Abstract
Spread of Streptococcus pneumoniae from the nasopharynx to other host tissues would require the organism to adapt to a variety of environmental conditions. Since heat shock proteins are induced by environmental stresses, we investigated the effect of heat shock on ClpL and ClpP synthesis and the effect of clpL and clpP mutations on the expression of key pneumococcal virulence genes. Pulse labeling with [35S]methionine and chase experiments as well as immunoblot analysis demonstrated that ClpL, DnaK, and GroEL were stable. Purified recombinant ClpL refolded urea-denatured rhodanese in a dose-dependent manner, demonstrating ClpL's chaperone activity. Although growth of the clpL mutant was not affected at 30 or 37°C, growth of the clpP mutant was severely affected at these temperatures. However, both clpL and clpP mutants were sensitive to 43°C. Although it was further induced by heat shock, the level of expression of ClpL in the clpP mutant was high at 30°C, suggesting that ClpP represses expression of ClpL. Furthermore, the clpP mutation significantly attenuated the virulence of S. pneumoniae in a murine intraperitoneal infection model, whereas the clpL mutation did not. Interestingly, immunoblot and real-time reverse transcription-PCR analysis demonstrated that pneumolysin and pneumococcal surface antigen A were induced by heat shock in wild-type S. pneumoniae. Other virulence genes were also affected by heat shock and clpL and clpP mutations. Virulence gene expression seems to be modulated not only by heat shock but also by the ClpL and ClpP proteases.
Streptococcus pneumoniae, a gram-positive and naturally transformable organism, causes bacterial pneumonia, otitis media, and meningitis (46). It is carried in the nasopharynx of healthy individuals, and this is a major reservoir for pneumococcal infections. Pneumococci are subject to a number of environmental stresses in vivo. Change of environmental niche in the host, such as penetration of pneumococci from the nasopharynx into the bloodstream, can trigger dramatic changes in morphology as well as gene expression. In fact, pneumococci in the nasopharynx have been shown to be predominantly of a transparent colony phenotype and tend to express less capsule and more choline binding protein A (CbpA), whereas pneumococci in the bloodstream are predominantly of the opaque colony morphology and tend to produce more capsule and less CbpA (24, 42).
S. pneumoniae encounters heat stress as a regular feature of its pathogenic life cycle after penetration from the nasal mucosa (30 to 34°C versus 28°C) into blood and/or meninges (37°C). The elevated temperatures they encounter within a mammalian host may serve as a key trigger for a rapid, transient increase in the synthesis of a highly conserved set of proteins referred to as heat shock proteins (HSPs). HSPs protect bacteria against such adverse effects as elevated temperatures, exposure to ethanol, and heavy metals, increasing their survival rate. Therefore, a thorough understanding of the heat shock response could provide useful information on adaptation of the pneumococcus to the hostile environment it encounters.
HSPs can be classified into Hsp100, Hsp70, Hsp60, and small Hsp families depending on molecular weight and are ubiquitously present in prokaryotes and eukaryotes (9, 21, 30). One of the HSPs, hsp100/Clp (caseinolytic protease) family, is present as a 104-kDa protein in eukaryotes but as an 80- to 95-kDa protein in prokaryotes. It carries out a chaperone function and is also involved in proteolysis, removing damaged and denatured proteins (39). Proteolysis by Clp requires a serine-type peptidase ClpP subunit and a regulatory ATPase subunit (39). Regulatory Clp subunit proteins can be assigned, in general, to two classes: class I, which comprises clpA, clpB, clpC, and clpD, contains two ATP-binding regions; class II, which comprises clpM, clpN, clpX, and clpY, contains only one ATP-binding region. Clps have been classified by the size of the central spacer segment, the need for gaps in aligning the overall sequences, and sequence similarities in the well-conserved regions and in the N- and C-terminal segments. The variable leader regions have very different sequences in each subfamily (39).
Although substantial progress has been made in understanding the mechanisms of action of the Clp family in gram-negative bacteria such as Escherichia coli (33, 39), little is known about Clp in gram-positive bacteria. The clpP gene and clpC operon are negatively regulated by CtsR, which recognizes a directly repeated operator sequence (A/GGT CAA ANA NA/GG TCA AA) (10), but clpX does not have this sequence (15), and their specific mechanisms of action have not been determined in detail (10, 11, 16).
Since a variety of environmental signals, including temperature and nutrient availability, can control the expression of virulence factors (31), we examined the protein profiles of the heat shock response in pneumococci after exposure of the cells to several stresses. The major proteins induced by heat shock were 62, 72, and 84 kDa in size, identified subsequently as GroEL, DnaK, and ClpL, respectively (8). However, pulse labeling of proteins with [35S]methionine revealed that certain conditions which are known to induce stress responses in E. coli and Bacillus subtilis failed to induce any high-molecular-weight HSPs such as GroEL and DnaK homologues. However, a temperature shift from 30 to 37°C in vitro, similar to that encountered by S. pneumoniae after translocation from the nasal mucosa to the lungs, triggered induction of DnaK and GroEL (8). Recently, other heat shock genes, clpC, clpX, clpE, and clpP, were identified, but their specific roles in virulence have not been fully elucidated (6, 7, 11, 41). In addition, the role of ClpC in autolysis, transformation, and virulence remains controversial (6, 7).
The nucleotide sequences of clpL from several gram-positive organisms are known (Lactococcus lactis [X62333]; Staphylococcus aureus [AP003365, AP003137]; Streptococcus pyogenes [AE006538 and AE004092]; and Lactobacillus rhamnosus [AF323526]), but functional studies on ClpL have been limited (22). To date, information on the role of clpL in pathogenesis is not available. Since HSPs are representative of proteins induced by other stresses such as oxidative stress and heavy metals (33), the effect of heat shock on ClpL and ClpP synthesis was investigated. In addition, the impact of clpL and clpP mutation on in vitro expression of key pneumococcal virulence genes was evaluated. Furthermore, the effect of clpL and clpP mutations on the virulence of S. pneumoniae was evaluated in a mouse intraperitoneal challenge model. Here we demonstrate that the heat shock process induced expression of pneumolysin and modulated the expression of other virulence factors in wild-type pneumococci. We also show that mutation in clpP resulted in an increase in mRNA expression but not in the activity of pneumolysin at elevated temperatures.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and transformation.
The bacterial strains used in this work are presented in Table 1. S. pneumoniae CP1200 (8), a derivative of Rx-1, was used in this study and was grown at 37°C in Casitone-tryptone (CAT)-based medium to the mid-exponential phase (8). Complete transformation medium was prepared from CAT broth by addition of (per liter) 147 mg of CaCl2 and 2 g of bovine serum albumin (fraction V; Sigma). Competence was controlled by appropriate addition of the competence-specific peptide and quantitated as novobiocin-resistant transformants obtained after exposure of cells to DNA in culture medium, as described previously (20). Encapsulated strain D39 (type 2) was grown in brain heart infusion broth or Todd-Hewitt broth and transformed as described (4). For selection of pneumococcal transformants, erythromycin or novobiocin was added to the growth medium at a concentration of 2.5 μg/ml or 10 μg/ml, respectively.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| E. coli strains | ||
| BL21(DE3) | gal (λcIts857 ind1 Sam7 nin5 lacUV5-T7 gene 1) | Novagen |
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) | BRL |
| XL1-Blue | relA1 lac [F′ proAB lacIqZ ΔM15 Tn10 (Tetr)] | Stratagene |
| S. pneumoniae strains | ||
| CP1200 | Nonencapsulated derivative of Rx1, malM511 str-1 | Choi et al.(8) |
| HYK1 | CP1200, ΔclpL::ermB | This study |
| HYK2 | CP1200, ΔclpP::ermB | This study |
| D39 | Encapsulated, type 2 | Avery et al. (1) |
| HYK302 | D39, ΔclpP::ermB | This study |
| HYK304 | D39, ΔclpL::ermB | This study |
| Plasmids | ||
| pET30(a) | 5.4 kb, Kmr | Novagen |
| pBluescript | 3.0 kb, Apr | Stratagene |
| pGEM-T | 3.0 kb, Apr, TA cloning vector | Promega |
| pG8413 | 1.3 kb, clpL PCR fragment in pBluescript | This study |
| pKHY004 | 7.5 kb, histidine-tagged clpL in pET30(a) | This study |
Escherichia coli strains were grown in Luria-Bertani (LB) broth or on LB agar. Plasmids were introduced into E. coli by transformation as described by Hanahan (19). For selection of E. coli transformants, ampicillin (100 μg/ml) was added to the growth medium. Plasmid vectors along with new recombinants generated in this study are listed in Table 1.
