Abstract
Pseudomonas aeruginosa is a gram-negative pathogen that infects immunocompromised and cystic fibrosis patients. The molecular basis of the host-P. aeruginosa interaction and the effect of specific P. aeruginosa virulence factors on various components of the innate immunity pathways are largely unknown. We examine interactions between P. aeruginosa virulence factors and components of innate immunity response in the Drosophila melanogaster model system to reveal the importance of the Toll signaling pathway in resistance to infection by the P. aeruginosa human isolate PA14. Using the two PA14-isogenic mutants plcS and dsbA, we show that Drosophila loss-of-function mutants of Spatzle, the extracellular ligand of Toll, and Dorsal and Dif, two NF-κB-like transcription factors, allow increased P. aeruginosa infectivity within fly tissues. In contrast, a constitutively active Toll mutant and a loss-of-function mutant of Cactus, an IκB-like factor that inhibits the Toll signaling, reduce infectivity. Our finding that Dorsal activity is required to restrict P. aeruginosa infectivity in Drosophila provides direct in vivo evidence for Dorsal function in adult fly immunity. Additionally, our results provide the basis for future studies into interactions between P. aeruginosa virulence factors and components of the Toll signaling pathway, which is functionally conserved between flies and humans.
Microbial pathogens use a variety of complex strategies to subvert host defenses to ensure their multiplication and survival. Pseudomonas aeruginosa is a medically important opportunistic human pathogen (10, 42) that, unlike most pathogens, exhibits an extremely broad host range that includes vertebrates (9), insects (3, 5, 7, 15), nematodes (22, 38), and plants (30). The virulence mechanisms used by P. aeruginosa to infect these phylogenetically diverse hosts (30) are remarkably conserved (30), suggesting that dissection of the immune strategies used by one model host system to subvert P. aeruginosa pathogenesis would provide an understanding of the strategies used by unrelated host organisms.
Drosophila melanogaster defends itself against pathogens via both humoral and cellular immune responses (1, 12). NF-κB-like transcription factors are thought to play critical roles in mediating the immune response of the fly, and at least two signaling pathways appear to trigger their activities. Relish (Rel) functions downstream of the immune deficiency gene imd (11), whereas Dorsal and Dif are controlled by the Toll pathway (24, 25, 35). Fungi and gram-positive, but not gram-negative, bacteria predominantly activate the Toll signaling pathway (20, 21, 26, 35) via the extracellular ligand, Spatzle. Toll pathway activation results in the proteolytic degradation of Cactus (27), a homologue of the mammalian I-κB inhibitor of Rel proteins, to mediate the nuclear import of Dorsal (18, 24, 33, 43) and Dif (13, 35, 43). Although both Dorsal and Dif are translocated to the host nuclei following microbial challenge, it is still unknown whether Dorsal participates in the adult fly immune response.
D'Argenio et al. recently reported that the gram-negative pathogen P. aeruginosa can infect D. melanogaster (5). In light of the striking evolutionary conservation of innate immunity defenses in insects and mammals (1, 12), we have utilized D. melanogaster to identify and dissect the disease response pathways involved in P. aeruginosa infection. Using virulence factors also required for mammalian pathogenesis, we show here that the P. aeruginosa human isolate PA14 establishes a progressive and lethal infection in Drosophila. We further show that the Toll signaling pathway, which is highly conserved between flies and mammals, is required to restrict the ability of P. aeruginosa to proliferate within fly tissues and be highly lethal. In addition, we present evidence that the Drosophila NF-κB-like factor Dorsal plays an important role during disease development and the immune response. These results offer insights into the interactions between P. aeruginosa virulence factors and components of the Toll signaling pathway during pathogenesis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
P. aeruginosa strain PA14 and the isogenic PA14 TnphoA mutants were previously described (22, 30, 31, 38). Other human isolates were obtained from Milton Schroth's UCPP strain collection (University of California at Berkeley). Bacteria were grown at 37°C in Luria-Bertani medium plus 100 μg of rifampin/ml for PA14 or 200 μg of kanamycin/ml for PA14 mutants. None of the P. aeruginosa strains used in this study exhibit any in vitro growth defects.
Fly stocks.
Fly stocks (obtained from Bloomington Fly Center unless specified) were maintained on standard yeast extract-agar-sucrose-cornmeal medium at 24°C. Tl10b flies carry a constitutively active Toll gene (37). Tl10b/+ flies were obtained by mating virgin Oregon-R (OR) females to Tl10b males. Loss-of-function trans-heterozygotes in Toll and Imd pathway mutants were obtained from the crosses TM3/spzrm7 × TM1/spz197, CyO/cactA2 × CyO/cactIIIG, and CyO/dl17 × CyO/dl1. F1 offspring that did not carry the balancer were used for infection assays. Bc1imd1 (19) and Rele20 (11) were homozygous for imd−/− and rel−/−, respectively. Homozygous recessive Dif1 (dif−/−) mutants were derived from ywDD1,cnbw (35).
Fly infection assays.
All experiments used healthy, 4- to 7-day-old adult male flies. Aseptic injuries were produced by pricking flies on the dorsal thorax with a 10-μm-diameter needle (Ernest F. Fullam, Inc., Latham, New York) which had previously been dipped into 10 mM MgSO4. After being dipped into a bacterial suspension containing 5 × 107 CFU/ml from an early stationary phase (optical density at 600 nm, 3.0) bacterial culture, the needle was used to infect flies with 10 to 100 P. aeruginosa cells/fly. Fly lethality was determined for groups of ≥25 infected flies for up to 72 h at 24°C. Flies that died within 12 h after infection were excluded. Lethality studies were repeated at least three times. For bacterial growth studies, infected flies were collected at indicated time intervals, ground in 10 mM MgSO4, and plated onto Luria-Bertani agar plates supplemented with appropriate antibiotics, with colony counts scored after 14 to 16 h at 37°C.
