Abstract
Target of rapamycin (TOR) is an evolutionally conserved protein kinase in eukaryotes and a central cell growth controller. TOR exists in two distinct complexes, termed TORC1 and TORC2. Mammalian TORC2 has recently been shown to possess kinase activity toward the C-terminal hydrophobic site of Akt/PKB. Here, we report that Sin1 is an essential component of TORC2 but not of TORC1, and functions similarly to Rictor, the defining member of TORC2, in complex formation and kinase activity. Knockdown of Sin1decreases Akt phosphorylation in both Drosophila and mammalian cells and diminishes Akt function in vivo. It also disrupts the interaction between Rictor and mTOR. Furthermore, Sin1 is required for TORC2 kinase activity in vitro. Disruption of the Rictor gene in mice results in embryonic lethality and ablates Akt phosphorylation. These data demonstrate that Sin1 together with Rictor are key components of mTORC2 and play an essential role in Akt phosphorylation and signaling.
Keywords: Sin1, TORC, TSC, Rictor, Akt, mTOR
Rapamycin is an Food and Drug Administration (FDA)-approved drug used for immunosuppression in organ transplants, prevention of restenosis post-angioplasty, and chemotherapy for soft tissue and bone sarcomas (Abraham and Wiederrecht 1996; Sawyers 2003). The target of rapamycin (TOR) is a highly conserved phosphatidylinositol kinaserelated serine/threonine protein kinase that regulates cell growth, proliferation, morphology, and survival (Inoki et al. 2005; Wullschleger et al. 2006). The functions of TOR in translation and cell size control have been well characterized due to the availability of rapamycin, a highly specific TOR inhibitor.
TOR forms two distinct structural and functional complexes (TORC1 and TORC2) (Wullschleger et al. 2006). TORC1 and TORC2 were initially identified in Saccharomyces cerevisiae, which has two TOR genes, while higher eukaryotes have only one TOR gene. Yeast TORC1 consists of either Tor1 or Tor2, Kog1, Lst8, and Tco89, while yeast TORC2 contains Tor2, Lst8, Avo1, Avo2, Avo3, and Bit61 (Loewith et al. 2002; Reinke et al. 2004). Biochemical studies show that TORC1 activity is inhibited by rapamycin. In contrast, TORC2 activity is insensitive to rapamycin inhibition. Recently, both TORC1 and TORC2 have also been identified in higher eukaryotes (Jacinto et al. 2004; Sarbassov et al. 2004). Mammalian TORC1 (mTORC1) consists of mTOR, Raptor (homolog of Kog1) and mLST8 (also known as GβL) (Hara et al. 2002; Kim et al. 2002, 2003). mTORC1 regulates translation and cell growth by phosphorylating ribosomal S6 kinase (S6K) and eukaryote initiation factor 4E-binding protein 1 (4EBP1) in a rapamycinsensitive manner (Hay and Sonenberg 2004). Much of the current knowledge about mTOR in literature is associated with mTORC1 because of the wide utilization of rapamycin in biochemical studies. The identification of these two distinct TOR complexes ties together many yeast and mammalian studies and has significantly advanced the understanding of TOR biology (Wullschleger et al. 2006).
Recent discoveries have highlighted the functional importance of TORC2 (Jacinto et al. 2004; Sarbassov et al. 2004, 2005). Sabatini's group (Sarbassov et al. 2005) has demonstrated that TORC2 directly phosphorylates Ser473 in the hydrophobic motif of Akt/PKB. In contrast, TORC2 does not phosphorylate S6K, a well-characterized mTOR substrate. The discovery that TORC2 is the long-sought PDK2 greatly widens the cellular functions of mTOR. Akt is a key signaling component downstream from PI3K (Brazil et al. 2004) and regulates a wide range of cellular activities including cell growth, metabolism, and survival. Akt controls these important cellular functions by phosphorylating and regulating a large number of cellular proteins such as GSK3 (Cross et al. 1995; Woodgett 2005). Akt activation is tightly controlled by PI3K signaling and requires both the phosphorylation of Thr308 in the activation loop and the phosphorylation of Ser473 in the hydrophobic motif. Phosphorylation of these two residues in Akt is accomplished by PDK1 and PDK2 (TORC2), respectively (Alessi et al. 1997). PDK1 has been well characterized as an essential link between PI3K and Akt by phosphorylating Thr308 in the activation loop of Akt (Mora et al. 2004). Similarly, the phosphorylation of Ser473 in Akt is stimulated by PI3K activation, though the molecular mechanism of TORC2 regulation is not clear.
mTORC2 consists of mTOR, Rictor (Avo3 homolog) and mLST8 (Jacinto et al. 2004; Sarbassov et al. 2004). It is also known as the “Rictor complex,” because Rictor is the defining component of mTORC2, while both mTOR and mLST8 are also present in mTORC1. Rictor is essential for both mTORC2 complex formation and its biological function. Rictor knockdown by RNA interference (RNAi) dramatically inhibits Ser473 phosphorylation of Akt in both Drosophila and mammalian cells. Moreover, TORC2 immunoprecipitated from mammalian cells using a Rictor antibody efficiently phosphorylates Akt on Ser473 in vitro (Sarbassov et al. 2005; Wullschleger et al. 2005). However, phosphorylation of Akt Ser473 is not completely abolished in Rictor knockdown cells. This might be due to the incomplete knockdown of Rictor by RNAi. Nevertheless, the existence of other PDK2 activities cannot be excluded by the current data, and TORC2 may represent only one of multiple PDK2 activities in the cell (Balendran et al. 1999; Toker and Newton 2000; Persad et al. 2001; Rane et al. 2001; Feng et al. 2004). The essential role of Rictor in mTORC2 is further supported by its function in actin cytoskeletal organization, which is also regulated by mTORC2 (Jacinto et al. 2004; Sarbassov et al. 2004). Excluding Rictor, no other protein has been identified as a component unique to mTORC2.
Avo1 was purified as a component of TORC2 in the budding yeast S. cerevisiae. Disruption of AVO1 results in phenotypes similar to those observed in TOR2 disruption, suggesting that Avo1 functions together with Tor2 in the yeast TORC2 (Loewith et al. 2002). Human Sin1 homolog (hSin1) was originally named based on its homology with Sin1, the Avo1 ortholog in the fission yeast Schizosaccharomyces pombe (Wilkinson et al. 1999). In fission yeast, Sin1 is involved in stress responses and interacts with Sty1/Spc1, a member of the mitogenactivated protein kinase family. However, the function of Sin1 in TOR signaling has not been investigated. Mammalian Sin1/Mip1 was isolated as a MEKK2-interacting protein (Cheng et al. 2005). MEKK2 is a member of the mitogenactivated protein (MAP) kinase kinase kinases and activates the JNK kinase pathway (Hagemann and Blank 2001). However, previous reports had suggested that hSin1 is not part of mTORC2, but the evidence was not extensive (Loewith et al. 2002). Since hSin1 is widely expressed in human tissue similar to the expression profile of mTOR (Loewith et al. 2002; Schroder et al. 2004), it is possible that hSin1 is a component of mTORC2.
In this report, we examined the function of hSin1 and its relationship with mTORC2. Our studies show that hSin1 specifically interacts with mTOR and Rictor, but not Raptor. Furthermore, the knockdown of hSin1 results in decrease of Rictor phosphorylation and protein levels as well as disrupting the binding between Rictor and mTOR. Consistent with a role of hSin1 in TORC2 activity, Rictor knockdown dramatically decreases hSin1 protein levels. Moreover, the knockdown of Sin1 in both Drosophila and mammalian cells diminishes Akt phosphorylation. hSin1 knockdown cells display less phosphorylation of Akt substrates and are more sensitive to apoptosis. We also demonstrate that TORC2 plays a pivotal role in Akt phosphorylation in mouse embryos by disruption of the Rictor gene. We found that Rictor disruption results in embryonic lethality. The Akt phosphorylation on Ser473 is abolished, whereas the decrease of Thr308 phosphorylation varies among different embryos. This study establishes that hSin1 is an essential component of TORC2 and plays a crucial role in Akt phosphorylation and signaling.
Results
dSin1 is essential for dAkt phosphorylation in Drosophila S2 cells
Previous studies have shown that Avo1 is an essential component of TORC2 in S. cerevisiae (Loewith et al. 2002; Wullschleger et al. 2005). Our initial sequence homology search by BLAST or PSIBLAST using the S. cerevisiae Avo1 sequence only identified the S. pombe Sin1 gene product as an obvious homolog. When the Sin1 sequence was used in a BLAST search, Sin1 homologs were found in higher eukaryotes, including Drosophila and human. However, whether this Avo1 ortholog (Sin1) functions in higher eukaryotic TORC2 was not clear. In fact, a previous report suggested that hSin1 does not interact with mTOR (Loewith et al. 2002).
