Abstract
Co-immunoprecipitation experiments in cell extract from cultured cells or target tissues indicated that estrogen receptor was complexed with the retinoblastoma binding protein RIZ in a ligand-dependent manner. Mapping of interaction sites indicated that in both proteins the same regions and motifs responsible for the interaction of transcriptional co-activator and nuclear receptors were involved. In cultured cells, estradiol induced a redistribution of RIZ protein within the nucleus and in the cytoplasm. A similar effect was produced in vivo, in prepuberal rat endometrium, by administration of a physiological dose of estradiol. Therefore, RIZ protein could be a specific effector of estrogen action downstream of the hormone-receptor interaction, presumably involved in proliferation control.
A mandatory step in the mechanism of action of nuclear receptor is the binding to specific sequences present in the regulatory region of controlled genes, the hormone responsive element, to achieve transcriptional regulation of target genes (1). However, binding of the estrogen receptor to the estrogen-responsive element is not ligand-dependent (2). Occupation of the hormone-binding site modulates the accessibility of the receptor DNA-binding domain to DNA and the transcriptional activation function (3). Protein-protein interaction with components of the transcription initiation complex and/or with adapters bridging receptor and transcription initiation complex mediates transcriptional activation (4–11). A subset of these proteins cooperating in the estrogen signaling transduction pathway contains a LXXLL motif that interacts with the AF-2 core sequence in the estrogen receptor hormone binding domain (12). In a recent report, we presented evidence that a protein was able to bestow estrogen-inducibility to a promoter containing an incomplete estrogen-responsive element (13). This protein, co-immunoprecipitated from MCF-7 cell extract by a monoclonal antibody to estrogen receptor, was identified as the retinoblastoma-interacting zinc-finger protein RIZ.
The RIZ protein has been identified for its ability to bind the retinoblastoma gene product (14). It is a 220-kDa protein, containing eight zinc finger motifs. RIZ is mainly expressed in neuroendocrine tissues, with a ubiquitous low level of expression. It has independently been recognized as G3B, a GATA-3 factor binding protein (15), or as MTB-Zf, a transcription factor for the heme-oxygenase-1 gene (16). The interaction with the retinoblastoma protein (Rb) suggests that RIZ could be involved in the control of proliferation by an alternative mechanism. RIZ protein binds Rb with a cr2 motif, containing the core sequence LXCXE, common to the Rb-binding domain of the oncogenic products of some DNA tumor viruses. The cr2 motif is located in an acidic region near the N terminus of RIZ protein and is responsible for the binding to the pocket domain of Rb. In addition, RIZ protein shares an antigenic epitope with the C terminus of adenovirus E1A protein, suggesting evolutionary relationship among these proteins (17). The structural and antigenic similarities shared by RIZ protein and E1A suggested a role for RIZ protein as endogenous ligand of Rb.
RIZ gene encodes two protein products, RIZ1 and RIZ2, that differ for a motif present in the N-terminal domain (18, 19). This motif, defined as PR domain and lacking in RIZ2 protein, is structurally related to a similar domain present in the myeloid leukemia gene MDS1-EVI1 and in the transcription repressor/differentiation factor PRDI-BF1/BLIMP1. These gene products appear to function as negative regulators of tumor growth. RIZ1 expression was reduced or absent in human malignant tissue and tumor cell lines whereas increased expression of RIZ2 was detected (20). This suggests that the function of RIZ protein is within the control mechanism of cell proliferation. In addition, RIZ gene was physically mapped to chromosome 1q36, a region that undergoes deletions, rearrangements, or loss-of-heterozygosity in a broad spectrum of human tumors, including breast cancer (16, 21). For these reasons, RIZ gene was suggested to be a tumor suppressor.
Aside of the functional control of target cells, estradiol is a potent mitogen for breast and uterine epithelia, and, in this way, it is responsible for tumor-promoting action on breast and endometrial neoplasm (22). The hypothesis that the mitogenic effect was solely mediated by hormone-regulated autocrine growth factor secretion (23) has been contradicted by the evidence that estrogens can directly promote G1 progression through direct activation of cell cycle genes (24) or through the ras-MAP kinase pathway (25). Here we show that a physiological concentration of estradiol is specifically able to induce the binding of estrogen receptor to RIZ protein both in a target tissue and in cultured cells. This interaction was reproducible in vitro and involved functional regions of both proteins. By this new property, RIZ protein becomes an important target of estradiol action, and, because of its ability to complex with Rb protein, it is reasonable to speculate that it could mediate the proliferation and growth induced by estradiol in some target tissues.
Materials and Methods
Antibodies.
Preparations of monoclonal antibodies AER314, AER 311, and AER317 to estrogen receptor (26) or RZ1745 and RZ1802 to RIZ protein (13) are described elsewhere. Hybridoma cell supernatant contained 20–30 μg/ml of specific antibody. Rabbit polyclonal antibodies aER (amino acids 154–171) to estrogen receptor (amino acids 154–171) and the monoclonal antibody to proliferating cell nuclear antigen were from Sigma. Antibodies to Rb were 14001a (monoclonal) from PharMingen and C-15 (rabbit polyclonal) from Santa Cruz Biotechnology.
Preparation of Cell and Tissue Extracts.
