Abstract
The functions of most of the 12 subunits of the RNA polymerase II (Pol II) enzyme are unknown. In this study, we demonstrate that two of the subunits, hRPB2 and hRPB10α, mediate the regulated stimulation of transcription. We find that the transcriptional coactivator BRCA1 interacts directly with the core Pol II complex in vitro. We tested whether single subunits from Pol II would compete with the intact Pol II complex to inhibit transcription stimulated by BRCA1. Excess purified Pol II subunits hRPB2 or hRPB10α blocked BRCA1- and VP16-dependent transcriptional activation in vitro with minimal effect on basal transcription. No other Pol II subunits tested inhibited activated transcription in these assays. Furthermore, hRPB10α, but not hRPB2, blocked Sp1-dependent activation.
Regulation of class II gene expression in eukaryotes involves a myriad of transcription factors and protein complexes that ultimately exert control over the enzymatic activity of the multisubunit RNA polymerase II enzyme (Pol II). Pol II and the general transcription factors bind to template DNA in a sequential fashion before the initiation of transcription to form a preinitiation complex (1–3). The process is more complex because Pol II also exists in a preformed complex known as the Pol II holoenzyme, which contains general transcription factors, mediator and SRB proteins, chromatin remodeling factors, and coactivators such as BRCA1 and CBP (4). Further, regulated transcription requires promoter-specific DNA-binding factors to target stimulation of specific genes and multiple other factors that function in the gene activation process (5, 6).
Although hundreds of polypeptides orchestrate gene expression in the nucleus, the mass of a protein required to catalyze a DNA-dependent RNA synthesis is not excessive. Indeed, a single 99-kDa polypeptide, such as the bacteriophage T7 RNA polymerase, is sufficient to catalyze promoter-dependent RNA polymerization (7). Bacterial RNA polymerase consists of an enzymatic core of three subunits (α2ββ′) and a variable specificity factor (σ) that selects for various classes of promoters. Promoter-specific activators primarily interact with the α and σ subunits, although all four subunits can be targets (8–12). By analogy, some of the many subunits of the eukaryotic RNA polymerase would also be targets for activators.
Human Pol II consists of 12 subunits (see Table 1 and ref. 14). Several subunits are homologous to prokaryotic subunits, and presumably function similarly. The two largest subunits, hRPB1 and hRPB2, are homologues of the prokaryotic β′ and β subunits that bind DNA and nucleotide triphosphate substrate, respectively (15). hRPB3 contains some sequence similarity to α, and likewise contributes to core Pol II assembly. Analysis of individual Pol II subunits in yeast has revealed a role for a number of Pol II subunits in polymerase assembly (16–20), elongation (21, 22), and accurate initiation of transcription (23–25). However, only the C-terminal domain of RPB1 and the full-length polypeptides of RPB5 and RPB7 have been shown to function in the stimulation of transcription from specific promoters (26–29).
Table 1.
Human Pol II subunits
| Subunit | Mass, kDa | Saccharomyces cerevisiae |
|---|---|---|
| hRPB1 | 220 | B220 |
| hRPB2 | 140 | B150 |
| hRPB3 | 33 | B44 |
| hRPB4 | 16.3 | B32 |
| hRPB5 | 25 | ABC27 |
| hRPB6 | 14.4 | ABC23 |
| hRPB7 | 19 | B16 |
| hRPB8 | 17 | ABC14.5 |
| hRPB9 | 14.5 | B12.6 |
| hRPB10α | 7.0 | ABC10α |
| hRPB10β | 7.6 | ABC10β |
| hRPB11 | 14 | B12.5 |
Here we demonstrate that two of the subunits of Pol II mediate the stimulation of transcription from a subset of gene activators. We find a direct interaction between the transcriptional coactivator BRCA1 and core Pol II, and we demonstrate a role for two subunits, hRPB2 and hRPB10α, in BRCA1- and VP16-dependent stimulation of transcription. In contrast, hRPB10α, but not hRPB2, mediates Sp1-dependent activation in vitro.
Materials and Methods
In Vitro Binding Assay.
