Abstract
Cytadherence-related molecules of Mycoplasma gallisepticum strain R-low were identified by Tn4001 transposon mutagenesis with the hemadsorption (HA) assay as an indicator for cytadherence. Three Gmr HA-negative (HA−) colonies displaying a stable HA− phenotype through several successive generations in which gentamicin selection was maintained were isolated from four independent transformation experiments and characterized. Southern blot analysis showed that the transposon was inserted as a single copy within the genome of each of the HA− mutants, suggesting that the transposon insertion was directly responsible for their inability to attach to erythrocytes. Sequence analysis of the transposon insertion sites revealed that in two mutants, the transposon was inserted at two distinct sites within the gapA structural gene. In the third mutant, the insertion was mapped within the crmA gene, which is located immediately downstream of the gapA gene as part of the same operon. In vitro attachment experiments with the MRC-5 human lung fibroblast cell line showed that the cytadherence capabilities of the HA− mutants were less than 25% those of original strain R. Experimental infection of chickens, the natural host of M. gallisepticum, with each of the three mutants demonstrated significantly impaired colonization and host responses. These data demonstrate conclusively the role of both GapA and CrmA proteins in the adherence of M. gallisepticum to host cells in model systems and in vivo colonization. Furthermore, these results underscore the relevance of in vitro cytadherence model systems for studying the pathogenesis of natural infections in chickens.
Mycoplasma gallisepticum is an important pathogen of chickens and turkeys and is of considerable economic importance to poultry producers throughout the world (26). M. gallisepticum infections have a wide variety of clinical manifestations, the most significant of which is chronic respiratory disease of chickens, causing pathology in the form of tracheitis and air sacculitis (27). Like that of the human pathogen Mycoplasma pneumoniae, the morphology of M. gallisepticum is characterized by a specialized tip-like organelle which appears to mediate cytadherence to tracheal epithelial cells (19, 33, 34). Extensive studies on the molecular mechanisms of M. pneumoniae cytadherence have revealed a complex process involving the coordinated action of the primary cytadhesin molecule, P1, in concert with an array of high-molecular-weight accessory proteins (17, 19, 20). The emerging scenario from several studies of M. gallisepticum surface molecules that may be involved in cytadherence to host cells indicates a complex and multifactorial process that, in principle, may be analogous to that in M. pneumoniae (3, 8, 10, 12, 29, 43).
Several putative M. gallisepticum cytadherence proteins have been identified, mostly on the basis of sequence homology with cytadhesins from pathogenic human mycoplasmas. In some cases, their putative roles as adhesins have been supported by in vitro cytadherence models and by electron microscopy. These include MGC2, a 32-kDa protein homologous to the M. pneumoniae P30 and M. genitalium P32 cytadhesins (12), and PvpA, a 55-kDa protein which shows homology to accessory molecule HMW3 of M. pneumoniae as well as to the P30 protein of M. genitalium (3). In addition, major surface lipoproteins pMGA (29) and lp64 (8, 14) have been implicated in cytadherence, mainly on the basis of the inhibition of cytadherence or hemagglutinin by specific antibodies. However, GapA, a 105-kDa protein, is considered the primary cytadhesin molecule on the basis of its homology to the P1 protein of M. pneumoniae and the fact that anti-GapA Fab fragments significantly inhibit M. gallisepticum cytadherence (10, 13). CrmA, a 116-kDa protein, shows significant sequence homology to the M. pneumoniae open reading frame (ORF) 6 (ORF6) gene product, which has been shown to play a role as an accessory protein in P1-mediated cytadherence (30, 43). The crmA gene is located downstream of the gapA gene as part of the same operon (30). Notably, while both GapA and CrmA proteins are expressed in virulent M. gallisepticum strain R-low (35), they are absent from avirulent M. gallisepticum strain R-high (passage 164) (25, 30). Complementation of strain R-high with the wild-type gapA gene alone did not restore cytadherence capability, and such a strain was not virulent for chickens (30, 31). However, in a recent study, complementation of M. gallisepticum R-high with both gapA and crmA wild-type alleles restored cytadherence to the level of wild-type strain R-low, indicating that both GapA and CrmA are essential for M. gallisepticum cytadherence (31).
In the present study, the chromosome of M. gallisepticum strain R-low was subjected to random transposon mutagenesis, and transformants were screened for hemadsorption (HA)-negative (HA−) mutants with the goal of identifying and characterizing insertions within genomic regions that include putative cytadherence genes. Three HA− transformants from four independent transformation experiments were shown to possess insertions within the gapA and crmA genes. The role of the three M. gallisepticum R-low mutants in cytadherence was evaluated in an in vitro model system and in vivo in chickens, the natural hosts.
MATERIALS AND METHODS
Mycoplasma strains.
The origins and properties of M. gallisepticum R-low, a widely used prototype pathogenic strain used at passage 9, and vaccine strain F were previously described (25, 35, 42). The properties of M. gallisepticum strain R-high (passage 164) were described previously (25, 30). Commercial live vaccine strains ts-11 (developed by Bioproperties, North Ringwood, Victoria, Australia, and marketed in the United States by Merial Select, Gainesville, Ga.) and 6/85 (Intervet America, Millsboro, Del.) were obtained from S. H. Kleven (University of Georgia, Athens). Mycoplasma imitans type strain 4229, an avian mycoplasma species phylogenically closely related to M. gallisepticum (4), was obtained from J. M. Bradbury (University of Liverpool, Liverpool, United Kingdom).
Plasmids and chemicals.
Transposon Tn4001mod (pISM2062) (16) was kindly provided by Duncan Kraus (University of Georgia, Athens). Escherichia coli strain DH5αMCR (Gibco BRL Life Technologies, Inc., Gaithersburg, Md.) was used as a host. Recombinant clones were constructed in plasmid vector pKS (Strategene, La Jolla, Calif.). E. coli cultures for plasmid isolation were grown in Luria-Bertani broth (39). Restriction enzymes, T4 ligase, and T4 polynucleotide kinase were purchased from MBI Fermentas, Vilnius, Lithuania. 5-Bromo-4-chloro-3 indolyl-β-d-galactopyranoside (X-Gal), isopropyl-β-d-thiogalactopyranoside (IPTG), gentamicin, and ampicillin were purchased from Sigma Chemical Co. (St. Louis, Mo.). [α-32P]CTP was purchased from Amersham, Little Chalfont, United Kingdom.
DNA extractions, labeling, and manipulations.
Genomic DNA was extracted from M. gallisepticum cultures and purified as previously described (3). The DNA was digested to completion with restriction enzymes, electrophoresed, and subjected to Southern blot hybridization as previously described (3). Labeling of DNA probes was performed by using a HexaLabel DNA labeling kit (MBI Fermentas). DNA was extracted from tracheal cell suspensions or broth cultures of isolates from infected chickens as follows. A 500-μl tracheal swab suspension sample or a 100-μl culture sample was mixed with 100 μl of lysing solution (5% sodium dodecyl sulfate [SDS], 0.05 M EDTA, 1 μg of proteinase K/ml) and incubated for 10 min at 60°C. An equal volume of phenol-chloroform (volume/volume) was added, and the suspension was vortexed vigorously and centrifuged for 5 min at 15,000 × g and room temperature. The top layer was removed and extracted with an equal volume of chloroform. DNA was precipitated by adding an equal volume of isopropanol containing 10% 3 M sodium acetate, followed by incubation for 1 h at −20°C.
