Abstract
Negative feedback regulation of the proopiomelanocortin (POMC) gene by the glucocorticoid (Gc) receptor (GR) is a critical feature of the hypothalamo–pituitary–adrenal axis, and it is in part exerted by trans-repression between GR and the orphan nuclear receptors related to NGFI-B. We now show that Brg1, the ATPase subunit of the Swi/Snf complex, is essential for this trans-repression and that Brg1 is required in vivo to stabilize interactions between GR and NGFI-B as well as between GR and HDAC2. Whereas Brg1 is constitutively present at the POMC promoter, recruitment of GR and HDAC2 is ligand-dependent and results in histone H4 deacetylation of the POMC locus. In addition, GR-dependent repression inhibits promoter clearance by RNA polymerase II. Thus, corecruitment of repressor and activator at the promoter and chromatin modification jointly contribute to trans-repression initiated by direct interactions between GR and NGFI-B. Loss of Brg1 or HDAC2 should therefore produce Gc resistance, and we show that ∼50% of Gc-resistant human and dog corticotroph adenomas, which are the hallmark of Cushing disease, are deficient in nuclear expression of either protein. In addition to providing a molecular basis for Gc resistance, these deficiencies may also contribute to the tumorigenic process.
Keywords: Repression, nuclear receptor, Swi/Snf, pituitary tumors, POMC, Cushing
Important aspects of glucocorticoid (Gc) action are exerted through repression of transcription (Helmberg et al. 1995; Reichardt et al. 1998), such as their anti-inflammatory action (Wintermantel et al. 2004). Contrary to activation of transcription resulting from DNA binding of the glucocorticoid receptor (GR), repression by GR is often achieved through protein:protein interactions and mutual antagonism with other transcription factors. The first proposal of a mechanism for this type of transcriptional repression, now known as trans-repression, involved GR antagonism of AP-1-dependent transcription on the collagenase 1 gene (Jonat et al. 1990; Schüle et al. 1990; Yang-Yen et al. 1990). Many features of this mechanism of repression were revealed in this early work. Mainly, Gc-dependent repression is mediated by GR, but without direct GR:DNA interactions as for GR- activated transcription (McKenna and O'Malley 2002). Rather, GR represses transcription through protein:protein interactions with DNA-bound AP-1. In contrast to GR activation of transcription, Gc-dependent trans-repression by GR is exerted by monomers (Heck et al. 1994) and, consequently, is independent of dimerization (Reichardt et al. 1998; Martens et al. 2005). Direct protein interactions between GR and AP-1 (jun/fos) were observed and initially led to the suggestion that complex formation between these factors titrated AP-1 away from transcription targets, thus resulting in apparent repression. It was later shown by in vivo footprinting that pro-moter occupancy is not altered in the repressed state, and thus a model of repressors interacting with promoter-bound activators was proposed (König et al. 1992). This mechanism was shown to be reciprocal, such that activators (such as GR, AP-1, NFκB, or NGFI-B) can behave as either activator or repressor, with the DNA- bound factor acting as activator. Trans-repression was also shown to occur between AP-1 and different nuclear receptors (NR) in addition to GR (for review, see De Bosscher et al. 2003).
The anti-inflammatory action of Gc is largely exerted by its repressor activity (Hayashi et al. 2004). Indeed, repression by GR of NFκB action on genes encoding pro- inflammatory cytokines such as interleukin 8 (IL-8) (Wintermantel et al. 2004) was suggested to use similar mechanisms of trans-repression as those between GR and AP-1 (De Bosscher et al. 2003). Recent insights into the molecular mechanism of GR trans-repression of the NFκB-activated IL-8 gene followed the introduction of the chromatin immunoprecipitation (ChIP) technique, which confirmed and extended the trans-repression model: In this case, GR does not inhibit formation of a preinitiation complex, but rather interferes with phosphorylation of the RNA polymerase II (Pol II) C-terminal repeat (CTD) at Ser2 (Nissen and Yamamoto 2000) and with recruitment of p-TEFb, the Ser2 CTD kinase. In this system, failure to recruit p-TEFb appeared to decrease gene expression at a post-initiation step (Luecke and Yamamoto 2005). More recently, trans-repression initiated by another NR, PPARγ, was shown to depend on ligand-induced SUMOylation of the PPARγ ligand- binding domain and the resulting stabilization of its interaction with the corepressor NCor (Pascual et al. 2005).
We have investigated the mechanism of Gc repression of the pituitary proopiomelanocortin (POMC) gene. Pituitary POMC is at the center of the hypothalamo–pituitary–adrenal (HPA) axis that ultimately controls Gc synthesis and modulates the stress response, energy metabolism, and immune response. Central activation of the HPA axis is mediated through secretion of hypothalamic CRH into the pituitary portal system, where CRH stimulates secretion of presynthesized POMC-derived ACTH and transcription of the POMC gene (Philips et al. 1997b). Blood-borne ACTH is the major stimulus of adrenal Gc synthesis, and maintenance of physiological levels of cortisol requires adequate negative feedback regulation by Gc of pituitary ACTH secretion and POMC gene transcription. Disregulation of this negative feedback loop has severe metabolic consequences that characterize Cushing disease.
Cushing syndrome, or hypercortisolism, is characterized by upper body obesity (moon face and buffalo hump), muscle weakness, high blood pressure, and glucose intolerance. This condition is caused by elevated blood Gc that may result from high-dose Gc treatment, excessive cortisol production by adrenal tumors, and, in most noniatrogenic cases, overproduction of ACTH by ectopic or pituitary tumors (Arnaldi et al. 2003). Typically, ACTH-producing pituitary tumors are micro- adenomas that are not malignant, produce excessive amounts of ACTH, and are resistant to Gc negative feedback. These tumors define Cushing disease (as opposed to syndrome), and they appear to be due to tumorigenic transformation of anterior pituitary corticotroph cells (Vallette-Kasic et al. 2003). The corticotrophs are one of two proopiomelanocortin (POMC)-expressing pituitary lineages (Pulichino et al. 2003), and they are at the center of the HPA axis. The development of Gc resistance in corticotroph adenomas may be a critical (and possibly primary) step in tumorigenesis.
Although various human pathological conditions, such as Cushing disease and depressive illness, have been associated with deficient Gc feedback, little mechanistic insight exists into the molecular defects causing Gc resistance, except for a few rare mutations in GR itself (Lamberts 2002). We have used Gc/GR repression of POMC gene transcription to gain insight into mechanisms of Gc resistance. Repression of POMC gene transcription by Gc results, at least in part, from trans-repression exerted by GR on the activity of orphan NRs related to NGFI-B (Philips et al. 1997b). Indeed, transcription elicited by NGFI-B (Nur77) and by the closely related orphan NRs, Nurr1 and NOR1, is subject to GR repression by a mechanism that is very similar to trans-repression between GR and AP-1 and between GR and NFκB (Philips et al. 1997b; Martens et al. 2005). In pituitary corticotroph cells that produce ACTH, NGFI-B, and its related NRs are mediators of the stimulatory signals elicited by CRH (Philips et al. 1997a; Maira et al. 1999). Acting through its membrane receptors, CRH leads to activation of protein kinase A (PKA) and mitogen- activated protein kinase (MAPK) pathways that quickly result in: (1) dephosphorylation of the NGFI-B DNA- binding domain (DBD), which is required for NGFI-B interaction with DNA; (2) formation of Nur factor dimers that recognize the POMC promoter NurRE sequence; and (3) recruitment of the NR coactivator SRC-2 (TIF2) to the AF-1 domain of NGFI-B (Maira et al. 2003b). CRH signals also act on the POMC promoter through SRC-2 coactivation of Tpit (Maira et al. 2003a), a highly cell-restricted T-box transcription factor (Lamolet et al. 2001). Although both NGFI-B and Tpit activities are enhanced in response to SRC2 and CRH signals, only NurRE-dependent activity is subject to Gc repression (Martens et al. 2005).
We now report on the molecular mechanism of trans-repression between GR and NGFI-B, and in particular on the requirement for the Swi/Snf chromatin remodeling protein Brg1 and its ATPase activity in this mechanism. Brg1 is critical for formation of stable in vivo complexes between GR and NGFI-B, and between GR and HDAC2. Promoter recruitment of both GR and HDAC2 are Gc- dependent and are associated with decreased acetylated histone H4 at the promoter and throughout the gene. Assembly of a trans-repression machinery at the POMC promoter also appears to hamper initiation of transcription by blocking Pol II at the promoter. Protein:protein interactions at the promoter between activator (NGFI-B) and repressor (GR) together with histone deacetylation result in inhibition of transcription initiation. Further, we found that ∼50% of pituitary adenomas from Cushing disease patients or dogs have misexpression of either Brg1 or HDAC2 in tumor tissue but not in adjacent normal pituitary tissue. The high frequency of misexpression of these proteins in corticotroph adenomas clearly supports their importance in Gc negative feedback regulation and in Gc resistance syndromes.
