Abstract
We describe a novel requirement for the condensin complex in sister chromatid cohesion in Saccharomyces cerevisiae. Strikingly, condensin-dependent cohesion can be distinguished from cohesin-based pairing by a number of criteria. First, condensin is required to maintain cohesion at several chromosomal arm sites but, in contrast to cohesin, is not required at either centromere or telomere-proximal loci. Second, condensin-dependent interlinks are established during mitosis independently of DNA replication and are reversible within a single cell cycle. Third, the loss of condensin-dependent linkages occurs without affecting cohesin levels at the separated URA3 locus. We propose that, during mitosis, robust sister chromatid cohesion along chromosome arms requires both condensinand cohesin-dependent mechanisms, which function independently of each other. We discuss the implications of our results for current models of sister chromatid cohesion.
Keywords: Condensin complex, cohesin complex, sister chromatid cohesion, chromosome segregation, mitotic chromosome structure
The fidelity of genetic inheritance is orchestrated by a number of evolutionarily conserved mechanisms. In eukaryotic cells, two such mechanisms are sister chromatid cohesion (SCC) and mitotic chromosome condensation (for recent reviews, see Hagstrom and Meyer 2003; Nasmyth and Haering 2005). Cohesion holds newly replicated sisters in close proximity from S phase until the initiation of chromosome segregation in anaphase, whereas condensation organizes mitotic chromosomes into compact, discrete segregatable units. While both the cohesion and condensation machineries—called cohesin and condensin, respectively—are essential for accurate chromosome transmission and are conserved from yeast to man, their molecular mechanisms of action remain poorly understood (see Yokomori 2003; Hirano 2005a; Nasmyth and Haering 2005).
In Saccharomyces cerevisiae, the cohesin complex consists of two structural maintenance of chromosome (SMC) proteins, termed Smc1 and Smc3, plus two additional non-SMC subunits called Mcd1/Scc1 and Scc3. SMCs are ubiquitous in nature and function in numerous aspects of DNA metabolism including SCC, DNA repair, chromosome condensation, and transcriptional regulation (Hirano 2005b). While the precise mechanism(s) of SMC function in these processes is unknown, the structural features of SMC proteins have inspired a number of hypothetical models (Haering et al. 2002; Huang et al. 2005). SMCs consist of hinge and head (ATPase) domains separated by large coiled-coil regions (Strunnikov et al. 1993), which fold back upon themselves to form compact monomers (Haering et al. 2002). Upon dimerization of monomers through both hinge and head regions, ring structures can be observed by electron microscopy, consistent with biochemical experiments (Anderson et al. 2002; Haering et al. 2002; Gruber et al. 2003; Ivanov and Nasmyth 2005). The existence of a cohesin ring led to the proposal that cohesion between sister chromatids could be mediated through the entrapment of both sister chromatids within a single cohesin ring, which would occur as a consequence of replication (Haering et al. 2002; Gruber et al. 2003; Ivanov and Nasmyth 2005). While this “replication through a ring” model is attractive, a number of alternative models posit that the formation of SCC involves the multimerization of cohesin complexes (for review, see Huang et al. 2005; Skibbens 2005). Until cohesed sister chromatids are amenable to direct experimentation however, the molecular nature of cohesin-based cohesion will remain hotly debated.
A second abundant SMC complex conserved in eukaryotes is condensin (for review, see Hirano 2005a). In budding and fission yeasts as well as in Xenopus laevis egg extracts, condensin is the major chromosome condensation activity (Hirano et al. 1997; Sutani et al. 1999; Freeman et al. 2000). The budding yeast condensin subunits are Smc2, Smc4, Ycs4, Ycg1, and Brn1, and mutants in any of these cause dramatic chromosome condensation and segregation defects (Strunnikov et al. 1995; Freeman et al. 2000; Ouspenski et al. 2000; Bhalla et al. 2002; Lavoie et al. 2002). While the precise activities underlying mitotic chromosome folding remain mysterious, condensation involves both the resolution and compaction of sister chromatids into discrete segregatable units. Interestingly, significant in vivo metaphase chromosome compaction is still observed in condensin mutants/knockdowns in metazoans (Hirano et al. 1997; Kimura et al. 1998, 2001; Schmiesing et al. 2000; Giet and Glover 2001; Hagstrom et al. 2002; Stear and Roth 2002), indicating that additional compaction activities are likely to exist in these cells. Nonetheless, anaphase bridges are observed in all cells lacking condensin, indicating a fundamental and conserved role in chromosome transmission throughout evolution. While condensin is known to be required for higher-order chromosome structure at all genomic regions, recent work in budding yeast has demonstrated a specialized role for condensin in resolving the cohesin-independent linkages observed at the rDNA during anaphase (D'Amours et al. 2004; Sullivan et al. 2004). Strikingly, this rDNA resolution step is temporally distinct from the Ipl1 kinase-dependent (and cohesin-independent) condensation that occurs during anaphase (D'Amours et al. 2004; Lavoie et al. 2004; Machin et al. 2005), suggesting that condensin has additional and possibly compaction-independent functions in mitosis.
While much emphasis has been placed on the molecular mechanism of cohesin-based cohesion, the recent description of cohesin-independent linkages at repetitive loci suggests that additional mechanisms for cohesion exist in both yeast and higher eukaryotes (D'Amours et al. 2004; Dynek and Smith 2004; Sullivan et al. 2004). Whether cohesin-independent interlinks exist at nonrepetitive DNA is an important question, particularly since in metazoans most cohesin complexes are removed from chromosomes long before the onset of anaphase (Losada et al. 1998; Waizenegger et al. 2000; Warren et al. 2000; Gimenez-Abian et al. 2004). Despite tremendous progress in identifying proteins required for cohesion and condensation, the molecular mechanisms underlying each of these essential cellular processes remain poorly understood. In this work, we describe a novel role for the condensin complex in sister chromatid cohesion. We show that during mitosis, condensin is required for sister chromatid cohesion at multiple loci on chromosome arms, but not at centromeric nor telomeric proximal loci. This cohesion is distinguishable from that deriving from the cohesin complex by several criteria. We discuss our results in the context of current models for sister chromatid cohesion.
Results
Condensin is required for sister chromatid cohesion
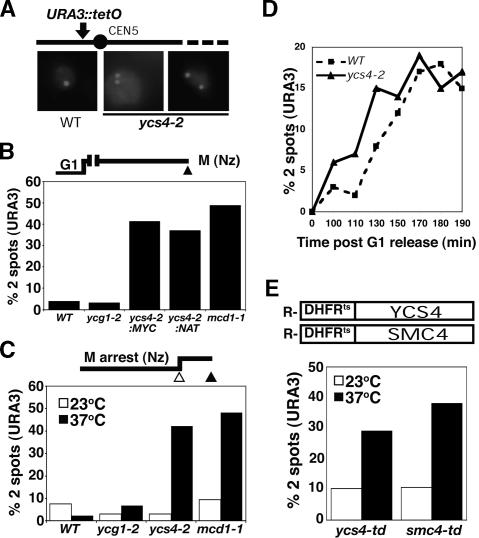
To address whether condensins play a role in sister chromatid pairing at nonrepetitive loci, we monitored mitotic cohesion in the absence of functional condensin complex using previously described temperature-sensitive (ts) alleles of two condensin subunits: ycg1-2 and ycs4-2 (Lavoie et al. 2004). A GFP-Tet repressor fusion protein bound to an array of Tet operators integrated at the URA3 locus on chromosome V was used to visualize sister chromatid cohesion in mitotic cells (Fig. 1A; Michaelis et al. 1997). Because sister cohesion exists from S phase through metaphase, each mitotic cell possesses one fluorescent signal, whereas cells with precociously separated sisters contain two signals (Guacci et al. 1994). We used two different experimental regimens to inactivate condensins during different phases of the cell cycle. In both regimens, exponentially growing cells were arrested in G1 phase using α-factor at the permissive temperature (23°C) to generate a uniform starting population. Cells were either shifted to the restrictive temperature immediately (i.e., from S through M phase) or allowed to proceed into a metaphase arrest by treatment with the microtubule-depolymerizing drug nocodazole prior to the temperature shift. The number of GFP spots per large budded cell was then determined (Fig. 1). In both cases, ycs4-2 cells showed a dramatic increase in precocious sister separation at the URA3 locus (37%–42%, two spots) (Fig. 1A–C). Similar levels of precocious separation were observed in two independent ycs4-2 strains (plus or minus a 12XMYC tag) as well as in the well-characterized cohesin mutant mcd1-1 (Guacci et al. 1997), which showed two spots in 49% of metaphase cells. In contrast, inactivation of another condensin subunit, Ycg1, caused little or no precocious sister separation (2%–4%, two spots). These data are in keeping with previous studies that showed that the abrogation of chromosome condensation per se does not induce precocious sister separation (Strunnikov et al. 1995; Freeman et al. 2000; Lavoie et al. 2000; Ouspenski et al. 2000; Bhalla et al. 2002). Since ycs4-2 cells traverse mitosis without triggering a checkpoint, it was possible to assess whether Ycs4 is required for URA3 cohesion in cycling cells (Fig. 1D). Cultures were synchronized in G1 at the permissive temperature, shifted to the restrictive temperature, and then released into the cell cycle at 37°C. While both wild-type and ycs4-2 cultures proceeded through the cell cycle at similar rates (as determined by flow cytometry and microscopy) (data not shown), precocious separation at the URA3 locus was observed in the ycs4-2 mutant, indicating that Ycs4 function is required for cohesion in cycling cells.
