Abstract
The mechanism of lineage specification in multipotent stem cells has not been fully understood. We recently isolated progenitors with the eosinophil, basophil, or mast cell lineage potential, all of which originate from granulocyte/monocyte progenitors (GMPs). By using these prospectively purified progenitors, we show here that the expression timing of GATA-2 and CCAAT enhancer-binding protein α (C/EBPα) can differentially control their lineage commitment. The expression of GATA-2 instructed C/EBPα-expressing GMPs to commit exclusively into the eosinophil lineage, while it induced basophil and/or mast cell lineage commitment if C/EBPα was suppressed at the GMP stage. Furthermore, simply by switching the order of C/EBPα and GATA-2 transduction, even lymphoid-committed progenitors recaptured these developmental processes to be reprogrammed into each of these lineages. We propose that the order of expression of key transcription factors is critical for their interplay to selectively drive lineage specification programs, by which stem cells could generate multiple lineage cells in a hierarchical manner.
Keywords: Hematopoiesis, hematopoietic progenitors, transcription factor
It has been proposed that antagonistic or cooperative effects of multiple transcription factors, called a “transcription factor network” (Sieweke and Graf 1998; Orkin 2000), play a pivotal role in establishment of lineage diversities from stem cells. In this context, multiple transcription factors can antagonize to specify cell fates. For example, in hematopoiesis, GATA-1 and GATA-2, major transcription factors for megakaryocyte/erythrocyte (MegE) development inhibit the expression and transactivation functions of PU.1, a transcription factor indispensable for myeloid or lymphoid development (Zhang et al. 1999; Nerlov et al. 2000), and vice versa (Nerlov and Graf 1998; Rekhtman et al. 1999; Zhang et al. 1999, 2000; Walsh et al. 2002), suggesting that these antagonistic effects may be critical at least for the MegE versus the granulocyte/monocyte (GM) lineage commitment. It has also been proposed that CCAAT enhancer-binding protein α (C/EBPα) and PU.1 mutually antagonize their expression and/or functions to decide the neutrophil versus the monocyte fates (Reddy et al. 2002; Dahl et al. 2003). Thus, mutually exclusive competition among more than two transcription factor functions could be a major mechanism for a single lineage program to be activated at a bipotent progenitor stage. However, for a variety of hematopoietic lineages to be generated from single multipotent hematopoietic stem cells (HSCs), there should be some other form of interplay among major transcription factors.
Eosinophils, basophils, and mast cells are multifunctional hematopoietic effectors that cooperate to mount a variety of allergic and innate immune responses (Rothenberg 1998; Galli 2000; Wedemeyer et al. 2000). Eosinophils and basophils normally constitute only 1%–2% of circulating white blood cells, and mast cells are scattered mainly in the skin and the mucosa. Recently, we have identified the bipotent basophil/mast cell progenitor (BMCP) in the adult mouse spleen, providing formal evidence that basophils and mast cells share their origin (Arinobu et al. 2005). We have also identified the eosinophil lineage-committed progenitor (EoP) in the bone marrow (Iwasaki et al. 2005). BMCPs and EoPs were found as distinct populations downstream from GM progenitors (GMPs), suggesting that developmental pathways for the eosinophil and the basophil/mast cell lineages might be separable. On the other hand, eosinophils and basophils possess morphological similarities such as a polymorphic nucleus like neutrophils, and cells possessing both basophilic and eosinophilic granules have been found in some leukemia patients (Weil and Hrisinko 1987; Boyce et al. 1995), suggesting that certain developmental programs could be commonly used in the eosinophil and the basophil lineages. In fact, all three cell types express GATA-1 and GATA-2, major transcription factors also for MegE development (Martin et al. 1990; Zon et al. 1993). GATA-1-deficient mice displayed the impaired maturation of mast cells (Harigae et al. 1998) and eosinophils (Hirasawa et al. 2002) as well as MegE cells (Fujiwara et al. 1996) that could be rescued by GATA-2 to a certain degree (Tsai et al. 1998; Takahashi et al. 2000; Hirasawa et al. 2002). The decreased expression of GATA-1 caused the impaired mast cell maturation as well as the frequent development of mast cells with MegE components in single colonies (Migliaccio et al. 2003), and GATA-2-deficient mice lacked mast cells (Jippo et al. 1996; Tsai and Orkin 1997; Walsh et al. 2002). When enforced in CD34+ cord blood progenitor cells, both GATA-1 and GATA-2 stimulated the formation of eosinophil colonies at the expense of GM colonies (Hirasawa et al. 2002), and enforced GATA-1 converted GM cells into the MegE and the eosinophil lineages (Kulessa et al. 1995; Heyworth et al. 2002; Iwasaki et al. 2003).
In addition to these GATA factors, GM-related C/EBP transcription factors are critical for eosinophil and basophil development. C/EBPα-deficient mice lacked eosinophils as well as neutrophils, although their basophil development was not tested (Zhang et al. 1997), and a chicken cell line expressing GATA-1 could differentiate into eosinophils by the enforced expression of C/EBPα or C/EBPβ (Nerlov et al. 1998). We have reported that C/EBPα is necessary for the late basophil development after the BMCP stage (Arinobu et al. 2005), and that enforced C/EBPα plays an instructive role in commitment into the basophil lineage excluding the mast cell fate (Arinobu et al. 2005). These forced expression experiments show that GATA factors and C/EBPα can cooperatively instruct commitment for the eosinophil or the basophil lineage (McNagny and Graf 2002).
The question is how GATA or C/EBP transcription factors regulate the development of rare eosinophils, basophils, or mast cells differentially from that of neutrophils and monocytes. In the present paper, by analyzing the development of eosinophils and basophils from purified GMPs, we found that although either cell type requires both GATA-2 and C/EBPα for their development, the order of expression of these transcription factors is critical to selectively activate the program for each of these lineages. We propose that hierarchical developmental programs in hematopoiesis might be achieved at least by the ordered expression pattern of lineage instructive transcription factors.
Results
Expression profiles of transcription factors and lineage-related genes in progenitor populations restricted to each granulocytic lineage
We tested the development of eosinophils, basophils, and mast cells from purified lineage-restricted progenitors such as common myeloid progenitors (CMPs), GMPs, megakaryocyte/erythrocyte progenitors (MEPs) (Akashi et al. 2000), and common lymphoid progenitors (CLPs) (Kondo et al. 1997), and found that these cell types develop along the myeloid lineage differentiation pathway (Arinobu et al. 2005; Iwasaki et al. 2005). Based on these data, we have also identified EoPs, BMCPs, basophil progenitors (BaPs), and mast cell progenitors (MCPs) in normal murine hematopoiesis (Arinobu et al. 2005; Iwasaki et al. 2005). The developmental scheme of these lineage-restricted progenitor populations is shown in Figure 1A.
Figure 1.
Expression profiles of transcription factors and lineage-related genes in purified progenitor populations. (A) A developmental scheme of lineage-restricted progenitor populations used in this study. (HSC) Hematopoietic stem cell; (CLP) common lymphoid progenitor; (CMP) common myeloid progenitor; (MEP) megakaryocyte/erythrocyte progenitor; (GMP) granulocyte/monocyte progenitor; (BMCP) basophil/mast cell progenitor; (MCP) mast cell progenitor; (BaP) basophil progenitor; (EoP) eosinophil progenitor. (B) Semiquantitative RT–PCR analyses of transcription factors and lineage-related genes in purified progenitor populations. (mMCP-5) Mouse mast cell protease-5; (MBP) major basic protein (EoPO) eosinophil peroxidase; (HPRT) hypoxanthine–guanine phosphoribosyl transferase. (*) MCPs were purified from GMP cultures on day 3. (C) Real-time PCR assays of transcription factor expression in purified stem and progenitor cells. ΔCt values for each sample were standardized by GAPDH Ct values. PCR assays were performed using three sets of cDNA samples prepared independently, resulting in the same pattern of relative expression.
