Abstract
Dendrites exhibit a wide range of morphological diversity, and their arborization patterns are critical determinants of proper neural connectivity. How different neurons acquire their distinct dendritic branching patterns during development is not well understood. Here we report that Spineless (Ss), the Drosophila homolog of the mammalian aryl hydrocarbon (dioxin) receptor (Ahr), regulates dendrite diversity in the dendritic arborization (da) sensory neurons. In loss-of-function ss mutants, class I and II da neurons, which are normally characterized by their simple dendrite morphologies, elaborate more complex arbors, whereas the normally complex class III and IV da neurons develop simpler dendritic arbors. Consequently, different classes of da neurons elaborate dendrites with similar morphologies. In its control of dendritic diversity among da neurons, ss likely acts independently of its known cofactor tango and through a regulatory program distinct from those involving cut and abrupt. These findings suggest that one evolutionarily conserved role for Ahr in neuronal development concerns the diversification of dendrite morphology.
Keywords: Drosophila, neuron, dendrite, axon, aryl hydrocarbon receptor, dioxin
Each neuronal cell type is endowed with a unique dendritic architecture, from the relatively simple arbors of a bipolar neuron to the elaborate and expansive dendritic tree of a cerebellar Purkinje cell. Critical for proper neural connectivity, how a neuron elaborates its dendritic arbor in large part determines the way it integrates and processes synaptic or sensory inputs (Hausser et al. 2000; Jan and Jan 2001; Scott and Luo 2001; Wong and Ghosh 2002). Furthermore, the morphological properties of dendrites can influence the activity, and hence the function, of a neuron (Hausser et al. 2000; Wong and Ghosh 2002). Developmental programs that regulate dendrite morpho- genesis, by controlling such distinct modes as dendrite outgrowth, branching, stabilization, and maintenance, require the concerted actions of various extrinsic and intrinsic factors (Scott and Luo 2001; Whitford et al. 2002; Jan and Jan 2003). However, relatively little is known about how different types of neurons acquire their distinct dendrite arborization patterns.
The Drosophila peripheral nervous system (PNS) provides an ideal in vivo model system to investigate the molecular mechanisms that regulate dendrite patterning. The Drosophila PNS is comprised of many different cell types including the dendritic arborization (da) sensory neurons (Bodmer and Jan 1987). The da neurons are further subdivided into four classes (I–IV) based on their increasing dendritic branching complexity, which is defined by branch number and branch order as assessed by Strahler analysis (Grueber et al. 2002). The da neurons are born during mid-embryogenesis, and each neuron elaborates a stereotypical dendritic arbor that innervates the epidermis (Bodmer and Jan 1987). Different classes of da neurons are thought to function in different sensory modalities such as nociception, thermosensation, and locomotion (Ainsley et al. 2003; Liu et al. 2003; Tracey et al. 2003; Xu et al. 2004), suggesting that their distinct dendrite arborization patterns are important for specific behaviors. This classification of the da neurons, initially based primarily on their dendrite morphologies, is corroborated by more recent evidence demonstrating molecular differences among these four classes of neurons (Grueber et al. 2003a; Li et al. 2004; Sugimura et al. 2004). Each of the 15 da neurons in an abdominal hemisegment exhibits stereotypical dendritic arbors, thereby allowing systematic analysis of dendritic morphogenesis tailored for individual cell types.
Previous studies in Drosophila have shown that transcriptional regulation plays an integral role in the proper development of da neuron dendrites (Brenman et al. 2001; Grueber et al. 2003a; Li et al. 2004; Sugimura et al. 2004; Parrish et al. 2006). These studies have provided insight into the mechanisms that different neurons use to establish their distinct dendrite morphologies. Neurons may act through transcriptional programs that directly promote dendrite outgrowth and branching. For example, the homeodomain protein Cut promotes the formation of complex dendritic arbors, and increasing levels of Cut expression correlate with greater dendritic complexity of da neurons (Grueber et al. 2003a). Neurons may also use cell-type-specific transcriptional programs to regulate dendritic branching. Thus, class I da neurons express the BTB-zinc finger protein Abrupt, but not Cut, to limit dendrite complexity (Li et al. 2004; Sugimura et al. 2004). Whether there are genetic programs that specifically govern the dendritic complexity unique to each of the four classes of da neurons, however, is not known.
Through a mutagenesis screen, we found the spineless (ss) gene to be critical for the diversification of dendrite complexity. Since the discovery of the first ss mutant >80 yr ago by Bridges and Morgan (1923), three classes of phenotypes have been associated with ss mutants: (1) transformation of the distal part of the antenna to the distal part of the second leg (mutants with this pheno-type are known as spineless-aristapedia mutants), (2) deletion of most of the tarsal regions of the legs, and (3) reduction of bristle size. In addition, a recent study reports that stochastic ss expression enables R7 photoreceptors to express a particular form of rhodopsin for color vision (Wernet et al. 2006). Although ss has been previously shown to be expressed in the PNS (Duncan et al. 1998), no PNS phenotype has been reported with then available markers such as the widely used monoclonal 22C10 antibody that labels the cell body and axons of sensory neurons (Zipursky et al. 1984). Here we report that ss mutants exhibit a striking dendrite phenotype that only becomes apparent with suitable dendrite markers.
Spineless (Ss) was previously identified as a member of a family of transcription factors that contain the basic helix–loop–helix (bHLH)-PAS domain (Duncan et al. 1998). The PAS domain mediates dimerization and is named after the founding members of the family: Drosophila Period, mammalian Ahr, and Drosophila Single- minded (Huang et al. 1993; Gu et al. 2000). The Ss protein is most similar to the mammalian aryl hydrocarbon (dioxin) receptor (Ahr), as ss and Ahr share almost twice as much sequence identity as either shares with its next closest relative (Duncan et al. 1998). Ss can heterodimerize with the bHLH-PAS protein Tango (Tgo), the Drosophila homolog of the aryl hydrocarbon receptor nuclear translocator (Arnt), to initiate transcription of downstream target genes (Emmons et al. 1999). Mammalian Ahr binds the environmental toxin 2,3,7,8-tetra- chlorodibenzo-p-dioxin (TCDD, or dioxin), which induces a broad range of toxic effects including teratogenesis, immunosuppression, and tumor promotion (Schmidt and Bradfield 1996; Puga et al. 2005). Fetal exposure to dioxin can cause a range of cognitive and behavioral defects (ten Tusscher and Koppe 2004), possibly due to dioxin action via Ahr in the brain. Previous reports have shown that Ahr and Arnt are widely distributed throughout the rat brain including several hypothalamic and brainstem regions involved in the regulation of appetite and circadian rhythm, functions that are disrupted by dioxin exposure (Petersen et al. 2000). Although invertebrate homologs of Ahr do not bind dioxin (Powell-Coffman et al. 1998; Butler et al. 2001), recent studies in Caenorhabditis elegans support an evolution- arily conserved role for Ahr in neuronal development (Huang et al. 2004; Qin and Powell-Coffman 2004).
In this study, we report that ss acts cell-autonomously to regulate the morphological diversity of dendrites in all classes of Drosophila da sensory neurons. Remarkably, ss exerts diametrically opposing functions in different classes of da neurons to limit branching complexity in neurons that normally possess simple dendrites but to promote the formation of higher-order branches in neurons that normally elaborate more complex dendritic arbors. Furthermore, we show that ss is essential for the formation of distinct morphological features of dendrites in different classes of da neurons. In the absence of ss function, different classes of da neurons elaborate dendrites with similar morphologies, although these da neurons still project their axons to the appropriate location within the ventral nerve cord (VNC). We further show that ss likely functions independently of its known heterodimerization partner tango, via a novel regulatory program separate from the actions of cut and abrupt, to specify the branching patterns of sensory neuron dendrites. Our findings thus reveal a novel role for ss in generating dendritic diversity.
Results
spineless is required for proper dendrite morphogenesis in Drosophila da neurons
Drosophila class I da neurons elaborate their dendritic arbor in a simple and highly stereotypical pattern, projecting a primary branch then secondary branches that are typically straight and perpendicular to the primary branch (Fig. 1A; Sugimura et al. 2004). To identify genes important for class I dendrite morphogenesis, we conducted an EMS mutagenesis screen using the Gal4221 enhancer trap line to drive GFP expression in all class I neurons (Fig. 1A,C; Grueber et al. 2003a). We found a lethal mutant line (line 134) that showed primary branch overgrowth and increased terminal branching of class I dendrites in late-stage embryos (18–20 h after egg laying [AEL]) (Fig. 1B), a defect that persisted throughout the third instar larval stage (Fig. 1D). When we introduced the mutant allele into the Gal4109(2)80 reporter line, which labels all sensory neurons (Gao et al. 1999), we observed widespread dendrite defects in the da neurons, but we did not observe any obvious defects in the development of neurons without dendritic arbors, including the bipolar dendrite (bd) neurons, the tracheal dendrite (td) neurons, external sensory (es) organs, and chordotonal organs (data not shown). Immunostaining with 22C10 antibody revealed no obvious defects in cell migration or cell number in the PNS of embryos as well as third instar larvae (Supplementary Fig. S1), consistent with previous reports (Duncan et al. 1998). Furthermore, the mutant did not exhibit any axon guidance defects in the VNC as revealed by immunostaining with Fasciclin II (FasII) antibody (Supplementary Fig. S1; Lin et al. 1994).
