Abstract
Citrobacter spp., Enterobacter spp., and Klebsiella spp. isolated from the human gut were investigated for the biosynthesis of cellulose and curli fimbriae (csg). While Citrobacter spp. produced curli fimbriae and cellulose and Enterobacter spp. produced cellulose with various temperature-regulatory programs, Klebsiella spp. did not show pronounced expression of those extracellular matrix components. Investigation of multicellular behavior in two Citrobacter species and Enterobacter sakazakii showed an extracellular matrix, cell clumping, pellicle formation, and biofilm formation associated with the expression of cellulose and curli fimbriae. In those three strains, the csgD-csgBA region and the cellulose synthase gene bcsA were conserved. PCR screening for the presence of csgD, csgA and bcsA revealed that besides Klebsiella pneumoniae and Klebsiella oxytoca, all species investigated harbored the genetic information for expression of curli fimbriae and cellulose. Since Citrobacter spp., Enterobacter spp., and Klebsiella spp. are frequently found to cause biofilm-related infections such as catheter-associated urinary tract infections, the human gut could serve as a reservoir for dissemination of biofilm-forming isolates.
Besides Escherichia coli, other genera of the family Enterobacteriaceae, mainly Citrobacter, Enterobacter, Klebsiella, and Proteus spp., are regularly recovered from the human gastrointestinal tract, where they are members of the normal fecal floras or transient colonizers (14). Besides inhabitants of the human gastrointestinal tract, the Enterobacteriaceae, and primarily the above-listed genera, are the most prominent family of gram-negative bacteria that cause biofilm-related infections such as biliary tract infections, bacterial prostatitis, and catheter-associated urinary tract infections, the most common type of nosocomial infection (4). The gut is considered a primary source for dissemination and transmission of those potential pathogens to susceptible sites.
The rdar morphotype is a multicellular behavior commonly expressed by Salmonella enterica serotype Typhimurium isolates (26) and certain isolates of Escherichia coli (2, 24). The rdar morphotype mediates different types of multicellular behavior, for example, cell aggregation in liquid culture, pellicle formation at the air-liquid interface, and biofilm formation at liquid-solid interfaces (17). The rdar morphotype produces an extracellular matrix consisting of cellulose and curli fimbriae, which is the major determinant of cell-cell interactions and cell adherence to hydrophilic and hydrophobic abiotic surfaces. Besides their distinct roles in bacterial self-organization, features related to virulence and transmission such as adherence and invasion of epithelial cells and chlorine resistance have been assigned to curli fimbriae and cellulose, respectively (3, 8, 13, 21, 22).
Gene required for cellulose biosynthesis are encoded by the bcsABZC operon. While bcsA encodes the cellulose synthase, the exact functions of the other genes remain to be determined. Structural genes for curli fimbriae are encoded by the csgBA(C) operon. Cellulose and curli fimbria biosynthesis is commonly regulated, either through adrA or directly by csgD, a transcriptional activator, divergently transcribed to csgBA(C).
Performing this study, we wanted to gain insights into the ability of clinically important Enterobacteriaceae besides E. coli and serotype Typhimurium to express cellulose and curli fimbriae. The isolates were collected from the gastrointestinal tract as the major human reservoir for dissemination of isolates.
Isolation and phenotypic characterization of bacterial strains.
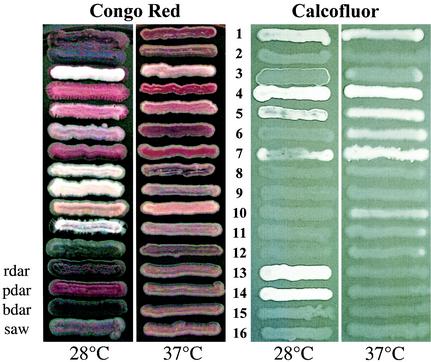
Since we suspected that cellulose and curli fimbria expression are unstable phenotypes upon subculture, we collected primary isolates of bacterial strains from 21 volunteers who provided fecal samples. Fecal swabs were immediately inoculated on MacConkey agar plates (Oxoid) and incubated overnight at 37°C. The confluent microbial outgrowth was collected and resuspended, and appropriate dilutions were plated on MacConkey agar plates for single colonies. A total of 25 non-E. coli isolates which belonged to 13 species were identified with the API 20E kit (BioMerieux, Nürtingen, Germany): Citrobacter sp. (isolated independently two times), Citrobacter freundii (n = 3), Citrobacter koseri/farmeri (n = 1), Enterobacter sp. (n = 1), Enterobacter aerogenes (n = 2), Enterobacter cloacae (n = 3), Enterobacter sakazakii (n = 2), Klebsiella sp. (n = 2), K. oxytoca (n = 2), K. pneumoniae (n = 2), Klyvera sp. (n = 2), Proteus mirabilis (n = 1), and Raoultella ornithinolytica (n = 2). When the fecal strains were grown on Congo Red (CR) plates (17), we observed that several of the isolates selectively bound CR, thereby resembling the rdar, bdar, or pdar morphotype of serotype Typhimurium reference strains (Fig. 1). Serotype Typhimurium colonies which show the rdar morphotype express cellulose and curli fimbriae, while colonies which show the pdar morphotype express cellulose and colonies which show the bdar morphotype express curli fimbriae (17, 26).