Results
Brg1 or Brm is required for trans-repression between GR and NGFI-B
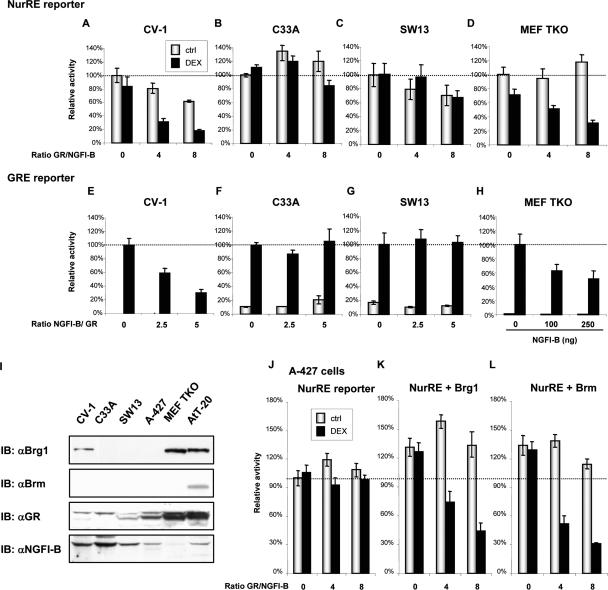
We have identified the POMC gene promoter NurRE element and its cognate transcription factors, NGFI-B and the related NRs, as the primary target for repression of POMC transcription by GR (Philips et al. 1997b; Martens et al. 2005). Trans-repression between NGFI-B and GR can be monitored using simple reporter plasmids containing the NurRE or GRE. When we assessed the sensitivity of the NurRE reporter to Gc repression in a panel of cell lines, we realized that a subset of cells does not support transcriptional antagonism between NGFI-B and GR. Indeed, while CV-1 cells are subject to Dex- and GR-dependent repression of NGFI-B activity (Fig. 1A), C33A, SW13, and A-427 cells are insensitive to Gc/GR action (Fig. 1B,C,J). Conversely, CV-1 cells exhibit antagonism of GR-dependent transcription by NGFI-B (Fig. 1E), while C33A (Fig. 1F) and SW13 cells (Fig. 1G) do not support this antagonism. Expression of GR and NGFI-B was similar in cells that support antagonism or not (Supplementary Fig. S1). Since C33A and SW13 cells are deficient in Rb expression (Wong et al. 2000), and given the role of Rb in CRH responsiveness (Batsche et al. 2005a), we assessed the importance of Rb and the related pocket proteins p107 and p130 in trans-repression. To this end, we used mouse embryo fibroblasts (MEF) that are deficient in all three pocket proteins (TKO) (Sage et al. 2000). These MEF TKO cells appeared to be fully capable of supporting trans-repression by GR of NGFI-B- dependent activity (Fig. 1D) as well as NGFI-B repression of GR-dependent activity (Fig. 1H). These results clearly indicate that Rb-related proteins are not required for trans-repression between GR and NGFI-B.
Figure 1.
Cell lines can/cannot support trans-repression between GR and NGFI-B. Assessment of trans-repression between GR and NGFI-B using either NurRE (A–D) or GRE (E–H) reporters and expression vectors for NGFI-B (25 ng of expression vector in A–D, resulting in eightfold to 10-fold activation) and GR (100 ng of expression plasmid in E–H). CV-1 (A,E) and MEFs from Rb−/−, p107−/−, and p130−/− mouse (MEF TKO) cells (D,H) support trans-repression, whereas the Brg1/Brm-deficient C33A (B,F) and SW13 (C,G) cells do not. (H) GR is expressed at significant levels in MEF TKO cells and was not overexpressed in these experiments. (I) Western blot analysis of cellular proteins showing that the ability for trans-repression correlates with expression of Brg1 or Brm. Whereas GR and NGFI-B are expressed at various levels in all six cell lines, only the trans-repression-proficient CV-1, AtT-20, and MEF TKO cells express Brg1 and/or Brm. (J) Brg1/Brm-deficient A-427 cells do not support trans-repression, which can be reconstituted by expression of Brg1 (K) or Brm (L) as assessed using NurRE reporter and expression vectors for GR and NGFI-B (25 ng of expression vector resulting in eightfold activation). Total DNA is kept constant in all transfections, and data represent the means ± SEM of three experiments, each performed in duplicate.
Another common property of C33A, SW13, and A-427 cells is that they do not express significant levels of the SWI/SNF proteins Brg1 or Brm (Wong et al. 2000) as verified by Western blot, whereas AtT-20, MEF TKO, and CV-1 cells express at least one of these proteins (Fig. 1I). Since the A-427 lung adenocarcinoma cells express Rb- related proteins (Wong et al. 2000), we used them to assess the importance of Brg1 or Brm in GR repression. Expression of either Brg1 (Fig. 1K) or Brm (Fig. 1L) is sufficient to fully restore GR- and Gc-dependent repression of NGFI-B-dependent activity. It is noteworthy that expression of either Brg1 or Brm slightly but significantly increased basal transcriptional activity (Fig. 1, cf. K,L and J). These experiments suggest that Brg1 is essential for GR repression of NGFI-B-dependent transcription.
Brg 1 is essential for Gc repression of POMC gene transcription
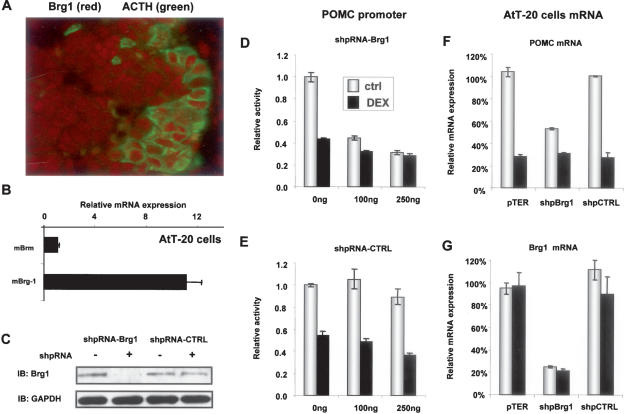
Brg1 is expressed by most cells and particularly by POMC (ACTH)-positive cells as revealed by immunohistochemistry (Fig. 2A). In AtT-20 cells, a model of POMC- expressing pituitary corticotroph cells, Brg1 appears to be expressed at ∼10 times higher levels than Brm as assessed by RT–QPCR (Fig. 2B).
Figure 2.
Brg1 is required for POMC promoter activity and trans-repression by GR. (A) Colocalization of Brg1 (red) with the corticotroph marker ACTH (green) in mouse pituitary section. (B) Relative mRNA expression (RT–QPCR) of Brg1 and Brm in corticotroph AtT-20 cells showing a preponderance of Brg1. (C) Western blot analysis of Brg1 expression in AtT20 cells transfected with expression plasmids for Brg1 or control shRNA; GAPDH was measured as loading control. (D,E) Effects of Brg1 (D) or control (E) shRNA knockdown on the activity (ctrl) and Gc (Dex) repression of a POMC promoter luciferase reporter assessed in transfected AtT-20 cells. (F,G) Effects of the same shRNAs on endogenous POMC (F) and Brg1 (G) mRNAs in transfected and FACS-sorted AtT-20 cells. Endogenous mRNAs were quantitated by RT–QPCR. Data of three experiments, each performed in duplicate, are shown as means ± SEM.
As these data suggest that Brg1 is predominantly expressed in POMC cells, we directly tested its role in Gc feedback repression of POMC using small hairpin RNA (shRNA) expression plasmids in AtT-20 cells to knock down Brg1 expression as ascertained by Western blot (Fig. 2C). The importance of Brg1 was assessed by cotransfection of a POMC-luciferase reporter together with shRNA expression plasmids against Brg1 (Fig. 2D) or a random sequence (Fig. 2E). Knockdown of Brg1 expression decreased basal POMC promoter activity by almost 50%, and the residual (but significant) activity was no longer repressible by Dex. It thus appears that part of the basal POMC promoter activity is dependent on Brg1, and that it is this activity that is targeted by Dex repression. When similarly transfected and FACS-sorted AtT-20 cells were assessed for endogenous POMC (Fig. 2F) and Brg1 (Fig. 2G) mRNA levels, it was also found that constitutive POMC mRNA levels are reduced by ∼50% and that the remaining expression is relatively resistant to Dex repression. Thus in the absence of Brg1, POMC expression and promoter activity appear to become Dex- resistant, in agreement with the importance of this protein for trans-repression between GR and NGFI-B (Fig. 1).
Brg1 interacts with both NGFI-B and GR
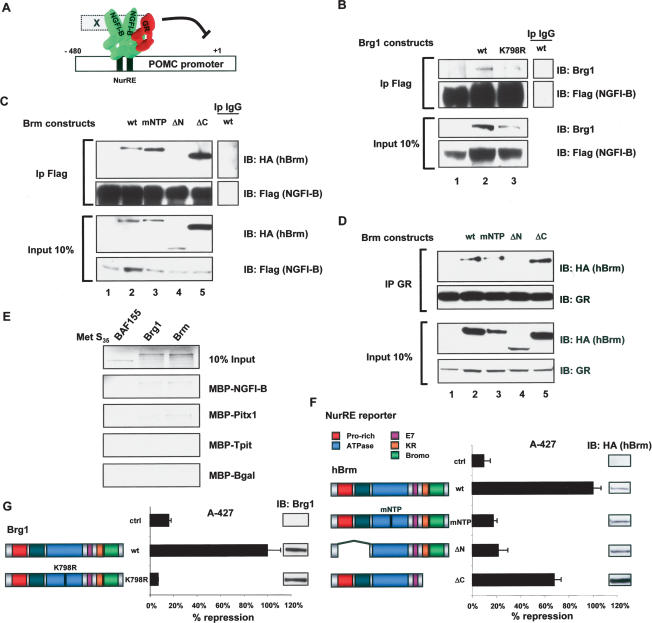
We have previously shown that Gc repression of POMC promoter activity is exerted through trans-repression involving direct protein–protein interactions between GR and NGFI-B (Fig. 3A; Martens et al. 2005). The involvement of Brg1 in Gc repression of NGFI-B activity may result from interactions between Brg1 and NGFI-B and/ or GR. In order to assess these putative interactions, we used immunoprecipitation of Flag-tagged NGFI-B from extracts of transfected C33A cells. This resulted in co- precipitation of Brg1 (Fig. 3B, lane 2), and similar results were obtained for Brm (Fig. 3C, lane 2). It was previously shown that GR interacts physically with Brg1 (Fryer and Archer 1998), and in similar coimmunoprecipitations we showed coprecipitation of HA-tagged Brm with GR (Fig. 3D, lane 2). We then used available mutants of Brg1 and Brm to first assess the structural requirements for interaction with NGFI-B or GR. For Brg1, the ATPase-deficient mutant K798R was as efficiently coprecipitated with NGFI-B as the wild-type protein (Fig. 3B, lane 3). Similarly, the mNTP mutant of Brm was also efficiently coimmunoprecipitated with NGFI-B (Fig. 3C, lane 3) and with GR (Fig. 3D, lane 3). Using deletion mutants of Brm, we could show that both NGFI-B and GR interactions required the Brm N terminus (ΔN) (Fig. 3C,D, lanes 4), but not the Brm C terminus (Fig. 3C,D, lanes 5). We then tested whether Brg1 and Brm could interact directly with NGFI-B. A pull-down assay using a MBP–NGFI-B fusion protein was used to show interactions with Brg1 and Brm, but less so with another Swi/Snf protein, BAF155 (Fig. 3E).