Statistical analysis of fly survival kinetics.
Statistical analysis was performed using R project software (www.r-project.org/). To test the null hypothesis that the survival kinetics of the mutant is equivalent to that of the wild type, each mutant survival curve was analyzed by two methods: (i) the log-rank test (Mantel-Haenszel) of the Kaplan-Meier estimate of survival (16) and (ii) the Cox proportional hazards regression model (4). The former method is nonparametric, while the latter utilizes a popular parametric model in the field of survival analysis. In each case, a P value was calculated from an analysis of the difference of the overall time course, representing the probability of randomly selecting subjects, from those predicted by the null hypothesis, whose survival curves are as different as (or more different than) those actually observed.
Fly cytological studies.
Male OR flies infected with PA14 were collected at time intervals, embedded and sectioned in paraffin, stained with hematoxylin and eosin, and examined by light microscopy.
RESULTS
Human isolates of P. aeruginosa cause lethal infections in D. melanogaster.
To determine whether Drosophila can serve as a model host for the determination of innate immune responses to P. aeruginosa infection, we assessed the viability of male adult OR flies infected with 10 human isolates of P. aeruginosa (Table 1). Each fly was infected with 10 to 100 bacterial cells. While control flies, pricked with a sterile needle, quickly recovered from injury (Fig. 1A), human isolates of P. aeruginosa caused differing degrees of lethality. In Table 1, these isolates are grouped into three lethality classes (high, medium, and low), members of which showed 93 to 100% lethality (strains PA14, PA37, and PA8); 73 to 84% lethality (strains PA12, PA15, PA38, PA46, PA4, and PA13); and 47% lethality (strain PA2), respectively. Previous studies have shown that other P. aeruginosa strains, including the well-characterized PAO1, cause highly lethal Drosophila infections (5, 7).
TABLE 1.
Human isolates of P. aeruginosa cause varying degrees of lethality in D. melanogaster
| Lethality group | P. aeruginosa human isolate | % Fly mortalitya |
|---|---|---|
| High | PA14 | 100 |
| PA37 | 96 | |
| PA8 | 93 | |
| Moderate | PA12 | 84 |
| PA15 | 83 | |
| PA38 | 83 | |
| PA46 | 81 | |
| PA4 | 76 | |
| PA13 | 73 | |
| Low | PA2 | 47 |
Percent mortality was determined in adult D. melanogaster OR flies at 72 h postinoculation. A minimum of 25 flies were infected with each P. aeruginosa strain. The results reflect data from four independent experiments. Human isolates are classified based on the levels of lethality produced in Drosophila.
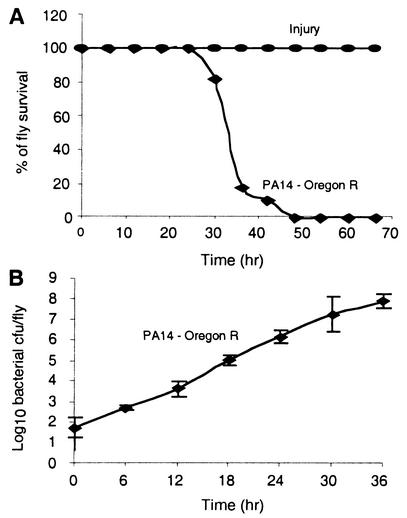
FIG. 1.
P. aeruginosa proliferates within D. melanogaster tissues with a lethal outcome. (A) Percent survival of adult OR flies following injury and infection. Control flies were injured with a needle that had been dipped into 10 mM MgSO4. Infected flies received 10 to 100 bacterial cells/fly. Approximately 40 flies were used for each experiment. The data reflect results from five independent experiments. (B) Proliferation of P. aeruginosa strain PA14 in OR flies. Means ± standard deviations (SD) of results for five flies per time point are shown. The data reflect results from four independent experiments.
We chose the highly lethal strain PA14 to study the antagonistic interactions of P. aeruginosa with Drosophila. PA14 is also highly virulent in other model host systems such as plants (30, 31), nematodes (22, 38), the insect Galleria mellonella (15), and mice (30). We assessed the resistance of OR flies to PA14 infection by monitoring percent survival and bacterial proliferation over time. Figure 1A shows that PA14-infected flies began to die at 28 h postinoculation and exhibited 0% survival by 48 h. Figure 1B shows that the degree of proliferation of PA14 within fly tissues correlated with the kinetics of mortality. The number of viable bacteria in PA14-infected flies rapidly proliferated, increasing 5 logs within 24 h postinoculation, and bacterial titers continued to increase over time and reached a maximum of 7 logs by 48 h (data not shown). These results indicate that the fly mortality produced by PA14 correlates with the ability of the bacteria to proliferate and establish a lethal infection.
P. aeruginosa strain PA14 invades, proliferates in, and colonizes fly tissues.
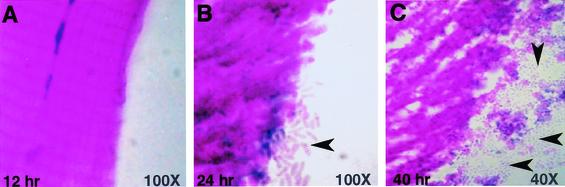
Following infection in the dorsal thorax, we evaluated stained sections of the infected flies at consecutive time points (Fig. 2). Between 0 and 12 h postinoculation, the infection was restricted to the focus of inoculation, with the greater portion of the thoracic muscles unaffected (Fig. 2A). By 24 h postinfection and beyond, widespread bacterial invasion was seen and the thoracic muscles began to show muscle fiber disruption (Fig. 2B). By 40 h, significant tissue consumption and complete disruption of striated muscle morphology were observed (Fig. 2C) and bacteria were observed throughout every body organ of the fly (data not shown). These findings demonstrate a progressive invasive process and systemic spread of the pathogen that correlates with PA14 cell proliferation over time and indicate that PA14 is able to invade, colonize, and utilize fly tissues as a nutrient source.