Electroporation and isolation of HA− transformants.
A 100-μl aliquot containing 107 to 108 CFU of mid-logarithmic-phase M. gallisepticum strain R-low was mixed with 5 μg of Tn4001mod (pISM2062) (16), and electroporation was performed with a 0.2-cm Gene Pulser cuvette (Bio-Rad Laboratories, Hercules, Calif.) at 2.5 kV, 100-Ω resistance, and 25-μF capacitance (11). The cell suspension was added to 1 ml of Edward broth medium (32) and incubated at 37°C for 1 h. The mycoplasma suspension was plated on agar plates containing 25 μg of gentamicin/ml and incubated for 4 to 6 days at 37°C. Transformants growing in the presence of gentamicin (Gmr) were screened microscopically for HA− by using a previously described method (1, 11, 37). Gmr HA− colonies were picked by using sterile Pasteur pipettes, filter cloned, and rescreened for HA. Each HA− mutant was plated (with gentamicin selection), and the resultant colonies were assayed for the HA− phenotype. One colony (HA−) was picked, replated, and assayed for its HA phenotype. This was done three times to ensure the stability of the HA− phenotype. Three Gmr HA− mutants that exhibited a stable HA− phenotype during three successive generations were chosen for further study.
DNA sequence analysis.
DNA sequence analysis of both strands was performed by the dideoxy chain termination method (40). Synthetic oligonucleotides were synthesized on a model 380B DNA synthesizer (Applied Biosystems, Inc., Foster City, Calif). A primer, designated ISb1, that represents an 18-bp sequence between nucleotides 79 and 98 from within the bottom strand of the IS256 arm of Tn4001 (5, 18) was used to determine the junction between the Tn4001 arm and the flanking mycoplasma DNA. Sequencing was done by using an ABI PRISMA 377 automatic sequencer for dye terminator cycle sequencing (Perkin-Elmer, Foster City, Calif). Sequence data were analyzed by using AssemblyLIGN and MacVector 6.0 software.
Electrophoresis and Western immunoblotting.
SDS-polyacrylamide gel electrophoresis (PAGE) was performed by the method of Laemmli (21). Samples were prepared by heating at 100°C for 5 min in sample buffer (2% SDS, 5% [vol/vol] 2-mercaptoethanol, 10% [vol/vol] glycerol, 62.5 mM Tris [pH 6.8]). Proteins were separated in 9% acrylamide gels and transferred to nitrocellulose membrane filters (0.45-μm-pore size; Schleicher & Schuell, Dassel, Germany) by the method of Towbin et al. (41). Blot contents were incubated for 1 h at room temperature with phosphate-buffered saline (PBS) containing 3% bovine serum albumin (Sigma, St. Louis, Mo.) and then incubated overnight at 4°C with primary antibodies diluted in PBS containing 20% (vol/vol) fetal calf serum. Monospecific polyclonal anti-GapA or anti-CrmA antibodies prepared in rabbits (30) were used as primary antibodies. After three washes in PBS, blots were incubated for 2 h at room temperature with peroxidase-conjugated goat antiserum to rabbit immunoglobulin G (Jackson ImmunoResearch Laboratories, West Grove, Pa., and Nordic, Tilburg, The Netherlands). For detection, the enzyme substrate o-dianisidine (Sigma) was used as previously described (36).
Mycoplasma cultivation.
M. gallisepticum was cultivated in Edward modified medium (32) with the addition of gentamicin as needed for the selection and maintenance of transposon mutants. For experimental infection, M. gallisepticum was grown in Edward’s modified medium with 25 μg of gentamicin/ml but without thallium acetate. Isolation from chickens was carried out with Edward medium in the absence of gentamicin. CFUs were determined by plating aliquots of 10-fold dilutions of mycoplasma cultures on agar and counting the colonies, whereas color-changing units (CCU) were determined with 10-fold dilutions of a tracheal swab suspension in mycoplasma broth medium. Broth cultures showing an acid color change were plated on mycoplasma agar medium (32), and selected representative samples were tested by a direct immunofluorescence assay with M. gallisepticum-specific fluorescein-conjugated antiserum by using a modification of the method described by Baas and Jasper (2). For monitoring of chickens prior to infection or chickens in the noninfected control group, tracheal swab samples were inoculated in Frey’s medium by standard methods for the isolation of avian mycoplasmas (15).
In vitro attachment of M. gallisepticum to MRC-5 cells.
Attachment of M. gallisepticum was evaluated in an in vitro attachment model system by using a modification of the method described by Geary and Gabridge (9) and MRC-5 human embryo lung fibroblasts. A cell suspension of MRC-5 cells (ATCC CCL-171) was inoculated in a 24-well cell culture plate and incubated at 37°C in the presence of 5% CO2 up to the confluence of the cells in the wells. The plate containing adherent MRC-5 cells was washed three times with prechilled PBS prior to the addition of 1 ml of 3H-palmitic acid-labeled M. gallisepticum (107 to 108 CFU), washed three times with PBS-0.5% bovine serum albumin, and incubated for 1 h at 37°C. The wells were washed gently three times with 2 ml of prechilled PBS. MRC-5 and attached mycoplasma cells were detached from the plate by the addition of 200 μl of trypsin to each well, followed by 5 min of incubation at 37°C. The contents of the wells were transferred to scintillation vials for counting. The attachment values in each experiment were calculated as total MRC-5-associated counts per minute/total counts per minute in the M. gallisepticum culture, averaged for three replicate wells of each strain. Average results from three separate experiments are presented.
PCR for species and transposon identification.
