Abstract
We have previously shown that the unique vimA (virulence-modulating) gene could modulate proteolytic activity in Porphyromomas gingivalis. Although a reduction in cysteine protease activity was observed in the vimA-defective mutant, P. gingivalis FLL92, compared to that of the wild-type strain, no changes were seen in the expression of the gingipain genes. This result might suggest posttranscriptional regulation of protease expression. To determine whether there was a defect in the translation, transport, or maturation of the gingipains, P. gingivalis FLL92 was further characterized. In contrast to the wild-type strain, a 90% reduction was seen in both Rgp and Kgp protease activities in strain FLL92 during the exponential growth phase. These activities, however, increased to approximately 60% of that of the wild-type strain during stationary phase. Throughout all the growth phases, Rgp and Kgp activities were mostly soluble, in contrast to those of the wild-type strain. Western blot analyses identified unique Rgp- and Kgp-immunoreactive bands in extracellular protein fractions from FLL92 grown to late exponential phase. Also, the RgpB proenzyme was identified in this fraction by mass spectrometry. In addition, in vitro protease activity could be induced by a urea denaturation-renaturation cycle in this fraction. These results indicate that protease activity in P. gingivalis may be growth phase regulated, possibly by multiple mechanisms. Furthermore, the gingipain RgpB is excreted in an inactive form in the vimA mutant. In addition, these results provide the first evidence of posttranslational regulation of protease activity in P. gingivalis and may suggest an important role for the vimA gene in protease activation in this organism.
Porphyromonas gingivalis, a black-pigmented, gram-negative anaerobic bacterium, is an important etiological agent of chronic adult periodontitis (reviewed in references 23, 25, and 43) and is associated with other systemic diseases, including atherosclerosis (reviewed in reference 11). While several virulence factors have been implicated in the pathogenicity of P. gingivalis, the high-level proteolytic abilities of this organism have been the focus of much attention and appear to play an important role in its virulence. The major proteases, called gingipains, are both extracellular and cell associated. They consist of arginine-specific protease (Arg-gingipain [Rgp]) and lysine-specific protease (Lys-gingipain [Kgp]) (35). The Rgp is encoded by two genes, rgpA and rgpB, while Kgp is encoded by a single gene, kgp (28, 33). In addition to being essential for growth, proteases have other functions. Several reports (reviewed in reference 22) have documented the multiple effects of proteases, including degradation of complement and immunoglobulin, inactivation of cytokines and their receptors, aggregation of platelets, attenuation of neutrophil antibacterial activities, and increases in vascular permeability and blood clotting prevention. Furthermore, these gingipains, which have been suggested to be not coordinately regulated (44), have also been shown to act as major processing enzymes for various cell surface proteins, including individual proteases (3, 4, 17).
Previously, it has been reported that the recA locus can affect the phenotypic expression and distribution of the gingipains in P. gingivalis (1, 2, 10). A defective mutant was constructed by allelic exchange (1) by using the cloned vimA gene, which is downstream of the recA gene and appears to be part of the same transcriptional unit. The mutant strain, designated FLL92, was not black pigmented and showed increased autoaggregration and significant reductions in proteolytic, hemolytic, and hemagglutinating activities. In in vivo experiments using a mouse model, the virulence of P. gingivalis FLL92 was dramatically reduced in comparison to that of the wild-type W83 strain. While reductions in Arg-X- and Lys-X-specific proteolytic activities were observed in P. gingivalis FLL92, transcription of the gingipain genes was unaltered in this mutant compared to that of the wild-type strain (1). A similar unaltered level of expression of the gingipain genes was also observed in P. gingivalis FLL32, a recA-defective isogenic mutant that had reduced Arg-X- and Lys-X-specific proteolytic activities (2). The detection of immunoreactive bands in an extracellular fraction of P. gingivalis FLL32 by using anti-Rgp or anti-Kgp has raised the question of whether the reduced level of proteolytic activity in this mutant may be due to a defect in the processing of the proteases (2). While these data may suggest posttranscriptional regulation of these genes, we cannot rule out the possibility that altered translation and/or transport of the gingipains may contribute to the observed reduced protease activity in the vimA-defective mutant. Further, if it were shown that the gingipain proenzyme forms were detected extracellularly, it might suggest that gingipain biogenesis requires a host-specific factor(s).
In this report, we have further characterized the vimA-defective P. gingivalis FLL92 strain. A late onset of gingipain protease activity occurred during the stationary phase of growth. In the absence of significant proteolytic activity during exponential growth phase, Western blot analyses identified unique Rgp- and Kgp-immunoreactive bands in extracellular protein fractions from P. gingivalis FLL92. Moreover, the RgpB proenzyme was present in this reduced proteolytic extracellular fraction. These results indicate that the gingipain RgpB is excreted in an inactive form in the vimA-defective mutant. Furthermore, protease activity in P. gingivalis may be regulated by multiple mechanisms. Finally, this finding may suggest an important role for the vimA gene in protease activation in P. gingivalis.
MATERIALS AND METHODS
Bacterial strains.
P. gingivalis W83 and P. gingivalis FLL92 (vimA::ermF- ermAM) were grown in brain heart infusion (BHI) broth (Difco Laboratories, Detroit, Mich.) supplemented with hemin (5 μg/ml), vitamin K (0.5 μg/ml), and cysteine (1%). Unless otherwise stated, all cultures were incubated at 37°C and maintained in an anaerobic chamber (Coy Manufacturing, Ann Arbor, Mich.) in 10% H2, 10% CO2, and 80% N2. The growth rates for the P. gingivalis strains were determined spectrophotometrically (optical density at 660 nm [OD660]). Where appropriate, P. gingivalis was grown in the presence of clindamycin (0.5 μg/ml).
Cell fraction preparation and protease assay.
One-liter cultures of P. gingivalis strains FLL92 and W83 were grown from actively growing cells. Preparations of whole-cell culture, cell-free medium, cell suspension, vesicles, and vesicle-free medium were made as previously reported (36). The whole-cell culture fraction is a sample of the culture after the bacterium has been grown to a specific growth phase. This sample has the bacterial cells suspended in the growth medium, and the enzyme activity includes the gingipains that are attached to the bacterial cell surface plus those that are secreted in the culture medium. After centrifugation, the cell pellet is resuspended in a buffer, and the enzyme activity in this sample (the cell suspension fraction) represents the gingipains that are attached to the bacterial cell surface. The enzyme activity in the supernatant (the cell-free medium fraction) includes the gingipains that are secreted in the culture medium. Secreted gingipains can be either associated with vesicles or soluble in the culture medium; thus, ultracentrifugation of the cell-free supernatant pellets the vesicles, while the soluble gingipains remain suspended. The enzyme activity of this vesicle pellet resuspended in a buffer (the vesicle fraction) indicates the activity associated with the vesicles. The enzyme activity of the supernatant (the vesicle-free medium fraction) after ultracentrifugation represents the soluble gingipains.