FIG. 2.
P. aeruginosa invades and degrades D. melanogaster tissues. OR flies infected with PA14 were collected and sectioned at 12, 24, and 40 h postinoculation, stained with hematoxylin and eosin, and evaluated by light microscopy. (A) Intact striated flight muscle exhibited no sign of bacterial invasion at 12 h. (B) P. aeruginosa cells (arrowhead) associated with flight muscles at 24 h. (C) Flight muscle degradation with large numbers of P. aeruginosa cells (arrowheads) at 40 h. Original magnifications: ×100 for panels A and B and ×40 for panel C.
Flies and mammals are susceptible to overlapping subsets of P. aeruginosa virulence factors.
To determine whether D. melanogaster is a suitable model host for the study of specific interactions between P. aeruginosa virulence factors and mammalian host responses, we tested 11 PA14 mutants, previously shown to have reduced virulence in mice (23, 32), for their ability to infect flies (Table 2). Virulence genes mutated in these strains encode proteins involved in protein folding, quorum sensing, posttranscriptional control, efflux systems, biosynthesis of redox active compounds, toxins, and proteins of unknown function (23, 32). Some of these genes have previously been shown to be required for virulence in the insect Galleria mellonella (15). Table 2 shows that all 11 P. aeruginosa virulence-associated genes that are necessary for full mammalian pathogenesis are also essential for maximum pathogenicity in the fly. In addition, all mutants that produced reduced fly lethality also proliferated to lower levels within fly tissues (Table 2). These findings validate Drosophila as a host to study interactions between P. aeruginosa virulence factors and host innate immunity and suggest that insects and mammals share functionally conserved mechanisms of resistance.
TABLE 2.
P. aeruginosa infects D. melanogaster and mammals using shared subsets of virulence factors
| Mutant or gene | % Drosophila lethalitya | % Mouse lethalityb | Comments | Bacterial replication (log10 CFU/fly ± SD) |
|---|---|---|---|---|
| PA14 | 100 | 100 | Wild-type strain | 8.12 ± 0.32 |
| dsbA | 15 | 60 | Periplasmic thiol-disulfide oxidoreductase | 3.48 ± 1.20 |
| mtrR | 24 | 53 | Transcriptional regulator of multidrug transporter | 3.99 ± 2.09 |
| gacA | 28 | 50 | Two-component quorum-sensing regulator | 4.65 ± 1.95 |
| phzB | 28 | 18 | Phenazine biosynthesis | 3.46 ± 1.92 |
| 33C7 | 33 | 0 | Unknownc | 5.40 ± 1.24 |
| plcS | 44 | 40 | Phospolipase C, lyses eukaryotic cell | 4.00 ± 0.39 |
| toxA | 50 | 40 | Exotoxin A, inhibits mammalian protein synthesis | 4.67 ± 1.65 |
| 44B1 | 50 | 56 | Unknownc | 5.43 ± 1.84 |
| pstP | 56 | 0 | Transcriptional regulator of RpoN-dependent operons | 3.99 ± 2.44 |
| mvfR | 56 | 35 | Quorum-sensing regulator | 4.52 ± 2.15 |
| pqsB | 56 | 63 | Hydroxy-alkylquinoline synthesis-quorum-sensing signaling | 4.40 ± 1.77 |
The lethality rate was measured in adult D. melanogaster OR flies. At least 25 flies were inoculated with a needle previously dipped into a 5 × 107/ml bacterial suspension and incubated at 24°C. Similar lethality and proliferation rates were obtained from 3 independent experiments. Bacterial loads were determined 36 hours postinfection.
Mouse lethality rates were obtained using a thermally injured mouse model (30).
Unknown, genes which encode hypothetical proteins of unknown function.
The Toll signaling pathway is required to limit P. aeruginosa infection in flies.
Since the Toll signaling pathway is thought to mediate pathogen defense in Drosophila, we infected fly strains mutant for genetic components of the Toll signaling pathway with the highly virulent strain PA14 and with two PA14-isogenic mutant derivatives, dsbA and plcS, which display reduced morbidity and mortality in mice (30, 31). The dsbA gene encodes a periplasmic thiol-disulfide oxidoreductase that catalyzes proper folding of bacterial proteins, including virulence-associated proteins, such as extracellular proteases, toxins, and type III secretion apparatus components (32, 44). The plcS gene encodes a hemolytic phospholipase capable of degrading eukaryotic cell membrane phospholipids (28) and has been shown to suppress the respiratory burst by neutrophils in mammals (39) and to cause the release of inflammatory mediators from human granulocytes and monocytes in vitro (17).