PCR was used to monitor the presence of M. gallisepticum in experimentally infected chickens as well as the presence and position of the Tn4001 transposon within the mycoplasma genome. Amplifications were carried out by using a PC-Personal Cycler (Biometra, Gottingen, Germany). Primers for the M. gallisepticum gapA gene (gapA3F and gapA4R) were selected by M. Garcia (University of Georgia, Athens). Amplification was carried out with ABGene Reddy-Mix (ABgene, Epson, Surrey, United Kingdom), 0.5 μl of each primer (50 ng), and 10 ng of DNA template in a 25-μl total reaction mixture. Denaturation for 5 min at 95°C was followed by 40 cycles of 20 s of denaturation at 94°C, 40 s of annealing at 58°C, and 60 s of extension at 72°C and finally by extension at 72°C for 7 min in the last cycle. Primers GENTA5 and GENTA6 were used to amplify the gentamicin gene as previously described (6) with 1 U of Biotaq (Bioline, Herzilia, Israel). To monitor the position of the Tn4001 transposon within the gapA-crmA operon (Tn-position-PCR), primer ISb1 (5, 18) and primer GAf4, which represents a 21-bp sequence between nucleotides 1242 and 1263 of the gapA gene, were used for each mutant. The amplification program for Tn-position-PCR was as follows: denaturation for 5 min at 95°C; 35 cycles of 30 s of denaturation at 92°C, 30 s of annealing at 55°C, and 3 min of extension at 72°C; and finally extension at 72°C for 7 min in the last cycle. Primers for species-specific regions of the 16S rRNA genes of M. gallisepticum (MG13 and MG14) and Mycoplasma synoviae (MSL1 and MSL2), designed as described by Lauerman (22), were used to monitor the commercial breeding flock from which test birds were obtained as well as for testing of birds prior to infection and at the end of the experiment (noninfected control birds). The nucleotide sequences of all primers used in this study are listed in Table 1.
TABLE 1.
Primers used for PCR and to amplify Tn4001 insertion sites
| Primer designation | Sequence | Gene | Orientation | Reference(s) or source |
|---|---|---|---|---|
| ISb1 | 5′-AAGTCCTCCTGGGTATGT-3′ | IS256 arm of Tn4001 | Reverse | 5, 18 |
| GAf4 | 5′-GGTGGTAACGGTACATGAGTT-3′ | gapAa | Forward | This studyb |
| gapA3F | 5′-TTCTAGCGCTTTARCCCTAAACCC-3′ | gapAa | Forward | This studyb |
| gapA4R | 5′-CTTGTGGAACAGCAACGTATTCGC-3′ | gapAa | Reverse | This studyb |
| GENTA5 | 5′-TATGAAAAAGGTGATAAATAAATG-3′ | Gentamicin | Forward | 6 |
| GENTA6 | 5′-AATTTCTGGTGTTAAAAAAGTTCC-3′ | Gentamicin | Reverse | 6 |
| MG13 | 5′-GCTTCCTTGCGGTTAGCAAC-3′ | 16S rRNA of M. gallisepticum | Forward | 22 |
| MG14 | 5′-GAGCTAATCTGTAAAGTTGGTC-3′ | 16S rRNA of M. gallisepticum | Reverse | 22 |
| MSL1 | 5′-GAGAAGCAAAATAGTGATATCA-3′ | 16S rRNA of M. synoviae | Forward | 22 |
| MSL2 | 5′-CAGTCGTCTCCGAAGTAAACAA-3′ | 16S rRNA of M. synoviae | Reverse | 22 |
GenBank accession number AF214004.
M. Garcia, University of Georgia, personal communication.
Experimental infection.
In three separate trials (I, II, and III), 5-week-old Leghorn-type chickens from a mycoplasma-free commercial breeding flock were infected by intratracheal inoculation of 100 μl of mycoplasma culture by using a MicroAliquator (Scientific Manufacturing Industries). Logarithmic-phase cultures of M. gallisepticum strain R-low or transposon mutants that contained approximately 107 to 108 CFU were used. Chickens in the noninfected control group were inoculated with sterile mycoplasma broth medium. Chickens were maintained in positive-pressure Horsfal-Bauer isolation cells from time of hatching (experiments I and II) or from 4 weeks of age (experiment III) for the duration of the experiments (4 weeks postinfection [p.i.]). Each experimental group was comprised of 25 to 30 chickens, of which 4 or 5 birds were sacrificed at each sampling time (usually 3, 7, 11, 14, 21, and 28 days p.i.). At each sampling time, chickens were bled for serological testing, and tracheal swab samples were obtained from live birds (experiments I and II) or from excised tracheas (experiment III) for isolation, quantitation, and phenotypic analysis of M. gallisepticum. Tracheal swab suspensions were plated on mycoplasma agar medium and incubated at 37°C. The resulting colonies were examined for HA as previously described. Tracheal levels of M. gallisepticum were determined as CCU with 10-fold dilutions of tracheal swab suspensions in Edward medium in the absence of gentamicin. In parallel, DNA was prepared from tracheal swab suspensions for PCR testing with primers gapA3F, gapA4R, GENTA5, and GENTA6, and Tn-position-PCR was performed with DNA extracted from organisms grown in broth cultures inoculated with tracheal swab suspensions.
Serological tests.
Rapid slide agglutination (RSA) was performed with serum samples and commercial stained antigens for M. gallisepticum and M. synoviae (Noblis, Intervet Ltd., Boxmeer, The Netherlands) according to the manufacturer's instructions. Enzyme-linked immunosorbent assays (ELISAs) were performed by using ProFlok kits for M. gallisepticum and M. synoviae (Kirkegaard & Perry Laboratories, Gaithersburg, Md.).
Statistical analysis.
For comparison of the attachment of the mutants and the vaccine strains as a percentage of that of wild-type strain R-low in the MRC-5 cell tissue culture assay, a mixed linear model was adapted. The least-squares means (LSMeans) were computed, and multiple comparisons between the expected means were performed. Statistical analysis of the numbers of organisms isolated from the tracheas of experimentally infected chickens was performed with Duncan's multiple-range test. Tracheal levels were expressed as the mean of the log CCU for each group at each time p.i. Analysis was done with SAS software (version 8.02; SAS Institute, Cary, N.C.).
Nucleotide sequence accession numbers.
The nucleotide sequences of the gapA genes from strains F and ts-11 have been assigned GenBank accession numbers AY227006 and AY212515.
RESULTS
Isolation of M. gallisepticum HA− phenotype transformants.
Four independent transformations of M. gallisepticum strain R-low with transposon Tn4001mod (16) were performed and yielded between 800 and 1,000 Gmr transformants per transformation. The Gmr colonies were examined for HA capability (1), and colonies exhibiting the Gmr HA− phenotype were isolated. Notably, screening of the original M. gallisepticum R-low population did not reveal colonies displaying the HA− phenotype, despite several attempts to enrich the HA− phenotype. This was done by three cycles of incubation of the mycoplasma cells with a suspension of fresh erythrocytes, followed by centrifugation and screening of the supernatant fraction. Nevertheless, since the HA− phenotype could potentially arise due to spontaneous events that were not detected in our screenings and not as a result of the transposon insertion, each Gmr HA− transformant was filter cloned, plated again, and assayed for the HA− phenotype. This was done three times to ensure the stability of the HA− phenotype. In all cases, the three mutants exhibited an HA− phenotype. If the transposon insertion is responsible for the HA− phenotype, then the reversion rate should be several orders of magnitude lower than that normally seen with spontaneously arising variants. On the other hand, if the insertion is unrelated to the HA− phenotype, then the transformants should readily revert. Three distinct Gmr HA− transformants (designated E117, E325, and E345) displaying a stable HA− phenotype through several successive generations in which gentamicin selection was maintained were isolated for further study.