MCF-7 cells were grown in 140-mm diameter Petri dishes or in 175-cm2 flasks in DMEM supplemented with 5% FCS, 7 ng/ml (0.2 units/ml) bovine insulin, and 1 nM cortisol in a 5% CO2 environment. All tissue culture reagents were from GIBCO. Subconfluent cells were cultured for an additional 5 days in DMEM without phenol red, serum, and hormones. The medium was changed every day during this time. The cells were then treated with hormones as described in the legends to the figures. The total cellular lysate (27) and nuclear extract (28) were prepared as described elsewhere. Phosphocellulose chromatography was performed as described elsewhere (29). Alternatively, rat uteri were removed from rats killed by cervical dislocation, were incubated for 10 min in complete protease inhibitor mixture (Boehringer Mannheim), and were homogenized in 5 volumes of ice-cold buffer A (10 mM Hepes, pH 7.9/10 mM KCl/0.5 mM DTT/1.5 mM MgCl2) by 10 5-sec bursts of Ultraturrax homogenizer (Jane & Kunkel, Staufen, Germany). Nuclear extract was prepared as described (28). All nuclear extracts were dialyzed versus buffer D [20 mM Hepes, pH 7.8/100 mM KCl/0.5 mM DTT/0.1 mM EDTA/15% (vol/vol) glycerol/10% (wt/vol) sucrose/0.5 mM phenylmethylsulfonylfluoride] and were clarified at 14,000 × g for 15 min. Protein concentration was usually between 8 and 10 mg/ml.
Immunoprecipitation.
Purified monoclonal antibodies (24–60 ng/μg of resin) or hybridoma cell supernatants (40 μl/μg of resin) were incubated with CNBr-activated Sepharose coupled with anti-mouse IgG (Pharmacia Biotech, Uppsala). The samples (≈0.8 mg of protein from total cell extract, nuclear extract, or phosphocellulose column fractions) were incubated overnight at 4°C with 12.5 μg of the antibody-loaded resin. At the end of incubation, resins were washed with 40 volumes of 20 mM Hepes (pH 7.9), 2 mM MgCl2, 0.1 mM EDTA, 100 mM KCl, 1 mM DTT, 10% (vol/vol) glycerol, and 0.1% Nonidet P-40 (once for 10 min and three times for 2 min). Proteins were eluted with 2 volumes of Laemmli sample buffer additioned with 10 mM EDTA and analyzed by PAGE in the presence of SDS (SDS/PAGE).
Western Blot Analysis.
SDS/PAGE was performed according to published methods (30). Molecular weight markers were from Bio-Rad (Hercules, CA). Eight percent acrylamide gels were prepared with a constant 1/40 ratio mono-/bis-acrilammide. Western blot analysis was performed as reported (29).
Two-Hybrid Assay.
The two-hybrid assay (Matchmaker Two-Hybrid System) was purchased form CLONTECH. pGAD424(+1) was obtained by digestion of pGAD424 with EcoRI, followed by filling and ligation. pGEX-M1 and pGEX-M2 were obtained from pGEX-RIZ, introducing a frame-shift by opening the unique ClaI or NdeI sites, respectively, filling, and ligation. pGEX-M4 was obtained from pGEX-RIZ by deletion of a NgoM1 fragment, filling, and ligation. Filling was necessary to introduce a nearer stop codon, being the second NgoM1 site in the 3′ nontranslated region. pGAD-RIZwt, pGAD-M1, pGAD-M2, and pGAD-M10 were obtained by introducing in the BamHI site of pGAD424(+1) the BamHI insert obtained from pGEX-RIZ, pGEX-M1, pGEX-M2, and pGEX-M10, respectively. pGAD-M3 was obtained by introducing the EcoRI fragment of pGEX-M2 in the EcoRI site of pGAD424. pGAD-M5wt was obtained from pGAD-M3 by deletion of the SmaI fragment and ligation. Point mutations in pGAD-M5 were introduced in the EcoRI-SmaI insert. Two PCR reactions were performed by using Pfu polymerase and a partially degenerated oligonucleotide (GCTGCCTCCTGYCTTGATCCCCA) or its complement in combination with T3 or T7 primers. The amplification products were annealed and made double-stranded with Pfu polymerase. The product was digested with EcoRI and SmaI and was cloned in pGAD424. By sequence analysis of the clones, pGAD-M5Val965 and pGAD-M5Ala965 were selected. pGBT-HEG14 was obtained by cloning in the SmaI site of the pGBT9 plasmid the SmaI-EcoRI fragment, was digested from the pGEX/hER281-595 (29), and was filled. pGBT-M1HD536-547 and pGBT-M2H-ALA were obtained by cloning the BamHI fragments, digested from the pGST-ER(DEF)D536–547 and from pGST-ER(DEF)M543A/L544A, respectively (11), in the BamHI-SmaI site of the pGBT9(-1) plasmid.
Immunocytochemistry.
