Abstract
Leigh syndrome French Canadian variant (LSFC) is an autosomal recessive neurodegenerative disorder due to mutation in the LRP130 (leucine-rich protein 130 kDa) gene. Unlike classic Leigh syndrome, the French Canadian variant spares the heart, skeletal muscle, and kidneys, but severely affects the liver. The precise role of LRP130 in cytochrome c oxidase deficiency and hepatic lactic acidosis that accompanies this disorder is unknown. We show here that LRP130 is a component of the PGC-1α (peroxisome proliferator-activated receptor coactivator 1-α) transcriptional coactivator holocomplex and regulates expression of PEPCK (phosphoenolpyruvate carboxykinase), G6P (glucose-6-phosphatase), and certain mitochondrial genes through PGC-1α. Reduction of LRP130 in fasted mice via adenoviral RNA interference (RNAi) vector blocks the induction of PEPCK and G6P, and blunts hepatic glucose output. LRP130 is also necessary for PGC-1α-dependent transcription of several mitochondrial genes in vivo. These data link LRP130 and PGC-1α to defective hepatic energy homeostasis in LSFC, and reveal a novel regulatory mechanism of glucose homeostasis.
Keywords: PGC-1α, LRP130, LPRPRC, Leigh syndrome, gluconeogenesis
Leigh disease, also called subacute necrotizing encephalopathy or Leigh syndrome, is a progressive neurodegenerative disorder characterized by Denis Leigh in 1951 (Leigh 1951; V.A. McKusick, Online Mendelian Inheritance in Man (OMIM); http://www.ncbi.nlm.nih.gov/omim). Patients suffer progressive decline of their central nervous system due to focal necrotizing lesions of the brainstem, basal ganglia, and cerebellum, accompanied by capillary proliferation. Clinical hallmarks include psychomotor delay, truncal ataxia, intention tremor, lactic acidosis of the blood and cerebrospinal fluid, and death between ages 6 mo and 12 yr. Genetic and biochemical studies implicated several molecules and enzymes involved in ATP production as etiologic factors (Leigh 1951; Merante et al. 1993; Morin et al. 1993; Robinson 2000; V.A. McKusick, OMIM; http://www.ncbi.nlm.nih.gov/omim).
Cytochrome c oxidase deficiency is particularly common in Leigh disease. Cytochrome c oxidase (complex IV or COX) completes the terminal event of electron transfer by transferring electrons from cytochrome c to molecular oxygen. COX is comprised of 13 subunits, three of which (COX I, COX II, and COX III) are encoded by the mitochondrial genome. Mutations in nuclear-encoded assembly factors prove causative in some patients with Leigh syndrome (Stryer 1988; Robinson 2000; Shoubridge 2001). Patients with Leigh syndrome due to COX deficiency generally have COX activity <25% of normal in all tissues. In addition to neurological defects, these patients may exhibit cardiomyopathy, renal disease, and severe myopathy accompanied by profound weakness and moderate lactic acidosis (Morin et al. 1993; Robinson 2000). Leigh syndrome French Canadian variant (LSFC) is a notable exception. Besides the brain, only the liver exhibits marked reduction in COX activity. Therefore, these patients do not exhibit signs or symptoms of cardiomyopathy, renal disease, or severe myopathy (Merante et al. 1993; Morin et al. 1993). LSFC patients still exhibit chronic moderate lactic acidosis and episodes of fatal lactic acidosis during bouts of febrile illness, suggesting a defect in hepatic energy metabolism.
LSFC was identified in northeastern Quebec-Saguenay-Lac-Saint Jean and Charlevoix (Merante et al. 1993; Lin et al. 2004). It maps to chromosome 2p16 (Lee et al. 2001) and is a recessive mutation caused by a C ® T transition at 1119 base pairs (bp) of the LRP130 (leucine-rich protein 130 kDa) gene. This results in a missense mutation to valine from a highly conserved alanine at amino acid 354 (Mootha et al. 2003). LRP130, official nomenclature LRPPRC (leucine-rich pentatricopeptide repeat motifcontaining protein; OMIM 607544), was initially isolated as a lecitin-binding protein of unknown function from HepG2 cells (Hou et al. 1994) and contains 11 pentatricopeptide repeats, which have been implicated in mitochondrial RNA processing (Small and Peeters 2000; Mili and Pinol-Roma 2003; Tsuchiya et al. 2004). It is conserved from Drosophila to human and has homology with yeast PET309p, a molecule involved in mitochondrial metabolism (Mili and Pinol-Roma 2003). No studies have unraveled the biology and molecular physiology of LRP130 nor its function in patients with LSFC. One study suggests that the mutant protein is unstable and noted a specific decline in COX I and possibly COX III mRNA (Xu et al. 2004). This is in contrast to a report demonstrating normal levels of complex IV subcomponents, suggesting that LRP130 might be an assembly factor (Merante et al. 1993). Another study suggested that LRP130 binds a specific DNA element called inverted MED1 situated within two hepatic multidrug resistance gene promoters, MDR1 and MVP (Labialle et al. 2004). However, this study contradicted another study that suggests LRP130 only binds RNA and single-stranded DNA, not double-stranded DNA (Tsuchiya et al. 2004). Thus, no unifying molecular mechanism for the action of LRP130 can be readily discerned from available studies. In particular, no studies have addressed the unique and isolated hepatic phenotype of the patient population carrying mutations in this gene.
PGC-1α (peroxisome proliferator-activated receptor coactivator 1-α) was initially identified as a binding protein of PPARγ in brown fat and was shown to be sufficient to drive mitochondrial biogenesis in many tissues. It also regulates hepatic glucose homeostasis as well as many other aspects of energy metabolism in many other tissues (Puigserver et al. 1998; Wu et al. 1999; Scarpulla 2002a, b; St Pierre et al. 2003; Russell et al. 2004). Importantly, PGC-1α was shown to coactivate the broad family of nuclear receptors and many transcription factors outside the nuclear receptor superfamily. PGC-1α is induced in the liver by fasting and is elevated in murine models of both Type 1 and Type 2 diabetes milletus (Yoon et al. 2001). The rate of hepatic gluconeogenesis is established by the rate-limiting enzymes phosphoenolpyruvate carboxykinase (PEPCK) and glucose-6-phosphatase (G6P); both enzymes undergo transcriptional regulation following hormonal stimulation. The transcription factors FOXO1 and HNF4α are required for gluconeogenesis and transcriptionally activate PEPCK and G6P in an insulin-sensitive manner (Brunet et al. 1999; Nakae et al. 1999, 2002; Hall et al. 2000; Schmoll et al. 2000; Rhee et al. 2003). Moreover, PGC-1α potently coactivates both transcription factors (Puigserver et al. 2003; Rhee et al. 2003) and lies downstream from glucagon signaling. PGC-1α is both necessary and sufficient to drive gluconeogenic gene expression (Yoon et al. 2001; Puigserver et al. 2003; Koo et al. 2004; Lin et al. 2004), and PGC-1α is regulated by the transcription factor CREB in a fasting-dependent manner (Herzig et al. 2001).
Notably, PGC-1α functions as a transcriptional coactivator in the context of a large multiprotein complex. It has been previously shown that the N terminus binds to several other proteins having histone acetyltransferase activity, such as CBP/p300 and SRC-1 (Puigserver et al. 1999). Many components of the TRAP/DRIP/mediator complex bind near the C terminus and activate transcription via recruitment of RNA polymerase II (Wallberg et al. 2003) and perhaps via other mechanisms. To date, however, no biochemical purification of full-length PGC-1α with associated proteins has been reported. In this study, we have purified a complex of proteins assembled by PGC-1α from mammalian cell extracts and show that LRP130 constitutes a prominent component of this complex. Biochemical and functional analyses clearly indicate that LRP130 functions in mitochondrial and liver biology, at least in part, by modulating the function of PGC-1α.