EoPs were isolated as CD34+IL-5Rα+FcγRII/III+c-Kitlo cells in the bone marrow, and BMCPs were as CD34+β7hi FcγRII/ III+c-Kithi cells in the spleen (Arinobu et al. 2005; Iwasaki et al. 2005). EoPs and BMCPs were progeny of GMPs (CD34+IL- 5Rα−β7−FcγRII/III+c-Kit+), and BMCPs further generated monopotent BaPs (CD34+β7loFcεRIαhiFcγRII/III+c-Kit−) and MCPs (CD34+β7hiFcεRIαloc-Kit+) (Arinobu et al. 2005; Iwasaki et al. 2005). We purified these progenitors, and analyzed their expression profiles of lineage-related genes by RT–PCR (Fig. 1B). IL-5Rα was strongly expressed only in EoPs, whereas FcεRIα was expressed in BMCPs, BaPs, and MCPs but not in EoPs. Major basic protein (MBP) was expressed in BMCPs, BaPs, and EoPs but not in MCPs, whereas murine mast cell protease-5 (mMCP-5) was expressed in BMCPs and MCPs but not BaPs or EoPs. Eosinophil peroxidase (EoPO) was strongly expressed only in EoPs. GMPs had not expressed any of these lineage-related genes. Thus, the gene expression profile of each progenitor population appears to reflect that of their own mature progeny.
GATA-1 and GATA-2 are expressed in mature eosinophils, basophils, and mast cells (Zon et al. 1993; Harigae et al. 1998) as well as in MEPs (Akashi et al. 2000). C/EBPα is required for neutrophil and eosinophil development (Zhang et al. 1997), and PU.1 is essential for GM, B cell, and mast cell development (Scott et al. 1994; Walsh et al. 2002). As shown in Figure 1B, GATA-1 and GATA-2 were not expressed in GMPs, but were up-regulated in BMCPs, BaPs, MCPs, and EoPs. PU.1 was also expressed in all of these purified progenitors. Interestingly, C/EBPα was expressed in GMPs, BaPs, and EoPs, but it was suppressed in BMCPs or MCPs. Figure 1C shows the results of quantitative real-time PCR analyses for GATA-1, GATA-2, and C/EBPα mRNA expression in hematopoietic stem and progenitor populations. The expression level of GATA-1 was highest in MEPs, while GMPs did not express significant level of GATA-1. BMCPs, MCPs, BaPs, and EoPs expressed approximately three- to sixfold lower levels of GATA-1 as compared with that in MEPs (Fig. 1C, upper panel). In contrast, GATA-2 expression in BMCPs, MCPs, BaPs, and EoPs was significantly higher than that in MEPs (Fig. 1C, middle panel), suggesting the important role of GATA-2 in the development of basophil/mast cell/eosinophil lineages. C/EBPα expression was highest in BaPs followed by GMPs and EoPs, while BMCPs and MCPs possessed approximately eightfold decreased level of C/EBPα as compared with that in GMPs (Fig. 1C, bottom panel). In summary, BaPs and EoPs expressed GATA factors and C/EBPα, whereas BMCPs and MCPs expressed GATA factors but not C/EBPα.
The enforced expression of GATA-2 instructs GMPs to exclusively select the eosinophil fate
We have previously reported that GATA-1 is important for lineage instruction primarily for the MegE lineage commitment or conversion of HSCs, GMPs, and lymphoid-committed progenitors (Iwasaki et al. 2003). Consistent with this data, GATA-1 expression was highest in MEPs (Fig. 1C, upper panel). On the other hand, high levels of GATA-2 expression were observed in BMCPs, BaPs, MCPs, and EoPs, but not in MEPs (Fig. 1C, middle panel). These results led us to speculate that GATA-2 might be primarily important for eosinophil, basophil, and mast cell development.
Since EoPs and BaPs expressed both GATA-2 and C/EBPα while upstream GMPs expressed only C/EBPα (Fig. 1B,C), we first introduced GATA-2 into GMPs via a green fluorescence protein (GFP)-tagged retrovirus vector, and tested their differentiation activity. As shown in Figure 2A, although the vast majority of GMPs generated neutrophil/monocyte (NM) colonies in the presence of a cytokine cocktail containing Slf, IL-3, IL-5, GM-CSF, Epo, and Tpo, GMPs transduced with GATA-2 (GATA- 2+ GMPs) gave rise exclusively to pure eosinophil colonies at the expense of NM colonies (Fig. 2A,B). The formation of eosinophil colonies from GATA-2+ GMPs was not affected by removing IL-5, Epo, and Tpo from the culture. Progeny of GATA-2+ GMPs did not contain FcεRIα+c-Kit+ mast cells or FcεRIα+c-Kit− basophils (Fig. 2C). Figure 2D shows the change in expression pattern of eosinophil-related genes in GMPs immediately after the completion of GATA-2 transduction. GATA-2+ GMPs expressed GATA-1, IL-5Rα, and EoPO, maintaining the expression of PU.1 and C/EBPα, the patterns of which were identical to those of purified EoPs (Fig. 1B). Taken together, the ectopic expression of GATA-2 instructs GMPs to become EoPs but not BaPs.
Figure 2.
Enforced GATA-2 instructs GMPs to exclusively select the eosinophil fate. (A) Clonogenic analyses of GMPs with or without GATA-2 transduction. Purified GMPs were transduced with a GFP-tagged retrovirus carrying a murine GATA-2 cDNA or an empty control retro-virus. GFP-positive GMPs were isolated 36 h after the retroviral infection and subjected to methylcellulose assays. Types of colonies were evaluated at day 5 by May-Giemsa staining. Cytokines added are indicated. (B) Progeny of GMPs transduced with the GATA-2 retrovirus. An image of fluorescence microscopy (GFP filter, ×1000, upper panel), and that of May-Giemsa staining (×1000, bottom panel) are shown. Cells positive for GFP possessed eosinophilic granules. (C) The phenotypic analysis of progeny of GMPs transduced with GATA-2. Although BMCPs gave rise to FcεRIα+ basophils and mast cells in the culture, GATA-2+ GMPs did not give rise to FcεRIα+ cells. (D) Gene expression analyses of GMPs immediately after the completion of GATA-2 transduction. (E) Real-time PCR analysis of GATA-2 mRNA before and after GATA-2 transduction. ΔCt values for each samples were standardized by GAPDH Ct values. GATA-2 transduction experiment was performed three times using GMPs purified independently, and the same results were obtained.
Normal BaPs expressed approximately fivefold higher level of GATA-2 as compared with that in EoPs (Fig. 1C, middle panel), raising possibility that the expression of GATA-2 at a low level may be critical for eosinophil lineage commitment at the GMP stage. As shown in Figure 2E, the expression level of enforced GATA-2 in GATA-2+ GMPs was comparable to (1.5-fold higher than) that in normal BaPs, while it was 10-fold higher than that in normal EoPs. Because such a high level of enforced GATA-2 still instructed GMPs to become EoPs but not BaPs, these data collectively suggest that GATA-2 is likely to specifically instruct eosinophil commitment at least at the GMP stage.
The enforced expression of C/EBPα in GMPs blocked basophil/mast cell development
GMPs also give rise to BMCPs, BaPs, and MCPs (Fig. 1A; Arinobu et al. 2005). C/EBPα was highly expressed in GMPs but significantly decreased in BMCPs or MCPs (Fig. 1B,C), suggesting that the down-regulation of C/EBPα is critical for the development of basophil and mast cell lineages. Our previous data, however, showed that C/EBPα is indispensable for basophil development after the BMCP stage, and that the enforced expression of C/EBPα can convert MCPs into basophils (Arinobu et al. 2005). To test whether C/EBPα needs to be suppressed for GMPs to choose the basophil/mast cell fate, we constitutively expressed C/EBPα in GMPs via a human (h) CD4-tagged retrovirus vector. Interestingly, GMPs with transduced C/EBPα (C/EBPα+) could not generate basophils or mast cells efficiently, while they gave rise mainly to neutrophils and rare eosinophils (Fig. 3A). Thus, C/EBPα needs to be suppressed at the GMP stage for both basophil and mast cell development. These data suggest that C/EBPα is expressed in a biphasic manner for basophil development from GMPs through BMCPs, such as the down-regulation of C/EBPα at the GMP stage to proceed into BMCPs, and its reactivation at the BMCP stage to give rise to mature basophils.
Figure 3.