Preparation of antisera.
Production of HSP antibodies against S. pneumoniae DnaK and GroEL has been described previously (8). To prepare antibodies against ClpL, an exponential-phase culture of S. pneumoniae CP1200 was incubated at 42°C for 30 min; the cells were sonicated, and proteins were separated by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-10% PAGE) and lightly stained with Coomassie brilliant blue. The 84- and 21-kDa protein bands were cut out and electroeluted. One hundred micrograms of either protein per ml of saline was mixed with 1 ml of Freund's incomplete adjuvant. This mixture was then injected intramuscularly and subcutaneously into rabbits. Two booster doses were administered at 2-week intervals, and antiserum was collected after 6 weeks. The preparation of sera against CbpA, pneumococcal surface antigen A (PsaA), and pneumolysin was done essentially as described previously (34).
Protein labeling and gel electrophoresis.
For protein labeling experiments, cells were grown in CAT medium to an A550 of 0.2 and then divided into 2-ml aliquots. The cells were then harvested, resuspended in fresh warmed low-methionine labeling medium and equilibrated for 10 min at 30°C. To this was added 10 μCi of [35S]methionine (1,000 Ci/mmol, Amersham), and the culture was then transferred to 42°C for heat shock. The cells were harvested, resuspended in 20 μl of lysis buffer (5 mM Tris [pH 8.0], 30 mM EDTA, 0.1% Triton X-100, 0.025% [wt/vol] phenylmethylsulfonyl fluoride, 1 mM dithiothreitol), and then lysed completely by sonication (on ice) as described previously (8).
SDS-PAGE (either 10 or 15% polyacrylamide gel) was carried out as described by Laemmli (26), and the proteins were visualized with Coomassie brilliant blue staining. Polyacrylamide gels were exposed to a radiation-sensitive imaging plate for several days to obtain the images. The radiographic imaging data were quantitated with an image analysis system (Fujix Bio-imaging Analyzer BAS2500, Fuji Photo Film Co.).
Immunoblotting.
Proteins separated by SDS-10% PAGE were electroblotted onto a polyvinylidene difluoride membrane and then reacted with either 1:100 dilutions of a rabbit antiserum raised against heat shock proteins of S. pneumoniae or 1:5,000 dilutions of mouse antiserum raised against virulence proteins of S. pneumoniae (CbpA, PsaA, and pneumolysin) as the primary antibodies. The secondary antibody was a 1:2,000 dilution of goat anti-rabbit or goat anti-mouse immunoglobulin G conjugated to either horseradish peroxidase (Sigma) or alkaline phosphatase (Bio-Rad).
Real-time RT-PCR.
Total RNA was extracted with the hot acid phenol method, as described previously (35). Levels of mRNA for pneumolysin (ply), psaA, cbpA, and cps2A were quantitated by one-step real-time reverse transcription-PCR (RT-PCR) with the Promega Access RT-PCR system (Promega Biotech, catalog no. A1250). The specific primers used for the various RT-PCR assays have been described elsewhere (35) and were used at a final concentration of 50 nM per reaction. As an internal control, primers specific for the 16S rRNA were employed. Separate RT-PCRs (differing only in the constituent primers) were set up (on ice) from a master mix to which Sybr Green (Molecular Probes) had been added to a final concentration of 1:50,000. The mix was then aliquoted into tubes containing the respective upstream and downstream primers on ice and thoroughly mixed by gentle vortexing. Each mix was finally aliquoted into thin-walled 0.1-ml reaction tubes and placed in a Rotor-Gene 2000 Real-Time cycler (Corbett Research, Australia).
The RT-PCR cycling conditions comprised one cycle at 48°C for 39 min (for first-strand cDNA synthesis) and one cycle at 94°C for 2 min (for avian myeloblastosis virus reverse transcriptase inactivation and RNA/cDNA/primer denaturation), followed by 40 cycles of PCR amplification comprising denaturation (94°C for 30 s), primer annealing (60°C for 30 s), and extension (72°C for 39 s). Amplification data were acquired at the extension step and analyzed with the Corbett Research Software version 4.4 with the comparative critical threshold (ΔΔCT) values. Between RNA extracts, levels of target transcripts were normalized with reference to transcript levels obtained for the internal 16S rRNA control. All experiments were carried out in quadruplicate.
Construction of clpL and clpP deletion mutants.
To create an insertion-deletion mutation of clpL (ΔclpL::ermB) in S. pneumoniae, an 860-bp ermB cassette (43) was amplified with prs3 (5′-CCG GGC CCA AAA TTT GTT TGA T-3′) and prs4 (5′-AGT CGG CAG CGA CTC ATA GAA T-3′) from erythromycin-resistant E. coli chromosomal DNA and used to disrupt clpL. A 410-bp fragment (clpLup) containing part of both clpL and the 5′ end of ermB was amplified with hlp3 (5′-CGG TAC CAT GAA CAA TAA TTT TAA C-3′) and hlp1 (5′-ATC AAA CAA ATT TTG GGC CCG GTC AGA TGT TTC TTG AAT TTC C-3′) from CP1200 DNA. A 300-bp fragment (clpLdown) containing part of both the downstream clpL sequence and the 3′ terminus of ermB was amplified with hlp2 (5′-ATT CTA TGA GTC GCT GCC GAC TGT TCT AGA TGA TGG TCG TTT G-3′) and hlp4 (5′-GGC CGA GCT CTT AGA CTT TCT CAC GAA TAA C-3′) from CP1200 DNA.
The three PCR products were used as a mixed template for PCR with hlp3 and hlp4 to produce a 1.6-kb fragment with a 1,300-bp deletion of clpL (nucleotides 321 to 1620) that was replaced by the ermB gene. The tripartite 1.6-kb fragment was subsequently introduced into either S. pneumoniae CP1200 or D39 strains by transformation, and recipient bacteria that had integrated the recombinant fragment into the chromosome by homologous recombination were selected by resistance to erythromycin. Transformants were screened for the correct deletion by PCR and immunoblot analysis (not shown). CP1200 and D39 clpL mutants, HYK1 and HYK304, respectively, contained the correct deletion within clpL and were used for further studies. clpP mutants of either CP1200 (HYK2) or D39 (HYK302) were constructed with the same strategy except for the primers for clpPup (234 bp): hpp3 (5′-:CGA ATT CAT GAT TCC TGT AGT TAT-3′) and hpp11 (5′-ATT CTA TGA GTC GCT GCC GAC TCA GAA CCA CCT GGT GTA TTG A-3′) and clpPdown (319 bp): hpp10 (5′-ATC AAA CAA ATT TTG GGC CCG GAT CGC ATC AAG TGG AGC AAA A-3′) and hpp6 (5′-CGA GCT CTT AGT TCA ATG AAT TGT TG-3′) with a deletion of 95 bp (nucleotides 206 to 300).