MCF-7 cells were grown on slides for immunofluorescence (BioMerieux sa, Marcy l'Etoile, France) in DMEM supplemented with 5% FCS (heat inactivated), 2 mM l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 50 μg/ml gentamicin in a 5% CO2 environment. Cell growth in subconfluent cultures was arrested by replacing the growth medium with phenol red-free medium for 24 h. Cells were then incubated for 48 h in serum-free medium containing estradiol or other agents, as indicated. Cells were cultured for 5 further days in DMEM containing 5% charcoal-treated FCS. Cells were fixed with 4% (wt/vol) formaldehyde in PBS for 10 min, were permeabilized with 0.2% Triton X-100 in PBS for 10 min (where indicated), and were blocked with PBS containing 1% goat serum and 5% BSA for 1 h. Cells were then stained overnight with antibody RZ1802 to RIZ (diluted 1:20) or to proliferating cell nuclear antigen (diluted 1:3,000) and were washed twice with PBS. Cells were then incubated with secondary FITC-conjugated goat anti-mouse antibody (Ortho Diagnostics) (diluted 1:40) for 45 min, followed by two washes with PBS. Slides were mounted in Fluo prep mounting medium (BioMerieux).
Fragments of rat uteri were formalin-fixed and paraffine-embedded. Sections were deparaffined, then were treated with 0.1% hydrogen peroxide for 15 min at 4°C, were air-dried, and were blocked with PBS containing 2% normal goat serum and 5% BSA. After three washes with PBS, sections were incubated overnight at 4°C with RZ1723 hybridoma cell supernatant or control antibodies. After three washes with PBS, sections were incubated with biotinylated goat anti-mouse IgG (Boehringer Mannheim) for 1 h at room temperature and were further processed with ABC kit (Vector Laboratories), following the manufacturer's instructions. Slides were washed in water, were dehydrated with ethanol and xylene, and then were mounted with Eukitt mounting medium (O. Kindler GmbH, Freiburg, Germany).
Flow Cytometry.
Cell were harvested with PBS containing 1 mM EDTA and washed once with Cell Wash (Beckton Dickinson) at 1,200 rpm. Permeabilization was performed by incubation of 106 cell in 0.5 ml of FACS permeabilizing solution (Beckton Dickinson) for 20 min at 4°C in the dark. Cells were then washed with Cell Wash, were incubated for 30 min at 4°C with antibody RZ1802 to RIZ protein (diluted 1:100), and were washed twice with Cell Wash. Cells were then incubated with secondary FITC-conjugated goat anti-mouse antibody (Ortho Diagnostics) (diluted 1:40) for 30 min at 4°C, were washed again, and were resuspended in 0.1 ml of PBS. Flow cytometry data were collected by using a FACScalibur instrument and were analyzed by using cell quest software (Beckton Dickinson).
Results
Ligand Effect on RIZ Protein–Estrogen Receptor Interaction.
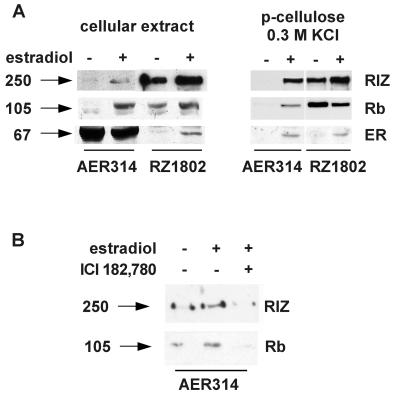
Proteins were immunoprecipitated from a total cell extract of MCF-7 cell with the AER317 antibody to estrogen receptor or with the RZ1802 antibody to RIZ protein and were analyzed by Western blot with the same antibodies. Cells were grown in a hormone-deprived medium or were treated with estradiol for 20 h. The result, presented in Fig. 1A, indicated that antibodies to estrogen receptor were able to co-immunoprecipitate RIZ protein and vice versa. A very faint band of RIZ protein was co-immunoprecipitated with estrogen receptor in the absence of hormone whereas in cells treated with estradiol the signal was much stronger. The variable ratio of the estrogen receptor/RIZ protein co-immunoprecipitated by the two different antibodies indicated that only a fraction of the receptor molecules present in the extract were complexed with RIZ protein and vice versa. Phosphocellulose chromatography of the nuclear extract from hormone-deprived cells separated RIZ protein, eluting at 0.3 M KCl, from estrogen receptor, eluting in the 0.5 M KCl fraction. However, in the 0.3 M KCl eluate of extract from cells treated with estradiol, part of estrogen receptor was detectable by Western blot analysis (data not presented). This was co-immunoprecipitated by antibodies to RIZ protein and antibodies to estrogen receptor co-immunoprecipitated RIZ protein (Fig. 1A). The same ratio of band intensity confirmed that all of the receptor molecules present in this fraction were complexed with RIZ protein, indicating, therefore, that estrogen receptor modified its elution properties being complexed to RIZ protein. Analysis of the same blots with the antibodies to Rb showed that it was co-immunoprecipitated by both antibodies and that estradiol treatment increased the amount of Rb co-immunoprecipitated with estrogen receptor. This evidence was consistent with the described Rb-binding property of RIZ protein, but it also indicated that Rb was present in multiprotein complexes with estrogen receptor in the extract of hormone-deprived cells. When cells were incubated in the presence of both estradiol and anti-estrogen ICI182,780 in the cell extract, RIZ protein was not co-immunoprecipitated with estrogen receptor, and neither was Rb (Fig. 1B).
Figure 1.