Figure 1.
Condensin is required for sister chromatid cohesion. (A) The GFP-TetR/TetO system for visualizing sister chromatids. The Tet operator (224 copies) are integrated at the URA3 locus, 35 kb upstream of CEN5. Representative cells containing one GFP spot (wild type) and two spots (ycs4-2 mutant) are shown. (B) Establishment of chromosome cohesion. Wild-type (WT) (YBL29-10), ycg1-2 (YBL44-9D), ycs4-2:MYC (YBL81-8B), ycs4-2:NAT (YBL146c), and mcd1-1 (YBL86-54D) cultures were synchronized in G1 at 23°C and allowed to proceed through S phase into an early mitotic arrest (M) at 37°C. The number of GFP signals per large budded cell was scored; ≥100 large budded cells were counted per sample. (C) Maintenance of chromosome cohesion. Strains, as in B excepting YBL146c, were synchronized in early M phase at 23°C before inactivation of mutants as indicated. Samples were taken before (23°C) and after (37°C) a 30-min temperature shift, fixed, and scored for GFP spots. (D) Precocious sister separation in cycling cells. YBL29-10 (wild type) and YBL146c (ycs4-2) cultures were synchronized in G1 phase with α-factor and released into fresh YPD medium containing 0.1 mg/mL pronase. The percent of cells with separated URA3 loci was monitored at various time points post-release. (E) Precocious sister separation in condensin-degron strains. Heat-inducible N-degron-tagged YCS4 (YBL310a) or SMC4 (YBL311) strains were synchronized in mitosis (nocodazole, Nz) at 23°C then shifted to the restrictive temperature (37°C) for 30 min to induce protein degradation. Sister chromatid cohesion was assessed at the URA3 locus.
The cohesion defect observed with ycs4-2 was a surprise since all condensin mutants tested to date caused death in mitosis due to chromosome condensation defects. Therefore, it was important to address whether the ycs4-2 mutation represents a true null or a gain-of-function allele. To do this, we generated ts degradable (degron) alleles (Dohmen et al. 1994; Labib et al. 2000) of two condensin subunits, Ycs4 and Smc4, and assayed sister chromatid cohesion following degradation of the degron-tagged proteins (Fig. 1E). Cultures were first grown at 23°C and synchronized in mitosis with nocodazole to allow the establishment of sister chromatid cohesion and condensation. Following a shift to 37°C for 30 min to induce protein degradation, cohesion was monitored at URA3. Comparable levels of precocious sister separation were observed (30%–40%, two spots) in both condensin mutants, indicating that (1) the ycs4-2 mutant is likely to be a null rather than a gain-of-function allele and (2) Ycs4 functions in the context of condensin, as both an SMC and non-SMC subunit similarly implicate condensin in sister chromatid cohesion.
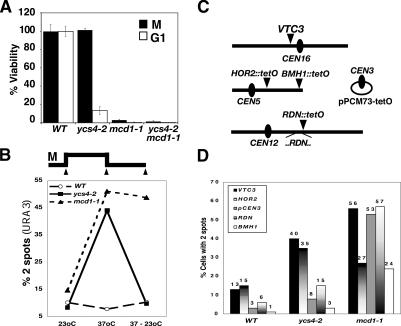
In cohesin mutants, a loss of sister chromatid cohesion in metaphase irreversibly compromises cell viability (Guacci et al. 1997; Michaelis et al. 1997; Uhlmann and Nasmyth 1998); therefore, we asked whether this would also be the case in the absence of Ycs4 function (Fig. 2A). G1-phase cells were shifted to the restrictive temperature, released, and rearrested in M phase to maximally disrupt cohesion, then an aliquot of cells was plated at 23°C to assess viability (Materials and Methods). The remaining culture was released from metaphase arrest at 37°C and allowed to proceed through mitosis to the subsequent G1 phase at 37°C, and was then plated at 23°C. As expected, when cells completed mitosis in the absence of cohesin or condensin function, rapid cell death ensued (Fig. 2A, open bars). Strikingly, however, when the ycs4-2 cells were returned to 23°C prior to the completion of mitosis, they remained viable—indicating that, in contrast to mcd1-1 cells, correct mitotic chromosome segregation occurred in these cells.
Figure 2.
Characterization of condensin-dependent cohesion. (A) Cell viability. G1-synchronized cultures of wild type (WT) (YBL29-10), ycs4-2 (YBL81-8B), mcd1-1 (YBL86-54D), and ycs4-2 mcd1-1 (YBL143a) were released at 37°C and split. One half was rearrested in M phase, sonicated, counted, and then plated at 23°C (M, black bars). The second half was allowed to proceed into the subsequent G1 at 37°C prior to plating (G1, open bars). Percent viability was determined as the number of viable colonies divided by the total number of cells plated (×100). Data were normalized to wild type. (B) Reversibility of Ycs4 cohesion in mitosis. Mitotically blocked (23°C) cultures of wild type (YBL29-10, open circles), ycs4-2 (YBL81-8B, closed squares), and mcd1-1 (YBL86-54D, closed circles) were shifted to 37°C to inactivate respective ts components, then brought back to the permissive temperature (37°C → 23°C). At each time point, the percent of large budded cells with two GFP spots was determined; ≥100 cells were scored per sample. (C) Schematic of probes used for cohesion assays. VTC3 is 40 kb from CEN16, HOR2::tetO is 129 kb from CEN5, and BMH1::tetO is 30 kb from TEL on ChrV. pPCM73-tetO is a reporter for CEN3 cohesion (Megee and Koshland 1999). RDN::tetO is as described (D'Amours et al. 2004). (D) Cohesion at multiple chromosomal loci. Synchronized mitotic cultures were shifted to 37°C to inactivate mutants and processed for fluorescence microscopy. Wild-type (YPH499), ycg1-2 (BLY07), ycs4-2 (ZW204), and mcd1-1 (VG955-7D) cells were processed for FISH (VTC3). Cohesion at other sites used a tetR-GFP reporter that binds to tetO arrays integrated at specific loci. HOR2::tetO.nat was integrated in wild-type (YBL196), ycs4-2 (YBL197a), and mcd1-1 (YBL198b) backgrounds. BMH1::tetO.URA3 was in wild-type (YBL125a), ycg1-2 (YBL132b), ycs4-2 (YBL126a), and mcd1-1 (YBL129a) strains. Centromeric cohesion was monitored using the pPCM73tetO plasmid in wild-type (2174-5C), ycg1-2 (YBL11-16C), ycs4-2 (YBL151-5C), and mcd1-1 (YBL150-2-14a) backgrounds. RDN::tetO were generated by integrating tet operators on the telomeric side of the rDNA array on chromosome XII in various backgrounds (wild type, YBL371a; ycs4-2, YBL372a; mcd1-1, YBL373a). Percent of large budded cells with two fluorescent signals was determined at each loci as indicated; ≥100 cells were scored per sample.
Condensin-dependent cohesion is reversible in M phase
Two mechanisms could account for the preservation of cell viability in ycs4-2 cells. First, sister cohesion could be re-established during mitosis, similar to the restoration of condensation observed in condensin mutants (Lavoie et al. 2002). Second, condensin could mediate cohesion at a restricted subset of loci. To test these possibilities, cohesion at URA3 was disrupted in the presence of nocodazole by a temperature shift, after which the cultures were returned to the permissive temperature for 0.5 h (Fig. 2B). Under these conditions, 44% of ycs4-2 cells had two URA3 spots at 37°C, versus 10% after recovery at 23°C, which is similar to background (8%). Cells remained arrested in metaphase and had not proceeded into telophase or the next G1 phase of the cell cycle as judged by Pds1 nuclear staining and nuclear morphology (data not shown). In contrast, the defect in cohesion in mcd1-1 cells remains after return to 23°C, consistent with previous observations that the loss of cohesin-based pairing is irreversible in mitotic cells (Guacci et al. 1997; Uhlmann and Nasmyth 1998; Lavoie et al. 2002). From these experiments we draw two conclusions: First, condensin-dependent cohesion is reversible in mitosis and, second, it can be restored in the absence of DNA replication.