Results
LRP130 in PGC-1α holocomplex and physical interaction between PGC-1α and LRP130
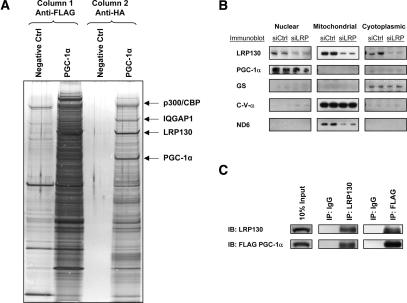
A biochemical approach was undertaken to isolate PGC-1α holocomplexes. PGC-1α protein complexes could not be purified directly from cells, since high ectopic expression of this coactivator interfered with the growth of the cells. We therefore devised a new method to purify PGC-1α protein complexes. Murine PGC-1α protein encoding N-terminal affinity tags HA and Flag was purified to homogeneity from bacteria. This preparation has been previously shown to support in vitro transcription (Wallberg et al. 2003). Purified PGC-1α protein (80 μg) was incubated in vitro with HeLa nuclear extract (100 mg) in high salt in order to disrupt protein interactions. The sample was slowly dialyzed to allow reassembly of specific complexes. PGC-1α protein complexes were then purified via tandem affinity chromatography over Flag and HA affinity columns, followed by native elution with respective peptides as previously described (Fig. 1A; Ikura et al. 2000). Bands from the purified complexes that underwent PAGE-SDS electrophoresis were visualized by Coomassie Brilliant Blue staining and were then subjected to MALDI-MS analysis.
Figure 1.
Purification of PGC-1α-specific protein complexes, and coimmunoprecipition in the nucleus. HeLa nuclear extract was incubated with purified PGC-1α protein in high salt. Following gradual dialysis and clarification, the complexes underwent sequential chromatography and native elution over Flag and HA affinity columns, respectively. (A) Silver staining showing aliquots of eluted complexes for negative control (no addition of purified protein) and PGC-1α samples following Flag chromatography (left two lanes) and HA chromatography (right two lanes). The arrows designate the positions of p300/CBP, LRP130, and PGC-1α.(B) Subcellular fractionation in H2.35 cells showing colocalization of LRP130 and PGC-1α to the nucleoplasmic fraction. Glycogen synthase (GS) is a cytoplasmic marker. C-V-α is a mitochondrial protein encoded in the nucleus. ND6 is a mitochondrial protein encoded in the mitochondrion. (C) Coimmunoprecipitation demonstrating interaction between LRP130 and PGC-1α in nucleus.
Three proteins were reproducibly and highly enriched by this procedure (Fig. 1A). CBP/p300, a bona fide binding partner for PGC-1α (Puigserver et al. 1999), was copurified, suggesting that the procedure afforded specificity. IQGAP1, a calmodulin-binding protein (Roy et al. 2004), was copurified, while LRP130 protein constituted a prominent band present in near-equimolar amount relative to PGC-1α. Both proteins share common expression in the nucleoplasmic fraction (Fig. 1B) and coimmunoprecipitate from the nuclear compartment (Fig. 1C). We concentrated on studies of LRP130, because of its known association with mitochondrial dysfunction in human patients and the well-established role of PGC-1α in the control of mitochondrial gene expression.
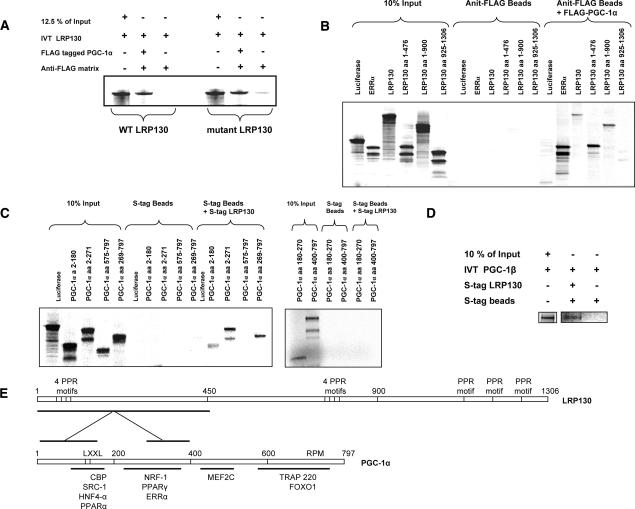
The interaction between full-length LRP130 and PGC-1α was studied using in vitro translated LRP130 incubated with purified PGC-1α. As shown in Figure 2A, the interaction between PGC-1α and LRP130 was robust. No discernable differences in binding were noted between wild-type and the mutant LRP130 protein present in LSFC patients. Truncations of LRP130 were used to map the regions of LRP130 that interact with PGC-1α (Fig. 2B). Three truncations of murine LRP130 were used— amino acids 1–476, 1–900, and 925–1306. Six PGC-1α truncations—amino acids 2–180, 2–271, 269–797, 575–797, 180–270, and 400–797—were used to define the regions of PGC-1α that interact with LRP130 (Fig. 2C). Based on the binding assays, a region within amino acids 1–476 of the N terminus of LRP130 binds most strongly with two regions of PGC-1α encompassing amino acids 1–180 and 270–400 (Fig. 2E). Additionally, LRP130 was shown to interact with PGC-1β, the closest homolog of PGC-1α (Fig. 2D). Homology comparison with the two interaction sites on PGC-1α was made to PGC-1β, and showed 49% and 46% homology, respectively (Supplementary Fig. 4).
Figure 2.
Physical interaction between LRP130 and PGC-1α. (A) Wild-type and mutant LRP130 were 35S-labeled by in vitro translation and incubated with purified Flag-tagged PGC-1α. (B) Truncations of LRP130 were 35S-labeled by in vitro translation and incubated with purified Flag-tagged PGC-1α. Anti-Flag matrix was used to immunopurify Flag-tagged PGC-1α protein bound to LRP130. (C) Truncations of PGC-1α or full-length PGC-1β were 35S-labeled and incubated with full-length LRP130. (D) PGC-1β was 35S-labeled and incubated with full-length LRP130. (E) Schematic of LRP130 and PGC-1α indicating regions of interaction.
Functional interaction between LRP130 and PGC-1α
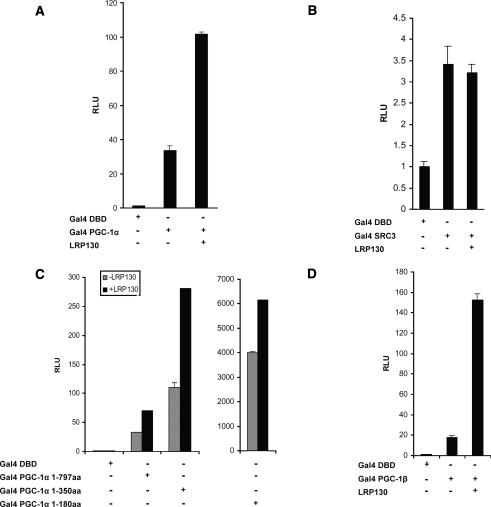
We first used luciferase reporter assays to investigate functional interactions between LRP130 and PGC-1α. The effect of LRP130 on the inherent transcriptional activity of PGC-1α was investigated using full-length PGC-1α fused to the DNA-binding domain of Gal4. LRP130 enhanced the transcriptional activity of PGC-1α fused to Gal4 DBD (DNA-binding domain) about two-to threefold. No such enhancement was noted when LRP130 was combined with the Gal4 DBD alone or SRC3 fused to Gal4 DBD (Fig. 3A,B). Similar findings were noted with truncated Gal4-PGC-1α constructs. We also examined the effect of LRP130 on PGC-1β, the closest homolog of PGC-1α. LRP130 increased the activity of Gal4-PGC-1β even more than that of Gal4-PGC-1α (eightfold to 10-fold for PGC-1β vs. two-to threefold for PGC-1α) (Fig. 3D). Thus, LRP130 increases the transcriptional activity of both PGC-1α and PGC-1β. Reciprocal effects were explored using LRP130-Gal4 fusions; however, no inherent transcription activity was noted. This may represent inherent lack of transcriptional activity or interference by the Gal4 fusion (data not shown).
Figure 3.
Transcription assays using either Gal4-PGC-1α or Gal4-PGC-1β with LRP130. The indicated constructs were transiently transfected into COS cells and analyzed for luciferase activity after 24 h. (A) LRP130 cotransfected with Gal4 PGC-1α. (B) LRP130 cotransfected with SRC3. (C) LRP130 cotransfected with Gal4-PGC-1α fragments 1–180 amino acids, 1–350 amino acids, and 1–797 amino acids. (D) LRP130 cotransfected with PGC-1β. Experiments were normalized to the Gal4 DNA-binding domain (Gal4-DBD). Each experiment was performed in duplicate and is representative of n ≥ 3 independent biological experiments. Error bars represent SEM.