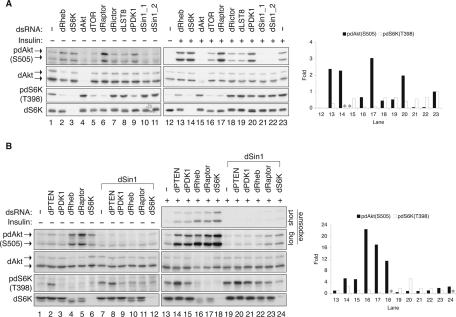
To test the function of Sin1 homologs in higher eukaryotes, we first performed RNAi experiments in Drosophila S2 cells, in which the function of TORC2 has been characterized. Two nonoverlapping dSin1 double stranded RNAs (dsRNA), targeting different dSin1 coding regions, were generated to knockdown dSin1 in cultured S2 cells. The phosphorylation of dAkt and dS6K, detected by phosphospecific antibodies, was used as a functional readout for the activities of dTORC2 and dTORC1, respectively. We found that down-regulation of dSin1 by RNAi decreased basal dAkt phosphorylation on Ser505, which is equivalent to Ser473 of the mammalian Akt1, the direct site of phosphorylation by TORC2 (Fig. 1A). Under basal conditions, the effects of dTOR, dRictor, and dLST8 RNAi were not obvious, probably due to the low levels of basal dAkt phosphorylation (Fig. 1A). Therefore, we examined these RNAi effects under insulin stimulation conditions, which stimulate basal dAkt phosphorylation (Fig. 1A, cf. lanes 12 and 23) and thus make the RNAi inhibitory effects more visible. We observed that knockdown of dSin1 ablated the insulin-induced dAkt phosphorylation. It is worth noting that the effect of dSin1 knockdown on dAkt phosphorylation was more dramatic than the effects of dTOR and dRictor knockdown. This could be partly due to a more efficient knockdown of dSin1 (Supplementary Fig. S1). We also observed that knockdown of dSin1 did not decrease dS6K phosphorylation, suggesting that dSin1 is not important for TORC1 activity. As expected, knockdown of dTOR strongly inhibited dS6K phosphorylation. dLST8 RNAi did not noticeably inhibit dS6K phosphorylation, probably due to an insufficient knockdown (Supplementary Fig. S1). Together, our data demonstrate that dSin1 is important for dTORC2 function in dAkt phosphorylation and is not required for dTORC1 function.
FIGURE 1.
dSin1 is essential for dAkt phosphorylation in Drosophila S2 cells. (A) dSin1 knockdown inhibits both basal and insulin-stimulated dAkt phosphorylation but not dS6K phosphorylation. dsRNA was used to target an individual dTOR pathway component as indicated. “dSin1_1” and “dSin1_2” denote two nonoverlapping dsRNAs targeting different regions of the dSin1 mRNA. Insulin stimulation (100 nM, 15 min) is indicated. Protein levels and phosphorylation were determined by Western blotting with indicated antibodies. The dAkt protein levels were detected by mammalian Akt antibody. The two dAkt isoforms generated by alternative splicing are indicated by the arrows. The column chart shows the quantification results (ImageJ) of the dAkt and dS6K phosphorylation under insulin stimulation condition. The lane numbers in the chart correspond to those labeled at the bottom of the Western blot panels. (*) Knockdown of either dAkt or dS6K that led to undetectable protein levels for quantification. The phosphorylation under basal condition was too low to be accurately quantified. (B) dSin1 knockdown blocks the effects caused by knockdown of other TOR pathway components on dAkt but not on dS6K phosphorylation. RNAi experiments, insulin stimulation, Western blotting, and quantification were performed the same as described in A. Where indicated, dSin1 dsRNA was used in combination and added to the cells at the same time with the other dsRNAs.
It has been reported that TORC1 and S6K exert an inhibitory effect on TORC2, possibly via the feedback inhibition mechanism involving S6K (Radimerski et al. 2002a, b; Stocker et al. 2003; Harrington et al. 2004; Shah et al. 2004; Um et al. 2004; Yang et al. 2006). Knockdown of dRaptor, dRheb, or dS6K increased both basal and insulin-induced dAkt phosphorylation (Fig. 1A,B). We examined the relationship between these genes and dSin1 in dAkt regulation. Knockdown of dSin1 completely blocked the positive effect on dAkt phosphorylation caused by knockdown of dRaptor, dRheb, or dS6K (Fig. 1B). dPTEN knockdown promoted phosphorylation of both dAkt and dS6K. Again, dSin1 was required for the dPTEN knockdown-induced dAkt phosphorylation, but not required for the enhancement of dS6K phosphorylation. Our data also confirm previous reports that dAkt is not required for dS6K phosphorylation in Droshophila cells (Radimerski et al. 2002b; Sarbassov et al. 2005; Yang et al. 2006). Together, these results indicate that dSin1 functions proximal to dAkt and are consistent with the hypothesis that dSin1 is a key component of dTORC2.
hSin1 plays a positive role in Akt phosphorylation in mammalian cells
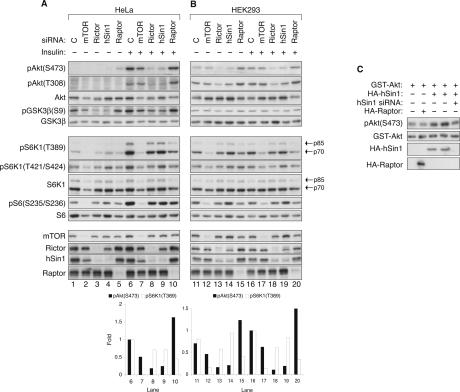
To further confirm the function of Sin1 in Akt phosphorylation, we next performed RNAi experiments in HeLa cells. We chose HeLa cells because RNAi is very efficient and TORC2 activity has been well characterized in this cell line (Sarbassov et al. 2005). The RNAi effects of mTOR (required for both mTORC1 and mTORC2), Rictor (mTORC2 specific), Raptor (mTORC1 specific), and hSin1 were analyzed by Western blotting. Phosphorylation of both Ser473 (the PDK2/TORC2 site) and Thr308 (the PDK1 site) of Akt was examined. Because the levels of basal Akt phosphorylation are low in HeLa cells, RNAi effects are more evident under insulin stimulation conditions (Fig. 2A). Similar to the observations made in Drosophila S2 cells, knockdown of both hSin1 and Rictor, but not Raptor, significantly reduced the phosphorylation of Akt on Ser473 and Thr308, and had a limited effect on S6K phosphorylation. The weak reduction in S6K phosphorylation could be due to a slight decrease of mTOR protein level (Figs. 2A,B, 4A, below) or an inhibition of Akt activity by the hSin1 and Rictor RNAi. Knockdown of mTOR also decreased Akt phosphorylation, although less dramatically than hSin1 knockdown. The relatively weak effect of mTOR RNAi could be due to a less-efficient knockdown of mTOR mRNA (Supplementary Fig. S2A), and/or a relief of the S6K-mediated feedback inhibition caused by mTOR knockdown. Supporting the latter possibility, Raptor RNAi caused a reproducible increase of Akt phosphorylation. The above observations are consistent with our data obtained in Drosophila S2 cells and suggest that hSin1 is required for proper Akt phosphorylation in mammalian cells.
FIGURE 2.
hSin1 positively regulates Akt phosphorylation in mammalian cells. (A) Knockdown of hSin1 inhibits Akt phosphorylation but not S6K phosphorylation in HeLa cells. Different siRNAs used in transfection are indicated. “C” denotes control siRNA targeting GFP. HeLa cells were treated with 100 nM insulin for 15 min before harvesting where indicated. Antibodies used for Western blotting were labeled next to each panel. The phospho-S6K1 (T389) and S6K1 antibodies detect both the p85 and the p70 isoforms that are indicated by the arrows. The column chart shows quantification results (ImageJ) of the Western blotting. The phosphorylation of Akt or S6K1 under basal conditions was too low to be accurately quantified. (B) Knockdown of hSin1 inhibits Akt phosphorylation in HEK293 cells. Experiments were performed similarly as those described in A. (C) Overexpression of HA-hSin1 promotes Akt phosphorylation. HA-hSin1 was cotransfected with GST-Akt into HEK293 cells. hSin1 siRNA and HA-Raptor were used as controls where indicated. Phosphorylation of cotransfected GST-Akt was monitored by phospho-specific antibody as indicated.
