Abstract
The cell wall lipids in Mycobacterium tuberculosis are probably involved in pathogenesis. The largest open reading frame in the genome of M. tuberculosis H37Rv, pks12, is unique in that it encodes two sets of domains needed to produce fatty acids. A pks12-disrupted mutant was produced, and disruption was confirmed by both PCR analysis and Southern blotting. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis showed that a 430-kDa protein band present in the wild type was missing in the mutant. Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MS) and liquid chromatography (LC)-MS analysis of tryptic peptides showed that 54 peptides distributed throughout this protein matched the pks12-encoded sequence. Biochemical analysis using [1-14C]propionate as the radiotracer showed that the pks12 mutant was deficient in the synthesis of dimycocerosyl phthiocerol (DIM). SDS-PAGE, immunoblot analysis of proteins, and analysis of fatty acids showed that the mutant can produce mycocerosic acids. Thus, the pks12 gene is probably involved in the synthesis of phthiocerol, the diol required for DIM synthesis. Growth of the pks12 mutant was attenuated in mouse alveolar macrophage cell line MH-S, and the virulence of the mutant in vivo was highly attenuated in a murine model. Thus, pks12 probably participates in DIM production and its expression is involved in pathogenesis.
Tuberculosis accounts for more than one-fourth of all preventable adult deaths in the world (Global tuberculosis control, World Health Organization report, http://www.who.int/gtb/publications/glovrep01/, 2001). Mycobacterium tuberculosis, the causative agent, can evade the defense mechanisms of the host and grow in macrophages that normally phagocytose and destroy most other bacterial pathogens. It has been widely recognized that the unusually complex cell wall of the organism plays a major role in the exceptional ability of the pathogen to be a successful pathogen (3, 8). The cell wall is rich (50 to 60%) in unusual lipids that are highly hydrophobic, and some of the cell wall components may also help the pathogen to enter macrophages (24) and suppress the defense mechanisms of the host (18, 22). Therefore, it is not surprising that cell wall synthesis has proved to be the target of some of the most successful antimycobacterial drugs. However, the pathogen has acquired resistance to several frontline antimycobacterial drugs, and the multidrug-resistant M. tuberculosis has been classified as a class C organism by the Centers for Disease Control and Prevention. The potential for such multidrug-resistant pathogens to cause major public health problems in highly populated areas makes identification of new targets for antimycobacterial therapy a critical need.
The presence of very-long-chain fatty acids with multiple methyl branches at alternate positions near the carboxyl end is a unique feature of mycobacterial cell wall lipids (17). Derivatives of such acids are virulence factors. For example, it was suggested that dimycocerosyl phthiocerol (DIM), composed of mycocerosic acids (2,4,6,8-tetramethyl C32 fatty acid and homologues) esterified to the long-chain diol phthiocerol, is a virulence factor because mutants that lack this compound were attenuated in human monocytes and in the murine lung (4, 7, 25). We cloned the mycocerosic acid synthase (MAS) gene, mas (19), and proved it to be the one responsible for the production of mycocerosic acids by gene disruption (1). The mycobacterial genome contains many polyketide synthase (PKS) genes (pks) (6), including seven mas-like (msl) genes (25). These genes usually encode one full complement of catalytic domains required to catalyze the synthesis of a fully saturated fatty acid (one module). These include the acyl transferase (AT) that transfers the reactants to the synthase ketoacyl synthase (KS), which catalyzes the condensation of the reactants to form the carbon-carbon bond generating the ketoacyl derivative ketoreductase, which reduces the carbonyl to the secondary hydroxyl dehydratase, which eliminates water to generate the unsaturated acyl group and enoyl reductase, which reduces the olefin to a fully saturated moiety. All of these reactions happen while the growing chain is still attached to the phosphopantetheine of the acyl carrier protein (ACP) domain of the synthase (17, 19). One of the msls (msl6, pks12), the largest open reading frame (ORF) in the mycobacterial genome (6), encodes two modules that can catalyze the synthesis of a saturated acid. Such large ORFs encoding multiple sets of modules have been previously found only in antibiotic-producing organisms (15, 16, 20).
Whether this gene is expressed in M. tuberculosis and, if it is, what the nature of the product and its biological function are and whether gene expression contributes to virulence remain unknown. In this paper we report that this largest mycobacterial ORF is expressed in M. tuberculosis and we identify the protein product by showing that the amino acid sequences of 54 peptides distributed throughout the 430-kDa protein in M. tuberculosis H37Rv matches with the sequences predicted from the nucleotide sequence of the gene. We also report disruption of this gene in M. tuberculosis and show that the msl6 mutant does not produce the 430-kDa protein. The msl6-disrupted mutant is defective in DIM synthesis and is highly attenuated.
MATERIALS AND METHODS
General DNA-handling techniques.
Molecular cloning and restriction enzyme digestions were performed by standard procedures (23). Cloning vectors used were pBluescript KS(−) (Stratagene, La Jolla, Calif.) and pYUB572 and phAE87 (kindly provided by Stoyan Bardarov, Albert Einstein College of Medicine, New York, N.Y.). Molecular cloning and restriction endonuclease digestions were performed by standard techniques (23).
Bacterial strains and culture conditions.
Escherichia coli DH5α (Life Technology) and HB101 were used as host strains for cloning experiments and were grown on Luria-Bertani (LB) broth or agar containing 100 μg of ampicillin (Sigma Chemical Co.)/ml or 150 μg of hygromycin B (Calbiochem)/ml. Mycobacterium smegmatis mc2155 was grown in liquid LB medium with 0.05% Tween 80 for competent-cell preparation and in Middlebrook 7H9 broth (Difco) with 0.05% Tween 80 for transduction. M. tuberculosis H37Rv (ATCC 25618) was grown in Middlebrook 7H9 broth supplemented with 10% oleic acid-albumin-dextrose (OADC) enrichment (7H9-OADC; BBL Microbiology Media) plus 0.05% Tween 80 in roller bottles or on Middlebrook 7H10-OADC agar plates. When required, hygromycin B was used at a concentration of 50 μg/ml.
Construction of mutant strains of M. tuberculosis.