FIG. 1.
Congo red and Calcofluor staining of morphotypes of representatives of enterobacterial species isolated from feces. The strains were grown for 48 h at 28°C and for 24 h at 37°C. Morphotypes were compared with the phenotype of serotype Typhimurium UMR1 (ATCC 14028-1s, Nal r) and its mutants, which have been streaked at the lower part of the panel. ROWS: 1, Citrobacter sp. strain Fec2; 2, C. freundii Fec4; 3, C. koseri/farmeri Fec157; 4, Enterobacter sp. strain Fec125; 5, E. aerogenes Fec135; 6, E. cloacae Fec36; 7, E. sakazakii Fec39; 8, Klebsiella sp. strain Fec164; 9, K. oxytoca Fec139; 10, K. pneumoniae Fec141; 11, R. ornithinolytica Fec153; 12, P. mirabilis Fec162; 13, serotype Typhimurium UMR1 (cellulose 28+, curli 28+); 14, serotype Typhimurium MAE1 (cellulose 28+, curli −); 15, serotype Typhimurium MAE222 (cellulose −, curli 28+); 16, serotype Typhimurium MAE51 (cellulose −, curli −).
Citrobacter spp. regularly expressed the rdar or the bdar morphotype at 28°C, while Enterobacter spp. commonly expressed the pdar morphotype at either 28°C and 37°C or only at 37°C. Klebsiella spp. did not show characteristic morphotypes that indicated pronounced expression of cellulose or curli fimbriae.
Expression of curli fimbriae and cellulose.
Three strains with characteristic morphotypes, Citrobacter sp. strain Fec2, C. freundii Fec4, and E. sakazakii Fec39, were chosen for more detailed analyses. Citrobacter sp. strain Fec2 expressed the bdar morphotype at 28°C, C. freundii Fec4 expressed the rdar morphotype at 28°C, and E. sakazakii Fec39 expressed the pdar morphotype at 28°C and 37°C (Fig. 1). However, as displayed in Fig. 1, during storage for several months at −70°C, Citrobacter sp. strain Fec2 changed morphotype to rdar at 28°C and pdar at 37°C, indicating enhanced cellulose biosynthesis. However, since those changes occurred after the molecular study of the isolates, they will not be considered here.
After an enrichment procedure employed for plate-grown cultures of Citrobacter sp. strain Fec2 (18a), the putative curli fimbria subunit was detected as a prominent band at ∼16 kDa that was cut out, trypsin digested, and subjected to sequence analysis by quadruple time-of-flight mass spectrometry (20). Sequence comparison of three peptides with the database indicated that the protein was homologous to CsgA, the structural subunit of curli fimbriae expressed by E. coli and serotype Typhimurium. Western blot analysis detected curli fimbriae after formic acid treatment (5) produced by Citrobacter sp. strain Fec2 and C. freundii Fec4 at 28°C, while E. sakazakii Fec39 did not produce any curli fimbriae (data not shown).