Genomic DNAs from wild-type M. gallisepticum strain R-low and the three HA− transformants were isolated, digested with restriction endonuclease EcoRI, and subjected to Southern blot hybridization with a 2.5-kb HindIII fragment carrying the gentamicin gene (5) as a probe (Fig. 1A). Notably, one EcoRI site is present at the end of the left Tn4001 arm. In two mutants (E117 and E345), a single hybridization band of 6.1 kb was observed, while in mutant E325, a single fragment of 5.6 kb was detected (Fig. 1A, lanes 2 to 4, respectively). No hybridization signal was detected in the genome of wild-type M. gallisepticum strain R-low (Fig. 1A, lane 1). These results confirm that transposon Tn4001 was inserted as a single copy within the genome of each of the M. gallisepticum HA− mutants and suggest that the transposon insertion is directly responsible for their inability to attach to erythrocytes.
FIG. 1.
Identification of Tn4001 insertions within the genomes of M. gallisepticum HA− transformants. Lanes: 1, wild-type M. gallisepticum R-low; 2 to 4, M. gallisepticum mutants E117, E345, and E325, respectively. In each lane, 2 to 4 μg of chromosomal DNA was digested with EcoRI (A) or XbaI (B and C) restriction enzymes, subjected to Southern blot hybridization, and probed with the 32P-labeled gentamicin gene (A), gp-1 fragment (B), or cm-1 fragment (C). The sizes of the hybridization bands are shown on the left.
Identification of Tn4001 insertion sites.
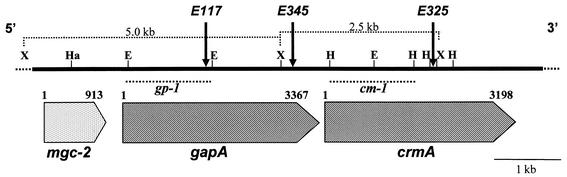
The site of the Tn4001 insertion in each HA− mutant was first mapped by Southern blot hybridization to a HindIII-restricted digest of genomic DNA with Tn4001 as a probe. Two HindIII sites flanking the gentamicin gene within Tn4001 (5, 16) generate a 2.5-kb HindIII fragment bearing the gentamicin gene and two additional fragments that contain the left or the right arm of the transposon as well as a flanking mycoplasmal genomic region (data not shown). Tn4001 arm-bearing fragments of each mutant were excised from the gel and subcloned into plasmid vector pKS. To precisely determine the site of the transposon insertion in each HA− mutant, the junction between the transposon and the mycoplasma DNA was sequenced by using primer ISb1 (5, 18). Finally, a genomic region of about 8 kb of M. gallisepticum R-low was cloned and sequenced (Fig. 2). A comparison of the resultant mycoplasmal nucleotide sequences to existing sequences in GenBank revealed that in two mutants, E117 and E345, the Tn4001 transposon was inserted at two distinct nucleotide positions (1423 and 2981, respectively) within the same gene, designated gapA (Fig. 2) (10, 30). In the third mutant, E325, the insertion was mapped at nucleotide position 2011 of a gene, designated crmA, which is located immediately downstream of the gapA gene in the same operon (30) (Fig. 2). In addition, upstream of the gapA gene but not in the gapA operon, another previously described putative adhesion gene, designated mgc-2, was identified (Fig. 2) (12). Localization of the transposon insertion in each of the three mutants was confirmed by Southern blot hybridization of XbaI-restricted genomic DNA with a 1.6-kb EcoRI fragment or a 1.2-kb HindIII fragment, each spanning the 5′ region of the gapA or the crmA gene, as a probe (designated gp-1 or cm-1, respectively) (Fig. 2). Since insertion in mutant E117 was within a 5.0-kb XbaI genomic fragment, the gp-1 probe identified the transposon insertion on a 9.8-kb XbaI fragment (Fig. 1B, lane 2). In mutants E345 and E325, the transposon insertion occurred within a 2.5-kb XbaI genomic fragment. Thus, the cm-1 probe recognized a 7.3-kb XbaI fragment in both mutant E345 and mutant E325 (Fig. 1C, lanes 3 and 4, respectively).
FIG. 2.
Identification of Tn4001 insertion sites within the M. gallisepticum R-low gapA-crmA operon. The solid line represents an 8-kb genomic fragment of M. gallisepticum R-low that was cloned and sequenced. The locations and directions of the mgc-2, gapA, and crmA genes are indicated by large labeled arrows. The positions of HaeIII (Ha), HindIII (H), XbaI (X), and EcoRI (E) restriction sites are indicated. The positions of Tn4001 insertions in mutants E117, E345, and E325 are indicated by labeled arrows. The locations of the gp-1 and cm-1 genomic fragments used as probes are indicated by broken lines. Two XbaI genomic fragments (shown in Fig. 1) are indicated by dotted brackets along with their corresponding sizes.
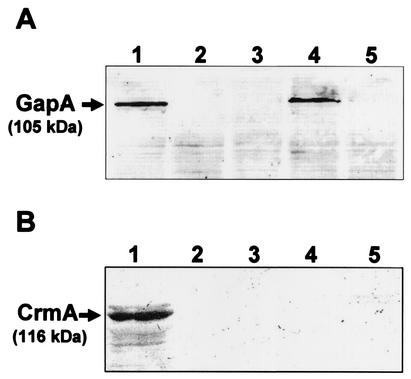
Western blot analysis of total cell proteins with monospecific anti-GapA and anti-CrmA antibodies (30) demonstrated the presence of reactive 105-kDa GapA and 116-kDa CrmA protein bands in M. gallisepticum R-low (Fig. 3, lanes 1). These antibodies, moreover, confirmed the lack of expression of the GapA protein in mutants E117 and E345 (Fig. 3A, lanes 2 and 3, respectively), the presence of the GapA protein in mutant E325 (Fig. 3A, lane 4), and the absence of the CrmA protein in all three mutants (Fig. 3B, lanes 2 to 4). Collectively, these data confirm previous findings indicating that the two genes represent an operon (30).
FIG. 3.
Western blot analysis of M. gallisepticum R-low Tn4001 mutants. Total cell proteins from wild-type M. gallisepticum R-low (lane 1), mutants E117, E345, and 325 (lanes 2 to 4, respectively), and M. gallisepticum R-high (lane 5) were subjected to SDS-PAGE and immunoblotted with monospecific anti-GapA (A) or anti-CrmA (B) antibodies. The 105-kDa GapA and 116-kDa CrmA protein bands are indicated.
Assessing the adherence capability of M. gallisepticum strain R-low HA− mutants by an in vitro attachment assay.