The M. tuberculosis msl6 (pks12, Rv2048c) mutant was constructed by allelic exchange using the previously described specialized transducing phage system (9) in a manner similar to that we used before (25). Part of the msl6 gene (bp 4101 to 8201 of the coding sequence; bp 49385 to 53485 of the M. tuberculosis genome) was amplified from genomic DNA with sense primer 5′-GGAAGCTTCGAAAATCTGCGGCTCGA-3′ (A) and antisense primer 5′-GGAAGCTTGACCGCAGCGATGTCAAC-3′ (B), introducing HindIII sites at the 5′ and 3′ ends of the sequence. The 4,100-bp PCR product was cloned into a HindIII-digested pUC19 vector, and a 2,193-bp PmlI fragment was replaced by a hygromycin resistance gene cassette (hyg). The resulting disrupted msl6 gene and flanking regions were cloned into the cos vector pYUB572. The resulting recombinant cosmid was digested with PacI and ligated into PacI-digested phAE87 DNA. The ligation mixture was packaged in the lambda in vitro packaging mixture (Gigapack III; Stratagene), transduced into E. coli HB101, and plated on LB plates with hygromycin. DNA from several phasmid clones was isolated, confirmed by restriction digestion, and electroporated into M. smegmatis strain mc2155, and the strain was plated for plaques at 30°C. Individual plaques were tested for thermosensitivity, amplified, and used to infect M. tuberculosis H37Rv. Colonies grown at 37°C on Middlebrook 7H10-OADC agar containing hygromycin (50 μg/ml) were screened by PCR for disruption of the msl6 gene. PCR amplification, performed directly on cell lysate obtained by boiling the cells by standard protocols (23), was performed with Platinum Taq polymerase (Life Technology) and sense primer 5′-CGCACTGCGAGCCCATGCGGT-3′ (E) and antisense primer 5′-AAGCCTTCTACCGGCTCGGCG-3′ (F). Positive clones were verified by Southern blot analysis and by further PCR analysis using two other sets of primers, each containing a hygromycin primer and a primer in the mycobacterial genome directly outside the sequences used to make the disruption construct: sense primer 5′-ACCGACCATGAATCCGGGGTGCTG-3′ (C) and antisense primer 5′-TGGACCTCGACGACCTGCAGGCAT-3′ (H1) for amplification of the 5′-flanking region and 5′-GACGTCGCCAGTAGGCCGCTGATC-3′ (D) and 5′-GGAACTGGCGCAGTTCCTCTGGGG-3′ (H2) for amplification of the 3′-flanking region. Primer pair E-F was used in reverse transcription-PCR (RT-PCR) analysis.
Genomic DNA isolation and Southern blotting.
M. tuberculosis genomic DNA was isolated by the GTC method using guanidine thiocyanate, Tris-HCl, and sarcosyl solution (17a). DNA samples were digested with pstI, transferred to nylon membranes (Nytran Plus; Schleicher & Schuell, Keene, N.H.), and hybridized with labeled probes generated with [α-32P]dCTP by using the random-prime labeling system Rediprime II (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom).
Analysis of the msl6 gene product.
Cells of M. tuberculosis H37Rv and its msl6-disrupted mutant, collected by centrifugation, were suspended in 0.1 M potassium phosphate buffer, pH 7.2, containing 1 mM dithioerythritol, 1 mM EDTA, 10% glycerol, and 1 mM phenylmethylsulfonyl fluoride and disrupted by being shaken six times, 20 s each time with a 2-min cooling on ice, with a Fast Prep F120 instrument (Q. BIOgene). The extract was centrifuged in a Microfuge, and the supernatant was filter sterilized (0.2 μm-pore-size filter). The extract was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 4% separating gel and stained with Coomassie blue.
Immunoblot analysis was done with anti-MAS antibodies as described before (13). For protein digestion, the 430-kDa band that was present in the wild type but not in the mutant was cut from the gel as closely as possible, divided into a number of smaller pieces, and washed and destained in two aliquots of 50% ethanol-5% acetic acid for 1 h each. The gel pieces were washed, reduced in dithiothreitol, and alkylated in iodoacetamide. The gel pieces were then washed in 0.1 M ammonium bicarbonate, dehydrated in 200 μl acetonitrile, and dried in a Speed-vac. The protease was driven into the gel pieces by rehydrating them in 50 μl of 50 mM ammonium bicarbonate containing 1 mg of trypsin on ice for 10 min. Any excess trypsin solution was removed, and 20 μl of 50 mM ammonium bicarbonate was added. The sample was incubated overnight at room temperature. The peptides were extracted from the polyacrylamide in two aliquots of 30 μl of 50% acetonitrile-5% formic acid, and the extracts were combined and evaporated to <20 μl for liquid chromatography-mass spectrometry (LC-MS) analysis and analyzed on an LC-MS system (Finnigan LCQ-Deca ion trap mass spectrometer system with a Protana microelectrospray ion source interfaced to a self-packed 10-cm by 75-μm [inside diameter] Phenomenex Jupiter C18 reverse-phase capillary chromatography column). One microliter of the peptides was injected, and the peptides were eluted from the column by an acetonitrile-0.05 M acetic acid gradient at a flow rate of 0.2 μl/min. The microelectrospray ion source was operated at 2.5 kV. The digest was analyzed by using the data-dependent multitask capability of the instrument, acquiring full-scan mass spectra to determine peptide molecular weights and product ion spectra to determine amino acid sequences in successive instrument scans. The data were analyzed by using the National Center for Biotechnology Information nonredundant database with the search program TurboSequest. All matching spectra were verified by manual interpretation. The interpretation process was also aided by additional searches using the programs Mascot and Fasta, which were carried out as needed.
The peptides were also analyzed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) MS. For these analyses, the digest was desalted with a Millipore C18 ZipTip column that was eluted in the MALDI matrix (5 mg of α-cyano-4-hydroxycinnamic acid/ml in 50% acetonitrile-2.5% trifluoroacetic acid). Aliquots of the extract (1.5 μl) were spotted on a sample plate and analyzed with a Micromass Tof Spec 2E MS system. The spectra were internally calibrated with trypsin autolysis peptides, giving mass accuracies that were generally better than 25 ppm.
Test for expression of mas, msl, and pps genes.
RNA was isolated from the M. tuberculosis cells grown to mid-exponential phase. Chilled cells isolated by centrifugation were resuspended in RNeasy lysis buffer (Qiagen), transferred to a 2-ml tube containing ceramic and silica beads (FastRNA Blue), and disrupted with a FastPrep F120 instrument (Q. BIOgene). The extract collected by centrifugation was used to isolate total RNA with an RNeasy kit (Qiagen) according to the protocol provided by the manufacturer. Reverse transcription was performed with random primers and SuperScript RNase H reverse transcriptase (Life Technologies). PCR on the cDNA was done with Platinum Taq DNA polymerase (Invitrogen) and the primers shown in Table 1. A control without the reverse transcriptase verified the absence of DNA contamination. msl6 expression in Mycobacterium bovis BCG was also tested as described here.