Extracellular proteases were prepared from the culture supernatant (500 ml) of cells grown to late log phase (OD660 of 0.8) or stationary phase (OD660 of 1.5). The cells were harvested by centrifugation (10,000 × g, 30 min), and the supernatant was further clarified by filtration through a 0.45-μm-pore-size membrane (Millipore Corporation, Bedford, Mass.). The cell-free culture fluid was precipitated with 37.5 or 60% acetone (−20°C), and the protein pellet was resuspended in 7 ml of 100 mM Tris-HCl buffer (pH 7.4), dialyzed for 24 h against the same buffer, and then stored on ice or at 0°C. The presence of Arg-X- and Lys-X-specific cysteine protease activities was determined with a microplate reader (Bio-Rad Laboratories, Hercules, Calif.) according to methods reported elsewhere (1).
Enzyme purification.
Cell-free culture fluid was obtained by centrifugation (10, 000 × g, 30 min) of the harvested whole-cell culture. The supernatant was further clarified by vacuum filtration with a 0.45-mm-pore-size membrane (Millipore Corporation). The cell-free culture fluid was precipitated with 60 or 37.5% acetone at −20°C and applied to a Sephadex G-150 column as described by Pike et al. (34). Nine-milliliter fractions were collected and pooled according to N-α-benzoyl-dl-arginine-p-nitroanilide (BAPNA; for Rgp activity) and acetyl-lysine-p-nitroanilide (Ac-Lys-pNA; for Kgp activity) activities and reactivities to RgpA-, RgpB-, and Kgp-specific antibodies (36). Three peaks of activity against the two substrates and reactivity to specific antibodies were found. The Rgp-specific peak fractions were pooled, concentrated, and applied to an arginine-Sepharose column according to the protocol described by Otsuka et al. (30), with modified elution volumes of 75 ml. The fractions were pooled and used for further analysis.
SDS-PAGE and Western blot analysis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed with a 4 to 12% bis-Tris separating gel in MOPS (morpholinepropanesulfonic acid)-SDS running buffer (NuPAGE Novex gels; Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions. Samples were prepared (65% sample, 25% 4× NuPAGE LDS sample buffer, 10% NuPAGE reducing agent), heated at 72°C for 10 min, and then electrophoresed at 200 V for 65 min with the XCell SureLock Mini-Cell (Invitrogen). The protein bands were visualized by staining with Simply Blue Safe stain (Invitrogen). The separated proteins were then transferred to nitrocellulose membranes (Schleicher & Schuell, Keene, N.H.) and processed at 15 V for 25 min with a Semi-Dry Trans-blot apparatus (Bio-Rad). The blots were probed with antibodies against specific protease domains. Immunoreactive proteins were detected by the procedure described in the Western Lightning Chemiluminescence Reagent Plus kit (Perkin-Elmer Life Sciences, Boston, Mass.). The secondary antibody was immunoglobulin G (heavy plus light chains)-horseradish peroxidase conjugate (Zymed Laboratories, Inc., South San Francisco, Calif.).
Denaturation-renaturation procedure for in vitro protease activation.
Extracellular protein fractions from P. gingivalis were mixed with 8 M urea and incubated at 4°C for 1 h. The urea was slowly removed from the mixture by centrifugation (10,000 × g) in a Millipore filtration unit (Biomax 10K NMWL membrane, 0.5-ml volume) with the addition of increasing volumes of 100 mM Tris-HCl buffer (pH 7.4).
Mass spectrometry sequence analysis.
Sequence analysis of the proteins was performed in the Mass Spectrometry Core Facility at the Beckman Research Institute of the City of Hope (Duarte, Calif.). The gel-separated proteins were reduced, alkylated, and digested in the gel as described in published procedures (9). Tryptic peptides were extracted and analyzed with a custom-built nano flow high-performance liquid chromatography system (8) coupled to an LCQ Classic Ion Trap mass spectrometer (ThermoFinnigan, San Jose, Calif.). Peptide tandem mass spectra were automatically screened to remove low-quality spectra (26) and compared to spectra in the National Center for Biotechnology Information nonredundant database (http://ncbi.nlm.nih.gov/blast/db) by using TurboSequest (ThermoFinnigan). Individual peptide matches were confirmed, and proteins were considered to be identified if at least two different peptides were identified.
Fluorescence labeling of proteases by using DNS-EGR-CK.
Fluorescence labeling of proteases in the extracellular protein fraction from P. gingivalis was done as previously reported (4). Briefly, solutions of extracellular protein fractions from P. gingivalis were treated at 4°C for 10 min with equal volumes of 0.2 M Tris-HCl (pH 8.4) containing 20 mM CaCl2 and 20 mM 2-mercaptoethanol. Dansyl-glutamyl-glycyl-arginyl chloromethyl ketone (DNS-EGR-CK) (0.25 mg; Calbiochem, San Diego, Calif.) was dissolved in 600 μl of 95% (vol/vol) aqueous ethanol just before use. Fifty microliters was added to the reduced protease solution, and the reaction was allowed to proceed at 4°C until the enzyme activity (monitored by BAPNA hydrolysis) was abolished. Samples of the protein solution were then dried in a SpeedVac concentrator (Thermo Savant, Holbrook, N.Y.). To identify the protease band unambiguously after SDS-PAGE, labeling was also performed in the presence of 50 μM leupeptin. The dried fluorescence-labeled proteases were then treated with SDS-PAGE sample buffer and subjected to electrophoresis.
Casein substrate zymography.
Novex Zymogram gels (12% Tris-glycine gel with β-casein incorporated as a substrate) were obtained from Invitrogen and used according to the manufacturer's instructions. Proteolytic activity was visualized as a clear band against a blue background.
RESULTS
The onset of proteolytic activity in P. gingivalis FLL92 occurs during stationary phase.