Overexpression of ClpL in E. coli.
To overexpress His6-tagged ClpL in E. coli, the clpL open reading frame was amplified with prs3 and HYG4 (5′-GGC CGA GCT CTT AGA CTT TCT CAC GAA TAA C-3′, which incorporates KpnI and SacI sites, respectively) from CP1200 DNA. The fragment was digested with KpnI and SacI and cloned into the KpnI and SacI sites of pET30(a) (Novagen) to generate plasmid pKHY004. His6-tagged protein was expressed in E. coli and subjected to DEAE-Sepharose fast flow chromatography (Amersham Pharmacia) eluted with a 0.1 to 0.4 M NaCl gradient. The fractions containing ClpL were pooled and purified on a nickel-nitriloacetic acid column according to the manufacturer's instructions (Novagen) with minor modifications. Bound His6-tagged protein was washed with 40 mM imidazole buffer, eluted with 0.4 M imidazole buffer (pH 7.9), and dialyzed against 20 mM Tris-HCl (pH 7.8)-5 mM MgCl2. The protein was >95% pure as judged by SDS-PAGE and staining with Coomassie brilliant blue R250 (data not shown).
Determination of chaperone activity.
The chaperone activity of ClpL was determined as described previously (25) with a modification as follows. Rhodanese (9 μM) was denatured in 200 mM potassium phosphate buffer (pH 7.6) containing 1 mM β-mercaptoethanol and 8 M urea for 1 h at 25°C. Spontaneous and ClpL-assisted refolding was initiated by diluting 2.5 μl of denatured enzyme in 8 M urea to a final volume of 250 μl of a solution containing 50 mM Tris-HCl (pH 7.8), 200 mM β-mercaptoethanol, 5 mM sodium thiosulfate, 10 mM MgCl2, and 10 mM KCl. The final concentration of rhodanese in the refolding reaction was 90 nM. The refolding reaction was carried out for 30 min at 25°C. The chaperone activity of ClpL was measured by refolding of rhodanese into its native conformation. The enzyme activity of rhodanese was determined as described by Sorbo (40).
Virulence studies.
Intraperitoneal challenge with a highly virulent capsular type 2 strain (D39) and its isogenic clpP and clpL mutants (HYK302 and HYK304, respectively) was performed to evaluate the effect, if any, of mutating clpL or clpP on the virulence of S. pneumoniae. Bacteria were cultured at 37°C overnight on blood agar (supplemented with erythromycin as required) and then grown in serum broth (10% [vol/vol] horse serum in meat extract broth) for 3 h at 37°C to give ca. 108 CFU/ml (34). Each bacterial culture was then diluted in serum broth to ca. 106 CFU/ml, and groups of 10 BALB/c mice were infected intraperitoneally with 0.1-ml volumes of either D39, HYK302, or HYK304. The survival of the challenged mice was monitored four times daily for the first 5 days, twice daily for the following 5 days, and daily until 21 days postchallenge.
Pneumolysin assay.
Hemolytic activity was determined as previously described (29) with a minor modification. Briefly, pneumococci grown in THY broth to early to mid-log phase (absorbance at 600 nm = 0.05 to 0.1) were harvested by centrifugation at 3,900 × g for 10 min at 4°C and resuspended in phosphate-buffered saline. Sodium deoxycholate was added to a final concentration of 0.1% and then incubated at 37°C for 10 min. After centrifugation of the samples, the supernatant was withdrawn and serially diluted. Hemolytic activity was determined by incubation with an equal volume of 1.5% washed human red blood cells in 96-well microtiter plates. Hemolytic titer was determined as the reciprocal of the estimated dilution at which 50% of erythrocytes were lysed at A540.
Statistics.
Statistical analysis was performed with an unpaired Student's t test. Data presented are means ± standard deviation of the mean for two to four independent experiments.
Differences in median survival times between groups were analyzed by the Mann-Whitney U test (two-tailed), and differences in overall survival rate between groups were analyzed by the Fisher exact test.
RESULTS
Characterization of ClpL.
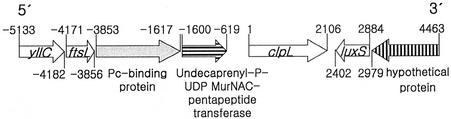
Previously, an 84-kDa HSP was identified as ClpL by N-terminal amino acid sequencing (8). Members of the Clp family contain two highly conserved ATP-binding regions (ATP-1 and ATP-2), each of which contains a consensus sequence for adenine nucleotide binding (17). To confirm that putative pneumococcal ClpL is indeed a member of the Clp family, degenerate oligonucleotides from the N-terminal amino acid sequence (5′-GAT GAA YAA YAA YTT YAA YAA YTT YAA-3′) and the second ATP-binding site for Clp members (5′-GTY TTN CCN CAN CCN GYN GG-3′, where Y is T or C and N is A, C, G, or T), which was the most conserved amino acid sequence (PTGVGKT) of the clp family, were used for PCR amplification of CP1200 chromosomal DNA. PCR yielded a 1.37-kb DNA fragment expected from the size of L. lactis ClpL. This was cloned into pGEM-T to generate plasmid pG8403. Sequence analysis of the cloned fragment demonstrated homology to the L. lactis clpL and bovine clp genes (data not shown). The complete clpL gene was then identified in the TIGR S. pneumoniae type 4 genome with Blast analysis. Moreover, the clpL homologue in S. pneumoniae R6 showed 98% identity with that of the type 4 clpL homologue, and CP1200 clpL revealed high sequence homology with R6 clpL (data not shown). The organization of this region of the genome is shown in Fig. 1.
FIG. 1.
Physical map of S. pneumoniae clpL locus.
A detailed analysis of the sequence of clpL showed an open reading frame of 2,106 bp encoding a putative polypeptide of 701 amino acids with a molecular mass of 77,699 Da and a pI of 4.99. Analysis of the nucleotide sequence showed that S. pneumoniae clpL has a sigma A type promoter (TTGACC-17 bp-TATATT) 240 bp upstream of the ATG codon. Upstream of clpL there is a CtsR repressor binding sequence, GTC AAA NAN RGT CAA A (R is A or G), which has been found adjacent to clp genes in several organisms (10). Thus, clpL may be regulated by CtsR. A gene 619 bp upstream from clpL encodes a putative undecaprenyl-p-UDP-MurNAC-pentapeptide transferase and is in the same orientation. The gene downstream of clpL, encoding LuxS, is in the opposite orientation (Fig. 1), suggesting that clpL is organized as a monocistronic transcription unit.
Blast analysis indicated that pneumococcal ClpL has high homology to all members of the Clp family in the two conserved ATP-binding regions (p-loops) at amino acids 121 to 128 (GDAGVGKT) and 391 to 398 (GSTGVGKT). Eight amino acids (MDDLFNQL) at positions 11 to 18 in the hydrophilic N-terminal region were also absolutely conserved with the bovine Clp-like protein and L. lactis ClpL. The pneumococcal ClpL ATPase shows strongest homology with a bovine Clp-like protein (76% identity and 88% similarity) and L. lactis ClpL (59% identity and 76% similarity). It also shows high homology to that of other species (data not shown).
Transient induction but high stability of ClpL after heat shock.