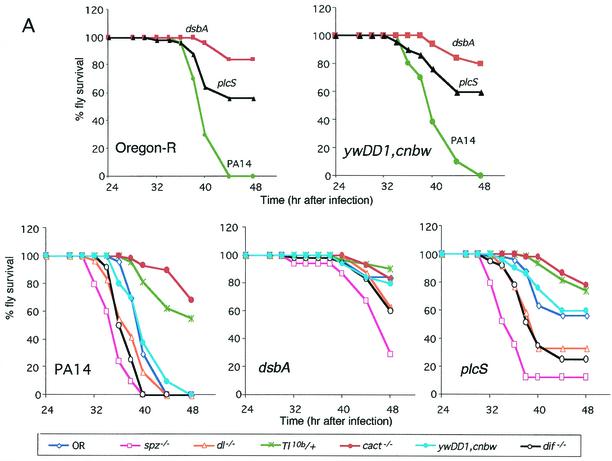
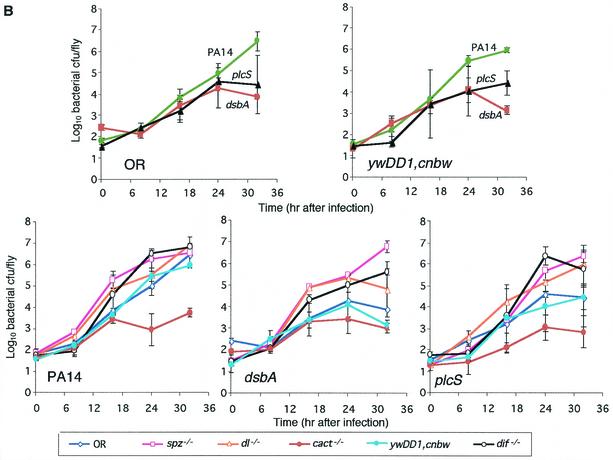
Figure 3A shows that PA14 caused 100% lethality in wild-type OR and ywDD1,cnbw flies while dsbA and plcS mutant bacteria produced much lower levels of lethality of 20 and 45%, respectively. In contrast, flies that carried loss-of-function mutations in the Toll ligand spatzle (spz−/−) and the NF-κB-like transcription factors dorsal (dl−/−) and Dorsal-related immunity factor Dif (dif−/−) were more susceptible to infection by dsbA and plcS, suggesting the importance of the Toll signaling pathway in limiting P. aeruginosa lethality (Fig. 3). Specifically, at 48 h postinfection, spz−/−, dl−/−, and dif−/− flies infected with dsbA exhibited 29, 62, and 60% survival, respectively, compared to dsbA survival levels for wild-type OR and ywDD1,cnbw flies of 85 and 80%, respectively. Similarly, 12, 33, and 25% of spz−/−, dl−>/−, and dif−/− flies, respectively, survived plcS infection versus 56 and 60% survival with this strain for the wild-type OR and ywDD1,cnbw flies, respectively.
FIG. 3.
Mutations in genetic components of Toll signaling alter P. aeruginosa lethality and proliferation. (A) Comparative survival analysis of adult flies infected with P. aeruginosa strain PA14 or its isogenic mutant derivatives dsbA and plcS. Survival in wild-type flies (OR and ywDD1,cnbw) or Toll component-mutated flies (spz−/−, dl−/−, cact−/−, dif−/−, and Tl10b/+) was assessed. A total of 100 to 150 flies were infected and monitored for survival at 48 h postinoculation. The results reflect data from three independent experiments. (B) Comparative analysis of bacterial proliferation in adult wild-type flies (OR and ywDD1,cnbw) or Toll component-mutated flies (spz−/−, dl−/−, cact−/−, dif−/−, and Tl10b/+). Flies were infected with P. aeruginosa strain PA14 or its isogenic mutants dsbA and plcS. Five flies per time point were used for CFU determination. Means ± SD of results for five flies per time point are shown. The results reflect data from three independent experiments.
As presented in Table 3, statistical analysis shows that the survival kinetics (Fig. 3A) of the various loss-of-function mutant flies infected with dsbA or the plcS mutant was indeed affected. Table 3 lists P values resulting from comparisons of survival curves from Mantel-Haenszel tests using the Kaplan-Meier estimate of survival (16) and likelihood ratio tests using the Cox proportional hazards regression model (4). As is presented in Table 3, the two tests give similar results. The spz−/− flies began to die at least 5 h earlier than wild-type OR flies when infected by PA14 and plcS, indicating increased susceptibility of these mutant flies to the bacterial strains. Although the survival kinetics of dl−/− and dif−/− flies infected with PA14 or plcS shows only a slight time difference (1 to 2 h) compared to the OR flies, this difference is statistically significant (Table 3).
TABLE 3.
Statistical analysis of the survival of wild-type and mutant flies after infection with the wild-type P. aeruginosa strain PA14 and isogenic mutants plcS and dsbA using the Kaplan-Meier (16) and Cox (4) modelsa
| Strains |
P by:
|
|
|---|---|---|
| Kaplan-Meier | Cox proportional hazards model | |
| PA14-OR vs dsbA-OR | 0 | 0 |
| PA14-OR vs plcS-OR | 4.11e−15 | 2.22e−16 |
| PA14-YWDD1 vs dsbA-YWDD1 | 0 | 0 |
| PA14-YWDD1 vs plcS-YWDD1 | 1.48e−14 | 8.88e−16 |
| PA14-OR vs PA14-spz | 0 | 0 |
| PA14-OR vs PA14-dl | 1.03e−05 | 0.000862 |
| PA14-OR vs PA14-dif | 1.11e−16 | 4.44e−16 |
| PA14-OR vs PA14-Tl10b | 0 | 0 |
| PA14-OR vs PA14-cact | 0 | 0 |
| dsbA-OR vs dsbA-spz | 3.84e−13 | 2.68e−14 |
| dsbA-OR vs dsbA-dl | 0.00328 | 0.00237 |
| dsbA-OR vs dsbA-dif | 0.000566 | 0.000418 |
| dsbA-OR vs dsbA-Tl10b | 0.21 | 0.197 |
| dsbA-OR vs dsbA-cact | 0.985 | 0.992 |
| plcS-OR vs plcS-spz | 0 | 0 |
| plcS-OR vs plcS-dl | 2.53e−05 | 3.56e−05 |
| plcS-OR vs plcS-dif | 1.23e−07 | 3.01e−07 |
| plcS-OR vs plcS-Tl10b | 0.0013 | 0.00124 |
| plcS-OR vs plcS-cact | 0.000156 | 0.000131 |
Further experiments with both Cactus loss-of-function (cact−/−) and Toll gain-of-function (Tl10b/+) flies provided additional evidence for the role of the Toll pathway in mediating resistance. The Cactus protein is an inhibitor of Toll signaling, and therefore, if the Toll pathway plays a role in mediating resistance, then cact−/− mutant flies should exhibit (versus wild-type flies) heightened levels of resistance to P. aeruginosa pathogenesis. This result was indeed observed, as shown in Fig. 3A. The cact−/− flies were (versus wild-type OR flies) significantly more resistant to infection by the mutant plcS and parental strain PA14 (Table 3). Significantly, 68% of the cact−/− flies infected with PA14 survived compared to 0% survival in the PA14-infected OR flies. Similarly, gain-of-function Tl10b/+ flies, which constitutively expressed activated Toll signaling, should have exhibited increased resistance to lethality. As predicted, 55% of infected Tl10b/+ flies survived PA14 infection compared to the 0% survival of infected OR flies. Furthermore, both Tl10b/+ and cact−/− flies exhibited delayed rates of lethality when infected by either PA14 or the plcS mutant strain. Interestingly, a delay in death (versus wild-type flies) was not observed in Tl10b/+ and cact−/− flies or in any other mutant flies when infected with the dsbA mutant strain. This result might have been expected, since the dsbA mutant strain exhibited highly attenuated virulence in most fly strains (Fig. 3), likely due to the inability of the dsbA mutant to properly fold multiple virulence-associated proteins. It is possible that the low level of virulence obtained with dsbA could be mediated via the gradual accumulation of non-dsbA-dependent factors.