Previous literature reported that the A354V LRP130 mutant protein is unstable and present at reduced levels in both the mitochondrial and nucleoplasmic fractions of patients carrying the homozygous mutation (Xu et al. 2004). To investigate the levels of the wild-type and mutant proteins, we first depleted the H2.35 hepatoma cell line of LRP130 using retroviral small interfering RNA (siRNA) (Supplementary Fig. 1), in order to minimize any effects of the wild-type protein on the expressed proteins. As shown in Supplementary Figure 2A, there was significantly less mutant protein 72 h after adenoviral infection relative to the wild-type protein. This is despite similar levels of wild-type and mutant LRP130 mRNA (Supplementary Fig. 2B). We revisited the Gal4-PGC-1α experiments using these conditions and found a 50% reduction in Gal-4-PGC-1α activity when comparing mutant to wild-type protein (Supplementary Fig. 2C).
LRP130 functions with PGC-1α in the regulation of gluconeogenic and mitochondrial genes
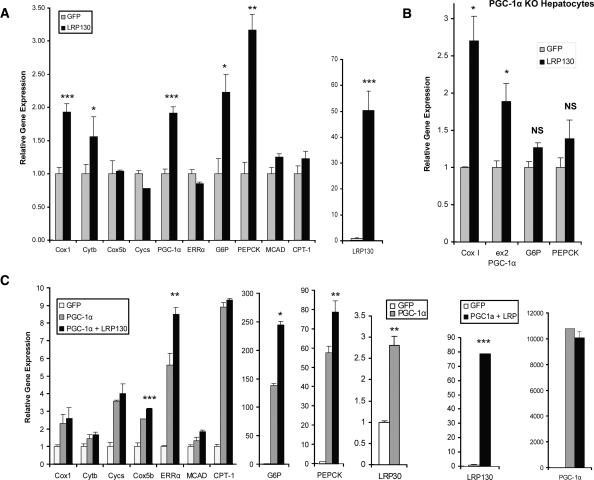
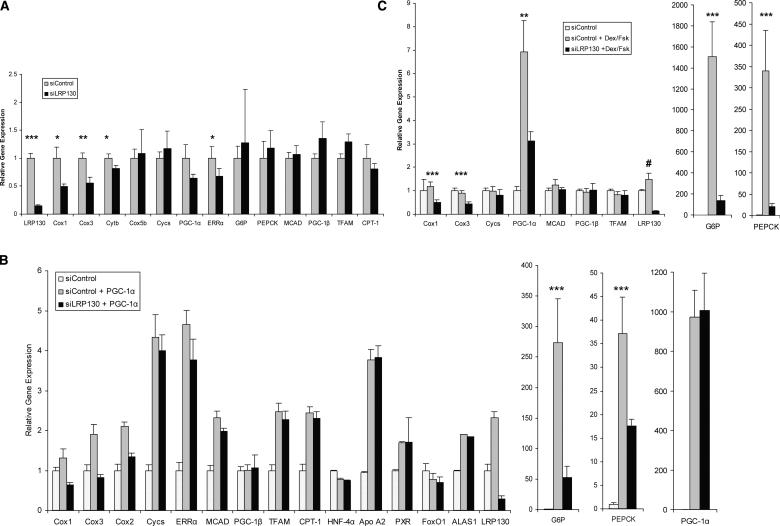
In order to ascertain the effects of LRP130 on endogenous PGC-1α target genes, we expressed LRP130 in primary hepatocytes using adenoviral vectors (Fig. 4A). Forced expression of LRP130 minimally impacted mRNA levels of nuclear-encoded genes involved in mitochondrial respiration and fatty acid oxidation such as cyctochrome c, Cox5b, MCAD, or CPT-1. Expression of ERRα, a potent transcription factor involved in mitochondrial biogenesis, was unaltered. Forced expression of LPR130 induced PGC-1α, PEPCK, and G6P mRNAs. Cytochrome b and Cox1 are encoded by the mitochondrial genome and are induced by LRP130 at the mRNA level as well. Although PGC-1α mRNA was induced 1.9-fold, this apparently was not sufficient to drive other known targets of PGC-1α. The positive effect of forced LRP130 expression on Cox I, PGC-1α, G6P, and PEPCK might be the result of modest PGC-1α induction or represent a PGC-1α-independent process. We addressed this using forced expression of LRP130 in primary hepatocytes, isolated from PGC-1α knockout mice. Remnant PGC-1α transcript was monitored by assaying exon 2. Notably, induction of gluconeogenic genes completely requires PGC-1α protein, while the induction of Cox I and PGC-1α gene expression does not require PGC-1α protein (Fig. 4B). Overall, these results demonstrate that LRP130 regulates a subset of PGC-1α target genes.
Figure 4.
Gene expression induced by LRP130 in primary hepatocytes. Transcription of genes responsive to PGC-1α coactivation and implicated in gluconeogenesis, mitochondrial respiration, and fatty acid oxidation were measured using RT quantitative PCR. (A) Forced expression of LRP130 in primary hepatocytes. Statistical comparisons are made between GFP and LRP130. (B) Effect of forced expression of LRP130 in primary knockout (KO) hepatocytes. Residual PGC-1α transcript was detected in the knockout animals using primers for exon 2 (ex2). (C) Coforced expression of LRP130 and PGC-1α in primary hepatocytes. Statistical comparisons are made between GFP coexpressed with PGC-1α and LRP130 coexpressed with PGC-1α. Experiments were performed in triplicate and are representative of n = 3 independent biological experiments. (*) p < 0.05; (**) p < 0.01; (***) p < 0.001. Error bars represent standard deviation.
We next examined possible cooperative effects between LRP130 and PGC-1α on endogenous genes by expressing PGC-1α, with or without exogenously expressed LRP130. Comparisons were made to a GFP control adenoviral vector. In the presence of exogenously expressed LRP130/PGC-1αα, PEPCK mRNA expression increased 37% (p < 0.05) and G6P mRNA 78% (p < 0.01), compared with PGC-1α expression without exogenously expressed LRP130 (Fig. 4C). Cox1 and Cytb are mitochondrially encoded genes, and neither showed further enhancement by coexpression of LRP130 and PGC-1α, compared with PGC-1α expression alone. Similarly, cytochrome c, MCAD, and CPT-1 exhibited no further enhancement. In the presence of coexpressed LRP130 and PGC-1α, Cox5b showed a small, but statistically significant change. Coexpression of LRP130 and PGC-1α stimulated gene expression of ERRα by 51%, compared with PGC-1α expression alone (p < 0.01) (Fig. 4C). Thus, LRP130 cooperates with PGC-1α to preferentially govern certain PGC-1α target genes.
LRP130 is a rather abundant mRNA and protein, so the effects of LRP130 reduction were studied through the use of an RNA interference (RNAi)-expressing adenovirus. Compared with an siControl, adenoviral expression of siLRP130 depleted >85% of endogenous LRP130 mRNA in primary murine hepatocytes (Fig. 5A). Depletion of LRP130 had no effect on nuclear-encoded mitochondrial genes such as cytochrome c, Cox5b, and ATP5o (Fig. 5B). ERRα (p < 0.05) and PGC-1α (trend p = 0.10) underwent reductions of ∼40% due to a reduction in LRP130. However, mitochondrial-encoded genes of the electron transport chain exhibited a much greater reduction—Cox1 (∼70%), Cox2, Cox3, Cytb (∼60%) (Fig. 5A). A second siRNA construct against LRP130 was designed and exhibited similar specificity (Supplementary Fig. 3). Additionally, the second RNAi construct was stably introduced into a nonhepatic cell line, and exhibited identical specificity for mitochondrion-encoded and nuclear encoded genes (data not shown), compared with our primary RNAi construct.
Figure 5.