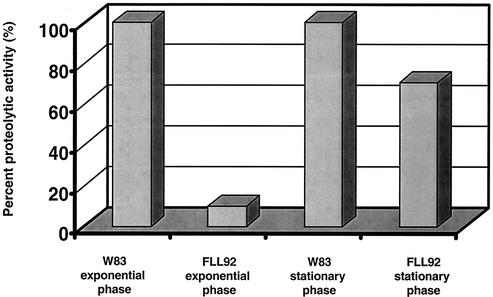
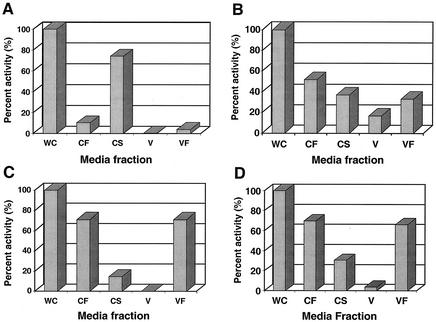
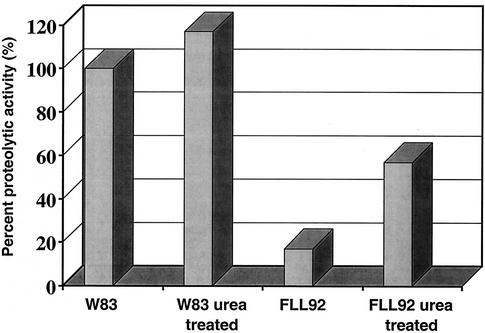
P. gingivalis strains W83 and FLL92 were assayed for proteolytic activity by using BAPNA and Ac-Lys-pNA. In late-exponential-growth-phase cultures, proteolytic activity in P. gingivalis FLL92 was reduced by 90% compared to that of the wild-type strain (Fig. 1). In the stationary phase of growth, the proteolytic activity of P. gingivalis FLL92 showed a dramatic increase in contrast to the proteolytic activity of the wild-type strain, which remained constant throughout this growth phase (Fig. 1). In addition, most of the Arg-X-specific activity present in P. gingivalis FLL92 was found in the vesicle-free fraction, while that of W83 was mostly membrane bound (Fig. 2). No change was observed in the distribution of the proteolytic activity of P. gingivalis FLL92 in stationary phase compared to that in exponential phase (Fig. 2). Similar results were observed for the Lys-X-specific activity profile in P. gingivalis FLL92 (data not shown). Taken together, these data suggest that under the same physiological conditions there is a late onset of proteolytic activity in the vimA mutant compared to that of the wild-type strain.
FIG. 1.
Proteolytic activity of P. gingivalis mutants. P. gingivalis was grown to late log phase (OD660 of 0.8) or stationary phase (OD660 of 1.5) in 1 liter of BHI broth supplemented with hemin and vitamin K. Activity against BAPNA was tested in whole-cell culture according to methods reported elsewhere (1). The results shown are representative of four independent experiments.
FIG. 2.
Distribution of Rgp protease activity in P. gingivalis. Activities against BAPNA were tested in whole-cell culture (WC), cell-free medium (CF), cell suspension (CS), vesicles (V), and vesicle-free medium (VF). The fractions were prepared according to the methods of Potempa et al. (36). One hundred percent of the activities in the whole-cell fraction were assumed to be produced by that strain. The activities measured in P. gingivalis FLL92 represent 10 and 60% of the activities measured in the wild-type strain W83 in exponential (OD660 = 0.8) and stationary (OD660 = 1.5) growth phases, respectively. CF + CS = WC; VF + V = CF.
Extracellular proteins from P. gingivalis FLL92.
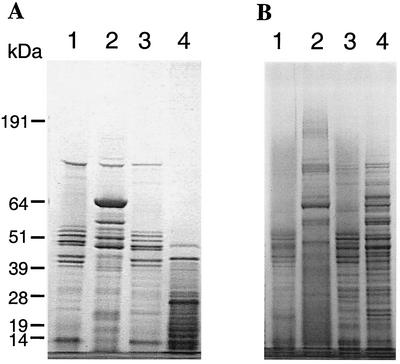
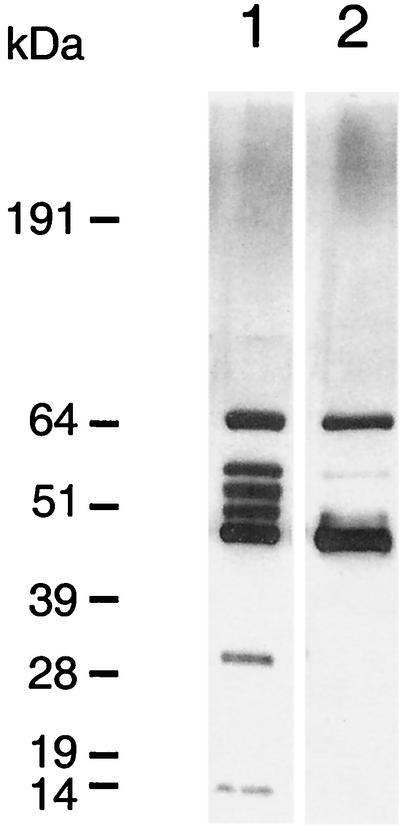
Although the proteolytic activity was reduced in P. gingivalis FLL92, no change in the expression of the gingipain genes was observed (1). This lack of change may suggest a defect in the translation, transport, or maturation of the protease. To examine the profile of the extracellular proteins from P. gingivalis FLL92, two acetone-precipitated protein preparations of the culture supernatant were subjected to SDS-PAGE. As shown in Fig. 3, several protein bands observed in the extracellular protein fraction from P. gingivalis FLL92 were missing in similar preparations of the wild type. High-molecular-weight protein bands of over 100 kDa were more prominent in extracellular protein fractions from P. gingivalis FLL92. The 37.5% acetone selectively precipitated a 64-kDa protein that was present only in P. gingivalis FLL92 grown to exponential phase. There were no changes in the protein profile when the extracellular protein fractions were prepared in the presence of Nα-p-tosyl-l-lysine chloromethyl ketone (data not shown).
FIG. 3.