Expression of BRCA1(1,560–1,863) protein fused to the biotin-binding PinPoint domain has been described (30). Bacterial lysates were incubated with 10 μl of Dynabeads M-280 Streptavidin for 3 h at 4°C. The beads were washed three times in 0.25 ml of buffer H (20 mM Tris⋅OAc, pH 7.9/1 mM EDTA/20% glycerol) containing 750 mM KOAc, 0.5% Nonidet P-40, 0.2 mg/ml BSA, and 1 mM DTT. Binding of equal amounts of PinPoint fusion proteins was verified by immunoblot analysis using a streptavidin-horseradish peroxidase conjugate. Purified calf thymus Pol II (31) was added in the same buffer except containing 75 mM KOAc and was incubated overnight at 4°C. The beads were washed again in the same buffer containing 750 mM KOAc and were resolved by SDS/PAGE. Samples were transferred to nitrocellulose, were probed with a Pol II-specific antibody (31), and were visualized by chemiluminescence.
Plasmid Templates.
G-less cassette construct G5-E4 contains five GAL4 DNA-binding sites and the adenoviral E4 promoter upstream of a 380-base DNA sequence that lacks guanosine in the coding strand from the start site. SV-MLP contains the SV40 21-bp repeat region found in the 40–117 sequence of the SV40 promoter (GenBank accession no. J02400; gift of M. Timmers; Utrecht University, Utrecht, The Netherlands) and the adenoviral major late promoter upstream of the same G-less cassette. Reactions including BRCA1 or VP16 contained the G5-E4 template; Sp1 reactions contained the SV-MLP template. All reactions contained the internal control template pΔML-200, which consists of core adenoviral major late promoter upstream of a shortened 210-base pair G-less cassette.
Cloning, Mutagenesis, and Expression of hRPB Subunits.
For nomenclature of the RPB subunits, refer to Table 1. The cDNAs for the hRPB7 and hRPB4 were kindly provided by E. Golemis (Fox Chase Cancer Center, Philadelphia). The cDNAs for the remaining nine human RNA polymerase II subunits were cloned from human cDNA libraries by using PCR and appropriate primers based on the published sequences, as described previously for hRPB5, hRPB8, and hRPB6 (32). PCR-generated fragments encoding the full-length subunits or their deletion mutants were cloned into pGEX-2TJL1 (a derivative of the pGEX-2T vector containing a modified polylinker). Mutants hRPB10α (19, 22)A, hRPB10α (34, 38)A, and hRPB10α (36)A were created by site-directed mutagenesis (GeneEditor, Promega). hRPB10α truncation mutants were generated by PCR and were cloned into pGEX-2TK. All constructs were verified by DNA sequencing. Glutathione S-transferase (GST)–hRPB fusion proteins were expressed in BL21 (DE3) cells (Novagen), were purified on glutathione-agarose beads, and were eluted with glutathione according to the manufacturer's protocol (Amersham Pharmacia).
Transcription Reactions.