The transposon mutagenesis results strengthened our postulate that HA can serve as an efficient indicator for the identification of M. gallisepticum cytadherence-related genes. To directly demonstrate that disruption of the gapA or crmA gene has an impact on the cytadherence of M. gallisepticum, the attachment capabilities of strain R-low and the three HA− mutants (E117, E345, and E325) were evaluated in an vitro attachment model with the MRC-5 human lung fibroblast cell line (9). To assess attachment, MRC-5 cells immobilized in multiwell dishes were incubated with 3H-labeled M. gallisepticum cells from a mid-logarithmic-phase culture. After static incubation at 37°C for 60 min, MRC-5 cells were rinsed and assayed for the uptake of radioactivity (see Materials and Methods). As shown in Fig. 4, the attachment efficiency of all three mutants with transposon insertions within the gapA or crmA gene was significantly lower (<25%) than that of wild-type R-low (statistically significant at a P value of <0.0001). No significant difference was found among the three mutants. Notably, several mutants in which Tn4001 was inserted in different locations within the chromosome showed no decrease in attachment relative to that of the wild type (data not shown).
FIG. 4.
In vitro attachment of M. gallisepticum to MRC-5 cells. Hatched and stippled bars indicate the attachment of M. gallisepticum mutants and strains relative to that of wild-type M. gallisepticum R-low (data are presented relative to the R-low cytadherence value of 100%). The Tn4001 transposon mutants and M. gallisepticum strains used are indicated below the bars. Division of the attachment values for the strains by multiple comparisons of the LSMeans into three significantly different clusters is indicated by letters above the bars. Error bars represent standard deviations.
The MRC-5 model was also used to test the attachment capabilities of several other M. gallisepticum strains. M. gallisepticum R-high is a strain derived from R-low by 164 passages in vitro. This strain, which was previously reported to possess a nonsense mutation within the gapA coding region (30) and which displayed a GapA− CrmA− phenotype (Fig. 3, lanes 5), also exhibited a low level of attachment to MRC-5 cells (Fig. 4). Two M. gallisepticum strains that are used as live vaccines (ts-11 and 6/85) exhibited attachment levels similar to those of the transposon mutants, according to statistical analysis by multiple comparisons of the LSMeans. Vaccine strain F also showed a significantly lower level of attachment in the MRC-5 model, relative to that of M. gallisepticum R-low, but in a cluster separate from the mutants and other vaccines (Fig. 4).
Western blot analysis of total cell proteins with anti-GapA and anti-CrmA antibodies (Fig. 5A and B, respectively) revealed that CrmA is expressed in all strains tested (Fig. 5B, lanes 1 to 3), while GapA is absent only from strain ts-11 (Fig. 5A, lane 1). Sequence analysis revealed that a change of a cytosine to a thymidine at nucleotide position 143 of the gapA gene in strain ts-11 resulted in premature termination of translation (TAA) (Fig. 5C). Western blot analysis with anti-GapA and anti-CrmA antibodies also revealed the presence of the cognate proteins in M. imitans, an avian mycoplasma phylogenetically, biologically, and morphologically closely related to M. gallisepticum (4, 38) (Fig. 5A and B, lanes 4).
FIG. 5.
Western blot analysis of M. gallisepticum strains. (A and B) Total cell proteins from M. gallisepticum strains were subjected to SDS-PAGE and immunoblotted with monospecific anti-GapA (A) or anti-CrmA (B) antibodies. The GapA and CrmA protein bands are indicated. M. gallisepticum strains included ts-11, F, and 6/85 (lanes 1 to 3, respectively). M. imitans is shown in lane 4. (C) A base substitution at position 139 (boxed) in the gapA structural gene of M. gallisepticum ts-11, generating a TAA termination codon, is shown in comparison to the corresponding region in strain R-low.
Analysis of M. gallisepticum strain R-low HA− mutants in experimentally infected chickens.
Three experimental infection trials (I to III) comparing the behavior in vivo of M. gallisepticum R-low and the derivative transposon mutants were performed with mycoplasma-free 5-week-old Leghorn-type chickens as described in Materials and Methods. Tracheal infection was monitored by gapA PCR and by isolation of M. gallisepticum from tracheal swab suspensions. In order to compare the abilities of the strains to colonize the respiratory tract, the numbers of CCU of M. gallisepticum present in the tracheas of individual infected birds were determined at designated times p.i. The M. gallisepticum strain used for infection was recovered from infected birds in each experimental group, but M. gallisepticum R-low and the transposon mutants showed markedly different behavior in vivo. The recovery of each of the inoculated transposon mutants was significantly lower than that of the original strain R-low at each time of sampling after infection. For example, the numbers of M. gallisepticum cells recovered in each of the experimental groups in experiment III (expressed as mean log CCU) are presented in Table 2. All birds received the same inoculum dose of M. gallisepticum R-low or mutant E117, E345, or E325 (about 107 CFU inoculated into the trachea). As shown in Table 2, at each sampling time, the numbers of M. gallisepticum organisms recovered from the tracheas of the chickens infected with each of the transposon mutants were 3 to 4 log units lower than the numbers of M. gallisepticum organisms recovered from chickens infected with M. gallisepticum R-low (significantly different at a P value of <0.001). The numbers of organisms recovered from the transposon mutant-infected chickens at the first time of testing (3 days p.i.) were markedly lower than the numbers of organisms introduced (102 to 103 CCU recovered compared to 107 CCU introduced). Tracheal levels in all groups tended to increase between 3 and 6 days p.i., indicating colonization in the trachea, but decreased at later times of testing, although the differences with time within a group were not statistically significant (Table 2). The relatively high standard deviations of the means for the log values within groups reflect the biological differences in the tracheal levels of M. gallisepticum between birds. The differences in efficiency of tracheal infection seen in this experiment (III) were also found in experiments I and II, although initial infection levels and absolute tracheal levels over time differed among the experiments (data not shown).
TABLE 2.
Tracheal levels of cytadherence-deficient mutants of M. gallisepticum in experimentally infected chickens
| Day p.i. | CCU for M. gallisepticuma:
|
|||
|---|---|---|---|---|
| E117 | E345 | E325 | R-low | |
| 3 | 2.0 ± 0.82 A | 1.2 ± 0 A | 1.2 ± 0.96 A | 4.7 ± 0.58 B |
| 6 | 2.5 ± 0.58 A | 1.2 ± 0.50 A | 1.7 ± 1.3 A | 6.3 ± 0.58 B |
| 10 | 1.3 ± 1.15 A | 0.2 ± 0.50 A | 1.2 ± 0.86 A | 5.3 ± 0.58 B |
| 14 | 1.0 ± 0.82 A | 1.0 ± 0.82 A | 1.2 ± 0.96 A | 4.7 ± 2.38 B |
CCU for M. gallisepticum organisms isolated in mycoplasma broth cultures from tracheal swab samples. The inoculum was 107 CFU. For each sampling time, four birds were used for E117, E345, and E325, and three birds were used for R-low. Statistical analysis was done by Duncan's multiple-range test for mean and standard deviation log tracheal levels of M. gallisepticum. Comparisons were made at each sampling time among all four groups. Different letters following data indicate significant differences at a P value of P < 0.001.