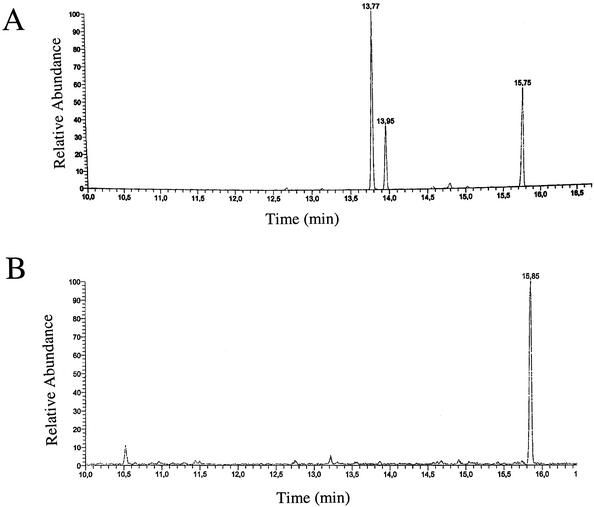
Calcofluor binding indicated variable production of cellulose, a 1,4-β-glucan, in the three strains (Fig. 1). Since Calcofluor binding is not absolutely specific for cellulose, we confirmed cellulose production by isolating crystalline cellulose by the Updegraff method with a mixture of hot 58% acetic acid and 19% nitric acid (25, 26). After hydrolysis of the isolated polymer, the glucose monomers detected by high-pressure liquid chromatography (HPLC) (Fig. 2) were indicative of cellulose production. While glucose was detected when samples from E. sakazakii Fec39 grown at 28°C and 37°C and C. freundii Fec4 grown at 28°C were analyzed, Citrobacter sp. strain Fec2 samples did not show any significant glucose peak.
FIG. 2.
Detection of glucose monomers by HPLC after isolation of crystalline cellulose. Results for (A) E. sakazakii Fec39 and (B) serotype Typhimurium MAE51 are shown as examples of positive and negative outcomes, respectively. Crystalline cellulose was hydrolyzed and sugar monomers were detected by HPLC. Glucose shows two peaks representing the α- and β-anomers (retention time, 13.77 and 13.95 min, respectively); 1 μg of myoinositol was used as an internal standard (retention time, 15.75 min).
In conclusion, consistent with the expression of the pdar morphotype expressed at 28°C and 37°C, E. sakazakii Fec39 produced cellulose but not curli fimbriae at both temperatures. C. freundii Fec4, which showed the rdar morphotype at 28°C, produced cellulose and curli fimbriae at 28°C, while Citrobacter sp. strain Fec2, which showed the bdar morphotype, produced only curli fimbriae.
Multicellular behavior of isolates.
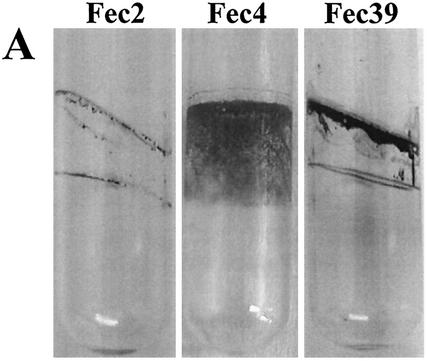
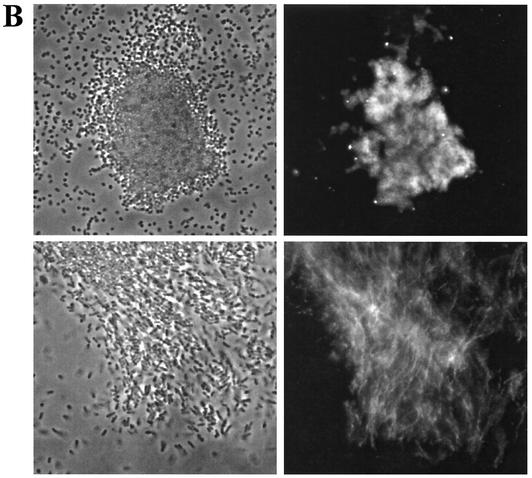
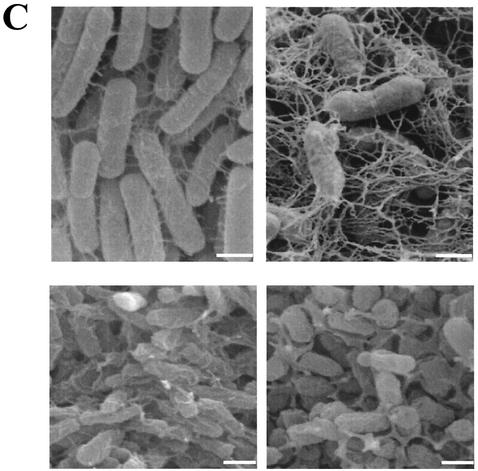
The expression of the extracellular matrix components cellulose and curli fimbriae is associated with biofilm formation and other modes of multicellular behavior (17). We used a steady-state model, incubation of the strains in glass tubes (17), to test biofilm formation by Citrobacter sp. strain Fec2, C. freundii Fec4, and E. sakazakii Fec39. Optimal biofilm formation was achieved with different incubation conditions for each strain (Fig. 3A and data not shown), which indicated that the regulatory patterns of biofilm formation with respect to oxygen tensions were different between the strains. Pellicle formation was observed for all three strains (data not shown). While E. sakazakii Fec39 formed a pellicle after just 24 h of incubation at 28°C in standing cultures, pellicle formation in cultures of Citrobacter sp. strain Fec2 and C. freundii Fec4 required 48 h to develop. In liquid culture at 28°C after incubation for 24 h, all three strains formed clumps, although the extent of clumping and the consistency of the clumps varied between the strains (Fig. 3B and data not shown). When observed by fluorescence microscopy, Calcofluor-stained cellulose fibrils could be observed for E. sakazakii Fec39 and C. freundii Fec4 (Fig. 3B). Free-floating cellulose fibrils associated with cell clumps were observed for E. sakazakii Fec39, as in serotype Typhimurium when cellulose is the sole extracellular matrix component (26). On the other hand, C. freundii Fec4 produced cellulose fibrils which were tightly wrapped around the cells, as in serotype Typhimurium when cellulose is coexpressed with curli fimbriae (17). Electron microscopy studies of the extracellular matrix of plate-grown colonies of E. sakazakii Fec39 and C. freundii Fec4 further supported the view that the extracellular matrix components cellulose and curli formed structures similar to those in serotype Typhimurium (Fig. 3C).