Figure 3.
Brg1 and Brm ATPase activity is required for repression but not for interaction with NGFI-B or GR. (A) Standard model of trans-repression by GR. On the POMC promoter, dimers of NGFI-B or heterodimers between NGFI-B and the related orphan NRs Nurr1 and NOR1 bind the NurRE; their activity is enhanced by CRH signaling. Gc repression is thought to be initiated through protein:protein interactions between GR and promoter-bound NGFI-B. (B) Coimmunoprecipitation of Brg1 or its ATPase-deficient K798R mutant with Flag-tagged NGFI-B. GR- and Brg1-deficient C33A cells were transfected with expression vectors as indicated, and relevant proteins were revealed by immunoblotting (IB) after immunoprecipitation (IP) of Flag-NGFI-B. (C) Coimmunoprecipitation of Brm or its indicated mutants following immunoprecipitation of Flag-NGFI-B. (D) Coimmunoprecipitation of Brm or its indicated mutants following immunoprecipitation of GR. (E) In vitro pulldown of [35S]-labeled Brg1 or Brm with MBP–NGFI-B, MBP-Pitx1, MBP-Tpit, or MBP-βGal as control. The Swi/Snf protein BAF155 was used as control. (F,G) Reconstitution of trans-repression using a NurRE reporter in A-427 cells with hBrm and mutants of this protein (F) as well as with Brg1 and the ATPase deficiency Brg1 mutant K798R (G). Band reporter activity was similar for all Brg1 and Brm proteins. (Insets) The relative expression levels of each Brm or Brg1 protein were assessed by Western blot. Data represent the means ± SEM. of three experiments, each performed in duplicate.
In order to test the importance of Brg1 or Brm domains in trans-repression, we used the reconstituted system in A-427 cells in which trans-repression is dependent on ectopic expression of Brg1 or Brm (Fig. 1J–L). Interestingly, the ATPase-deficient Brg1 K798R and Brm mNTP mutants that retain the ability to interact with NGFI-B and GR are both unable to reconstitute trans-repression (Fig. 3F,G). This result clearly indicates a dependence on ATPase activity for trans-repression. In agreement with the deletion analysis for protein interactions, deletion of the Brm N terminus (ΔN) prevented reconstitution of trans-repression (Fig. 3F). These analyses clearly implicate many domains of Brg1/Brm in the process of trans-repression, and they corroborate the coimmunoprecipitation analyses.
Brg1 is required in vivo for complex formation between GR and NGFI-B
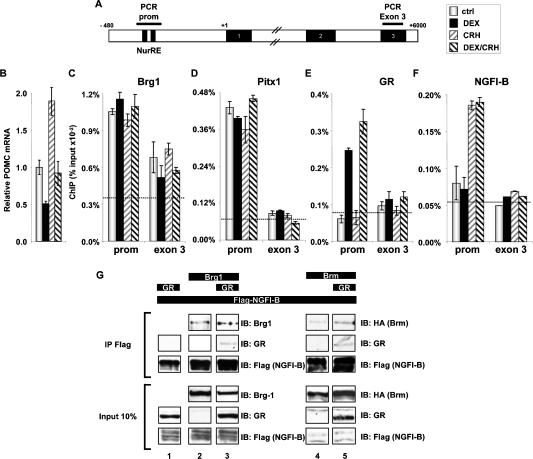
We next wanted to verify that Brg1, GR, and NGFI-B are present at the POMC promoter, and that their recruitment coincides with trans-repression. Using ChIP, we first assessed recruitment of Brg1 to the POMC promoter. In these experiments, promoter recruitment was compared with the presence of proteins over exon 3 of the POMC gene, which lies 6 kb downstream (Fig. 4A). Further, promoter recruitment was assessed in basal conditions as well as in activated and repressed states (Fig. 4B). It is noteworthy that Brg1 is present at the POMC promoter in basal conditions and that promoter occupancy is not altered by either activation (CRH) or repression (Dex) of transcription (Fig. 4C). Brg1 also exhibits significant presence at the 3′-end of the gene (Corey et al. 2003), and although differences are not significant, the amount of Brg1 over exon 3 has a tendency to follow the effects of hormone treatment on transcription. The recruitment of Pitx1, a factor involved in basal POMC transcription (Lamonerie et al. 1996) and showing in vitro interaction with Brg1 (Fig. 3E), is also constant and unchanged by hormonal stimulation (Fig. 4D). Promoter presence of Brg1 in basal conditions is consistent with its purported role in basal POMC transcription, as suggested by the shRNA experiments (Fig. 2D). In agreement with a model of Gc repression that depends on nuclear trans- location and promoter recruitment of ligand-activated GR, we observed recruitment of GR at the POMC promoter in the presence but not in the absence of Dex; in contrast, no GR recruitment was observed over POMC exon 3 sequences (Fig. 4E). Similarly, POMC promoter recruitment of NGFI-B was observed upon CRH activation, and NGFI-B recruitment was not observed over exon 3 (Fig. 4F). Corecruitment of GR with NGFI-B in Dex/CRH-treated cells is consistent with the protein: protein interaction model of trans-repression (Fig. 3A).
Figure 4.
Brg1 is required for complex formation between NGFI-B and GR. (A) Schematic representation of the mouse POMC gene indicating the position of PCR-amplified regions in ChIP experiments. (B) Abundance of POMC mRNA measured by RT–QPCR in control and AtT-20 cells treated as indicated. Both the synthetic Gc dexamethasone (Dex) and the hypothalamic hormone CRH were used at 10−7 M. (C) ChIP analysis of Brg1 recruitment at the POMC promoter and exon 3 of the POMC gene in AtT-20 cells. The dashed line indicates background ChIP signal observed with control IgG. (D) ChIP analysis of Pitx1 recruitment to the POMC promoter and exon 3. (E) ChIP analysis of GR recruitment to the POMC promoter and exon 3. (F) ChIP analysis of NGFI-B recruitment to the POMC promoter and exon 3. (G) Coimmunoprecipitation of GR requires Brg1 or Brm with Flag-NGFI-B in cotransfected C33A cells.
These data indicate that involvement of Brg1 in a trans-repression complex would presumably involve de novo association of GR with Brg1 that is already present at the promoter.
Previous work had shown direct in vitro interaction between GR and NGFI-B (Martens et al. 2005), and thus these data did not indicate a requirement for another protein such as Brg1. In order to define the role of Brg1, we first assessed the putative interaction of GR and NGFI-B in vivo using coimmunoprecipitation in Brg1- deficient C33A cells. When Flag-NGFI-B and GR were coexpressed, immunoprecipitation of NGFI-B was not sufficient to coprecipitate GR (Fig. 4G, lane 1). On the other hand, NGFI-B was sufficient for in vivo interaction with Brg1 (Fig. 4G, lane 2) or Brm (Fig. 4G, lane 4). In presence of Brg1 (Fig. 4G, lane 3) or Brm (Fig. 4G, lane 5), GR was coimmunoprecipitated with Flag-NGFI-B, thus showing the in vivo dependence on Brg1/Brm for GR recruitment to a trans-repression complex. It thus appears that the requirements for GR/NGFI-B interactions in vivo are more stringent than in vitro and that Brg1 or Brm may play an essential role as a bridge to stabilize interactions between the two proteins, either directly (Fig. 3E; Fryer and Archer 1998) or indirectly.
Brg1- and GR-dependent HDAC2 recruitment
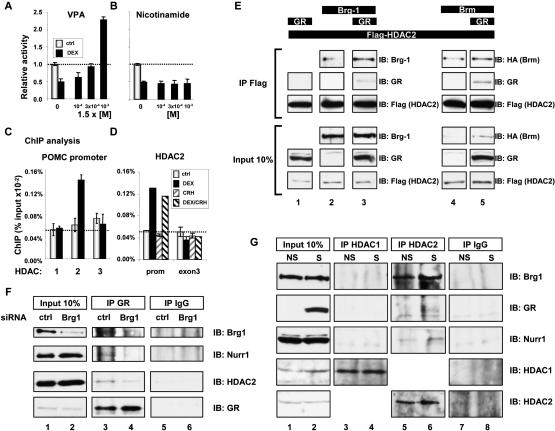
The formation of a putative trans-repression complex raises the question of how such a complex may repress transcription. A first possibility would be that recruitment of GR passively lowers transcription by preventing NGFI-B-dependent activation; alternatively, the Brg1/ NGFI-B/GR complex may also recruit corepressors such as histone deacetylases (HDAC). To test a putative role of HDAC activity in repression of POMC transcription by Gc/GR, we used valproic acid (VPA), an anticonvulsant used in clinical practice that was reported to be a class I and II HDAC inhibitor, and nicotinamide, an inhibitor of NAD-dependent class III HDACs. While nicotinamide had no effect (Fig. 5B), VPA treatment prevented Gc repression of POMC promoter activity at low concentration and even reversed the normally Gc-re- pressed POMC promoter into a Gc-inducible promoter at high VPA concentration (Fig. 5A). Similar results were obtained for trichostatin A and sodium butyrate (data not shown). All three HDAC inhibitors share the ability not only to prevent, but to reverse the sensitivity of POMC transcription to Gc. These results indicate an essential role of histone deacetylases in Gc repression of POMC transcription.
Figure 5.
Brg1-dependent recruitment of HDAC2 to the trans-repression complex. AtT-20 cells were treated with valproate (VPA) (A) or nicotinamide (B) at the indicated concentrations and assessed for Dex repression of POMC-luciferase activity. Class I and II (A), but not class III (B), HDAC inhibitors show a complete loss of Dex repression. (C) ChIP analysis of the POMC promoter showing Dex-dependent recruitment of HDAC2 to the promoter, but not HDAC1 or HDAC3. (D) ChIP analysis of HDAC2 recruitment to the POMC promoter, but not to POMC exon 3, following treatment of AtT-20 cells with Dex. (E) Coimmunoprecipitation of GR with Flag-HDAC2 requires the presence of Brg1 or Brm in transfected C33A cells. (F) Coimmunoprecipitation of Brg1, the Nur-related factor Nurr1, and HDAC2 with endogenous GR in AtT-20 cell nuclear extracts (lane 3) is impaired following siRNA knockdown (lane 2) of Brg1 (lane 4). (G) Coimmunoprecipitation of endogenous Brg1, GR, and Nurr1 in AtT-20 cells stimulated (S) or not (NS) with CRH and Dex following HDAC2 immunoprecipitation. Neither immunoprecipitation with antibody against HDAC1 nor control IgG brought down these proteins.