Not only were the tracheal levels of M. gallisepticum transposon mutants lower than that of M. gallisepticum R-low in infected birds, but also the elicitation of specific anti-M. gallisepticum antibodies by the host was affected. Serological reactions in M. gallisepticum R-low-infected chickens, as measured by commercial RSA and ELISA diagnostic tests, were detected from about 6 days p.i. (RSA) and 10 days p.i. (ELISA) (data not shown). In contrast, no significant serological reactions were detected in transposon mutant-infected chickens in any of the three experiments. These data support the notion of a key role for the GapA and CrmA proteins in the colonization of M. gallisepticum in natural infections.
It is important to note that in general, M. gallisepticum isolates from the transposon mutant-infected chickens retained the intact Tn4001 transposon during colonization in vivo. This finding was determined by testing DNA extracted from broth cultures of M. gallisepticum strains isolated from experimentally infected chickens at each time of testing. The presence of the transposon was indicated by amplification of an 880-bp product with the GENTA5 and GENTA6 PCR primers. In addition, the genomic position of the transposon in isolates from each mutant-infected group was verified by performing Tn-position-PCR (see Materials and Methods). Figure 6A illustrates the presence and the distinct size of the Tn4001 transposon in each mutant (E117, E345, and E325). The PCRs confirmed the presence and position of the transposon in isolates from chickens infected with each of the mutants for at least 14 days p.i.. These data clearly indicate that multiplication of M. gallisepticum in the trachea does occur in the absence of detectable GapA and CrmA functions, although at an efficiency much lower than that found with the original strain.
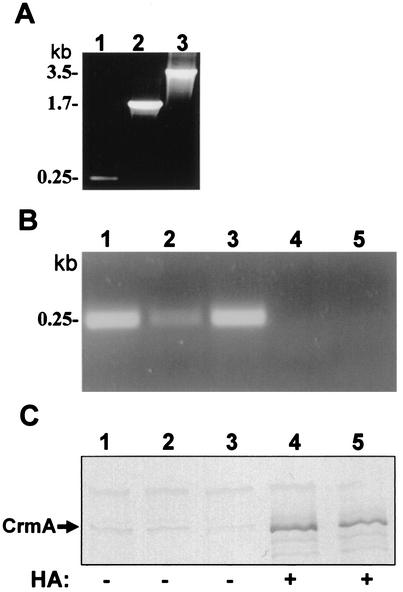
FIG. 6.
Monitoring the presence and genomic location of the Tn4001 transposon in M. gallisepticum R-low mutants. (A) Tn-position-PCR of various mutants. Primers ISb1 and GAf4 were used to amplify the junction between the IS256 arm of the Tn4001 transposon and the gapA gene in M. gallisepticum R-low mutants E117, E345, and E325 (lanes 1 to 3, respectively). The sizes of the PCR products corresponding to the mutants are shown on the left. (B) Tn-position-PCR of mutant E117 isolated directly from the trachea at 3, 7, 11, 21, and 28 days p.i. (lanes 1 to 5, respectively) with primers ISb1 and GAf4. The 0.25-kb PCR product is indicated. (C). Western blot analysis of total cell proteins from mutant E117 (depicted in panel B) with anti-CrmA antibodies. The 116-kDa CrmA protein band is indicated. The HA phenotype of colonies from each isolate on agar plates (positive or negative, + or −, respectively) is shown at the bottom.
Nonetheless, it should be noted in a few isolates from transposon mutant-infected chickens, excision of the transposon was detected, but only at times later than 14 days p.i. and not in all groups. For example, in experiment II, M. gallisepticum isolated from E117-infected chickens at 3, 7, and 11 days p.i. manifested the E117 phenotype, as shown by Tn-position-PCR yielding a 0.25-kb fragment (Fig. 6B, lanes 1 to 3), by the nonexpression of CrmA (Fig. 6C, lanes 1 to 3), and by the HA− phenotype on agar plates. In contrast, two isolates from chickens in this group at 21 and 28 days p.i. manifested the original R-low phenotype, as shown by negative Tn-position-PCR results, positive results for CrmA expression, and an HA-positive (HA+) phenotype on agar plates (Fig. 6B and C, lanes 4 and 5).
DISCUSSION
M. gallisepticum strain R-low is known to be pathogenic for chickens, capable of colonizing the trachea, and capable of producing air sac and trachea lesions (7, 35). It was shown that multiple in vitro passages (164 passages) of this strain (designated R-high) resulted in a decrease in the ability to colonize the tracheal mucosa and produce pathological changes in the respiratory tract (25), suggesting that during multiple in vitro passages, the proper function of certain components was impaired. In this study, we randomly mutated the chromosome of strain R-low (passage 9) by Tn4001 transposon mutagenesis and screened for cytadherence-deficient mutants by using an HA assay as an indicator for cytadherence (11). Four independent transformation experiments in which 106 to 107 cells per transformation were used yielded a total of about 3,500 Gmr transformants, but only 5 colonies exhibited an HA− phenotype. It should be noted that the original M. gallisepticum R-low population displayed an HA+ phenotype and that colonies displaying the HA− phenotype, which may have represented spontaneously arising HA− variants, were not detected. Moreover, in several passages of filter-cloned HA− transformants, the HA− phenotype did not reveal any HA+ revertants within the variant population. We concluded, therefore, that the transposon insertion is directly responsible for the inability of strain R-low to hemadsorb.
Mapping of the Tn4001 insertion site of the M. gallisepticum R-low Gmr HA− mutants identified three in which the insertion was found to be within the coding regions of two adjacent genes, gapA and crmA (10, 13, 30, 43) (Fig. 2). One insertion was identified within a gene encoding an ATP or GTP binding protein, and one was identified within a gene with an unknown function. The gapA and crmA gene products expressed in strain R-low have been shown by Papazisi et al. to be missing in highly passaged strain R-high (30). The gapA and crmA genes are part of a single operon encoding two proteins that belong to a conserved mycoplasma adhesion family (ADP1 family) (31, 34) and that may play a role in M. gallisepticum cytadherence. The coordinated loss of the M. gallisepticum GapA and CrmA proteins in highly passaged strain R-high, together with its reduced cytadherence capability, provides further support for the key role of this operon in cytadherence.
Our in vitro attachment experiments with the MRC-5 model system (Fig. 4) showed a marked decreased in the efficiency of attachment of each of the gapA and crmA mutants (E117, E345, and E325) in comparison to that of wild-type strain R-low. In other words, inactivation of either the gapA or the crmA gene alone is sufficient to abolish attachment, implying that the coordinated action of the GapA and CrmA proteins is necessary for M. gallisepticum cytadherence. Indeed, independent experiments in which R-high was complemented with wild-type alleles of gapA or crmA demonstrated that neither GapA nor CrmA alone is sufficient to mediate efficient M. gallisepticum cytadherence (30). However, a recent study in which R-high was complemented with both wild-type gapA and wild-type crmA alleles demonstrated that their coexpression is necessary for efficient cytadherence and virulence (31). It is important to note, moreover, that two M. gallisepticum strains used as live vaccines, 6/85 and F, exhibited significantly less attachment than did strain R-low in the MRC-5 assay despite the fact that both CrmA and GapA are expressed in these strains. On the other hand, in another vaccine strain, ts-11, only the expression of GapA was not detected. Overall, these findings further indicate that the cytadherence of M. gallisepticum is not solely mediated by the GapA and CrmA proteins but reflects a multifactorial process that involves an array of accessory proteins and that may be analogous to the process that has been intensively studied in M. pneumoniae (17, 19). Indeed, other putative cytadherence molecules have been identified in M. gallisepticum, but their precise roles in cytadherence have not yet been determined (3, 8, 12, 14, 28, 29).