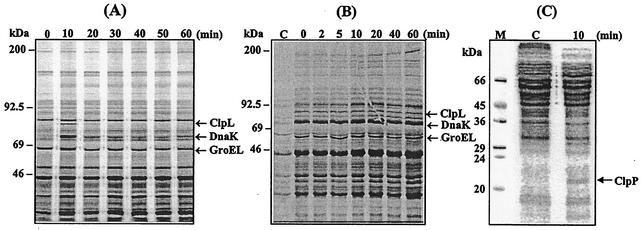
HSPs are induced upon infection and under environmental stresses and serve as antigens in some pathogens (5, 13, 23, 33). Major HSPs, ClpL, DnaK, and GroEL, which have molecular masses of 84, 73, and 65 kDa, respectively, have been identified by N-terminal amino acid sequencing of corresponding S. pneumoniae proteins after heat shock (8). The coordinate or independent control of HSP expression has not been determined; hence, we examined the kinetics of HSP synthesis by pulse labeling with [35S]methionine. Cells grown at 30°C to the mid-exponential phase were heat shocked by shifting the growth temperature to 42°C and then labeled for 10 min with [35S]methionine. Cells were harvested, and proteins were separated by SDS-PAGE followed by autoradiography.
The result revealed that the induction of HSPs peaked at 10 min after the upshift in temperature and then rapidly diminished to baseline levels (Fig. 2A). After incubating the cells at 42°C for 10 min, synthesis of ClpL, DnaK, and GroEL, was increased 11.3- ± 0.8-, 5.0- ± 0.3-, and 2.7- ± 0.2-fold, respectively, relative to the control. Although the GroEL band was very close to the nearby protein band, a higher magnification of the autoradiogram clearly showed that GroEL was induced (see Fig. 2B). However, synthesis of the major HSPs after the initial 10-min exposure at 42°C rapidly leveled off to 2.0- ± 0.2-, 2.2- ± 0.3-, and 1.2- ± 0.1-fold, respectively, relative to the non-heat-shocked control, suggesting that synthesis of HSPs reached a new steady-state level. Similar to the results presented in Fig. 2 (A), pulse labeling for 2.5 min with [35S]methionine from 0 to 15 min showed that GroEL, DnaK, and ClpL were made early, and the induction of HSPs peaked at around 5 min after the temperature upshift but fell off to the steady state after 7.5 min and resulted in net 1.5- to 2-fold increases relative to the control (data not shown). These results indicate that these HSPs, although in different classes, have the same kinetics of induction. Also, the increase in the rate of synthesis upon heat shock is similar to the increase in mRNA level of clpL and groEL in the stationary growth phase of the pneumococcus (38).
FIG. 2.
Transient induction and stability of S. pneumoniae ClpL after heat shock. (A) To determine induction kinetics, exponentially growing CP1200 cells (A550 = 0.2) were pulse labeled for 10 min with [35S]methionine, starting from the indicated time after the shift to 42°C. Two milliliters of cultures was harvested, and the cells were lysed by sonication in lysis buffer. The cell lysates were then analyzed by SDS-PAGE, and protein bands were visualized by autoradiography. (B) To determine the stability of heat shock proteins, exponentially growing CP1200 cells (A550 = 0.2) were stressed at 42°C for 10 min and pulse labeled with [35S]methionine at that time, and then the cell cultures were returned to 30°C, followed by chasing with excess nonradioactive methionine for the indicated times. Two milliliters of cultures was harvested, and the cells were lysed by sonication. The cell lysates were then analyzed by SDS-PAGE, and protein bands were visualized by autoradiography. (C) To determine induction of ClpP, exponentially growing CP1200 cells (A550 = 0.2) were pulse labeled for 10 min with [35S]methionine, starting from the indicated time after a shift to 42°C. The proteins from 2 ml of culture were separated by SDS-15% PAGE and visualized by autoradiography. A representative of duplicate experiments is shown. Lane C, not stressed. Numbers on top show time elapsed (minutes) after a return to the nonstress condition. The heavy arrows indicate major HSPs. Molecular sizes are indicated on the left.
Since the ATPase subunit of Clp members forms a complex with the ClpP serine protease (39), induction of ClpP after heat shock was examined by pulse labeling of proteins for 10 min with [35S]methionine. The result revealed induction of a 21-kDa HSP after heat shock, which was subsequently identified as by immunoblot analysis with ClpP antibodies (Fig. 2C).
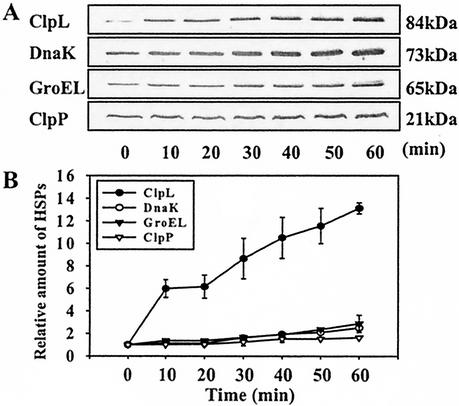
HSPs are immunogenic in some pathogens (23), and the persistence of HSPs may help in the survival of the pathogens in the host. Therefore, the stability of HSPs was examined. Bacteria were heat shocked at 42°C for 10 min, pulse labeled at that time, returned to 30°C, and then chased with nonradioactive methionine for various lengths of time. When we examined for HSPs after 1 to 60 min, there was no detectable decrease in the amount of radioactive ClpL, DnaK, or GroEL, i.e., the HSPs produced during heat shock persisted through the temperature downshift for 60 min (Fig. 2B). Interestingly, immunoblot analysis with pneumococcal HSP antibody revealed that the absolute amount of ClpL increased significantly and steadily during sustained heat shock (up to 14-fold by 60 min). However, the amount of DnaK and GroEL was increased by 2.4- and 3.4-fold, respectively, during a 60-min period (Fig. 3), but thereafter there was a reduction in the amounts of all the HSPs (data not shown). These results indicate that ClpL is fairly stable in S. pneumoniae.
FIG. 3.
Steady accumulation of ClpL after heat shock. (A) Immunoblot analysis of whole-cell lysates of exponentially growing S. pneumoniae CP1200 cells exposed to 42°C. S. pneumoniae cells grown at 30°C to an A550 = 0.3 were heat shocked at 42°C for the indicated times. The culture was harvested and resuspended in lysis buffer. The cells were lysed by sonication. Then 10 μg of proteins was separated by SDS-PAGE and reacted with antisera to ClpL, DnaK, and GroEL. In the case of ClpP, 30 μg of proteins was used for SDS-PAGE, followed by immunoblot analysis. (B) Densitometric analysis of relative levels of ClpL, ClpP, DnaK, and GroEL after heat shock, as shown in panel A. The figure shows the standard deviation from three independent experiments.
Phenotype of clpL and clpP mutants.