The effect of enforcement or reduction of C/EBPα expression on the basophil/mast cell development from myeloid progenitors. (A) FACS analyses of day 3 progeny of GMPs with or without C/EBPα transduction. Purified GMPs were transduced with a hCD4-tagged retrovirus carrying a murine C/EBPα cDNA or an empty retrovirus as control. hCD4-positive GMPs were isolated 36 h after the retroviral infection and cultured for an additional 72 h in the presence of Slf, IL-3, and IL-6. (Right) By maintaining C/EBPα expression, GMPs became incapable of producing FcεRIαloc-Kithi MCPs or FcεRIαhic-Kit− BaPs. FACS plots are representative of three independent experiments. (B) Limiting dilution analysis of mast cell or basophil development from C/EBPαΔ/Δ GMPs. GMPs were purified from C/EBPαF/F mice (Zhang et al. 2004) and transduced with a GFP-tagged retrovirus carrying a Cre cDNA or an empty retrovirus as control. GFP-positive GMPs were isolated 36 h after the retroviral infection and subjected to limiting dilution assays in the presence of Slf, IL-3, and IL-6. A complete excision of floxed C/EBPα alleles after Cre transduction was determined by PCR assay as previously reported (Zhang et al. 2004) (not shown). The development of basophils and/or mast cells in each cultures was determined by May-Giemsa and Toluidine Blue staining. The frequency of mast cell read-out from C/EBPαΔ/Δ GMPs was fourfold higher than that from control C/EBPαF/F GMPs (left), whereas mature basophil development was blocked completely by the C/EBPα disruption (right). (C) Reduction of C/EBPα by an RNAi technique using shRNA expression vector by retrovirus transduction. GMPs transduced with three different shRNAs targeting C/EBPα (see Materials and Methods) were measured for their expression of C/EBPα by a real-time PCR analysis standardized by a GAPDH expression level (left), and also analyzed for their frequency of mast cell readout by a limiting dilution assay (right). The expression level of C/EBPα in GMPs was inversely correlated with the frequency of mast cell lineage readout.
The expression level of C/EBPα in GMPs is inversely correlated with the frequency of mast cell lineage readout
The fact that the sustained C/EBPα expression blocked basophil/mast cell development from GMPs led us to predict that the decreased C/EBPα expression would reciprocally increase their development. Since we have shown that C/EBPα is required for the late basophil development after the BMCP stage (Arinobu et al. 2005), the suppression of C/EBPα should inhibit the development of mature basophils. Therefore, this experiment was focused mainly on mast cell development. First, we disrupted C/EBPα in GMPs by using a conditional knockout system (Zhang et al. 2004). We purified GMPs from mice in which the C/EBPα gene is flanked by the loxP sequences (floxed, F), and C/EBPαF/F GMPs were transduced with the Cre recombinase via a GFP-tagged retrovirus vector. C/EBPαF/F GMPs with the Cre transduction displayed almost complete excision of floxed C/EBPα alleles determined by PCR assays (data not shown), and therefore we refer to Cre-transduced C/EBPαF/F cells as C/EBPαΔ/Δ. C/EBPαΔ/Δ GMPs gave rise to neutrophils and monocytes (data not shown) as reported (Zhang et al. 2004) but not to basophils (Fig. 3B, right panel), which is consistent with our previous observation that C/EBPα is essential for BMCPs to develop mature basophils (Arinobu et al. 2005).
In contrast, the frequency of mast cell lineage readout from C/EBPαΔ/Δ GMPs increased by fourfold compared with that in C/EBPαF/F GMPs transduced with a control (empty) retrovirus vector (Fig. 3B, left panel). We further found that the frequency of mast cell lineage readout from GMPs was gradually enhanced by the graded reduction of C/EBPα expression. To suppress C/EBPα expression at the GMP stage, we employed an RNA interference (RNAi) technique using a short hairpin RNA (shRNA) expression retrovirus vector. Transduction of three separate C/EBPα shRNAs into GMPs resulted in a graded reduction of C/EBPα mRNA by ∼30%, ∼50%, or ∼70% of the normal level (Fig. 3C, left panel), and the frequency of mast cell readout was enhanced according to the level of C/EBPα mRNA suppression induced by each shRNA (Fig. 3C, right panel). Thus, the mast cell lineage-commitment from GMPs was inversely correlated with the expression level of C/EBPα. These data collectively suggest that C/EBPα needs to be down-regulated for GMPs to become BMCPs, but it is required for the later development of basophils but not mast cells after the BMCP stage.
The enforced expression of GATA-2 instructs the mast cell fate in C/EBPα−/− myeloid progenitors
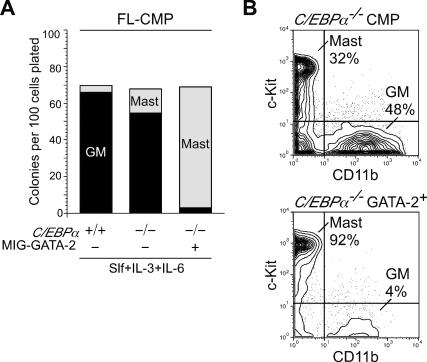
The enforcement of GATA-2 expression in C/EBPα-expressing GMPs resulted in their exclusive differentiation into eosinophils (Fig. 2). Since the differentiation of GMPs into BMCPs and mast cells was inversely correlated with the expression level of C/EBPα, we tested the effect of GATA-2 expression on the lineage fate decision of C/EBPα-deficient myeloid progenitors. To this end, we used embryonic day 14 C/EBPα−/− fetal liver (FL) progenitor cells. We have reported that C/EBPα−/− FL does not have GMPs, but have Lin−Sca-1−c-Kit+CD34+FcγRII/ IIIlo myeloid progenitors whose phenotype corresponds to normal CMPs (Zhang et al. 2004). As shown in Figure 4A, C/EBPα−/− FL-CMPs formed approximately fourfold more number of pure mast cell colonies as compared with wild-type FL-CMPs. More interestingly, upon GATA-2 transduction via a GFP-tagged retrovirus vector, the vast majority of GATA-2/GFP+ C/EBPα−/− FLCMPs formed pure mast cell colonies (Fig. 4A,B). Thus, when C/EBPα is absent, GATA-2 instructs lineage specification into the mast cell but not the eosinophil lineage.
Figure 4.
Enforced expression of GATA-2 in C/EBPα−/− myeloid progenitors CMPs purified from E14 C/EBPα−/− FL were transduced with a GFP-tagged retrovirus carrying a murine GATA-2 cDNA or an empty retrovirus as control. GFP-positive CMPs were isolated 36 h after the retroviral infection and cultured for 7 d on methylcellulose (A) or in suspension (B) in the presence of Slf, IL-3, and IL-6. Types of colonies were determined by May-Giemsa and Toluidine Blue staining. The enforced expression of GATA-2 instructed C/EBPα−/− FL-CMPs to select the mast cell fate. Representative data of three independent experiments are shown.
The enforced expression of C/EBPα and/or GATA-2 can convert lymphoid-committed progenitors into either GMPs, EoPs, BMCPs, or BaPs simply by switching the order of their expression
Data described above collectively suggest that the order of expression of C/EBPα and GATA-2 should be critical for lineage specification in eosinophil versus basophil/ mast cell development. CLPs, which give rise to all lymphoid but not myeloid components (Kondo et al. 1997; Traver et al. 2000), still possess plasticity that can be triggered by lineage instructive signals including ectopic cytokines and transcription factors (Kondo et al. 2000; Iwasaki et al. 2003; Iwasaki-Arai et al. 2003; Baba et al. 2005). Thus, the introduction of ectopic myeloid-related genes into CLPs should be a good tool to analyze myeloid lineage instructive signals without being affected by preexisting myeloid programs. CLPs do not express myeloid-related genes including C/EBPα and GATA-2 (Akashi et al. 2000). We thus transduced CLPs with these transcription factors in a different order to test its impact on myeloid lineage specification.