FIG. 3.
Phenotypes of Citrobacter sp. strain Fec2, C. freundii Fec4, and E. sakazakii Fec39. (A) Biofilm formation. Cultures were grown at 28°C for 48 h in Luria broth without salt and with shaking. Optimal biofilm formation was achieved at 180 and 210 rpm for Citrobacter sp. strain Fec2, C. freundii Fec4, and E. sakazakii Fec39, respectively. (B) Cell clumping and arrangement of Calcofluor-stained cellulose fibrils in C. freundii Fec4 (top) and E. sakazakii Fec39 (bottom). Note that C. freundii Fec4, due to the expression of cellulose and curli fimbriae, formed tighter clumps than E. sakazakii Fec39, which only expressed cellulose. Left, phase contrast; right, fluorescence microscopy. Magnification, ×600. (C) Scanning electron microscopy of plate-grown cells. Upper panel: C. freundii Fec4 (left) in comparison with serotype Typhimurium MAE52 (right; cellulose +, curli +). Lower panel: E. sakazakii Fec39 (left) in comparison with serotype Typhimurium MAE97 (right; cellulose +, curli −). Bars, 1 μm.
Sequence analysis of curli and cellulose biosynthesis genes.
Using conserved and strain-specific primers, we sequenced structural genes and regulatory regions involved in cellulose and curli fimbria production in the two Citrobacter strains and E. sakazakii. In particular, csgA was amplified with primers AGFA66 (ATGATGTTAACAATACTGGGTGC) and AGFA60 (CGGCCATTGTTGTGATAAATG), csgD was amplified with primers EC-AGFD1 (CAGCAGTGCAACATCTGTCAG) and EC-AGFD2 (AAAGTCTGGAAAATAACGTCCTG), and the intergenic region between csgA and csgD was amplified with primers FEC-IRD (GATCAACAATAATGTATGACCA) and FEC-IRB (GCTGCCTGATTAAATGAAGAC).
For bcsA, the following consensus primers were used: BCSA74A (CTTCCGTATTGGCAGTCAGGTTCAGGACG) and BCSA70 (GCGCCAGCGGGTTAAACGGCTG) for the N terminus, BCSA74 (GCAACAGATTCAATTTCTGCCCTTC) and BCSA86 (GCACCCGC-GCTGGCAGCGTATTCG) for the middle part of the gene, and BCSA62 (TGGGTCTTCTACAACCTGATTA) and BCSA62A (GCGGCGGTGCAATTTGCGCAAAGGT) for the C terminus.
Sequence data were submitted to the EMBL data library under the following accession numbers: Citrobacter sp. strain Fec2, csgD-csgBA, AJ515700; bcsA, AJ515698; C. freundii Fec4, csgD-csgBA, AJ515701; E. sakazakii Fec39, csgD-csgBA, AJ515702; bcsA, AJ515699. Although E. sakazakii Fec39 did not express curli fimbriae, it contained the csgD-csgBA region with intact genes.