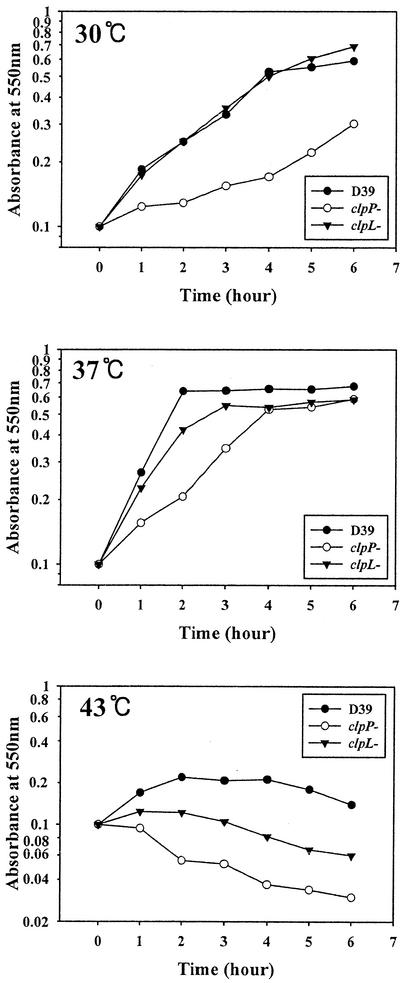
To construct clpL and clpP mutants, a DNA fragment containing either a ΔclpL::ermB or ΔclpP::ermB insertion was amplified by PCR and incorporated into the chromosome by transformation as described in Materials and Methods. The insertion mutation was confirmed by PCR and by immunoblot analysis to demonstrate the absence of ClpL or ClpP, respectively. The growth rate of the D39 derivative HYK304 carrying ΔclpL::ermB was similar to that of the parent at 30°C, but it grew slowly at 37°C, with a doubling time of 55 min, compared to about 40 min for the parent (Fig. 4). Thus, ClpL does not seem to be essential for growth of S. pneumoniae at 30°C and 37°C (Fig. 4). In contrast, HYK302 carrying the ΔclpP::ermB mutation, showed severely impaired growth at both 30°C (doubling time = 270 min) and 37°C (doubling time = 100 min) compared to the parent strain (100 and 40 min, respectively) (Fig. 4), as described previously (7). At 43°C, growth of D39 increased for the first 2 h but decreased thereafter (Fig. 4). The viability of the parent strain was maintained over a 45-min period at 42°C; however, beyond 45 min, the viability started to drop (data not shown). At 43°C, the growth of clpL and clpP mutants (HYK304 and HYK302, respectively) was impaired (Fig. 4). Furthermore, isogenic CP1200 derivatives HYK1 and HYK2 showed growth patterns similar to those of HYK304 and HYK302, respectively (data not shown).
FIG. 4.
Growth of D39 and its clpL and clpP mutants. Cultures of D39 and its isogenic clpL (HYK304) and clpP (HYK302) mutants were grown to an absorbance at 550 nm of 0.1. The temperature was then shifted from 37°C to 43°C, and the cultures were incubated for the indicated times.
Induction of ClpL in clpP mutant.
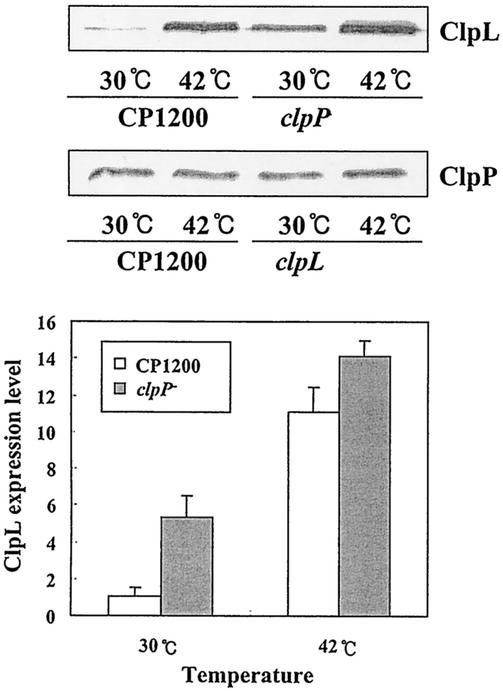
The facts that ClpL and ClpP seem to be regulated by the same repressor, CtsR (7), and that ClpP is involved in CtsR degradation (10) make it likely that ClpL might be controlled by ClpP. To examine this possibility, we determined the amounts of ClpL and ClpP with either CP1200 or its clpL or clpP mutant. Exponentially growing S. pneumoniae cells were heat shocked at 42°C for 30 min, and cellular proteins were subjected to immunoblot analysis with either polyclonal anti-ClpL or anti-ClpP antibody. In wild-type cells (CP1200) and the clpL mutant (HYK1), ClpP was detected at 30°C, but after the cells were heat shocked for 30 min, the amount of ClpP was marginally increased (Fig. 5), as shown in Fig. 3. Although ClpL was induced, the amount of ClpL in the uninduced culture was greater in a clpP mutant (HYK2) than in the wild type, suggesting that ClpP represses expression of ClpL.
FIG. 5.
Induction of ClpL in clpP mutant. Exponentially growing S. pneumoniae CP1200 (A550 = 0.3) and its isogenic clpL and clpP derivatives were heat shocked at 42°C for 30 min. Proteins from 3 ml of culture were subjected to immunoblot analysis with either anti-ClpL or anti-ClpP polyclonal serum. The positions of ClpL and ClpP are indicated.
Chaperone function of ClpL.
Since HSPs promote secretion and assist in the proper folding and translocation of proteins (9, 21, 33), the chaperone activity of ClpL in S. pneumoniae was examined. To measure chaperone activity quantitatively, refolding of a denatured protein into its native conformation is used (32). Since unassisted refolding of rhodanese occurs relatively slowly (32) and rhodanese activity can be determined by a simple and sensitive assay (40), refolding of denatured rhodanese has been used extensively to study protein folding. Histidine-tagged ClpL (pKHY004) was overexpressed in E. coli, purified, and used for determination of refolding activity. Under the test conditions, denatured rhodanese showed only 2.8 to 7.7% of the native rhodanese activity, as shown previously (32), indicating that spontaneous refolding of denatured rhodanese occurs inefficiently when diluted 100-fold from an 8 M urea solution. This activity is expressed as a percentage of the activity of native rhodanese carried through the same procedure. Inclusion of ClpL in the refolding reaction mixture in an approximately threefold molar excess to denatured rhodanese increased renaturation to almost 10% of the native rhodanese activity. However, when a 12-fold excess of ClpL was added to denatured rhodanese, it increased activity to 30% of the native level. Increasing the amount of ClpL added above this point yielded little further renaturation in the presence of ATP (Table 2). These results demonstrated that ClpL could function independently as a chaperone to refold the denatured protein, as shown previously for ClpA in E. coli (45).
TABLE 2.
ClpL-dependent in vitro refolding of denatured rhodanesea
| ClpL concn | Rhodanese activity (% of control) |
|---|---|
| None (control) | 4.3 ± 1.8 |
| 90 nM | 9.0 ± 4.2* |
| 270 nM | 9.5 ± 3.2** |
| 541 nM | 15.2 ± 7.2** |
| 1.08 μM | 30.1 ± 14.0** |
| 1.6 μM | 35.7 ± 14.0** |
| 2.2 μM | 35.9 ± 6.5** |
Denatured rhodanese (90 nM final concentration) was incubated at 37°C for 1 h alone or together with ClpL in the presence of 2 mM ATP. The activity of the refolded enzyme was measured after 30 min of incubation at 25°C and is expressed as a percentage of the activity of the same amount of native enzyme incubated at 25°C under the same conditions. The mean value and standard deviation of five independent experiments are shown. *, significantly different from the control (no ClpL), P < 0.05; **, significantly different from the control, P < 0.001.
Modulation of expression of virulence-associated factors by heat shock.
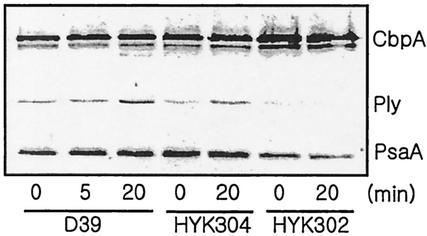
Environmental stress, including heat shock and starvation, can affect the expression of virulence factors (31). Hence, the effect of heat shock on the expression of virulence-associated factors in the encapsulated strain D39 and its clpP (HYK302) and clpL (HYK304) mutants was determined by immunoblot analysis with antibodies against choline-binding protein A (CbpA), PsaA, pneumococcal surface protein A (PspA), pneumolysin, and autolysin (LytA). Unexpectedly, pneumolysin was induced after heat shock in the wild-type D39 as well as in the clpL mutant. PsaA was also induced slightly in D39 after heat shock, but it was not induced in the clpL mutant. In contrast, in the clpP mutant, CbpA was induced, but expression of pneumolysin and PsaA was decreased (Fig. 6). PspA and LytA levels did not change after heat shock regardless of genetic background (result not shown). To confirm the increase of pneumolysin expression after heat shock, the hemolytic activity of the pneumolysin in cell lysates was determined. Although pneumolysin activity was increased 1.8-fold in D39 after heat shock, it was not increased in the clpP mutant (Table 3). These results demonstrated that heat shock increased pneumolysin expression in the wild type and the clpL mutant, possibly contributing to a potential gain in virulence upon stress challenge, but this did not occur in the clpP mutant.