Experimental tracheal infection of birds with M. gallisepticum mutants E117, E345, and E325 in comparison to infection with wild-type R-low resulted in a significant decrease in the number of M. gallisepticum organisms recovered from the trachea at each sampling time and a marked decreased in serological reactions, as measured by commercial RSA and ELISA diagnostic tests. The finding that the inactivation of either the gapA or the crmA gene in strain R-low by transposon mutagenesis hampered its capability to colonize the upper respiratory tracts of experimentally infected chickens (Table 2) provides strong evidence for the coordinated role of the GapA and CrmA proteins in M. gallisepticum cytadherence and pathogenicity in vivo. Moreover, these studies indicate the relevance of in vitro cytadherence models, such as those using HA and MRC-5 cells, to infection in the natural animal host.
It should be noted that the loss of the Tn4001 transposon was detected during some infection experiments but was confined to times later than 14 day p.i. and did not occur in every case. With the loss of the transposon, the wild-type phenotype was restored (Fig. 6). Since M. gallisepticum with the wild-type phenotype would be predicted to have a selective advantage in colonization and transmission, it would readily multiply in birds and spread within the group. Thus, under our experimental conditions, in which chickens in each group were maintained in close contact in isolation cages, it was not possible to determine the frequency of the loss of the transposon.
CrmA displays significant homology to the precursor of M. pneumoniae ORF6 gene products, which play a role in P1 (ADP1)-mediated cytadherence (20, 30, 31). M. pneumoniae ORF6 gene products are found in close proximity to ADP1 and other M. pneumoniae cytadherence-related molecules, such as HMW1, HMW3, p65, and p30 (23, 24). Interestingly, upstream of the gapA operon (Fig. 2) there exists an ORF, designated MGC2, that is predicted to encode a protein with homology to the M. pneumoniae p30 and M. genitalium p32 cytadhesins (12). Double-sided immunogold labeling localized MGC2 on the terminal bleb, and anti-MGC2 antiserum inhibited attachment. The relationship of the MGC2 protein to the GapA and CrmA proteins in mediating M. gallisepticum cytadherence is not yet known, but the location of the MGC2 ORF adjacent to the gapA operon may suggest such a role.
Collectively, our findings support previous data and provide compelling additional independent evidence indicating significant roles for both the GapA and the CrmA proteins in the adherence of M. gallisepticum to host cells in model systems as well as in colonization in vivo in the natural animal host. This study also underscores the efficiency of combining Tn4001 transposon mutagenesis and HA capability as efficient tools for identifying mycoplasmal molecules involved in pathogenesis.
Acknowledgments
This work was supported by grant 1999127 from the United States-Israel Binational Science Foundation (BSF) to D.Y. and S.J.G. and by grant IS-3126-99 from United States-Israel Binational Agricultural Research and Development (BARD) to S.L. and D.Y.
We gratefully acknowledge the assistance of A. Lublin in the statistical analysis of quantitative infection data.
Editor: V. J. DiRita
REFERENCES
- 1.Athamna, A., R. Rosengarten, S. Levisohn, I. Kahane, and D. Yogev. 1997. Adherence of Mycoplasma gallisepticum involves variable surface membrane proteins. Infect. Immun. 65:2468-2471. [DOI] [PMC free article] [PubMed]
- 2.Baas, E. J., and D. E. Jasper. 1972. Agar block technique for identification of mycoplasmas by use of fluorescent antibody. Appl. Microbiol. 23:1097-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boguslavsky, S., D. Menaker, I. Lysnyansky, T. Liu, S. Levisohn, R. Rosengarten, M. Garcia, and D. Yogev. 2000. Molecular characterization of the Mycoplasma gallisepticum pvpA gene, which encodes a putative variable cytadhesin protein. Infect. Immun. 68:3956-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradbury, J. M., O. M. Saed, C. A. Yavari, J. P. Dupiellet, and J. M. Bove. 1993. Mycoplasma imitans sp. nov. is related to Mycoplasma gallisepticum and found in birds. Int. J. Syst. Bacteriol. 43:721-728. [DOI] [PubMed] [Google Scholar]
- 5.Byrne, M. E., D. A. Rouch, and R. A. Skurray. 1989. Nucleotide sequence analysis of IS256 from the Staphylococcus aureus gentamicin-tobramicin-kanamicin-resistance transposon Tn4001. Gene 81:361-367. [DOI] [PubMed] [Google Scholar]
- 6.Dhandayuthapani, S., W. G. Rasmussen, and J. B. Baseman. 1999. Disruption of gene mg218 of Mycoplasma genitalium through homologous recombination leads to an adherence-deficient phenotype. Proc. Natl. Acad. Sci. USA 96:5227-5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dykstra, M. J., S. Levisohn, O. J. Fletcher, and S. H. Kleven. 1985. Evaluation of cytopathologic changes induced in chicken tracheal epithelium by Mycoplasma gallisepticum in vivo and in vitro. Am. J. Vet. Res. 46:116-122. [PubMed] [Google Scholar]
- 8.Forsyth, M. H., M. E. Tourtellotte, and S. J. Geary. 1992. Localization of an immunodominant 64kDa lipoprotein (LP64) in the membrane of Mycoplasma gallisepticum and its role in cytoadherence. Mol. Microbiol. 6:2099-2106. [DOI] [PubMed] [Google Scholar]
- 9.Geary, S. J., and M. G. Gabridge. 1987. Characterization of a human lung fibroblast receptor site for Mycoplasma pneumoniae. Isr. J. Med. Sci. 23:462-468. [PubMed] [Google Scholar]
- 10.Goh, M. S., T. S. Gorton, M. H. Forsyth, K. E. Troy, and S. J. Geary. 1998. Molecular and biochemical analysis of a 105 kDa Mycoplasma gallisepticum cytadhesin (GapA). Microbiology 144:2971-2978. [DOI] [PubMed] [Google Scholar]
- 11.Hedreyda, C. T., and D. C. Krause. 1995. Identification of a possible cytadherence regulatory locus in Mycoplasma pneumoniae. Infect. Immun. 63:3479-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hnatow, L. L., C. L. Keeler, Jr., L. L. Tessmer, K. Czymmek, and J. E. Dohms. 1998. Characterization of MGC2, a Mycoplasma gallisepticum cytadhesin with homology to the Mycoplasma pneumoniae30-kilodalton protein P30 and Mycoplasma genitalium P32. Infect. Immun. 66:3436-3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keeler, L. L., Jr., L. L. Hnatow, P. L. Whetzel, and J. E. Dohms. 1996. Cloning and characterization of a putative cytadhesin gene (mgcl) from Mycoplasma gallisepticum. Infect. Immun. 64:1541-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kheyar, A., S. K. Reddy, and A. Silim. 1995. The 64 kDa lipoprotein of Mycoplasma gallisepticum has two distinct epitopes responsible for haemagglutination and growth inhibition. Avian Pathol. 24:55-68. [DOI] [PubMed] [Google Scholar]
- 15.Kleven, S. H., and S. Levisohn. 1996. Mycoplasma infections of poultry, p. 283-292. In J. G. Tully and S. Razin (ed.), Molecular and diagnostic procedures in mycoplasmology, vol. II. Academic Press, Inc., New York, N.Y.