We next assessed the recruitment of different HDACs to the POMC promoter by ChIP (Fig. 5C). These experiments clearly indicated recruitment of HDAC2 but not HDAC1 or HDAC3. Further, comparison of cells treated or not with CRH and/or Dex indicated that HDAC2 is recruited to the promoter only in Dex-treated cells (Fig. 5D). It is thus possible that HDAC2 is directly associated with GR and Brg1. In order to assess this possibility, we performed coimmunoprecipitation using Flag-tagged HDAC2 (Fig. 5E). Whereas HDAC2 was not sufficient for interaction with GR in these experiments (Fig. 5E, lane 1), it was found that HDAC2 could interact in vivo with Brg1 (Fig. 5E, lane 2) or Brm (Fig. 5E, lane 4), but, most significantly, Brg1 or Brm appeared necessary for corecruitment of GR with HDAC2 (Fig. 5E, lanes 3,5).
So far, we have shown in reconstituted systems corecruitment of Brg1 with NGFI-B, GR, and HDAC2 (Figs. 3–5). In order to show similar interactions between endogenous proteins, we used immunoprecipitation in nuclear extract of AtT-20 cells. Immunoprecipitation of GR led to the coimmunoprecipitation of Brg1, HDAC2, and the NGFI-B-related factor, Nurr1 (Fig. 5F, lane 3). In previous work, we have shown enhanced activity of NGFI-B and the related NRs Nurr1 and NOR1 in response to CRH (Maira et al. 1999), and we have also shown that GR antagonism is equally exerted on the three Nur-related factors (Martens et al. 2005); we used Nurr1 antibodies in the present experiments because available NGFI-B antibodies are not adequate. In order to test the importance of Brg1 for complex formation, we used small interfering RNAs (siRNAs) to knock down Brg1 (Fig. 5F, lane 2), and this led to decreased coimmunoprecipitation of Nurr1 and HDAC2 with GR (Fig. 5F, cf. lanes 4 and 3).
Since ChIP analyses revealed a preference for HDAC2 in GR repression of POMC, we also assessed HDAC specificity in complex formation. AtT-20 cell nuclear extracts were immunoprecipitated with antibodies against either HDAC1 or HDAC2 and revealed for coimmuno- precipitation of Brg1, GR, and Nurr1. As shown in Figure 5G, all three proteins were coimmunoprecipitated with HDAC2 (lane 6) but not with HDAC1 (lane 4), in close agreement with promoter recruitment data (Fig. 5C). Taken together, these data are consistent with the idea of trans-repression protein complexes formed in the presence of ligand-activated GR: These complexes appear to contain Brg1, GR, Nurr1, and HDAC2. Although we cannot formally exclude the possibility that some of these proteins are present in separate complexes, the recruitment of all these proteins to the POMC promoter in response to cognate signals strongly argues for their joint presence and their association on the promoter.
Trans-repression reduces histone acetylation and promoter clearance
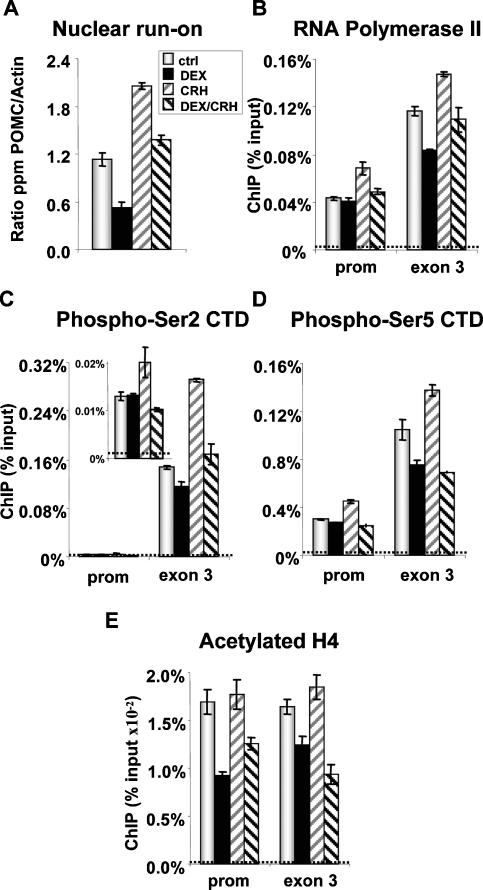
Previous work had indicated that the effects of CRH and Dex on POMC transcription are essentially transcriptional, and they are due to changes in initiation rate (Gagner and Drouin 1985, 1987). Indeed, the rate of POMC transcription as measured in nuclear run-on as- says was decreased by Dex and increased by CRH (Fig. 6A). These changes in transcription initiation should be reflected in promoter recruitment of Pol II, and this was assessed using ChIP (Fig. 6B). Contrary to expectation, Dex inhibition of POMC initiation (Fig. 6A) was not paralleled by a decrease in Pol II recruitment at the promoter, since promoter occupancy was unchanged by Dex treatment (Fig. 6B). As expected, CRH stimulation enhanced Pol II promoter recruitment, and this recruitment was reversed by Dex (Fig. 6B). It is thus noteworthy that Dex repression has different effects on basal and CRH-stimulated transcription: Whereas in both conditions Dex decreases transcription rates (Fig. 6A), only CRH-induced Pol II recruitment is reversed by Dex. This suggests that another mechanism of repression is triggered by Dex and GR, in addition to modulation of Pol II promoter recruitment. The abundance of Pol II over exon 3 sequences was modulated in the same way as transcriptional activity, consistent with a correlation between the number of polymerases over the gene and the level of transcripts (Figs. 4B, 6A). The discrepancy in Pol II abundance at the promoter versus exon 3 in Dex-treated cells suggests reduced promoter clearance by Pol II.
Figure 6.
POMC promoter function in repressed (Dex) and activated (CRH) conditions. (A) Initiation of POMC transcription as measured in nuclear run-on assays in pituitary primary culture treated with Dex and/or CRH. (B) ChIP analyses of RNA Pol II recruitment to the POMC promoter and exon 3 in control and treated AtT-20 cells. (C) ChIP analysis of POMC gene occupancy by Pol II with phospho-Ser2 CTD. (D) ChIP analysis of POMC gene occupancy by Pol II with phospho-Ser5 CTD. (E) ChIP analysis of acetylated histone H4 over the POMC promoter and exon 3. Dashed lines indicate background ChIP signals.
Previous work showed differential Pol II CTD Ser2 and Ser5 phosphorylation on the Gc-repressed NFκ-B-activated IL-8 gene (Nissen and Yamamoto 2000). We therefore assessed the presence of Pol II phospho-Ser2 (Fig. 6C) and phospho-Ser5 (Fig. 6D) forms at the POMC gene and found similar recruitment of both forms of phospho CTD as well as total Pol II at the POMC promoter and exon 3 in basal, stimulated, and repressed conditions. This discrepancy likely reflects differences in mechanisms of trans-repression between GR and NGFI-B (pre- sent work), which occurs at the level of transcription initiation (Fig. 6A), and between GR and NFκB, which occurs at a post-initiation step. Indeed, Luecke and Yamamoto (2005) showed a decrease of phospho-Ser2 Pol II at the IL-8 promoter following Dex, and this was correlated with reduced elongation rates; in the case of POMC, we did not observe Dex-dependent changes in phospho-Ser2 Pol II at the promoter. In agreement with elongation-dependent increases in phospho-Ser2 Pol II (Cho et al. 2001), we found much higher levels of this Pol II form at exon 3 compared with the promoter (Fig. 6C).
The relative accumulation of Pol II at the POMC promoter in Dex-repressed cells may be due to a sequestering effect of the trans-repression complex; alternatively and not mutually exclusively, it may result from modification of chromatin structure through acetylation since HDAC2 is corecruited with GR. We therefore assessed the level of acetylated histone H4 over the promoter and exon 3 of the POMC gene using ChIP (Fig. 6E). A significant reduction of acetylated H4 was observed over the POMC promoter after Dex repression in both basal and CRH-stimulated conditions. It is noteworthy that the level of acetylated H4 is not increased by CRH, which enhances transcription (Fig. 6A), but it should be remembered that constitutive/basal activity of the POMC promoter corresponds to significant transcription, and this may be correlated with significant acetylation of the promoter. Similar (but of lesser magnitude) changes in the level of acetylated H4 were observed downstream over exon 3 (Fig. 6E). These data are entirely consistent with the recruitment of HDAC2 to the POMC promoter and clearly support a model of trans-repression in which active rather than passive repression is exerted through chromatin modification, together with the possibility that the trans-repression complex itself may contribute to retain Pol II at the promoter or prevent its recruitment in the CRH-stimulated condition.
In summary, two complementary mechanisms appear to account for GR trans-repression of the POMC gene. First, recruitment of HDAC2 to the promoter would lead to histone deacetylation, and this appears to inhibit promoter clearance in both basal and CRH-stimulated conditions. Second, the CRH-stimulated enhancement of Pol II recruitment to the promoter is prevented under Dex repression, suggesting that further Pol II recruitment is incompatible with the corecruitment of Brg1, NGFI-B, GR, and HDAC2.