SDS-PAGE of acetone-precipitated P. gingivalis extracellular proteins. Proteins in the supernatant fractions of cultures at different growth phases in BHI medium were precipitated with 37.5% (A) or 60% (B) acetone. The fractions were solubilized at 72°C in reducing buffer. NuPAGE bis-Tris gels (4 to 12%) were stained with Simply Blue Safe stain. Lanes: 1, W83 at exponential phase; 2, FLL92 at exponential phase; 3, W83 at stationary phase; and 4, FLL92 at stationary phase. The molecular mass markers (in kilodaltons) are indicated on the left. Each lane contains 20 μg of protein. It is noteworthy that the 64-kDa band was observed in abundance when 37.5% acetone instead of 60% acetone was used for the precipitation.
Analysis of the extracellular proteins from P. gingivalis FLL92 by using specific antiprotease antibodies as probes.
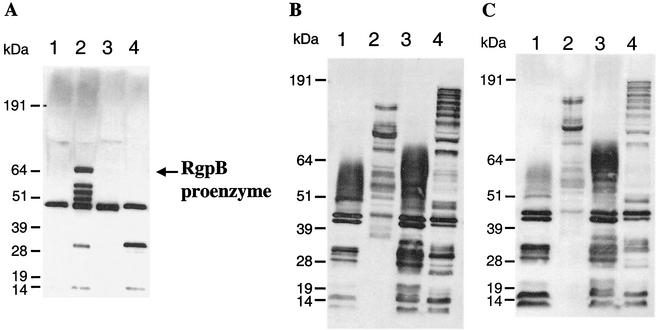
Similar to the recA-defective mutant P. gingivalis FLL32, the transcription of the major protease genes in P. gingivalis FLL92 (1, 2) showed no changes. Further, in P. gingivalis FLL32, immunoreactive bands to antibodies against the major proteases were detected in the extracellular fractions, suggesting the presence of a product encoded by the protease genes (2). Thus, the presence of an mRNA transcript for the major protease genes in P. gingivalis FLL92 may suggest a defect in protease biogenesis. To determine the presence of the major proteases in P. gingivalis FLL92, culture supernatants from cells grown to late log and stationary growth phases were tested by Western blot analysis by using antibodies against RgpB, RgpA, and Kgp. As shown in Fig. 4, the extracellular proteins produced during exponential and stationary growth phases by P. gingivalis FLL92 showed multiple unique immunoreactive bands to RgpB, RgpA, and Kgp antibodies, compared to those of the wild-type strain. During the stationary phase, several of the immunoreactive bands in P. gingivalis FLL92 were similar in size to those representing the catalytic and hemagglutinin domains in RgpA and Kgp in the wild-type strain. It is noteworthy that the unique 64-kDa protein band present in P. gingivalis FLL92 in exponential growth phase (Fig. 3A) immunoreacted only with anti-RgpB-specific antibodies (Fig. 4A). This immunoreactive band was not detected in stationary phase; however, a band similar in size to the band representing the catalytic domain in the wild-type strain was observed in both growth phases. When gel filtration and arginine-Sepharose gel affinity chromatography were used to fractionate the extracellular proteins from P. gingivalis FLL92, antibodies against RgpA, RgpB, and Kgp immunoreacted with the fractions that contained high-molecular-weight proteins with no detectable proteolytic activity (data not shown). Taken together, these data suggest that the protein products of the protease genes are excreted in P. gingivalis FLL92. Further, in the absence of proteolytic activity, protein species that are recognized by anti-gingipain antibodies are present in the extracellular medium.
FIG. 4.
Western immunoblot analysis of the extracellular proteins from P. gingivalis by using specific anti-Rgp and anti-Kgp antibodies as probes. All lanes contained 20 μg of acetone (37.5%)-precipitated proteins from the supernatant fractions of cultures in BHI medium at different growth phases. The membrane was reacted with antiserum raised in rabbits or chickens against the Arg-X- and Lys-X-specific protease from P. gingivalis. The secondary antibody was goat anti-rabbit or anti-chicken immunoglobulin G-horseradish peroxidase conjugate.(Zymed Laboratories Inc.). Reactions were done with rabbit anti-RgpB (A), rabbit anti-RgpA (B), and chicken anti-Kgp (C). Lanes: 1, W83 at exponential phase; 2, FLL92 at exponential phase; 3, W83 at stationary phase; 4, FLL92 at stationary phase. It is noteworthy that the 64-kDa band was identified as the RgpB proenzyme by mass spectrometry (see the text).
Identification of the RgpB proenzyme.
The presence of unique high-molecular-weight protein species that immunoreact with antibodies against the gingipains may suggest that the gingipains are secreted in an inactive form in the vimA-defective mutant during exponential growth phase. The prominent, unique 64-kDa protein band present in P. gingivalis FLL92 in exponential growth phase that immunoreacted only with anti-RgpB-specific antibodies was isolated by SDS-PAGE. Mass spectrometry of a total of 15 tryptic peptides constituting 29% of the total protein revealed the identity of the 64-kDa protein to be RgpB with its attached propeptide (GenBank accession number AF007124). This result confirms that RgpB is translated and excreted.
Protease activation.
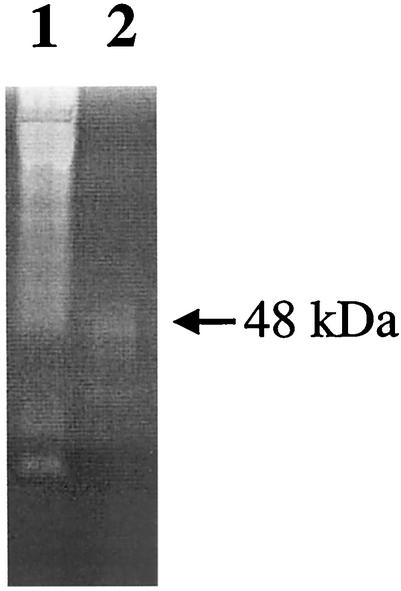
Proteolytic cleavage is a common mechanism that can generate an active enzyme from a larger proenzyme. While activation can be achieved by the action of other proteolytic enzymes (5, 18), we cannot rule out the possibility that the tertiary structure of the putative proenzyme may be incompatible for activation or that an inhibitor may be present at its catalytic site. Since most proteins are denatured in the presence of high concentrations of urea and some will renature if the high concentration is slowly reduced (16), extracellular proteins from P. gingivalis FLL92 grown to late exponential phase were subjected to a urea denaturation-renaturation cycle. Figure 5 shows proteolytic activation that is approximately fourfold greater in the extracellular protein fraction from P. gingivalis FLL92 grown to late exponential phase than that of the wild-type strain. Western blot analysis of the urea-activated fraction using antibodies against RgpB shows the continued presence of the 64-kDa band; however, immunoreactive bands in the 50- to 57-kDa range were missing (Fig. 6). Taken together, these findings suggest that the decrease in the 50- to 57-kDa immunoreactive bands accompanied by an increase in the intensity of the 48-kDa band may explain the increase in Rgp proteolytic activity. The 48-kDa band is similar in size to the band representing the catalytic domain in the wild-type strain (Fig. 4A).