We infected CLPs with the hCD4-tagged C/EBPα and the GFP-tagged GATA-2 retroviruses in a reciprocal order. Figure 5A shows the expression pattern of hCD4 and GFP after the sequential transduction. The expression levels of hCD4 and GFP were almost identical in all experiments. In the first experiment, C/EBPα was transduced into CLPs, and 24 h after the C/EBPα transduction, GATA-2 was additionally transduced. Purified hCD4+ and/or GFP+ cells were cultured in vitro in the presence of Slf, IL-3, IL-5, IL-6, IL-7, IL-9, and Epo. Control (empty) hCD4+ and GFP+ CLPs (Fig. 5A, upper panel, Control) always gave rise to CD19+B220+ preB cells in vitro reflecting their natural lineage potential (Fig. 5B,C; Kondo et al. 1997). Interestingly, C/EBPα+GATA-2− CLPs (Fig. 5A, middle panel, R1) gave rise only to GM colonies at the expense of preB colonies, indicating that enforced C/EBPα is sufficient for CLPs to be reprogrammed into the GM lineage. Consistent with the data that GATA-2+ GMPs became EoPs (Fig. 2), C/EBPα+GATA-2+ CLPs (Fig. 5A, middle panel, R2) exclusively formed pure eosinophil colonies (Fig. 5B,C). In the second experiment, we switched the order of C/EBPα and GATA-2 transduction; GATA-2 was first transduced into CLPs, and 24 h after the GATA-2 transduction, C/EBPα was additionally transduced. Strikingly, GATA-2+C/EBPα− CLPs (Fig. 5A, bottom panel, R3) gave rise mainly to basophils and mast cells (Fig. 5B,C), indicating that GATA-2 converts CLPs into BMCPs. Additional C/EBPα transduction (Fig. 5A, bottom panel, R4) instructed GATA-2+ CLPs to only generate basophil colonies (Fig. 5B,C).
Figure 5.
The ordered expression of C/EBPα and/or GATA-2 can convert CLPs to either GMPs, EoPs, BMCPs, or BaPs. (A) Expression profile of hCD4 and GFP reporters after the two-step transduction. Note that the expression level of GFP and hCD4 was consistent irrespective of the order of transduction. (B) Methylcellulose assays for lineage outcomes of CLPs transduced with C/EBPα and/or GATA-2. R1–R4 represent sorting regions defined in A. Cytokines added were Slf, IL-3, IL-5, IL-6, IL-7, GM-CSF, Epo, and Tpo. Types of colonies were determined at day 7 by May-Giemsa and Toluidine Blue staining. (C) Phenotypic and morphological analyses of progeny of CLPs transduced with C/EBPα and/or GATA-2 in a different order. CLPs were purified according to sorting regions (R1–R4) defined in A and cultured in suspension for 7 d in the presence of Slf, IL-3, IL-5, IL-6, IL-7, GM-CSF, Epo, and Tpo. FACS analyses and morphologies (May-Giemsa staining, ×1000) of day 7 progeny are shown. Representative data of three independent experiments are shown.
The expression patterns of lineage-affiliated genes were evaluated by RT–PCR immediately after the completion of the second transduction. Consistent with their functional activities, C/EBPα+GATA-2− (R1), C/EBPα+GATA-2+ (R2), GATA-2+C/EBPα− (R3), and GATA-2+C/EBPα+ (R4) CLPs displayed the expression profile of genes almost identical to freshly isolated GMPs, EoPs, BMCPs, and BaPs, respectively (Figs. 1B, 6A). In contrast, CLPs doubly infected with empty hCD4 and GFP viruses expressed a series of lymphoid but not myeloid genes (Fig. 6A). Lymphoid-affiliated genes including EBF, Pax-5, and Notch-1 were down-regulated in all fractions of CLPs transduced with C/EBPα and/or GATA-2 (Fig. 6A). These data strongly suggest that although EoPs and BaPs coexpress C/EBPα and GATA-2, the order of their expression is critical for their lineage decision.
Figure 6.
Expression profiles of transcription factors and lineage-related genes in CLPs after two-step retroviral transduction of C/EBPα and GATA-2. (A)RT–PCR analyses of lineage-related gene expression in CLPs purified immediately after the completion of two-step retroviral transduction. R1–R4 represent sorting regions defined in Figure 5. (B) Change in the expression level of C/EBPα and GATA-2 mRNA in CLPs after tranduction of control-GFP, C/EBPα, or GATA-2, determined by a real-time PCR assay. ΔCt values for each samples were standardized by GAPDH Ct values. (C) Change in the expression level of lymphoid-affiliated EBF, Pax-5, and GATA-3 in CLPs after tranduction of control-GFP, C/EBPα, or GATA-2. Representative data of three independent experiments are shown.
We evaluated the expression level of enforced C/EBPα and GATA-2 in this experiment. The quantification of C/EBPα and GATA-2 mRNA after the completion of the first transduction of each transcription factor revealed that their expression levels were only slightly higher than those used in normal hematopoiesis (Fig. 6B). C/EBPα+ CLPs expressed almost 1.1- and 1.8-fold higher level of C/EBPα as compared with that in normal BaPs and GMPs, respectively, while the GATA-2 expression remained undetectable. GATA-2+ CLPs expressed GATA-2 at a level of ∼1.4-fold higher than normal BaPs, while the C/EBPα level was as low as that of control CLPs. Since the expression level of the hCD4 or the GFP reporter was not altered by double transduction procedures (Fig. 5A), expression levels of transduced C/EBPα and GATA-2 should be stable in this set of experiments. These data collectively suggest that the order of expression of transcription factors is critical for CLPs to select eosinophil versus basophil lineage fate decision.
We further measured EBF, Pax-5, and GATA-3 mRNA in CLPs after the first transduction of either C/EBPα or GATA-2. The quantitative real-time PCR experiments showed that all of these lymphoid-affiliated transcription factors were profoundly suppressed by the enforced expression of either C/EBPα or GATA-2 (Fig. 6C). These data suggest that C/EBPα or GATA-2 alone can reprogram CLPs into the myeloid lineage, at least by suppressing the lymphoid developmental program in CLPs.
Discussion
HSCs can give rise to multiple lineage cells, but the underlying mechanisms for this lineage specification process have not been fully understood. By using prospectively purified progenitor populations, we show that the order of expression of key transcription factors can differentially regulate fate decisions of multiple hematopoietic lineages including neutrophils/monocytes, eosinophils, basophils, and mast cells.
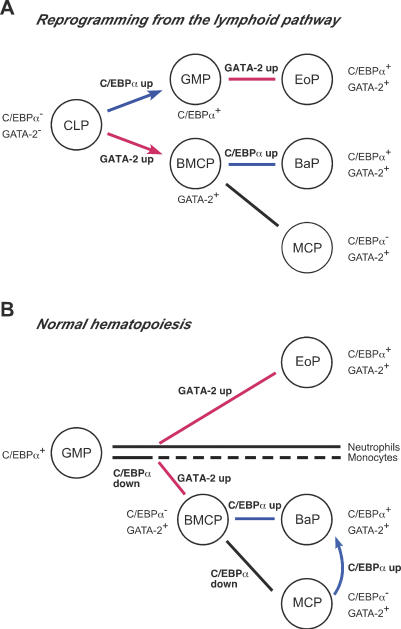
It is important to note that the enforced expression of GATA-2 instructed GMPs to differentiate into EoPs, while it converted CLPs into BMCPs. This interesting data suggests that GATA-2 can induce two different fate outcomes in the presence or absence of ongoing GM developmental program activated in GMPs. This then raises the question as to what signal within the GM program is critical for GATA-2 to instruct eosinophil versus basophil/mast cell lineage commitment. C/EBPα is one of key transcription factors for GM development (Tenen et al. 1997; Nerlov 2004), since C/EBPαΔ/Δ animals do not possess GMPs (Zhang et al. 2004). CLPs are lymphoid lineage-restricted progenitors, but they still possess multipotentiality when triggered by the enforced lineage-instructive signals (Kondo et al. 2000; Iwasaki et al. 2003; Iwasaki-Arai et al. 2003). We found that virtually all CLPs were converted into GMPs by the enforced expression of C/EBPα, and that the sequential expression of GATA-2 instructed them to differentiate further into the eosinophil lineage (Fig. 7A). Consistent with a recent report (Xie et al. 2004), these data indicate that C/EBPα plays a lineage-instructive role in GM commitment, and that the expression of C/EBPα is sufficient for GATA-2 to activate the eosinophil differentiation program.
Figure 7.
Schematic presentation of roles of transcription factors in lineage specification to eosinophils, basophils, and mast cells during reprogramming of CLPs and in normal hematopoiesis. (A) Lineage-instructive signals for each myeloid lineage on the basis of reprogramming of CLPs. CLPs can be converted to GMPs and BMCPs by the enforced expression of C/EBPα and GATA-2, respectively. In this model, the order of C/EBPα and GATA-2 expression is critical for CLPs to differentiate into EoPs via GMPs, or into BaPs via BMCPs. (B) Hematopoietic progenitor development downstream from GMPs in a physiological setting. Up-regulation of GATA-2 instructs GMPs to differentiate into eosinophil, basophil, and mast cell lineages. Upon GATA-2 up-regulation, if GMPs maintain C/EBPα expression, they become EoPs, whereas if GMPs down-regulate C/EBPα, they become BMCPs. For BaP development, C/EBPα needs to be reactivated after the BMCP stage. The mechanism of C/EBPα down-regulation in GMPs is not clear at this time, but since C/EBPα is not required for generation of mature neutrophils and monocytes after the GMP stage (Zhang et al. 2004), a fraction of GMPs may naturally down-regulate C/EBPα. (Red lines) Up-regulation of GATA-2; (blue lines) up-regulation of C/EBPα; (arrows) reprogramming of progenitors by enforced expression of transcription factors.