Co-immunoprecipitation of estrogen receptor and RIZ protein from cell extracts. Arrows on the left side indicate the molecular mass (kDa) of bands calculated from migration of standard proteins. (A) MCF-7 cells were grown in charcoal-treated medium in the absence or presence of 10 nM estradiol for 20 h (lanes marked by minus or plus signs, respectively) before harvesting. Aliquots of cell extracts or of the 0.3 M KCl eluate from a phosphocellulose chromatography, as indicated on the top, were incubated with the monoclonal antibody AER314 to estrogen receptor or RZ1802 to RIZ protein, as indicated by the underlining bar. Immunoprecipitated proteins were analyzed by SDS/PAGE, followed by Western blot analysis with the antibodies RZ1802 to RIZ protein (RIZ), 14001A (cellular extract) or C-15 (p-cellulose) to Rb protein (Rb), and AER317 (cellular extract) or rabbit polyclonal antibodies αER (154–171) (p-cellulose) to estrogen receptor (ER), as indicated on the side. (B) MCF-7 cell were grown in charcoal-treated medium in the absence or presence of 10 nM estradiol without or with 30 nM ICI182,780 (lanes marked by minus or plus signs, respectively) for 20 h before harvesting. Aliquots of cell extracts were incubated with the monoclonal antibody to estrogen receptor AER314, and immunoprecipitated proteins were analyzed by SDS/PAGE followed by Western blot analysis with the antibody RZ1802 to RIZ protein (RIZ) or 14001A to Rb protein (Rb).
Identification of the Domains Involved in the Interaction.
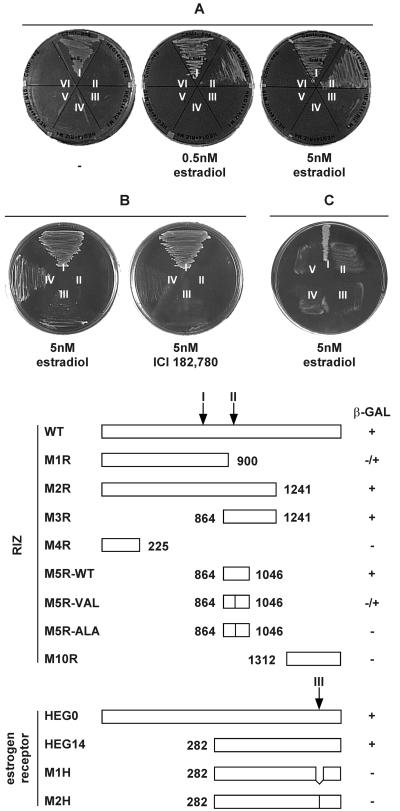
To evaluate whether estrogen receptor was involved in a direct interaction with RIZ protein, a yeast two-hybrid assay was implemented, using the full RIZ cDNA and a cDNA fragment encoding the hormone-binding domain of the human estrogen receptor (amino acids 281–595). Yeast cells only grew in the presence of estradiol, in a dose-dependent manner, but not in the presence of the anti-estrogen ICI182,780 (Fig. 2 A and B) or tamoxifen (not presented), thus confirming that the interaction was not mediated by other protein moieties and that, even in vitro, the estradiol effect was specific. By using RIZ deletion mutants in the same experimental design, the site of interaction was mapped between amino acids 864 and 1,046 of RIZ protein. In this region, there is a LXXLL motif, identified in transcriptional co-activators interacting with nuclear receptors (12). Site-directed mutagenesis of the first leucine codon in the M5 deletion fragment (amino acids 864–1,046) produced a partial (M5R-VAL, L→V mutation) or complete (M5R-ALA, L→A mutation) loss of binding (Fig. 2B). This result indicated that the LXXLL motif was involved in estrogen receptor binding and that hydrophobicity alone was not sufficient for this interaction. A second LXXLL motif at position 695–699 was apparently not involved in the interaction with the estrogen receptor, but it was presumably responsible of the weak growth of yeast cells co-transfected with the pGAD-M1R plasmid, encoding the first 900 amino acids of RIZ protein. Estrogen receptor mutants that were unable to activate transcription were tested with full RIZ cDNA in the same assay. Deletion of amino acids 535–547 (M1HD536–547) or substitution of amino acids 543 and 544 (M2H-ALA, M→A and L→A mutations) disrupted the ability of HEG14 to interact with RIZ (Fig. 2C). A graphic representation of the tested mutants is reported on the bottom part of Fig. 2. These results confirmed that estrogen receptor region interacting with RIZ protein was the same involved in the interaction with transcriptional co-activators (4, 11).
Figure 2.
Interaction of estrogen receptor and RIZ protein in yeast two-hybrid system. (Upper) Growth of yeast cell on plates lacking leucine, tryptophan, and histidine and incubated in the presence of the estradiol or ICI182,780 at the concentration indicated below. (A) Cells were transfected with: I, pVA3 and pVT1; II, pGAD-M2R and pGBT-HEG14; III, pGAD-M1R and pGBT-HEG14; IV, pGAD-M4R and pGBT-HEG14; V, pGAD-M10 and pGBT-HEG14; VI, pGAD424 and pGBT-HEG14. (B) Cells were transfected with: I, pVA3 and pVT1; II, pGAD-M5RAla965 and pGBT-HEG14; III, pGAD-M5RVal965 and pGBT-HEG14; IV, pGAD-M5R and pGBT-HEG14. (C) Cell were transfected with: I, pGAD-M2R and pGBT-HEG14; II, pGAD and pGBT-M1HD536–547; III, pGAD and pGBT-M2H-ALA; IV, pGAD-M2R and pGBT-M1HD536–547; V, pGAD-M2R and pGBT-M2H-ALA. (Lower) Graphic representation of the mutants of RIZ protein co-transfected in yeast cells with the plasmid pHEG14 coding for a fragment of human estrogen receptor (marked as RIZ on the left) or of the mutants of estrogen receptor co-transfected in yeast cells with the plasmid coding for RIZ protein (marked as estrogen receptor on the left); β-galactosidase activity is reported on the right; arrows marked I and II indicate the position of the LXXLL motifs in the RIZ protein primary sequence; the arrow marked III indicates the position of the region interacting with transcriptional co-activators on estrogen receptor primary sequence; numbers indicate the first or last amino acids of the mutant.