- 16.Knudson, K. L., and F. C. Minion. 1993. Construction of Tn4001lac derivatives to be used as promoter probe vectors in mycoplasmas. Gene 137:217-222. [DOI] [PubMed] [Google Scholar]
- 17.Krause, D. C. 1996. Mycoplasma pneumoniae cytadherence: unravelling the tie that binds. Mol. Microbiol. 20:247-253. [DOI] [PubMed] [Google Scholar]
- 18.Krause, D. C., T. Proft, C. T. Hedreyda, H. Hilbert, H. Plagens, and R. Herrmann. 1997. Transposon mutagenesis reinforces the correlation between Mycoplasma pneumoniae cytoskeletal protein HMW2 and cytadherence. J. Bacteriol. 179:2668-2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krause, D. C. 1998. Mycoplasma pneumoniae cytadherence: organization and assembly of the attachment organelle. Trends Microbiol. 6:15-18. [DOI] [PubMed] [Google Scholar]
- 20.Krause, D. C., and M. F. Balish. 2001. Structure, function, and assembly of the terminal organelle of Mycoplasma pneumoniae. FEMS Microbiol. Lett. 198:1-7. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 22.Lauerman, L. H. 1998. Nucleic acid amplification assays for diagnosis of animal diseases. American Association of Veterinary Laboratory Diagnosticians, Turlock, Calif.
- 23.Layh-Schmitt, G., and R. Herrmann. 1994. Spatial arrangement of gene products of the P1 operon in the membrane of Mycoplasma pneumoniae. Infect. Immun. 62:974-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Layh-Schmitt, G., A. Podtelejnikov, and M. Mann. 2000. Proteins complexed to the P1 adhesin of Mycoplasma pneumoniae. Microbiology 146:741-747. [DOI] [PubMed] [Google Scholar]
- 25.Levisohn, S., M. J. Dykstra, M. Y. Lin, and S. H. Kleven. 1986. Comparison of in vivo and in vitro methods for pathogenicity evaluation for Mycoplasma gallisepticum in respiratory infection. Avian Pathol. 15:233-246. [DOI] [PubMed] [Google Scholar]
- 26.Levisohn, S., and S. H. Kleven. 2000. Avian mycoplasmosis (Mycoplasma gallisepticum), p. 425-442. In C. W. Beard and S. McNulty (ed.), Diseases of poultry: world trade and public health implications, vol. 19. Office International des Epizooties, Paris, France. [PubMed]
- 27.Ley, D. H., and H. W. Yoder, Jr. 1997. Mycoplasma gallisepticum infection, p. 194-207. In B. W. Calnek, H. J. Barnes, C. Beard, L. R. MacDougald, and Y. M. Saif (ed.), Diseases of poultry, 10th ed. Iowa State University Press, Ames.
- 28.Liu, L., D. M. Payne, V. L. van Santen, K. Dybvig, and V. S. Panangala. 1998. A protein (M9) associated with monoclonal antibody-mediated agglutination of Mycoplasma gallisepticum is a member of the pMGA family. Infect. Immun. 66:5570-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Markham, P. F., M. D. Glew, J. E. Sykes, T. R. Bowden, T. D. Pollocks, G. F. Browning, K. G. Whithear, and I. D. Walker. 1994. The organization of the multigene family which encodes the major cell surface protein, pMGA, a hemagglutinin of Mycoplasma gallisepticum. FEBS Lett. 352:347-352. [DOI] [PubMed] [Google Scholar]
- 30.Papazisi, L., K. E. Troy, T. S. Gorton, X. Liao, and S. J. Geary. 2000. Analysis of cytadherence-deficient, GapA-negative Mycoplasma gallisepticum strain R. Infect. Immun. 68:6643-6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Papazisi, L., S. Frasca, Jr., M. Gladd, X. Liao, D. Yogev, and S. J. Geary. 2002. GapA and CrmA coexpression is essential for Mycoplasma gallisepticum cytadherence and virulence. Infect. Immun. 70:6839-6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Razin, S. 1963. Osmotic lysis of mycoplasma. J. Gen. Microbiol. 33:471-475. [DOI] [PubMed] [Google Scholar]
- 33.Razin, S., and E. Jacobs. 1992. Mycoplasma adhesion. J. Gen. Microbiol. 138:407-422. [DOI] [PubMed] [Google Scholar]
- 34.Razin, S., D. Yogev, and Y. Naot. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62:1094-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodriguez, R., and S. H. Kleven. 1980. Pathogenicity of two strains of Mycoplasma gallisepticum in broilers chickens. Avian Dis. 24:801-807. [PubMed] [Google Scholar]
- 36.Rosengarten, R., A. Behrens, A. Stetefeld, M. Heller, M. Ahrens, K. Sachse, D. Yogev, and H. Kirchhoff. 1994. Antigen heterogeneity among isolates of Mycoplasma bovis is generated by high-frequency variation of diverse membrane surface proteins. Infect. Immun. 62:5066-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosengarten, R., and D. Yogev. 1996. Variant colony surface antigenic phenotypes within mycoplasma strain populations: implications for species diagnosis and strain standardization. J. Clin. Microbiol. 34:149-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saed, O. M., G. Ross, and J. M. Bradbury. 1996. Pathogenicity and cytadherence of Mycoplasma imitans in chick and duck embryo tracheal organ cultures. Infect. Immun. 64:563-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 40.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yogev, D., S. Levisohn, S. H. Kleven, D. Halachmi, and S. Razin. 1988. Ribosomal RNA gene probes detect intraspecies heterogeneity in Mycoplasma gallisepticum and Mycoplasma synoviae. Avian Dis. 32:220-231. [PubMed] [Google Scholar]
- 43.Yoshida, S., A. Fujisawa, Y. Tsuzaki, and S. Saitoh. 2000. Identification and expression of a Mycoplasma gallisepticum surface antigen recognized by a monoclonal antibody capable of inhibition of both growth and metabolism. Infect. Immun. 68:3186-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]