Conversely, the sustained expression of retrovirally transduced C/EBPα in GMPs blocked the basophil/mast cell lineage (Fig. 3A) but not the eosinophil lineage readout. The graded suppression of C/EBPα expression in GMPs progressively increased the frequency of mast cell readout (Fig. 3C), but more interestingly, C/EBPαΔ/Δ GMPs became incapable of differentiation into basophils (Fig. 3B). Given that down-regulation of C/EBPα is necessary for GMPs to develop into BMCPs, C/EBPα reactivation should be required for further basophil development after the BMCP stage. We have reported that when C/EBPα was transduced into MCPs, C/EBPα+ MCPs converted into BaPs, giving rise to mature basophils (Arinobu et al. 2005). Therefore, BMCPs can develop into MCPs only if C/EBPα remains suppressed, while C/EBPα should be reactivated for BMCPs to develop into BaPs. For GMPs to proceed into the basophil/mast cell pathway (including the BMCP, BaP, and MCP stages), down-regulation of C/EBPα might be the initial event (Fig. 7B), although this itself does not affect NM potential of GMPs as C/EBPαΔ/Δ GMPs could generate normal numbers of neutrophils and macrophages (Zhang et al. 2004).
Interestingly, development of basophils and mast cells via these pathways was recaptured again in CLPs that do not express either C/EBPα or GATA-2 in normal hematopoiesis. We showed that transduction of GATA-2 alone converted CLPs into BMCPs, and the sequential transduction of C/EBPα further converted them into BaPs (Fig. 5). These data suggest that simply by switching the order of C/EBPα and GATA-2 expression, CLPs can be reprogrammed into any of myeloid components such as neutrophils, monocytes, eosinophils, basophils, and mast cells. Thus, in normal hematopoiesis, EoP and BMCP development might be instructed by GATA-2 up-regulation depending on the presence and absence of C/EBPα expression, respectively.
Since the graded expression of transcription factors can affect lineage outcomes (Kulessa et al. 1995; DeKoter and Singh 2000), it is important to consider the expression level of enforced C/EBPα and GATA-2 used in this study. As shown in Figure 6B, the level of enforced C/EBPα and GATA-2 might not be unphysiologically high because the expression level of C/EBPα and GATA-2 was only 1.1- and 1.4-fold higher than that in normal BaPs (Fig. 6B). Furthermore, in all of these experiments, the expression level of C/EBPα and GATA-2 was stable, not being affected by double transduction procedures (Fig. 5A). Thus, these data collectively suggest that the order but not the level of transcription factor expression controls eosinophil versus basophil lineage commitment. We, however, did not analyze the transcription factor expression at a protein level in this study because of a technical limitation as to quantification of proteins in rare progenitor populations. To formally prove such action of transcription factors, the change of their protein expression levels during each developmental stage should be evaluated in future studies.
Transduction of either C/EBPα or GATA-2 in CLPs immediately induced the significant reduction of lymphoid-affiliated transcription factors such as EBF, Pax-5, and GATA-3 (Fig. 6C), suggesting that each of them alone can convert CLPs to the myeloid lineage. C/EBPα and GATA-2 do not appear to antagonize each other's function; CLPs with enforced C/EBPα converted into GMPs that developed normal numbers of GATA-2-ex pressing eosinophils, while CLPs with enforced GATA-2 became BMCPs that generated normal numbers of C/EBPα-expressing basophils (Fig. 5). Furthermore, we showed that enforced C/EBPα did not affect the expression level of GATA-2 in CLPs, and vice versa (Fig. 6B). Therefore, although both EoPs and BaPs coexpress C/EBPα and GATA-2, they might develop via independent developmental pathways guided by the order of C/EBPα and GATA-2 expression.
Previous studies have shown that GATA-1 and GATA-2 are mutually redundant to some extent (Takahashi et al. 2000). By enforcing the expression of GATA-1 and GATA-2 at identical stages of hematopoietic development, we found clear functional differences between GATA-1 and GATA-2. We have reported that the enforced expression of GATA-1 using the identical retroviral transduction system can induce MegE commitment excluding other fate outcomes in HSCs, and supports their differentiation into mature MegE cells in the absence of Epo (Iwasaki et al. 2003). In the present study, enforced GATA-2 in HSCs did not increase the frequency of their MegE lineage readout, nor did it permit MegE differentiation in the absence of Epo (data not shown). The enforced expression of GATA-1 mainly induced conversion into the MegE lineage in CLPs and GMPs (Iwasaki et al. 2003). In contrast, enforced GATA-2 specified GMPs into EoPs, while it converted CLPs into BMCPs (Figs. 2, 5). The marked contrast between GATA-1 and GATA-2 functions in our system suggests that it is critical to use a homogenous cell population at a defined developmental stage to correctly evaluate transcription factor functions. These are interesting phenomena because in all of these cases, enforced GATA-1 or GATA-2 up-regulated the expression of the other after transduction (Figs. 2D, 6A; Iwasaki et al. 2003). Since GATA-1 and GATA-2 have some redundant properties (Takahashi et al. 2000), it is important to test the absolute requirement of GATA-1 or GATA-2 at the GMP stage for eosinophil or basophil development by utilizing conditional knockout systems, for example. It is also possible that another MegE/eosinophil-related transcription factor, Friend of GATA-1 (FOG-1) (Tsang et al. 1997, 1998), may be involved in GATA-1- or GATA- 2-dependent lineage specification. FOG-1 is required for normal MegE development, but in turn, exerts inhibitory effects on eosinophil development from a multipotent cell line (Querfurth et al. 2000). FOG-1 is not expressed in EoPs, BMCPs, BaPs, and MCPs (data not shown). FOG-1 was transactivated in GATA-1-transduced GMPs and CLPs (Iwasaki et al. 2003) but not in GATA-2-transduced GMPs (Fig. 2D). Thus, it is tempting to speculate that FOG-1 can be transactivated only when the GATA-1 expression precedes GATA-2, or when GATA-1 is predominant (by a retroviral transduction in our experiments) as compared with GATA-2. These possible interplays of GATA factors and FOG-1, dependent upon their timing or levels of expression, should also be evaluated in future studies.
Our data regarding GATA-1 and GATA-2 effects on lineage instruction are not entirely consistent with previous papers using different systems. A previous report showed that a Myb-Ets-transformed chicken myelo-blasts converted into thromboblasts by the enforced expression of GATA-1, while they differentiated into eosinophils when GATA-1 was expressed at a lower level (Kulessa et al. 1995). In our study, enforced GATA-2 in CLPs and GMPs was associated with GATA-1 up-regulation: The expression level of GATA-1 in GATA-2+ CLPs was ∼1.2- and twofold higher than that in normal BaPs and EoPs, respectively, while approximately twofold less than that in normal MEPs on a quantitative real-time PCR assay (data not shown). Although it is difficult to judge whether such level of GATA-1 reflects its physiological expression level at the EoP, BaP, or MEP stages, the principal mechanism of GATA-2-induced eosinophil lineage instruction in GMPs and CLPs could be dependent upon a combinatory action of C/EBPα and a medium level of secondary GATA-1. This possibility should also be tested in a future study by the enforced expression of graded levels of GATA-1 at the GMP or CLP stage.
Another report showed that human CD34+ cord blood cells mainly generated eosinophil colonies by the enforced expression of either murine GATA-1 or GATA-2 (Hirasawa et al. 2002). This difference could be due to heterogeneity of the CD34+ cord blood population that contains HSCs as well as all myelo-erythroid progenitors (Manz et al. 2002). It is also important to note that there may be a significant difference in roles of GATA factors in human and mouse hematopoiesis: We found that in human hematopoiesis, mature eosinophils expressed GATA-1 but not GATA-2, while basophils expressed only GATA-2 (Y. Mori and H. Iwasaki, unpubl.). This suggests that one should be careful to interpret data of transcription factor expression and functions generated in different species.