Figure 1.
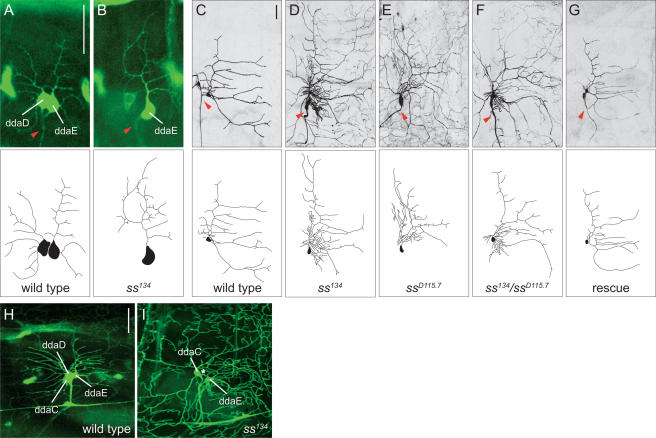
spineless regulates the dendritic morphogenesis of class I da neurons. (A) Wild-type dorsal class I neurons ddaD and ddaE in a stage 17 (18–20 h AEL) embryo visualized by the Gal4221 reporter. (B) ddaE neuron in a homozygous ss134 mutant exhibits overgrowth of the primary branch and increased terminal branching. (C) Wild-type ddaE neuron in a third instar larva. (D–F) ddaE neurons in a homozygous ss134 mutant (D), ssD115.7 mutant (E), and ss134/ssD115.7 transheterozygous mutant (F) exhibit overgrowth of the primary branch and increased terminal branching. (G) Expressing UAS-ss with Gal4221 in ss mutants rescues the primary branch overgrowth and increased terminal branching phenotype. Red arrowheads indicate axons. (H) Expression of GFP in the dorsal class I neurons (ddaD and ddaE) in a second instar larva as visualized by the Gal4221 reporter. GFP is weakly expressed in the dorsal class IV neuron ddaC at this developmental stage. (I) Expression of GFP in a homozygous ss134 mutant as visualized by the Gal4221 reporter. GFP is highly expressed in ddaE and ddaC, while absent in ddaD (normal position is indicated by a white asterisk). The terminal branches of ddaC are clearly visible. Anterior is to the left, and dorsal is up in this and all subsequent figures. Bar, 25 μm.
Based on genetic mapping via recombination and complementation tests with available deficiency lines and ss mutants, we identified the mutant line 134 as a new allele of spineless (hereafter referred to as ss134). A trans-heteroallelic combination of ss134 with the ssaBR30-71 loss-of-function allele (McMillan and McGuire 1992) recapitulated the ss adult phenotype including a reduction in the size of bristles and an antenna-to-leg transformation. By conducting complementation tests with a deficiency line that disrupts the ss gene [Df(3R)ssD114.4] and a lethal ss-null allele (ssD115.7) (Duncan et al. 1998), we further showed that a mutation in the ss gene was responsible for both lethality and the dendrite phenotype (Fig. 1E,F). Moreover, class I da neuron-specific expression of Ss in transgenic flies bearing the ss134/ssD115.7 mutation fully suppressed the primary branch over- growth and increased terminal branching phenotype (Fig. 1G), suggesting that ss functions in class I da neurons to limit their dendritic arborization.
Spineless is expressed in all sensory neurons
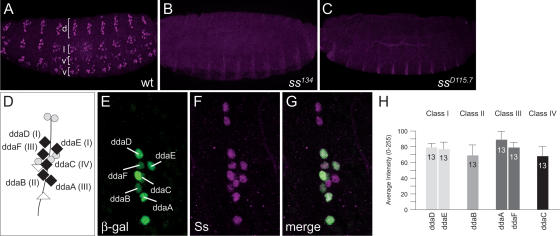
A prerequisite for a cell-autonomous action of Ss in class I neurons is the expression of Ss in these neurons. Indeed, in our immunostaining analysis using a polyclonal antibody raised against a C-terminal fragment of Ss (see Materials and Methods), we found that Ss is expressed in all sensory neurons in the embryonic PNS (Fig. 2A), consistent with previous in situ hybridization data (Duncan et al. 1998). Ss expression is largely restricted to the PNS in embryos, although Ss was also detected in four unidentified cells per segment within the VNC (data not shown). Ss was detected in the PNS as early as stage 13 (9–10 h AEL), and expression persisted throughout late embryogenesis (20–22 h AEL). Ss is expressed in the soma of all da neurons at similar levels (Fig. 2D–H), as well as the soma of bd and es neurons (Fig. 2D–G) and td neurons and chordotonal organs (data not shown). Ss expression is absent in both ss134 and ssD115.7 mutant lines, confirming that both are null alleles (Fig. 2B,C), and thus verifying the specificity of the antibody.
Figure 2.
Spineless expression in the embryonic PNS. (A) Expression of Ss protein in stage 16 (16–18 h AEL) embryo. Ss is expressed in all sensory neurons in the dorsal (d), lateral (l), ventral prime (v′), and ventral (v) clusters. (B,C) Ss expression is absent in homozygous ss134 (B) and ssD115.7 (C) mutants. (D) Schematic depiction of sensory neurons in the dorsal cluster. Black diamonds represent da neurons, gray circles represent es organs, and white triangles represent bd neurons. (E) All multidendritic (md) neurons in the dorsal cluster in a stage 16 embryo labeled by the E7-2-36 lacZ enhancer trap line as revealed by staining with β-gal. (F) Expression of Ss in the dorsal cluster. (G) Merged image of E and F showing expression of Ss in the da neurons. (H) Quantification of Spineless immunoreactivity (measured as mean pixel intensity) in dorsal da neurons.
Simple dendritic arbors become more complex in spineless mutants
To further test whether ss functions cell-autonomously to regulate dendrite morphogenesis, we performed mosaic analysis with a repressible cell marker (MARCM) experiments with both ss134- and ssD115.7-null alleles to examine single cell mutant clones in third instar larvae (Lee and Luo 1999). Consistent with the ss134 homozygous mutant phenotype (Fig. 1D), dorsal ss134 and ssD115.7 MARCM clones of the class I neurons ddaE (Fig. 3B,C) and ddaD (Fig. 3F,G) showed a dramatic over- branching phenotype. ddaE normally elaborates very few branches and typically projects a single primary dendrite followed by secondary branches that project toward the posterior segment border (Fig. 3A). In contrast, the overall number of branchpoints in ss ddaE mutant clones was 245% greater than that in wild-type clones (Fig. 3D). Unlike the primary branch overgrowth phenotype in homo- zygous ss mutants (Fig. 1D), however, the outgrowth of primary dendrites was mostly unaffected in ss MARCM clones (Fig. 3B,C).
Figure 3.
spineless functions cell-autonomously to regulate dendritic branching in class I and II da neurons. (A) Normal morphology of class I neuron ddaE dendrites in a wild-type third instar larva MARCM clone. ddaE normally projects its secondary dendrites toward the posterior segment border and elaborates few terminal branches. (B,C) ddaE neurons exhibit increased terminal branching in ss134 (B) and ssD115.7 (C) MARCM clones. (D) Quantification of the total number of branchpoints for ddaE in wild-type and ssD115.7 clones. Error bars represent standard deviation in this and all subsequent figures (asterisk; p < 0.001, two- tailed t-test). (E) Normal morphology of class I neuron ddaD dendrites in a wild-type MARCM clone. ddaD normally projects its secondary dendrites toward the anterior segment border. (F,G) ddaD neurons show increased branching and dendrite routing defects in ss134 (F) and ssD115.7 (G) MARCM clones. Secondary dendrites are misoriented, and branches improperly terminate posterior to the cell body. (H) Quantification of total number of branchpoints for ddaD in wild- type and ssD115.7 clones (asterisk; p < 0.001, two-tailed t-test). (I) Normal morphology of dorsal class II neuron ddaB dendrites in a wild-type MARCM clone. ddaB extends long primary and secondary dendrites with few terminal branches. (J,K) ddaB neurons exhibit increased dendritic branching at the distal tips of primary and secondary dendrites in ss134 (J) and ssD115.7 (K) MARCM clones. (L) Quantification of total number of branchpoints for ddaB in wild-type and ssD115.7 clones (asterisk; p < 0.001, two-tailed t-test). Arrowheads indicate axons. Bar, 50 μm.