TABLE 1.
Primers used to test for the expression of mas, msl, and pps genes by RT-PCRa
| ORF(s) | Primer sequences (5′→3′) |
|---|---|
| ppsA and -B | CGCAGTGCCCGACGACATCA, GTCGAGTGAGTTCCGGGCAC |
| ppsB and -C | GCCGAGAGGTCGGTGTGATGC, AGCATTCGCTGCTGCGGGTCC |
| ppsC and -D | AATGACCGCAGCGACACCAGATCG, TAGCCGGCCCGCCAGCGTGAGCAT |
| ppsD and -E | ATGACAAGTCTGGCGGAGCGCGCG, TCCGGTGCTCAGGTACGGGTCGAT |
| mas | GCCGGATCTGCTCGGCGAAC, ATCGTGTATGGGCCTCGCGT |
| msl1 | TACCGTCGCCGTACACCCAT, TGAACGGTGCGGCCACCAAA |
| msl2 | GCCGAGCACCCTCACCTGCG, GAAGATCTCAGCGCCCGCCG |
| msl3 | TGGAGATCCGGTCGAATACGC, GTCAAAACTGCTCGCCGTCAC |
| msl4 | TACCGAAGCATCCGCCAGCCG, ACAACCCACCTCGTCACCGGC |
| msl5 | GCGCGGGCGCCCTTGAGGAT, GTCCGGCCTCCATCAAGTCG |
| msl6 | CGCACTGCGAGCCCATGCGGT, AAGCCTTCTACCGGCTCGGCG |
| msl7 | GTATGCGGCCACCCACAC, CGCGCAGCGTGTCCCCAC |
For pps, the primers used amplified the junctions between two consecutive ORFs.
Biochemical analysis of cell wall lipids in the wild-type M. tuberculosis and the mutant with msl6 disrupted.
Sodium [1-14C]propionate (50 μCi; specific activity, 55 Ci/mol) (American Radiolabeled Chemicals, St. Louis, Mo.) was added to 30 ml of 12-day-old cultures of M. tuberculosis H37Rv and the msl6 mutant (optical density at 600 nm, 1.7 to 1.8), and incubation was continued at 37°C in roller bottles for a further 24 h. Cells were collected by centrifugation at 6,000 × g for 10 min and autoclaved. The cells were extracted with an excess of chloroform-methanol (2:1 [vol/vol]) with constant stirring at room temperature for several hours, and total cellular lipids were extracted and assayed for total 14C with Scintiverse BD scintillation fluid (Fisher Scientific, Pittsburgh, Pa.) in a Beckman LA3801 liquid scintillation counter (25). Total cellular lipids were separated on silica gel G plates (20 by 20 cm, 0.5 mm thick) with hexane-ethyl ether (9:1 [vol/vol]), and the polar lipids that remained at the origin in this solvent system were recovered from silica gel by elution with chloroform-methanol (2:1 [vol/vol]) and subjected to thin-layer chromatography (TLC) with chloroform-methanol (9:1 [vol/vol]) as the developing solvent. The lipids were visualized under UV light after the plates were sprayed with a 0.1% ethanolic solution of 2,7-dichlorofluorescein or with 5% dichromate in 50% H2SO4 followed by heating at 180°C. The 14C-labeled lipids were detected by scanning the chromatograms in a Berthold Tracemaster 20 automatic TLC linear analyzer and by autoradiography. Silica gel containing 14C was recovered from TLC plates, and the lipids were eluted with chloroform-methanol (2:1 [vol/vol]). The recovered lipids were subjected to exhaustive alkaline hydrolysis followed by methylation as described previously (25). The products recovered after methylation were subjected to TLC on silica gel G plates by using hexane-diethyl ether (9:1 [vol/vol]) as the developing solvent, and both nonhydroxylated and hydroxy fatty acid methyl ester fractions were recovered from the silica gel. The hydroxy fatty acid methyl esters were subjected to acetylation with acetic anhydride-pyridine (2:1) at room temperature overnight. The acetyl derivatives recovered from the reaction mixture were isolated by TLC, with hexane-ethyl ether (9:1 [vol/vol]) as the developing solvent; fatty acid methyl esters and acetylated hydroxyacid methyl esters were analyzed by radio gas chromatography (radio-GC). To examine the composition of the fatty acids from the total lipids, these lipids were subjected to the same procedures as those described above and analyzed by radio-GC. The methyl esters of fatty acids from the total lipids of both the wild type and the mutant were also separated by argentation-TLC (5% AgNO3 in silica gel) using hexane-ethyl ether (9:1 [vol/vol]) as the developing solvent (10), and each separated fatty acid methyl ester fraction was analyzed by radio-GC using a Varian model 3300 gas chromatograph with a coiled stainless steel column (3.2 mm by 2 m) packed with 3% (wt/wt) OV-1 on Chrom W-HP 80/100 (Varian, Inc.) and a Lablogic GC-RAM radioactivity monitor using Winflow (IN/US Systems, Tampa, Fla.) software. For analysis of fatty acid methyl esters and acetylated mycolipanolic acid methyl esters, a 180 to 300°C program at 15°C/min was used. For acetylated hydroxyphthioceranic acid methyl esters, isothermal analysis at 290°C was done with a carrier gas (He) flow of 30 ml/min.
Bacterial growth in an alveolar macrophage cell line.
Mouse alveolar macrophage cell line MH-S (ATCC CRL-2019) was obtained from the American Type Culture Collection and propagated in RPMI 1640 medium (Gibco-BRL) supplemented with 10% heat-inactivated fetal bovine serum (FBS). Bacteria were grown in Middlebrook 7H9 medium to an optical density at 600 nm of 0.4 and diluted 1:400 in RPMI 1640-FBS. Two days before infection, 24-well plates were seeded with 2 × 105 MH-S cells and cultures were infected in triplicate at 37°C with 0.1 ml of a single-cell suspension of M. tuberculosis H37Rv or the msl6 gene-disrupted mutant (0.5 bacterial cell/macrophage; multiplicity of infection, 0.5). The bacteria were allowed to adsorb for 4 h, and extracellular bacteria were removed by four successive washings with warm RPMI 1640. The infected cells were lysed in 7H9 medium containing 200 μl of 0.067% SDS for 30 min at 37°C. Serial 10-fold dilutions of the lysates were plated on 7H10 Middlebrook agar medium supplemented with 10% OADC, 0.5% Tween 80, and hygromycin wherever needed. Colonies were counted after 4 weeks of incubation at 37°C.