FIGURE 4.
hSin1 is important for TORC2 kinase activity. (A) hSin1 is indispensable for TORC2 kinase activity in vitro. HeLa cells were transfected with each indicated siRNA and lysed in CHAPS buffer. “C” denotes control siRNA targeting GFP. mTOR and Rictor IPs were performed as indicated. Dephosphorylated Akt (Upstate Biotechnology, from insect cells) was used as a substrate for the in vitro TORC2 kinase assay. Phosphorylation of Akt was first determined by the phospho-Akt(S473) antibody, and then reprobed by the phospho-Akt(T308) antibody after low pH stripping. Note that the pAkt(T308) blot had a very long exposure time and only shows the background signal. A small fraction of the kinase reactions was blotted with Akt antibody for protein levels. (B) hSin1 directly precipitates active TORC2 kinase. HEK293 cells were transfected with indicated plasmids, lysed in CHAPS buffer, and immunoprecipitated with HA antibody. Purified GST-Akt (from HEK293 cells) and the dephosphorylated Akt (Upstate Biotechnology) were used as substrates for the kinase assays. The phosphorylation of the Akt substrates was determined by Western blotting with phospho-specific antibody as indicated. Protein levels were determined by HA, MYC, GST, and Akt antibodies. (C) hSin1 is required for insulin-stimulated TORC2 activity. HeLa cells were transfected with various siRNAs as indicated. Cells were stimulated with insulin (100 nM, 15 min) and lysed in CHAPS buffer. In vitro kinase assay of immunoprecipitated mTOR was performed using dephosphorylated Akt as a substrate. (D) Insulin stimulation does not alter TORC2 integrity. HeLa cells were serum-starved overnight before being treated with insulin (100 nM, 15 min). Cells were harvested with CHAPS buffer and the lysates were subjected to Rictor immunoprecipitation. TORC2 kinase activity was assayed using dephosphorylated Akt as a substrate.
To determine whether hSin1 RNAi decreases Akt activity in vivo, we examined the phosphorylation of GSK3β Ser9, which is a physiological substrate of Akt (Cross et al. 1995). In agreement with decreased Akt phosphorylation, both basal and insulin-stimulated GSK3β Ser9 phosphorylation were inhibited by knockdown of either hSin1 or Rictor (Fig. 2A). The results indicate that both hSin1 and Rictor are required for basal and insulin-stimulated Akt activity. It is worth noting that the decrease of GSK3 phosphorylation caused by hSin1 knockdown is less dramatic than the decrease of Akt phosphorylation. This is likely due to the fact that Akt is not the only kinase responsible for GSK3 Ser9 phosphorylation (McManus et al. 2005; Woodgett 2005). In the same experiments, phosphorylation of the S6 protein, a physiological substrate of S6K, was not significantly affected by knockdown of either hSin1 or Rictor. Therefore, hSin1, similar to Rictor, is selectively required for the phosphorylation and activation of the TORC2 substrate Akt, but not the TORC1 substrate S6K. Similar experiments were performed in HEK293 cells, which have much higher basal levels of Akt phosphorylation and display a weaker response to insulin stimulation. Still, knockdown of either hSin1 or Rictor decreased both basal and insulin-stimulated Akt phosphorylation (Fig. 2B). Therefore, these data further support that hSin1 functions similarly to Rictor and plays an essential role in Akt phosphorylation.
We also examined the effects of mLST8 RNAi, a component known to be present in both TORC1 and TORC2. Knockdown of mLST8 showed only minor effects on phosphorylation of both Akt and S6K (Supplementary Fig. S3A). It is possible that the mLST8 protein is more abundant than the other TORC components, and therefore is less sensitive to knockdown. Alternatively, mLST8 may have a less critical function in TORC1 and TORC2.
The effects of PDK1 knockdown were also examined. Knockdown of PDK1 decreased S6K phosphorylation as expected. Surprisingly, it increased Akt phosphorylation on Ser473 and had no visible effect on Thr308 phosphorylation (Supplementary Fig. S3A). The increase of Akt phosphorylation on Ser473 could be due to the PDK1 knockdown-induced inhibition of S6K, therefore relieving the inhibitory effect of S6K on TORC2 (Radimerski et al. 2002a, b; Stocker et al. 2003; Harrington et al. 2004; Shah et al. 2004; Um et al. 2004; Yang et al. 2006). The lack of PDK1 RNAi effect on the Thr308 phosphorylation of Akt can be explained by the excessive PDK1 activity needed for Akt phosphorylation in the HeLa cells. It is consistent with a previous report that a 90% reduction of PDK1 does not affect Akt phosphorylation in response to insulin (Lawlor et al. 2002).
In the RNAi experiments, we observed that knockdown of one target decreased the protein level of another TORC2 component (Figs. 2A,B, 4A, below). For example, Rictor knockdown significantly decreased hSin1 protein levels. One may question the specificity of the SMARTpool small interfering RNA (siRNA), which is composed of four individual siRNAs for each mRNA target. However, several lines of evidence argue against the possibility of nonspecific targeting by the SMARTpool siRNA. First, Rictor knockdown did not affect the protein levels of Raptor, Akt, GSK3β, S6K, and S6. Second, we performed quantitative real-time PCR (QRT-PCR) to determine mRNA levels. Our data showed that the SMART pool siRNAs were indeed specifically targeting their respective targets (Supplementary Fig. S2A–C). Furthermore, we examined four different siRNA duplexes, targeting either hSin1 or Rictor (Supplementary Fig. S3B). The results demonstrated that independent knockdown of Rictor by four different siRNAs also decreased hSin1 protein levels. Similarly, hSin1 knockdown by four different siRNAs decreased Rictor protein levels. These data not only demonstrate the specificity of the RNAi experiments, but also further support that hSin1 is a component of TORC2 and is stabilized by complex formation with Rictor.
To complement the hSin1 RNAi experiments, we examined the effects of hSin1 overexpression on Akt phosphorylation. Overexpression of HA-hSin1 reproducibly promoted the phosphorylation of the cotransfected GSTAkt on Ser473 (Fig. 2C). As a negative control, overexpression of HA-Raptor had no effect on the phosphorylation of GST-Akt. These results further demonstrate a positive role of hSin1 in Akt phosphorylation. Akt is an important cellular regulator known to phosphorylate multiple substrates. We used a phospho-Akt substrate antibody, which recognizes RXRXXS*/T* (X for any residue, * for phosphorylation) motif (Alessi et al. 1996), to determine whether down-regulation of hSin1 affects the phosphorylation of Akt substrates. It is important to note that the “phospho-Akt substrate antibody” also recognizes proteins that are phosphorylated by other kinases as long as the protein sequence matches to the antibody recognition motif. The phospho-Akt substrate antibody detected multiple proteins in HeLa cell lysates, many of which were stimulated by insulin and inhibited by LY249002, a PI3K-specific inhibitor (Supplementary Fig. S4). Knockdown of either hSin1 or Rictor caused a reduction in phosphorylation of several proteins as detected by the phospho-Akt substrate antibody. In contrast, knockdown of Raptor had little effect on the phosphorylation of those putative Akt substrates. Therefore, hSin1 plays an important role in the regulation of Akt activity and the phosphorylation of Akt substrates in vivo.
hSin1 is an essential component of TORC2 required for complex formation
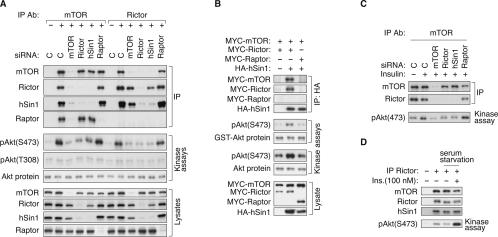
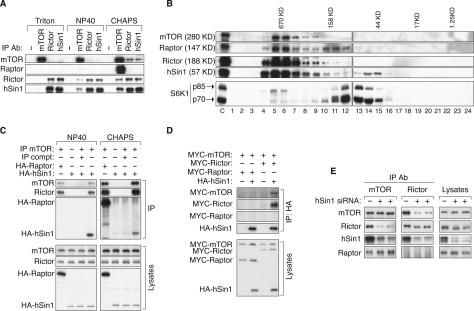
Based on the functional similarities between hSin1 and Rictor, as well as the limited sequence similarity between hSin1 and Avo1, we hypothesize that hSin1 is a component of TORC2. To test this model, we immunoprecipitated endogenous TOR complexes under different buffer conditions and examined the interactions between hSin1 and TORC2 components (Fig. 3A). HEK293 cells were lysed in buffers containing either 1% Triton X-100, 1% NP40, or 0.3% CHAPS. Consistent with previous findings (Kim et al. 2002; Sarbassov et al. 2004), both TORC1 and TORC2 were maintained in CHAPS buffer. We found that TORC1 was sensitive to both NP40 and Triton X-100, while TORC2 was less sensitive to NP40 but was disrupted by Triton X-100. Interestingly, the interaction between Rictor and hSin1 was not disrupted by any of the detergents tested, indicating that hSin1 and Rictor form a tight complex. This result is consistent with the observation that the knockdown of either protein results in the destabilization of the other.
