone. Microbiology 148:909-922. [DOI] [PubMed] [Google Scholar]
- 102.Yamanaka, K., and M. Inouye. 2001. Selective mRNA degradation by polynucleotide phosphorylase in cold shock adaptation in Escherichia coli. J. Bacteriol. 183:2808-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yi, K., and T. F. Murphy. 1997. Importance of an immunodominant surface-exposed loop on outer membrane protein P2 of nontypeable Haemophilus influenzae. Infect. Immun. 65:150-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yu, J., and J. S. Kroll. 1999. DsbA: a protein-folding catalyst contributing to bacterial virulence. Microbes Infect. 1:1221-1228. [DOI] [PubMed] [Google Scholar]
- 105.Zhao, Q., and K. Poole. 2000. A second tonB gene in Pseudomonas aeruginosa is linked to the exbB and exbD genes. FEMS Microbiol. Lett. 184:127-132. [DOI] [PubMed] [Google Scholar]