Survival of Toll pathway mutant flies correlates with P. aeruginosa proliferation.
The enhanced susceptibility of spz−/−, dl−/−, and dif−/− flies and the increased resistance of cact−/− and Tl10b/+ flies to the PA14 mutants plcS and dsbA implicate the Toll signaling pathway in mediating resistance to P. aeruginosa. To determine whether the increased fly mortality seen in flies with mutations in components of the Toll and Imd pathways corresponds to an increased ability of dsbA and plcS bacteria to multiply within these immune-deficient hosts, we profiled the kinetics of bacterial replication. Figure 3B demonstrates that both dsbA and plcS mutant bacteria displayed decreased growth versus wild-type PA14 bacteria in OR and ywDD1,cnbw wild-type flies, correlating with the higher postinfection survival observed with these strains. This is particularly evident at 32 h postinoculation, when the bacterial loads in PA14-infected OR flies were 2.6 and 2.0 logs higher than those seen in OR flies infected with dsbA and plcS, respectively. In contrast, dsbA and plcS bacteria replicated to higher loads in spz−/−, dl−/−, and dif−/− mutant flies (Fig. 3B). Specifically, by 32 h postinoculation, the dsbA and plcS bacterial loads were 2.9 and 1.9 logs higher in spz−/− than in OR flies, respectively. Similarly, the bacterial loads of dsbA and plcS in dl−/− flies at 24 and 32 h were 1.1 and 0.9 logs higher and 0.6 and 1.5 logs higher, respectively, than those seen with the wild-type OR flies. Furthermore, by 32 h postinfection, the bacterial loads of dsbA and plcS in dif−/− flies were 2.5 and 1.3 logs higher than those seen with the wild-type parent ywDD1,cnbw flies. These data strengthen the idea of a correspondence between fly survival and decreased bacterial proliferation and suggest that both Dorsal and Dif participate in host immune defense to limit P. aeruginosa infection.
Bacterial proliferation was also scored in cact−/− and Tl10b/+ flies, which exhibited constitutive Toll pathway activity. As predicted, increased survival of both these flies to bacterial infection correlated with decreased bacterial proliferation over the course of infection. Figure 3B shows that viable PA14 counts for cact−/− flies were approximately 3 logs lower than for OR flies at 32 h, and similar data (data not shown) were obtained for Tl10b/+ flies. Furthermore, both dsbA and plcS strains proliferate to lower bacterial loads throughout the course of infection in cact−/− versus OR flies (Fig. 3B).
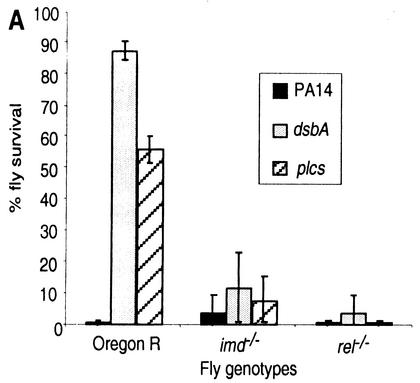
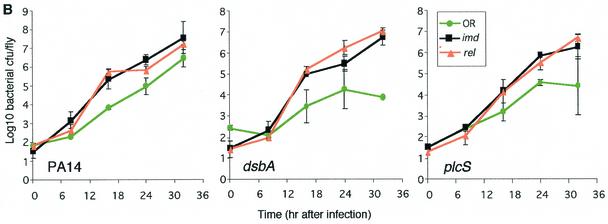
P. aeruginosa virulence is elevated in imd and rel mutant flies.
imd and rel mutations disable the Imd pathway that functions in the Drosophila immune response. To ascertain that this pathway also plays a role against P. aeruginosa infection, we infected imd−/− and rel−/− flies with PA14, dsbA, and plcS bacteria. Figure 4A shows that, as seen with Toll pathway mutant flies, dsbA and plcS lethality was greatly increased in the imd−/− and rel−/− mutants to levels similar to those observed for dsbA and plcS bacteria in spz−/− flies. These results agree with previous studies showing a requirement for the Imd pathway in fighting gram-negative bacterial infections (6, 19). Figure 4B demonstrates that as seen with the Toll pathway-deficient flies, PA14, dsbA, and plcS bacteria replicated to higher densities in the imd−/− and rel−/− versus wild-type OR flies, reaching ≥2 logs higher at 32 h postinfection. These results indicate that both the Toll and Imd pathways are required to confer maximum resistance to P. aeruginosa in the adult fly, presumably by generating a fully competent immune system.