Similar to ss ddaE clones, ss ddaD clones showed a 285% increase in total dendritic branch number when compared with control clones (Fig. 3H). However, unlike ss ddaE clones, the dendrite phenotypes were more variable as we also observed overall patterning defects in ss ddaD clones, likely due to misrouting of the dendritic branches (Fig. 3F,G). Normally, secondary branches from ddaD and ddaE rarely occupy the space between the two neurons, suggesting that there is a mechanism to restrict the respective target fields of ddaD and ddaE (Parrish et al. 2006). Compared with secondary branches in control ddaD clones, which typically project toward the anterior segment border and rarely ever invade the space posterior to the cell body (<0.5% of the total number of branches in all control clones) (Fig. 3E), many more branches in ss ddaD clones (29% of the branches in all ss ddaD clones) occupied territories posterior to the cell body (Fig. 3F,G). Presumably, these branches are unresponsive to repulsive cues that normally prevent ddaD secondary branches from invading the receptive field of ddaE. Dendritic branches from ss ddaE clones occasionally terminated improperly; however, this occurred at a much lower frequency than in ss ddaD clones. Thus, in addition to regulating dendritic branching, ss may also play a role in dendrite routing.
The increase in terminal branching in class I ss MARCM clones suggested that ss normally functions cell-autonomously to limit higher-order branching indendrites of class I neurons. Given that Ss is also expressed in class II neurons, which also elaborate simple dendritic arbors with branching complexity comparable with that of class I dendrites as assessed by Strahler analysis (Grueber et al. 2002), we wondered whether ss acts to limit dendritic branching in class II neurons as well. The dorsal class II neuron ddaB typically extends long primary and secondary dendrites, but has very few higher-order branches (Fig. 3I). In contrast, ss ddaB clones showed a significant increase in overall branch number (183%) (Fig. 3J–L). Taken together, these findings suggest that ss restricts the morphological complexity of the class I and class II neurons by limiting the overall number of their dendritic branches.
Complex dendritic arbors become simpler in spineless mutants
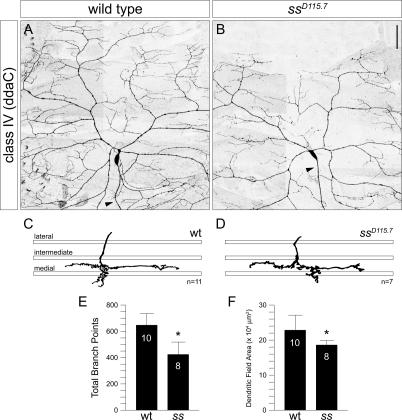
Class IV neurons differ from class I and class II neurons in that they project very elaborate dendritic arbors that cover the entire body wall (Grueber et al. 2002, 2003b). To test the function of Ss expressed in these class IV neurons, we examined MARCM clones of the dorsal class IV neuron ddaC, which is very highly branched and extends its arbor to the dorsal midline and both the anterior and posterior segment borders (Fig. 4A). In contrast to the significant increase in dendritic branching in class I and class II ss MARCM clones, ss ddaC clones exhibited a significant reduction (34%) in the total number of terminal branches when compared with wild-type clones (Fig. 4B,E). Furthermore, there was a significant reduction (19%) in the total dendritic field size of ss ddaC clones (Fig. 4F). Consequently, in most cases the primary dendrites failed to reach the dorsal midline (Fig. 4B). Not surprisingly, there was no overlap of the dendrites between adjacent mutant clones or in the null mutant as revealed by the class IV-specific ppk::EGFP reporter (data not shown). Thus, ss is required for terminal branching in class IV neurons. Without ss function, the complexity of class IV dendritic arbors is significantly reduced.
Figure 4.
Loss of spineless function reduces the complexity of class IV dendrites. (A) Normal morphology of class IV neuron ddaC dendrites in a wild-type MARCM clone. ddaC extends its dendrites to the dorsal midline and anterior and posterior segment borders. (B) A ddaC ssD115.7 MARCM clone shows reduced terminal branching and failure of dendrites to reach the dorsal midline. Arrowheads indicate axons. (C,D) Tracing of representative ddaC axon terminals in wild-type (C) and ssD115.7 (D) MARCM clones. The three bars represent the lateral, intermediate, and medial fascicles of the VNC as revealed by FasII immunostaining. ddaC axon terminals in wild-type (C) and ssD115.7 (D) MARCM clones invariably terminate near the medial fascicle. (E) Quantification of total number of branchpoints for ddaC in wild-type and ssD115.7 clones (asterisk; p < 0.001, two- tailed t-test). (F) Quantification of total dendritic field area for ddaC in wild-type and ssD115.7 clones (asterisk; p < 0.02, two- tailed t-test). Bar, 50 μm.
Whereas ss ddaC clones exhibited severe defects in dendrite morphology, there was no obvious abnormality in the axon projections of individual ss ddaC clones (Fig. 4C,D). The ddaC axons normally terminate near the me- dial fascicle of the VNC, as revealed by FasII immuno- staining, and the ddaC axon terminals typically extend an anterior- and posterior-directed branch, although their lengths are highly variable, and often, but not always, extend a protruding branch toward the ventral midline (Fig. 4C). Similar to axons from control clones, axon terminals from ss ddaC clones exhibited variable morphology but invariably terminated near the medial fascicle (Fig. 4D). It thus appears that ss may function preferentially in controlling the morphogenesis of the dendrites of da neurons.
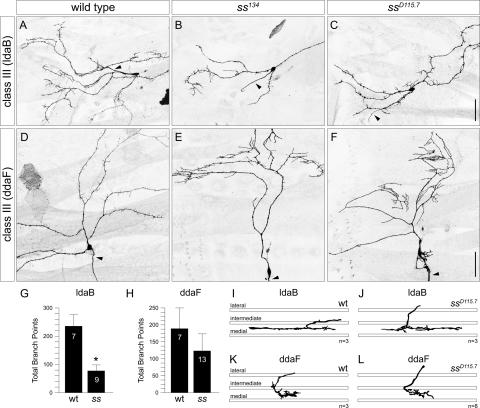
The Ss protein is present at similar levels in neurons of class III as well as classes I, II, and IV (Fig. 2H). However, class III neurons are unique among the da neurons in that they possess characteristic fine actin-based protrusions of 2–20 μm in length along their dendrites that are known as dendritic spikes (Grueber et al. 2002). In ss MARCM clones, these spiky protrusions were largely absent in all class III neurons. Compared with wild-type clones, where the lateral class III neuron ldaB showed a high density of dendritic spikes all along its primary and secondary branches (Fig. 5A), ss134 and ssD115.7 ldaB clones showed a significant decrease in overall branch number, which reflected an obvious reduction in dendritic spikes along the primary and secondary dendrites (Fig. 5B,C,G). The dorsal class III neuron ddaA also showed a significant reduction (55%) in the total number of branches and dendritic spikes in ss clones (Table 1). The other dorsal class III neuron, ddaF, normally extends long primary and secondary dendrites that are decorated with dendritic spikes (Fig. 5D). Although the dendrite phenotypes were variable from clone to clone, the most common defects were the absence of some primary and secondary branches and primary branch overgrowth (Fig. 5E,F). In many cases, the primary branch extended beyond the dorsal midline. Although there was not a significant change in the total branch number due to the high variability of the phenotype (Fig. 5H), dendritic spikes were mostly eliminated from the primary dendrites and replaced by clusters of longer terminal branches (Fig. 5E,F). Therefore, similar to loss of ss function in class IV dendrites, loss of ss function in class III neurons resulted in reduced dendritic complexity, and more specifically, in the loss of dendritic spikes.
Figure 5.
spineless regulates the number of dendritic spikes in the class III neurons. (A) Normal morphology of lateral class III neuron ldaB dendrites in a wild-type MARCM clone. Dendritic spikes are found along all major branches. (B,C) ldaB neurons show a reduction of dendritic spikes along all major branches in ss134 (B) and ssD115.7 (C) MARCM clones. (D) Normal morphology of dorsal class III neuron ddaF dendrites in a wild-type MARCM clone. ddaF extends long primary and secondary dendrites that are decorated with dendritic spikes. (E,F) ddaF neurons show a reduction in the number of dendritic spikes and exhibit clusters of longer terminal branches along the primary dendrites in ss134 (E) and ssD115.7 (F) MARCM clones. Arrowheads indicate axons. (G) Quantification of total number of branchpoints for ldaB in wild-type and ssD115.7 clones (asterisk; p < 0.001, two-tailed t-test). (H) Quantification of total number of branchpoints for ddaF indicates no significant difference between wild-type and ssD115.7 clones. (I,J) Tracing of representative ldaB axon terminals in wild-type (I) and ssD115.7 (J) MARCM clones. ldaB axons always terminate between the intermediate and medial fascicles in wild-type (I) and ssD115.7 (J) MARCM clones. (K,L) Tracing of representative ddaF axon terminals in wild-type (K) and ssD115.7 (L) MARCM clones. ddaF axons project into the VNC and curve posteriorly between the intermediate and medial fascicles in wild-type (K) and ssD115.7 (L) MARCM clones. Bar, 50 μm.