Human recombinant Sp1 was obtained from Promega. The purification of all other transcription factors used in this study has been described previously (33). Reactions were performed in 25-μl volumes containing 20 mM Hepes⋅NaOH (pH 7.9), 20% glycerol, 1 mM EDTA, 6 mM MgOAc, 90 mM KOAc, 3 mM DTT, 4 μM ZnSO4, 0.2 mg/ml BSA, 100 μM each ATP and UTP, 2.5 μM CTP, 0.5 μM α32P-CTP, 50 μM 3′-OMe-GTP, and 30 ng of each template. Proteins included were 100 ng of TFIIA (34, 35), 60 ng of TFIIB, 4 ng of TFIIE, 100 ng of TFIIF, 40 ng Pol II, 0.25 μl of immunoaffinity-purified TFIID (36), and 0.5 μl of TFIIH fraction. Twenty nanograms of GAL4-BRCA1 or thirty nanograms of GAL4-VP16 were added as indicated, with one hundred nanograms of PC4 (37, 38). Reactions containing Sp1 (1 μl) were supplemented with 0.5 μl of USA fraction derived from the high salt elution of a nuclear extract that was applied to a phosphocellulose column (5). Three hundred nanograms each of GST-hRPB3 to hRPB11 and twelve hundred of GST-hRPB2 polypeptides were added as indicated. The reactions were incubated at 30°C for 90 min and were terminated by the addition of 0.2 ml of solution containing 7 M urea, 1% SDS, 10 mM EDTA, 0.35 M ammonium acetate, 0.1 M LiCl, 0.1 mg/ml tRNA, and a radioactive 550-nucleotide RNA recovery control (not shown in figures). Reactions were extracted in phenol, were precipitated in ethanol as per standard procedures, and were subjected to electrophoresis on 6% polyacrylamide gels containing 8.3 M urea. Dried gels were exposed to film, generally for 16–24 h, and were quantified by using a PhosphorImager (Molecular Dynamics). Activation was measured as the ratio of RNA product of the G5-E4 or SV-MLP template to the RNA product of the ΔML-200 template, all divided by the same ratio in control reactions that lacked activator. Staged initiation reactions were performed as described above except nucleotide triphosphate concentrations were 100 μM ATP and 0.5 μM α32P-CTP. This permits synthesis of only the first seven phosphodiester bonds at the G5-E4 start site. After incubation at 30°C for 45 min, the reactions were supplemented with an additional 100 μM each ATP, UTP, and CTP, 50 μM 3′-OMe-GTP, and 6 mM MgOAc and incubated for another 45 min. Addition of excess CTP at this step prevents labeling of newly initiated transcripts.
Results
BRCA1 Binds Core Pol II Enzyme in Vitro.
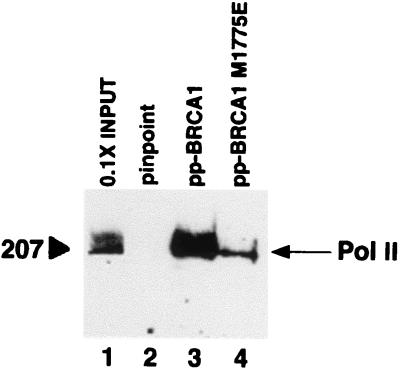
The carboxy-terminal domain of BRCA1 (amino acids 1,560–1,863) functions as a transcriptional activator in vivo when fused to a GAL4 DNA-binding domain (39–41). Recently, we reconstituted activation of transcription in vitro by GAL4-BRCA1 in reactions containing purified transcription factors (33). Whereas we have shown that BRCA1 interacts with the Pol II holoenzyme in cells (41, 42), this cell-free transcriptional regulation by BRCA1 functions with core Pol II. We examined, using an in vitro binding assay, whether BRCA1 interacts directly with the core Pol II enzyme using a fusion protein of BRCA1(1,560–1,863) and purified calf thymus Pol II. Pol II bound directly to BRCA1(1,560–1,863), but not to the fusion partner alone (Fig. 1). Binding of core Pol II to the inactivating point mutant BRCA1(1,560–1,863) M1775E was diminished by about 10-fold (Fig. 1), suggesting that the interaction between the BRCA1 carboxy terminus and core Pol II is specific.
Figure 1.
BRCA1 binds core Pol II in vitro. Purified calf thymus Pol II was incubated with PinPoint protein (pp; lane 2) or PinPoint-BRCA1 fusion proteins (lanes 3 and 4) immobilized on streptavidin beads. After washing in buffer containing 750 mM KOAc and 0.5% Nonidet P-40, bound proteins were resolved by SDS/PAGE and were visualized by immunoblot analysis using a Pol II RPB1-specific antibody. Lane 1 represents 10% of total Pol II in binding reactions.
Pol II Subunits hRPB10α and hRPB2 Function in BRCA1-Dependent Transcriptional Activation.