Estrogen-Induced Redistribution of RIZ Protein in Cultured Cells.
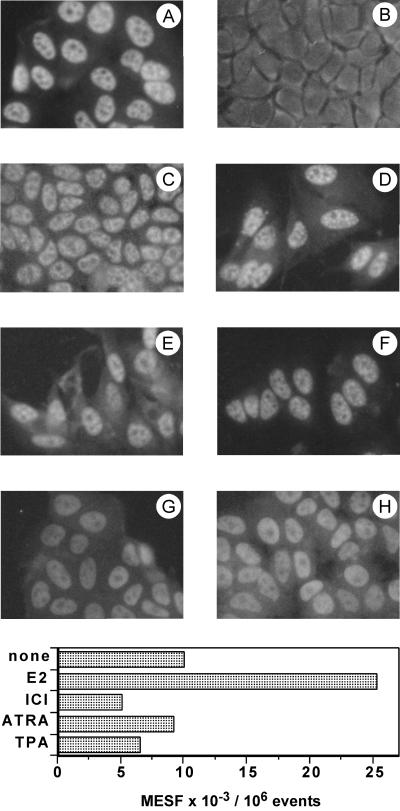
Immunofluorescence staining of serum-starved MCF-7 cells with the antibody RZ1802 indicated that RIZ was mostly localized in the nucleus (Fig. 3A). On estradiol treatment, the nuclear staining was dramatically reduced, and a moderate florescence appeared in the cytoplasm (Fig. 3B). The loss of nuclear staining was mainly attributable to epitope masking because detergent treatment restored the immunoreactivity as a patchy nuclear staining (Fig. 3C). No effect was visible in the estrogen-independent MDA cell line (not presented). Simultaneous treatment with estradiol and the anti-estrogen ICI182,780 prevented the effect (Fig. 3D). Another ligand of a nuclear receptor, retinoic acid, or 12-O-tetradecanoylphorbol-13-acetate did not modify the nuclear staining of RIZ (Fig. 3 E and F). Both molecules have an antiproliferative effect on MCF-7 cells (31–33). Localization of a different nuclear protein, proliferating cell nuclear antigen, was not modified by estradiol treatment (Fig. 3 G and H). Pretreatment of cells with a permeabilizing agent allowed the access of fluorescent antibodies to the antigen present in the cytoplasm for flow cytometry analysis. Cell treatment with the antiproliferative agents (12-O-tetradecanoylphorbol-13-acetate and retinoic acid) that preserved the nuclear localization of RIZ did not affect or slightly reduced the amount of measured fluorescence. On the other hand, the appearance of cytoplasmic immunoreactivity of RIZ protein on estradiol treatment was confirmed by a specific 2.5-fold increase of fluorescence in estradiol-treated cells that was prevented by the anti-estrogen (Fig. 3 Lower). The presented data are from a single experiment, but the increase of fluorescence caused by the presence of RIZ protein in the cytoplasm was repeatable and ranged between 2- and 5-fold.
Figure 3.
Immunofluorescence analysis of RIZ protein in MCF-7 cells. (Upper) Cells were incubated without hormones (A and G), with 50 nM estradiol (B, C, and H), with 10 nM estradiol and 10 μm ICI182,780 (D), with 1 μM all-trans-retinoic acid (E), or with 100 ng/ml 12-O-tetradecanoylphorbol-13-acetate (F). (A, B, and D–F) Staining with antibody to RIZ protein RZ1802. (C) Permeabilization with Triton X-100 followed by staining with antibody to RIZ protein RZ1802. (G and H) Staining with antibody to proliferating cell nuclear antigen. (Lower) Cells were grown in Petri dishes in hormone-free medium and, subsequently, were incubated with 50 nM estradiol (E2), with 10 nM estradiol and 10 μm ICI182,780 (ICI), with 1 μM all-trans-retinoic acid (ATRA), or with 100 ng/ml 12-O-tetradecanoylphorbol-13-acetate (TPA) or kept in hormone free-medium (none). Harvested cells were permeabilized and processed for flow cytometry as described in Materials and Methods. Results were measured as number of molecules of equivalent soluble fluorochrome (MESF).
In Vivo Effect of Estrogens on RIZ Protein.