CsgD, the central regulator for multicellular behavior, was highly conserved among the species (data not shown). Most diverse were the CsgD proteins from E. sakazakii Fec39 and serotype Typhimurium ATCC 14028, with a homology of 94%. While several amino acid substitutions were found in the N-terminal domain, a single amino acid substitution was found in the DNA binding domain.
csgD expression is highly regulated by environmental conditions mediated by response regulators and DNA architectural proteins (9, 12), which bind in the intergenic region. Surprisingly, binding sites for OmpR and IHF are not located in the most conserved regions (data not shown). This finding suggested that OmpR and IHF binding might differ among the species and consequently the response to environmental conditions. The 521-bp csgD-csgBA intergenic region can be divided into four regions, IR1 to IR4 (17). The sequence of the IR3 region is highly diverse among the species, but its length is conserved in all strains, suggesting structural importance.
CsgA and CsgB, the structural proteins of curli fimbriae, were also highly conserved (Fig. 4A and data not shown). Most diverse were the CsgA and CsgB proteins from C. freundii Fec4 and E. sakazakii Fec39 with a homology of 80% and 78%, respectively. Polymerized CsgA of Salmonella enteritidis possesses two domains, an N-terminal domain of 22 residues, which is proteinase K susceptible, and a C-terminal core domain, which is proteinase K resistant (6). The highest sequence diversity among the CsgA proteins was found in the glycine-rich N-terminal domain, which is proteinase K susceptible. The C-terminal domain of each protein consisted of five tandemly arranged sequences each containing the SX5QXGX2NXAX3Q consensus sequence (6).
FIG. 4.
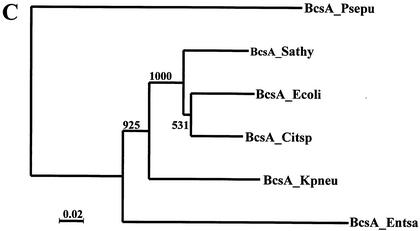
Comparative analysis of structural genes required for curli and cellulose biosynthesis. (A) Multiple alignment of CsgA sequences from different Enterobacteriaceae. The alignment was carried out with ClustalX in default mode (23). Amino acids identical in all the proteins are displayed in bold. An arrow indicates the cleavage site for the signaling peptide. The five internal repeats (C5a to C5e) are boxed. Proteinase-sensitive (light bar) and proteinase-resistant (dark bar) regions are indicated above the alignment. N- and C-terminal amino acid stretches identified in E. coli to mediate protein binding (15) are indicated by bars under the alignment. Three tryptic peptides of CsgA from Citrobacter sp. strain Fec2 were subjected to mass spectrometric sequence analysis, and the partial sequences (underlined), identical within the limits of technical accuracy to the translated csgA gene product, were detected. From the molecular mass of tryptic peptide 1, M+H: 3,990.7 (theoretic mass: 3,990.9), the amino terminus of the protein could be deduced as GSVP. CsgA sequences of E. coli K-12 (P28307) and serotype Typhimurium ATCC 14028 (P55225) were taken from the Swiss Prot database. Abbrevations: Sathy, serotype Typhimurium UMR1; Ecoli, E. coli K-12; Citsp, Citrobacter sp. strain Fec2; Citfr, C. freundii Fec4; Entsa, E. sakazakii. (B) Multiple alignment of the core region of enterobacterial BcsA, the catalytic subunit of the cellulose synthase. Conserved amino acids are displayed as described for panel A. Conserved motifs are indicated by boxes above and below the sequence. Abbrevations: Kpneu, K. pneumoniae; Psepu, Pseudomonas putida KT2440. BcsA sequences of E. coli K-12 (P37653) and serotype Typhimurium ATCC14028 (Q93IN2) were taken from the Swiss Prot database. BcsA from P. putida (http://www.tigr.org) and K. pneumoniae (http://genome.wustl.edu/projects/bacterial/?kpneumoniae=1) were from unfinished genome projects. (C) Phylogeny for BcsA sequences with BcsA from P. putida as an outgroup. After alignment of sequence, the tree was constructed by the neighbor-joining method subjected to 1,000 bootstrap trials. The tree was drawn with TreeView.
Phylogenetic analysis of the CsgD, CsgB, and CsgA proteins showed that, for all three proteins, the E. sakazakii Fec39 sequences developed fastest (Fig. 4 and data not shown). Protein sequences from Citrobacter sp. strain Fec2 and C. freundii Fec4 were usually more closely related to other species than to each other (Fig. 4 and data not shown).