FIG. 4.
Mutations in genetic components of the Drosophila Imd signaling pathway increase P. aeruginosa lethality and proliferation. (A) Survival analysis of wild-type OR and loss-of-function imd−/− and rel−/− (homozygous) mutant flies following infection with strain PA14 and its isogenic derivatives dsbA and plcS. A total of 100 to 150 flies were infected, and survival was scored at 48 h postinoculation. (B) Comparative analysis of bacterial proliferation in adult wild-type OR flies and imd−/− and rel−/− mutant flies infected with P. aeruginosa strain PA14 or its isogenic mutants dsbA and plcS. Five flies per time point were used for CFU determination. Means ± SD for five flies per time point are shown. The results reflect data from three independent experiments.
DISCUSSION
The results presented in this report provide several significant conclusions with regard to the progression of the host-pathogen interaction between D. melanogaster and the human opportunistic pathogen P. aeruginosa. These results also provide novel insights into the identity of both pathogen and host genetic factors that enhance or restrict pathogenesis. In addition, these results further establish Drosophila as a model system to gain future insights into the mechanisms of P. aeruginosa pathogenesis and host resistance.
With regard to the bacterial side of this interaction, we have shown that different P. aeruginosa isolates elicit infections in Drosophila of differing degrees of lethality and that strain PA14 produces 100% mortality at 36 to 48 h postinfection from an inoculum as low as 10 bacteria. We note that Drosophila has previously been reported to be susceptible to other human P. aeruginosa isolates (5, 7). We have further shown that the bacteria invade and proliferate in adult fly tissue and that the ability of PA14 and its derivatives to cause maximum lethality correlates with the kinetics of invasion and proliferation. Significantly, PA14 utilizes many of the same virulence factors to infect the fly that it uses to mediate pathogenesis in mammalian hosts and mutations in virulence factors that give decreased mortality exhibit restricted kinetics of proliferation. Additionally, we demonstrated that the dsbA and plcS virulence gene functions are required for P. aeruginosa to produce full disease symptoms (invasion, proliferation, and lethality) in flies.
With regard to the host side of this interaction, we have shown using different fly mutant lines that both the Toll and Imd signaling pathways function to limit the severity of pathogenesis both in the degree of lethality and in the kinetics of bacterial invasion and proliferation. Indeed, the activities of all three Drosophila NF-κB-like factors are required to fully restrict infection, as the PA14-isogenic mutants dsbA and plcS are significantly more virulent in flies that carry mutations in Dorsal, Dif, or Relish. Interestingly, our results on Toll contradict previous reports that the Drosophila Toll pathway is not involved in resistance to gram-negative bacterial infections (2, 19, 21, 35, 36). However, the recent microarray analysis by De Gregorio et al. provides evidence that the Toll signaling pathway functions in combating gram-negative infections (6). Here, we functionally demonstrate the importance of the Toll pathway by showing that the two PA14-isogenic mutants dsbA and plcS, which produce attenuated virulence in wild-type flies, are significantly more virulent in flies that carry mutations in the Toll pathway genes, spatzle, dif, and dorsal.
That the Toll pathway contributes resistance to P. aeruginosa infection is further supported by our findings that the gain-of-function Toll pathway mutant Tl10b/+, a constitutively active allele, and the loss of function in negative regulator Cactus (cact−/−) exhibit heightened resistance to P. aeruginosa strain PA14, as manifested by lower fly lethality and in vivo bacterial proliferation. Unchallenged Tl10b/+ flies have previously been shown to exhibit strong constitutive expression of the antimicrobial peptides Drosomycin and Metchnikowin, as well as increased levels (two- to fivefold) of other antibacterial peptides, versus unchallenged wild-type OR flies (14, 20). Because Tl10b/+ and cact−/− larvae also exhibit increased numbers of hemocytes (29), we cannot rule out the possibility that the enhanced resistance to P. aeruginosa infection observed in Tl10b/+ and cact−/− flies results from more phagocytosis and that a collective effort by both humoral (8, 40, 41) and cellular (29, 34) responses underlies the increased survival of Tl10b/+ and cact−/− flies to PA14 infections. Future studies using other fly mutants should help to determine the importance of these two classes of responses against P. aeruginosa.
Previous studies have indicated that Dorsal, Dif, and Relish play distinct roles in immunity and development (1, 12). To date, no in vivo study has demonstrated the involvement of Dorsal, which controls dorsal-ventral patterning in the developing embryo, in the adult fly immune response. Using genetic data, we provide such evidence, showing that Dorsal is required to mediate a maximal immune response: dl−/− flies infected with dsbA and plcS exhibit significantly greater susceptibility than do wild-type flies. Interestingly, dsbA and plcS give decreased levels of lethality and proliferation in dl−/− and dif −/− versus spz−/− flies. One explanation for this result is that Dif and Dorsal functions are partially redundant in the adult fly immune system in a manner similar to that previously suggested to occur for these two activities in Drosophila larvae (24).
In conclusion, we show that study of the pathogenic interaction between P. aeruginosa and Drosophila allows for the dissection of both the pathogenicity of, and the resistance to, this important human pathogen. The P. aeruginosa-Drosophila interaction permits the genetic manipulation of both the pathogen and host partners, which is not possible when studying pathogenesis in humans. Significantly, P. aeruginosa utilizes many of the same virulence factors to infect insects as it does to infect mammals, including humans, and the Drosophila Toll signaling pathway, which is required for full immunity to P. aeruginosa, shares striking molecular and functional conservation with the human IL-1/NF-κB signaling pathways. These findings demonstrate the utility of Drosophila for probing the mechanistic interactions between P. aeruginosa virulence factors and components of the Toll signaling pathway, which should give novel insights into how these components function to provide resistance.