Misexpression of Brg1 or HDAC2 in Cushing disease
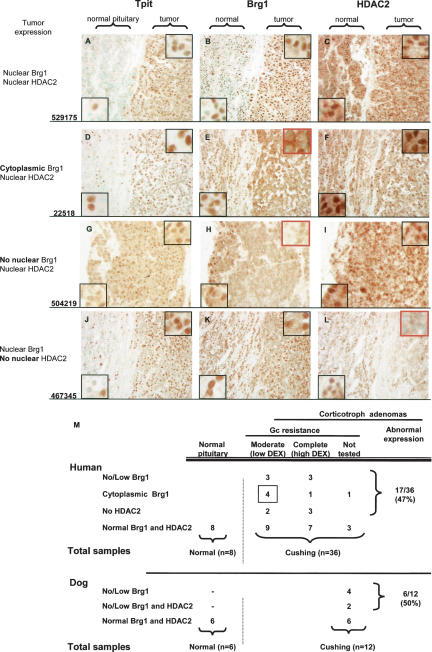
Pituitary corticotroph adenomas that define Cushing disease (Cushing 1932) are typically resistant to Gc feedback and secrete the excessive levels of ACTH that are responsible for the associated hypercortisolism. Gc resistance, or the loss of Gc feedback inhibition, is observed in all Cushing disease patients when assessed by the Dex suppression test. All patients show no response to a low dose of Dex, and a subset of patients exhibits a complete resistance to Gc as revealed by challenge with a high dose of Dex. In view of the importance of Brg1 and HDAC2 for Gc repression of POMC transcription, we assessed expression of these proteins in corticotroph adenomas that had been classified as either moderate or complete Gc-resistant in the Dex suppression test (Fig. 7M). The Tpit transcription factor is present only in nuclei of corticotroph cells, and this factor has been shown to be an excellent marker of corticotroph adenomas (Vallette-Kasic et al. 2003). For many pituitary surgery samples, we had some normal as well as tumor tissue, the latter being marked by extensive Tpit-positive nuclei as opposed to dispersed corticotrophs in the normal tissue (Fig. 7A). Some tumors did not show any change of either Brg1 (Fig. 7B) or HDAC2 (Fig. 7C) expression in comparison with normal pituitary. In these tumors, as in all normal pituitary samples, Tpit, Brg1, and HDAC2 were found to be exclusively nuclear. A subset of tumors presented with a striking change in the distribution of Brg1 within the tumor but not within the adjacent normal pituitary tissue; indeed, these tumor cells have cytoplasmic, rather than nuclear, localization of Brg1 (Fig. 7E). Six tumors of this type were found (Fig. 7M), and in each case tumor expression of HDAC2 was not altered (Fig. 7F). Another six tumor samples were found to have low/undetectable Brg1 (Fig. 7H) with normal HDAC2 (Fig. 7I), whereas five tumors were found with normal Brg1 (Fig. 7K) and no HDAC2 (Fig. 7L). None of these tumors had altered GR expression as assessed by immunohistochemistry (data not shown).
Figure 7.
Deficient Brg1 or HDAC2 expression in Cushing disease corticotroph adenomas. Immunohistochemical analysis of the corticotroph marker TPIT (A,D,G,J), Brg1 (B,E,H,K), and HDAC2 (C,F,I,L) in representative corticotroph adenomas. (A–C) Tumor revealed by nuclear TPIT staining (A) with normal nuclear Brg1 (B) and nuclear HDAC2 (C). (D–F) Tumor with cytoplasmic Brg1 (E) and normal nuclear HDAC2 (F). Note nuclear Brg1 in contiguous normal tissue. (G–I) Tumor with no nuclear Brg1 (H) and normal nuclear HDAC2 (I). (J–L) Tumor with normal nuclear Brg1 (K) and no nuclear HDAC2 (L). Each row presents data from one patient who is representative of the others in the same group as summarized in M. Human patients were subdivided into moderate or complete Gc resistance groups depending on their response to low or high doses of dexamethasone. In 19 cases, expression of Brg1 and HDAC2 were similar in tumor and normal pituitary tissues. Overall, 17 of 36 patients (47%) present with abnormal expression of Brg1 or HDAC2. The panel of 12 dog corticotroph adenomas (Supplementary Fig. S2) had four with no or low Brg1 expression and two with no tumor Brg1 and low HDAC2.
Cushing disease is more prevalent in dogs than humans (Meij et al. 2002), and we also studied Brg1 and HDAC2 expression in a panel of dog tumors (Supplementary Fig. S2). Whereas four out of 12 tumors were found to have low or undectectable Brg1 together with normal HDAC2 expression, no dog tumor was found to have cytoplasmic Brg1 or isolated HDAC2 deficiency (Fig. 7M). In contrast, we found two tumors with both undetectable Brg1 and low HDAC2, an association that was not observed in the panel of human tumors. This may reflect species differences in the requirement (or redundancy) for HDAC2.
The compilation of human tumor data according to the level of Gc resistance (Fig. 7M) revealed a higher prevalence of cytoplasmic Brg1 in tumors from patients with moderate Gc resistance. Although the number of patients is low, one may speculate that in tumors with abundant cytoplasmic Brg1, there is still enough nuclear Brg1 for a moderate Gc response. Overall, 17 of the 36 human corticotroph adenomas studied (47%) have mis- expression of either Brg1 or HDAC2. Similarly, six out of 12 (50%) dog tumors have misexpression of Brg1 with or without deficient HDAC2. Consistent with the essential roles of Brg1 and HDAC2 for Gc repression of POMC transcription, the loss of nuclear expression of either protein in corticotroph adenomas may account for the associated Gc resistance.
Discussion
The present work has revealed a critical role for Brg1 (Figs. 1, 2) in trans-repression involving GR, NGFI-B, and HDAC2. Brg1 is required for in vivo recruitment of GR and NGFI-B to the same complex (Figs. 4G, 5F) and also to bring HDAC2 to GR (Fig. 5E). We propose that trans-repression complexes containing Brg1, GR, Nur factors, and HDAC2 (Fig. 5F,G) would assemble on the POMC promoter (Figs. 4C–F, 5D) and lead to repression of transcription (Figs. 4B, 6A) by blockade of Pol II release from the promoter (Fig. 6B–D). This active repression may result from histone deacetylation (Fig. 6E) and stands in contrast to the CRH-induced increase in POMC transcription (Figs. 4B, 6A) that occurs through promoter recruitment of Pol II (Fig. 6B–D). Thus, Brg1 behaves as a scaffold required to form a ligand- and GR-dependent repression machinery that includes HDAC2 and leads to histone deacetylation and repression by blockade of promoter clearance. The in vivo relevance of these proteins was shown in Gc-resistant corticotroph adenomas that characterize Cushing disease (Fig. 7).
Role of Brg1 in POMC transcription
Brg1 and Brm, the ATPase subunits of the Swi/Snf chromatin remodeling complexes, have been associated with the control of transcription in many different ways (Narlikar et al. 2002). Some genes preferentially recruit Brg1 or Brm, whereas other genes do not show specificity or a need for either protein (Kadam and Emerson 2003). They interact directly with a large array of transcription factors belonging to almost all structural classes of DNA- binding proteins. The actions of Brg1 on transcription are thought to occur through chromatin remodeling. Some Brg1-containing complexes also contain HDACs, and it may be this association that determines whether activation or repression of transcription takes place (Pal et al. 2003). The existence of HDAC-containing Brg1 complexes does not, however, in itself explain how such complexes are recruited to specific genes.
Transcription of the POMC gene and of NGFI-B-dependent reporters relies in part on Brg1, and this activity appears to be the target of GR trans-repression. This was suggested by the gain of basal activity in complementation experiments (Fig. 1K,L) and by the loss of POMC promoter activity following Brg1 knockdown (Fig. 2). We currently do not know how and in particular through which transcription factors(s) Brg1 is recruited to the POMC promoter. There are many candidates for this since it was already shown that Brg1 interacts with NeuroD1 (Seo et al. 2005), a basic helix–loop–helix (bHLH) factor contributing to cell specificity of POMC transcription (Poulin et al. 2000). We have also detected in vitro interaction of Brg1 with Pitx1, but not with the cell- restricted T-box factor Tpit (Fig. 3E). The constant presence of Brg1 at the POMC promoter (Fig. 4C) together with Pitx1 (Fig. 4D) may be taken to argue for de novo recruitment to the promoter of Brg1-interacting proteins in response to signals rather than for replacement of complexes; however, it is equally possible that the highly dynamic nature of protein recruitment to promoters (Metivier et al. 2002) allows for replacement of different Brg1-containing complexes in response to activating or repressing signals.
Brg1, a scaffold for recruitment of GR and corepressors
Although activation of POMC transcription in response to CRH may be mediated through different regulatory elements and their cognate transcription factors NGFI-B and Tpit (Philips et al. 1997a; Maira et al. 2003a), trans-repression by GR specifically requires the NurRE and Nur factors (Martens et al. 2005). This strict requirement for Nur factors is a reflection of the specificity of the trans-repression mechanism that relies in some part at least on direct interactions between GR and NGFI-B. Those direct interactions were documented in vitro (Philips et al. 1997b; Martens et al. 2005), but they appear insufficient in vivo to readily allow complex formation between GR and NGFI-B (Fig. 4G, lane 1); the in vivo requirement on Brg1 (Fig. 4G, lane 3) clearly indicates that additional interactions between GR and Brg1 (Fryer and Archer 1998) and between Brg1 and NGFI-B (Fig. 3B) are required in order to form a stable tripartite in vivo complex. Thus, Brg1 plays an essential role as a scaffold for formation of this complex, and presumably for the in vivo recruitment of GR to the promoter (Fig. 4E). Although our data clearly argue for an essential role of Brg1, they do not exclude the presence or involvement of other proteins, such as those of the Swi/Snf complex in trans-repression. Indeed, whereas the ATPase activity of Brg1 is required for trans-repression (Fig. 3G), an activity that may involve the Swi/Snf complex, this same activity is not required for interaction of Brg1/Brm with GR or NGFI-B (Fig. 3B–D). Thus, Brg1 may play its scaffolding role on its own whereas its action in repression depends on chromatin remodeling.
The trans-repression machinery also involves HDAC2 (Fig. 5E–G), and since HDAC2 recruitment to the promoter (Fig. 5C,D) follows that of GR (Fig. 4E), it is likely that GR and HDAC2 are corecruited. We have observed direct interactions between HDAC2 and GR in pull- down assays (data not shown), but, obviously, in vivo interactions of HDAC2 with GR require Brg1 or Brm (Fig. 5E). Again, these data highlight the essential role played by Brg1 in allowing association of the different proteins that constitute the trans-repression machinery.