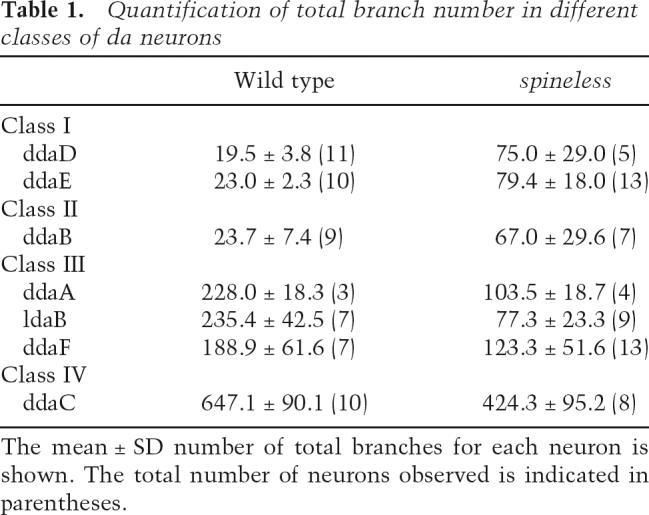
Table 1.
Quantification of total branch number in different classes of da neurons
Like ss ddaC clones, ss ldaB and ddaF clones exhibited apparently normal axon projections (Fig. 5I–L). The ldaB axons normally terminate between the intermediate and medial fascicles, and their axon terminals project both anterior- and posterior-directed branches of varying lengths (Fig. 5I), whereas the ddaF axons project into the VNC and curve toward the posterior then terminate between the intermediate and medial fascicles (Fig. 5K). Axon projections from both ss ldaB and ddaF clones invariably terminated between the intermediate and me- dial fascicles similarly to wild-type clones (Fig. 5J,L). These observations reinforce the notion that ss function is likely restricted to the dendrites of the da neurons.
Loss of spineless function reduces dendritic diversity
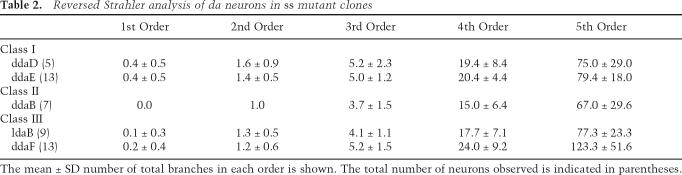
From our MARCM analysis, we noticed that the dendritic patterns of different classes of da neurons in ss mutants became more homogeneous. For example, in the absence of ss function, the dendritic arbor of the class III neuron ddaF lost its characteristic dendritic spikes and resembled the dendrites with supernumerary branching of the class II neuron ddaB in ss MARCM clones (Supplementary Fig. S2). The loss of ss function also caused ddaF dendrites to resemble dendrites from the class I neuron ddaD (Supplementary Fig. S2). Indeed, with the exception of the class IV neurons, the overall branch number was comparable for dendrites of different classes of da neurons in ss mutants, in contrast to the very significant differences in wild-type controls (Table 1). For example, the overall number of dendritic branches in the class III neuron ldaB was typically 10-fold higher than that of the class I neurons ddaD and ddaE in wild- type controls (Table 1). In ss mutants, however, ldaB elaborated, on average, the same number of branches as ddaD and ddaE (Table 1). Furthermore, different classes of da neurons exhibited similar degrees of complexity as assessed by Strahler analysis (Table 2). Therefore, as a result of the opposite dendrite phenotypes in class I and II versus class III and IV da neurons, loss of ss function led to a dramatic reduction in the overall dendritic diversity of the da neurons and, as a consequence, different classes of da neurons (except for class IV) elaborated morphologically similar dendrites.
Table 2.
Reversed Strahler analysis of da neurons in ss mutant clones
Overexpression of spineless results in opposite dendrite phenotypes in different types of sensory neurons
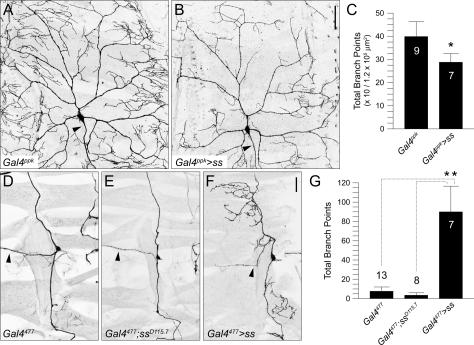
Our data showed that all sensory neurons express Ss at similar levels (Fig. 2H), yet loss of ss function resulted in different phenotypes in different classes of sensory neurons. We therefore wanted to test whether altering the expression level of Ss in the sensory neurons would result in any dendrite phenotype. Using available Gal4 drivers, we first overexpressed Ss in the different classes of da neurons and found no noticeable phenotype in class I or class II dendrites (data not shown). Class III neurons that overexpressed Ss also did not exhibit any detectable defects (data not shown). However, overexpression of Ss in class IV neurons via the Gal4ppk driver (Kuo et al. 2005) resulted in a significant reduction in overall branch number (Fig. 6A-C), similar to the ss loss-of-function phenotype (Fig. 4B). Having found that class IV, but not class I, II, and III da neurons are sensitive to the level of Ss expression, we went on to test whether overexpression of Ss could affect the morphology of sensory neurons that did not exhibit an ss loss-of-function pheno-type. Overexpression of Ss did not generate dendritic phenotypes in bd neurons, es organs, and chordotonal organs (data not shown). In contrast, expression of Ss in the td neuron via the Gal4477 driver (Grueber et al. 2003a) resulted in a very striking phenotype (Fig. 6F). The td neuron normally extends bipolar dendrites ventrally and dorsally with little or no branching (Fig. 6D), as did td neurons in ss mutants (Fig. 6E). However, with overexpression of Ss, the td neuron exhibited a significant increase in ectopic dendritic branching in 100% of the cases observed (Fig. 6G). Taken together, these observations suggest different sensory neurons are sensitive to different ranges of Ss expression. Like bd neurons, es organs, and chordotonal organs, the simple dendritic pattern of the td neuron persists even in the absence of ss function. However, unlike in these neurons, td neuron dendrites can be converted to complex dendritic patterns characteristic of da neurons by Ss overexpression. Similarly, while all classes of da neurons normally require ss function, class I–III da neurons apparently tolerate a wider range of Ss expression levels compared with the class IV da neuron. Thus, class IV dendrites are more sensitive to changes in Ss expression, and the more elaborate dendritic branching pattern of class IV neurons can only be achieved with the appropriate level of Ss expression.
Figure 6.
Overexpression of Spineless leads to opposite phenotypes in different sensory neurons. (A) Wild- type morphology of the class IV neuron ddaC as revealed by the Gal4ppk reporter line. (B) Overexpression of Ss in ddaC causes a severe reduction in terminal branching. (C) Quantification of total number of branchpoints for ddaC in wild type (Gal4ppk) and over-expressing Ss (Gal4ppk>ss) (asterisk; p < 0.001, two-tailed t-test). (D) Wild-type morphology of the td neuron as revealed by the Gal4477 reporter line. (E) The td neuron does not show any dendrite defects in ssD115.7 mutants. (F) Overexpression of Ss in the td neuron causes ectopic dendritic branches to form. (G) Quantification of total number of branchpoints for the td neuron in wild-type (Gal4477), ssD115.7 mutant (Gal4477; ssD115.7), and overexpressing Ss (Gal4477>ss) (asterisks; p < 0.001, two tailed t-test). Arrowheads indicate axons. Bar, 50 μm.
Loss of spineless function does not alter Abrupt or Cut expression in the da neurons
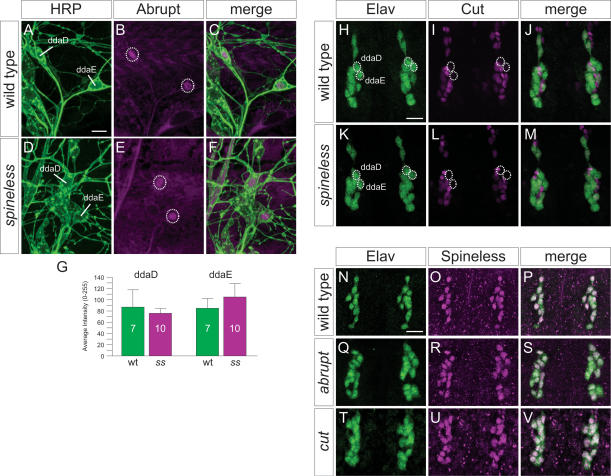
Previous reports have established the role of the BTB- zinc finger protein Abrupt and the homeodomain protein Cut in specifying the distinct dendritic branching patterns of class I and class II–IV da neurons, respectively (Grueber et al. 2003a; Li et al. 2004; Sugimura et al. 2004). Thus, Ab is only expressed in class I of the da neurons (Fig. 7A–C) and regulates its dendritic development (Li et al. 2004; Sugimura et al. 2004). Loss of ab function in class I neurons leads to a significant increase in overall branch number (Li et al. 2004; Sugimura et al. 2004), similar to class I dendrites in ss loss-of-function mutants. To test whether ss regulates Ab expression in the class I neurons and whether the class I dendrite phenotype could be attributed to a reduction in the levels of Ab expression, we examined Ab expression in ss mutants and found that Ab was expressed in class I neurons at comparable levels to wild-type controls (Fig. 7D–G); the slight increase in Ab expression in ddaE neurons of ss mutants was not statistically significant (p = 0.08, two- tailed t-test). Furthermore, we did not observe Ab expression in any of the other da neurons. As Ab can also serve as a molecular marker for class I da neurons, it appears that class I neurons maintained their identity in ss mutants and the dendritic phenotypes of class II–IV da neurons could not be attributed to their transformation to class I neurons.