Ycs4 is not required for cohesion at centromeric or telomeric loci
Given that cohesion can be restored in ycs4-2 cells, we asked how sister loci find each other. One simple mechanism would be that some pairing between sister chromatids remains intact, thus maintaining chromatid alignment. To address this, we assessed the requirement for condensin for maintaining cohesion at other chromosomal loci: two arm loci (VTC3, 30 kb from CEN16 and HOR2, 129 kb from CEN5), as well as centromeric (CEN3) and telomeric regions (BHM1) (Fig. 2C,D). Cells were arrested in M phase and shifted to 37°C to inactivate condensin or cohesin components. Cohesion at VTC3 was examined by fluorescence in situ hybridization (FISH) using a probe that hybridizes near the VTC3 locus, and the number of fluorescent signals within each cell nucleus was scored as described previously (Guacci et al. 1994). Cohesion at HOR2 was monitored using the GFP-TetR/TetO system as before, where the Tet operators were integrated at HOR2. Inactivation of ycs4-2 showed ∼40% precocious sister separation at both the VTC3 and HOR2 arm loci, indicating that precocious sister separation occurs at multiple loci and does not result from perturbations of chromosome structure incurred by the integration of the large Tet operator array. To monitor centromeric cohesion under conditions that minimize contributions of arm cohesion, we used a reporter plasmid (pPCM73-tetO) that is dependent on its centromere for cohesion (Megee and Koshland 1999). pPCM73-tetO, which contains CEN3 sequences and an array of Tet operators, was transformed into strains expressing the TetR-GFP fusion construct. Cohesion near telomeres was monitored using tet operators integrated at BMH1 (30 kb from ChrV TEL) (Alexandru et al. 2001). Contrary to what we observed for arm sites, cohesion at both CEN3 and BMH1 was maintained in the absence of Ycs4 (Fig. 2D) while, as expected, cohesin complex disruption (mcd1-1) caused precocious sister separation at all loci tested. Thus condensin-dependent cohesion, unlike cohesin complex-dependent pairing, is specific to chromosomal arm sites.
Condensin-based cohesion is distinguishable from cohesin-mediated pairing
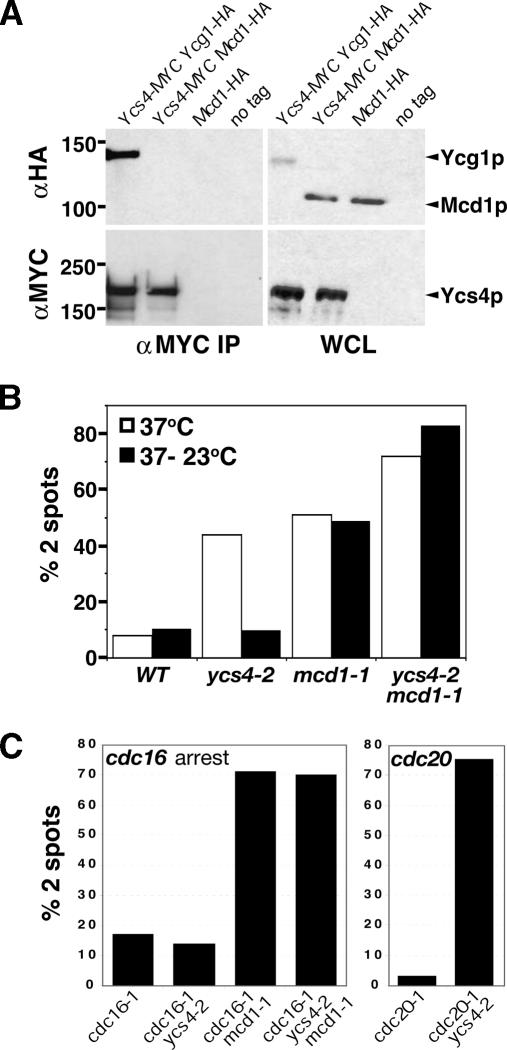
Condensin could modulate sister chromatid cohesion through the known cohesin machinery or through a distinct, cohesin-independent mechanism. We first asked whether Ycs4 and cohesin interact in cell extracts. While Ycs4 efficiently coimmunoprecipitates the condensin subunit Ycg1, no stable association with cohesin complex (Mcd1) could be detected (Fig. 3A). Consistent with this observation, no detectable change was observed for the bulk chromatin association of cohesin, as judged by chromatin spreads using an epitope-tagged Mcd1-HA6p in a ycs4-2 background, and vice versa (data not shown). These data are consistent with previous studies and suggest that cohesins and condensins do not stably interact (Strunnikov et al. 1995; Toth et al. 1999; Bhalla et al. 2002).
Figure 3.
Distinct requirements for Mcd1 and Ycs4 in mitotic cohesion. (A) Ycs4-MYC12 coimmunoprecipitation. YCG1-HA (YBL37-4c) and MCD1-HA (YBL96-1) strains containing a 12XMYC tag integrated at endogenous YCS4, and MCD1-HA (YBL95-1), and wild-type (YPH499) strains containing untagged YCS4 were used to immunoprecipitate condensin (Ycg1) and/or cohesin (Mcd1) subunits. Western blots are shown. (B) Genetic analysis. Cohesion defects at the URA3 locus in wild type (WT) (YBL29-10), ycs4-2 (YBL81-8B), and mcd1-1 (YBL86-54D), vs. a ycs4-2 mcd1-1 dm (YBL143a). Samples were taken after inactivation (37°C) and recovery (37°C → 23°C) of ts mutants. The number of GFP signals per large budded cell was scored. (C) Cohesion defects in cdc16-1 and cdc20-1-arrested cells (URA3). cdc16-1 (YBL251-7-11B), cdc16-1 ycs4-2 (YBL249-3-10b), cdc16-1 mcd1-1 (YBL250-5-6a), cdc16-1 ycs4-2 mcd1-1 (YBL252), cdc20-1 (YBL388-18d), and cdc20-1 ycs4-2 (YBL388-11c) strains were arrested in G2/M phase by temperature shift (3 h at 37°C) and the percent of cells with two GFP spots was determined. In all experiments, ≥100 large budded cells were scored per sample.
If condensin and cohesin promote cohesion through independent mechanisms, then two simple predictions should be fulfilled. First, a condensin–cohesin double mutant (dm) should grow robustly at 23°C but exhibit additive cohesion defects at 37°C. Second, each mechanism should be separately regulated during the cell cycle. As predicted, no synthetic growth defects were observed in ycs4-2 mcd1-1 cells at 23°C, in contrast to the synthetic lethality or sickness observed in every condensin–condensin or cohesin–cohesin dm we constructed (data not shown). More importantly, ycs4-2 mcd1-1 dm cells showed an enhanced cohesion defect at 37°C, where 72%–85% of large budded cells possessed two URA3 spots, in comparison with 44% in ycs4-2 (37°C) and 50% in mcd1-1 (Fig. 3B). These data suggest that cohesins and condensins both contribute to robust arm cohesion, and that the reversibility of condensin-based pairing is dependent on the close proximity of sisters maintained by residual cohesin.
To uncouple cohesin and condensin function during mitosis, we next made use of a conditional mutant in the essential APC subunit CDC16. The cdc16-1 mutant causes a cell cycle arrest in G2/M phase, after the establishment of sister chromatid cohesion but before full chromosome condensation (Guacci et al. 1994). As expected, mcd1-1 cdc16-1 cells showed precocious sister separation at URA3 in 71% of large budded cells (Fig. 3C). This increase over the ∼50% two spots consistently obser ved in nocodazole-arrested cells likely results from the spindle tension in cdc16 cells and is reflected by the localization of marked loci at opposite ends of the nucleus (data not shown). In contrast, only background levels of sister separation were observed in ycs4-2 cdc16-1 cells (14% vs. 17% in wild type), unlike the 75% detected in a ycs4-2 cdc20-1 background where chromosomes are fully condensed (Guacci et al. 1994). Taken together, our results suggest that while early in mitosis cohesin appears predominantly responsible for cohesion, robust arm cohesion requires condensin as the cells approach the metaphase-to-anaphase transition.