Effect of depletion of LRP130 on endogenous gene expression. Primary hepatocytes were depleted of endogenous LRP130 using an RNAi construct designated siLRP130 or a scrambled control, siControl. Transcription of genes responsive to PGC-1α coactivation and implicated in gluconeogenesis, heme metabolism, mitochondrial respiration, and fatty acid oxidation was measured using RT quantitative PCR. (A) Forced expression of siControl or siLRP130. (B) Coforced expression of either siControl/GFP (siControl), siControl + PGC-1α, or siLRP130 + PGC-1α. (C) Forced expression of siControl or siLRP130 in the presence or absence of dexamethasone and forskolin (Dex/Fsk). Experiments were performed in triplicate and are respresentative of n = 3 independent biological experiments. (*) p < 0.05; (**) p < 0.01; (***) p < 0.001. Asterisks designate statistical comparisons made between siControl and siLRP130 (A) siControl + PGC-1α and siLRP130 + PGC-1α (B), and siControl + Dex/Fsk and siLRP130 + Dex/Fsk (C). (#) p < 0.01; stastistical comparisons are made between siControl and siControl + PGC-1α (B), and siControl and siControl + Dex/Fsk (C). The control for Dex/Fsk contained an appropriate vehicle. Error bars represent standard deviation.
Depletion of LRP130 in the presence of forced PGC-1α expression dramatically reduced the ability of this coactivator to increase transcription of both the PEPCK and G6P genes compared with siControl (Fig. 5B). Similar findings were noted for mitochondrial-encoded genes, including Cox1, Cox2, and Cox3. The effect was specific for this subset of genes in that no significant effect was noted on nuclear-encoded mitochondrial genes, controlled by PGC-1α, such as cytochrome c and CPT-1. Importantly, the PGC-1α mRNA level was identical for both siControl and siLRP130 groups, and HNF-4α and FoxO1 mRNA levels were similar as well (Fig. 5B).
Dexamethasone and forskolin treatment mimic the hormonal milieu present in the fasted state of mammals, and have been shown to induce endogenous PGC-1α (Lin et al. 2004). Depletion of LRP130 by RNAi profoundly reduced the transcription of endogenous PEPCK and G6P gene expression. Interestingly, PGC-1α induction was also significantly suppressed. However, the reduction in PGC-1α gene expression here was considerably less than the reduction in PEPCK and G6P mRNA gene expression (Fig. 5B). Again, the results appeared specific in that no such reduction in nuclear-encoded mitochondrial genes was noted, while reductions in mitochondrial-encoded genes were present (Fig. 5B). These data indicate that LRP130 and PGC-1α cooperate to regulate expression of certain mitochondrially encoded genes and gluconeogenic genes.
LRP130 augments the recruitment of PGC-1α to a FoxO1-binding site on chromatin
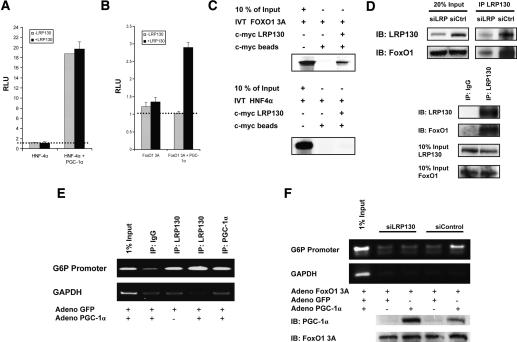
The data above illustrate that LRP130 is required for some of the actions of PGC-1α, but not for others. To address how LRP130 might augment or modify the actions of PGC-1α, we first compared the ability of LRP130 to modify the coactivation of two liver-enriched transcription factors that are targets of PGC-1α: HNF4α and FoxO1. As was originally shown by Yoon et al. (2001), PGC-1α coactivates HNF4α robustly. This occurs in cells depleted of LRP130 (Fig. 6A), and addition of exogenous LRP130 does not quantitatively influence the co-activation. In contrast, PGC-1α has lost its ability to coactivate FoxO1 in cells depleted of LRP130, while exogenous expression of LRP130 restores the coactivation function of PGC-1α (Fig. 6B).
Figure 6.
Effect of LRP130 on coactivation and docking of PGC-1α. H2.35 cells stably depleted of LRP130 were used for the coactivation studies. Exogenous LRP130 was used to restore LRP130 where indicated. (A) LRP130 was cotransfected with PGC-1α and HNF-4α as indicated, and a multimerized AF-1-binding element reporter construct was used. (B) LRP130 was cotransfected with PGC-1α and FoxO1 3A (constitutively active mutant) as indicated. A multimerized IRS-1-binding element reporter construct was used. The dotted line represents reporter construct alone. No statistical difference was observed between reporter alone versus reporter plus transcription factor, or for LRP130 and PGC-1α plus reporter. (C) Assessment of interactions between LRP130 and FoxO1 or HNF4α. FoxO1 or HNF4α was 35S-labeled and incubated with purified full-length S-tagged LRP130 protein. (D) Specific interaction of LRP130 with FoxO1 in liver and cells. (Top panel) Coimmunoprecipitation of LRP130 and FoxO1 from liver whole-cell extract showing dose effect of interaction. (Bottom panel) Coimmunoprecipitation from nuclear extract using H2.35 cells. (E) ChIP showing recruitment of LRP130 to the FoxO1-binding site in PGC-1α knockout (KO) primary hepatocytes. (Top panel) Radioactive ChIP processed by PhosphorImager. (F) ChIP in H2.35 cells stably depleted of LRP130 using an RNAi construct compared with a siControl cell line. The panels below indicate protein levels of PGC-1α and FoxO1 3A. Note, the upper specific band migrates in close proximity to the lower single-peak unreacted primer. Experiments were performed in n ≥ 3 independent biological experiments.
PGC-1α is a unique coactivator, and functions by bridging the recruitment of general coactivators to transcription factors, in a cellular and promoter-specific manner. Based on the coactivation studies in Figure 6, we reasoned that LRP130 might mediate the effects of PGC-1α on certain transcription factors by performing a similar role of bridging. In vitro assays demonstrated that LRP130 and FoxO1 interact (Fig. 6C). Coimmunoprecipitations using whole-cell extract from mouse liver showed a dose-dependent enrichment of FoxO1, when comparing siControl to siLRP130. Furthermore, we documented that this interaction takes place in the nucleus, using nuclear extracts from H2.35 cells (Fig. 6D). No interaction with HNF4α was noted (Fig. 6C), and this may explain why LRP130 is not required for apoliprotein A2 and PXR expression (Fig. 5B), two genes under the control of HNF4α and PGC-1α, and may also explain LRP130-independent coactivation of HNF4α by PGC-1α in Figure 6A.
In principle, LPR130 might have this regulatory effect on PGC-1α function through FoxO1 by several mechanisms, including direct and indirect modulation of transcriptional activation, mRNA processing, or docking of PGC-1α on this transcription factor. LRP130 interacts with both PGC-1α and FoxO1; therefore, we used chromatin immunoprecipitation (ChIP) to examine whether LRP130 is recruited to the FoxO1-binding site of the G6P promoter, and if it influences the docking of PGC-1α on the well-characterized FoxO1-binding site of the G6P promoter (Puigserver et al. 2003). ChIP in primary hepatocytes isolated from PGC-1α knockout mice shows recruitment of LRP130 to the FoxO1-binding sites, and notably, recruitment does not require PGC-1α (Fig. 6E). However, as seen in Figure 6F, depletion of LRP130 almost completely ablates the docking of PGC-1α onto the FoxO1-binding site of the G6P promoter, compared with the level of docking observed in the siControl cells. This finding is apparent despite slightly less expression of PGC-1α protein in the sample that was not depleted of LRP130—that is, the siControl cells. These data strongly suggest that LRP130 alters PGC-1α function, at least in part, through regulation of the docking of this coactivator on certain transcription factor targets. We did not assay whether LRP130 occupied the HNF4α-binding site; however, the totality of the genetic, coactivation, and interaction studies make it highly unlikely that LRP130 is required for PGC-1α-dependent coactivation of HNF4α.
LRP130 regulates the gluconeogenic program and the expression of mitochondrial-encoded genes in vivo
We used tail vein delivery of siRNA adenoviral constructs into mice in order to determine the effect of LRP130 depletion in vivo. The liver is the predominant organ susceptible to adenovirus infection and provides a convenient method to assess specific hepatic effects. Four to six days post-inoculation, the mice were fed ad libitum or fasted. Genetic and serologic data were subsequently assessed.