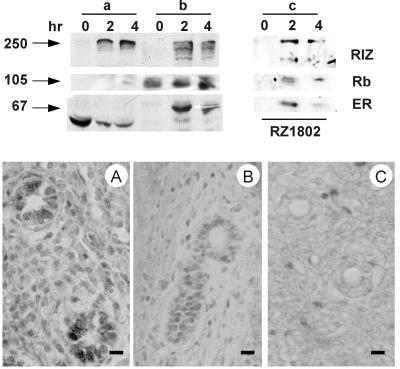
Estrogen treatment of prepuberal rat elicits a dramatic proliferative response in the uterus. As expected, the hormone treatment rapidly induced the partitioning of estrogen receptor from the cytosol to the nucleus (Fig. 4, a and b lanes). In the untreated control, RIZ protein was undetectable by Western blot analysis, in both cytosol and nuclear extract, whereas immunohistochemical analysis showed an intense nuclear staining. The discrepancy could be explained as a consequence of relative insolubility of RIZ protein present in the nuclei of unstimulated tissue. On hormone treatment, RIZ protein became detectable, by Western blot analysis, both in the cytosol and in the nuclear extract (Fig. 4, a and b lanes). Estrogen receptor was co-immunoprecipitated with RIZ protein in the nuclear extract after 2 and 4 h from estradiol injection (Fig. 4, c lanes). Western blot analysis of the same samples with an antibody to Rb protein indicated that Rb was always present in the nuclear extract and that it was co-immunoprecipitated by the antibody to RIZ protein. At 2 h after hormone injection, immunohistochemical staining with a monoclonal antibody to RIZ protein of endometrial cell nuclei was reduced, and it disappeared, in a time-dependent manner, after 4 h (Fig. 4, Lower). Taken together, the rat uterus and MCF-7 cell results indicated that the interaction between RIZ protein and estrogen receptor was induced by a physiological concentration of estradiol and mostly involved molecules present in the nuclear compartment.
Figure 4.
Co-immunoprecipitation of estrogen receptor and RIZ protein from nuclear extracts of immature rat uterus after estrogen treatment and immunohistochemical localization. (Upper) Arrows on the left side indicate the molecular mass (kDa) of bands calculated from migration of standard proteins; immature (21 days old) female Wistar rats were killed at the indicated times after the i.p. injection of 0.5 μg of estradiol in 50 μl of saline; the uterus was removed, and aliquots of the cytosol (a lanes) or of nuclear extract (b and c lanes) were either loaded onto the gel (a and b lanes) or were incubated with the monoclonal antibody to RIZ protein RZ1802 (c lanes). The samples were analyzed by SDS/PAGE followed by Western blot analysis with the antibodies RZ1802 to RIZ protein (RIZ), 14001A to Rb protein (Rb), or AER317 to estrogen receptor (ER), as indicated on the side. The deformed appearance of estrogen receptor bands in lanes a was caused by the presence of a large band of serum albumin migrating in the same region. (Lower) Immature (21 days old) female Wistar rats were killed after 4 h of the i.p. injection of 0 (A), 0.5 (B) ,or 2.5 (C) μg of estradiol in 50 μl of saline. Uterine tissue was treated as described in Materials and Methods. (Bar = 10 μm.)
Discussion
The ability of RIZ protein to complex with estrogen receptor was discovered by screening a HeLa cell expression library with a probe designed around a six-base motif (TTGGC). This sequence was recurrent in fragments selected from a random pool by repeated cycles of estrogen receptor-specific immunoprecipitation in the presence of a nuclear extract and PCR amplification [cyclic amplification and selection of target (CAST) for multiple elements] (13). Furthermore, in transfection experiments, RIZ protein was able to bestow estrogen-inducibility to a promoter containing an incomplete estrogen responsive element and the identified TTGGC motif (13). This interference might reflect a physiological function in the context of complex promoters containing incomplete estrogen responsive elements, establishing the point of integration of various intra or extra-cellular signals.
Data presented in this report indicate that interaction of estrogen receptor with RIZ gene product was ligand-dependent at physiological concentrations of hormone and was observable both in intact cells and in vitro. The hormone-binding domain of estrogen receptor binds the central domain of RIZ. The interaction involved the AF-2 domain of estrogen receptor. This is the same domain binding other transcriptional co-activators (4). RIZ protein binds estrogen receptors with a LXXLL motif, which is also present on many transcriptional co-activators (12).
Morphological observation of cultured cells and target tissue indicated that estradiol was able to specifically induce a redistribution of RIZ protein within the nucleus. In MCF-7 cells, the antigen clustered and became inaccessible to the antibody, unless the fixed slide was treated with a detergent before antibody incubation. This effect was estradiol-specific and receptor-mediated because it was prevented by an anti-hormone. It was specific for RIZ protein because it was not shared by another nuclear protein. A specific effect of estradiol on the nuclear staining of RIZ protein was observed in immature rat uterus in a dose-dependent manner at physiological concentrations of hormone. Furthermore, both immunofluorescence staining and flow cytometry indicated that estradiol treatment induced the appearance of RIZ protein in the cytoplasm of MCF-7 cells. We have no evidence indicating whether this effect relays on nuclear export or on de novo synthesis, but it supports the hypothesis that RIZ protein, being a tumor suppressor, needs to be removed from the nucleus, or sequestered within it, to allow cell proliferation. This could be a common feature of PR-domain containing proteins. In fact, the protein SC-1, another member of the same family, moves from the cytoplasm to the nucleus of COS cells transfected with p75 neurotrophyn receptor on differentiating stimuli, such as nerve growth factor treatment or serum deprivation (34). It is known that neurotrophyns promote cell differentiation and survival through the p75 receptor pathway. The intracellular distribution of PR-domain containing proteins might, therefore, be correlated to the proliferative state of the cell. The same consideration could be supported by the observed insolubility of RIZ protein from the nuclei of immature rat uterine cells. The behavior of RIZ in these cells is different from hormone-deprived MCF-7 cells, where RIZ protein was detectable in the nuclear extract. This difference might dwell on the different proliferative status of transformed cells arrested by hormone withdrawal compared with normal cells never “primed” by estradiol, where RIZ protein, being a negative regulator of proliferation, could be more tenaciously bound to its nuclear target.