FIG. 6.
Induction of virulence-associated genes by heat shock. Exponentially growing encapsulated S. pneumoniae D39 (A600 = 0.1) and its isogenic clpP (HYK302) and clpL (HYK304) derivatives were heat shocked at 42°C for 20 min. Then 0.6 ml of culture was centrifuged, and the cell pellets were resuspended in lysis buffer, followed by boiling for 3 min. Subsequently, cell lysates were subjected to immunoblot analysis with a mixture of polyclonal antisera raised against CbpA, pneumolysin, and PsaA. The relative positions of CbpA, pneumolysin (Ply), and PsaA are indicated.
TABLE 3.
Effect of heat shock on hemolytic activity of pneumolysina
| Strain | Hemolytic units
|
% increase after heat shock | |
|---|---|---|---|
| 30°C | 42°C | ||
| D39 | 9,331 ± 2,347 | 16,515 ± 4,592 | 177* |
| HYK304 (clpL mutant) | 10,210 ± 1,429 | 13,477 ± 3,155 | 132 |
| HYK302 (clpP mutant) | 13,063 ± 2,859 | 15,414 ± 2,489 | 118 |
Hemolytic activity in cultures equivalent to an A600 of 1. Fifty microliters of cell lysate was serially diluted (1:1) into 50 μl of phosphate-buffered saline. Then 50 μl of a 1.5% suspension of human red blood cells was added to each well. The plate was incubated for 30 min at 37°C. The hemolytic units were calculated from the well in which 50% hemolysis had occurred. Values are the means ± standard deviation of three independent experiments. *, P < 0.05.
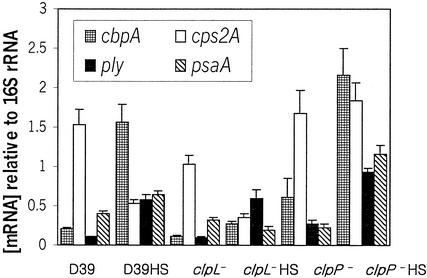
In Yersinia enterocolitica, the ClpP protease repressed the expression of both ail transcript level and cell surface-expressed Ail protein (36). This prompted us to examine the modulation of virulence gene expression at the mRNA level in S. pneumoniae. RNA was prepared from cultures, and the mRNA levels of ply, cbpA, psaA, and the capsule synthesis gene cps2A were determined by RT-PCR (Fig. 7). At 30°C, expression of cbpA in the clpL mutant was decreased relative to that of D39 (P = 0.001) but increased in the clpP mutant (P = 0.01). Although no significant changes in expression of ply and psaA in the wild-type and clpL mutant strains were detected, in the clpP mutant, expression of ply was increased 2.5-fold (P < 0.01), but expression of psaA was decreased by half (P < 0.01). After heat shock, cbpA mRNA levels were increased 7.48-, 2.39-, and 3.48-fold (P < 0.001, P < 0.001, P = 0.001, respectively) relative to 30°C levels in D39 and the clpL and clpP mutants, respectively. Similarly, the mRNA levels of ply were increased 5.27-, 6.0-, and 3.48-fold relative to 30°C in D39 and the clpL and clpP mutants, respectively (P < 0.001 in all cases).
FIG. 7.
Relative mRNA concentrations of cbpA, cps2A, ply, and psaA in D39 and the clpL and clpP mutants before and after heat shock as determined by real-time RT-PCR. Between RNA extracts, levels of individual mRNA species were corrected with reference to that obtained for the internal 16S rRNA control. Data points represent means ± standard deviation of quadruplicate samples from each RNA extract.
After heat shock, expression of cps2A was significantly decreased relative to the 30°C levels in both D39 and the clpL mutant (P = 0.001 in both cases). The expression of cps2A in the clpP mutant was increased after heat shock, but the increase was not statistically significant. In contrast, heat shock increased the mRNA level of psaA 1.6- and 5.04-fold in D39 and the clpP mutant, respectively (P < 0.01 and P = 0.001, respectively), but it was decreased in the clpL mutant (P < 0.01; Fig. 7). These results suggest that the clpL mutation may negatively affect the expression of psaA, whereas the clpP mutation may positively affect the expression of cps2A in some unknown way. These findings provide evidence that clpL and clpP as well as heat shock modulate a variety of virulence-associated genes to cope with new environmental challenges.
Effect of clpL and clpP mutations on virulence.
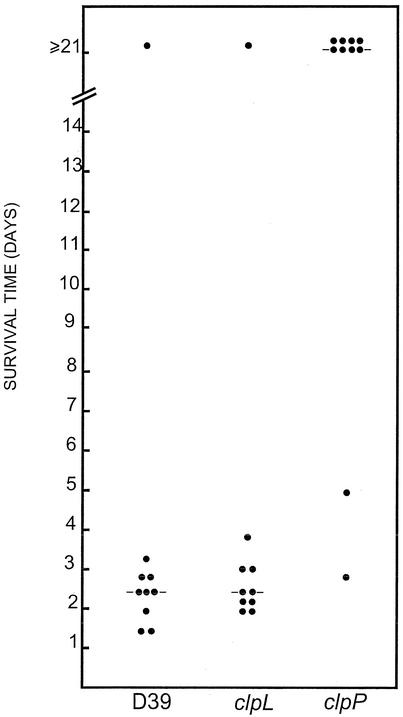
To further investigate the effect of clpL and clpP mutations on the virulence of D39, the survival time of mice after intraperitoneal infection with ca. 105 CFU of pneumococci was measured. The median survival time for mice in the groups infected with the parent strain (D39) and the clpL mutant was 55 h and 60 h, respectively. This difference was not statistically significant. However, the group of mice infected with the clpP mutant became sick 2 to 3 days postinfection, but most gradually recovered 4 to 5 days postinfection. Only two mice challenged with the clpP mutant died after 67 and 119 h (Fig. 8). The differences in median survival time and overall survival between the group infected with the clpP mutant and the groups infected with either D39 or the clpL mutant were highly significant (P ≪ 0.001 in all cases). This result indicates that ClpP function is critical for virulence factor expression in S. pneumoniae.
FIG. 8.
Survival times of mice after intraperitoneal challenge. Groups of 10 BALB/c mice were infected with approximately 105 CFU of D39 or its clpP (HYK302) or clpL (HYK304) derivative. Each point represents one mouse. The horizontal lines denote the median survival time for each group.
DISCUSSION
In this study, we identified an ATP-dependent Clp protease, AAK74513, in S. pneumoniae as the clpL homologue. ClpL homologues have been identified in several gram-positive organisms (L. lactis X62333, S. aureus AP003365, AP003137, S. pyogenes AE006538 and AE004092, and Lactobacillus rhamnosus AF323526) but not in gram-negative organisms, and so ClpL, like ClpE, seems to be specific to gram-positive organisms (11).