FIGURE 3.
hSin1 is a genuine component of TORC2 and is essential for the complex formation. (A) Effect of different detergents on the integrity of TORC1 and TORC2. HEK293 cell were lysed in buffers containing either Triton X-100 (1%), NP40 (1%), or CHAPS (0.3%) as indicated. Each lysate was equally divided into four aliquots and subjected to immunoprecipitation with the indicated antibodies. (−) The immunoprecipitation control (no antibody was used). Coimmunoprecipitation of the TORC components was detected by specific antibodies as indicated. (B) hSin1 is present in a large-molecular-weight complex including mTOR and Rictor. HeLa cells were lysed in CHAPS buffer. Total cell lysates were fractionated by a Superose 12 Prep Grade Column. “C” indicates the whole-cell lysates before fractionation as a control. Numbers (1–24) at the bottom of the panel indicate fractionation numbers. Elution peaks of molecular weight standard are labeled at top of the panels. (C) mTOR interacts with transfected HA-hSin1. HA-hSin1 or HA-Raptor was transfected into HEK293 cells, and the transfected cells were lysed in either NP40 (1%) or CHAPS (0.3%) buffer as indicated. Samples were immunoprecipitated by an mTOR antibody (IP mTOR) as indicated. For the immunoprecipitation competition (compt), 1 μg of mTOR-peptide was added to the lysates prior to the addition of the mTOR antibody. The immunoprecipitated samples were probed with mTOR, Rictor, and HA antibodies. (D) The interaction between hSin1 and mTOR requires Rictor. MYC-mTOR, MYC-Rictor, MYC-Raptor, and HA-hSin1 were cotransfected into HEK293 cells as indicated. Cells were lysed in CHAPS buffer and immunoprecipitated with HA antibody. HA and MYC antibodies were used to determine the protein levels. (E) hSin1 is required for TORC2 integrity. HEK293 cells were transfected with hSin1 siRNA in duplicate where indicated. After harvesting, cell lysates were subjected to immunoprecipitation with either mTOR or Rictor antibody, followed by Western blotting with antibodies against TORC components.
Coimmunoprecipitation of hSin1 in TORC2 was also observed in HeLa cells (Supplementary Fig. S5A). Endogenous mTOR, Rictor, and hSin1 were coimmunoprecipitated with each other. In contrast, neither Rictor nor hSin1 could coprecipitate Raptor, while mTOR could. It is worth noting that the amount of mTOR coprecipitated by hSin1 was comparable to that coprecipitated by Rictor (Fig. 3A; Supplementary Fig. S5A). Furthermore, the amount of endogenous hSin1 coprecipitated by the endogenous Rictor was not much less than that precipitated by the hSin1 antibody itself (Fig. 3A; Supplementary Fig. S5A). Similarly, the amount of Rictor coprecipitated by hSin1 was not much less than that precipitated by the Rictor antibody itself. However, the amount of hSin1 and Rictor coprecipitated by mTOR were less than that precipitated by their respective antibodies. This could be due to less-efficient immunoprecipitation by the mTOR antibody or a relatively weak interaction between mTOR and hSin1 or Rictor. Based on these coimmunoprecipitation data, we conclude that hSin1 is a genuine component of TORC2, and is not present in TORC1.
To further support the notion that hSin1 is a genuine component of TOR complex, gel-filtration experiments using both HeLa and HEK293 cell lysates were performed to examine the association of hSin1 with the other TORC components. We found that the majority of endogenous hSin1 protein in HeLa cells coeluted with mTOR and Rictor as a high-molecular complex, while a small portion of hSin1 eluted as a monomer in the predicted 57-KD fraction (Fig. 3B). Similarly, mTOR, Rictor, and Raptor were coeluted as a high-molecular complex. As a control, most of the S6K1 protein in the lysates was detected in the fractions around 70 KD. Interestingly, a small amount of S6K coeluted with mTOR, consistent with an association between S6K and TORC1. We obtained similar results using HEK293 cell lysates (data not shown). Together, these data support that hSin1 is a subunit of the TOR complex under physiological conditions.
We performed additional transfection experiments to verify the interaction between hSin1 and mTOR. HA-hSin1 and HA-Raptor were transfected into HEK293 cells. Immunoprecipitation of endogenous mTOR was performed with an mTOR antibody in two different lysis buffers, NP40 and CHAPS buffer. HA-hSin1 was specifically coimmunoprecipitated by mTOR regardless of the detergent conditions (Fig. 3C). As expected, mTOR also coprecipitated both HA-Raptor and endogenous Rictor in the CHAPS buffer. Antibody competition by the mTOR peptide in the immunoprecipitations completely abolished the mTOR immunoprecipitation as well as the coprecipitated HA-hSin1, Rictor, and HA-Raptor, demonstrating the specificity of the coimmunoprecipitations. It is worth noting that the amount of HA-hSin1 coprecipitated with mTOR was significantly less than the HARaptor control. However, this difference was likely due to the relatively low level of HA-hSin1 expression, as indicated by the HA Western blot of the cell lysates (Fig. 3C).
Endogenous mTOR immunoprecipitates hSin1 (Fig. 3A,C; Supplementary Fig. S5A), but it is not clear whether Rictor is required for the interaction between mTOR and hSin1. To address this question, we performed HA-hSin1 immunoprecipitations when TORC components were overexpressed in HEK293 cells. HA-hSin1 coprecipitated MYC-Rictor/MYC-mTOR, but not MYC-Raptor/MYC-mTOR (Fig. 3D). In the absence of MYC-Rictor coexpression, HA-hSin1 coprecipitated little MYC-mTOR. These data indicate that Rictor is important for the interaction between hSin1 and mTOR, consistent with the observed strong interaction between hSin1 and Rictor (Fig. 3A; Supplementary Fig. S5A). The lack of interaction between HA-hSin1 and MYC-mTOR in the absence of MYC-Rictor cotransfection could be due to the fact that the endogenous Rictor level is limited.
To determine the function of hSin1 in TORC2 complex formation, hSin1 was knocked-down by RNAi in HEK293 cells. Endogenous mTOR or Rictor was immunoprecipitated, and the coprecipitated TORC components were analyzed by Western blotting. We found that knockdown of hSin1 significantly decreased the coprecipitation between Rictor and mTOR but had no effect on the coprecipitation between Raptor and mTOR (Fig. 3E). These data support that hSin1 is essential for the integrity of TORC2. In other words, when hSin1 protein is reduced, the interaction between mTOR and Rictor is significantly compromised.
Interestingly, hSin1 knockdown caused a significant increase of Rictor electrophoretic mobility, indicative of Rictor dephosphorylation (Fig. 3E; Supplementary Fig. S5B), suggesting that hSin1 may modulate Rictor phosphorylation. λ-phosphatase treatment increased the mobility of both hSin1and Rictor on SDS-PAGE (Supplementary Fig. S5C), demonstrating that the mobility shift was indeed caused by phosphorylation. Our results also indicate that the dephosphorylated Rictor may have a weaker affinity for mTOR binding. To test this hypothesis, we examined the Rictor protein precipitated by the Rictor antibody and those coprecipitated by the mTOR antibody. A gradient gel was used to better resolve the difference (Supplementary Fig. S5B). Our data clearly showed that the Rictor antibody precipitated both the hypo-and hyperphosphorylated Rictor proteins (Fig. 3E; Supplementary Fig. S5B). In contrast, the mTOR antibody preferentially coprecipitated the hyperphosphorylated Rictor (the slow migrating form). These data suggest that Rictor phosphorylation may modulate its interaction with mTOR and, more importantly, hSin1 may facilitate TORC2 complex formation by stimulating Rictor phosphorylation. In addition, we also observed that mTOR preferentially coprecipitated the slow migrating hSin1 (Fig. 3E; Supplementary Fig. S5B), indicating that the phosphorylation of hSin1 may also modulate its interaction with mTOR. Alternatively, it is possible that both hSin1 and Rictor are phosphorylated by mTOR when they are in the TORC2 complex.
QRT-PCR demonstrated that RNAi decreased hSin1 mRNA >90% (Supplementary Fig. S2C). Western blotting with the hSin1 antibody confirmed the effectiveness of the hSin1 RNAi (Figs. 2A,B, 3E, 4A). Interestingly, hSin1 protein levels were also dramatically decreased in Rictor knockdown cells. In fact, the reduction of hSin1 protein by Rictor RNAi (with no significant change of the hSin1 mRNA level) (Supplementary Fig. S2B) was comparable to the reduction caused by hSin1 RNAi. mTOR knockdown also decreased hSin1 protein levels, although to a lesser degree. These results are consistent with the notion that hSin1 protein levels depend on Rictor and mTOR, suggesting that hSin1 is stabilized by being in the TORC2 complex. The decrease of hSin1 protein caused by Rictor and mTOR knockdown further supports our model that hSin1 is an integral subunit of mTORC2 and functions mainly in TORC2 in vivo.
hSin1 is important for TORC2 kinase activity
It has been reported that TORC2 directly phosphorylates Ser473 of Akt (Sarbassov et al. 2005). We tested the effects of hSin1 knockdown on the TORC2 kinase activity in vitro. HeLa cells were treated with siRNAs targeting GFP (control), mTOR, Rictor, hSin1, and Raptor. mTOR was immunoprecipitated and in vitro kinase assays were performed using purified Akt as a substrate. We found that immunoprecipitated mTOR phosphorylated Akt on Ser473 but not on Thr308 (Fig. 4A). Knockdown of mTOR, Rictor, and hSin1 decreased the TORC2 kinase activity toward Ser473 of Akt. In contrast, knockdown of Raptor had no effect on the kinase activity toward Ser473 of Akt. Similar experiments were performed with Rictor immunoprecipitation. hSin1 knockdown almost completely eliminated the Rictor-associated kinase activity as did the mTOR and Rictor knockdown (Fig. 4A). Parallel experiments were performed using dephosphorylated recombinant S6K as a substrate to assay TORC1 activity. As expected, immunoprecipitated mTOR phosphorylated S6K in vitro and this kinase activity was diminished by mTOR knockdown (Supplementary Fig. S6). However, knockdown of neither Rictor nor hSin1 inhibited the mTOR kinase activity toward S6K (TORC1 activity) in vitro, consistent with the fact that they are not components of TORC1. These results provide strong in vitro biochemical evidence to support the idea that hSin1 is required for TORC2 to phosphorylate Ser473 of Akt, but it is dispensable for TORC1 to phosphorylate Thr389 of S6K.