FIG. 5.
Protease activation in the extracellular protein fraction from P. gingivalis FLL92. P. gingivalis was grown to late log phase (OD660 of 0.8) in 1 liter of BHI broth supplemented with hemin and vitamin K. Acetone (37.5%)-precipitated proteins were mixed with 8 M urea and incubated at 4°C for 1 h. The urea was slowly removed from the mixture by centrifugation (10,000 × g) in a Millipore filtration unit with the addition of increasing volumes of 100 mM Tris-HCl buffer (pH 7.4). Activities against BAPNA were tested according to the methods of Potempa et al. (36). The results shown are representative of four independent experiments.
FIG. 6.
Western blot analysis of the urea-activated fraction using antibodies against RgpB. All lanes contained 20 μg of acetone (37.5%)-precipitated proteins from the supernatant fractions of P. gingivalis FLL92 cultures grown in BHI medium at exponential growth phases. The membrane was reacted with antiserum raised in rabbits against RgpB. Lanes: 1, FLL92 exponential-growth-phase extracellular proteins; 2, FLL92 exponential-growth-phase extracellular proteins treated with urea (see the text).
Active-site labeling of the RgpB proenzyme.
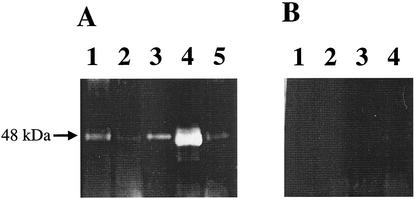
DNS-EGR-CK binds irreversibly to the active site of the arginine-specific proteases of P. gingivalis and can be detected by UV illumination after electrophoresis (4). As expected, DNS-EGR-CK-treated extracellular proteins from the wild-type strain grown to both late exponential and stationary phases generate fluorescent bands characteristic of the catalytic domains of the arginine-specific proteases on SDS-PAGE gels (Fig. 7). DNS-EGR-CK treatment of extracellular proteins from P. gingivalis FLL92 grown to late exponential or stationary phase showed a 48-kDa fluorescent band (Fig. 7). However, there was no detectable specifically labeled protein that was similar in size to the 64-kDa RgpB proenzyme in the late-exponential- or stationary-phase extracellular protein fraction (Fig. 7). Extracellular proteins from P. gingivalis FLL92, grown to late exponential phase and subjected to a urea denaturation-renaturation cycle, showed a fluorescent band that was indistinguishable from that for the catalytic domain of the wild-type strain on SDS-PAGE gels. It is noteworthy that the DNS-EGR-CK specifically labeled protein band was more intense in the increased proteolytic extracellular fraction from P. gingivalis FLL92 grown to stationary phase than in the late-exponential-phase extracellular fraction (Fig. 7). Moreover, and similar to what was observed for the wild-type strain, labeling with DNS-EGR-CK was inhibited by leupeptin, thereby confirming the specificity of the reaction to the protease-active site. Deficiency in protease activity of the 64-kDa RgpB proenzyme was further confirmed by casein substrate zymography. As shown in Fig. 8, only a proteolytic band at approximately 48 kDa was present in the extracellular proteins from P. gingivalis FLL92 grown to late exponential phase. Taken together, these results suggest that the 64-kDa protein band identified as RgpB with its attached propeptide is missing proteolytic activity and that its active site is not accessible to DNS-EGR-CK even after a urea denaturation-renaturation cycle.
FIG. 7.
Gel electrophoresis of DNS-EGR-CK-labeled extracellular protein from P. gingivalis. Acetone-precipitated extracellular proteins from P. gingivalis W83 and P. gingivalis FLL92 were labeled with the fluorescent irreversible inhibitor DNS-EGR-CK as described in the text and subjected to SDS-PAGE on 4 to 12% gels as described for Fig. 3A. The gels were viewed under UV light to visualize labeled proteins. (A) Lanes: 1, W83 at exponential phase; 2, FLL92 at exponential phase; 3, W83 at stationary phase; 4, FLL92 at stationary phase; 5, FLL92 at exponential phase and treated with urea. (B) Lanes: 1, W83 at exponential phase; 2, FLL92 at exponential phase; 3, W83 at stationary phase; and 4, FLL92 at stationary phase. All the lanes in panel B were labeled with DNS-EGR-CK in the presence of 50 μM leupeptin. Each lane contains 20 μg of protein. The arrow indicates the DNS-EGR-CK-labeled protein band.
FIG. 8.
Detection of proteolytic activity in casein-conjugated polyacrylamide gel. All lanes contained 20 μg of acetone (37.5%)-precipitated proteins from the supernatant fractions of cultures in BHI medium at exponential growth phases. Samples were solubilized at room temperature for 10 min in a Tris-glycine SDS sample buffer (Invitrogen) prior to electrophoresis. Lanes: 1, W83; 2, FLL92. The arrow indicates a 48-kDa proteolytic band present in the FLL92 exponential-growth-phase extracellular proteins.
DISCUSSION
Several studies have suggested that the phenotypic expression of gingipains in P. gingivalis is involved in iron utilization and virulence (20, 21, 24, 27, 29). Specifically, a defect in the gingipain Kgp results in a non-black-pigmented mutant, thus suggesting that the accumulation of iron on the cell surface has been affected. While other nongingipain genes, including the vimA gene, can also affect this phenotypic property, the mechanism is still unclear. We have further characterized the non-black-pigmented vimA-defective mutant of P. gingivalis that was demonstrated previously to have reduced proteolytic activity and virulence (1). In contrast to the wild-type strain, the vimA-defective mutant, P. gingivalis FLL92, has mostly soluble proteolytic activity and little or no cell-associated activity. It is noteworthy, however, that in stationary phase, P. gingivalis FLL92 demonstrated a unique late onset of Arg-X- and Lys-X-specific proteolytic activity. This finding may suggest that in P. gingivalis, proteolytic activity is regulated by multiple mechanisms. During exponential growth phase, proteolytic activity in P. gingivalis may be regulated in a vimA-dependent manner. However, during stationary phase, an alternative pathway for protease activation may be upregulated to overcome or bypass the vimA mutation or it may be activated in the absence of VimA. Because activation of proteolytic activity between the log and stationary growth phases was not observed in the wild-type strain, it is unclear if the mechanisms of activation are complementary or growth phase synchronized. These mechanisms are being further investigated in the laboratory.