Using scanning densitometry of immunoblots, we found that S. pneumoniae expressed high basal levels of DnaK, GroEL, and ClpP but not ClpL at 30°C. These levels increased up to twofold upon exposure of the organism to heat shock over a 40-min period. However, pulse labeling of proteins for 10 min with [35S]methionine demonstrated rapid and transient induction of all the HSPs, indicating that DnaK, GroEL, and ClpP were expressed constitutively in large amounts at 30°C. Moreover, persistence of ClpL, DnaK, and GroEL upon return to 30°C indicates that HSPs do not appear to be actively degraded upon return to normal culture conditions. Since HSPs function as chaperones and promote renaturation of unfolded proteins (21) and are induced during infection in a wide variety of bacterial pathogens (5, 12, 13), survival in vivo could be enhanced by the stabilizing effect of HSPs on bacterial macromolecular complexes in hostile environments (5). Therefore, persistence of the HSPs upon return to normal conditions and induction of virulence proteins such as PsaA and pneumolysin by heat shock might contribute to or enhance the virulence of the pneumococcus. The major HSP, DnaK, is highly immunogenic in S. pneumoniae (18), and there is substantial evidence in the literature that HSPs are immunodominant antigens in infections by various pathogens (23). Whether the pathogenic life style of S. pneumoniae necessitates high levels of DnaK and ClpL and whether ClpL associates with the specific substrate and forms a complex with ClpP for proteolysis is the subject of an ongoing study with recombinant proteins.
It is well documented that mutation in HSP genes impacts on adherence and virulence in many pathogens. The stress-induced ClpP serine protease contributes to virulence in Salmonella enterica serovar Typhimurium (44) and modulates adhesion invasion locus (ail) gene expression in Yersinia enterocolitica (36). In Listeria monocytogenes, ClpP is essential for intracellular parasitism and virulence (14). Our results indicate that ClpP also plays an essential role in the virulence of S. pneumoniae and support the recent finding of Robertson et al. (37).
In this study, we demonstrated that the mRNAs for virulence associated genes such as cbpA, ply, and psaA were upregulated by heat shock. When gene expression in the wild type and the clp mutants at 30°C was compared, the clpL mutant exhibited almost the same expression pattern as the wild type for cbpA, ply, psaA, and cps2A, whereas the clpP mutant showed increased expression of cbpA but decreased expression of ply and psaA. Thus, clpP seems to act as a negative regulator for cbpA expression but a positive regulator for ply expression. Contrary to our observation, Chastanet et al. (7) reported that pneumolysin production was not affected by clpP mutation. This discrepancy might be due to a difference in measurement method for pneumolysin activity, as they assessed this qualitatively by observing hemolytic halos on blood agar plates, whereas we employed a quantitative hemolysis assay. Since pneumolysin is a proven virulence factor in pneumococcal bacteremia (3), increased expression after heat shock may be a contributing factor in pathogenesis. Although thermoregulation of virulence genes in the food-borne pathogen Listeria monocytogenes has been demonstrated (27), this is the first report of regulation of virulence genes by heat shock in the respiratory pathogen S. pneumoniae.
After heat shock, real-time RT-PCR data demonstrated an increase in ply expression in the clpP mutant, whereas immunoblot analysis and pneumolysin activity measurements revealed no increase. This inconsistency could be attributed to instability of ply mRNA at high temperatures in the clpP mutant. It is also conceivable that ClpP might act in activating nascent pneumolysin directly. Our immunoblot data also demonstrated that the clpP mutation resulted in high-level expression of ClpL regardless of heat shock, suggesting that ClpP may negatively regulate ClpL. This result corroborates a recent microarray study which also showed high induction of clpL at 37°C in a clpP mutant (37). Additionally, after heat shock, the level of expression of cps2A, the first gene in the capsule biosynthesis locus, was reduced in the wild type and clpL mutant, implying potentially lower resistance to the host immune system. In contrast, there was no reduction in the level of expression of cps2A in the clpP mutant. This result suggests that the clpP mutant ought to exhibit a wild-type level of resistance to host macrophages upon stress challenge, even though overall virulence was decreased. This may lead to the establishment of chronic bacteremia, in which the bacteria are able to evade the host immune system and survive in the host but unable to cause fulminant disease, a phenomenon that has been previously demonstrated for a pneumolysin-negative mutant of D39 (2, 3).
Virulence gene regulation could be modulated not only by heat shock but also by ClpL and ClpP proteases. The thermosensitivity of the clpL mutant as well as the refolding activity of denatured rhodanese by the recombinant ClpL provide evidence for a chaperone function of ClpL. Furthermore, clpP was demonstrated to play an essential role in regulation of ply and cbpA expression.
Acknowledgments
This work was supported by grants from the Korea Research Foundation (2000-015-FP0032) and the National Health and Medical Research Council of Australia.
Editor: A. D. O'Brien
REFERENCES
- 1.Avery, O. T., C. M. MacLeod, and M. McCarty. 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. Induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J. Exp. Med. 79:137-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benton, K. A., M. P. Everson, and D. E. Briles. 1995. A pneumolysin-negative mutant of Streptococcus pneumoniae causes chronic bacteremia rather than acute sepsis in mice. Infect. Immun. 63:448-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berry, A. M., J. Yother, D. E. Briles, D. Hansman, and J. C. Paton. 1989. Reduced virulence of a defined pneumolysin negative mutant of Streptococcus pneumoniae. Infect. Immun. 57:2037-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bricker, A. L., and A. Camilli. 1999. Transformation of a type 4 encapsulated strain of Streptococcus pneumoniae. FEMS Microbiol. Lett. 172:131-135. [DOI] [PubMed] [Google Scholar]
- 5.Buchmeier, N. A., and F. Heffron. 1990. Induction of Salmonella stress proteins upon infection of macrophages. Science 248:730-732. [DOI] [PubMed] [Google Scholar]
- 6.Charpentier, E., R. Novak, and E. Tuomanen. 2000. Regulation of growth inhibition at high temperature, autolysis, transformation and adherence in Streptococcus pneumoniae by clpC. Mol. Microbiol. 37:717-726. [DOI] [PubMed] [Google Scholar]
- 7.Chastanet, A., M. Prudhomme, J. P. Claverys, and T. Msadek. 2001. Regulation of Streptococcus pneumoniae clp genes and their role in competence development and stress survival. J. Bacteriol. 183:7295-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi, I. H., J. H. Shim, S. W. Kim, S. N. Kim, S. N. Pyo, and D. K. Rhee. 1999. Limited stress response in Streptococcus pneumoniae. Microbiol. Immunol. 43:807-812. [DOI] [PubMed] [Google Scholar]
- 9.Craig, E. A., B. D. Gambill, and R. J. Nelson. 1993. Heat shock proteins: Molecular chaperones of protein biogenesis. Microbiol. Rev. 57:402-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derre, I., G. Rapoport, and T. Msadek. 2000. The CtsR regulator of stress response is active as a dimer and specifically degraded in vivo at 37°C. Mol. Microbiol. 38:335-347. [DOI] [PubMed] [Google Scholar]