To directly demonstrate that hSin1 associates with TORC2 kinase activity, we assayed the HA-hSin1 immunoprecipitation from HEK293 cells. Both purified GST-Akt from HEK293 cells and Akt from insect cells were used as substrates in the in vitro kinase reactions. In the presence of MYC-mTOR and MYC-Rictor, immunoprecipitated HA-hSin1 complex phosphorylated Ser473 of both Akt substrates (Fig. 4B). When MYC-Rictor was replaced by MYC-Raptor in the transfection, HA-hSin1 failed to precipitate kinase activity toward Ser473 of Akt.
We next examined the effects of Rictor and hSin1 on TORC2 activity in response to insulin stimulation. HeLa cells were treated with insulin and endogenous mTOR (Fig. 4C) or Rictor (Fig. 4D) was immunoprecipitated, and the kinase activity was assayed in vitro. We observed that insulin increased TORC2 kinase activity toward Akt, whereas the interaction among the TORC2 components was not significantly affected by insulin stimulation (Fig. 4C,D). Knockdown of mTOR, Rictor, or hSin1 significantly attenuated the insulin-stimulated TORC2 kinase activity assayed in vitro. In contrast, knockdown of Raptor did not decrease TORC2 kinase activity.
Functional role of hSin1 in cytoskeletal organization and apoptosis
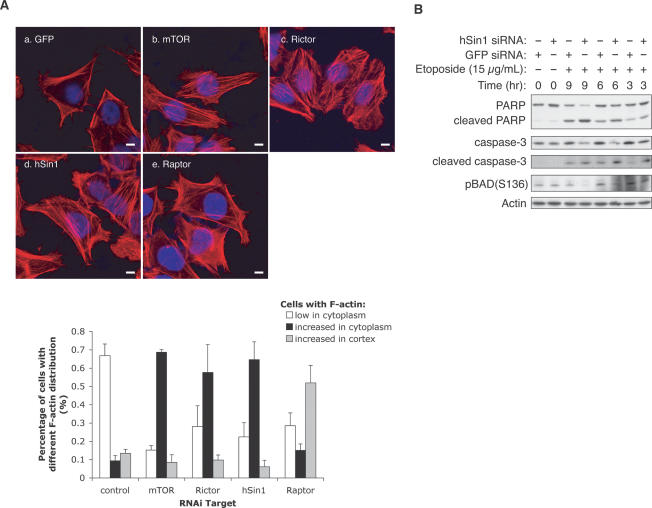
It has been well established that the yeast TOR genes are involved in the regulation of actin organization (Schmidt et al. 1996). Recently, the mammalian TORC2 has also been implicated in the regulation of cytoskeletal structure and actin organization (Jacinto et al. 2004; Sarbassov et al. 2004). To further support the role of hSin1 in TORC2 function, we stained F-actin (with rhodamineconjugated phalloidin) in HeLa cells with knockdown of different TORC components. As previously reported (Sarbassov et al. 2004), in mTOR knockdown cells actin fibers were present in the whole cell, while in control RNAi-treated cells, they were mainly localized in the cell cortex (Fig. 5A). We discovered that knockdown of hSin1 increased actin fibers in the cytoplasm. Similarly, Rictor knockdown also increased actin fibers in the cytoplasm. In cells with knockdown of mTOR, Rictor, or hSin1, the actin fibers were no longer restricted to the cell cortex. In contrast, Raptor knockdown promoted actin fibers accumulation in the cell cortex, but the cells clearly showed fewer actin fibers throughout the cytoplasm than those with either hSin1 or Rictor knockdown. Our results are consistent with the observations reported previously (Sarbassov et al. 2004) and further support our model that hSin1 is a genuine component of TORC2.
FIGURE 5.
hSin1 plays a role in actin organization and cell survival. (A) hSin1 RNAi displays similar actin fiber organization as Rictor RNAi. HeLa cells were transfected with siRNA against GFP control (a), mTOR (b), Rictor (c), hSin1 (d), and Raptor (e). Thirty-six hours post-transfection, cells were serum-starved for 12 h, followed by insulin stimulation (200 nM, 30 min) before fixation. F-actin (red) and nucleus (blue) were visualized under confocal microscope (bars, 10 μm). The column chart shows the quantification results of the change on cellular F-actin organization. For each knockdown, three sample groups were counted. Cells were divided into three categories based on F-actin staining: those with few stress fibers (<20% of the cell area, open bars), those with intense cytoplasmic stress fibers (filled bars), and those with strong Factin in the cortex (hatched bars). Quantification was determined by visual counting of confocal images. (B) hSin1 knockdown sensitizes cells to apoptosis. HeLa cells were transfected with either hSin1 or GFP (control) siRNA. Cells were treated with 15 μg/mL etoposide at 9, 6, and 3 h prior to harvesting; lysed in NP40 buffer; and analyzed by Western blotting. PARP antibody was used to detect both full-length and cleaved PARP. Caspase-3 antibody and caspase-3 (Asp175) antibody were used to detect procaspase-3 (full length) and cleaved caspase-3, respectively. Phosphorylation of BAD was determined by phospho-specific antibody as indicated. Note that the hSin1 RNAi treatment at T = 0 is shorter than the other experiments because the cells were treated with etopside for additional hours. This may explain a lack of effect of hSin1 RNAi on BAD phosphorylation. Additionally, BAD can be phosphorylated by other kinases besides Akt.
Akt activity is known to play an important role in inhibiting apoptosis (Brazil et al. 2004). Cells with reduced Akt activity often display high sensitivity to apoptosis inducers. We examined the effect of hSin1 knockdown on apoptosis in HeLa cells in response to etopo-side, a potent apoptosis inducer. We observed that hSin1 knockdown increased the sensitivity to etoposide, as judged by the increase of caspase-3 cleavage and PARP cleavage, both of which are well-characterized apoptosis markers (Fig. 5B). Since the basal level of Akt phosphorylation could not be accurately detected (Fig. 2A), we decided to examine the phosphorylation of BAD on Ser136. It is well established that Akt phosphorylates BAD on Ser136 to inhibit apoptosis. Phosphorylation of BAD was indeed decreased by hSin1 knockdown (Fig. 5B). These observations are consistent with the decreased Akt activity observed in hSin1 knockdown cells and support a positive role of hSin1 in Akt activation.
Rictor knockout diminishes Akt phosphorylation
The evidence for TORC2 involvement in Akt regulation is largely based on RNAi-mediated knockdown in cultured cells and in vitro biochemical studies. Phosphorylation of Akt on Ser473 is reduced, but not eliminated, in Rictor knockdown cells. We created Rictor knockout mice to determine the function of Rictor in Akt activation in vivo. The Rictor knockouts were derived from an embryonic stem (ES) cell that contains an insertion of both β-galactosidase and neomycin genes in intron 3 of the Rictor gene (Supplementary Fig. S7A). This insertion truncates Rictor protein at amino acid residue 65, and therefore inactivates Rictor.
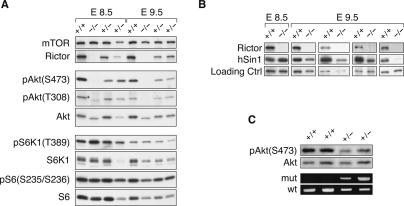
We failed to obtain homozygous Rictor−/− mice, demonstrating that Rictor knockout is embryonic lethal. We found that Rictor−/− embryos die around embryonic day 10.5–12.5 (E10.5–E12.5) (detailed analysis of Rictor knockout requires further studies and will be described elsewhere). To determine the critical role of Rictor in Akt phosphorylation, we examined Rictor−/− embryos. We found that the Rictor−/− embryos had no detectable Akt phosphorylation on Ser473 (Fig. 6A). As expected, phosphorylation of S6K and S6 was not decreased in Rictor−/− embryos. These results unequivocally demonstrate that Rictor plays an essential role in Akt phosphorylation and is the long-sought kinase responsible for Ser473 phosphorylation in the hydrophobic region. Interestingly, phosphorylation of Akt on Thr308 in E8.5 Rictor−/− embryos was reduced but not eliminated. In E9.5 Rictor−/− embryos, the Thr308 phosphorylation of Akt was greatly decreased, an observation consistent with our in vivo RNAi knockdown data in cultured cells. However, the decrease in Akt Thr308 phosphorylation was less dramatic in later-stage embryos (data not shown). These data indicate that Rictor may have a positive, though not direct role in the phosphorylation of Thr308. Sin1 protein levels were unchanged in the E8.5 Rictor knockout embryos. However, in Rictor−/− E9.5 embryos, Sin1 protein levels varied from a slight decrease to a dramatic reduction (Fig. 6B). Possible explanations for the variations could be that the embryos were different in genetic background and/or the degradation machinery for Sin1 develops around E9.5. As a consequence, a small change of the embryo age may result in a dramatic effect on Sin1 protein levels. Together, these data support our observations in the cultured cell systems that Rictor stabilizes Sin1 protein.