In a recent study, a porR-defective, non-black-pigmented mutant of P. gingivalis also showed a late onset of soluble proteolytic activity (42). It is unclear, however, if both the vimA and porR genes are part of a common pathway for protease activation and distribution. The porR gene product is part of the polysaccharide biosynthetic pathway which can affect gingipain biogenesis, its anchorage, and its possible distribution on the cell surface (42). While the porR gene does not regulate the transcription of the gingipain genes, we cannot rule out the possibility that the posttranscriptional-posttranslational regulation of gingipain biogenesis may involve the indirect effect of the porR gene product on the cell membrane. The generation and maintenance of subcellular organization in bacteria are critical for many cell processes and properties; thus, it is possible that the polysaccharide effect on the fluidity of the cell membrane could affect the transport, maturation, or distribution of the gingipains. In Shigella flexneri, distribution of the virulence protein IcsA (VirG) on the bacterial cell surface can be altered by modification of the lipopolysaccharide in the outer membrane (12, 39, 40). It is unclear if the vimA gene affects polysaccharide synthesis in P. gingivalis and thus could use a similar mechanism like S. flexneri to affect gingipain biogenesis and distribution. Preliminary reverse transcriptase PCR experiments show that the vimA mutation does not affect the expression of the porR gene at the transcriptional level. A relationship between polysaccharide biosynthesis and VimA is being further investigated.
The late onset of proteolytic activity in P. gingivalis FLL92 in stationary phase may be consistent with a stringent response, although we cannot rule out the possibility that other protease genes (e.g., tpr) (31, 32) might be upregulated to compensate for the gingipain deficiency. Bacteria undergoing nutritional stress, such as amino acid deprivation, synthesize highly phosphorylated guanosine nucleotides (ppGpp and pppGpp) (6). As part of this phenomenon, termed the stringent response, these nucleotides have been shown to play a role in growth-related gene expression during nutrient deprivation in Escherichia coli (6) or virulence gene expression in stationary phase in Legionella pneumophila (14, 15). A homologue of the relA gene in P. gingivalis has been identified (41). Construction of the relA mutant P. gingivalis KS7 revealed that it was defective in ribosome-mediated ppGpp formation and in the stringent response (41); however, it is unclear if this gene plays a role in the modulation of proteolytic activity in this organism.
P. gingivalis FLL92 released soluble proteins that reacted with gingipain-specific antibodies into the media. These soluble proteins appear to contain most of the total proteolytic activity in this strain. If this is so, it would be consistent with the DNS-EGR-CK specifically labeled protein band, representing the catalytic domain, being more intense in the increased proteolytic, extracellular fraction from P. gingivalis FLL92 grown to stationary phase. The high-molecular-weight immunoreactive proteins could represent the unprocessed or partially processed proteases. A unique 64-kDa protein was selectively precipitated from exponential-phase culture fluid by using approximately 38% acetone. This protein was definitively confirmed in this study to be RgpB with its profragment. The intermediate species in gingipain biogenesis may have their catalytic sites blocked, but the site can be activated by a urea denaturation-renaturation cycle. All the anti-RgpB immunoreactive bands between 55 and 67 kDa were no longer observed after a urea denaturation-renaturation cycle of a nonproteolytic fraction that resulted in a fourfold increase in protease activity. It is noteworthy that the 64-kDa protein band was observed in the urea-treated fraction; however, the inability to label this protein with DNS-EGR-CK may suggest that the active site is not accessible. Casein substrate zymography revealed that the 64-kDa band did not display any proteolytic activity. Thus, taken together, these results may indicate that the proenzyme is not responsible for the increased activity observed after urea activation.
An active enzyme can be generated from a larger polypeptide by autoprocessing or by the action of other proteolytic enzymes (19). Several cysteine proteases are converted to their active forms by removal of the prosegment by an autocatalytic mechanism (19). There is accumulating evidence that a multicomponent maturation pathway(s), perhaps including an autolytic mechanism, may be involved in the production of Arg-X- and Lys-X-specific proteases in P. gingivalis (4, 37). While it is clear that proteolytic processing of the full-length gingipain precursors in P. gingivalis is required to produce the isoforms detected (4, 37, 38), the continued presence of the RgpB proenzyme in the urea-activated fraction may suggest that the mechanism of protease activation in P. gingivalis requires a host-specific factor. It is unclear whether a factor, perhaps the vimA gene product or a product regulated by the vimA gene, is involved in the maturation process. It is also unclear if the vimA gene may affect the phenotypic expression of other proteases in P. gingivalis. The multiple proteolytic bands which may represent different isoforms of gingipain R and other proteases (7, 13) were observed only in assays of the wild-type strain. The effects of VimA on other proteases are currently under investigation.
Finally, it is noteworthy that the gingipain genes are not clustered on the P. gingivalis chromosome (oral pathogen sequence databases are available at http://www.oralgen.lanl.gov). The results of this study, therefore, would support the hypothesis that inactivation of the vimA gene may affect a common pathway for protease biogenesis. Collectively, our data suggest that the protease genes are regulated differently during different growth phases. While vimA may affect regulation of proteolytic activity during exponential growth phase, it is apparent that the onset of proteolytic activity in stationary phase may be under the control of a different gene(s). We have presented evidence for posttranslational regulation of proteolytic activity in P. gingivalis. Furthermore, to our knowledge, this is the first report of the identification of a gingipain proenzyme from P. gingivalis. This model system will facilitate a more careful study of gingipain biogenesis in P. gingivalis.
Acknowledgments
We thank Roger Moore for technical assistance with mass spectrometry. We thank Jan Potempa for the gift of the antigingipain antibodies.