In conclusion, our data suggest that the order of activation or inactivation of C/EBPα and GATA-2 plays a pivotal role in lineage specification into myeloid cells. Presumably, induction of conformational changes of chromatin by a preceding transcription factor alters accessibility to differentiation programs induced by a subsequently activated one. This timing-based lineage instruction by multiple transcription factors should lead to formation of hematopoietic hierarchy through step-wise lineage choices, which could be one of the critical mechanisms for lineage fate decisions in a variety of stem cell systems.
Materials and methods
Mice
C57BL/6J mice were purchased from The Jackson Laboratory. Generation of C/EBPα−/+ and C/EBPαF/F mice was previously reported (Zhang et al. 1997, 2004). All mice were bred and maintained in the Research Animal Facility at Dana-Farber Cancer Institute in accordance with the guidelines.
Antibodies, cell staining, and sorting
Sorting of HSCs and CLPs was accomplished by staining bone marrow cells with biotinylated anti-IL-7Rα chain (A7R34) monoclonal antibodies (eBioscience), FITC-conjugated anti-Sca-1 (E13-161-7) (Pharmingen), APC-conjugated anti-c-Kit (2B8) (Pharmingen), and PE-Cy5-conjugated rat antibodies specific for the following lineage markers: CD3 (CT-CD3), CD4 (RM4-5), CD8 (5H10), B220 (6B2), Gr-1 (8C5), and CD19 (6D5) (Caltag), followed by avidin-PE (Caltag). HSCs and CLPs were sorted as IL-7Rα−Lin−Sca-1hic-Kithi and IL-7Rα+Lin−Sca-1loc-Kitlo populations, respectively (Kondo et al. 1997). For myeloid progenitor sorting, bone marrow and FL cells were stained with purified rat anti-IL-7Rα chain monoclonal antibodies (A7R34) (eBioscience) and the lineage cocktail described above. IL-7Rα−Lin− cells were negatively selected by using sheep antirat IgG-conjugated magnetic beads (Dynabeads M-450; Dynal A.S.), and the remaining IL-7Rα+Lin+ cells were visualized by staining with PE-Cy5-conjugated goat anti-rat IgG antibodies (Caltag). After blocking with purified rat IgG (Sigma), cells were stained with PE-conjugated anti-FcγRII/III (2.4G2), FITC-conjugated anti-CD34 (RAM34), APC-conjugated anti-c-Kit (2B8), and biotinylated anti-Sca-1 (E13-161-7) monoclonal antibodies (Pharmingen), followed by avidin-APC/Cy7 (Caltag). Myeloid progenitors were sorted as IL-7Rα−Lin−Sca-1−c-Kit+CD34+FcγRII/IIIlo (CMPs), IL-7Rα−Lin−Sca-1−c-Kit+CD34+FcγRII/IIIhi (GMPs), and IL-7Rα−Lin−Sca-1−c-Kit+CD34−FcγRII/IIIlo (MEPs) as described previously (Akashi et al. 2000). To sort EoPs, pre-enriched IL-7Rα−Lin− bone marrow cells were stained with biotinylated anti-IL-5Rα chain (H7) (Yamaguchi et al. 1990), FITC-conjugated anti-CD34 (RAM34), APC-conjugated anti-c-Kit (2B8), and PE-Cy5-conjugated anti-Sca-1 (D7) (eBioscience) monoclonal antibodies, followed by avidin-PE. EoPs were purified as IL-7Rα−Lin−Sca-1−CD34+IL-5Rα+c-Kitlo cells with a low level of side scatter profile as described (Iwasaki et al. 2005). For BMCP sorting, Lin− spleen cells were stained with FITC-conjugated anti-T1/ST2, PE-conjugated anti-β7 integrin, APC-conjugated anti-c-Kit (2B8), and biotinylated anti-FcγRII/III (2.4G2) (Pharmingen), followed by avidin-APC/Cy7 (Caltag). BMCPs were sorted as Lin−c-Kit+FcγRII/IIIhiβ7hi cells as described (Arinobu et al. 2005). For BaP or MCP sorting, bone marrow or intestinal cells were stained with FITC-conjugated anti-CD34 (RAM34), PE-conjugated anti-FcεRIα (MAR-1), APC-conjugated anti-c-Kit (2B8), and/or APC/Cy7-conjugated anti-CD45.2. BaPs and MCPs were purified as Lin−CD34+FcεRIαhic-Kit− bone marrow cells and CD45.2+Lin−CD34+β7hiFcεRIαlo intestinal cells, respectively (Arinobu et al. 2005). All of these stem and progenitor cells were double-sorted using a highly modified double laser (488 nm/350 nm Enterprise II + 647 nm Spectrum) high-speed FACS (Moflo-MLS, Cytomation). For all analyses and sorts, dead cells were excluded by propidium iodide staining. For single-cell and limiting dilution assays, cells were directly sorted into 60-well Terasaki plates or 96-well plates by using an automatic cell deposition unit (ACDU) system. Data were analyzed with FlowJo software (Treestar, Inc.).
Cell cultures
Cells were cultured in Iscove's Modified Dulbecco's Medium (IMDM; Invitrogen) supplemented with 20% fetal calf serum (FCS), 50 μM 2-mercaptoethanol, 100 U penicillin, and 100 μg/ mL streptomycin (Invitrogen). For clonogenic analyses, cells were cultured in methylcellulose medium (Methocult H4100, Stem Cell Technologies) supplemented with 30% FCS, 1% bovine serum albumin, and 2 mM L-glutamine (Stem Cell Technologies). Cytokines such as murine Slf (20 ng/mL), IL-3 (20 ng/mL), IL-5 (50 ng/mL), IL-6 (20 ng/mL), IL-7 (20 ng/mL), IL-11 (10 ng/mL), GM-CSF (10 ng/mL), Epo (2 U/mL), and Tpo (10 ng/mL) (R&D Systems) were added at the initiation of cultures. All cultures were incubated at 37°C in a humidified chamber under 5% CO2. Cell components of each colonies or cultures were determined morphologically and cytochemically by May-Giemsa and Toluidine Blue staining.
Retroviral transduction of stem and progenitor cells
The MSCV-Flag-mGATA-2-ires-EGFP retrovirus vector was provided by Dr. Atsushi Iwama. The MSCV-mC/EBPα-ireshCD4 retrovirus vector was kindly provided by Dr. Thomas Graf. A Cre cDNA was subcloned into the EcoRI site of the MSCV-ires-EGFP vector. The virus supernatant was obtained from cultures of 293T cells cotransfected with the target retro-virus vector, and gag-pol and VSV-G expression plasmids using a standard CaPO4 coprecipitation method. FACS-purified progenitors were plated onto a recombinant fibronectin fragment-coated culture dish (RetroNectin dish; Takara) with 1 mL of the virus supernatant containing the respective cytokine cocktail (for CMPs and GMPs, Slf [20 ng/mL] and IL-11 [10 ng/mL]; for MCPs, Slf [20 ng/mL], IL-3 [20 ng/mL], and IL-6 [20 ng/mL]; for CLPs, Slf [20 ng/mL] and IL-7 [20 ng/mL]), and cultured for 24–48 h. In the case of CLPs, prior to virus infection, FACS-purified cells were stimulated for 18 h in the presence of Slf (20 ng/mL) and IL-7 (20 ng/mL). At the completion of transduction, cells positive for GFP or hCD4 were purified by FACS, and were subsequently subjected to further analyses.
Conditional C/EBPα disruption in GMPs
GMPs were purified from C/EBPαF/F mice bone marrow and infected with MSCV-Cre-ires-EGFP retroviruses as described above. Following transduction, cells were isolated into EGFP− and EGFP+ populations and assessed for a recombination by PCR using the following primers sets (5′-TGGCCTGGAGAC GCAATGA-3′ and 5′-CGCAGAGATTGTGCGTCTTT-3′ for the C/EBPαF allele [269-base-pair {bp} product]; 5′-GCCTGG TAAGCCTAGCAATCCT-3′ and 5′-TGGAAACTTGGGT TGGGTGT-3′ for the C/EBPαΔ allele [400-bp product]). Isolated EGFP− (C/EBPαF/F) and EGFP+ (C/EBPαΔ/Δ) GMPs were subjected to limiting dilution assays for their lineage readouts.