Figure 7.
spineless acts through a regulatory program distinct from cut and abrupt to control dendritic diversity. (A) Wild-type third instar larva stained with the pan-neuronal marker horseradish peroxidase (HRP) indicating positions of class I neurons in dorsal cluster (ddaD and ddaE). (B) Wild-type expression pattern of Ab in the dorsal cluster. (C) Merged image of A and B showing that Ab is only expressed in the class I neurons. (D) HRP stain of homozygous ssD115.7 mutant third instar larva indicating the positions of ddaD and ddaE. (E) Expression pattern of Ab in the dorsal cluster in a homozygous ssD115.7 mutant. (F) Merged image of D and E showing Ab expression in the dorsal class I neurons in the ssD115.7 mutant. (G) Quantification of Ab immunoreactivity in class I neurons ddaD and ddaE in wild type and ss mutants reveals similar levels of Ab expression. (H) Wild-type embryo (18–20 h AEL) stained with the pan-neuronal marker Elav. (I) Wild-type expression pattern of Cut in the dorsal cluster. Cut is not expressed in the class I neurons ddaD and ddaE (circles indicate positions). (J) Merged image of H and I showing different levels of Cut expression in the da neurons. (K) Homozygous ssD115.7 mutant embryo stained with Elav. (L) Expression pattern of Cut in the dorsal cluster in a homozygous ssD115.7 mutant. Cut is not expressed in ddaD and ddaE (circles). (M) Merged image of K and L showing that Cut expression is normal in the ssD115.7 mutant. (N) Wild-type expression pattern of Elav in the dorsal cluster. (O) Wild-type expression pattern of Ss. (P) Merged image of N and O showing Ss expression in all Elav-positive neurons. (Q) Expression pattern of Elav in abK02807 loss-of-function mutant. (R) Expression pattern of Ss in the abK02807 mutant. (S) Merged image of Q and R showing that Ss expression is not regulated by ab in the sensory neurons. (T) Expression pattern of Elav in cutc145 loss-of-function mutant. (U) Expression pattern of Ss in the cutc145 mutant. (V) Merged image of T and U showing that Ss expression is not regulated by cut in the sensory neurons. Bar, 10 μm.
Different levels of Cut expression contribute to the branching diversity of class II–IV da neurons (Grueber et al. 2003a). Cut expression is highest in the class III neurons, at intermediate levels in the class IV neurons, at low levels in the class II neurons, and largely undetectable in the class I neurons (Fig. 7H–J; Grueber et al. 2003a). Previous studies have shown that ectopic expression of ss can suppress Cut expression in antennal discs (Emerald et al. 2003). We therefore examined the possibility that ss may regulate cut in the da neurons to control dendritic branching. However, there was no obvious alteration of Cut expression in any of the da neurons in ss mutants when compared with wild-type controls (Fig. 7K–M). Cut expression was absent in the class I neurons and was always detected in the other classes. These observations thus rendered it unlikely that ss acts upstream of ab or cut. Furthermore, ectopic expression of Ss did not alter Cut expression levels in the da neurons, nor did it suppress the Cut overexpression phenotype in class I dendrites (data not shown). The inability of ss to modulate Cut expression in any of the four classes of da neurons further suggests that these neurons maintain their identity in ss loss-of-function mutants.
Having found no evidence that ss acts upstream of cut or ab, we then asked whether the dendrite phenotypes associated with loss of ab or cut function are associated with changes in Ss expression. However, there was no detectable alteration of Ss expression in any of the da neurons or other sensory neurons in the PNS of abk02807 and cutc145 loss-of-function mutants (Fig. 7N–V). Thus, ss is neither upstream of nor downstream from ab and cut in its regulation of dendritic complexity. Most likely, ss acts in a pathway independent of cut and ab to regulate dendritic branching.
spineless likely acts independently of tango to regulate dendrite morphogenesis
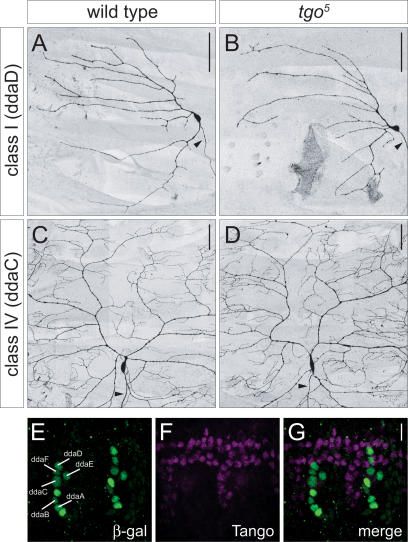
Previous reports have shown that Ss can heterodimerize with the bHLH-PAS protein Tgo in the nucleus and that Ss and Tgo act in concert to regulate the transcription of downstream target genes (Emmons et al. 1999). In Caenorhabditis elegans, both the Ahr (ahr-1) and Arnt (aha- 1) homologs are involved in cell fate specification and neuronal development in specific subclasses of neurons (Huang et al. 2004; Qin and Powell-Coffman 2004). To determine if the regulation of dendritic morphogenesis by ss requires tgo, we first examined MARCM clones with a lethal tgo allele, tgo5 (Emmons et al. 1999). Surprisingly, the tgo clones showed no obvious dendrite defects in any of the classes of da neurons and, hence, no phenotype that resembled the ss dendrite phenotype (Fig. 8A–D). Although it is possible that a maternal contribution of tgo may be masking a dendrite phenotype, we did not detect nuclear accumulation of Tgo in any of the daneurons at any stage during embryonic development (Fig. 8E–G). It remains possible that tgo may have non- cell-autonomous functions in dendrite development as RNA interference (RNAi) experiments with tgo dsRNA resulted in abnormal dendrites of class I da neurons (Parrish et al. 2006; data not shown). However, loss of tgo resulted in reduced arborization of class I dendrites (Parrish et al. 2006), while class I dendrites are more highly branched in ss mutants. Although we cannot completely rule out the possible involvement of tgo, we favor a scenario in which ss acts independently of tgo to control dendrite morphogenesis in Drosophila da neurons. It remains possible that ss acts together with another cofactor to regulate transcription within the da neurons.
Figure 8.
tgo does not act cell-autonomously to regulate the development of da neuron dendrites. (A) Wild-type MARCM clone of the class I neuron ddaD. (B) The ddaD tgo5 MARCM clone does not exhibit any dendrite branching or routing defects. (C) Wild-type MARCM clone of the class IV neuron ddaC. (D) The ddaC tgo5 MARCM clone does not show a reduction in terminal branching or dendritic field size. Arrowheads indicate axons. Bar, 50 μm. (E) Multidendritic (md) neurons in the dorsal cluster labeled by the E7- 2-36 lacZ enhancer trap line as revealed by staining with β-gal. (F) Wild-type expression pattern of Tgo. Tgo is largely accumulated in tracheal cells. (G) Merged image of E and F showing that Tgo is not accumulated in the nuclei of da neurons. Bar, 10 μm.
spineless differentially regulates gene expression in the da neurons
Although ss has been previously shown to activate the transcription of target genes in vitro (Emmons et al. 1999; Brown et al. 2005), little is known of how ss regulates gene expression in Drosophila. In the Gal4221 reporter line, Gal4 is expressed at high levels in all class I neurons, but is also expressed at lower levels in all class IV neurons, although expression is most evident in later developmental stages (Fig. 1H). In addition to class I dendrite defects revealed by the Gal4221 reporter line in a ss134 or ssD115.7 mutant background, we saw an elevated level of GFP signal in all class IV neurons and the absence of GFP expression in ddaD and the ventral class I neuron vpda (Fig. 1I; data not shown). Expression of GFP in the class IV neurons was seen as early as 15 h AEL and persisted throughout late larval stages. In contrast, GFP expression never reached such high levels in class IV neurons in wild-type embryos or larvae (Fig. 1H). Furthermore, in ss mutants, GFP expression was only visible in ddaE among the class I neurons in later stages when all class I neurons normally express GFP at high levels (Fig. 1I). The ability of ss to modulate the expression of the Gal4221 reporter in different da neurons is one indication that ss can differentially regulate gene expression in the da neurons.