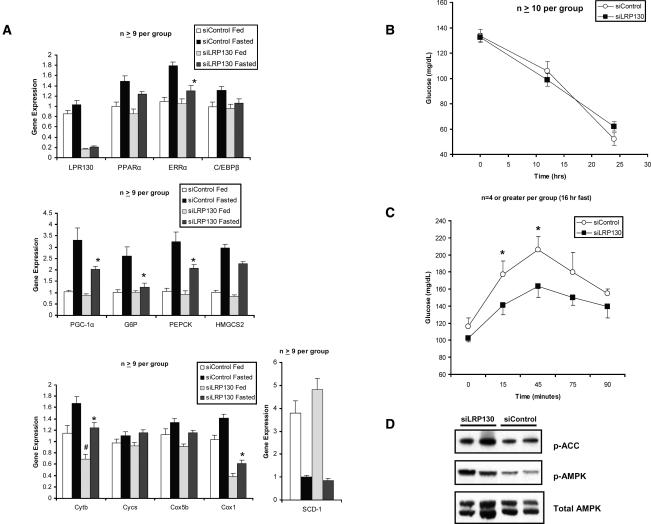
PGC-1α, PEPCK, and G6P gene expression was strongly induced by fasting in control animals as previously shown (Herzig et al. 2001; Yoon et al. 2001). Genetic profiling of liver tissue using quantitative PCR demonstrated blunted induction of PEPCK (23% reduction) and G6P (80% reduction) by siLRP130, compared with siControl following a 24-h fast (Fig. 7A). Gene expression of PGC-1α and ERRα was also blunted as well. None of these genes exhibited a decrement in their basal levels. In contrast, mitochondrial-encoded genes underwent a decline in both the fed basal state and the fasted state. The effects were specific as cytochrome c, Cox5b, and SCD-1 showed no perturbations in response to siLRP130 (Fig. 7A). Our findings are in agreement with our cellular data and indicate the importance of LRP130 in specifically modulating the normal fasted response.
Figure 7.
Effect of depletion of LRP130 in the hepatic fasting response in vivo. (A) Relative gene expression of several genes involved in energy homeostasis. Asterisks (*) indicate that statistical comparisons are made between siControl and siLRP130 in fasted animals. Number signs (#) indicate that statistical comparisons are made between siControl and siLRP130 in fed animals. (*) p < 0.05; (**) p < 0.01; (***) p < 0.001; (#) p < 0.05. Error bars represent standard deviation. (B) Comparison of glucose levels at 0, 12, and 24 h. (C) Comparison of hepatic glucose output as assessed by pyruvate tolerance test following a 16-h fast. (D) Immunoblot for AMPK, phosphorylated-AMPK (p-AMPK), and phosphorylated-acetyl CoA carboxylase (p-ACC) from the fasted animals analyzed in A. Two representative animals are shown.
We assessed glucose homeostasis in vivo with two methods: the measurement of fasted glucose and a pyruvate tolerance test. Fasting glucose levels reflect hepatic gluconeogenesis, but also reflect many other systemic parameters. Despite blunted transcription of PEPCK and G6P in response to fasting, glucose levels were normal at 12, 16, and 24 h of fasting (Fig. 7B). Pyruvate tolerance tests look more specifically at the process of gluconeo-genesis, as pyruvate is converted to glucose in gluconeo-genesis. Pyruvate tolerance testing revealed that hepatic glucose output was significantly reduced in animals treated with RNAi against LRP130 compared with control RNAi (Fig. 7C). This reduction likely reflects suppression of PEPCK and G6P during fasting, but may also reflect depressed ATP pools, since our in vitro and in vivo data demonstrated depression of several mitochondrial-encoded genes involved in electron transport. AMP kinase (AMPK) senses the energy status of the cell in the form of a perturbed ATP:AMP ratio. We assessed the activation of AMPK in fasted animals and found that AMPK and ACC underwent increased phosphorylation in the livers of siLRP130 animals compared with siControl, suggesting a perturbed ATP:AMP ratio (Fig. 7D). Thus, reduced hepatic glucose output likely results from both depressed activation of gluconeogenic genes and failure to generate sufficient ATP for gluconeogenesis. These data indicate that LRP130 is required in vivo for both hepatic gluconeogenesis and expression of several mitochondrial-encoded genes.
Discussion
LSFC is a disease characterized by severe neurodegeneration and liver defects. However, the molecular mechanisms for either clinical phenotype are unknown. We show here that LRP130 is a significant component of the PGC-1α holocomplex and regulates PGC-1α activity in cells and animals. Notably, cooperation between LRP130 and PGC-1α is required for two programs relevant to glucose homeostasis: gluconeogenesis and mitochondrial electron transport. Both processes are critical for the conversion of lactate and other substrates into glucose. Indeed, patients with LSFC develop striking lactic acidosis during bouts of septic illness. Increased lactate during sepsis occurs secondary to hypoperfusion of tissues. The liver, via gluconeogenesis, rapidly clears the lactate in order to maintain a steady-state level of lactate and normal blood pH. We observed specific defects in hepatic gluconeogenesis in mouse liver depleted of LRP130 protein, and experiments in primary hepatocytes clearly indicate that this is through defective PGC-1α action. These observed defects likely constitute a major contributory factor for the diathesis toward lactic acidosis in patients with LSFC, in which LRP130 protein is severely reduced.
Neurodegenerative defects are prominent features of LSFC. These patients have lesions of the basal ganglia, cerebellum, and thalamus. Although we show here that LRP130 and PGC-1α cooperate in the liver to maintain glucose homeostasis, it is highly plausible that LRP130 and PGC-1α also cooperate to regulate energy homeostasis in the brain. In fact, recent data have demonstrated that PGC-1α-null mice have neurological defects in the striatum, which is situated in the basal ganglia, consisting of spongiform lesions and reactive astrocytes. PGC-1α-null mice also have a striking neurological phenotype characterized by hyperactivity reminiscent of movement disorders like Huntington's disease. LRP130 and PGC-1α may cooperate to regulate mitochondrial gene expression encoded by the mitochondrial genome and other genes important for neuronal development and function. Future studies are under active investigation to unravel this potential biology.
Finally, one very interesting issue that has emerged from these studies is the gene specificity that LRP130 imposes on PGC-1α function. Previous work has shown that PGC-1α induces mitochondrial gene expression, regardless of whether the genes are encoded within the mitochondrion or the nucleus. In contrast, LRP130 modulates PGC-1α action on several mitochondrial subunits encoded only within the mitochondrion, but not those encoded by the nuclear genome. Additionally, LRP130 modulates gluconeogenic genes, which are encoded by the nuclear genome. How LRP130 specifically modulates the expression of certain gene targets of PGC-1α, but not others, is not known. While there are several possibilities, two seem most likely. First, LRP130 may influence the interaction between PGC-1α and certain transcription factors. This is an intriguing notion as it suggests that LRP130 may be required to increase or decrease the docking of PGC-1α, and its concomitant transcriptional activity, on specific transcription factors and promoters. The data presented in Figure 6, showing that LRP130 augments docking on a FoxO1-binding site, are consistent with this model. On the other hand, the ability of LRP130 to increase the transcriptional activity of PGC-1α tethered to the Gal4-DBD strongly suggests that the promotion of selective docking of PGC-1α on target transcription factors cannot be its exclusive function in PGC-1α-linked gene regulation. A second possible mechanism might relate to mRNA metabolism. PGC-1α is involved in mRNA processing following the initiation of transcription (Monsalve et al. 2000). It is possible that LRP130 participates in these post-transcriptional processes, as it also contains RNA-binding motifs (Mili and Pinol-Roma 2003). Different gene targets of PGC-1α may have different requirements for processing following initiation of transcription, and some tissues may have special requirements for LRP130. Importantly, the two possible mechanisms discussed here are not mutually exclusive.
In conclusion, these studies illustrate a novel function for LRP130, the gene that is defective in a subset of patients with Leigh syndrome. While this protein is partly resident in mitochondria (Xu et al. 2004) and may directly affect mitochondrial function, we show here that LRP130 is a component of the PGC-1α complex, and affects numerous processes related to energy homeostasis via PGC-1α.