Design of retrovirus vectors for shRNA expression directed against C/EBPα
To suppress the expression level of C/EBPα to various extents, we prepared a series of retrovirus vectors that possess expression cassettes of shRNA against C/EBPα (GenBank accession no. BC058161). Target sequences were selected according to rules proposed by Tuschl's group (Elbashir et al. 2001), using small interfering RNA (siRNA) Selection Program of Whitehead Institute for Biomedical Research (Yuan et al. 2004). Oligo-nucleotides were synthesized to form shRNA on transcription, in order of sense, loop (lower case), and antisense sequences (Brummelkamp et al. 2002) as follows: C118, 5′-TGGAGTCG GCCGACTTCTAttcaagagaTAGAAGTCGGCCGACTCCA-3′; C939, 5′-GCCAAGAAGTCGGTGGACAttcaagagaTGTCCAC CGACTTCTTGGC-3′; C1015, 5′-GCCGAGATAAAGCCAA ACAttcaagagaTGTTTGGCTTTATCTCGGC-3′. These oligo-nucleotides were annealed with their complementary ones and cloned subsequently into pSUPER vector (kindly provided by Dr. R. Agami) carrying an shRNA expression cassette driven by an H1 promoter with some modifications at the promoter site. After confirming their sequences, these shRNA cassettes were introduced into a 3′LTR region of the pQCXIN retrovirus vector (Clontech), in which a neomycin-resistant gene was replaced with an EGFP gene.
Analysis of gene expression from total RNA
Total RNA extracted from 2000 cells for each population was subjected to semiquantitative RT–PCR or real-time PCR analyses as described previously (Iwasaki et al. 2003; Arinobu et al. 2005). Primer and probe sequences and PCR protocols for each specific gene are shown in Supplementary Table 1.
Acknowledgments
We thank A.B. Cantor, M.F. Gurish, and K.F. Austen for helpful discussions. We thank Izumi Noguchi for expert assistance with preparation of the manuscript. This work was supported in part by grants from NIH (DK050654, DK061320, and CA072009 to K.A.; HL56745 to D.G.T.), the Ministry of Education, Culture, Sports, Science, and Technology, Japan (no. 18604006 to H.I.; a Special Research Grant-In-Aid for Development of Characteristic Education to K.A.), Damon-Runyon Cancer Research to K.A., Takeda Science Foundation to H.I., and Uehara Memorial Foundation to S.M.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1493506
References
- Akashi, K., Traver, D., Miyamoto, T., Weissman, I.L. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- Arinobu, Y., Iwasaki, H., Gurish, M.F., Mizuno, S.I., Shigematsu, H., Ozawa, H., Tenen, D.G., Austen, K.F., Akashi, K. Developmental checkpoints of the basophil/ mast cell lineages in adult murine hematopoiesis. Proc. Natl. Acad. Sci. 2005;102:18105–18110. doi: 10.1073/pnas.0509148102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba, Y., Garrett, K.P., Kincade, P.W. Constitutively active β-catenin confers multilineage differentiation potential on lymphoid and myeloid progenitors. Immunity. 2005;23:599–609. doi: 10.1016/j.immuni.2005.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce, J.A., Friend, D., Matsumoto, R., Austen, K.F., Owen, W.F. Differentiation in vitro of hybrid eosinophil/basophil granulocytes: Autocrine function of an eosinophil developmental intermediate. J. Exp. Med. 1995;182:49–57. doi: 10.1084/jem.182.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp, T.R., Bernards, R., Agami, R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Dahl, R., Walsh, J.C., Lancki, D., Laslo, P., Iyer, S.R., Singh, H., Simon, M.C. Regulation of macrophage and neutrophil cell fates by the PU.1:C/EBPα ratio and granulocyte colony-stimulating factor. Nat. Immunol. 2003;4:1029–1036. doi: 10.1038/ni973. [DOI] [PubMed] [Google Scholar]
- DeKoter, R.P., Singh, H. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science. 2000;288:1439–1441. doi: 10.1126/science.288.5470.1439. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Lendeckel, W., Tuschl, T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes & Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara, Y., Browne, C.P., Cunniff, K., Goff, S.C., Orkin, S.H. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc. Natl. Acad. Sci. 1996;93:12355–12358. doi: 10.1073/pnas.93.22.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli, S.J. Mast cells and basophils. Curr. Opin. Hematol. 2000;7:32–39. doi: 10.1097/00062752-200001000-00007. [DOI] [PubMed] [Google Scholar]
- Harigae, H., Takahashi, S., Suwabe, N., Ohtsu, H., Gu, L., Yang, Z., Tsai, F.Y., Kitamura, Y., Engel, J.D., Yamamoto, M. Differential roles of GATA-1 and GATA-2 in growth and differentiation of mast cells. Genes Cells. 1998;3:39–50. doi: 10.1046/j.1365-2443.1998.00166.x. [DOI] [PubMed] [Google Scholar]
- Heyworth, C., Pearson, S., May, G., Enver, T. Transcription factor-mediated lineage switching reveals plasticity in primary committed progenitor cells. EMBO J. 2002;21:3770–3781. doi: 10.1093/emboj/cdf368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirasawa, R., Shimizu, R., Takahashi, S., Osawa, M., Takayanagi, S., Kato, Y., Onodera, M., Minegishi, N., Yamamoto, M., Fukao, K., et al. Essential and instructive roles of GATA factors in eosinophil development. J. Exp. Med. 2002;195:1379–1386. doi: 10.1084/jem.20020170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki, H., Mizuno, S., Wells, R.A., Cantor, A.B., Watanabe, S., Akashi, K. GATA-1 converts lymphoid and myelomonocytic progenitors into the megakaryocyte/erythrocyte lineages. Immunity. 2003;19:451–462. doi: 10.1016/s1074-7613(03)00242-5. [DOI] [PubMed] [Google Scholar]
- Iwasaki, H., Mizuno, S., Mayfield, R., Shigematsu, H., Arinobu, Y., Seed, B., Gurish, M.F., Takatsu, K., Akashi, K. Identification of eosinophil lineage-committed progenitors in the murine bone marrow. J. Exp. Med. 2005;201:1891–1897. doi: 10.1084/jem.20050548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki-Arai, J., Iwasaki, H., Miyamoto, T., Watanabe, S., Akashi, K. Enforced granulocyte/macrophage colony-stimulating factor signals do not support lymphopoiesis, but instruct lymphoid to myelomonocytic lineage conversion. J. Exp. Med. 2003;197:1311–1322. doi: 10.1084/jem.20021843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jippo, T., Mizuno, H., Xu, Z., Nomura, S., Yamamoto, M., Kitamura, Y. Abundant expression of transcription factor GATA-2 in proliferating but not in differentiated mast cells in tissues of mice: Demonstration by in situ hybridization. Blood. 1996;87:993–998. [PubMed] [Google Scholar]
- Kondo, M., Weissman, I.L., Akashi, K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- Kondo, M., Scherer, D.C., Miyamoto, T., King, A.G., Akashi, K., Sugamura, K., Weissman, I.L. Cell-fate conversion of lymphoid-committed progenitors by instructive actions of cytokines. Nature. 2000;407:383–386. doi: 10.1038/35030112. [DOI] [PubMed] [Google Scholar]
- Kulessa, H., Frampton, J., Graf, T. GATA-1 reprograms avian myelomonocytic cell lines into eosinophils, thromboblasts, and erythroblasts. Genes & Dev. 1995;9:1250–1262. doi: 10.1101/gad.9.10.1250. [DOI] [PubMed] [Google Scholar]
- Manz, M.G., Miyamoto, T., Akashi, K., Weissman, I.L. Prospective isolation of human clonogenic common myeloid progenitors. Proc. Natl. Acad. Sci. 2002;99:11872–11877. doi: 10.1073/pnas.172384399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, D.I., Zon, L.I., Mutter, G., Orkin, S.H. Expression of an erythroid transcription factor in megakaryocytic and mast cell lineages. Nature. 1990;344:444–447. doi: 10.1038/344444a0. [DOI] [PubMed] [Google Scholar]