Discussion
Most vertebrate and invertebrate nervous systems contain a variety of neurons with distinct dendritic morphologies that determine the manner in which these neurons receive and process information. How such diversity develops is not well understood. In this study, we provide evidence that the Drosophila homolog of the dioxin receptor ss serves a novel function for diversification of Drosophila da sensory neurons. By executing diametrically opposing regulation to maintain the simple dendritic arbors of class I and class II da neurons but to enable class III and class IV da neurons to elaborate complex dendrites, ss is essential for these neurons to acquire their distinct patterns of dendritic branching. Loss of ss function resulted in increased dendritic complexity of the class I and class II da neurons that normally elaborate simple arbors. In contrast, loss of ss function led to a reduction in the total number of dendritic spikes and terminal branches, respectively, of the class III and class IV neurons, which normally display complex dendritic patterns. Consequently, the dendrite patterns of different classes of da neurons resemble one another, and there is a dramatic reduction in the overall dendritic diversity of the da neurons in loss-of-function ss mutants. Yet, despite the striking dendrite phenotype, the axon projections of da neurons are normal in ss mutants. Interestingly, ss likely acts independently of its cofactor tango and functions in a regulatory program that is distinct from cut and ab to control dendrite morphogenesis. The Ss protein is present at nearly the same level in all da neurons and acts cell-autonomously to dictate their dendritic complexity, while different da neurons exhibit different sensitivity to the level of Ss, and even the bipolar td neuron can respond to elevated Ss activity by increasing dendritic complexity.
Do mutations in spineless affect the cell fate of da neurons?
Previous studies in C. elegans have demonstrated essential roles for invertebrate homologs of Ahr in neuronal cell fate determination. For example, ahr-1 regulates the differentiation program of a subclass of neurons that contact the pseudocoelomic fluid, and both ahr-1 and aha-1 specify GABAergic neuron cell fate in C. elegans (Huang et al. 2004; Qin and Powell-Coffman 2004). The dramatic changes in the dendrite morphologies of the da neurons, however, are not due to an all-or-nothing change in cell fate because the da neurons in ss mutants displayed normal class-specific expression patterns of the molecular markers Ab and Cut (Fig. 7) and normal axon projection patterns characteristic of individual da neurons (Figs 4, 5; data not shown). However, this also does not assume that a partial cell fate change has not occurred. One reflection of the ss function as a transcription factor is the altered expression levels of GFP in the class I Gal4221 reporter, with increased levels of expression in all class IV neurons and essentially no expression in the dorsal class I neuron ddaD and the ventral class I neuron vpda in ss mutants (Fig. 1H,I). It will be of interest to further characterize the genetic basis for this Gal4 reporter, to determine whether this regulation constitutes a partial cell fate alteration or transcriptional regulation of genes downstream from ss in the execution of adjustment of dendritic complexity.
Do da neuron dendrites in spineless mutants assume a primordial pattern?
There is an emerging theme that ss functions to diversify neuronal differentiation by expanding the photopigment repertoire of R7 photoreceptors in the Drosophila eye (Wernet et al. 2006) and by diversifying da neuron dendritic morphologies (this study).
Recent studies have demonstrated that the entire retinal mosaic pattern required for color vision in Drosophila is regulated by ss (Wernet et al. 2006). In the Drosophila retina, two types of ommatidia form the wild-type retinal mosaic: “pale” and “yellow.” In ssD115.7 mutants, the yellow ommatidial subtype is lost and normally yellow R7 cells are misspecified into the pale subtype (Wernet et al. 2006). As a result, nearly all R7 cells adopt the pale subtype, leading to loss of the retinal mosaic pattern. Thus, the pale R7 subtype represents the R7 “default state” (Wernet et al. 2006).
The overall lack of dendritic diversity in the da neurons in ss mutants is suggestive of the hypothesis that ss, an ancient, evolutionarily conserved gene, may act to convert a primordial dendrite pattern (perhaps a default state) to different complexities for different neurons in the peripheral nervous system. The loss-of-function ss phenotype in the da neuron dendrites might reflect such a primordial pattern as the dendrites in the mutant are devoid of specific morphological features that define distinct neuronal subclasses. In support of this notion, dendrites of the different classes of da neurons share similar morphological characteristics and elaborate similar numbers of total branches in ss mutants.
The ability of ss to regulate the complexity and diversity of this dendrite pattern, by limiting dendritic branching to shape the simpler arbors of the class I and class II neurons and by promoting class-specific terminal branching to shape the more complex arbors of the class III and class IV neurons, is quite unique. Of the many mutants that affect multiple classes of da neurons, the great majority affect da neurons with simple or complex dendritic arbors the same way; that is, causing them to all become simpler or more complex. The ss phenotype of making simple dendritic arbors more complex and complex arbors simpler is very unusual among the many mutants affecting dendrite complexity (e.g., Gao et al. 1999). It thus seems likely that the distinct dendritic patterns rely not only on a cohort of gene activities specifying the mechanics of dendrite outgrowth and branching, but also a genetic program that diverts the generic primordial mode of dendritic formation to a diverse range of dendritic patterns.
How might spineless exert its functions?
Unlike the homeodomain protein Cut, which promotes dendritic complexity in a specific direction (Grueber et al. 2003a), ss functions in an opposing manner in different cell types to regulate dendritic diversity. How might ss function differently in different neuronal cell types? One possibility is that ss is activated by different ligands in different neurons. Ss is incapable of binding dioxin and other related compounds (Powell-Coffman et al. 1998; Butler et al. 2001), suggesting that other, as yet unidentified ligands are required for its activation. Previous reports have suggested that Ss and other invertebrate homologs of Ahr are activated by an endogenous ligand or that no ligand is required at all (Powell-Coffman et al. 1998; Emmons et al. 1999; Butler et al. 2001; Brown et al. 2005). Recent studies have shown that Ahr can accumulate in the nucleus upon activation by the second messenger cyclic AMP (cAMP) (Oesch-Bartlomowicz et al. 2005), although it is not yet known whether cAMP signaling can activate Ss in Drosophila. Thus, it is conceivable that Ss is activated by different upstream factors in different cell types. It will be of interest to test in future studies whether in different neuronal cell types Ss is activated by different ligands or upstream second messengers and whether Ss acts in concert with regulatory programs for cell fate specification to dictate dendritic complexity.
In the canonical Ahr signaling pathway, Ahr requires the appropriate cofactor for its proper function (Reyes et al. 1992; Probst et al. 1993; Hankinson 1995). Members of the bHLH-PAS protein family are able to heterodimerize with other bHLH-PAS proteins (Huang et al. 1993). Previous studies have shown that, upon ligand binding, Ahr is translocated to the nucleus, where it heterodimerizes with Arnt to form a transcriptionally active complex (Reyes et al. 1992; Probst et al. 1993; Hankinson 1995). However, we found that tango, the Drosophila homolog of Arnt, is likely not required for the regulation of dendritic morphogenesis, indicating that ss is probably not acting through its canonical signaling pathway to specify dendritic complexity. In Sf9 cells, ss can act independently of tgo to enhance expression of a reporter in the absence of a ligand (Brown et al. 2005). Furthermore, Ahr is unable to interact with Arnt upon activation by cAMP (Oesch-Bartlomowicz et al. 2005). Although Ahr, Arnt, and the Arnt homolog Arnt2 are widely distributed throughout the rat brain, Ahr does not preferentially co- localize with either Arnt or Arnt2 (Petersen et al. 2000). Ahr is also expressed in specific regions of the rat brain where neither Arnt nor Arnt2 is expressed (Petersen et al. 2000). These studies support the notion that ss can act independently of tgo in certain developmental contexts. Tgo can heterodimerize with other bHLH-PAS proteins in addition to Ss (Emmons et al. 1999). It is conceivable that ss may act with different heterodimerization partners to mediate its different functions in different cell types.
Might Ahr be involved in mammalian dendrite development?
Although the roles of Ahr in mediating the toxic effects of dioxin have been studied extensively, relatively little is known about the role of Ahr in development. Ahr is conserved in both vertebrates and invertebrates with their common ancestors dating back for >500 million yr (Hahn 2002). Invertebrate homologs of Ahr do not bind dioxin (Powell-Coffman et al. 1998; Butler et al. 2001), suggesting that the functional ability of mammalian Ahr to mediate the toxic response likely evolved after the divergence of vertebrates and invertebrates (Hahn 2002). Reports that Ahr is highly expressed in the rat brain (Petersen et al. 2000) and that Ahr can mediate TCDD-induced alterations in spatial learning (Powers et al. 2005) suggest important roles for Ahr in the nervous system. Here we demonstrate its critical role in dendrite development in the specification of morphological properties of distinct neuronal cell types in Drosophila. It will be of interest to determine whether this function of Ahr in governing dendritic diversity is conserved in other species and underlies neurological disorders caused by dioxin.