Materials and methods
Plasmids, recombinant PGC-1α protein, and antibodies
Full-length murine LRP130 was isolated from a mouse liver cDNA library by PCR amplification and cloned in-frame into the KpnI/NotI site of pcDNA 3.1 myc His B. The mouse homolog of the human A354V mutant LRP130 was created by site-directed mutagenesis of C ® T at 800 bp of the mouse mRNA clone for LRP130 using the Quick Change II XL site-directed mutagenesis kit (Stratagene). LRP130 clones expressing amino acids 1–475, 1–900, and 925–1306 were generated by restriction digest with BsrGI/XhoI, XhoI, and BspEI, respectively. The digests were blunted with Klenow fragments and religated, in order to remain in-frame with C-terminal c-myc tag. These clones were then subcloned into the NcoI/NotI sites of pET30a vector, containing an N-terminal S-tag. Full-length murine PGC-1α was cloned into the BamHI/XhoI sites of pET30a and contained an N-terminal Flag and HA tag in tandem. PGC-1α truncations encoding amino acids 2–180, 2–271, 575–797, and 269–797 were PCR-amplified and cloned into the BamHI/XhoI site of the pET30a. Gal4 constructs were generated as previously described (Puigserver et al. 1998, 1999, 2001; Lin et al. 2002). Transient transfections were performed using Lipofectamine 2000 (Invitrogen) per the manufacturer's recommendations. Briefly, 100 ng of reporter wwere used with 100 ng of Gal4 construct and 1–2 μg of LRP130 construct. Reactions were adjusted to a total of 4 μg using a nonspecific plasmid. Differences in transfection efficiency were normalized by inclusion of RSV β-galactosidase as an internal control. PGC-1α recombinant protein was generated as previously described (Wallberg et al. 2003).
Mouse polyclonal antibodies were generated using 100 μg of purified antigen, either denatured C-terminal PGC-1α (amino acids 575–797) or C-terminal LRP130 protein (amino acids 925–1306). The antigens were emulsified with adjuvant in PBS and injected intraperitoneally on day 0. Booster injections were administered at day 21 and day 28. Serum was collected from tail bleeds and used at 1:1000 for immunoblotting.
Protein identification
Gel-resolved proteins (1–16c) (see Fig. 1) were digested with trypsin and batch-purified on a reversed-phase microtip, and resulting peptide pools were individually analyzed by matrix-assisted laser desorption/ionization reflectron time-of-flight (MALDI-reTOF) mass spectrometry (MS) (UltraFlex TOF/TOF; BRUKER) for peptide mass fingerprinting (PMF), as described (Erdjument-Bromage et al. 1998; Sebastiaan et al. 2002). Selected peptide ions (m/z) were taken to search a “nonredundant” protein database (NR; 3,245,378 entries on January 28, 2006; National Center for Biotechnology Information, Bethesda, MD) using the PeptideSearch algorithm (Matthias Mann, Max-Planck Institute for Biochemistry, Martinsried, Germany; an updated version of this program is currently available as “Pep-Sea” from Applied Biosystems/MDS Sciex). A molecular mass range up to twice the apparent molecular weight (as estimated from electrophoretic relative mobility) was covered, with a mass accuracy restriction of <35 ppm, and maximum one missed cleavage site allowed per peptide. To confirm PMF results with scores ≤40, mass spectrometric sequencing of selected peptides was done by MALDI-TOF/TOF (MS/MS) analysis on the same prepared samples, using the UltraFlex instrument in “LIFT” mode. Fragment ion spectra were taken to search NR using the MASCOT MS/MS Ion Search program (Perkins et al. 1999), version 2.0.04 for Windows (Matrix Science Ltd.). Any tentative confirmation (Mascot score ≥ 30) of a PMF result thus obtained was verified by comparing the computer-generated fragment ion series of the predicted tryptic peptide with the experimental MS/MS data.
RNAi construct and retrovirus
RNAi sequence (5′-GAAGCTAGATGACCTGTTT-3′) against murine LRP130 was custom generated (Dharmacon). The DNA duplex sequence was cloned into pSuper Retro (Oligoengine) per the manufacturer's protocol. H2.35 cells were stably transduced with retrovirus using either control RNAi or RNAi against LRP130. Retrovirus was generated in phoenix cells. Briefly, cells 70%–80% confluent in a 10-cm dish were transiently transfected with 10 μg of pSuper-retro construct, 5 μg of gag-pol, and 5 μg of VSV-G using a calcium phosphate transfection kit (Promega). Twenty-four hours later, the media was replaced with 5 mL of fresh media. Forty-eight hours thereafter, an additional 5 mL of fresh media was added, polybrene was added to a final concentration of 8 μg/mL, and the retroviral mix was filtered through a 0.45-μm filter. The retroviral mix was incubated with H2.35 cells, ∼50% confluent, at 37°C. After 16–24 h of incubation, the retroviral mix was replaced with fresh media. Outgrowth proceeded for 48 h, after which the cells were selected with 1.0 μg/mL puromycin until all the cells in a control plate died (typically at 48–96 h). Specific knockdown was confirmed by Northern blotting (data not shown) and real-time PCR of LRP130 mRNA, and immunoblotting for LRP130 protein. On occasion, RNAi constructs may have unintended effects beyond specific gene modulation. We therefore generated (Dharmacon) a second RNAi (5′-GCAGTTAGGTGTCGTATAT-3′) against LRP130 (siLRP130 #2) that exhibited similar specificity (Supplementary Fig. 3) to the siLRP130 construct used throughout our work.
Adenoviruses
Insulin-insensitive FoxO1 adenovirus (3A mutant) was the kind gift of Dr. Pere Puigserver (The Johns Hopkins School of Medicine, Baltimore, MD). Full-length LRP130 in the pcDNA 3.1 myc His B/LRP130 construct described above was restriction-digested with NotI/PmeI and subcloned containing its c-myc tag, downstream from the CMV promotor into the KpnI/EcoRV site of pAdTrack. RNAi adenovirus was generated by cloning the H1 promoter-LRP130 RNAi cassette into a pAdTrack devoid of an upstream CMV promoter. Recombination was performed in BJD135 cells (Stratagene). Subsequent screening and amplification were performed per the manufacturer's protocol. Adenovirus was purified by CsCl banding as previously described. Tail vein injections were performed as previously described (Lin et al. 2005), on 12-wk-old male C57BL/6 mice. Assays were performed 5 d after inoculation. Primary hepatocytes were infected at an m.o.i. (multiplicity of infection) of 8–16 for 48 h, except for RNAi experiments, in which an m.o.i. of 4–8 was used for 72 h. In circumstances in which two adenoviruses were combined, total viral m.o.i. was maintained between all samples and treatments.
Cells
H2.35 cells were purchased from ATCC and maintained in 5% fetal bovine serum, DMEM (GIBCO), 4 mM L-glutamine, and 200 nM dexamethasone. Primary hepatocytes were isolated from mice ages 6–8 wk as previously described (Lin et al. 2004). Primary hepatocytes were maintained in DMEM (GIBCO) supplemented with 10% fetal bovine serum (GIBCO), 2 mM pyruvate, 1 nM insulin (Sigma), and 10 nM dexamethasone (Sigma). Primary hepatocytes were changed to starvation media (DMEM supplemented with 0.1% bovine serum albumin, fraction V (Sigma), and 2 mM pyruvate for 16–24 h, prior to stimulation with 10 μM forskolin (Calbiochem) and 1 μM dexamethasone. DMSO served as a vehicle control for reagents dissolved in DMSO.
Animal experiments
All animal experiments were performed in accordance with the procedures approved by the Institutional Animal Care and Use Committee. Mice were maintained on a standard rodent chow with 12-h light–dark cycles. The pyruvate challenge test (pyruvate tolerance test) was performed as previously described (Miyake et al. 2002). Briefly, 12-wk-old male mice were fasted for 16 h prior to intraperitoneal administration of pyruvate (2 g/kg freshly prepared in normal saline). Plasma glucose levels were then assessed at various time points.
In vitro binding assays
Clones containing full-length PGC-1α or LRP130 were subjected to in vitro transcription/translation using the TnT kit (Promega). The full-length proteins served as bait, and were first immobilized on either Flag matrix (Sigma) or S-tag matrix (Novex), and then extensively washed with 1× PBS/0.1% Triton X-100 containing protease inhibitors. Translation of the unlabeled bait proteins was confirmed by silver staining of SDS-PAGE gels (data not shown). Radiolabeled fragments were 35S-methionine labeled, and 20 μL of these translated products was incubated with nonradiolabeled protein, preimmobilized to affinity matrix. Following incubation for 4 h to overnight at 4°C, the complexes were washed extensively and developed as previously described (Lin et al. 2004).