Acknowledgments
We thank D. Morisato for Tl10b flies, D. Ferrandon for Dif1 flies, and M. Mindrinos and S. Stachel for helpful discussions and for reading the manuscript and John Storey for helpful discussion on the statistical analysis.
This work was partially supported by a Shriners Hospital for Children grant. B.C.G. was supported by a Shriners Research Fellowship, and L.A.P. was supported by the National Science Foundation.
Editor: A. D. O'Brien
REFERENCES
- 1.Anderson, K. V. 2000. Toll signaling pathways in the innate immune response. Curr. Opin. Immunol. 12:13-19. [DOI] [PubMed] [Google Scholar]
- 2.Basset, A., R. S. Khush, A. Braun, L. Gardan, F. Boccard, J. A. Hoffmann, and B. Lemaitre. 2000. The phytopathogenic bacteria Erwinia carotovora infects Drosophila and activates an immune response. Proc. Natl. Acad. Sci. USA 97:3376-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boman, H. G., I. Nilsson, and B. Rasmuson. 1972. Inducible antibacterial defense system in Drosophila. Nature 237:232-235. [DOI] [PubMed] [Google Scholar]
- 4.Cox, D. R. 1972. Regression models and life tables. J. Royal Stat. Soc. Ser. B 34:187-220. [Google Scholar]
- 5.D'Argenio, D. A., L. A. Gallagher, C. A. Berg, and C. Manoil. 2001. Drosophila as a model host for Pseudomonas aeruginosa infection. J. Bacteriol. 183:1466-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Gregorio, E., P. T. Spellman, P. Tzou, G. M. Rubin, and B. Lemaitre. 2002. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 21:2568-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fauvarque, M. O., E. Bergeret, J. Chabert, D. Dacheux, M. Satre, and I. Attree. 2002. Role and activation of type III secretion system genes in Pseudomonas aeruginosa-induced Drosophila killing. Microb. Pathog. 32:287-295. [DOI] [PubMed] [Google Scholar]
- 8.Ferrandon, D., A. C. Jung, M. Criqui, B. Lemaitre, S. Uttenweiler-Joseph, L. Michaut, J. M. Reichhart, and J. A. Hoffmann. 1998. A drosomycin-GFP reporter transgene reveals a local immune response in Drosophila that is not dependent on the Toll pathway. EMBO J. 17:1217-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fink, R. B., Jr. (ed.). 1993. Pseudomonas aeruginosa the opportunist: pathogenesis and disease. CRC Press, Boca Raton, Fla.
- 10.Gang, R. K., R. L. Bang, S. C. Sanyal, E. Mokaddas, and A. R. Lari. 1999. Pseudomonas aeruginosa septicemia in burns. Burns 25:611-616. [DOI] [PubMed] [Google Scholar]
- 11.Hedengren, M., B. Asling, M. Dushay, I. Ando, S. Ekengren, M. Wihlborg, and D. Hultmark. 1999. Relish, a central factor in the control of humoral but not cellular immunity in Drosophila. Mol. Cell 4:827-837. [DOI] [PubMed] [Google Scholar]
- 12.Hoffmann, J. A., F. C. Kafatos, C. A. Janeway, and R. A. Ezekowitz. 1999. Phylogenetic perspectives in innate immunity. Science 284:1313-1318. [DOI] [PubMed] [Google Scholar]
- 13.Ip, Y. T., M. Reach, Y. Engstrom, L. Kadalayil, H. Cai, S. Gonzalez-Crespo, K. Tatei, and M. Levine. 1993. Dif, a dorsal-related gene that mediates an immune response in Drosophila. Cell 75:753-763. [DOI] [PubMed] [Google Scholar]
- 14.Irving, P., L. Troxler, T. S. Heuer, M. Belvin, C. Kopczynski, J. M. Reichhart, J. A. Hoffmann, and C. Hetru. 2001. A genome-wide analysis of immune responses in Drosophila. Proc. Natl. Acad. Sci. USA 98:15119-15124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jander, G., L. G. Rahme, and F. M. Ausubel. 2000. Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J. Bacteriol. 182:3843-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaplan, E., and P. Meier. 1958. Nonparametric estimation from incomplete observations. J. Am. Stat. Assoc. 53:457-481, 562-563.