FIGURE 6.
Rictor knockout inhibits Akt phosphorylation. (A) Akt phosphorylation but not S6K1 phosphorylation is inhibited in Rictor knockout. Rictor knockout embryos from heterozygous intercrosses at E8.5E9.5 were dissected. At the age of either E8.5 or E9.5, one wild-type (+/+), one homozygous knockout (−/−), and two heterozygous (+/−) embryos were analyzed by Western blotting for phosphorylation and protein levels as indicated. (B) Decreased hSin1 protein levels in Rictor knockout embryos. Samples were prepared as described in A. Each group of wild-type and knockout samples was prepared at the same time and analyzed by Western blotting in the same panel. Different sample groups were collected at different times. (C) Decreased Akt phosphorylation in the liver tissues of Rictor+/− heterozygous knockout mice. The mice liver tissues were collected at 2 wk of age. Samples from two wild-type (+/+) and two heterozygous (+/−) mice were analyzed by both Western blotting (top two panels) and PCR genotyping (bottom two panels).
Phosphorylation of putative Akt substrates in Rictor−/− embryos was analyzed by Western blotting with the phospho-Akt substrate antibody (Supplementary Fig. S7B). Our data show that phosphorylation of multiple proteins was decreased in the Rictor−/− embryos, indicating that Akt kinase activity decreased in vivo. These observations further demonstrate that TORC2 plays a critical role in Akt activation in vivo.
To further determine the role of Rictor in Akt phosphorylation, we measured Akt phosphorylation in the liver of Rictor heterozygous mice. Western blotting of Rictor+/− tissues indicated that the heterozygotes had a detectable decrease in Akt phosphorylation on Ser473 (Fig. 6C). This observation indicates Rictor haploinsufficiency in Akt phosphorylation. The data obtained with Rictor knockout studies demonstrate that Rictor is essential for Akt Ser473 phosphorylation in vivo.
Discussion
The biological importance of mTORC2 is appreciated by the finding that it is the long-sought kinase that plays a critical role in Akt phosphorylation and activation (Sarbassov et al. 2005). Three components of mTORC2, including mTOR, Rictor, and mLST8, have been characterized and all of them are structurally and functionally conserved in eukaryotes from yeast to mammalian cells (Wullschleger et al. 2006). Hall's group (Loewith et al. 2002) originally identified more components in TORC2, including Avo1 and Avo2. In addition, Rip3, an Avo1 homolog in Dictyostelium, has recently been found to be an essential component of TORC2 (Lee et al. 2005). Bit61 was also reported as another component of TORC2 (Wullschleger et al. 2005). Therefore, it is reasonable to speculate that mTORC2 has other components in addition to the three known ones. The existence of higher eukaryotic homologs of Avo2 and Bit61 is not apparent based on sequence analysis. In contrast, hSin1 could be identified as a potential homolog of yeast Avo1, based on the very limited sequence similarity. However, previous studies had indicated that hSin1 is not a mammalian TORC2 component (Loewith et al. 2002).
In this study, we have presented convincing biochemical and cell biological data demonstrating that hSin1 is a genuine component of mTORC2 and is essential for the mTORC2 complex formation. This conclusion is supported by the following observations: Endogenous hSin1 can be readily coprecipitated by both mTOR and Rictor under physiological conditions. hSin1 is detected in mTORC2 but absent in mTORC1. hSin1 coelutes with mTOR in gel-filtration chromatography. Transfected HA-hSin1 shows an interaction with transfected mTOR in a Rictor-dependent manner. Knockdown of hSin1 disrupts the mTORC2 complex formation. In addition, knockdown of Rictor, but not Raptor, dramatically decreases hSin1 protein levels. Furthermore, hSin1 knockdown inhibits Rictor phosphorylation and also decreases Rictor protein levels. These data establish hSin1 as an integral subunit of TORC2 and support an important function of hSin1 in TORC2 complex formation. It is worth noting that both Avo1 (Sin1 homolog) and Avo3 (Rictor homolog) are important for TORC2 complex formation in yeast (Loewith et al. 2002; Wullschleger et al. 2005). The TORC2 complex is disrupted in either AVO1 or AVO3 knockout yeast, indicating structural and functional conservation of TORC2 components.
By examining Akt phosphorylation, we show that hSin1 is required for the phosphorylation of Akt on Ser 473, the TORC2 site, in cultured cells. In addition, hSin1 knockdown significantly decreases the ability of immunoprecipitated mTOR to phosphorylate Akt in vitro. Moreover, hSin1 can immunoprecipitate TORC2 kinase activity. These observations establish the functional importance of hSin1 in TORC2 activity and Akt phosphorylation. Finally, similar to the results observed with Rictor RNAi, hSin1 knockdown also inhibits the phosphorylation of Akt on the PDK1 site, Thr308; however, this site is not directly phosphorylated by TORC2. It is possible that the phosphorylation of Ser473 facilitates the phosphorylation of Thr308 in Akt. Alternatively, but not exclusively, TORC2 may positively contribute to PDK1 function. Future studies are necessary to clarify the relationship between Thr308 and Ser473 phosphorylation. By all the criteria examined, hSin1 functions similarly to Rictor as a key component of TORC2 that is essential for Akt phosphorylation on Ser473. Knockdown of hSin1 decreases Rictor phosphorylation and its association with mTOR. This observation suggests that phosphorylation of Rictor may depend on hSin1 and TORC2 activity. It is reasonable to speculate that hSin1 may serve as the signal-receiving component in TORC2.
We propose that the hSin1 protein is stabilized in TORC2, especially by its interaction with Rictor. Reciprocally, hSin1 also contributes to Rictor stability. In the absence of TORC2, such as in Rictor knockdown cells, hSin1 protein decreases dramatically. Knockdown of mTOR also decreases hSin1 protein levels, although the effect is much less dramatic than that caused by Rictor knockdown. Currently, we cannot formally exclude the possibility that TORC2 (or Rictor) modulates hSin1 translation, thereby affecting hSin1 protein levels. Interestingly, Sin1 protein levels are also decreased to varying degrees in E9.5 Rictor−/− embryos. It is possible that the hSin1 degradation mechanism/machinery is not present in early embryos and develops around E9.5 during embryogenesis. The Rictor knockout results further support our model that hSin1 is stabilized by complex formation with Rictor.
The importance of Rictor in Akt phosphorylation is confirmed by our Rictor knockout experiments. Our results show that the Rictor dependent-kinase is the primary (perhaps sole) hydrophobic kinase for Akt in developing embryos. Therefore, TORC2 may be the primary kinase responsible for the phosphorylation of Akt on the hydrophobic site. However, we cannot exclude the possibility that, in addition to mTOR, Rictor may associate with another kinase that can also phosphorylate Ser473 in Akt. It is also possible that in addition to TORC2, another kinase(s) may phosphorylate Ser473 during the late stages of development or in adult animals. Although our Rictor knockout confirms a critical role of Rictor in Akt phosphorylation on Ser473, the requirement of Rictor for Akt Thr308 phosphorylation is less clear. RNAi knockdown experiments in cultured mammalian cells show that Rictor is also important for Akt Thr308 phosphorylation. Identical results are obtained with hSin1 knockdown. However, the effect of Rictor knockout on Akt Thr308 phosphorylation may be developmental stage dependent. We propose that both Rictor and hSin1 play a physiological, though indirect role, in the phosphorylation of Thr308 in the activation loop of Akt. Also, TORC2 may positively contribute to PDK1 activity regulation. Based on the Thr308 phosphorylation results of the E8.5 embryos, one may speculate that compensation occurs in the early stage Rictor−/− knockout embryos, thus reducing the dependency of Akt Thr308 phosphorylation on TORC2.
In summary, this report establishes that both hSin1 and Rictor are essential components of TORC2 and are required for the assembly and function of TORC2. We also show that hSin1 plays an important role in Akt phosphorylation. Therefore, mutation or uncontrolled activation of hSin1 could result in dysregulation of Akt and possibly cause abnormalities in cell growth and apoptosis, leading to tumor formation. Finally, we conclude that Rictor, hence TORC2, plays an essential role in Akt Ser473 phosphorylation and activity during embryonic development.