This work was supported by the Loma Linda University Schools of Medicine and Dentistry, by University of California Tobacco Related Disease Research Program grant 10RT-0122 (to H.M.F.), by National Institute of Dental and Craniofacial Research grant R01 DE13664 (to H.M.F.), and by National Institutes of Health Cancer Center support grant CA33572 (to M.K.Y.).
Editor: V. J. DiRita
REFERENCES
- 1.Abaibou, H., Z. Chen, G. J. Olango, Y. Liu, J. Edwards, and H. M. Fletcher. 2001. vimA gene downstream of recA is involved in virulence modulation in Porphyromonas gingivalis W83. Infect. Immun. 69:325-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abaibou, H., Q. Ma, G. J. Olango, J. Potempa, J. Travis, and H. M. Fletcher. 2000. Unaltered expression of the major protease genes in a non-virulent recA-defective mutant of Porphyromonas gingivalis W83. Oral Microbiol. Immunol. 15:40-47. [DOI] [PubMed] [Google Scholar]
- 3.Abe, N., T. Kadowaki, K. Okamoto, K. Nakayama, M. Ohishi, and K. Yamamoto. 1998. Biochemical and functional properties of lysine-specific cysteine proteinase (Lys-gingipain) as a virulence factor of Porphyromonas gingivalis in periodontal disease. J. Biochem. (Tokyo) 123:305-312. [DOI] [PubMed] [Google Scholar]
- 4.Aduse-Opoku, J., M. Rangarajan, K. A. Young, and M. A. Curtis. 1998. Maturation of the arginine-specific proteases of Porphyromonas gingivalis W50 is dependent on a functional prR2 protease gene. Infect. Immun. 66:1594-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun, P., W. Bitter, and J. Tommassen. 2000. Activation of Pseudomonas aeruginosa elastase in Pseudomonas putida by triggering dissociation of the propeptide-enzyme complex. Microbiology 146:2565-2572. [DOI] [PubMed] [Google Scholar]
- 6.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: molecular and cellular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 7.Curtis, M. A., H. K. Kuramitsu, M. Lantz, F. L. Macrina, K. Nakayama, J. Potempa, E. C. Reynolds, and J. Aduse-Opoku. 1999. Molecular genetics and nomenclature of proteases of Porphyromonas gingivalis. J. Periodontal Res. 34:464-472. [DOI] [PubMed] [Google Scholar]
- 8.Davis, M. T., and T. D. Lee. 1998. Rapid protein identification using a microscale electrospray LC/MS system on an ion trap mass spectrometer. J. Am. Soc. Mass Spectrom. 9:194-201. [DOI] [PubMed] [Google Scholar]
- 9.Falany, M. L., A. M. Thames III, J. M. McDonald, H. C. Blair, M. A. McKenna, R. E. Moore, M. K. Young, and J. P. Williams. 2001. Osteoclasts secrete the chemotactic cytokine mim-1. Biochem. Biophys. Res. Commun. 281:180-185. [DOI] [PubMed] [Google Scholar]
- 10.Fletcher, H. M., R. M. Morgan, and F. L. Macrina. 1997. Nucleotide sequence of the Porphyromonas gingivalis W83 recA homolog and construction of a recA-deficient mutant. Infect. Immun. 65:4592-4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Genco, R., S. Offenbacher, and J. Beck. 2002. Periodontal disease and cardiovascular disease: epidemiology and possible mechanisms. J.Am. Dent. Assoc. 133(Suppl.):14S-22S. [DOI] [PubMed]
- 12.Goldberg, M. B., and J. A. Theriot. 1995. Shigella flexneri surface protein IcsA is sufficient to direct actin-based motility. Proc. Natl. Acad. Sci. USA 92:6572-6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grenier, D., G. Chao, and B. C. McBride. 1989. Characterization of sodium dodecyl sulfate-stable Bacteroides gingivalis proteases by polyacrylamide gel electrophoresis. Infect. Immun. 57:95-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammer, B. K., and M. S. Swanson. 1999. Co-ordination of Legionella pneumophila virulence with entry into stationary phase by ppGpp. Mol. Microbiol. 33:721-731. [DOI] [PubMed] [Google Scholar]
- 15.Hammer, B. K., E. S. Tateda, and M. S. Swanson. 2002. A two-component regulator induces the transmission phenotype of stationary-phase Legionella pneumophila. Mol. Microbiol. 44:107-118. [DOI] [PubMed] [Google Scholar]
- 16.Hobson, A. H., C. M. Buckley, J. L. Aamand, S. T. Jorgensen, B. Diderichsen, and D. J. McConnell. 1993. Activation of a bacterial lipase by its chaperone. Proc. Natl. Acad. Sci. USA 90:5682-5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kadowaki, T., K. Nakayama, F. Yoshimura, K. Okamoto, N. Abe, and K. Yamamoto. 1998. Arg-gingipain acts as a major processing enzyme for various cell surface proteins in Porphyromonas gingivalis. J. Biol. Chem. 273:29072-29076. [DOI] [PubMed] [Google Scholar]
- 18.Kessler, E., M. Safrin, J. K. Gustin, and D. E. Ohman. 1998. Elastase and the LasA protease of Pseudomonas aeruginosa are secreted with their propeptides. J. Biol. Chem. 273:30225-30231. [DOI] [PubMed] [Google Scholar]
- 19.Khan, A. R., and M. N. James. 1998. Molecular mechanisms for the conversion of zymogens to active proteolytic enzymes. Protein Sci. 7:815-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuboniwa, M., A. Amano, and S. Shizukuishi. 1998. Hemoglobin-binding protein purified from Porphyromonas gingivalis is identical to lysine-specific cysteine proteinase (Lys-gingipain). Biochem. Biophys. Res. Commun. 249:38-43. [DOI] [PubMed] [Google Scholar]
- 21.Kuboniwa, M., A. Amano, S. Shizukuishi, I. Nakagawa, and S. Hamada. 2001. Specific antibodies to Porphyromonas gingivalis Lys-gingipain by DNA vaccination inhibit bacterial binding to hemoglobin and protect mice from infection. Infect. Immun. 69:2972-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuramitsu, H. K. 1998. Protease of Porphyromonas gingivalis: what don't they do? Oral Microbiol. Immunol. 13:263-270. [DOI] [PubMed] [Google Scholar]