Condensin and cohesin bind chromatin independently
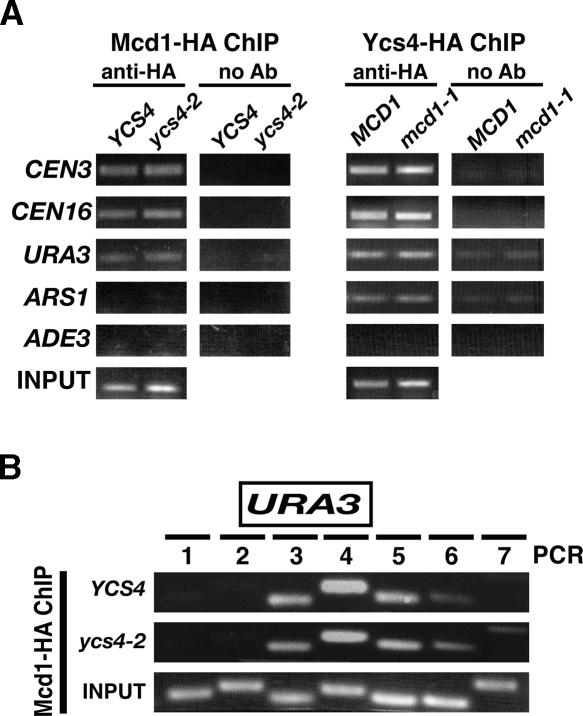
Previous work has shown that the bulk chromatin binding of cohesin and condensin are not interdependent (Strunnikov et al. 1995; Toth et al. 1999; Bhalla et al. 2002). To test whether condensin could affect local cohesin binding at loci where both are required for cohesion, we examined Mcd1 protein association at URA3. Chromatin immunoprecipitation (ChIP) was used to localize both Mcd1 and Ycs4 on mitotic chromosomes in vivo (Fig. 4). Wild-type and ycs4-2 mutant strains containing an epitope-tagged Mcd1 (MCD1-HA6) were arrested in M phase (nocodazole) at 37°C, followed by ChIP with anti-HA antibodies and PCR to detect enrichment of specific chromosomal DNA in the pellet fraction. As expected, Mcd1 was enriched at CEN3, CEN16, and URA3 sequences, but not at ARS1 or ADE3 (Megee et al. 1999; Glynn et al. 2004; Lengronne et al. 2004). Significantly, no change in Mcd1 binding was detected in the absence of condensin function at all loci tested, including URA3 where sister separation is observed (Fig. 4A). These data suggest that the loss of cohesion in ycs4-2 cells occurs despite the presence of cohesin. We next used ChIP to assess Ycs4 binding at these same loci (Fig. 4A). As predicted from its requirement for sister chromatid cohesion, Ycs4 was detected at the URA3 locus. In addition, Ycs4 was detected at CEN3, CEN16, and ARS1 but not at the ADE3 locus. This chromatin binding was independent of cohesin function, as no change in the IP signal was detected between MCD1 and mcd1-1 samples. Furthermore, the binding of Ycs4 at ARS1, which was not observed for Mcd1, indicates that the two proteins are likely to be enriched at different sites in mitotic chromatin, as recently indicated by genomic ChIP–chip studies (Wang et al. 2005).
Figure 4.
ChIP analysis of Ycs4 and cohesin at multiple loci. (A, left) ChIP of Mcd1-HA6 at indicated loci in the presence or absence of condensin function in YCS4 (YBL166-14D) vs. ycs4-2 (YBL169-9D) cells. (Right) ChIP of Ycs4-HA6 in MCD1 (YBL244) vs. mcd1-1 (YBL245c) strains. M-phase cultures where shifted to 37°C to inactivate mutants before proceeding with ChIP. PCR was used to detect protein association at indicated sequences. A no-antibody control is shown for each experiment. Input is the chromatin solution used to perform the ChIP. PCR reactions from each input chromatin solution at representative chromosome loci are as follows: ARS1 (Mcd1-HA ChIP, αHA), CEN16 (Mcd1-HA ChIP, no Ab), ADE3 (Ycs4-HA ChIP, αHA), and URA3 (Ycs4-HA ChIP, no Ab). (B) Chromatin-binding profiles at URA3. Seven PCR products designed to span the URA3 locus are shown. Mcd1-HA ChIP was in the presence (YCS4) or absence (ycs4-2) of functional Ycs4 as in A.
One possible explanation for the loss of cohesion at URA3 in ycs4-2 cells is that the distribution of Mcd1p could change within this locus, similar to that observed upon passage of the transcription machinery (Lengronne et al. 2004). To address this, a series of PCR primer pairs spanning the URA3 ORF were used to map Mcd1 association over the entire region (Fig. 4B). Mcd1-binding profiles in the presence (YCS4) and absence (ycs4-2) of condensin were indistinguishable: Mcd1 associates over an ∼1-kb region at URA3 (PCR 3–5). Therefore, while both Mcd1 and Ycs4 are detected at URA3, the premature separation observed in ycs4-2 cells does not result from any detectable loss or redistribution of cohesins.
Mcd1 levels at URA3 are independent of cohesion
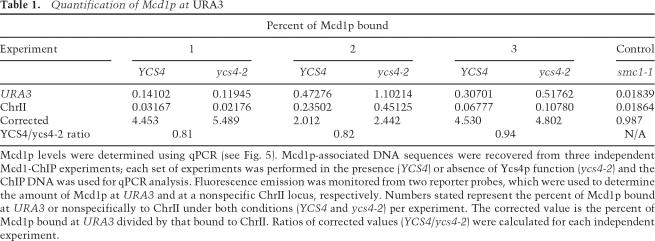
One concern from our ChIP assays is that a partial loss of cohesin complex might not be detectable within a cell population. Since 40%–50% of cells exhibit detectable sister separation at URA3 in the absence of Ycs4 (by microscopy), the signal observed by Mcd1-HA ChIP could result from cells within the population with intact cohesion. To quantitatively assess Mcd1 levels at paired (YCS4) versus separated (ycs4-2) URA3 loci, we performed quantitative real-time PCR (qPCR) on Mcd1bound DNA. Figure 5 shows the percent of Mcd1p bound either at URA3 or on ChrII (negative control) in the presence (YCS4) or absence of cohesion (ycs4-2 and smc1-1). In both YCS4 and ycs4-2 conditions, the amount of Mcd1 protein associated at URA3 (Fig. 5B, black bars) is approximately five times greater than that at the ChrII control locus (Fig. 5B, open bars). In contrast, disruption of the cohesin complex with smc1-1 resulted in only background levels of Mcd1 at URA3. Importantly, while ∼40%–50% sister separation is observed at URA3 upon ycs4-2 inactivation, no decrease in Mcd1 binding could be detected. Rather, the average ratio of Mcd1 at paired versus unpaired URA3 sites approached 1 (YCS4/ycs42 = 0.86 ± 0.06) (Table 1). Conservatively estimating that 30%–40% of the cells in the total population have unpaired URA3 loci (40% separated URA3 × 80% large budded cells), then the loss of Mcd1 would yield a ratio of >1.5 and possibly much larger, depending on the number of cohesins per URA3 site. The simplest interpretation then is that cohesin complexes mediate sister chromatid pairing through a mechanism that allows sisters to separate transiently in mitosis without a local loss in cohesin complex (Fig. 5C).
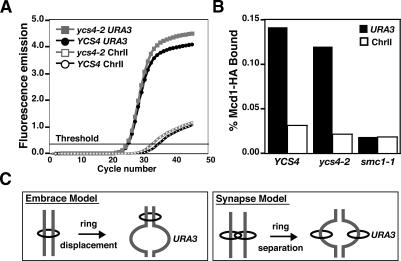
Figure 5.
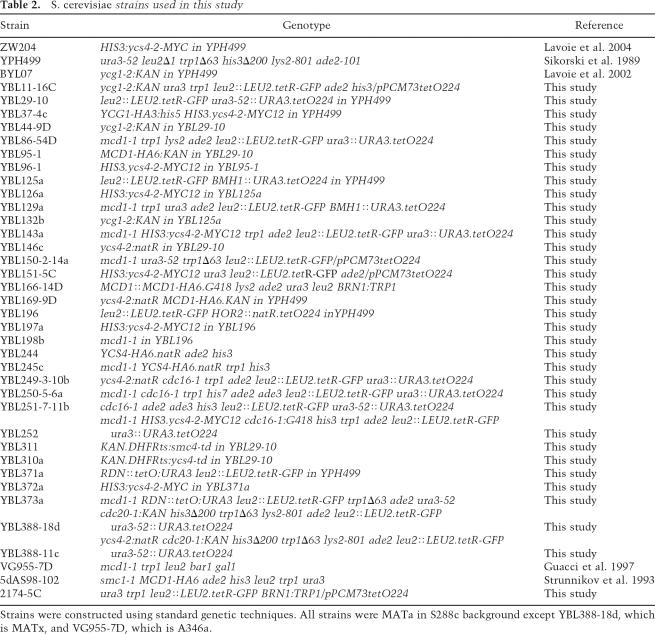
Quantitative determination of Mcd1 binding at URA3. (A) qPCR amplification plots. Curves show the accumulation of the locus-specific PCR products amplified from DNA pelleted from Mcd1-HA ChIP, performed in the presence (YCS4, black) or absence (ycs4-2, gray) of Ycs4 function, represented as a function of fluorescence over a total of 45 PCR cycles. Two fluorescent reporter probes were used. The URA3 probe monitors Mcd1 binding specifically at the URA3 locus (closed markers), while ChrII is a negative internal control probe that hybridizes to a Mcd1-free region, thereby detecting the nonspecific DNA (open markers). Threshold denotes the background noise in the system (see Materials and Methods). (B) Quantification of Mcd1 levels at URA3. Percent of Mcd1 bound to URA3 (black bars) and to the ChrII nonspecific site (open bars) were calculated from Mcd1-ChIP DNA done in YCS4 (YBL166-14D), ycs4-2 (YBL169-9D), and smc1-1 (5dAS98-102) cells. (C) Models for sister chromatid cohesion by cohesin. In the embrace model, cohesin complex (black ring) must be either cleaved or displaced in order for sister separation at URA3 to occur. Conversely, synapsis models allow for transient sister separation without the loss of cohesins at the URA3 locus.