Materials and methods
Plasmids, antibodies, and chemicals
pSRα-3HA-hSin1 was a kind gift from Dr. Bing Su (MD Anderson Cancer Center, Houston, TX). Other plasmid DNAs were described previously (Yang et al. 2006). Phospho-Drosophila Akt (Ser505), phospho-Drosophila S6K (Thr398), phospho-Akt (Ser473), phspho-Akt (Thr308), phospho-GSK3β (Ser9), phospho-S6K1 (Thr389), phospho-S6K1 (Thr421/Ser424), phospho-S6 (Ser235/Ser236), phospho-Akt substrate, Akt, S6K1, GSK3β, S6, mTOR (used for Western blotting), Raptor, PARP, caspase-3, cleaved caspase-3 (Asp175), and phospho-BAD (Ser136) antibodies were from Cell Signaling. Drosophila S6K antibody was kindly provided by Dr. Mary Stewart (North Dakota State University, Fargo, ND) and the hSin1 antibody was kindly provided by Dr. Bing Su. mTOR antibody, used in immunoprecipitation, was purchased from Santa Cruz Biotechnology. Rictor antibody was from Bethyl Laboratories. HA(HA.11) and MYC(9E10) antibodies were purchased from Covance. GST and actin antibodies were obtained from Molecular Probes and Sigma, respectively. LY294002 and etoposide were purchased from Calbiochem. Insulin was from Invitrogen.
Cell culture and transfection
HEK293 and HeLa cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM) containing 10% fetal bovine serum (FBS) with 50 U/mL penicillin/streptomycin (pen/strep). Drosophila S2 cells (Invitrogen) were cultured in Drosophila-SFM (Invitrogen) with 2.92 mg/mL L-glutamine and 50 U/mL pen/ strep. Unless specified, mammalian cells were cultured in six-well plates for all of the experiments, while S2 cells were all cultured in 12-well plates. Mammalian cell transfections, including siRNA transfections, were performed under serum-free conditions using lipofectamine reagent (Invitrogen), following the manufacturer's protocol.
RNAi
For each Drosophila S2 cell RNAi experiment, cells were plated in 12-well plates, with a starting density of 2 × 105 cells per well. 5′ primer GAGCCGGATCGCAACTACCC and 3′ primer GGCGTCGCTCCAAAATCTCG were used to generate dSin1_1 dsRNA, while 5′ primer TCGGGCGCTGGAGGAGAT and 3′ primer CCGTTTCGTTTGCATACCCACTTG were used to generate dSin1_2 dsRNA. Detailed protocols, including dsRNA synthesis procedure, RNAi experimental procedure, and additional information of primer sequences, were described previously (Yang et al. 2006). For HEK293 cell and HeLa cell RNAi, siGENOME SMARTpool siRNA targeting either mTOR (NM_004958), Rictor (NM_152756), Raptor (NM_020761), mLST8 (NM_022372), PDK1 (NM_002613), or hSin1 (NM_024117), and the single duplex siRNA targeting either Rictor or hSin1 were ordered from Dharmacon. For all mammalian cell RNAi experiments, cells were plated in six-well plates with a starting density of 3.6 × 105 cells per well and transfected with 80 nM siRNA for 4 h for each specific target. Cells were lysed 48 h post-transfection for immunoprecipitations and/or Western blotting analysis.
Gel filtration
Cells were cultured in a 10-cm plate. After reaching confluence, cells were lysed in 1 mL CHAPS buffer (see Immunoprecipitation). Cell lysate was centrifuged at 13,200 rpm for 15 min and the supernatant was concentrated to 250 μL using a Centriprep YM-10 KD (Millipore). Samples were loaded onto a Superose 12 Prep Grade Column connected to an FPLC system. Elution was collected in 400 μL per fraction.
Western blotting, immunoprecipitation, and kinase assay
For Western blot analysis, cells cultured in 12-and six-well plates were lysed in 100 and 250 μL of NP40 buffer (Mild Lysis buffer) (Yang et al. 2006), respectively. Rictor knockout mice embryos and liver tissues were also lysed in NP40 buffer. For imunoprecipitations, cells cultured in a six-well plate were lysed in 350 μL 1% Triton, 1% NP40, or 0.3% CHAPS buffer as previously described (Kim et al. 2002). One microgram of antibody was added to each of the cellular lysates and incubated for 90 min at 4°C. Ten microliters of protein G-sepharose slurry (50%) was added to the lysates and incubated for another hour. Immunoprecipitates were washed four times in the lysis buffer before Western blotting analysis. For kinase assays, after washing with lysis buffer, the immunoprecipitates were washed once more with kinase buffer and then incubated in 15 μL kinase assay reaction mix for 30 min at 37°C. Kinase assay reactions were designed as previously reported (Sarbassov et al. 2004). The dephosphorylated recombinant Akt substrate (Upstate Biotechnology), GST-S6K, and GST-Akt purified from HEK293 cells were used as substrates (Yang et al. 2006). To stop the reaction, 5 μL of 4× SDS sample buffer was added to each reaction, which was then boiled for 5 min.
Confocal microscopy
HeLa cells were grown and transfected on glass coverslips. Forty-eight hours after transfection, the cells were fixed with 4% paraformaldehyde/PBS, permeabilized with 0.1% Triton X-100/PBS, and blocked with 2% BSA/PBS. Cells were then incubated with rhodamine-phalloidin (1:500 dilution, Molecular Probes) and mounted with Vectashield H-1200 (with DAPI, Vector Laboratories). Signals were detected by using the Olympus FluoView 500 Laser Scanning Confocal Microscope at the Diabetes Center of the University of Michigan.
Generation of Rictor knockout mice
A mouse ES cell line (RRR347, strain 129/OlaHsd) containing an insertional mutation located in the third intron of Rictor was obtained from BayGenomics. The gene-trap vector used (pGT0Lxf) contains a splice-acceptor sequence upstream of the reporter gene β-geo (a fusion of lacZ and neomycin). The mutation results in the production of an in-frame fusion transcript consisting of exons 1–3 from Rictor (amino acids 1–65) and β-geo. The ES cells were microinjected into C57BL/6 blasto-cysts to produce chimeric male mice by Transgenic Animal Model Core Laboratory of the University of Michigan. Chimeric animals were bred to C57BL/6 mice (Jackson Laboratory) to produce heterozygous animals. The following primers were used for genotyping: Ric-F, 5′-GACCGATTTTCCACTTTCGTTG-3′; Ric-R, 5′-GACTTCTCTCTGCAACAGATGC-3′; and β-geo-R (from vector sequences), 5′-TCCAGACAAGTAGATCCCGG CG-3′. The wild-type allele was detected with primers Ric3-F and Ric3-R, yielding a 342-base-pair (bp) product, whereas the mutant allele was detected with primers Ric3-F and βgeo-R, yielding a 505-bp product.
Acknowlegments
We thank Drs. Bing Su (MD Anderson Cancer Center), David Sabatini (MIT), and Mary Stewart (North Dakota State University) for reagents; Drs. Yuan Zhu, Jiandie Lin, Jiahai Zhou, Zhaohui Xu, and Hiroyuki Mori for technical assistance; and Chung-Han Lee, Jean Guan, and Pamela Wong for critically reading the manuscript. This work is supported by grants from NIH and DOD (ot K.L.G); Q.Y. and I.T. are partially supported by the Anthony and Lilian Lu Fellowship and the Uehara Memorial Foundation and Mochida Memorial Foundation for Medical and Pharmaceutical Research, respectively.
Footnotes
Supplemental material is available at http://www.genesdev.org
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1461206.