- 23.Lantz, M. S. 1996. New insights into mechanisms of bacterial pathogenesis in periodontitis. Curr. Opin. Periodontol. 3:10-18. [PubMed] [Google Scholar]
- 24.Lewis, J. P., J. A. Dawson, J. C. Hannis, D. Muddiman, and F. L. Macrina. 1999. Hemoglobinase activity of the lysine gingipain protease (Kgp) of Porphyromonas gingivalis W83. J. Bacteriol. 181:4905-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayrand, D., and S. C. Holt. 1988. Biology of asaccharolytic black-pigmented Bacteroides species. Microbiol. Rev. 52:134-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore, R. E., M. K. Young, and T. D. Lee. 2000. Method for screening peptide fragment ion mass spectra prior to database searching. J. Am. Soc. Mass Spectrom. 11:422-426. [DOI] [PubMed] [Google Scholar]
- 27.Nakayama, K., D. B. Ratnayake, T. Tsukuba, T. Kadowaki, K. Yamamoto, and S. Fujimura. 1998. Haemoglobin receptor protein is intragenically encoded by the cysteine proteinase-encoding genes and the haemaglutinin-encoding gene of Porphyromonas gingivalis. Mol. Microbiol. 27:51-61. [DOI] [PubMed] [Google Scholar]
- 28.Okamoto, K., T. Kadowaki, K. Nakayama, and K. Yamamoto. 1996. Cloning and sequencing of the gene encoding a novel lysine-specific cysteine proteinase (Lys-gingipain) in Porphyromonas gingivalis: structural relationship with the arginine-specific cysteine proteinase (Arg-gingipain). J. Biochem. (Tokyo) 120:398-406. [DOI] [PubMed] [Google Scholar]
- 29.Okamoto, K., K. Nakayama, T. Kadowaki, N. Abe, D. B. Ratnayake, and K. Yamamoto. 1998. Involvement of a lysine-specific cysteine proteinase in hemoglobin adsorption and heme accumulation by Porphyromonas gingivalis. J. Biol. Chem. 273:21225-21231. [DOI] [PubMed] [Google Scholar]
- 30.Otsuka, M., J. Endo, D. Hinode, A. Nagata, R. Maehara, M. Sato, and R. Nakamura. 1987. Isolation and characterization of protease from culture supernatant of Bacteroides gingivalis. J. Periodontal Res. 22:491-498. [DOI] [PubMed] [Google Scholar]
- 31.Park, Y., B. Lu, C. Mazur, and B. C. McBride. 1997. Inducible expression of a Porphyromonas gingivalis W83 membrane-associated protease. Infect. Immun. 65:1101-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park, Y., and B. C. McBride. 1993. Characterization of the tpr gene product and isolation of a specific protease-deficient mutant of Porphyromonas gingivalis W83. Infect. Immun. 61:4139-4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pavloff, N., P. A. Pemberton, J. Potempa, W. C. Chen, R. N. Pike, V. Prochazka, M. C. Kiefer, J. Travis, and P. J. Barr. 1997. Molecular cloning and characterization of Porphyromonas gingivalis lysine-specific gingipain. A new member of an emerging family of pathogenic bacterial cysteine proteinases. J. Biol. Chem. 272:1595-1600. [DOI] [PubMed] [Google Scholar]
- 34.Pike, R., W. McGraw, J. Potempa, and J. Travis. 1994. Lysine- and arginine-specific proteinases from Porphyromonas gingivalis. Isolation, characterization, and evidence for the existence of complexes with hemagglutinins. J. Biol. Chem. 269:406-411. [PubMed] [Google Scholar]
- 35.Potempa, J., N. Pavloff, and J. Travis. 1995. Porphyromonas gingivalis: a proteinase/gene accounting audit. Trends Microbiol. 3:430-434. [DOI] [PubMed] [Google Scholar]
- 36.Potempa, J., R. Pike, and J. Travis. 1995. The multiple forms of trypsin-like activity present in various strains of Porphyromonas gingivalis are due to the presence of either Arg-gingipain or Lys-gingipain. Infect. Immun. 63:1176-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rangarajan, M., J. Aduse-Opoku, J. M. Slaney, K. A. Young, M. A. Curtis, and J. Aduse Opoku. 1997. The prpR1 and prR2 arginine-specific protease genes of Porphyromonas gingivalis W50 produce five biochemically distinct enzymes. Mol. Microbiol. 23:955-965. [DOI] [PubMed] [Google Scholar]
- 38.Rangarajan, M., S. J. Smith, S. U., and M. A. Curtis. 1997. Biochemical characterization of the arginine-specific proteases of Porphyromonas gingivalis W50 suggests a common precursor. Biochem. J. 323:701-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robbins, J. R., D. Monack, S. J. McCallum, A. Vegas, E. Pham, M. B. Goldberg, and J. A. Theriot. 2001. The making of a gradient: IcsA (VirG) polarity in Shigella flexneri. Mol. Microbiol. 41:861-872. [DOI] [PubMed] [Google Scholar]
- 40.Sandlin, R. C., K. A. Lampel, S. P. Keasler, M. B. Goldberg, A. L. Stolzer, and A. T. Maurelli. 1995. Avirulence of rough mutants of Shigella flexneri: requirement of O antigen for correct unipolar localization of IcsA in the bacterial outer membrane. Infect. Immun. 63:229-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sen, K., J.-I. Hayashi, and H. K. Kuramitsu. 2000. Characterization of the relA gene of Porphyromonas gingivalis. J. Bacteriol. 182:3302-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shoji, M., D. B. Ratnayake, Y. Shi, T. Kadowaki, K. Yamamoto, F. Yoshimura, A. Akamine, M. A. Curtis, and K. Nakayama. 2002. Construction and characterization of a nonpigmented mutant of Porphyromonas gingivalis: cell surface polysaccharide as an anchorage for gingipains. Microbiology 148:1183-1191. [DOI] [PubMed] [Google Scholar]
- 43.Sundqvist, G. 1993. Pathogenicity and virulence of black-pigmented gram-negative anaerobes. FEMS Immunol. Med. Microbiol. 6:125-137. [DOI] [PubMed] [Google Scholar]
- 44.Tokuda, M., W. Chen, T. Karunakaran, and H. K. Kuramitsu. 1998. Regulation of protease expression in Porphyromonas gingivalis. Infect. Immun. 66:5232-5237. [DOI] [PMC free article] [PubMed] [Google Scholar]