Assay for virulence in the murine model.
Aliquots of the M. tuberculosis H37Rv strain and its mutant strain were grown in modified 7H10 liquid media (7H10 agar formulations with agar and malachite green omitted) supplemented with 10% OADC for 1 week on a 37°C rotary shaker. Media for the mutant strains were supplemented with 50 μg of hygromycin/ml. At the end of the incubation period, culture growth was measured with a Klett-Summerson colorimeter (Klett Manufacturing, Brooklyn, N.Y.) and diluted to yield a final concentration of 1 Klett unit/ml or 5 × 105 CFU/ml. The inoculum size was determined by titration in triplicate on Middlebrook 7H10 (Difco Laboratories, Detroit, Mich.)-OADC agar plates.
Female C57BL6/J mice (Jackson Laboratories, Bar Harbor, Maine) were purchased at 6 weeks of age and allowed to acclimate in the facility for 1 week. Animals were housed in microisolator cages (Lab Products, Maywood, N.J.) and maintained with water and Prolab RMH 3000 rodent chow (Purina, St. Louis, Mo.) in a BSL-3 animal facility. Mice were randomly assigned to the following groups (four mice per group): day 1 postinfection, day 10 postinfection, and day 20 postinfection. Mice, anesthetized with telazol-xylazine, were infected intranasally with 20 μl of a suspension containing 104 CFU. At the time points indicated above, mice were euthanized by CO2 asphyxiation and their right lungs were sterilely removed. Lungs were homogenized in a 1-ml volume contained in an aerosol-resistant grinding assembly (Idea Works Laboratory Devices, Syracuse, N.Y.). Aliquots of the homogenate were serially diluted and titrated on 7H10 agar plates. Agar plates were incubated at 37°C in ambient air for 4 weeks, and viable-colony counts were enumerated. For the day 10 postinfection group infected with the wild type, there were only two animals because two died accidentally.
RESULTS
A search of the M. tuberculosis genome for ORFs with homology to mas revealed a large ORF of 12,455 bp, designated pks12, that could encode a protein containing 4,151 amino acids with a deduced molecular mass of 431 kDa, representing the largest ORF in the genome of M. tuberculosis. Analysis of the product of this ORF, which we designated msl6 (25), shows that it consists of a tandem repeat of two modules, each containing a full complement of catalytic domains necessary for fatty acid synthesis, in the same sequential order as in the product of mas. The two AT domains show more sequence variability (40% amino acid sequence identity, 55% similarity) than the KS domains (86% amino acid sequence identity and 91% similarity). A sequence comparison of AT domains for PKSs and fatty acid synthases revealed specific sequence motifs that distinguish between malonyl- and methylmalonyl-coenzyme A (CoA)-incorporating ATs (14). Analysis of the two AT domains encoded by msl6 indicates that AT1 falls in the group of ATs that use methylmalonyl-CoA as the substrate and that AT2 is homologous to those ATs that are specific for malonyl-CoA.
Disruption of the msl6 gene by homologous recombination.
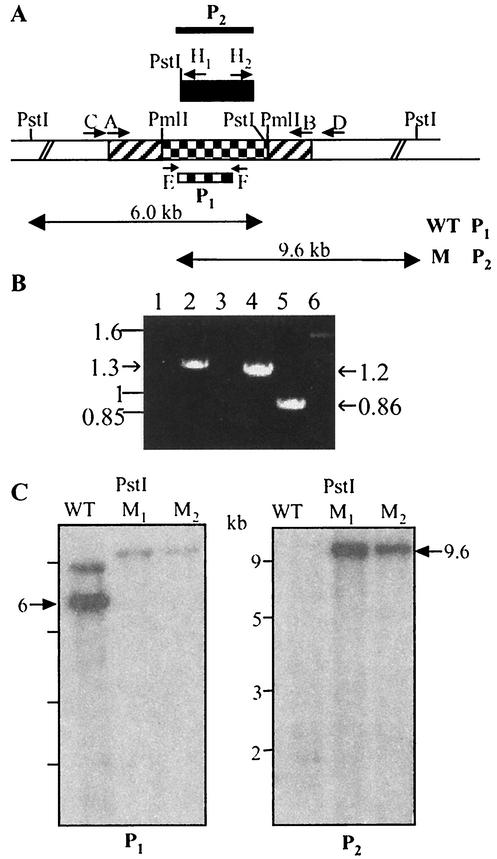
To investigate the function of the msl6 gene, we disrupted this gene by using a phage-mediated system to deliver the disruption construct into M. tuberculosis. A 4,100-bp PCR product containing part of msl6 (encoding the ER1, KR1, and ACP1 domains of the first module and the KS2 and AT2 domains of the second module) was amplified from M. tuberculosis genomic DNA by introducing HindIII sites at the 5′ and 3′ ends of the sequence. This product was subcloned into the HindIII-digested pUC19 vector, and a 2,193-bp internal PmlI fragment (encoding part of the ER1, ACP1, and KS2 domains) was replaced by the hyg gene and used to generate recombinant phages containing the disrupted copy of msl6 (Fig. 1A). After transduction of M. tuberculosis with these phages two hygromycin-resistant colonies were screened for allelic exchange with two primers specific for the deleted internal fragment that generate an 870-bp PCR product in the wild-type M. tuberculosis and that fail to amplify such a product from the mutant generated by allelic exchange. The two hygromycin-resistant colonies failed to amplify an 860-bp product specific for the deleted gene segment (Fig. 1B). Disruption of msl6 gene by allelic exchange was confirmed by further PCR analysis using two other sets of primers, each containing a hygromycin primer and a primer in the mycobacterial genome directly outside the msl6 sequences used to make the disruption construct. These primers generated the expected 1.3-kb 5′-flanking product and 1.2-kb 3′-flanking product (Fig. 1B). Southern blot analysis of the genes with disruption mutations was entirely consistent with the result of the PCR mapping (Fig. 1C). Genomic DNA samples from wild-type and mutant strains were digested with PstI. When an internal segment of the msl6 gene which was replaced by the hyg gene was used as a probe, the wild type showed the expected 6-kb hybridization band. The msl6-disrupted mutant did not show hybridization, confirming integration by double-crossover recombination. Analysis of the same blot with the hyg gene as a probe yielded a hybridization pattern consistent with replacement of the deleted msl6 segment with the hyg gene.
FIG. 1.