ChIP assays
Stably transduced siControl or siLRP130 H2.35 cells were infected for 48–72 h with FoxO1 (3A mutant) adenovirus and either GFP control adenovirus or PGC-1α adenovirus. Cells were trypsinized, counted, and subjected to ChIP using the EZ ChIP kit (Upstate Biotechnology). For the immunoprecipitations, 2.5 μL of anti-C-terminal PGC-1α mouse serum was used per reaction. Following processing, de-cross-linking, and column purification, the samples were normalized using either 18S or TBP by real-time PCR. In order to confirm linearity and acceptable primer efficiency (1.0 ± 0.1), calibration curves were generated by serial dilution. Variability was no greater than 10% between samples for each experiment. The region of the G6P promoter containing FoxO1-binding sites (IRS 1 and IRS 2) was PCR-amplified using forward primer, 5′-GTCAAGCAGTGTGCCCAAGTTA-3′, and reverse primer, 5′-ATAGCAAAACAGGCACACAAAAAC-3′. The linear region of PCR amplification was monitored by real-time assessment and resolved on a 2% agarose gel. Additionally, protein extract was isolated from a portion of the cells, in order to confirm similar protein expression for FoxO1 3A and PGC-1α.
Real-time RT–PCR
Total RNA was isolated from cells or tissues using Trizol reagent (Invitrogen) per the manufacturer's protocol. Reverse transcriptase reactions were performed using Iscript (Bio-Rad). Primers were designed using the Primer Express 2.0 (Applied Biosystems). The sequences of primers are shown in Supplementary Table 1. Real-time PCR was performed using SYBR green PCR mix (Applied Biosystems) with a primer concentration of 500 nM. TBP (TATA-binding protein) served as an internal control to normalize samples.
Protein immunoblot analyses
Protein extracts were performed on cells or tissues by homogenization in Flag lysis buffer. Samples were clarified at 14,000g for 30 min and quantified with Bradford reagent. Fifty micrograms of total protein was loaded onto a 4%–12% SDS-PAGE gels in MOPS buffer. Gels were transferred to PVDF (Immobilon) membranes in Towbin buffer and probed with primary antibodies per the manufacturer's protocol. Mouse polyclonal antibodies against the C terminus of LRP130 and the C terminus of PGC-1α were used at 1:1000. Anti-c-myc antibody (Santa Cruz Biotechnology) was used per the manufacturer's protocol. Immunoblots were then developed using the appropriate horse-radish peroxidase-conjugated secondary antibody (Zymed) and ECL detection reagent (Amersham). α-Tubulin served as the loading control.
Acknowledgments
We thank Dr. Vamsi K. Mootha for human LRPPRC constructs. We thank Dr. Pere Puigserver for FoxO1 3A adenovirus and Arpi Nazarian for help with mass spectrometric analysis. This work was supported by grants from the N.I.H., R01DK61562 (to B.M.S.), K08DK071017 (to M.P.C.), and NCI Cancer Center Support Grant P30 CA08748 (to P.T.).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1483906.
References
- Brunet, A., Bonni, A., Zigmond, M.J., Lin, M.Z., Juo, P., Hu, L.S., Anderson, M.J., Arden, K.C., Blenis, J., Greenberg, M.E. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- Erdjument-Bromage, H., Lui, M., Lacomis, L., Grewal, A., Annan, R.S., McNulty, D.E., Carr, S.A., Tempst, P. Examination of micro-tip reversed-phase liquid chromatographic extraction of peptide pools for mass spectrometric analysis. J. Chromatogr. A. 1998;826:167–181. doi: 10.1016/s0021-9673(98)00705-5. [DOI] [PubMed] [Google Scholar]
- Hall, R.K., Yamasaki, T., Kucera, T., Waltner-Law, M., O'Brien, R., Granner, D.K. Regulation of phosphoenolpyruvate carboxykinase and insulin-like growth factor-binding protein-1 gene expression by insulin. The role of winged helix/forkhead proteins. J. Biol. Chem. 2000;275:30169–30175. doi: 10.1074/jbc.M004898200. [DOI] [PubMed] [Google Scholar]
- Herzig, S., Long, F., Jhala, U.S., Hedrick, S., Quinn, R., Bauer, A., Rudolph, D., Schutz, G., Yoon, C., Puigserver, P., et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- Hou, J., Wang, F., McKeehan, W.L. Molecular cloning and expression of the gene for a major leucine-rich protein from human hepatoblastoma cells (HepG2) In Vitro Cell. Dev. Biol. Anim. 1994;30A:111–114. doi: 10.1007/BF02631402. [DOI] [PubMed] [Google Scholar]
- Ikura, T., Ogryzko, V.V., Grigoriev, M., Groisman, R., Wang, J., Horikoshi, M., Scully, R., Qin, J., Nakatani, Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- Koo, S.H., Satoh, H., Herzig, S., Lee, C.H., Hedrick, S., Kulkarni, R., Evans, R.M., Olefsky, J., Montminy, M. PGC-1 promotes insulin resistance in liver through PPAR-α-dependent induction of TRB-3. Nat. Med. 2004;10:530–534. doi: 10.1038/nm1044. [DOI] [PubMed] [Google Scholar]
- Labialle, S., Dayan, G., Gayet, L., Rigal, D., Gambrelle, J., Baggetto, L.G. New invMED1 element cis-activates human multidrug-related MDR1 and MVP genes, involving the LRP130 protein. Nucleic Acids Res. 2004;32:3864–3876. doi: 10.1093/nar/gkh722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, N., Daly, M.J., Delmonte, T., Lander, E.S., Xu, F., Hudson, T.J., Mitchell, G.A., Morin, C.C., Robinson, B.H., Rioux, J.D. A genomewide linkage-disequilibrium scan localizes the Saguenay-Lac-Saint-Jean cytochrome oxidase deficiency to 2p16. Am. J. Hum. Genet. 2001;68:397–409. doi: 10.1086/318197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh, D. Subacute necrotizing encephalomyelopathy in an infant. J. Neurol. Neurosurg. Psychiatr. 1951;14:216–221. doi: 10.1136/jnnp.14.3.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J., Puigserver, P., Donovan, J., Tarr, P., Spiegelman, B.M. Peroxisome proliferator-activated receptor γ co-activator 1β (PGC-1β), a novel PGC-1-related transcription coactivator associated with host cell factor. J. Biol. Chem. 2002;277:1645–1648. doi: 10.1074/jbc.C100631200. [DOI] [PubMed] [Google Scholar]
- Lin, J., Wu, P.H., Tarr, P.T., Lindenberg, K.S., St Pierre, J., Zhang, C.Y., Mootha, V.K., Jager, S., Vianna, C.R., Reznick, R.M., et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1α null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Lin, J., Yang, R., Tarr, P.T., Wu, P.H., Handschin, C., Li, S., Yang, W., Pei, L., Uldry, M., Tontonoz, P., et al. Hyperlipidemic effects of dietary saturated fats mediated through PGC-1β coactivation of SREBP. Cell. 2005;120:261–273. doi: 10.1016/j.cell.2004.11.043. [DOI] [PubMed] [Google Scholar]
- Merante, F., Petrova-Benedict, R., MacKay, N., Mitchell, G., Lambert, M., Morin, C., De Braekeleer, M., Laframboise, R., Gagne, R., Robinson, B.H. A biochemically distinct form of cytochrome oxidase (COX) deficiency in the Saguenay-Lac-Saint-Jean region of Quebec. Am. J. Hum. Genet. 1993;53:481–487. [Google Scholar]