Table 1.
Quantification of Mcd1p at URA3
Discussion
Condensin is required for arm cohesion
We show that the condensin complex is required for sister chromatid cohesion in budding yeast. Cells bearing null alleles of either YCS4 or SMC4, two condensin subunits, prematurely lose arm cohesion in metaphase-arrested cells at levels similar to those observed in the well-characterized cohesin mutant mcd1-1/scc1-73 (Guacci et al. 1997; Michaelis et al. 1997). This requirement was observed at multiple arm loci on different chromosomes (URA3, HOR2, and VTC3) and does not derive from the mitotic arrest, as ycs4-2 cells lose arm cohesion precociously in cycling cells. These data are the first to implicate condensin in sister chromatid cohesion in any organism. The loss of condensation per se cannot account for the precocious arm separation observed, since previously characterized alleles of condensin that abrogate condensation did not exhibit cohesion defects (Freeman et al. 2000; Lavoie et al. 2000, 2002; Ouspenski et al. 2000; Bhalla et al. 2002). Rather, we propose that condensin performs separable functions in both mitotic chromosome condensation and sister chromatid cohesion. How condensin mediates arm pairing remains unknown. One simple model is that condensin could promote both intermolecular and intramolecular interactions, which would result in condensation and cohesion, respectively. Consistent with this, condensin's role in cohesion can only be discerned during mitosis, when condensin is competent for condensation. Alternatively, condensin could impinge upon other essential aspects of mitotic chromosome cohesion, such as the cohesion maintenance machinery including PDS5 and SMT4 (see Hartman et al. 2000; Panizza et al. 2000; Bachant et al. 2002; Stead et al. 2003, and references therein).
Distinct mechanisms for chromosome arm cohesion
One possible model for condensin's involvement in sister chromatid cohesion is that it could modulate cohesin function at specific loci by perturbing the cohesin complex. Condensin-dependent cohesion can, however, be distinguished from cohesin-dependent pairing by a number of criteria. First, a role for condensin in sister cohesion could only be discerned along chromosome arms, and not centromeric or telomeric regions (this study), while the cohesin complex is required along the entire chromosome length (Guacci et al. 1997; Michaelis et al. 1997; Megee and Koshland 1999). Interestingly, condensin and cohesin both appeared equally important for cohesion at the URA3 locus, whereas condensin had a more pronounced effect at HOR2 (40% two spots in ycs4-2 vs. 25% in mcd1-1). These data suggest that condensinand cohesin-based cohesion mechanisms contribute differentially at distinct loci. Second, condensindependent cohesion, like mitotic chromosome condensation, is reversible in mitosis. This reversibility requires intact cohesin, however, presumably to maintain sister chromatid alignment via centromeric and telomeric sequences. In contrast, the inactivation of cohesin causes an irreversible defect in mitotic SCC, even when wild-type protein is provided (Uhlmann and Nasmyth 1998). The ability to generate condensin-dependent cohesion during M phase, independently of DNA replication, clearly distinguishes it from cohesin complex-based cohesion and supports a model where two distinct mechanisms collaborate to ensure robust pairing at arm loci. Consistent with this, additive cohesion defects were observed in a cohesin–condensin dm (85% two spots at URA3), indicating that maximal separation of arm loci in mitotic cells lacking spindle tension requires the inactivation of both mitotic mechanisms. Finally, the binding of the cohesin and condensin were not interdependent (Figs. 4, 5). At the URA3 locus, where both Ycs4 and cohesin function, no change in either Mcd1 levels or distribution was observed in the absence of condensin, and vice versa (this study). Since condensin and cohesin neither stably interact biochemically nor precisely colocalize on DNA (Strunnikov et al. 1995; Losada et al. 1998; Toth et al. 1999; Bhalla et al. 2002), it is likely that they function independently as well.
Cell cycle regulation of condensin-dependent linkages
We propose that at the onset of mitosis, condensin mediates cohesin-independent interlinks along chromosome arms. Cohesin-independent linkages have been previously described at heterochromatin-like loci such as centromeres, telomeres, and the repetitive rDNA (Bachant et al. 2002; D'Amours et al. 2004; Dynek and Smith 2004; Sullivan et al. 2004), and together these data indicate that cohesin-independent cohesion is widespread in the genome. While the mechanism of pairing is unknown at repetitive regions, our data indicate condensin is responsible for cohesin-independent interlinks along chromosome arms. Why, then, do cells require multiple cohesion mechanisms? One reason may be related to the cell cycle regulation of higher-order chromosome structure. In metazoans, cohesin is largely removed from chromosomes at the end of prophase, as chromosome condensation is initiated (Losada et al. 1998; Sumara et al. 2000, 2002; Waizenegger et al. 2000; Hauf et al. 2005). In yeast, the preanaphase regulation of cohesin function by Cdc5 (Alexandru et al. 2001) could necessitate additional cohesion mechanisms to maintain robust arm cohesion. Consistent with this, chromatin-bound cohesin levels decrease from S phase to metaphase in budding yeast (Hartman et al. 2000).
The existence of additional cohesion mechanisms, however, poses an additional challenge for the timely segregation of chromosomes in mitosis. At anaphase onset, Esp1-mediated proteolysis of cohesin is required for efficient chromosome segregation (Uhlmann et al. 1999). Given that Esp1 overexpression is sufficient for genome segregation in ∼20% of metaphase-arrested cells (Ciosk et al. 1998), it is tempting to speculate that condensin-based cohesion may also be directly or indirectly under separase control. The precise mechanism for dissolution of condensin-dependent arm cohesion remains to be determined, however, as condensin is not known to be proteolytically degraded during the metaphase-to-anaphase transition, and indeed remains functional during anaphase. One simple mechanism could involve the relocalization of condensin from arm to repetitive loci during anaphase, thereby releasing arm cohesion. In budding yeast, condensins are known to accumulate at the rDNA locus on ChrXII during anaphase (Freeman et al. 2000; Bhalla et al. 2002; D'Amours et al. 2004; Sullivan et al. 2004; Wang et al. 2004) where they promote the resolution, compaction, and segregation of the rDNA (D'Amours et al. 2004; Lavoie et al. 2004; Sullivan et al. 2004; Wang et al. 2004; Machin et al. 2005). At the 1-Mb RDN locus, condensin promotes the anaphase dissolution of as-yet ill-defined cohesin-independent interlinks (D'Amours et al. 2004; Sullivan et al. 2004; Wang et al. 2004). While it may seem counterintuitive that condensin could function in both promoting cohesion at arm loci during metaphase and resolving interlinks at repetitive loci in anaphase, it is noteworthy that these two events are temporally distinct and coincide with the onset of multiple mechanisms of condensin modification such as phosophorylation by the Ipl1/kinase (Lavoie et al. 2004), sumoylation (D'Amours et al. 2004), and the CDC14/FEAR pathway (D'Amours et al. 2004; Sullivan et al. 2004; Wang et al. 2004). Thus it is also plausible that changes in condensin's biochemical properties could underlie the transition between its pairing and resolution functions. Alternatively, condensin-dependent pairing could require additional factors (perhaps even specific to chromosome arms) that are themselves subject to regulation/degradation at the metaphase-to-anaphase transition. Further experiments will be required to distinguish between these possibilities.
Physical association of cohesin with chromosomes: rings versus snaps
Our observation that cohesins remain associated with separated loci has important implications for current models of cohesin function. Two types of cohesion models have been proposed, and each makes distinct predictions for cohesin localization when sisters are separated (Fig. 5C). The embrace model (Haering et al. 2002; Nasmyth and Haering 2005) predicts that a stable ∼40-nm ring of cohesin entraps both sister chromatids and must be displaced by sliding or cleavage to allow sister separation. In contrast, snap or bracelet models posit that intermolecular contacts between cohesin complexes could mediate cohesion (Huang et al. 2005; Skibbens 2005). In this scenario, precocious separation would occur by disrupting intermolecular cohesin contacts without necessarily causing dissociation of cohesin from chromatin. In this work we show that cohesin levels and distribution are unchanged at the URA3 arm locus regardless of whether the locus is paired or unpaired (in ycs4-2). Assuming that cohesin and condensin do not interact, then precocious sister separation along chromosome arms occurs in the presence of a presumably established cohesin complex, both in the absence of the mitotic spindle and in cycling cells. While transient and localized sister separation has previously been observed at or near centromeres under tension (He et al. 2000; Goshima and Yanagida 2001; Bachant et al. 2002), the mechanism is likely to be different than in condensin mutants since sister centromeres reassociated once tension was removed.