On the basis of our data, we can assume that RIZ protein is an effector of estrogen action downstream of the hormone-receptor interaction. Given the molecular mass of RIZ and the number of conserved motifs and domains present on its primary sequence, it is conceivable that this protein is the point of divergence for different signal transduction pathways activated by estradiol in the target cell.
It is an old observation that some target tissues, such as endometrium or mammary epithelium, respond to estradiol stimulation with a dramatic increase in number of actively proliferating cells. The cellular response to this stimulus includes the activation of different pathways leading to a G1-S transition and, therefore, to new DNA and RNA synthesis. Direct and indirect activation of cell cycle genes has been postulated (24, 25), but the rapid enhancement of DNA-dependent synthesis of RNA, preceding any increase in protein synthesis, suggests that other intracellular pathways might be triggered by estradiol. Evidences presented in this report suggest that RIZ protein could be involved in an alternative intracellular pathway mediating the mitogenic effect of estradiol.
It is not known how Rb function is modified by RIZ protein. The hormone-dependent interaction of estrogen receptor with RIZ protein allows a new conjecture, based on the presented evidence of co-immunoprecipitation of Rb with estrogen receptor. It is conceivable that they form tertiary complexes and that, in this way, the gatekeeping function of Rb could be modified by estradiol. Estrogen action on quiescent cell is dramatic and goes beyond the transcriptional activation of regulated genes containing the consensus estrogen-responsive element. Enhancement of RNA synthesis (rRNA and tRNAs) in the uterus has been detected within minutes after estradiol administration to immature or ovariectomized animals and before any detectable protein synthesis (35). This suggests a direct effect of estrogens even on DNA-dependent RNA polymerase I and III. In this respect, co-immunoprecipitation of RIZ and Rb with estrogen receptor might indicate that estrogen receptor could affect, by way of RIZ, Rb function as repressor of RNA polymerase I and III activities (36, 37). This is an important mechanism, alternative to E2F inhibition, by which Rb restrains cell growth (38).
Acknowledgments
We thank Dr. Pier Luigi Bontempo and Ms. Anna Maria Tonti and Pasqualina Contatore for histochemical analysis of uterine tissue. This investigation was supported by the Italian Ministry for University and Scientific and Technological Research and, partially, by the Italian Association for Cancer Research (AIRC).
Abbreviation
- Rb
retinoblastoma protein
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.050015697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.050015697
References
- 1.Beato M. Cell. 1989;56:335–344. doi: 10.1016/0092-8674(89)90237-7. [DOI] [PubMed] [Google Scholar]
- 2.Medici N, Nigro V, Abbondanza C, Moncharmont B, Molinari A M, Puca G A. Mol Endocrinol. 1991;5:555–563. doi: 10.1210/mend-5-4-555. [DOI] [PubMed] [Google Scholar]
- 3.Beato M, Herrlich P, Schutz G. Cell. 1995;83:851–857. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- 4.Cavailles V, Dauvois S, Danielian P S, Parker M G. Proc Natl Acad Sci USA. 1994;91:10009–10013. doi: 10.1073/pnas.91.21.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halachmi S, Marden E, Martin G, MacKay H, Abbondanza C, Brown M. Science. 1994;264:1455–1458. doi: 10.1126/science.8197458. [DOI] [PubMed] [Google Scholar]
- 6.Ing N H, Beekman J M, Tsai S Y, Tsai M J, O'Malley B W. J Biol Chem. 1992;267:17617–17623. [PubMed] [Google Scholar]
- 7.Jacq X, Brou C, Lutz J, Davidson I, Chambon P, Tora L. Cell. 1994;79:107–117. doi: 10.1016/0092-8674(94)90404-9. [DOI] [PubMed] [Google Scholar]
- 8.Le Douarin B, Zechel C, Garnier J M, Lutz Y, Tora L, Pierrat P, Heery D, Gronemeyer H, Chambon P, Losson R. EMBO J. 1995;14:2020–2033. doi: 10.1002/j.1460-2075.1995.tb07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oñate S A, Tsai S Y, Tsai M J, O'Malley B W. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 10.Voegel J J, Heine M J S, Zechel C, Gronemeyer H, Chambon P. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 11.vom Baur E, Zechel C, Heery D M, Heine M J S, Garnier J M, Vivat V, Le Douarin B, Gronemeyer H, Chambon P, Losson R. EMBO J. 1996;15:110–124. [PMC free article] [PubMed] [Google Scholar]