(A) Schematic representation of the construct used to disrupt the msl6 gene in M. tuberculosis. Hatched, checkered, and unshaded regions represent msl6 coding sequences, an internal segment replaced with the hyg gene (black box), and regions of the gene outside those used to make the disruption construct, respectively. Primer pair A-B was used to amplify the msl6 region chosen to generate the disruption construct. Primer pairs C-H1, D-H2, and E-F were used to test for allelic exchange by PCR analysis. P1 and P2, DNA segments used as probes in Southern blot analysis (P1, msl6 segment deleted in making the construct; P2, hyg gene). WT, wild type; M, mutant. (B) PCR analysis of internal and flanking regions of msl6 showing products consistent with allelic exchange. Lanes 1, 3, and 5, wild type; lanes 2, 4, and 6, mutant. Lane 1 and 2, 5′-flanking product with primers C and H1; lanes 3 and 4, 3′-flanking product with primers D and H2; lanes 5 and 6, internal deletion segment with primers E and F. (C) Southern blot analysis of M. tuberculosis H37Rv and msl6 mutants. Genomic DNA was digested with PstI. Left, DNA hybridized with an msl6 segment that was deleted in making the construct (probe P1); right, DNA probed with the hyg gene (probe P2). WT, wild type; M1 and M2, mutants.
Expression of mas, msl, and pps genes.
RT-PCR analysis of the msl6 mutant showed that the levels of transcripts of mas, msl1, msl2, msl3, msl4, msl5, msl7, ppsA, ppsB, ppsC, ppsD, and ppsE were very similar to those observed in the wild type (data not shown).
Analysis of total-protein extracts from wild-type M. tuberculosis and the msl6 mutant.
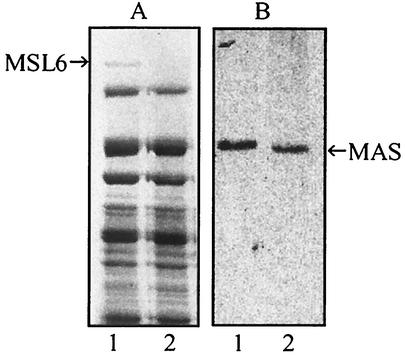
SDS-PAGE of total extracts from wild-type M. tuberculosis and the msl6 mutant showed a high-molecular-mass protein band of about 430 kDa that was missing in the mutant (Fig. 2A). In-gel reduction, carboxymethylation, and tryptic digestion, followed by analysis by MALDI and LC-MS, identified this protein as the product of msl6. The sequences for all of the 54 peptides, comprising 732 amino acid residues, whose coding sequences were distributed throughout the ORF of msl6 that we examined, matched exactly with those predicted from the msl6 sequence (data not shown).
FIG. 2.
SDS-PAGE (A) and immunoblot (B) analyses of total extracts from wild-type and msl6 mutant M. tuberculosis. The separated proteins were stained with Coomassie blue or analyzed by immunoblotting with anti-MAS antibodies. Lane 1, wild type; lane 2, msl6 mutant. The protein band at 430 kDa (MSL6) was used for amino acid sequence analysis.
Analysis of the lipids derived from [1-14C]propionic acid.
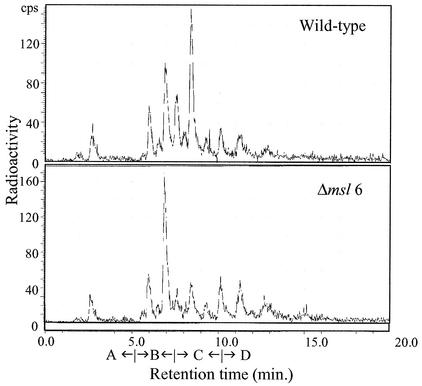
To investigate the biochemical consequences of msl6 disruption, we used [1-14C]propionate as the radiotracer to label the lipids because msl6 is expected to encode a methylmalonyl-CoA-utilizing synthase based on the sequence of one of the AT domains. The wild type and the mutant incorporated similar amounts (20 to 25%) of label into lipids. Radio-GC analysis of the methyl esters of fatty acids from the total lipids showed that the mutant generated all classes of fatty acids that are produced by the wild type. However, labeling of mycocerosic acids was much less in the msl6 mutant than in the wild type (Fig. 3). Argentation-TLC of fatty acids (as methyl esters) from the total lipids and radio-GC analysis of each fraction showed that the mutant generated all classes of fatty acids except that the labeling of the mycocerosic acid-containing fraction was much less in the mutant than in the wild type (data not shown).
FIG. 3.
Radio-GC analysis of the total fatty acid methyl esters derived from [1-14C]propionate in M. tuberculosis H37Rv (top) and its msl6 mutant (bottom). Retention time ranges for branched short-chain acids (A) mycolipanoic and mycolipenic acids (B), mycocerosic acids (C), and phthioceranic acids (D) are indicated.
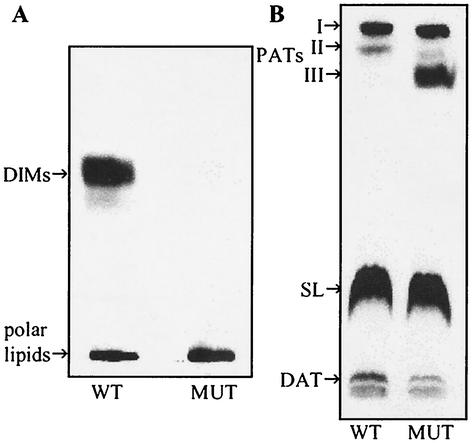
When the total lipids were separated by TLC using 10% ethyl ether in hexane, about 30% of the 14C was found in the DIM fraction in the wild type, whereas virtually no label was found in the DIM fraction in the msl6 mutant (Fig. 4A, Table 2). DIM was also not found in the culture medium. The absence of DIM in the msl6 mutant was confirmed by treatment of the plate with dichromate-H2SO4 followed by heating; the charred band at a retention factor of 0.6 found in the wild type was absent in the mutant. A detailed analysis of the more-polar lipids labeled by [1-14C]propionate showed that labeling of lipids other than DIM was not affected by msl6 disruption. The polar lipids that remained at the origin on the TLC plate developed in hexane-ethyl ether (9:1 [vol/vol]) as the solvent system were recovered and separated by TLC using 10% methanol in chloroform as the developing solvent (Fig. 4B). The distribution patterns of 14C among the polyacyl trehalose fractions, sulfolipids, and diacyl trehaloses for the wild type and the msl6 mutant were similar except for the heavier labeling in the mutant of a more-polar polyacyl trehalose that was barely labeled in the wild type (Fig. 4B, Table 2). A detailed analysis of fatty acids, obtained by hydrolysis and methyl esterification, present in individual lipid fractions was done. TLC analysis showed that there was increased amounts of 14C in the hydroxyphthioceranic acids in the msl6 mutant (Table 2). The high content of the hydroxyphthioceranic acids may explain the occurrence of a polar polyacyl trehalose component in the mutant (Fig. 4B). Radio-GC analysis of each fatty acid methyl ester fraction separated by argentation-TLC showed no significant differences in the fatty acid compositions of individual lipids between the wild type and msl6 mutant. Such detailed analysis revealed identical distributions of 14C among all classes of lipids and fatty acids (data not shown).