Together, these data pose interesting questions about how cohesin complex mediates transient sister separation. Clearly, the embrace model in which a single ∼40-nm cohesin ring entraps both sisters (Haering et al. 2002) is difficult to reconcile with the presence of cohesin at separated loci. It may be that cohesin rings open transiently, allowing one chromatid to escape but without dissociating from the other sister. The recent demonstration that cohesins lack chromatin-binding activity following ring opening argues against this model, however, especially given that no single domain within cohesin appears responsible for its stable association (Nasmyth and Haering 2005). Additional models posit that cohesin complexes bound to each sister chromatid could mediate cohesion through intercomplex interactions, possibly involving cohesion maintenance factors (Huang et al. 2005; Skibbens 2005). This type of mechanism would most easily explain sister separation in the presence of cohesin; however, experimental evidence for cohesin multimers has remained elusive (Ivanov and Nasmyth 2005). While currently available data cannot unambiguously distinguish between embrace and synapsis models, our observations provide additional experimental constraints in drafting future models of cohesin function.
Materials and methods
Reagents and media
Strains were grown in YPD media (1% yeast extract, 2% peptone, 2% dextrose ± 2% agar) (Guthrie and Fink 1991) with 10 mg/mL adenine. Yeast extract, peptone, agar, α-factor, and G418 were from Bioshop CA; dextrose was from EMD Pharmaceuticals; DAPI, pronase E, and nocodazole were from Sigma-Aldrich and clonNAT Werner BioAgents. Oligos were from IDT or Applied Biosystems (TaqMan probes and qPCR kit). Restriction enzymes and Taq DNA polymerase were from NEB. DIG-labeled FISH probes (VTC3 locus; CEN16 proximal Probe1) were as in Guacci et al. (1994). The antibodies used were mouse α-DIG, swine α-goat-FITC conjugate, mouse α-HA mAb 12CA5 (Roche); goat α-mouse-FITC, goat α-mouse-HRP (Jackson ImmunoResearch); and mouse α-MYC mAb 9E10 (Babco). Protein A Sepharose CL-4B beads (AmershamPharmacia) were swelled in water (0.2-g beads in 15 mL) for 2 h at room temperature, washed, and kept at 4°C as a 50% slurry in storage buffer (0.1% azide in TE at pH 8.0).
Yeast strain construction
Yeast strains (Table 2) were constructed using standard techniques (Guthrie and Fink 1991). Ts condensin alleles are as in Lavoie et al. (2002). To generate GFP reporter strains, tet operator arrays (224 copies) were integrated at specific loci: URA3 using pRS306-tetO224 (Michaelis et al. 1997), BMH1 with K3527 (Ciosk et al. 1998), and HOR2 with pWL9 in strains expressing tetR-GFP fusions (at LEU2). pWL9 was pFA6a-NATMX4 (Wach et al. 1994) into which both an 800-base-pair (bp) fragment of HOR2 (+SpeI linkers) and the 10.4-kb tetO array (BglII–BamHI) were cloned. MCD1-HA6-tagged strains were made by integrating NcoI-cut pPCM87 (P. Megee, University of Colorado Health Sciences Center, Aurora, CO) at URA3 or, alternatively, cotransforming a 1.9-kb mcd1-HA6 fragment (EcoRI–XbaI) along with the kanR gene (pFA6-kanMX4) containing 24-bp homology with HA and 50-bp homology with MCD1 3′ untranslated region. YCS4-HA6:natR-tagged strains were created by integrating a 7.1-kb fragment of pWL15 (HindIII–SpeI), a YCS4-HA6:natR construct in YCplac111. N-degron-tagged (Dohmen et al. 1994) YCS4 and SMC4 strains were created by integration of a KanMX–URA3pr–Ub-DHFRts cassette to replace the endogenous ATG of each gene.
Table 2.
S. cerevisiae strains used in this study
Cell culture, synchronization, and viability
Permissive and restrictive temperatures were 23°C and 37°C, respectively. Synchrony conditions were as described (Lavoie et al. 2002, 2004). Briefly, G1 or M arrests required 3 μM α-factor or 15–20 μg/mL nocodazole, respectively, for 3 h at 23°C. Arrests were monitored by microscopy, with 75%–90% of arrests being typical. For temperature shift experiments, G1 cells were shifted to 37°C for 30 min prior to their release into warmed medium containing 0.4 mg/mL pronase E plus nocodazole for 2.5–3 h. For cell viability experiments, following the inactivation regimen (S–M vs. S–G1 at 37°C), sonicated cells were counted in a hemocytometer and viability was determined after plating to rich media. Percent viability was calculated as the number of colony-forming units divided by the number of cells plated. Data were normalized to wild type.
Sister chromatid cohesion assays
G1-phase cultures (23°C) were released into a mitotic block induced by nocodazole (23°C or 37°C) or the cdc16-1 mutation (3hat37°C). For cells arrested at the permissive temperature, ts alleles were then inactivated by shifting to 37°C for at least 0.5 h. Cells were processed for GFP: One milliliter of culture was resuspended in 0.1 vol 4% paraformaldehyde (in 3.4% sucrose) and fixed for 15 min at room temperature, washed and resuspended in 0.4 mL of sorbitol buffer (2 M sorbitol/KHPO4 at pH 6.25), and sonicated briefly. The number of GFP spots per nucleus was determined by fluorescence microscopy using a Nikon Eclipse E1000 microscope (1000× magnification). Images were captured using a Hamamatsu ORCA-ER digital camera using the GFP filter (FITC-HYO, Nikon) with Nikon Simple PCI software (version 5.1.0.0110). Nuclear morphology was assessed by DAPI staining (45 μg/mL DAPI with 1 mg/mL phenylenediamine in 10% PBS/90% Glycerol) using the Nikon UV2A filter. FISH was performed exactly as described previously (Guacci et al. 1994; Lavoie et al. 2004). The DIG-labeled VTC3 probe hybridizing to a region 11–28 kb upstream of CEN16 was detected by IIF after sequential incubations with mouse α-DIG antibodies, goat α-mouse-FITC, and swine α-goat FITC antibodies.
ChIP
ChIP was as in Meluh and Koshland (1997) and Megee et al. (1999), with the following modifications: One-hundred-microliter cultures (YBL166-14D, YBL169-9D, YBL244, and/or YBL245c) OD600 ∼ 0.5 were arrested in M phase with 20 μg/mL nocodazole at 23°C for 3 h, then shifted to 37°C for 0.5 h. Crosslinking was with 1% formaldehyde for 2 h at 23°C. Harvested and washed cells (2 × 5 mL cold PBS) were transferred to 2-mL screw-cap tubes, and pellets were frozen in a dry ice/ethanol bath. Frozen cells were lysed by vigorously bead beating for 12–15 min of alternating 30-sec on/off cycles in 500 μL of lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl at pH 8.0 with protease inhibitors: 0.5 mM PMSF, 0.8 μL/mL pepstatin, 0.6 μg/mL leupeptin) with 500 μL of chilled 0.5-mm glass beads (Biospec Products) at 4°C using a Biospec Mini Bead Beater 8 (model #693) at the “homogenize” power setting. Whole-cell extracts were recovered and sonicated on ice for 20 × 30-sec on/off intervals on a Branson Sonifier 450 with a 0.125-in-diameter microtip at output setting 3, which gave 500–1500-bp DNA fragments. Lysates were spun at 15K rpm for 10 min at 4°C to obtain the soluble chromatin solution, which was diluted 1/10 in ice-cold IP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl at pH 8.0, 167 mM NaCl with protease inhibitors). ChIP reactions used 1.5 mL of the diluted chromatin solution with 2 μg of 12CA5 mouse α-HA antibody overnight at 4°C on a rotating wheel. Mock no-antibody IPs were also performed to assess nonspecific chromatin enrichment in the IPs. Immune complexes were recovered by adding 40 μL of 50% Sepharose-protein A slurry (3 h at 4°C). Beads were sequentially washed twice in 1 mL TSE-150 wash (0.1% SDS, 1% Triton X-100, 2 mM EDTA at pH 8.0, 20 mM Tris-HCl at pH 8.0, 150 mM NaCl), once in 1 mL of LiCl/ detergent wash (0.25 M LiCl, 1% NP-40, 1% deoxycholate [DOC], 1 mM EDTA at pH 8.0, 10 mM Tris-HCl at pH 8.0), and twice in 1 mL of TE (pH 8.0) for 3–5 min each at 4°C on rotating platform. During the second TE wash, the beads were transferred into a new microcentrifuge tube, and IP material was eluted twice with 250 μL of elution buffer (1% SDS, 0.1 M Na2CO3). The eluate, as well as a 300-μL aliquot of the input chromatin solution, were incubated at 65°C for 5 h to reverse cross-links, followed by treatment of 1.5 μL of proteinase K (15.3 mg/mL stock) for 2 h at 42°C. Organic extractions (for ChIP DNA, 300 μL of PCI [25:24:1], and 300 μL of CHCl3; for input chromatin, two with 400 μL of PCI and two with 400 μL of CHCl3) were performed. DNA was ethanol-precipitated and resuspended in 50 μL (ChIP) or 300 μL (input DNA solution) of TE.