References
- Abraham, R.T., Wiederrecht, G.J. Immunopharmacology of rapamycin. Annu. Rev. Immunol. 1996;14:483–510. doi: 10.1146/annurev.immunol.14.1.483. [DOI] [PubMed] [Google Scholar]
- Alessi, D.R., Caudwell, F.B., Andjelkovic, M., Hemmings, B.A., Cohen, P. Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 kinase. FEBS Lett. 1996;399:333–338. doi: 10.1016/s0014-5793(96)01370-1. [DOI] [PubMed] [Google Scholar]
- Alessi, D.R., James, S.R., Downes, C.P., Holmes, A.B., Gaffney, P.R., Reese, C.B., Cohen, P. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Bα . Curr. Biol. 1997;7:261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- Balendran, A., Casamayor, A., Deak, M., Paterson, A., Gaffney, P., Currie, R., Downes, C.P., Alessi, D.R. PDK1 acquires PDK2 activity in the presence of a synthetic peptide derived from the carboxyl terminus of PRK2. Curr. Biol. 1999;9:393–404. doi: 10.1016/s0960-9822(99)80186-9. [DOI] [PubMed] [Google Scholar]
- Brazil, D.P., Yang, Z.Z., Hemmings, B.A. Advances in protein kinase B signalling: AKTion on multiple fronts. Trends Biochem. Sci. 2004;29:233–242. doi: 10.1016/j.tibs.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Cheng, J., Zhang, D., Kim, K., Zhao, Y., Su, B. Mip1, an MEKK2-interacting protein, controls MEKK2 dimerization and activation. Mol. Cell. Biol. 2005;25:5955–5964. doi: 10.1128/MCB.25.14.5955-5964.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross, D.A., Alessi, D.R., Cohen, P., Andjelkovich, M., Hemmings, B.A. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- Feng, J., Park, J., Cron, P., Hess, D., Hemmings, B.A. Identification of a PKB/Akt hydrophobic motif Ser-473 kinase as DNA-dependent protein kinase. J. Biol. Chem. 2004;279:41189–41196. doi: 10.1074/jbc.M406731200. [DOI] [PubMed] [Google Scholar]
- Hagemann, C., Blank, J.L. The ups and downs of MEK kinase interactions. Cell. Signal. 2001;13:863–875. doi: 10.1016/s0898-6568(01)00220-0. [DOI] [PubMed] [Google Scholar]
- Hara, K., Maruki, Y., Long, X., Yoshino, K., Oshiro, N., Hidayat, S., Tokunaga, C., Avruch, J., Yonezawa, K. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–189. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- Harrington, L.S., Findlay, G.M., Gray, A., Tolkacheva, T., Wig-field, S., Rebholz, H., Barnett, J., Leslie, N.R., Cheng, S., Shepherd, P.R., et al. The TSC1-2 tumor suppressor controls insulin–PI3K signaling via regulation of IRS proteins. J. Cell Biol. 2004;166:213–223. doi: 10.1083/jcb.200403069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay, N., Sonenberg, N. Upstream and downstream of mTOR. Genes & Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- Inoki, K., Ouyang, H., Li, Y., Guan, K.L. Signaling by target of rapamycin proteins in cell growth control. Micro-biol. Mol. Biol. Rev. 2005;69:79–100. doi: 10.1128/MMBR.69.1.79-100.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto, E., Loewith, R., Schmidt, A., Lin, S., Ruegg, M.A., Hall, A., Hall, M.N. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 2004;6:1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- Kim, D.H., Sarbassov, D.D., Ali, S.M., King, J.E., Latek, R.R., Erdjument-Bromage, H., Tempst, P., Sabatini, D.M. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- Kim, D.H., Sarbassov, D.D., Ali, S.M., Latek, R.R., Guntur, K.V., Erdjument-Bromage, H., Tempst, P., Sabatini, D.M. GβL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol. Cell. 2003;11:895–904. doi: 10.1016/s1097-2765(03)00114-x. [DOI] [PubMed] [Google Scholar]
- Lawlor, M.A., Mora, A., Ashby, P.R., Williams, M.R., Murray-Tait, V., Malone, L., Prescott, A.R., Lucocq, J.M., Alessi, D.R. Essential role of PDK1 in regulating cell size and development in mice. EMBO J. 2002;21:3728–3738. doi: 10.1093/emboj/cdf387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S., Comer, F.I., Sasaki, A., McLeod, I.X., Duong, Y., Okumura, K., Yates, J.R., III, Parent, C.A., Firtel, R.A. TOR complex 2 integrates cell movement during chemo-taxis and signal relay in Dictyostelium . Mol. Biol. Cell. 2005;16:4572–4583. doi: 10.1091/mbc.E05-04-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith, R., Jacinto, E., Wullschleger, S., Lorberg, A., Crespo, J.L., Bonenfant, D., Oppliger, W., Jenoe, P., Hall, M.N. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- McManus, E.J., Sakamoto, K., Armit, L.J., Ronaldson, L., Shpiro, N., Marquez, R., Alessi, D.R. Role that phosphorylation of GSK3 plays in insulin and Wnt signalling defined by knockin analysis. EMBO J. 2005;24:1571–1583. doi: 10.1038/sj.emboj.7600633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora, A., Komander, D., van Aalten, D.M., Alessi, D.R. PDK1, the master regulator of AGC kinase signal transduction. Semin. Cell Dev. Biol. 2004;15:161–170. doi: 10.1016/j.semcdb.2003.12.022. [DOI] [PubMed] [Google Scholar]
- Persad, S., Attwell, S., Gray, V., Mawji, N., Deng, J.T., Leung, D., Yan, J., Sanghera, J., Walsh, M.P., Dedhar, S. Regulation of protein kinase B/Akt-serine 473 phosphorylation by integrin-linked kinase: Critical roles for kinase activity and amino acids arginine 211 and serine 343. J. Biol. Chem. 2001;276:27462–27469. doi: 10.1074/jbc.M102940200. [DOI] [PubMed] [Google Scholar]
- Radimerski, T., Montagne, J., Hemmings-Mieszczak, M., Thomas, G. Lethality of Drosophila lacking TSC tumor suppressor function rescued by reducing dS6K signaling. Genes & Dev. 2002a;16:2627–2632. doi: 10.1101/gad.239102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radimerski, T., Montagne, J., Rintelen, F., Stocker, H., van der Kaay, J., Downes, C.P., Hafen, E., Thomas, G. dS6K-regulated cell growth is dPKB/dPI(3)K-independent, but requires dPDK1. Nat Cell Biol. 2002b;4:251–255. doi: 10.1038/ncb763. [DOI] [PubMed] [Google Scholar]
- Rane, M.J., Coxon, P.Y., Powell, D.W., Webster, R., Klein, J.B., Pierce, W., Ping, P., McLeish, K.R. p38 kinase-dependent MAPKAPK-2 activation functions as 3-phosphoi-nositide-dependent kinase-2 for Akt in human neutrophils. J. Biol Chem. 2001;276:3517–3523. doi: 10.1074/jbc.M005953200. [DOI] [PubMed] [Google Scholar]
- Reinke, A., Anderson, S., McCaffery, J.M., Yates, J., III, Aronova, S., Chu, S., Fairclough, S., Iverson, C., Wedaman, K.P., Powers, T. TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae . J. Biol. Chem. 2004;279:14752–14762. doi: 10.1074/jbc.M313062200. [DOI] [PubMed] [Google Scholar]
- Sarbassov, D.D., Ali, S.M., Kim, D.H., Guertin, D.A., Latek, R.R., Erdjument-Bromage, H., Tempst, P., Sabatini, D.M. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- Sarbassov, D.D., Guertin, D.A., Ali, S.M., Sabatini, D.M. Phosphorylation and regulation of Akt/PKB by the Rictor–mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Sawyers, C.L. Will mTOR inhibitors make it as cancer drugs? Cancer Cell. 2003;4:343–348. doi: 10.1016/s1535-6108(03)00275-7. [DOI] [PubMed] [Google Scholar]
- Schmidt, A., Kunz, J., Hall, M.N. TOR2 is required for organization of the actin cytoskeleton in yeast. Proc. Natl. Acad. Sci. 1996;93:13780–13785. doi: 10.1073/pnas.93.24.13780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder, W., Cloonan, N., Bushell, G., Sculley, T. Alternative polyadenylation and splicing of mRNAs transcribed from the human Sin1 gene. Gene. 2004;339:17–23. doi: 10.1016/j.gene.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Shah, O.J., Wang, Z., Hunter, T. Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr. Biol. 2004;14:1650–1656. doi: 10.1016/j.cub.2004.08.026. [DOI] [PubMed] [Google Scholar]
- Stocker, H., Radimerski, T., Schindelholz, B., Wittwer, F., Bela-wat, P., Daram, P., Breuer, S., Thomas, G., Hafen, E. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila . Nat. Cell Biol. 2003;5:559–565. doi: 10.1038/ncb995. [DOI] [PubMed] [Google Scholar]
- Toker, A., Newton, A.C. Akt/protein kinase B is regulated by autophosphorylation at the hypothetical PDK-2 site. J. Biol. Chem. 2000;275:8271–8274. doi: 10.1074/jbc.275.12.8271. [DOI] [PubMed] [Google Scholar]
- Um, S.H., Frigerio, F., Watanabe, M., Picard, F., Joaquin, M., Sticker, M., Fumagalli, S., Allegrini, P.R., Kozma, S.C., Auwerx, J., et al. Absence of S6K1 protects against ageand diet-induced obesity while enhancing insulin sensitivity. Nature. 2004;431:200–205. doi: 10.1038/nature02866. [DOI] [PubMed] [Google Scholar]
- Wilkinson, M.G., Pino, T.S., Tournier, S., Buck, V., Martin, H., Christiansen, J., Wilkinson, D.G., Millar, J.B. Sin1: An evolutionarily conserved component of the eukaryotic SAPK pathway. EMBO J. 1999;18:4210–4221. doi: 10.1093/emboj/18.15.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodgett, J.R. Recent advances in the protein kinase B signaling pathway. Curr. Opin. Cell Biol. 2005;17:150–157. doi: 10.1016/j.ceb.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Wullschleger, S., Loewith, R., Oppliger, W., Hall, M.N. Molecular organization of target of rapamycin complex 2. J. Biol. Chem. 2005;280:30697–30704. doi: 10.1074/jbc.M505553200. [DOI] [PubMed] [Google Scholar]
- Wullschleger, S., Loewith, R., Hall, M.N. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Yang, Q., Inoki, K., Kim, E., Guan, K.L. TSC1/TSC2 and Rheb have different effects on TORC1 and TORC2 activity. Proc. Natl. Acad. Sci. 2006;103:6811–6816. doi: 10.1073/pnas.0602282103. [DOI] [PMC free article] [PubMed] [Google Scholar]