- 12.Heery D M, Kalkhovren E, Hoare S, Parker M G. Nature (London) 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 13.Medici N, Abbondanza C, Nigro V, Rossi V, Piluso G, Belsito A, Gallo L, Roscigno A, Bontempo P, Puca A A, et al. Biochem Biophys Res Commun. 1999;264:983–989. doi: 10.1006/bbrc.1999.1604. [DOI] [PubMed] [Google Scholar]
- 14.Buyse I M, Shao G, Huang S. Proc Natl Acad Sci USA. 1995;92:4467–4471. doi: 10.1073/pnas.92.10.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shapiro V S, Lee P, Winoto A. Gene. 1995;163:329–330. doi: 10.1016/0378-1119(95)00420-b. [DOI] [PubMed] [Google Scholar]
- 16.Muraosa Y, Takahashi K, Yoshizawa M, Shibahara S. Eur J Biochem. 1996;235:471–479. doi: 10.1111/j.1432-1033.1996.00471.x. [DOI] [PubMed] [Google Scholar]
- 17.Buyse I M, Huang S. J Virol. 1997;71:6200–6203. doi: 10.1128/jvi.71.8.6200-6203.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie M, Shao G, Buyse I M, Huang S. J Biol Chem. 1997;272:26360–26366. doi: 10.1074/jbc.272.42.26360. [DOI] [PubMed] [Google Scholar]
- 19.Liu L, Shao G, Steele-Perkins G, Huang S. J Biol Chem. 1997;272:2984–2991. doi: 10.1074/jbc.272.5.2984. [DOI] [PubMed] [Google Scholar]
- 20.He L, Yu J X, Liu L, Buyse I M, Wang M-S, Yang Q-C, Nakagawara A, Brodeur G M, Shi Y E, Huang S. Cancer Res. 1998;58:4238–4244. [PubMed] [Google Scholar]
- 21.Buyse I M, Takahashi E, Huang S. Genomics. 1996;34:119–121. doi: 10.1006/geno.1996.0249. [DOI] [PubMed] [Google Scholar]
- 22.Davidson N E, Lippman M E. CRC Crit Rev Oncogen. 1989;1:89–111. [PubMed] [Google Scholar]
- 23.Dickson R B, Lippman M E. Endocr Rev. 1995;16:559–589. doi: 10.1210/edrv-16-5-559. [DOI] [PubMed] [Google Scholar]
- 24.Altucci L, Cicatiello L, Dauvois S, Parker M G, Truss M, Beato M, Sica V, Bresciani F, Weisz A. Oncogene. 1996;12:2315–2324. [PubMed] [Google Scholar]
- 25.Migliaccio A, Di Domenico M, Castoria G, de Falco A, Bontempo P, Nola E, Auricchio F. EMBO J. 1996;15:1292–1300. [PMC free article] [PubMed] [Google Scholar]
- 26.Abbondanza C, De Falco A, Nigro V, Medici N, Armetta I, Molinari A M, Moncharmont B, Puca G A. Steroids. 1993;58:4–12. doi: 10.1016/0039-128x(93)90011-b. [DOI] [PubMed] [Google Scholar]
- 27.Lee W S, Kao C C, Bryant G O, Liu X, Berks A J. Cell. 1991;67:365–376. doi: 10.1016/0092-8674(91)90188-5. [DOI] [PubMed] [Google Scholar]
- 28.Dingam J D, Lebivitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abbondanza C, Rossi V, Roscigno A, Gallo L, Belsito A, Piluso G, Medici N, Nigro V, Molinari A M, Moncharmont B, Puca G A. J Cell Biol. 1998;141:1301–1310. doi: 10.1083/jcb.141.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 31.Dawson M I, Chau W-R, Pine P, Jong L, Hobbs P D, Rudd C K, Quick T C, Niles R M, Zhang X-K, Lombardo A, et al. Cancer Res. 1995;55:4446–4451. [PubMed] [Google Scholar]
- 32.Zhou W-Y, Jones C S, Kiss A, Matsukuma K, Amin S, De Luca L M. Exp Cell Res. 1997;234:293–299. doi: 10.1006/excr.1997.3589. [DOI] [PubMed] [Google Scholar]
- 33.Alblas J, Slager-Davidov R, Steenbergh P H, Sussenbach J S, van der Burg B. Oncogene. 1998;16:131–139. doi: 10.1038/sj.onc.1201485. [DOI] [PubMed] [Google Scholar]
- 34.Chittka A, Chao M V. Proc Natl Acad Sci USA. 1999;96:10705–10710. doi: 10.1073/pnas.96.19.10705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamilton T H, Means A R. Proc Natl Acad Sci USA. 1966;56:1594–1598. doi: 10.1073/pnas.56.5.1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cavanaugh A H, Hempel W M, Taylor L J, Rogalsky V, Todorov G, Rothblum L I. Nature (London) 1995;374:177–180. doi: 10.1038/374177a0. [DOI] [PubMed] [Google Scholar]
- 37.White R J, Trouche D, Martin K, Jackson S P, Kouzarides T. Nature (London) 1996;382:88–90. doi: 10.1038/382088a0. [DOI] [PubMed] [Google Scholar]
- 38.White R J. Trends Biochem Sci. 1997;22:77–80. doi: 10.1016/s0968-0004(96)10067-0. [DOI] [PubMed] [Google Scholar]