FIG. 4.
Autoradiograms of TLC of lipids derived from [1-14C]propionate in M. tuberculosis H37Rv (WT) and its msl6-disrupted mutant (MUT). (A) total lipids were subjected to TLC on silica gel G with 10% ethyl ether in n-hexane as the solvent. (B) The polar lipids remaining in the origin in panel A were recovered and subjected to TLC on silica gel G with 10% methanol in chloroform as the solvent. PATs, polyacyl trehaloses; SL, sulfolipids; DAT, diacyl trehaloses.
TABLE 2.
Relative percent distribution of 14C among the lipid classes derived from [1-14C]propionic acid in M. tuberculosis H37Rv (wild type) and the msl 6 mutanta
| Lipids | % 14C in:
|
|
|---|---|---|
| Wild type | msl6 mutant | |
| DIM | 30 | 0 |
| Fatty acids | 28 | 0 |
| Hydroxy fatty acids | 2 | 0 |
| PAT-I | 16 | 17 |
| Fatty acids | 14 | 13 |
| Mycolipanolic acids | 1.7 | 3 |
| Hydroxyphthioceranic acids | 0.3 | 1 |
| PAT-II | 4 | 3 |
| Fatty acids | 1 | 0.8 |
| Mycolipanolic acids | 1 | 0.6 |
| Hydroxyphthioceranic acids | 2 | 1.6 |
| PAT-III | ND | 7 |
| Fatty acids | ND | 1 |
| Mycolipanolic acids | ND | 0.6 |
| Hydroxyphthioceranic acids | ND | 5.4 |
| SL | 45 | 68 |
| Fatty acids | 10 | 10 |
| Mycolipanolic acids | 4 | 6 |
| Hydroxyphthioceranic acids | 31 | 52 |
| Very polar eipids | 5 | 4 |
| Fatty acids | 0.5 | 0.5 |
| Mycolipanolic acids | 0.3 | 0.2 |
| Hydroxyphthioceranic acids | 4.2 | 3.3 |
PAT, polyacyl trehalose; SL, sulfolipids; very polar lipids, lipids that remained in the origin and diacyl trehaloses that remained near the origin in TLC using 10% methanol in chloroform as the solvent system. ND, not detected.
To investigate whether mycocerosic acids, the acyl portion of DIM, can be generated by the mutants, the nonpolar lipids migrating near DIM in the TLC plate with 10% ethyl ether in hexane were recovered and hydrolyzed. Analysis of the fatty acids as their methyl esters by radio-GC showed low levels of 14C-labeled mycocerosic acids. Furthermore, radio-GC of the methyl esters from the sulfolipids also showed low levels of 14C-labeled mycocerosic acids in the msl6 mutant just as in the wild type. Coomassie staining of an SDS-PAGE gel of soluble protein and immunoblot analysis of the proteins from the wild type and the mutant showed MAS proteins of the expected sizes in the wild type and the msl6 mutant (Fig. 2B). Thus, the mutant was found to be capable of generating MAS and mycocerosic acids.
Attenuation of virulence.
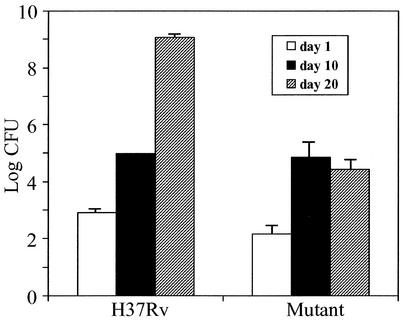
To test for the effect of msl6 disruption on virulence, we measured the growth of M. tuberculosis H37Rv in mouse alveolar macrophage cell line MH-S. We used a low inoculum level (0.5 bacteria per macrophage) to mimic natural infection and to avoid possible abnormal reactions. Starting with (2.3 ± 0.4) × 104 CFU, the wild type doubled in 2 days ([4.6 ± 0.44] × 104 CFU) and reached (4.3 ± 0.4) × 105 CFU in 5 days, and the msl6 mutant, starting with (2.0 ± 0.66) × 104 CFU, also doubled in 2 days ([4.3 ± 0.55] × 104 CFU) and reached (2.3 ± 0.6) × 105 CFU in 5 days. Thus, in a 5-day growth period the mutant showed considerable less growth than the wild type. Intranasal inoculation of C57BL6/J mice with M. tuberculosis H37Rv or its msl6 mutant followed by monitoring the growth of the pathogen in the lungs over a period of 20 days revealed a highly significant level of attenuation, especially after 20 days of growth. The growth of the mutant was several orders of magnitude less than that of the wild type (Fig. 5). The level of growth of the wild type found in these experiments was similar to those previously published (11). The animals appeared sick by day 20 after inoculation, especially those inoculated with the wild type, but there were no fatalities, even with 109 CFU/lung.
FIG. 5.
Growth of internasally administered M. tuberculosis H37Rv and its msl6-disrupted mutant in the lungs of C57BL6/J mice.