The trans-repression machinery actively represses transcription
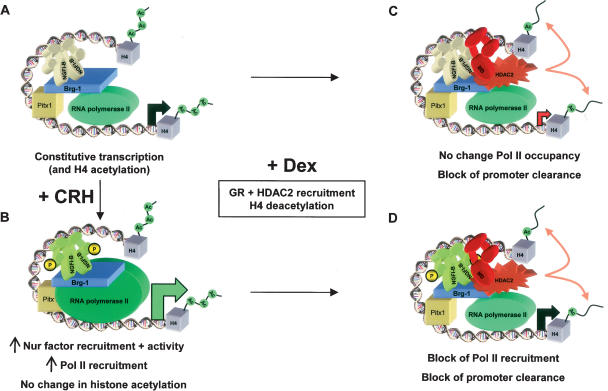
Taken collectively, our data lead to the proposal of different mechanisms for activation of POMC transcription in response to CRH and for repression as result of GR action (Fig. 8). In this model, basal (Fig. 8A) and CRH- activated (Fig. 8B) transcription are quite similar. This activity is associated with histone H4 acetylation of the promoter and gene (Fig. 8A). Stimulation of transcription by CRH leads to recruitment of more NGFI-B to the promoter as well as enhanced recruitment of Pol II (Fig. 8B). This enhanced Pol II recruitment is not associated with changes in acetylated histone H4 (Fig. 6E), but it results in enhanced transcription initiation (Figs. 4B, 6A).
Figure 8.
Model of Brg1-dependent trans-repression. (A) The pituitary POMC gene is constitutively expressed in corticotroph cells. This basal transcription relies on many ubiquitous and cell-restricted factors such as Pitx1, Tpit, and NeuroD1 (not shown), and on signal-regulated factors such as NGFI-B. This activity is partly dependent on Brg1 and is reflected by the presence of RNA Pol II at the promoter and by the presence of acetylated histone H4 over the promoter and gene. For the sake of clarity, nucleosomes are not shown in these diagrams and histones H4, which are always within nucleosomes, are portrayed as isolated entities in the promoter and gene regions. (B) CRH activation through the PKA and MAPK pathways enhances NGFI-B activity through phosphorylation of its AF-1 domain and its recruitment to the promoter. This leads to increased Pol II at the promoter and increased transcription initiation without changes in H4 acetylation. (C) Ligand (Dex) activation of GR initiates formation of a Brg1-dependent protein complex that also contains NGFI-B and HDAC2. Both GR and HDAC2 are recruited to the promoter, with neither contacting DNA directly. This results in deacetylation of H4 and inhibition of transcription initiation without changing promoter occupancy of Pol II; promoter clearance thus appears to be reduced. (D) Trans-repression of transcription initiation by GR is dominant over CRH activation, as GR and HDAC2 are still recruited in the presence of both regulators and H4 are deacetylated. The presence of the trans-repression complex containing Brg1, NGFI-B, GR, and HDAC2 may prevent promoter recruitment of Pol II by steric hindrance or through protein interactions.
Upon ligand (Dex) binding (Fig. 8C), GR is recruited to the POMC promoter (Fig. 4E) together with HDAC2 (Fig. 5D), and recruitment of both proteins to NGFI-B/Nurr1- containing complexes requires Brg1 (Figs. 4G, 5E–G). Consistent with a model of trans-repression in which GR is recruited to the promoter in part through interactions with NGFI-B (Philips et al. 1997b; Martens et al. 2005), promoter occupancy by NGFI-B is not modified in the presence of GR (Fig. 4F), and GR promoter recruitment is even slightly higher in CRH/Dex-treated cells compared with Dex alone (Fig. 4E). It is noteworthy that GR recruitment is not associated with a decrease of Pol II occupancy at the promoter, at least in the presence of Dex alone (Fig. 6B). Thus, the constant amount of Pol II associated with the promoter, presumably in a preinitiation complex, in the face of less Poll II reaching exon 3 of the gene (Fig. 6B), indicates a relative block of Pol II at the promoter in the presence of GR and the trans-repression machinery (Fig. 8C). This blockade of Pol II clearance is associated with a repressed transcription initiation rate (Fig. 6A) and, thus, this appears to be the major consequence of GR action. The hallmark of the action of Dex/GR is recruitment of HDAC2 to the promoter and the resulting decrease in histone acetylation. Promoter recruitment of HDAC activity was shown to be sufficient for inhibition of transcription initiation (Huang and Kadonaga 2001). Mediation of the repressor effect through HDAC2 recruitment and chromatin remodeling is consistent with the dependence on Brg1 ATPase activity (Fig. 3G). In contrast, a model of repression exerted by the trans-repression complex sequestering Pol II through protein interactions would be less likely to depend on the Brg1 ATPase activity, although such interactions may contribute directly or by steric hindrance to limit Pol II recruitment in CRH + Dex conditions (Fig. 8D) compared with CRH stimulation (Fig. 8B).
Gc resistance in Cushing disease
Hormone resistance is associated with many disease conditions. In some relatively rare cases of steroid hormone resistance, inherited or sporadic mutations of the gene encoding the cognate NR account for hormone insensitivity. More frequently, other and usually poorly defined mechanisms appear to be implicated. These mechanisms may include activation of cross-talk pathways that can alter the activity of NRs through phosphorylation of the receptors themselves or of coregula- tory molecules. In the case of Cushing disease, the development of Gc resistance appears to be an early, if not primary, event for pathogenesis. Indeed, ACTH-producing pituitary tumors are always more resistant to Gc than normal pituitary cells. Only rarely is the Gc resistance due to GR mutations (Lamberts 2002); accordingly, all tumors in our series had normal GR expression (data not shown).
Although there are numerous examples of the implication of Brg1 in NR-dependent transcription (Kadam and Emerson 2003), Brg1 has not been implicated previously in hormone resistance. The mislocalization of Brg1 to the cytoplasm may result from aberrant signaling. Alternatively, somatic mutations in Brg1 or HDAC2 may be responsible for altered targeting or loss of expression or function. The corticotroph adenomas being such small and infrequent tumors, it is very difficult to have enough material for biochemical investigation; all tumor sections used in the present study were from archival materials and, in some cases, only a few sections were still available. Thus, it is even difficult to undertake extensive genetic analysis of this material. The finding that human tumors were deficient in either Brg1 or HDAC2, but not both, suggests that both proteins are independently essential for Gc repression and that other HDACs cannot compensate for HDAC2 deficiency. This is consistent with our observations in mouse AtT-20 cells where Brg1 and HDAC activity are required (Figs. 1, 2, 5). The situation may be different in dogs since HDAC2 loss was observed only in association with Brg1 deficiency in this species.
Tumorigenic mechanisms in Cushing disease
The growth of AtT-20 cells is normally inhibited by Gc, and relief of this inhibition accelerates their growth (Svec 1984). Thus, derepressed corticotroph growth in the absence of Brg1 may create a hyperplastic state and predispose these cells to tumorigenesis. Loss of Brg1 expression has been observed in other tumors, including lung (Reisman et al. 2003) and oral (Gunduz et al. 2005) cancers, but it is not known whether this is associated with changes in hormone sensitivity.
Expression of HDACs was found to be either increased or decreased in different cancers. Like for Brg1, no relation with hormone sensitivity has been investigated. It is, however, noteworthy that HDAC inhibitors appear to have beneficial effects on progression of different tumors (Drummond et al. 2005). The mechanism(s) for this anti- cancer effect is not currently understood and may be multiple, as most HDAC inhibitors lack specificity and lead to genome-wide increases in histone acetylation and wide-scale changes in gene expression. Be that as it may, the possibility that long-term administration of such compounds may impair pituitary Gc feedback and pre- dispose to Cushing disease may have to be considered.
Materials and methods
Cell culture, transfection, and plasmids
CV-1, C33A, SW13, and MEF TKO cells were cultured in DMEM supplemented with 10% fetal calf serum and antibiotics. A-427 cells were cultured in MEM (Gibco) supplemented with 10% FBS, 1 mM L-glutamine, and 1 mM sodium pyruvate. CV-1, C33A, and MEF TKO were transfected as described (Batsche et al. 2005b; Martens et al. 2005). A-427 and SW13 were transfected using 500 ng of reporter plasmid and 0–250 ng of expression vector up to a total of 10 μg per quadriplicate. All cells were stimulated in duplicate for 24 h with dexamethasone 10−7M or vehicle with or without HDAC inhibitors (Sigma) as indicated. NurRE reporter and GRE reporters were described (Martens et al. 2005). All transfection data are shown in the figures as means ± SEM of three experiments, each performed in duplicate.
shRNA
AtT-20 cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) and antibiotics. The cells were transfected using 750 ng of POMC reporter plasmid and treated in duplicate for 24 h with dexamethasone 10−7 M. Oligonucleo- tides (63mer) directed against Brg1 or a random sequence were cloned in the pTER expression vector (van de Wetering et al. 2003). Cotransfection of 1 μg of CMV-eGFP (Clonetech) with 5 μg of shRNA plasmid was done using 3 × 106 cells and Lipofectamine (Invitrogen). Cells were then sorted by FACS, and 40,000 GFP-positive cells were loaded in each well. For luciferase assay, 0–250 ng of small hairpin plasmids were transfected in AtT-20 cells using Lipofectamine (Invitrogen) and 500 ng of POMC reporter plasmid. After SDS-PAGE, Western blots were revealed with antibodies against Brg1 (SNF2β, Chemicon) and GAPDH (Abcam).
Coimmunoprecipitation assays and Western blots
C33A cells (10-cm plate) were transfected with 2–20 μg of expression plasmids for tagged or native proteins and harvested 48 h later. Cells were harvested in cold PBS and extracted for 30 min at 4°C in TNEN250 with 1 mM PMSF and protease inhibitors. After centrifugation, supernatants were immunoprecipitated for2hat4°C with αFlag M2 (Sigma), and isotype-matched nonimmune IgG (Sigma) as control. Immunoprecipitates were washed three times with TNEN125. After SDS-PAGE, Western blots were revealed with antibodies against Flag M2, HA (SC- 805, Santa Cruz Biotechnology), Brg1 (H-88, Santa Cruz Bio- technology), Brm (Muchardt and Yaniv 1993), NGFI-B (Maira et al. 2003b), Nurr1 (N83220, BD Bioscience), and GR (P-20, Santa Cruz Biotechnology). For coimmunoprecipitations in AtT-20 cells, these cells were double-transfected with 50 nM Brg1 siRNA (ON-TARGETplus SMARTpool, Dharmacon) or random siRNA using lipofectamine. Briefly, the cells were transfected for 18 h, trypsinized, and retransfected after 8 h for 48 h. Cells were then stimulated for 30 min with CRH and Dex 10−7 M before harvest and coimmunoprecipitations performed as above.