ChIP DNA sequences were detected by PCR. Oligos (20–23 mers) were chosen to amplify the following genomic fragments (coordinates are SGD): CEN3 (ChrIII 114318–114561), CEN16 (ChrXVI 555847–556192), URA3 (ChrV 116617–116958), ARS1 (ChrIV 462831–463159), and ADE3 (ChrVII 906018–906323). Tiling of the URA3 locus in seven fragments corresponding to #1: 115431–115718; #2: 115860–116208; #3: 116346–116606; #4: 116617–116958; #5: 117051–117342; #6: 117372–117671; and #7: 117776–118246. For qPCR, the URA3 fragment amplified was ChrV 116897–116962 with the TaqMan probe hybridizing to 116940–116927; for ChrII (418224–418285), the TaqMan probe was ChrII 418247–418262.
PCR and qPCR
ChIP PCR reactions (29 cycles) were performed using 1 μL of ChIP DNA (equivalent to 30 μL of input chromatin solution) or 1 μL of input DNA (diluted 1/300 relative to ChIP DNA) in 15 μL of final volume in 1× PCR buffer (50 mM KCl, 10 mM TrisHCl at pH 9.0, 0.1% Triton X-100), 1.5 mM MgCl2, 0.2 mM dNTPs, and 0.05 U Taq. Annealing temperatures were optimized for each primer set. ChIP experiments were done at least three times. qPCR reactions were carried out using 2 mL of Mcd1-ChIP DNA template per reaction at the following cycling conditions: 15 min at 95°C (15 sec at 95°C, 90 sec at 56°C) for 45 cycles. The final volume of each qPCR reaction was 25 mL (1× PCR A buffer, 3.5 mM MgCl2, 200 mM dNTPs, 300 nM forward and reverse primers, 100–150 nM fluorescent probe, 0.625 U AmpliGold Taq polymerase). Quantification was done using the Applied Biosystems’ 7500 real-time PCR system as described in Costanzo et al. (2004). TaqMan chemistry probes were from Applied Biosystems and contained a 5′ fluorescent probe + 3′ black hole quencher. Levels of Mcd1p at URA3 versus a control region of ChrII were determined using two probes: URA3, which had a 5′ 6-carboxyfluorescein reporter (FAM), and ChrII, which used VIC. Fluorescence emitted upon removal of the 3′ quencher is dependent on the amount of PCR product and was detected at the end of each PCR cycle, up to a total of 45 cycles. Amount of PCR product in unknowns was determined based on the Ct value, defined by the cycle at which the fluorescence signal rises above background noise (threshold) (Fig. 5B), relative to standard curves generated for each probe in each experiment. Quantities stated in Table 1 were determined using three independent Mcd1-HA ChIP experiments. The percent of Mcd1p bound was calculated as the ratio of ChIP DNA versus input DNA. Nonspecific enrichment in the ChIP was corrected by dividing the amount of DNA by that obtained from the ChrII probe (Table 1, Corrected).
Acknowledgments
We are indebted to Lori Frappier, Andy Wilde, Bob Skibbens, and Vinny Guacci for critical reading of the manuscript, Mike Costanzo and Brenda Andrews for help with real-time PCR, and Paul Megee for advice with ChIP. This work was supported by a CIHR operating and salary grants to B.D.L.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1468806.
References
- Alexandru, G., Uhlmann, F., Mechtler, K., Poupart, M., Nasmyth, K. Phosphorylation of the cohesin subunit scc1 by polo/cdc5 kinase regulates sister chromatid separation in yeast. Cell. 2001;105:459–472. doi: 10.1016/s0092-8674(01)00362-2. [DOI] [PubMed] [Google Scholar]
- Anderson, D.E., Losada, A., Erickson, H.P., Hirano, T. Condensin and cohesin display different arm conformations with characteristic hinge angles. J. Cell Biol. 2002;156:419–424. doi: 10.1083/jcb.200111002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachant, J., Alcasabas, A., Blat, Y., Kleckner, N., Elledge, S.J. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol. Cell. 2002;9:1169–1182. doi: 10.1016/s1097-2765(02)00543-9. [DOI] [PubMed] [Google Scholar]
- Bhalla, N., Biggins, S., Murray, A.W. Mutation of YCS4, a budding yeast condensin subunit, affects mitotic and nonmitotic chromosome behavior. Mol. Biol. Cell. 2002;13:632–645. doi: 10.1091/mbc.01-05-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk, R., Zachariae, W., Michaelis, C., Shevchenko, A., Mann, M., Nasmyth, K. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 1998;93:1067–1076. doi: 10.1016/s0092-8674(00)81211-8. [DOI] [PubMed] [Google Scholar]
- Costanzo, M., Nishikawa, J.L., Tang, X., Millman, J.S., Schub, O., Breitkreuz, K., Dewar, D., Rupes, I., Andrews, B., Tyers, M. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell. 2004;117:899–913. doi: 10.1016/j.cell.2004.05.024. [DOI] [PubMed] [Google Scholar]
- D'Amours, D., Stegmeier, F., Amon, A. Cdc14 and condensin control the dissolution of cohesin-independent chromosome linkages at repeated DNA. Cell. 2004;117:455–469. doi: 10.1016/s0092-8674(04)00413-1. [DOI] [PubMed] [Google Scholar]
- Dohmen, R.J., Wu, P., Varshavsky, A. Heat-inducible degron: A method for constructing temperature-sensitive mutants. Science. 1994;263:1273–1276. doi: 10.1126/science.8122109. [DOI] [PubMed] [Google Scholar]
- Dynek, J.N., Smith, S. Resolution of sister telomere association is required for progression through mitosis. Science. 2004;304:97–100. doi: 10.1126/science.1094754. [DOI] [PubMed] [Google Scholar]
- Freeman, L., Aragon-Alcaide, L., Strunnikov, A. The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J. Cell Biol. 2000;149:811–824. doi: 10.1083/jcb.149.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giet, R., Glover, D.M. Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J. Cell Biol. 2001;152:669–682. doi: 10.1083/jcb.152.4.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez-Abian, J.F., Sumara, I., Hirota, T., Hauf, S., Gerlich, D., de la Torre, C., Ellenberg, J., Peters, J.M. Regulation of sister chromatid cohesion between chromosome arms. Curr. Biol. 2004;14:1187–1193. doi: 10.1016/j.cub.2004.06.052. [DOI] [PubMed] [Google Scholar]
- Glynn, E.F., Megee, P.C., Yu, H.G., Mistrot, C., Unal, E., Koshland, D.E., DeRisi, J.L., Gerton, J.L. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae . PLoS Biol. 2004;2:E259. doi: 10.1371/journal.pbio.0020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G., Yanagida, M. Time course analysis of precocious separation of sister centromeres in budding yeast: Continuously separated or frequently reassociated? Genes Cells. 2001;6:765–773. doi: 10.1046/j.1365-2443.2001.00464.x. [DOI] [PubMed] [Google Scholar]
- Gruber, S., Haering, C.H., Nasmyth, K. Chromosomal cohesin forms a ring. Cell. 2003;112:765–777. doi: 10.1016/s0092-8674(03)00162-4. [DOI] [PubMed] [Google Scholar]
- Guacci, V., Hogan, E., Koshland, D. Chromosome condensation and sister chromatid pairing in budding yeast. J. Cell Biol. 1994;125:517–530. doi: 10.1083/jcb.125.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci, V., Koshland, D., Strunnikov, A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae . Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C., Fink G.R., editors. Guide to yeast genetics and molecular biology. Academic Press; New York: 1991. [Google Scholar]
- Haering, C.H., Lowe, J., Hochwagen, A., Nasmyth, K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell. 2002;9:773–788. doi: 10.1016/s1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- Hagstrom, K.A., Meyer, B.J. Condensin and cohesin: More than chromosome compactor and glue. Nat. Rev. Genet. 2003;4:520–534. doi: 10.1038/nrg1110. [DOI] [PubMed] [Google Scholar]
- Hagstrom, K.A., Holmes, V.F., Cozzarelli, N.R., Meyer, B.J. C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes & Dev. 2002;16:729–742. doi: 10.1101/gad.968302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman, T., Stead, K., Koshland, D., Guacci, V. Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae . J. Cell. Biol. 2000;151:613–626. doi: 10.1083/jcb.151.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf, S., Roitinger, E., Koch, B., Dittrich, C.M., Mechtler, K., Peters, J.M. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 2005;3:e69. doi: 10.1371/journal.pbio.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, X., Asthana, S., Sorger, P.K. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell. 2000;101:763–775. doi: 10.1016/s0092-8674(00)80888-0. [DOI] [PubMed] [Google Scholar]
- Hirano, T. Condensins: Organizing and segregating the genome. Curr. Biol. 2005a;15:R265–R275. doi: 10.1016/j.cub.2005.03.037. [DOI] [PubMed] [Google Scholar]
- Hirano, T. SMC proteins and chromosome mechanics: From bacteria to humans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005b;360:507–514. doi: 10.1098/rstb.2004.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano, T., Kobayashi, R., Hirano, M. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell. 1997;89:511–521. doi: 10.1016/s0092-8674(00)80233-0. [DOI] [PubMed] [Google Scholar]