Sequence alignment of BcsA, the cellulose synthase, is shown in Fig. 4B. Previously, sequence comparison of all available bacterial cellulose synthases detected a highly conserved core region of 350 residues which contains the D3D2D35QRXRWA motif, common to processive β-glycosyltransferases, and five additional motifs (16). Although the overall sequence was highly conserved among the species over the whole length of the protein, again BcsA of E. sakazakii Fec39 showed the highest sequence diversity (Fig. 4C). Most remarkable, the PVDPYE and the FFCGS motif were replaced by a different sequence (Fig. 4B).
Detection of csgA, csgD, and bcsA in other enterobacterial species isolated from feces.
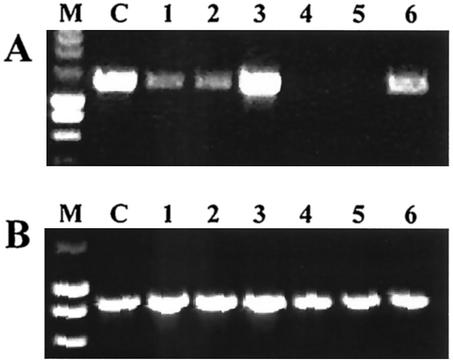
Conserved primers were used to detect the presence of csgA, csgD, and bcsA genes in other species isolated from feces (Fig. 5 and data not shown). bcsA was present in all species. In E. aerogenes Fec135 and R. ornithinolytica Fec153, csgD could not be detected, while csgA was present. We expect csgD to be present but not detectable by the primer pair used, because csgA is present. In summary, the genes required for the biosynthesis of curli fimbriae were present in all species investigated besides K. pneumoniae and K. oxytoca. Klebsiella ornithinolytica, which has recently been shown to belong to a phylogenetic line distinct from K. pneumoniae and K. oxytoca and therefore has been renamed R. ornithinolytica (10), harbors the gene cluster for curli biosynthesis.
FIG. 5.
Amplification of csgA (A) and bcsA (B) genes by PCR in enterobacterial species isolated from feces. PCR products of 1,301 bp and 860 bp were detected for csgA and bcsA, respectively. Lanes: M, size markers (SmartLadder), 200 to 10,000 kbp (Eurogentec); C, control serotype Typhimurium MAE52; 1, E. cloacae Fec36; 2, E. aerogenes Fec135; 3, C. koseri/farmeri Fec157; 4, K. oxytoca Fec139; 5, K. pneumoniae Fec141; 6, R. ornithinolytica Fec153. Primers for csgA were AGFA66 and AGFA60. bcsA primers were ES-BCSA162 (GACGATCTCTACCAGGTCTGG) and ES-BCSA681 (GTAACGCCACCAGATATAGCGG).
Temperature-regulated expression of rdar morphotype.
In this work, we showed that several non-E. coli enterobacterial isolates recovered from the gastrointestinal tract of humans produced either cellulose or curli fimbriae or both compounds under various temperature regulation programs. Different temperature regulation patterns of the rdar morphotype had been observed within clonal variants of serotype Typhimurium strains (17). Citrobacter species produced curli fimbriae at ambient temperature, but we did not investigate whether expression of curli fimbriae at 37°C might be triggered by iron depletion, as has been reported for serotype Typhimurium (17).
In the family Enterobacteriaceae, cellulose biosynthesis has been shown to occur in Salmonella spp., E. coli, and K. pneumoniae (21, 26). In this study, we found that most of the Enterobacter isolates produced cellulose. In particular, two unrelated isolates of E. sakazakii showed pronounced cellulose production at 28°C and 37°C (Fig. 1 and data not shown). Clinical isolates of E. sakazakii have been reported to display a peculiar wrinkled, leathery colony morphology, which, upon storage as agar stocks, readily dissociated into the standard smooth colony morphology (11). Based on the results from this study, this colony morphology is due to cellulose production, which is retained by E. sakazakii upon infection. Species from the genus Klebsiella and R. ornithinolytica isolated from feces did not show elevated expression of cellulose (Fig. 1), although the genetic information for the biosynthesis of cellulose is present (Fig. 5 and data not shown). However, as a possibility, cellulose biosynthesis might be induced in response to specific environmental signals, as shown for the plant symbiont Rhizobium leguminosarum bv. trifolii by contact with roots (1, 7). The observed species-specific expression pattern of curli fimbriae and cellulose in fecal strains raises the question of whether species-specific and consequently morphotype-specific niches exist in the gastrointestinal tract. Alternatively, a habitat-specific morphotype for each species is already determined outside the human body and retained when a strain passes through the gastrointestinal tract.