Pull-down assays
MBP fusion proteins were produced and [35S]-labeled BAF155, Brm, and Brg1 were synthesized in vitro as described (Batsche et al. 2005a). Labeled proteins were incubated with immobilized MBP-lacZ, MBP–NGFI-B, MBP-Pitx1, or MBP-Tpit in 350 μL of TNEN50 (50 mM Tris at pH 7.5, 5 mM EDTA, 50 mM NaCl, 0.1% NP-40) with 1 mM PMSF and 2% BSA for2hat4°C. Beads were washed at room temperature twice in TNEN250 and twice in TNEN125. Bound proteins were resolved on SDS-PAGE, stained with Coomassie blue to ensure that similar amounts of fusion proteins were recovered, and then autoradiographed.
ChIP, QPCR, and nuclear run-on assays
AtT-20 cells were treated or not for 30 min with 10−7 M CRH and/or 10−7M dexamethasone, and were then prepared for ChIP as described (Batsche et al. 2005a). Supernatants corresponding to 107 cells were subjected to overnight immunoprecipitation at 4°C with antibodies against Brg1 (Wang et al. 1996; Kadam and Emerson 2003), GR (M-20, Santa Cruz Biotechnology), NGFI-B (Maira et al. 2003b), RNA Pol II (N-20, Santa Cruz Biotechnology), phospho-Ser2 CTD (MMS-129R, Covance), phospho-Ser5 CTD (MMS-134R, Covance), HDAC1 (06-720, Upstate Biotechnology), HDAC2 (07-222, Upstate Biotechnology), HDAC3 (06- 890, Upstate Biotechnology), and acetyl-histone H4 (06-866, Upstate Biotechnology). Immunoprecipitates were collected with protein A/G agarose beads (Santa Cruz Biotechnology) saturated with tRNA. QPCR was as previously described (Batsche et al. 2005b). For quantitative RT–QPCR in AtT-20 cells, mRNA was prepared using RNAeasy (Qiagen), and quantitative real-time PCR was done using a OneStep RT–PCR Kit (Qiagen). The nuclear run-on experiments were performed as described (Gagner and Drouin 1985).
Immunofluorescence
For αACTH/Brg1 colocalization, anti-rabbit-Brg1 1:100 (H-88,Santa Cruz Biotechnology) was incubated overnight, antimouse-POMC 1:200 (Cortex Biochemicals) and anti-rabbit-biotinylated 1:200 (Vector Laboratories) were added next and, finally, anti mouse-fluorescein 1:200 (ImmunoPure Antibody) and avidin-rodhamine 1:200 (Vector Laboratories).
Clinical samples and immunohistochemestry
Corticotroph adenomas were selected from archival materials based on the extent of clinical investigation prior to surgery, the availability of data from a Dex suppression (Liddle) test, and the quantity and quality of available tissue sections. Immunohisto- chemistry was performed as described (Lanctôt et al. 1997). Antibodies were used as follows: rabbit anti-Tpit 1:25 (Lamolet et al. 2001), rabbit anti-Brg1 1:100 (H-88, Santa Cruz Biotechnology), and rabbit anti-GR 1:250 (M-20, Santa Cruz Biotechnology). All secondary antibodies were used 1:150 (Vector Laboratories). HDAC2 was detected with a rabbit anti-HDAC2 1:10 (Upstate Biotechnology, no. 07-222), and the signal was amplified with the TSA biotin system (PerkinElmer Life Sciences, Inc.).
Acknowledgments
We are grateful to many colleagues from the laboratory for comments on this work and manuscript and, in particular, we thank Eric Batsché, François Robert, Luc Gaudreau, and Jacques Côté for helpful comments. We thank Jean-Pierre Gagner for sharing his experience in nuclear run-on assays. We thank Moshe Yaniv and Christian Muchardt for Brm expression plasmids and antibodies, Weidang Wang for Brg1 expression plasmid and antibody, and Julien Sage for MEF TKO cells. We thank Hans Clevers for the pTER expression plasmids The expert secretarial assistance of Lise Laroche was greatly appreciated. This grant was supported by a fellowship from Fonds des chercheurs et aide à la recherche-Fonds de la recherché en santé du Québec (FCARFRSQ) and Canadian Institutes of Health Research (CIHR) to S.B., and by research grants to J.D. from the Canadian Institutes of Health Research (CIHR) and National Cancer Institute of Canada (NCIC).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1444606.
References
- Arnaldi, G., Angeli, A., Atkinson, A.B., Bertagna, X., Cavagnini, F., Chrousos, G.P., Fava, G.A., Findling, J.W., Gaillard, R.C., Grossman, A.B., et al. Diagnosis and complications of Cushing's syndrome: A consensus statement. J. Clin. Endocrinol. Metab. 2003;88:5593–5602. doi: 10.1210/jc.2003-030871. [DOI] [PubMed] [Google Scholar]
- Batsche, E., Desroches, J., Bilodeau, S., Gauthier, Y., Drouin, J. Rb enhances p160/SRC coactivator-depen- dent activity of nuclear receptors and hormone responsiveness. J. Biol. Chem. 2005a;280:19746–19756. doi: 10.1074/jbc.M413428200. [DOI] [PubMed] [Google Scholar]
- Batsche, E., Moschopoulos, P., Desroches, J., Bilodeau, S., Drouin, J. Retinoblastoma and the related pocket protein p107 act as coactivators of NeuroD1 to enhance gene transcription. J. Biol. Chem. 2005b;280:16088–16095. doi: 10.1074/jbc.M413427200. [DOI] [PubMed] [Google Scholar]
- Cho, E.J., Kobor, M.S., Kim, M., Greenblatt, J., Buratowski, S. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes & Dev. 2001;15:3319–3329. doi: 10.1101/gad.935901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey, L.L., Weirich, C.S., Benjamin, I.J., Kingston, R.E. Localized recruitment of a chromatin-remodeling activity by an activator in vivo drives transcriptional elongation. Genes & Dev. 2003;17:1392–1401. doi: 10.1101/gad.1071803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushing, H. The basophil adenomas of the pituitary body and their clinical manifestations (pituitary basophilism) Bull. Johns Hopkins Hosp. 1932;50:137–195. [Google Scholar]
- De Bosscher, K., Vanden Berghe, W., Haegeman, G. The interplay between the glucocorticoid receptor and nuclear factor-κB or activator protein-1: Molecular mechanisms for gene repression. Endocr. Rev. 2003;24:488–522. doi: 10.1210/er.2002-0006. [DOI] [PubMed] [Google Scholar]
- Drummond, D.C., Noble, C.O., Kirpotin, D.B., Guo, Z.X., Scott, G.K., Benz, C.C. Clinical development of histone deacetylase inhibitors as anticancer agents. Annu. Rev. Pharmacol. Toxicol. 2005;45:495–528. doi: 10.1146/annurev.pharmtox.45.120403.095825. [DOI] [PubMed] [Google Scholar]
- Fryer, C.J., Archer, T.K. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature. 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- Gagner, J.-P., Drouin, J. Opposite regulation of proopiomelanocortin gene transcription by glucocorticoids and CRH. Mol. Cell. Endocrinol. 1985;40:25–32. doi: 10.1016/0303-7207(85)90154-6. [DOI] [PubMed] [Google Scholar]
- Gagner, J.-P., Drouin, J. Tissue-specific regulation of pituitary proopiomelanocortin gene transcription by corticotropin-releasing hormone, 3′, 5′-cyclic adenosine monophosphate, and glucocorticoids. Mol. Endocrinol. 1987;1:677–682. doi: 10.1210/mend-1-10-677. [DOI] [PubMed] [Google Scholar]
- Gunduz, E., Gunduz, M., Ouchida, M., Nagatsuka, H., Beder, L., Tsujigiwa, H., Fukushima, K., Nishizaki, K., Shimizu, K., Nagai, N. Genetic and epigenetic alterations of BRG1 promote oral cancer development. Int. J. Oncol. 2005;26:201–210. [PubMed] [Google Scholar]
- Hayashi, R., Wada, H., Ito, K., Adcock, I.M. Effects of glucocorticoids on gene transcription. Eur. J. Pharmacol. 2004;500:51–62. doi: 10.1016/j.ejphar.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Heck, S., Kullmann, M., Gast, A., Ponta, H., Rahmsdorf, H.J., Herrlich, P., Cato, A.C. A distinct modulating domain in glucocorticoid receptor monomers in the repression of activity of the transcription factor AP-1. EMBO J. 1994;13:4087–4095. doi: 10.1002/j.1460-2075.1994.tb06726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmberg, A., Auphan, N., Caelles, C., Karin, M. Glucocorticoid-induced apoptosis of human leukemic cells is caused by the repressive function of the glucocorticoid receptor. EMBO J. 1995;14:452–460. doi: 10.1002/j.1460-2075.1995.tb07021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X., Kadonaga, J.T. Biochemical analysis of transcriptional repression by Drosophila histone deacetylase 1. J. Biol. Chem. 2001;276:12497–12500. doi: 10.1074/jbc.C100034200. [DOI] [PubMed] [Google Scholar]
- Jonat, C., Rahmsdorf, H.J., Park, K.-K., Cato, A.C.B., Gebel, S., Ponta, H., Herrlich, P. Antitumor promotion and antiinflammation: Down-modulation of AP-1 (fos/jun) activity by glucocorticoid hormone. Cell. 1990;62:1189–1204. doi: 10.1016/0092-8674(90)90395-u. [DOI] [PubMed] [Google Scholar]
- Kadam, S., Emerson, B.M. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol. Cell. 2003;11:377–389. doi: 10.1016/s1097-2765(03)00034-0. [DOI] [PubMed] [Google Scholar]
- König, H., Ponta, H., Rahmsdorf, H.J., Herrlich, P. Interference between pathway-specific transcription factors: Glucocorticoids antagonize phorbol ester-induced AP-1 activity without altering AP-1 site occupation in vivo. EMBO J. 1992;11:2241–2246. doi: 10.1002/j.1460-2075.1992.tb05283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamberts, S.W.J. Glucocorticoid receptors and Cushing's disease. Mol. Cell. Endocrinol. 2002;197:69–72. doi: 10.1016/s0303-7207(02)00280-0. [DOI] [PubMed] [Google Scholar]