DISCUSSION
The cell wall lipids of M. tuberculosis belong to many classes that have straight and methyl-branched carbon chains (17). The genome of this organism contains many PKS genes (pks) (6). One of the synthases that has been purified and characterized catalyzes repeated elongation of an n-C20 alkanoyl chain by using methylmalonyl-CoA as the substrate to generate tetramethyl-branched acids, the major component of a class of acids called mycocerosic acids, that are esterified to phthiocerol to generate DIM (21); it has been suggested that DIM plays roles in the permeability of the cell wall and in virulence (4, 5, 7, 25). The AT and KS domains of this synthase (MAS) showed high degrees of similarity to similar domains in the PKSs of Streptomyces spp., which use methylmalonyl-CoA to generate multiple methyl-branched macrocyclic lactone antibiotics (19). Methylmalonyl-CoA selectivity of these domains was confirmed by analyzing domains expressed in E. coli (12). The sequences of many other PKSs revealed certain features characteristic of malonyl-CoA-utilizing synthases and methylmalonyl-CoA utilizing synthases (14). The pks genes in the genome of M. tuberculosis encode proteins containing one module required to make either a branched acid or a straight-chain acid. The largest ORF in the M. tuberculosis genome, however, is unique in that it encodes two modules, one with an apparent selectivity for malonyl-CoA and the other with a selectivity for methylmalonyl-CoA. In the present report we show that disruption of this gene causes a drastic decrease in the synthesis of DIM. Radio-GC analysis of the fatty acids produced by the mutant shows that it is capable of producing mycocerosic acids although the absence of DIM results in a decreased level of total mycocerosic acids. Detailed analysis of the labeled fatty acids and hydroxy fatty acids produced from [1-14C]propionic acid showed that the msl6 mutant produces all classes of branched acids that are normally produced by the wild type although the level of mycocerosic acids was drastically reduced in the msl6 mutant. All classes of acyl lipids normally found in M. tuberculosis H37Rv are found in the msl6 mutant except DIM. When the flow of methylmalonyl-CoA into mycocerosic acids of DIM is blocked by the absence of DIM synthesis, this substrate is probably channeled into the other major products normally generated from methylmalonyl-CoA in this organism. The major part of methylmalonyl-CoA that would have normally gone into the production of mycocerosic acids appears to have gone into the synthesis of hydroxyphthioceranic acids. These hydroxyacids get incorporated into polyacyl trehaloses, generating a more-polar class of polyacyl trehaloses in the msl6 mutant.
Analysis of the total proteins of M. tuberculosis by SDS-PAGE clearly showed a protein band at about 430 kDa, consistent with the size of the protein predicted to be produced by the ORF of msl6. A proteomics approach showed that the amino acid sequences of 54 peptides distributed throughout the length of this protein matched with those predicted from the nucleotide sequence of the gene, confirming the identity of this protein as the msl6 product. Since the msl6 mutant did not show any band at 430 kDa, the msl6 product accounts for the entire protein represented by the band in the wild type. Since this band is of significant intensity, the msl6 product constitutes a significant component of the soluble proteins in M. tuberculosis. RT-PCR could readily detect msl6 transcripts not only in M. tuberculosis but also in M. bovis BCG (data not shown), indicating that msl6 expression may be common to the organisms in the tuberculosis complex.
The precise biochemical role of the msl6 gene product is not clear. The msl6 mutant produces MAS as shown by SDS-PAGE, Coomassie staining, and immunoblot analysis of the cellular proteins; radio-GC analysis of lipids derived from [1-14C]propionic acid showed that mycocerosic acid synthesis is not prevented by msl6 disruption. RT-PCR analysis showed msl6 disruption did not significantly affect the transcript levels of mas or any mas-like genes. The observed overall decrease in propionate incorporation into mycocerosic acids is probably due to deficiency in the sites for its esterification caused by the defect in phthiocerol synthesis. In M. bovis BCG, disruption of certain genes postulated to be involved in phthiocerol and phenolphthiocerol production caused a virtual absence of mycocerosic acid production, because these diols constitute the sole sites of esterification for mycocerosic acids in this organism, although enzymatically active MAS was present (2). In M. tuberculosis H37Rv some mycocerosic acids can also be esterified to other lipids, and thus some labeling of mycocerosic acids is observed in the mutant. However, DIM production is prevented by msl6 disruption.
Phthiocerol, a long-chain diol, is derived from malonyl-CoA and methylmalonyl-CoA. Its synthesis would involve many enzymes. Based on biochemical reasoning a pathway for the biosynthesis of phthiocerol has been postulated (2, 17). From the nature of the reactions to be catalyzed by the enzymes postulated to be involved in this process, the participation of a series of hypothetical enzymes with appropriate substrate specificities was hypothesized. Based on the postulated requirements for the appropriate combination of domains, it was suggested that a set of genes, designated the pps genes, were involved in phthiocerol synthesis. Although results of gene disruption studies of M. bovis BCG supported these postulates (2) and although the proposed involvement of these pps genes in DIM synthesis was further supported by transposon mutagenesis studies of M. tuberculosis (4, 5, 7), the enzymes involved in this process have not been demonstrated. RT-PCR analysis indicated that msl6 disruption did not affect the expression of the pps genes previously postulated to be involved in the latter stages of DIM synthesis. It is possible that the product of msl6, which has one module with postulated selectivity for malonyl-CoA and another module with postulated selectivity for methylmalonyl-CoA, is involved in the synthesis of phthiocerol, which requires both of these substrates. The pps gene products were postulated to catalyze the synthesis of phthiocerol and phenolphthiocerol starting with elongated products. The production of these starting materials would involve additional pks genes, which remain obscure. msl6 may be one such gene. Until the enzymology of phthiocerol synthesis is elucidated, the exact role of msl6 in the production of phthiocerol remains uncertain.
Disruption of msl6 in M. tuberculosis results in attenuation in a murine macrophage cell line and in the murine model in which the pathogen is introduced intranasally, presumably because of the absence of DIM. DIM-defective mutants have been generated previously by transposon mutagenesis, and they were found to be attenuated (4, 7). Since DIM production involves many proteins, mutations that affect the synthesis of any of these proteins can generate DIM-deficient mutants, as already seen with the present results and the previously described transposon insertion mutagenesis. Even though how the absence of DIM causes attenuation remains unclear, DIM synthesis might be an appropriate target for antimycobacterial therapy. Since DIM production involves many enzymes, this process offers multiple ways to intervene in DIM production and thus opens many possibilities for designing antimycobacterial agents.
Acknowledgments
We thank Mike Kinter for assistance with the amino acid sequence analysis.
This work was supported in part by grants AI46582 and AI35272 from the National Institutes of Health.