Multicellular behavior, including biofilm formation, is a fundamental life style of bacteria. It is reasonable to assume that such ancient behavior has common principles among bacteria. The rdar morphotype, defined as the expression of cellulose and curli fimbriae, represents a basic multicellular behavior of Enterobacteriaceae closely related to serotype Typhimurium and E. coli, but individual components of the rdar morphotype and its expression pattern are subject to species-specific adaptation for which no simple pathogen-commensal soil organism classification scheme can be proposed. For example, although semiconstitutive expression of the rdar morphotype is observed (17), the vast majority of pathogenic serovar Typhimurium and serotype Enteritidis strains express the rdar morphotype highly regulated by environmental conditions and only at ambient temperature (18, 18a). Otherwise, Shigella spp. and enteroinvasive E. coli, which cause invasive gastrointestinal disease (19), have lost the rdar morphotype and multicellular behavior. The rdar morphotype was also not found in the Klebsiella species analyzed in this study. However, constitutive expression of cellulose was observed in E. sakazakii, which can cause newborn meningitis, and the majority of E. coli strains from sepsis produce curli fimbriae or the rdar morphotype constitutively (2). Therefore, the expression of cellulose and curli fimbriae is complex and might be highly determined by the environmental microniche of the organism under study. In any case, the capacity to produce cellulose and curli fimbriae in Citrobacter, Enterobacter, and potentially Klebsiella species might contribute to the significant role those species play in biofilm-related infections.
Acknowledgments
This work was supported in part by grants from the Deutsche Forschungsgemeinschaft (RO2023/3-2), the Swedish Natural Science Research Council, and the Karolinska Institute (Elitforkartjänst to U.R.). During part of this work, U.R. was the recipient of a fellowship from the program “Infektionsbiologie” from the Bundesministerium für Forschung und Technologie (BMFT).
The excellent technical assistance of J. Majewski and A. Tiepold in mass spectrometric analysis is gratefully acknowledged. We thank Anne von Euler-Matell for electron microscopy of strains.
Editor: J. N. Weiser
REFERENCES
- 1.Ausmees, N., H. Jonsson, S. Hoglund, H. Ljunggren, and M. Lindberg. 1999. Structural and putative regulatory genes involved in cellulose synthesis in Rhizobium leguminosarum bv. trifolii. Microbiology 145:1253-1262. [DOI] [PubMed] [Google Scholar]
- 2.Bian, Z., A. Brauner, Y. Li, and S. Normark. 2000. Expression of and cytokine activation by Escherichia coli curli fibers in human sepsis. J. Infect. Dis. 181:602-612. [DOI] [PubMed] [Google Scholar]
- 3.Bian, Z., Z. Q. Yan, G. K. Hansson, P. Thoren, and S. Normark. 2001. Activation of inducible nitric oxide synthase/nitric oxide by curli fibers leads to a fall in blood pressure during systemic Escherichia coli infection in mice. J. Infect. Dis. 183:612-69. [DOI] [PubMed] [Google Scholar]
- 4.Bouza, E., R. San Juan, P. Munoz, A. Voss, and J. Kluytmans. 2001. A European perspective on nosocomial urinary tract infections II. Report on incidence, clinical characteristics and outcome (ESGNI-004 study). European Study Group on Nosocomial Infection. Clin. Microbiol. Infect. 7:532-542. [DOI] [PubMed] [Google Scholar]
- 5.Collinson, S. K., L. Emody, K. H. Muller, T. J. Trust, and W. W. Kay. 1991. Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. J. Bacteriol. 173:4773-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collinson, S. K., J. M. Parker, R. S. Hodges, and W. W. Kay. 1999. Structural predictions of AgfA, the insoluble fimbrial subunit of Salmonella thin aggregative fimbriae. J. Mol. Biol. 290:741-756. [DOI] [PubMed] [Google Scholar]
- 7.Dazzo, F. B., G. L. Truchet, J. E. Sherwood, E. M. Hrabak, M. Abe, and S. H. Pankratz. 1984. Specific phases of root hair attachment in the Rhizobium trifolii-clover symbiosis. Appl. Environ. Microbiol. 48:1140-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dibb-Fuller, M. P., E. Allen-Vercoe, C. J. Thorns, and M. J. Woodward. 1999. Fimbria- and flagella-mediated association with and invasion of cultured epithelial cells by Salmonella enteritidis. Microbiology 145:1023-1031. [DOI] [PubMed] [Google Scholar]