- Lamolet, B., Pulichino, A.M., Lamonerie, T., Gauthier, Y., Brue, T., Enjalbert, A., Drouin, J. A pituitary cell-restricted T-box factor, Tpit, activates POMC transcription in cooperation with Pitx homeoproteins. Cell. 2001;104:849–859. doi: 10.1016/s0092-8674(01)00282-3. [DOI] [PubMed] [Google Scholar]
- Lamonerie, T., Tremblay, J.J., Lanctôt, C., Therrien, M., Gauthier, Y., Drouin, J. PTX1, a bicoid-related homeobox transcription factor involved in transcription of pro-opiomelanocortin (POMC) gene. Genes & Dev. 1996;10:1284–1295. doi: 10.1101/gad.10.10.1284. [DOI] [PubMed] [Google Scholar]
- Lanctôt, C., Lamolet, B., Drouin, J. The bicoid-related homeoprotein Ptx1 defines the most anterior domain of theembryo and differentiates posterior from anterior lateral mesoderm. Development. 1997;124:2807–2817. doi: 10.1242/dev.124.14.2807. [DOI] [PubMed] [Google Scholar]
- Luecke, H.F., Yamamoto, K.R. The glucocorticoid receptor blocks P-TEFb recruitment by NFκB to effect promoter-specific transcriptional repression. Genes & Dev. 2005;19:1116–1127. doi: 10.1101/gad.1297105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maira, M.H., Martens, C., Philips, A., Drouin, J. Heterodimerization between members of the Nur subfamily of orphan nuclear receptors as a novel mechanism for gene activation. Mol. Cell. Biol. 1999;19:7549–7557. doi: 10.1128/mcb.19.11.7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maira, M.H., Couture, C., Le Martelot, G., Pulichino, A.M., Bilodeau, S., Drouin, J. The T-box factor Tpit recruits SRC/p160 coactivators and mediates hormone action. J. Biol. Chem. 2003a;278:46523–46532. doi: 10.1074/jbc.M305626200. [DOI] [PubMed] [Google Scholar]
- Maira, M.H., Martens, C., Batsche, E., Gauthier, Y., Drouin, J. Dimer-specific potentiation of NGFI-B (Nur77) transcriptional activity by the protein kinase A pathway and AF-1-dependent coactivator recruitment. Mol. Cell. Biol. 2003b;23:763–776. doi: 10.1128/MCB.23.3.763-776.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens, C., Bilodeau, S., Maira, M., Gauthier, Y., Drouin, J. Protein:protein interactions and transcriptional antagonism between the subfamily of NGFI-B/Nur77 orphan nuclear receptors and glucocorticoid receptor. Mol. Endocrinol. 2005;19:885–897. doi: 10.1210/me.2004-0333. [DOI] [PubMed] [Google Scholar]
- McKenna, N.J., O'Malley, B.W. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- Meij, B., Voorhout, G., Rijnberk, A. Progress in transsphenoidal hypophysectomy for treatment of pituitary-dependent hyperadrenocorticism in dogs and cats. Mol. Cell. Endocrinol. 2002;197:89–96. doi: 10.1016/s0303-7207(02)00283-6. [DOI] [PubMed] [Google Scholar]
- Metivier, R., Stark, A., Flouriot, G., Hubner, M.R., Brand, H., Penot, G., Manu, D., Denger, S., Reid, G., Kos, M., et al. A dynamic structural model for estrogen receptor-α activation by ligands, emphasizing the role of interactions between distant A and E domains. Mol. Cell. 2002;10:1019–1032. doi: 10.1016/s1097-2765(02)00746-3. [DOI] [PubMed] [Google Scholar]
- Muchardt, C., Yaniv, M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narlikar, G.J., Fan, H.Y., Kingston, R.E. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- Nissen, R.M., Yamamoto, K.R. The glucocorticoid receptor inhibits NFκB by interfering with serine-2 phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes & Dev. 2000;14:2314–2329. doi: 10.1101/gad.827900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal, S., Yun, R., Datta, A., Lacomis, L., Erdjument-Bromage, H., Kumar, J., Tempst, P., Sif, S. mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol. Cell Biol. 2003;23:7475–7487. doi: 10.1128/MCB.23.21.7475-7487.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual, G., Fong, A.L., Ogawa, S., Gamliel, A., Li, A.C., Perissi, V., Rose, D.W., Willson, T.M., Rosenfeld, M.G., Glass, C.K. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-γ . Nature. 2005;437:759–763. doi: 10.1038/nature03988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips, A., Lesage, S., Gingras, R., Maira, M.H., Gauthier, Y., Hugo, P., Drouin, J. Novel dimeric Nur77 signaling mechanisms in endocrine and lymphoid cells. Mol. Cell. Biol. 1997a;17:5946–5951. doi: 10.1128/mcb.17.10.5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips, A., Maira, M.H., Mullick, A., Chamberland, M., Lesage, S., Hugo, P., Drouin, J. Antagonism between Nur77 and glucocorticoid receptor for control of transcription. Mol. Cell. Biol. 1997b;17:5952–5959. doi: 10.1128/mcb.17.10.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin, G., Lebel, M., Chamberland, M., Paradis, F.W., Drouin, J. Specific protein:protein interaction between basic helix–loop–helix transcription factors and homeoproteins of the Pitx family. Mol. Cell. Biol. 2000;20:4826–4837. doi: 10.1128/mcb.20.13.4826-4837.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulichino, A.M., Vallette-Kasic, S., Tsai, J.P.Y., Couture, C., Gauthier, Y., Drouin, J. Tpit determines alternate fates during pituitary cell differentiation. Genes & Dev. 2003;17:738–747. doi: 10.1101/gad.1065703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt, H.M., Kaestner, K.H., Tuckermann, J., Kretz, O., Wessely, O., Bock, R., Gass, P., Schmid, W., Herrlich, P., Angel, P., et al. DNA binding of the glucocorticoid receptor is not essential for survival. Cell. 1998;93:531–541. doi: 10.1016/s0092-8674(00)81183-6. [DOI] [PubMed] [Google Scholar]
- Reisman, D.N., Sciarrotta, J., Wang, W.D., Funkhouser, W.K., Weissman, B.E. Loss of BRG1/BRM in human lung cancer cell lines and primary lung cancers: Correlation with poor prognosis. Cancer Res. 2003;63:560–566. [PubMed] [Google Scholar]
- Sage, J., Mulligan, G.J., Attardi, L.D., Miller, A., Chen, S., Williams, B., Theodorou, E., Jacks, T. Targeted disruption of the three Rb-related genes leads to loss of G(1) control and immortalization. Genes & Dev. 2000;14:3037–3050. doi: 10.1101/gad.843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüle, R., Rangarajan, P., Kliewer, S., Ransone, L.J., Bolado, J., Yang, N., Verma, I.M., Evans, R.M. Functional antagonism between oncoprotein c-jun and the glucocorticoid receptor. Cell. 1990;62:1217–1226. doi: 10.1016/0092-8674(90)90397-w. [DOI] [PubMed] [Google Scholar]
- Seo, S., Richardson, G.A., Kroll, K.L. The SWI/SNF chromatin remodeling protein Brg1 is required for vertebrate neurogenesis and mediates transactivation of Ngn and NeuroD. Development. 2005;132:105–115. doi: 10.1242/dev.01548. [DOI] [PubMed] [Google Scholar]
- Svec, F. Glucocorticoids inhibit the growth of AtT-20 mouse pituitary tumor cells. Mol. Cell. Endocrinol. 1984;35:33–40. doi: 10.1016/0303-7207(84)90027-3. [DOI] [PubMed] [Google Scholar]
- Vallette-Kasic, S., Figarella-Branger, D., Grino, M., Pulichino, A.M., Dufour, H., Grisoli, F., Enjalbert, A., Drouin, J., Brue, T. Differential regulation of proopiomelanocortin and pituitary-restricted transcription factor (TPIT), a new marker of normal and adenomatous human corticotrophs. J. Clin. Endocrinol. Metab. 2003;88:3050–3056. doi: 10.1210/jc.2002-021934. [DOI] [PubMed] [Google Scholar]
- van de Wetering, M., Oving, I., Muncan, V., Pon Fong, M.T., Brantjes, H., van Leenen, D., Holstege, F.C., Brummelkamp, T.R., Agami, R., Clevers, H. Specific inhibition of gene expression using a stably integrated, inducible small- interfering-RNA vector. EMBO Rep. 2003;4:609–615. doi: 10.1038/sj.embor.embor865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W., Xue, Y., Zhou, S., Kuo, A., Cairns, B.R., Crabtree, G.R. Diversity and specialization of mammalian SWI/ SNF complexes. Genes & Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- Wintermantel, T.M., Berger, S., Greiner, E.F., Schutz, G. Genetic dissection of corticosteroid receptor function in mice. Horm. Metab. Res. 2004;36:387–391. doi: 10.1055/s-2004-814567. [DOI] [PubMed] [Google Scholar]
- Wong, A.K.C., Shanahan, F., Chen, Y., Lian, L.B., Ha, P., Hendricks, K., Ghaffari, S., Iliev, D., Penn, B., Woodland, A.M., et al. BRG1, a component of the SWI–SNF complex, is mutated in multiple human tumor cell lines. Cancer Res. 2000;60:6171. [PubMed] [Google Scholar]
- Yang-Yen, H.F., Chambard, J.C., Sun, Y.L., Smeal, T., Schmidt, T.J., Drouin, J., Karin, M. Transcriptional interference between c-jun and the glucocorticoid receptor: Mutual inhibition of DNA binding due to direct protein–protein interaction. Cell. 1990;62:1205–1215. doi: 10.1016/0092-8674(90)90396-v. [DOI] [PubMed] [Google Scholar]