- 11.Derre, I., G. Rapoport, K. Devine, M. Rose, and T. Msadek. 1999. ClpE, a novel type of HSP100 ATPase, is part of the CtsR heat shock regulon of Bacillus subtilis. Mol. Microbiol. 32:581-593. [DOI] [PubMed] [Google Scholar]
- 12.Dorman, C. J., N. N. Bhriain, and C. F. Higgins. 1990. DNA supercoiling and environmental regulation of virulence gene expression in Shigella flexneri. Nature 344:789-792. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez, R. C., S. M. Logan, S. H. Lee, and P. S. Hoffman. 1996. Elevated levels of Legionella pneumophila stress protein Hsp60 early in infection of human monocytes and L929 cells correlate with virulence. Infect. Immun. 64:1968-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaillot, O., E. Pellegrini, S. Bregenholt, S. Nair, and P. Berche. 2000. The ClpP serine protease is essential for the intracellular parasitism and virulence of Listeria monocytogenes. Mol. Microbiol. 35:1286-1294. [DOI] [PubMed] [Google Scholar]
- 15.Gerth, U., A. Wipat, C. R. Harwood, N. Carter, P. T. Emmerson, and M. Hecker. 1996. Sequence and transcriptional analysis of clpX, a class-III heat-shock gene of Bacillus subtilis. Gene 181:77-83. [DOI] [PubMed] [Google Scholar]
- 16.Gerth, U., E. Kruger, I. Derre, T. Msadek, and M. Hecker. 1998. Stress induction of the Bacillus subtilis clpP gene encoding a homologue of the proteolytic component of the Clp protease and the involvement of ClpP and ClpX in stress tolerance. Mol. Microbiol. 28:787-802. [DOI] [PubMed] [Google Scholar]
- 17.Gottesman, S., C. Squires, E. Pichersky, M. Carrigton, M. Hobbs, J. S. Mattick, B. Dalrymple, H. Kuramitsu, T. Shiroza, T. Foster, W. P. Clark, B. Ross, C. L. Squires, and M. R. Maurizi. 1990. Conservation of the regulatory subunit for the Clp ATP-dependent protease in prokaryotes and eukaryotes. Proc. Natl. Acad. Sci. USA 87:3513-3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamel, J., D. Martin, and B. B. Brodeur. 1997. Heat shock response of Streptococcus pneumoniae: identification of immunoreactive stress proteins. Microb. Pathog. 23:11-21. [DOI] [PubMed] [Google Scholar]
- 19.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 20.Havarstein, L. S., G. Coomaraswamy, and D. A. Morrison. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 92:11140-11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hendrix, J. P., and F.-U. Hartl. 1993. Molecular chaperone functions of heat shock proteins. Annu. Rev. Biochem. 62:349-384. [DOI] [PubMed] [Google Scholar]
- 22.Huang, D. C., X. F. Huang, G. Novel, and M. Novel. 1993. Two genes present on a transposon-like structure in Lactococcus lactis are involved in a Clp-family proteolytic activity. Mol. Microbiol. 7:957-965. [DOI] [PubMed] [Google Scholar]
- 23.Kaufmann, S. H. E., and B. Schoel. 1994. Heat shock proteins as antigens in immunity against infection and self, p. 495-532. In R. I. Morimoto, A. Tissieres, and C. Georgopoulos (ed.), Biology of heat shock proteins and molecular chaperones. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Kim, J. O., and J. N. Weiser. 1998. Association of intrastrain phase variation in quantity of capsular polysaccharide and teichoic acid with the virulence of Streptococcus pneumoniae. J. Infect. Dis. 177:368-377. [DOI] [PubMed] [Google Scholar]
- 25.Kudlicki, W., A. Coffman, G. Kramer, and B. Hardesty. 1997. Renaturation of rhodanese by translational elongation factor (EF) Tu. Protein refolding by EF-Tu flexing. J. Biol. Chem. 272:32206-32210. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 27.Leimeister-Wachter, M., E. Domann, and T. Chakraborty. 1992. The expression of virulence genes in Listeria monocytogenes is thermoregulated. J. Bacteriol. 174:947-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindemann, J., R. Leiacker, G. Rettinger, and T. Keck. 2002. Nasal mucosal temperature during respiration. Clin. Otolaryngol. 27:135-139. [DOI] [PubMed] [Google Scholar]
- 29.Lock, R. A., Q. Y. Zhang, A. M. Berry, and J. C. Paton. 1996. Sequence variation in the Streptococcus pneumoniae pneumolysin gene affecting haemolytic activity and electrophoretic mobility of the toxin. Microb. Pathog. 21:71-83. [DOI] [PubMed] [Google Scholar]
- 30.Lund, P. A. 2001. Microbial molecular chaperones. Adv. Microb. Physiol. 44:93-140. [DOI] [PubMed] [Google Scholar]
- 31.Mekalanos, J. J. 1992. Environmental signals controlling expression of virulence genes determinants in bacteria. J. Bacteriol. 174:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mendoza, J. A., E. Rogers, G. H. Lorimer, and P. M. Horowitz. 1991. Unassisted refolding of urea unfolded rhodanese. J. Biol. Chem. 266:13587-13591. [PubMed] [Google Scholar]
- 33.Neidhardt, F. C., and R. A. VanBogelen. 1987. Heat shock response, p. 1334-11345. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. ASM Press, Washington, D.C.
- 34.Ogunniyi, A. D., R. L. Folland, D. E. Briles, S. K. Hollingshead, and J. C. Paton. 2000. Immunization of mice with combinations of pneumococcal virulence proteins elicits enhanced protection against challenge with Streptococcus pneumoniae. Infect. Immun. 68:3028-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogunniyi, A. D., P. Giammarinaro, and J. C. Paton. 2002. The genes encoding virulence-associated proteins and the capsule of Streptococcus pneumoniae are upregulated and differentially expressed in vivo. Microbiology 148:2045-2053. [DOI] [PubMed] [Google Scholar]
- 36.Pederson, K. J., S. Carlson, and D. E. Pierson. 1997. The ClpP protein, a subunit of the Clp protease, modulates ail gene expression in Yersinia enterocolitica. Mol. Microbiol. 26:99-107. [DOI] [PubMed] [Google Scholar]
- 37.Robertson, G. T., W. L. Ng, J. Foley, R. Gilmour, and M. E. Winkler. 2002. Global transcriptional analysis of clpP mutations of type 2 Streptococcus pneumoniae and their effects on physiology and virulence. J. Bacteriol. 184:3508-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saizieu, A., U. Certa, J. Warrington, C. Gray, W. Keck, and J. Mous. 1998. Bacterial transcript imaging by hybridization of total RNA to oligonucleotide arrays. Nat. Biotechnol. 16:45-48. [DOI] [PubMed] [Google Scholar]
- 39.Schirmer, E. C., J. R. Glover, M. A. Singer, and S. Lindquist. 1996. HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem. Sci. 21:289-296. [PubMed] [Google Scholar]
- 40.Sorbo, B. H. 1953. Crystalline rhodanese. I. Purification and physicochemical examination. Acta Chem. Scand. 7:1129-1136. [Google Scholar]
- 41.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple. H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 42.Tuomanen, E. 1999. Molecular and cellular biology of pneumococcal infection. Curr. Opin. Microbiol. 2:35-39. [DOI] [PubMed] [Google Scholar]
- 43.Vasseghi, H., and J. P. Claverys. 1983. Amplification of a chimeric plasmid carrying an erythromycin-resistance determinant introduced into the genome of Streptococcus pneumoniae. Gene 21:285-292. [DOI] [PubMed] [Google Scholar]
- 44.Webb, C., M. Moreno, M. Wilmes-Riesenberg, R. Curtiss III, and J. W. Foster. 1999. Effects of DksA and ClpP protease on sigma S production and virulence in Salmonella typhimurium. Mol. Microbiol. 34:112-123. [DOI] [PubMed] [Google Scholar]
- 45.Wickner, S., S. Gottesman, D. Skowyra, J. Hoskins, K. McKenney, and M. R. Maurizi. 1994. A molecular chaperone, ClpA, functions like DnaK and DnaJ. Proc. Natl. Acad. Sci. USA 91:12218-12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Willett, H. P. 1992. Streptococcus pneumoniae, p. 432-442. In W. K. Joklik, H. P. Willet, D. B. Amos, and C. M. Wilfert (ed.), Zinsser's microbiology. Prentice-Hall International, London, England.