- 9.Dorel, C., O. Vidal, C. Prigent-Combaret, I. Vallet, and P. Lejeune. 1999. Involvement of the Cpx signal transduction pathway of E. coli in biofilm formation. FEMS Microbiol. Lett. 178:169-175. [DOI] [PubMed] [Google Scholar]
- 10.Drancourt, M., C. Bollet, A. Carta, and P. Rousselier. 2001. Phylogenetic analyses of Klebsiella species delineate Klebsiella and Raoultella gen. nov., with description of Raoultella ornithinolytica comb. nov., Raoultella terrigena comb. nov. and Raoultella planticola comb. nov. Int. J. Syst. E vol. Microbiol. 51:925-932. [DOI] [PubMed] [Google Scholar]
- 11.Farmer, J. J., III, M. A. Asbury, F. W. Hickman, D. J. Brenner, and the Enterobacteriaceae Study Group. 1980. Enterobacter sakazakii: a new species of “Enterobacteriaceae” isolated from clinical specimens. Int. J. Syst. Bacteriol. 30:569-584. [Google Scholar]
- 12.Gerstel, U., and U. Römling. 2001. Oxygen tension and nutrient starvation are major signals that regulate agfD promoter activity and expression of the multicellular morphotype in Salmonella typhimurium. Environ. Microbiol. 3:638-648. [DOI] [PubMed] [Google Scholar]
- 13.Gophna, U., M. Barlev, R. Seijffers, T. A. Oelschlager, J. Hacker, and E. Z. Ron. 2001. Curli fibers mediate internalization of Escherichia coli by eukaryotic cells. Infect. Immun. 69:2659-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leclerc, H., D. A. Mossel, S. C. Edberg, and C. B. Struijk. 2001. Advances in the bacteriology of the coliform group: their suitability as markers of microbial water safety. Annu. Rev. Microbiol. 55:201-234. [DOI] [PubMed] [Google Scholar]
- 15.Olsen, A., H. Herwald, M. Wikstrom, K. Persson, E. Mattsson, and L. Bjorck. 2002. Identification of two protein-binding and functional regions of curli, a surface organelle and virulence determinant of Escherichia coli. J. Biol. Chem. 277:34568-34572. [DOI] [PubMed] [Google Scholar]
- 16.Römling, U. 2002. Molecular biology of cellulose production in bacteria. Res. Microbiol. 153:205-212. [DOI] [PubMed] [Google Scholar]
- 17.Römling, U., W. D. Sierralta, K. Eriksson, and S. Normark. 1998. Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol. Microbiol. 28:249-264. [DOI] [PubMed] [Google Scholar]
- 18.Römling, U., W. Bokranz, U. Gerstel, H. Lünsdorf, M. Nimtz, W. Rabsch, H. Tschäpe, and X. Zogaj. 2002. Dissection of the genetic pathway leading to multicellular behaviour in Salmonella typhimurium and other Enterobacteriaceae, p. 231-261. In M. Wilson and D. Devine (ed.), Medical implications of biolfilms. Cambridge University Press, Cambridge, England.
- 18a.Römling, U., W. Bokranz, W. Rabsch, X. Zogaj, M. Nimtz, and H. Tschäpe. Occurrence and regulation of the multicellular morphotype in Salmonella serovars important in human disease. Int. J. Med. Microbiol., in press. [DOI] [PubMed]
- 19.Sakellaris, H., N. K. Hannink, K. Rajakumar, D. Bulach, M. Hunt, C. Sasakawa, and B. Adler. 2000. Curli loci of Shigella spp. Infect. Immun. 68:3780-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 21.Solano, C., B. Garcia, J. Valle, C. Berasain, J. M. Ghigo, C. Gamazo, and I. Lasa. 2002. Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol. Microbiol. 43:793-808. [DOI] [PubMed] [Google Scholar]
- 22.Sukupolvi, S., R. G. Lorenz, J. I. Gordon, Z. Bian, J. D. Pfeifer, S. J. Normark, and M. Rhen. 1997. Expression of thin aggregative fimbriae promotes interaction of Salmonella typhimurium SR-11 with mouse small intestinal epithelial cells. Infect. Immun. 65:5320-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uhlich, G. A., J. E. Keen, and R. O. Elder. 2001. Mutations in the csgD promoter associated with variations in curli expression in certain strains of Escherichia coli O157:H7. Appl. Environ. Microbiol. 67:2367-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Updegraff, D. M. 1969. Semimicro determination of cellulose in biological materials. Anal. Biochem. 32:420-424. [DOI] [PubMed] [Google Scholar]
- 26.Zogaj, X., M. Nimtz, M. Rohde, W. Bokranz, and U. Römling. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 39:1452-1463. [DOI] [PubMed] [Google Scholar]