Editor: S. H. E. Kaufmann
REFERENCES
- 1.Azad, A. K., T. D. Sirakova, L. M. Rogers, and P. E. Kolattukudy. 1996. Targeted replacement of the mycocerosic acid synthase gene in Mycobacterium bovis BCG produces a mutant that lacks mycosides. Proc. Natl. Acad. Sci. USA 93:4787-4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azad, A. K., T. D. Sirakova, N. D. Fernandes, and P. E. Kolattukudy. 1997. Gene knock-out reveals a novel gene cluster for the synthesis of a class of cell wall lipids unique to pathogenic mycobacteria. J. Biol. Chem. 272:16741-16745. [DOI] [PubMed] [Google Scholar]
- 3.Brennan, P. J., and H. Nikaido. 1995. The envelope of mycobacteria. Annu. Rev. Biochem. 64:29-63. [DOI] [PubMed] [Google Scholar]
- 4.Camacho, L. R., D. Enserqueix, E. Perez, B. Gicquel, and C. Guilhot. 1999. Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol. Microbiol. 34:257-267. [DOI] [PubMed] [Google Scholar]
- 5.Camacho, L. R., P. Constant, C. Raynaud, M. A. Lanéelle, J. A. Triccas, B. Gicquel, M. Daffe, and C. J. Guilhot. 2001. Analysis of the phthiocerol dimycocerosate locus of Mycobacterium tuberculosis. Evidence that this lipid is involved in the cell wall permeability barrier. J. Biol. Chem. 276:19845-19854. [DOI] [PubMed] [Google Scholar]
- 6.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 7.Cox, J. S., B. Chen, M. McNeil, and W. R. Jacobs. 1999. Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature 402:79-83. [DOI] [PubMed] [Google Scholar]
- 8.Daffe, M., and P. Draper. 1998. The envelope layers of mycobacteria with reference to their pathogenicity. Adv. Microb. Physiol. 39:131-203. [DOI] [PubMed] [Google Scholar]
- 9.Derbyshire, K. M., and S. Bardarov. 2000. DNA transfer in Mycobacteria: conjugation and transduction, p. 93-107. In G. F. Hatfull and W. R. Jacobs, Jr., (ed.), Molecular genetics of Mycobacteria. ASM Press, Washington, D.C.
- 10.Dubey, V. S., T. D. Sirakova, and P. E. Kolattukudy. 2002. Disruption of msl 3 abolishes the synthesis of mycolipanoic and mycolipenic acids required for polyacyltrehalose synthesis in Mycobacterium tuberculosis H37Rv and causes cell aggregation. Mol. Microbiol. 45:1451-1459. [DOI] [PubMed] [Google Scholar]
- 11.Edwards, K. M., M. H. Cynamon, R. K. Voladri, C. C. Hager, M. S. DeStefano, K. T. Tham, D. L. Lakey, M. R. Bochan, and D. S. Kernodle. 2001. Iron-cofactored superoxide dismutase inhibits host responses to Mycobacterium tuberculosis. Am. J. Respir. Crit. Care Med. 164:2213-2219. [DOI] [PubMed] [Google Scholar]
- 12.Fernandes, N. D., and P. E. Kolattukudy. 1997. Methylmalonyl coenzyme A selectivity of cloned and expressed acyltransferase and β-ketoacyl synthase domains of mycocerosic acid synthase from Mycobacterium bovis BCG. J. Bacteriol. 179:7538-7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fitzmaurice, A. M., and P. E. Kolattukudy. 1998. An acyl-CoA synthase (acoas) gene adjacent to the mycocerosic acid synthase (mas) locus is necessary for mycocerosyl lipid synthesis in Mycobacterium tuberculosis var. bovis BCG. J. Biol. Chem. 273:8033-8039. [DOI] [PubMed] [Google Scholar]
- 14.Haydock, S. F., J. F. Aparicio, I. Molnar, T. Schwecke, L. E. Khaw, A. Konig, A. F. Marsden, I. S. Galloway, J. Staunton, and P. F. Leadlay. 1995. Divergent sequence motifs correlated with the substrate specificity of (methyl)malonyl-CoA:acyl carrier protein transacylase domains in modular polyketide synthases. FEBS Lett. 374:246-248. [DOI] [PubMed] [Google Scholar]
- 15.Hopwood, D. A. 1997. Genetic contributions to understanding polyketide synthases. Chem. Rev. 97:2465-2498. [DOI] [PubMed] [Google Scholar]
- 16.Katz, L. 1997. Manipulation of modular polyketide synthases. Chem. Rev. 97:2557-2576. [DOI] [PubMed] [Google Scholar]
- 17.Kolattukudy, P. E., N. D. Fernandes, A. K. Azad, A. M. Fitzmaurice, and T. D. Sirakova. 1997. Biochemistry and molecular genetics of cell wall lipid biosynthesis in mycobacteria. Mol. Microbiol. 24:263-270. [DOI] [PubMed] [Google Scholar]
- 17a.Larsen, M. H. 2000. Some common methods in mycobacterial genetics, p. 316. In G. F. Hatfull and W. R. Jacobs, Jr. (ed.), Molecular genetics of mycobacteria. ASM Press, Washington, D.C.
- 18.Liu, J., C. E. Barry III, and H. Nikaido. 1999. Cell wall: physical structure and permeability, p. 220-239. In C. Ratledge and J. Dale (ed.), Mycobacteria: molecular biology and virulence. Blackwell Science, Malden, Mass.
- 19.Mathur, M., and P. E. Kolattukudy. 1992. Molecular cloning and sequencing of the gene for mycocerosic acid synthase, a novel fatty acid elongating multi-functional enzyme, from Mycobacterium tuberculosis var. bovis bacillus Calmette-Guerin. J. Biol. Chem. 267:19388-19395. [PubMed] [Google Scholar]
- 20.Pfeifer, B. A., and C. Khosla. 2001. Biosynthesis of polyketides in heterologous hosts. Micobiol. Mol. Biol. Rev. 65:106-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rainwater, D. L., and P. E. Kolattukudy. 1985. Fatty acid biosynthesis in Mycobacterium tuberculosis var. bovis bacillus Calmette-Guerin. Purification and characterization of a novel fatty acid synthase, mycocerosic acid synthase, which elongates n-fatty acyl-CoA with methylmalonyl-CoA. J. Biol. Chem. 260:616-623. [PubMed] [Google Scholar]
- 22.Rhoades, E. R., and H. J. Ullrich. 2000. How to establish a lasting relationship with your host: lessons learned from Mycobacterium spp. Immunol. Cell Biol. 78:301-310. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 24.Schorey, J. S., M. C. Carrol, and E. J. Brown. 1997. A macrophage invasion mechanism of pathogenic mycobacteria. Science 277:1091-1093. [DOI] [PubMed] [Google Scholar]
- 25.Sirakova, T. D., A. K. Thirumala, V. S. Dubey, H. Sprecher, and P. E. Kolattukudy. 2001. The Mycobacterium tuberculosis pks2 gene encodes the synthase for the hepta- and octamethyl-branched fatty acids required for sulfolipid synthesis. J. Biol. Chem. 276:16833-16839. [DOI] [PubMed] [Google Scholar]