- McNagny, K., Graf, T. Making eosinophils through subtle shifts in transcription factor expression. J. Exp. Med. 2002;195:F43–F47. doi: 10.1084/jem.20020636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliaccio, A.R., Rana, R.A., Sanchez, M., Lorenzini, R., Centurione, L., Bianchi, L., Vannucchi, A.M., Migliaccio, G., Orkin, S.H. GATA-1 as a regulator of mast cell differentiation revealed by the phenotype of the GATA-1low mouse mutant. J. Exp. Med. 2003;197:281–296. doi: 10.1084/jem.20021149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerlov, C. C/EBPα mutations in acute myeloid leukaemias. Nat. Rev. Cancer. 2004;4:394–400. doi: 10.1038/nrc1363. [DOI] [PubMed] [Google Scholar]
- Nerlov, C., Graf, T. PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes & Dev. 1998;12:2403–2412. doi: 10.1101/gad.12.15.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerlov, C., McNagny, K.M., Doderlein, G., Kowenz-Leutz, E., Graf, T. Distinct C/EBP functions are required for eosinophil lineage commitment and maturation. Genes & Dev. 1998;12:2413–2423. doi: 10.1101/gad.12.15.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerlov, C., Querfurth, E., Kulessa, H., Graf, T. GATA-1 interacts with the myeloid PU.1 transcription factor and represses PU.1-dependent transcription. Blood. 2000;95:2543–2551. [PubMed] [Google Scholar]
- Orkin, S.H. Diversification of haematopoietic stem cells to specific lineages. Nat. Rev. Genet. 2000;1:57–64. doi: 10.1038/35049577. [DOI] [PubMed] [Google Scholar]
- Querfurth, E., Schuster, M., Kulessa, H., Crispino, J.D., Doderlein, G., Orkin, S.H., Graf, T., Nerlov, C. Antagonism between C/EBPβ and FOG in eosinophil lineage commitment of multipotent hematopoietic progenitors. Genes & Dev. 2000;14:2515–2525. doi: 10.1101/gad.177200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, V.A., Iwama, A., Iotzova, G., Schulz, M., Elsasser, A., Vangala, R.K., Tenen, D.G., Hiddemann, W., Behre, G. Granulocyte inducer C/EBPα inactivates the myeloid master regulator PU.1: Possible role in lineage commitment decisions. Blood. 2002;100:483–490. doi: 10.1182/blood.v100.2.483. [DOI] [PubMed] [Google Scholar]
- Rekhtman, N., Radparvar, F., Evans, T., Skoultchi, A.I. Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: Functional antagonism in erythroid cells. Genes & Dev. 1999;13:1398–1411. doi: 10.1101/gad.13.11.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg, M.E. Eosinophilia. N. Engl. J. Med. 1998;338:1592–1600. doi: 10.1056/NEJM199805283382206. [DOI] [PubMed] [Google Scholar]
- Scott, E.W., Simon, M.C., Anastasi, J., Singh, H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- Sieweke, M.H., Graf, T. A transcription factor party during blood cell differentiation. Curr. Opin. Genet. Dev. 1998;8:545–551. doi: 10.1016/s0959-437x(98)80009-9. [DOI] [PubMed] [Google Scholar]
- Takahashi, S., Shimizu, R., Suwabe, N., Kuroha, T., Yoh, K., Ohta, J., Nishimura, S., Lim, K.C., Engel, J.D., Yamamoto, M. GATA factor transgenes under GATA-1 locus control rescue germline GATA-1 mutant deficiencies. Blood. 2000;96:910–916. [PubMed] [Google Scholar]
- Tenen, D.G., Hromas, R., Licht, J.D., Zhang, D.E. Transcription factors, normal myeloid development, and leukemia. Blood. 1997;90:489–519. [PubMed] [Google Scholar]
- Traver, D., Akashi, K., Manz, M., Merad, M., Miyamoto, T., Engleman, E.G., Weissman, I.L. Development of CD8α-positive dendritic cells from a common myeloid progenitor. Science. 2000;290:2152–2154. doi: 10.1126/science.290.5499.2152. [DOI] [PubMed] [Google Scholar]
- Tsai, F.Y., Orkin, S.H. Transcription factor GATA-2 is required for proliferation/survival of early hematopoietic cells and mast cell formation, but not for erythroid and myeloid terminal differentiation. Blood. 1997;89:3636–3643. [PubMed] [Google Scholar]
- Tsai, F.Y., Browne, C.P., Orkin, S.H. Knock-in mutation of transcription factor GATA-3 into the GATA-1 locus: Partial rescue of GATA-1 loss of function in erythroid cells. Dev. Biol. 1998;196:218–227. doi: 10.1006/dbio.1997.8842. [DOI] [PubMed] [Google Scholar]
- Tsang, A.P., Visvader, J.E., Turner, C.A., Fujiwara, Y., Yu, C., Weiss, M.J., Crossley, M., Orkin, S.H. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell. 1997;90:109–119. doi: 10.1016/s0092-8674(00)80318-9. [DOI] [PubMed] [Google Scholar]
- Tsang, A.P., Fujiwara, Y., Hom, D.B., Orkin, S.H. Failure of megakaryopoiesis and arrested erythropoiesis in mice lacking the GATA-1 transcriptional cofactor FOG. Genes & Dev. 1998;12:1176–1188. doi: 10.1101/gad.12.8.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, J.C., DeKoter, R.P., Lee, H.J., Smith, E.D., Lancki, D.W., Gurish, M.F., Friend, D.S., Stevens, R.L., Anastasi, J., Singh, H. Cooperative and antagonistic interplay between PU.1 and GATA-2 in the specification of myeloid cell fates. Immunity. 2002;17:665–676. doi: 10.1016/s1074-7613(02)00452-1. [DOI] [PubMed] [Google Scholar]
- Wedemeyer, J., Tsai, M., Galli, S.J. Roles of mast cells and basophils in innate and acquired immunity. Curr. Opin. Immunol. 2000;12:624–631. doi: 10.1016/s0952-7915(00)00154-0. [DOI] [PubMed] [Google Scholar]
- Weil, S.C., Hrisinko, M.A. A hybrid eosinophilic–basophilic granulocyte in chronic granulocytic leukemia. Am. J. Clin. Pathol. 1987;87:66–70. doi: 10.1093/ajcp/87.1.66. [DOI] [PubMed] [Google Scholar]
- Xie, H., Ye, M., Feng, R., Graf, T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117:663–676. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, N., Hitoshi, Y., Mita, S., Hosoya, Y., Murata, Y., Kikuchi, Y., Tominaga, A., Takatsu, K. Characterization of the murine interleukin 5 receptor by using a monoclonal antibody. Int. Immunol. 1990;2:181–187. doi: 10.1093/intimm/2.2.181. [DOI] [PubMed] [Google Scholar]
- Yuan, B., Latek, R., Hossbach, M., Tuschl, T., Lewitter, F. siRNA Selection Server: An automated siRNA oligo-nucleotide prediction server. Nucleic Acids Res. 2004;32:W130–W134. doi: 10.1093/nar/gkh366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D.E., Zhang, P., Wang, N.D., Hetherington, C.J., Darlington, G.J., Tenen, D.G. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein α-deficient mice. Proc. Natl. Acad. Sci. 1997;94:569–574. doi: 10.1073/pnas.94.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, P., Behre, G., Pan, J., Iwama, A., Wara-Aswapati, N., Radomska, H.S., Auron, P.E., Tenen, D.G., Sun, Z. Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1. Proc. Natl. Acad. Sci. 1999;96:8705–8710. doi: 10.1073/pnas.96.15.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, P., Zhang, X., Iwama, A., Yu, C., Smith, K.A., Mueller, B.U., Narravula, S., Torbett, B.E., Orkin, S.H., Tenen, D.G. PU.1 inhibits GATA-1 function and erythroid differentiation by blocking GATA-1 DNA binding. Blood. 2000;96:2641–2648. [PubMed] [Google Scholar]
- Zhang, P., Iwasaki-Arai, J., Iwasaki, H., Fenyus, M.L., Dayaram, T., Owens, B.M., Shigematsu, H., Levantini, E., Huettner, C.S., Lekstrom-Himes, J.A., et al. Enhancement of hematopoietic stem cell repopulating capacity and self-renewal in the absence of the transcription factor C/EBP α. Immunity. 2004;21:853–863. doi: 10.1016/j.immuni.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Zon, L.I., Yamaguchi, Y., Yee, K., Albee, E.A., Kimura, A., Bennett, J.C., Orkin, S.H., Ackerman, S.J. Expression of mRNA for the GATA-binding proteins in human eosinophils and basophils: Potential role in gene transcription. Blood. 1993;81:3234–3241. [PubMed] [Google Scholar]