- Huang, C.E., Milutinovich, M., Koshland, D. Rings, bracelet or snaps: Fashionable alternatives for Smc complexes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005;360:537–542. doi: 10.1098/rstb.2004.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov, D., Nasmyth, K. A topological interaction between cohesin rings and a circular minichromosome. Cell. 2005;122:849–860. doi: 10.1016/j.cell.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Kimura, K., Hirano, M., Kobayashi, R., Hirano, T. Phosphorylation and activation of 13S condensin by Cdc2 in vitro. Science. 1998;282:487–490. doi: 10.1126/science.282.5388.487. [DOI] [PubMed] [Google Scholar]
- Kimura, K., Cuvier, O., Hirano, T. Chromosome condensation by a human condensin complex in Xenopus egg extracts. J. Biol. Chem. 2001;276:5417–5420. doi: 10.1074/jbc.C000873200. [DOI] [PubMed] [Google Scholar]
- Labib, K., Tercero, J.A., Diffley, J.F. Uninterrupted MCM2-7 function required for DNA replication fork progression. Science. 2000;288:1643–1647. doi: 10.1126/science.288.5471.1643. [DOI] [PubMed] [Google Scholar]
- Lavoie, B.D., Tuffo, K.M., Oh, S., Koshland, D., Holm, C. Mitotic chromosome condensation requires Brn1p, the yeast homologue of Barren. Mol. Biol. Cell. 2000;11:1293–1304. doi: 10.1091/mbc.11.4.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie, B.D., Hogan, E., Koshland, D. In vivo dissection of the chromosome condensation machinery: Reversibility of condensation distinguishes contributions of condensin and cohesin. J. Cell Biol. 2002;156:805–815. doi: 10.1083/jcb.200109056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie, B.D., Hogan, E., Koshland, D. In vivo requirements for rDNA chromosome condensation reveal two cellcycle-regulated pathways for mitotic chromosome folding. Genes & Dev. 2004;18:76–87. doi: 10.1101/gad.1150404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne, A., Katou, Y., Mori, S., Yokobayashi, S., Kelly, G.P., Itoh, T., Watanabe, Y., Shirahige, K., Uhlmann, F. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–578. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada, A., Hirano, M., Hirano, T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes & Dev. 1998;12:1986–1997. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machin, F., Torres-Rosell, J., Jarmuz, A., Aragon, L. Spindle-independent condensation-mediated segregation of yeast ribosomal DNA in late anaphase. J. Cell Biol. 2005;168:209–219. doi: 10.1083/jcb.200408087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megee, P.C., Koshland, D. A functional assay for centromere-associated sister chromatid cohesion. Science. 1999;285:254–257. doi: 10.1126/science.285.5425.254. [DOI] [PubMed] [Google Scholar]
- Megee, P.C., Mistrot, C., Guacci, V., Koshland, D. The centromeric sister chromatid cohesion site directs Mcd1p binding to adjacent sequences. Mol. Cell. 1999;4:445–450. doi: 10.1016/s1097-2765(00)80347-0. [DOI] [PubMed] [Google Scholar]
- Meluh, P.B., Koshland, D. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes & Dev. 1997;11:3401–3412. doi: 10.1101/gad.11.24.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis, C., Ciosk, R., Nasmyth, K. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K., Haering, C.H. The structure and function of smc and kleisin complexes. Annu. Rev. Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- Ouspenski, I.I., Cabello, O.A., Brinkley, B.R. Chromosome condensation factor Brn1p is required for chromatid separation in mitosis. Mol. Biol. Cell. 2000;11:1305–1313. doi: 10.1091/mbc.11.4.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panizza, S., Tanaka, T., Hochwagen, A., Eisenhaber, F., Nasmyth, K. Pds5 cooperates with cohesin in maintaining sister chromatid cohesion. Curr. Biol. 2000;10:1557–1564. doi: 10.1016/s0960-9822(00)00854-x. [DOI] [PubMed] [Google Scholar]
- Schmiesing, J.A., Gregson, H.C., Zhou, S., Yokomori, K. A human condensin complex containing hCAP-C–hCAP-E and CNAP1, a homolog of Xenopus XCAP-D2, colocalizes with phosphorylated histone H3 during the early stage of mitotic chromosome condensation. Mol. Cell. Biol. 2000;20:6996–7006. doi: 10.1128/mcb.20.18.6996-7006.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R.S., Hieter, P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens, R.V. Unzipped and loaded: The role of DNA helicases and RFC clamp-loading complexes in sister chromatid cohesion. J. Cell Biol. 2005;169:841–846. doi: 10.1083/jcb.200503129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead, K., Aguilar, C., Hartman, T., Drexel, M., Meluh, P., Guacci, V. Pds5p regulates the maintenance of sister chromatid cohesion and is sumoylated to promote the dissolution of cohesion. J. Cell Biol. 2003;163:729–741. doi: 10.1083/jcb.200305080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stear, J.H., Roth, M.B. Characterization of HCP-6, a C. elegans protein required to prevent chromosome twisting and merotelic attachment. Genes & Dev. 2002;16:1498–1508. doi: 10.1101/gad.989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunnikov, A.V., Larionov, V.L., Koshland, D. SMC1: An essential yeast gene encoding a putative head–rod–tail protein is required for nuclear division and defines a new ubiquitous protein family. J. Cell Biol. 1993;123:1635–1648. doi: 10.1083/jcb.123.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunnikov, A.V., Hogan, E., Koshland, D. SMC2, a Saccharomyces cerevisiae gene essential for chromosome segregation and condensation, defines a subgroup within the SMC family. Genes & Dev. 1995;9:587–599. doi: 10.1101/gad.9.5.587. [DOI] [PubMed] [Google Scholar]
- Sullivan, M., Higuchi, T., Katis, V.L., Uhlmann, F. Cdc14 phosphatase induces rDNA condensation and resolves cohesin-independent cohesion during budding yeast anaphase. Cell. 2004;117:471–482. doi: 10.1016/s0092-8674(04)00415-5. [DOI] [PubMed] [Google Scholar]
- Sumara, I., Vorlaufer, E., Gieffers, C., Peters, B.H., Peters, J.M. Characterization of vertebrate cohesin complexes and their regulation in prophase. J. Cell Biol. 2000;151:749–762. doi: 10.1083/jcb.151.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumara, I., Vorlaufer, E., Stukenberg, P.T., Kelm, O., Rede-mann, N., Nigg, E.A., Peters, J.M. The dissociation of cohesin from chromosomes in prophase is regulated by Polo-like kinase. Mol. Cell. 2002;9:515–525. doi: 10.1016/s1097-2765(02)00473-2. [DOI] [PubMed] [Google Scholar]
- Sutani, T., Yuasa, T., Tomonaga, T., Dohmae, N., Takio, K., Yanagida, M. Fission yeast condensin complex: Essential roles of non-SMC subunits for condensation and Cdc2 phosphorylation of Cut3/SMC4. Genes & Dev. 1999;13:2271–2283. doi: 10.1101/gad.13.17.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth, A., Ciosk, R., Uhlmann, F., Galova, M., Schleiffer, A., Nasmyth, K. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes & Dev. 1999;13:320–333. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann, F., Nasmyth, K. Cohesion between sister chromatids must be established during DNA replication. Curr. Biol. 1998;8:1095–1101. doi: 10.1016/s0960-9822(98)70463-4. [DOI] [PubMed] [Google Scholar]
- Uhlmann, F., Lottspeich, F., Nasmyth, K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- Wach, A., Brachat, A., Pohlmann, R., Philippsen, P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae . Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Waizenegger, I.C., Hauf, S., Meinke, A., Peters, J.M. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 2000;103:399–410. doi: 10.1016/s0092-8674(00)00132-x. [DOI] [PubMed] [Google Scholar]
- Wang, B.D., Yong-Gonzalez, V., Strunnikov, A.V. Cdc14p/FEAR pathway controls segregation of nucleolus in S. cerevisiae by facilitating condensin targeting to rDNA chromatin in anaphase. Cell Cycle. 2004;3:960–967. doi: 10.4161/cc.3.7.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, B.D., Eyre, D., Basrai, M., Lichten, M., Strunnikov, A. Condensin binding at distinct and specific chromosomal sites in the Saccharomyces cerevisiae genome. Mol. Cell. Biol. 2005;25:7216–7225. doi: 10.1128/MCB.25.16.7216-7225.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren, W.D., Lin, E., Nheu, T.V., Hime, G.R., McKay, M.J. Drad21, a Drosophila rad21 homologue expressed in S-phase cells. Gene. 2000;250:77–84. doi: 10.1016/s0378-1119(00)00166-9. [DOI] [PubMed] [Google Scholar]
- Yokomori, K. SMC protein complexes and the maintenance of chromosome integrity. Curr. Top. Microbiol. Immunol. 2003;274:79–112. doi: 10.1007/978-3-642-55747-7_4. [DOI] [PubMed] [Google Scholar]