Materials and methods
Drosophila stocks
To visualize da neuron dendrites, we used the following reporter lines: Gal4221, UAS-mCD8::GFP and Gal4477, UAS- mCD8::GFP (Grueber et al. 2003a); Gal4109(2)80, UAS- mCD8::GFP (Gao et al. 1999); ppk::EGFP (Grueber et al. 2003b); and Gal4ppk, UAS-mCD8::GFP (Kuo et al. 2005). The following spineless lines were used in this study: ssD115.7, Df(3R)ssD114.4, and UAS-ss (Duncan et al. 1998) and ssaBR30-71 (McMillan and McGuire 1992). The ss134 allele was uncovered in an EMS mutagenesis screen and newly described here. tgo5 (Emmons et al. 1999), cutc145 (Jack 1985), and abk02807 (Johannes and Preiss 2002) alleles are as described.
MARCM analysis
To generate mosaic mutant clones, FRT82B, ss134/TM6B or FRT82B, ssD115.7/TM6B flies were crossed to w; elav-Gal4, UAS- mCD8::GFP, hsFLP; FRT82B, tub-Gal80 flies. In brief, embryos were collected for 2 h and allowed to develop for 3–5 h at 25°C before heat shock. Embryos were heat-shocked for 30 min at 38°C, allowed to recover at room temperature for 30 min, then heat-shocked for an additional 45 min at 38°C. Heat-shocked embryos were allowed to develop at 25°C to the third instar larval stage. Larvae with mutant clones were then dissected, fixed, and costained with rat anti-mCD8 antibody at 1:200 dilution (Caltag) and monoclonal 22C10 antibody at 1:500 dilution (Developmental Studies Hybridoma Bank, DSHB). Fixed preps were mounted on poly-L-lysine-coated coverslips, dehydrated in an ethanol series, cleared in xylenes, then mounted in DPX medium. Confocal image stacks were obtained using a Leica TCS SP2 confocal microscope and reconstructed into single collapsed images using ImageJ software (NIH).
Immunocytochemistry
Anti-Ss antibody was generated by immunization of guinea pigs with a GST-Ss fusion protein containing the C-terminal 150 amino acids of Ss and used at 1:1000 dilution. For whole-mount embryo and third instar larvae immunolabeling, the following primary antibodies were used: mouse anti-Cut (2B10; DSHB) at 1:20 dilution, mouse anti-Abrupt (DSHB) at 1:5 dilution, mouse anti-Tango (DSHB) undiluted, rat anti-Elav (7E8A10; DSHB) at 1:1000 dilution, mouse anti-Fasciclin II (1D4; DSHB) at 1:50 dilution, mouse anti-22C10 (DSHB) at 1:200 dilution, rabbit anti-GFP at 1:2000 dilution, rabbit anti-β-gal at 1:10,000 dilution, and Cy2- or Cy5-conjugated HRP at 1:200 dilution. Cy2-, Cy3-, or Cy5-conjugated secondary antibodies (Jackson Laboratories) were used at 1:200 dilution.
In situ hybridization
DIG-labeled DNA probes were prepared using the PCR DIG Probe Synthesis Kit (Roche Applied Science). In situ hybridization of whole-mount embryos was essentially performed as described in Tautz and Pfeifle (1989).
ACKNOWLEDGMENTS
We thank Ian Duncan (Washington University, St. Louis, MO), Stephen Crews (University of North Carolina, Chapel Hill, NC), the Bloomington Stock Center, and the Developmental Studies Hybridoma Bank for fly stocks and antibodies; Denan Wang and Jay Parrish for help with in situ hybridization; Kazuo Emoto for advice on generating Ss antibody; Susan Younger for advice on Drosophila genetics; and members of the Jan laboratory for discussions and comments on the manuscript. This work was supported by National Institutes of Health Grant R01 NS40929 (to Y.N.J.). Y.N.J. and L.Y.J. are Investigators of the Howard Hughes Medical Institute.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1459706.
REFERENCES
- Ainsley, J.A., Pettus, J.M., Bosenko, D., Gerstein, C.E., Zinkevich, N., Anderson, M.G., Adams, C.M., Welsh, M.J., Johnson, W.A. Enhanced locomotion caused by loss of the Drosophila DEG/ENaC protein Pickpocket1. Curr. Biol. 2003;13:1557–1563. doi: 10.1016/s0960-9822(03)00596-7. [DOI] [PubMed] [Google Scholar]
- Bodmer, R., Jan, Y.N. Morphological differentiation of the embryonic peripheral neurons in. Drosophila. Rouxs Arch.Dev. Biol. 1987;196:69–77. doi: 10.1007/BF00402027. [DOI] [PubMed] [Google Scholar]
- Brenman, J.E., Gao, F.B., Jan, L.Y., Jan, Y.N. Sequoia, a tramtrack-related zinc finger protein, functions as a panneural regulator for dendrite and axon morphogenesis in. Drosophila. Dev. Cell. 2001;1:667–677. doi: 10.1016/s1534-5807(01)00072-7. [DOI] [PubMed] [Google Scholar]
- Bridges, C.B., Morgan, T.H. Carnegie Institute of Washington; Washington DC: 1923. The third-chromosome group of mutant characters of Drosophila melanogaster; p. Publ. No. 327. [Google Scholar]
- Brown, R.P., McDonnell, C.M., Berenbaum, M.R., Schuler, M.A. Regulation of an insect cytochrome P450 monooxygenase gene (CYP6B1) by aryl hydrocarbon and xanthotoxin response cascades. Gene. 2005;358:39–52. doi: 10.1016/j.gene.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Butler, R.A., Kelley, M.L., Powell, W.H., Hahn, M.E., Van Beneden, R.J. An aryl hydrocarbon receptor (AHR) homologue from the soft-shell clam Mya arenaria: Evidence that invertebrate AHR homologues lack 2,3,7,8-tetrachlorodibenzo-p-dioxin and β-naphthoflavone binding. Gene. 2001;278:223–234. doi: 10.1016/s0378-1119(01)00724-7. [DOI] [PubMed] [Google Scholar]
- Duncan, D.M., Burgess, E.A., Duncan, I. Control of distal antennal identity and tarsal development in Drosophila by spineless-aristapedia, a homolog of the mammalian dioxin receptor. Genes & Dev. 1998;12:1290–1303. doi: 10.1101/gad.12.9.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerald, B.S., Curtiss, J., Mlodzik, M., Cohen, S.M. distal antenna and distal antenna related encode nuclear proteins containing pipsqueak motifs involved in antenna development in. Drosophila. Development. 2003;130:1171–1180. doi: 10.1242/dev.00323. [DOI] [PubMed] [Google Scholar]
- Emmons, R.B., Duncan, D., Estes, P.A., Kiefel, P., Mosher, J.T., Sonnenfeld, M., Ward, M.P., Duncan, I., Crews, S.T. The Spineless-Aristapedia and Tango bHLH-PAS proteins interact to control antennal and tarsal development in. Drosophila. Development. 1999;126:3937–3945. doi: 10.1242/dev.126.17.3937. [DOI] [PubMed] [Google Scholar]
- Gao, F.B., Brenman, J.E., Jan, L.Y., Jan, Y.N. Genes regulating dendritic outgrowth, branching, and routing in Drosophila . Genes&. Dev. 1999;13:2549–2561. doi: 10.1101/gad.13.19.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueber, W.B., Jan, L.Y., Jan, Y.N. Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development. 2002;129:2867–2878. doi: 10.1242/dev.129.12.2867. [DOI] [PubMed] [Google Scholar]
- Grueber, W.B., Jan, L.Y., Jan, Y.N. Different levels of the homeodomain protein Cut regulate distinct dendrite branching patterns of Drosophila multidendritic neurons. Cell. 2003a;112:805–818. doi: 10.1016/s0092-8674(03)00160-0. [DOI] [PubMed] [Google Scholar]
- Grueber, W.B., Ye, B., Moore, A.W., Jan, L.Y., Jan, Y.N. Dendrites of distinct classes of Drosophila sensory neurons show different capacities for homotypic repulsion. Curr. Biol. 2003b;13:618–626. doi: 10.1016/s0960-9822(03)00207-0. [DOI] [PubMed] [Google Scholar]
- Gu, Y.Z., Hogenesch, J.B., Bradfield, C.A. The PAS superfamily: Sensors of environmental and developmental signals. Annu. Rev. Pharmacol. Toxicol. 2000;40:519–561. doi: 10.1146/annurev.pharmtox.40.1.519. [DOI] [PubMed] [Google Scholar]
- Hahn, M.E. Aryl hydrocarbon receptors: Diversity and evolution. Chem. Biol. Interact. 2002;141:131–160. doi: 10.1016/s0009-2797(02)00070-4. [DOI] [PubMed] [Google Scholar]
- Hankinson, O. The aryl hydrocarbon receptor complex. Annu. Rev. Pharmacol. Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- Hausser, M., Spruston, N., Stuart, G.J. Diversity and dynamics of dendritic signaling. Science. 2000;290:739–744. doi: 10.1126/science.290.5492.739. [DOI] [PubMed] [Google Scholar]