- 17.Konig, B., K. E. Jaeger, A. E. Sage, M. L. Vasil, and W. Konig. 1996. Role of Pseudomonas aeruginosa lipase in inflammatory mediator release from human inflammatory effector cells (platelets, granulocytes, and monocytes). Infect. Immun. 64:3252-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemaitre, B., M. Meister, S. Govind, P. Georgel, R. Steward, J. M. Reichhart, and J. A. Hoffmann. 1995. Functional analysis and regulation of nuclear import of dorsal during the immune response in Drosophila. EMBO J. 14:536-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemaitre, B., E. Kromer-Metzger, L. Michaut, E. Nicolas, M. Meister, P. Georgel, J. M. Reichhart, and J. A. Hoffmann. 1995. A recessive mutation, immune deficiency (imd), defines two distinct control pathways in the Drosophila host defense. Proc. Natl. Acad. Sci. USA 92:9465-9469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemaitre, B., E. Nicolas, L. Michaut, J. M. Reichhart, and J. A. Hoffmann. 1996. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 86:973-983. [DOI] [PubMed] [Google Scholar]
- 21.Lemaitre, B., J. M. Reichhart, and J. A. Hoffmann. 1997. Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc. Natl. Acad. Sci. USA 94:14614-14619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahajan-Miklos, S., M.-W. Tan, L. G. Rahme, and F. M. Ausubel. 1999. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 96:47-56. [DOI] [PubMed] [Google Scholar]
- 23.Mahajan-Miklos, S., L. G. Rahme, and F. M. Ausubel. 2000. Elucidating the molecular mechanisms of bacterial virulence using non-mammalian hosts. Mol. Microbiol. 37:981-988. [DOI] [PubMed] [Google Scholar]
- 24.Manfruelli, P., J. M. Reichhart, R. Steward, J. A. Hoffmann, and B. Lemaitre. 1999. A mosaic analysis in Drosophila fat body cells of the control of antimicrobial peptide genes by the Rel proteins Dorsal and Dif. EMBO J. 18:3380-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meng, X., B. S. Khanuja, and Y. T. Ip. 1999. Toll receptor-mediated Drosophila immune response requires Dif, an NF-κB factor. Genes Dev. 13:792-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michel, T., J. M. Reichhart, J. A. Hoffmann, and J. Royet. 2001. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature 414:756-759. [DOI] [PubMed] [Google Scholar]
- 27.Nicolas, E., J. M. Reichhart, J. A. Hoffmann, and B. Lemaitre. 1998. In vivo regulation of the I-κB homologue Cactus during the immune response of Drosophila. J. Biol. Chem. 273:10463-10469. [DOI] [PubMed] [Google Scholar]
- 28.Ostroff, R. M., B. Wretlind, and M. L. Vasil. 1989. Mutations in the hemolytic-phospholipase C operon result in decreased virulence of Pseudomonas aeruginosa PAO1 grown under phosphate-limiting conditions. Infect. Immun. 57:1369-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiu, P., P. C. Pan, and S. Govind. 1998. A role for the Drosophila Toll/Cactus pathway in larval hematopoiesis. Development 125:1909-1920. [DOI] [PubMed] [Google Scholar]
- 30.Rahme, L. G., E. J. Stevens, S. F. Wolfort, J. Shao, R. G. Tompkins, and F. M. Ausubel. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899-1902. [DOI] [PubMed] [Google Scholar]
- 31.Rahme, L. G., M.-W. Tan, L. Le, S. M. Wong, R. G. Tompkins, S. B. Calderwood, and F. M. Ausubel. 1997. Use of model plant hosts to identify Pseudomonas aeruginosa virulence factors. Proc. Natl. Acad. Sci. USA 94:13245-13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rahme, L. G., F. M. Ausubel, H. Cao, E. Drenkard, B. C. Goumnerov, G. W. Lau, S. Mahajan-Miklos, J. Plotnikova, M.-W. Tan, J. Tsongalis, C. L. Walendziewicz., and R. G. Tompkins. 2000. Plants and animals share functionally common bacterial virulence factors. Proc. Natl. Acad. Sci. USA 97:8815-8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reichhart, J. M., P. Georgel, M. Meister, B. Lemaitre, C. Kappler, and J. A. Hoffmann. 1993. Expression and nuclear translocation of the rel/NF-kappa B-related morphogen dorsal during the immune response of Drosophila. C. R. Acad. Sci. Ser. III 316:1218-1224. [PubMed] [Google Scholar]
- 34.Rosetto, M., Y. Engstrom, C. T. Baldari, J. L. Telford, and D. Hultmark. 1995. Signals from the IL-1 receptor homolog, Toll, can activate an immune response in a Drosophila hemocyte cell line. Biochem. Biophys. Res. Commun. 209:111-116. [DOI] [PubMed] [Google Scholar]
- 35.Rutschmann, S., A. C. Jung, C. Hetru, J. M. Reichhart, J. A. Hoffmann, and D. Ferrandon. 2000. The Rel protein DIF mediates the antifungal but not the antibacterial host defense in Drosophila. Immunity 12:569-580. [DOI] [PubMed] [Google Scholar]
- 36.Rutschmann, S., A. Kilinc, and D. Ferrandon. 2002. The toll pathway is required for resistance to gram-positive bacterial infections in Drosophila. J. Immunol. 168:1542-1546. [DOI] [PubMed] [Google Scholar]
- 37.Schneider, D. S., K. L. Hudson, T. Y. Lin, and K. V. Anderson. 1991. Dominant and recessive mutations define functional domains of Toll, a transmembrane protein required for dorsal-ventral polarity in the Drosophila embryo. Genes Dev. 5:797-807. [DOI] [PubMed] [Google Scholar]
- 38.Tan, M.-W., L. G. Rahme, J. A. Sternberg, R. G. Tompkins, and F. M. Ausubel. 1999. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify Pseudomonas aeruginosa virulence factors. Proc. Natl. Acad. Sci. USA 96:2408-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Terada, L. S., K. A. Johansen, S. Nowbar, A. I. Vasil, and M. L. Vasil. 1999. Pseudomonas aeruginosa hemolytic phospholipase C suppresses neutrophil respiratory burst activity. Infect. Immun. 67:2371-2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tzou, P., S. Ohresser, D. Ferrandon, M. Capovilla, J. M. Reichhart, B. Lemaitre, J. A. Hoffmann, and J. L. Imler. 2000. Tissue-specific inducible expression of antimicrobial peptide genes in Drosophila surface epithelia. Immunity 13:737-748. [DOI] [PubMed] [Google Scholar]
- 41.Tzou, P., J. M. Reichhart, and B. Lemaitre. 2002. Constitutive expression of a single antimicrobial peptide can restore wild-type resistance to infection in immunodeficient Drosophila mutants. Proc. Natl. Acad. Sci. USA 99:2152-2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wood, R. E. 1976. Pseudomonas: the compromised host. Hosp. Pract. 11:91-100. [DOI] [PubMed] [Google Scholar]
- 43.Wu, L. P., and K. V. Anderson. 1998. Regulated nuclear import of Rel proteins in the Drosophila immune response. Nature 392:93-97. [DOI] [PubMed] [Google Scholar]
- 44.Yu, J., and J. S. Kroll. 1999. DsbA: a protein-folding catalyst contributing to bacterial virulence. Microbes Infect. 1:1221-1228. [DOI] [PubMed] [Google Scholar]