- Mili, S., Pinol-Roma, S. LRP130, a pentatricopeptide motif protein with a noncanonical RNA-binding domain, is bound in vivo to mitochondrial and nuclear RNAs. Mol. Cell. Biol. 2003;23:4972–4982. doi: 10.1128/MCB.23.14.4972-4982.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake, K., Ogawa, W., Matsumoto, M., Nakamura, T., Sakaue, H., Kasuga, M. Hyperinsulinemia, glucose intolerance, and dyslipidemia induced by acute inhibition of phosphoinositide 3-kinase signaling in the liver. J. Clin. Invest. 2002;110:1483–1491. doi: 10.1172/JCI15880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsalve, M., Wu, Z., Adelmant, G., Puigserver, P., Fan, M., Spiegelman, B.M. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol. Cell. 2000;6:307–316. doi: 10.1016/s1097-2765(00)00031-9. [DOI] [PubMed] [Google Scholar]
- Mootha, V.K., Lepage, P., Miller, K., Bunkenborg, J., Reich, M., Hjerrild, M., Delmonte, T., Villeneuve, A., Sladek, R., Xu, F., et al. Identification of a gene causing human cyto-chrome c oxidase deficiency by integrative genomics. Proc. Natl. Acad. Sci. 2003;100:605–610. doi: 10.1073/pnas.242716699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin, C., Mitchell, G., Larochelle, J., Lambert, M., Ogier, H., Robinson, B.H., De Braekeleer, M. Clinical, metabolic, and genetic aspects of cytochrome c oxidase deficiency in Saguenay-Lac-Saint-Jean. Am. J. Hum. Genet. 1993;53:488–496. [Google Scholar]
- Nakae, J., Park, B.C., Accili, D. Insulin stimulates phosphorylation of the forkhead transcription factor FKHR on serine 253 through a Wortmannin-sensitive pathway. J. Biol. Chem. 1999;274:15982–15985. doi: 10.1074/jbc.274.23.15982. [DOI] [PubMed] [Google Scholar]
- Nakae, J., Biggs, W.H., III, Kitamura, T., Cavenee, W.K., Wright, C.V., Arden, K.C., Accili, D. Regulation of insulin action and pancreatic β-cell function by mutated alleles of the gene encoding forkhead transcription factor Foxo1. Nat. Genet. 2002;32:245–253. doi: 10.1038/ng890. [DOI] [PubMed] [Google Scholar]
- Perkins, D.N., Pappin, D.J., Creasy, D.M., Cottrell, J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Puigserver, P., Wu, Z., Park, C.W., Graves, R., Wright, M., Spiegelman, B.M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- Puigserver, P., Adelmant, G., Wu, Z., Fan, M., Xu, J., O'Malley, B., Spiegelman, B.M. Activation of PPARγ coactivator-1 through transcription factor docking. Science. 1999;286:1368–1371. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- Puigserver, P., Rhee, J., Lin, J., Wu, Z., Yoon, J.C., Zhang, C.Y., Krauss, S., Mootha, V.K., Lowell, B.B., Spiegelman, B.M. Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARγ coactivator-1. Mol. Cell. 2001;8:971–982. doi: 10.1016/s1097-2765(01)00390-2. [DOI] [PubMed] [Google Scholar]
- Puigserver, P., Rhee, J., Donovan, J., Walkey, C.J., Yoon, J.C., Oriente, F., Kitamura, Y., Altomonte, J., Dong, H., Accili, D., et al. Insulin-regulated hepatic gluconeogenesis through FOXO1–PGC-1α interaction. Nature. 2003;423:550–555. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- Rhee, J., Inoue, Y., Yoon, J.C., Puigserver, P., Fan, M., Gonzalez, F.J., Spiegelman, B.M. Regulation of hepatic fasting response by PPARγ coactivator-1α (PGC-1): Requirement for hepatocyte nuclear factor 4α in gluconeogenesis. Proc. Natl. Acad. Sci. 2003;100:4012–4017. doi: 10.1073/pnas.0730870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, B.H. Human cytochrome oxidase deficiency. Pediatr. Res. 2000;48:581–585. doi: 10.1203/00006450-200011000-00004. [DOI] [PubMed] [Google Scholar]
- Roy, M., Li, Z., Sacks, D.B. IQGAP1 binds ERK2 and modulates its activity. J. Biol. Chem. 2004;279:17329–17337. doi: 10.1074/jbc.M308405200. [DOI] [PubMed] [Google Scholar]
- Russell, L.K., Mansfield, C.M., Lehman, J.J., Kovacs, A., Courtois, M., Saffitz, J.E., Medeiros, D.M., Valencik, M.L., McDonald, J.A., Kelly, D.P. Cardiac-specific induction of the transcriptional coactivator peroxisome proliferator-activated receptor γ coactivator-1α promotes mitochondrial biogenesis and reversible cardiomyopathy in a developmental stage-dependent manner. Circ. Res. 2004;94:525–533. doi: 10.1161/01.RES.0000117088.36577.EB. [DOI] [PubMed] [Google Scholar]
- Scarpulla, R.C. Nuclear activators and coactivators in mammalian mitochondrial biogenesis. Biochim. Biophys. Acta. 2002a;1576:1–14. doi: 10.1016/s0167-4781(02)00343-3. [DOI] [PubMed] [Google Scholar]
- Scarpulla, R.C. Transcriptional activators and coactivators in the nuclear control of mitochondrial function in mammalian cells. Gene. 2002b;286:81–89. doi: 10.1016/s0378-1119(01)00809-5. [DOI] [PubMed] [Google Scholar]
- Schmoll, D., Walker, K.S., Alessi, D.R., Grempler, R., Burchell, A., Guo, S., Walther, R., Unterman, T.G. Regulation of glucose-6-phosphatase gene expression by protein kinase Bα and the forkhead transcription factor FKHR. Evidence for insulin response unit-dependent and -independent effects of insulin on promoter activity. J. Biol. Chem. 2000;275:36324–36333. doi: 10.1074/jbc.M003616200. [DOI] [PubMed] [Google Scholar]
- Sebastiaan, W.G., Lacomis, L., Philip, J., Erdjument-Bromage, H., Svejstrup, J.Q., Tempst, P. Isolation and mass spectrometry of transcription factor complexes. Methods. 2002;26:260–269. doi: 10.1016/S1046-2023(02)00030-0. [DOI] [PubMed] [Google Scholar]
- Shoubridge, E.A. Cytochrome c oxidase deficiency. Am. J. Med. Genet. 2001;106:46–52. doi: 10.1002/ajmg.1378. [DOI] [PubMed] [Google Scholar]
- Small, I.D., Peeters, N. The PPR motif—A TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 2000;25:46–47. doi: 10.1016/s0968-0004(99)01520-0. [DOI] [PubMed] [Google Scholar]
- St Pierre, J., Lin, J., Krauss, S., Tarr, P.T., Yang, R., Newgard, C.B., Spiegelman, B.M. Bioenergetic analysis of peroxisome proliferator-activated receptor γ coactivators 1α and 1β (PGC-1α and PGC-1β) in muscle cells. J. Biol. Chem. 2003;278:26597–26603. doi: 10.1074/jbc.M301850200. [DOI] [PubMed] [Google Scholar]
- Stryer, L. Biochemistry. W.H. Freeman and Company; New York: 1988. [Google Scholar]
- Tsuchiya, N., Fukuda, H., Nakashima, K., Nagao, M., Sugimura, T., Nakagama, H. LRP130, a single-stranded DNA/RNA-binding protein, localizes at the outer nuclear and endoplasmic reticulum membrane, and interacts with mRNA in vivo. Biochem. Biophys. Res. Commun. 2004;317:736–743. doi: 10.1016/j.bbrc.2004.03.103. [DOI] [PubMed] [Google Scholar]
- Wallberg, A.E., Yamamura, S., Malik, S., Spiegelman, B.M., Roeder, R.G. Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1α. Mol. Cell. 2003;12:1137–1149. doi: 10.1016/s1097-2765(03)00391-5. [DOI] [PubMed] [Google Scholar]
- Wu, Z., Puigserver, P., Andersson, U., Zhang, C., Adelmant, G., Mootha, V., Troy, A., Cinti, S., Lowell, B., Scarpulla, R.C., et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- Xu, F., Morin, C., Mitchell, G., Ackerley, C., Robinson, B.H. The role of the LRPPRC (leucine-rich pentatricopeptide repeat cassette) gene in cytochrome oxidase assembly: Mutation causes lowered levels of COX (cytochrome c oxidase) I and COX III mRNA. Biochem J. 2004;382:331–336. doi: 10.1042/BJ20040469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, J.C., Puigserver, P., Chen, G., Donovan, J., Wu, Z., Rhee, J., Adelmant, G., Stafford, J., Kahn, C.R., Granner, D.K., et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413:131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]