- Huang, Z.J., Edery, I., Rosbash, M. PAS is a dimerization domain common to Drosophila Period and several transcription factors. Nature. 1993;364:259–262. doi: 10.1038/364259a0. [DOI] [PubMed] [Google Scholar]
- Huang, X., Powell-Coffman, J.A., Jin, Y. The AHR-1 aryl hydrocarbon receptor and its co-factor the AHA-1 aryl hydrocarbon receptor nuclear translocator specify GABAergic neuron cell fate in C. elegans . Development. 2004;131:819–828. doi: 10.1242/dev.00959. [DOI] [PubMed] [Google Scholar]
- Jack, J.W. Molecular organization of the cut locus of Drosophila melanogaster . Cell. 1985;42:869–876. doi: 10.1016/0092-8674(85)90283-1. [DOI] [PubMed] [Google Scholar]
- Jan, Y.N., Jan, L.Y. Dendrites. Genes & Dev. 2001;15:2627–2641. doi: 10.1101/gad.916501. [DOI] [PubMed] [Google Scholar]
- Jan, Y.N., Jan, L.Y. The control of dendrite development. Neuron. 2003;40:229–242. doi: 10.1016/s0896-6273(03)00631-7. [DOI] [PubMed] [Google Scholar]
- Johannes, B., Preiss, A. Wing vein formation inDrosophila melanogasterHairless is involved in the cross-talk between Notch and EGF signaling pathways. Mech. Dev. 2002;115:3–14. doi: 10.1016/s0925-4773(02)00083-7. [DOI] [PubMed] [Google Scholar]
- Kuo, C.T., Jan, L.Y., Jan, Y.N. Dendrite-specific remodeling of Drosophila sensory neurons requires matrix metalloproteases, ubiquitin–proteasome, and ecdysone signaling. Proc. Natl. Acad. Sci. 2005;102:15230–15235. doi: 10.1073/pnas.0507393102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, T., Luo, L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Li, W., Wang, F., Menut, L., Gao, F.B. BTB/POZ-zinc finger protein Abrupt suppresses dendritic branching in a neuronal subtype-specific and dosage-dependent manner. Neuron. 2004;43:823–834. doi: 10.1016/j.neuron.2004.08.040. [DOI] [PubMed] [Google Scholar]
- Lin, D.M., Fetter, R.D., Kopczynski, C., Grenningloh, G., Goodman, C.S. Genetic analysis of Fasciclin II in Drosophila Defasciculation, refasciculation, and altered fasciculation. Neuron. 1994;13:1055–1069. doi: 10.1016/0896-6273(94)90045-0. [DOI] [PubMed] [Google Scholar]
- Liu, L., Yermolaieva, O., Johnson, W.A., Abboud, F.M., Welsh, M.J. Identification and function of thermosensory neurons in Drosophila larvae. Nat. Neurosci. 2003;6:267–273. doi: 10.1038/nn1009. [DOI] [PubMed] [Google Scholar]
- McMillan, P.A., McGuire, T.R. The homeotic gene spineless-aristapedia affects geotaxis in Drosophila melanogaster . Behav. Genet. 1992;22:557–573. doi: 10.1007/BF01074308. [DOI] [PubMed] [Google Scholar]
- Oesch-Bartlomowicz, B., Huelster, A., Wiss, O., Antoniou-Lipfert, P., Dietrich, C., Arand, M., Weiss, C., Bockamp, E., Oesch, F. Aryl hydrocarbon receptor activation by cAMP vs. dioxin: Divergent signaling pathways. Proc. Natl. Acad. Sci. 2005;102:9218–9223. doi: 10.1073/pnas.0503488102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish, J.Z., Kim, M.D., Jan, L.Y., Jan, Y.N. Genome- wide analyses identify transcription factors required for proper morphogenesis of Drosophila sensory neuron dendrites. Genes & Dev. 2006;20:820–835. doi: 10.1101/gad.1391006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, S.L., Curran, M.A., Marconi, S.A., Carpenter, C.D., Lubbers, L.S., McAbee, M.D. Distribution of mRNAs encoding the arylhydrocarbon receptor, arylhydro- carbon receptor nuclear translocator, and arylhydrocarbon receptor nuclear translocator-2 in the rat brain and brainstem. J. Comp. Neurol. 2000;427:428–439. doi: 10.1002/1096-9861(20001120)427:3<428::aid-cne9>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Powell-Coffman, J.A., Bradfield, C.A., Wood, W.B. Caenorhabditis elegans orthologs of the aryl hydrocarbon receptor and its heterodimerization partner the aryl hydro- carbon receptor nuclear translocator. Proc. Natl. Acad. Sci. 1998;95:2844–2849. doi: 10.1073/pnas.95.6.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers, B.E., Lin, T.M., Vanka, A., Peterson, R.E., Juraska, J.M., Schantz, S.L. Tetrachlorodibenzo-p-dioxin exposure alters radial arm maze performance and hippocampal morphology in female AhR mice. Genes Brain Behav. 2005;4:51–59. doi: 10.1111/j.1601-183X.2004.00098.x. [DOI] [PubMed] [Google Scholar]
- Probst, M.R., Reisz-Porszasz, S., Agbunag, R.V., Ong, M.S., Hankinson, O. Role of the aryl hydrocarbon receptor nuclear translocator protein in aryl hydrocarbon (dioxin) receptor action. Mol. Pharmacol. 1993;44:511–518. [PubMed] [Google Scholar]
- Puga, A., Tomlinson, C.R., Xia, Y. Ah receptor signals cross-talk with multiple developmental pathways. Biochem. Pharmacol. 2005;69:199–207. doi: 10.1016/j.bcp.2004.06.043. [DOI] [PubMed] [Google Scholar]
- Qin, H., Powell-Coffman, J.A. The Caenorhabditis elegans aryl hydrocarbon receptor, AHR-1, regulates neuronal development. Dev. Biol. 2004;270:64–75. doi: 10.1016/j.ydbio.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Reyes, H., Reisz-Porszasz, S., Hankinson, O. Identification of the Ah receptor nuclear translocator protein (Arnt) as a component of the DNA binding form of the Ah receptor. Science. 1992;256:1193–1195. doi: 10.1126/science.256.5060.1193. [DOI] [PubMed] [Google Scholar]
- Schmidt, J.V., Bradfield, C.A. Ah receptor signaling pathways. Annu. Rev. Cell Dev. Biol. 1996;12:55–89. doi: 10.1146/annurev.cellbio.12.1.55. [DOI] [PubMed] [Google Scholar]
- Scott, E.K., Luo, L. How do dendrites take their shape? Nat. Neurosci. 2001;4:359–365. doi: 10.1038/86006. [DOI] [PubMed] [Google Scholar]
- Sugimura, K., Satoh, D., Estes, P., Crews, S., Uemura, T. Development of morphological diversity of dendrites in Drosophila by the BTB-zinc finger protein Abrupt. Neuron. 2004;43:809–822. doi: 10.1016/j.neuron.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Tautz, D., Pfeifle, C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene. hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- ten Tusscher, G.W., Koppe, J.G. Perinatal dioxin exposure and later effects—A review. Chemosphere. 2004;54:1329–1336. doi: 10.1016/S0045-6535(03)00254-6. [DOI] [PubMed] [Google Scholar]
- Tracey, W.D., Jr., Wilson, R.I., Laurent, G., Benzer, S. painless, a Drosophila gene essential for nociception. Cell. 2003;113:261–273. doi: 10.1016/s0092-8674(03)00272-1. [DOI] [PubMed] [Google Scholar]
- Wernet, M.F., Mazzoni, E.O., Celik, A., Duncan, D.M., Duncan, I., Desplan, C. Stochastic spineless expression creates the retinal mosaic for colour vision. Nature. 2006;440:174–180. doi: 10.1038/nature04615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford, K.L., Marillat, V., Stein, E., Goodman, C.S., Tessier-Lavigne, M., Chedotal, A., Ghosh, A. Regulation of cortical dendrite development by Slit–Robo interactions. Neuron. 2002;33:47–61. doi: 10.1016/s0896-6273(01)00566-9. [DOI] [PubMed] [Google Scholar]
- Wong, R.O., Ghosh, A. Activity-dependent regulation of dendritic growth and patterning. Nat. Rev. Neurosci. 2002;3:803–812. doi: 10.1038/nrn941. [DOI] [PubMed] [Google Scholar]
- Xu, K., Bogert, B.A., Li, W., Su, K., Lee, A., Gao, F.B. The fragile X-related gene affects the crawling behavior of Drosophila larvae by regulating the mRNA level of the DEG/ENaC protein Pickpocket1. Curr. Biol. 2004;14:1025–1034. doi: 10.1016/j.cub.2004.05.055. [DOI] [PubMed] [Google Scholar]
- Zipursky, S.L., Venkatesh, T.R., Teplow, D.B., Benzer, S. Neuronal development in the Drosophila retina: Monoclonal antibodies as molecular probes. Cell. 1984;36:15–26. doi: 10.1016/0092-8674(84)90069-2. [DOI] [PubMed] [Google Scholar]