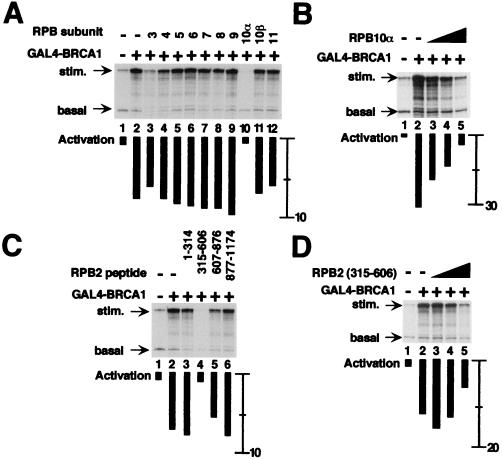
We hypothesized that, if the activation of transcription by BRCA1 depended on contacts with the 12-subunit core Pol II complex, activated transcription will be specifically inhibited by the addition in excess of single Pol II subunits. The standard transcription assay includes two templates, one that encodes a 380-nt RNA and contains GAL4 DNA binding elements and a second template lacking GAL4 sites that generates a 210-nt RNA that controls for nonspecific effects on transcription. In the absence of added Pol II subunit, the BRCA1 fusion protein activated transcription of the 380 nt RNA about eight-fold (Fig. 2A, compare lanes 1 and 2). Purified human Pol II subunits hRPB3 to hRPB11 fused to glutathione S-transferase (GST) were added in excess in Fig. 2A, lanes 3–12. Most subunits, when added to reactions in excess, had no effect on either activated or basal transcription. In contrast, addition of hRPB10α selectively inhibited BRCA1-activated transcription, with a minimal effect on basal transcription (Fig. 2A, lane 10). hRPB3 inhibited both stimulated and basal transcription, resulting in no net change in activation (Fig. 2A, lane 3). The effect of inhibition by GST-hRPB10α could be titrated with little effect on the accumulation of basal transcript (Fig. 2B). Full inhibition of activated transcription was observed using 300 ng of GST-hRPB10α, a 125-fold molar excess relative to Pol II.
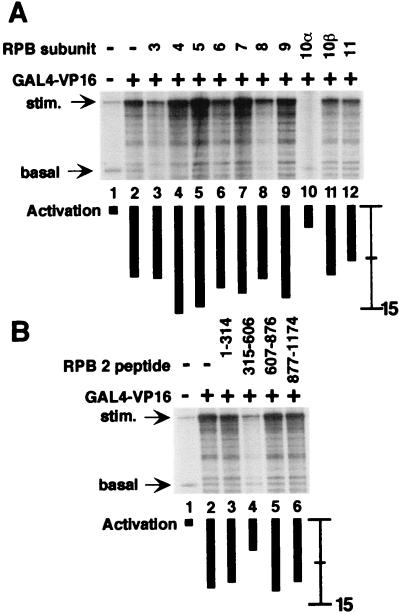
Figure 2.
Pol II subunits hRPB10α and hRPB2 mediate BRCA1-dependent transcriptional activation. All reactions contain purified basal transcription factors, the coactivator PC4, and core Pol II in the absence or presence of GAL4-BRCA1. Individual Pol II subunits fused to GST were added as indicated. Transcription from the GAL4-dependent template resulted in the accumulation of 380-nucleotide RNA (stim.), and transcription from a control template lacking GAL4 response elements resulted in accumulation of 210-nucleotide RNA (basal). The relative increase in transcription upon addition of activator, quantified by using PhosphorImager analysis, is indicated by black bars under each lane. For comparison, the activation in lane 2 of A (absence of added Pol II subunit) is eight-fold. (A) Inhibition of BRCA1-dependent stimulation of transcription by 300 ng of GST fusion of each hRPB3 to hRPB11 (lanes 3–12). (B) Titration of RPB10α results in an incremental decrease in BRCA1-stimulated transcription. GST-hRPB10α was included in reactions at 75 ng (lane 3), 150 ng (lane 4), and 300 ng (lane 5). (C) Inhibition of BRCA1-dependent transcriptional activation by hRPB2 peptides. Twelve hundred nanograms of each fragment were added as indicated. (D) Titration of hRPB2(315–606) results in an incremental decrease in BRCA1-stimulated transcription. GST-hRPB2(315–606) was included in reactions at 300 ng (lane 3), 600 ng (lane 4), and 1,200 ng (lane 5).
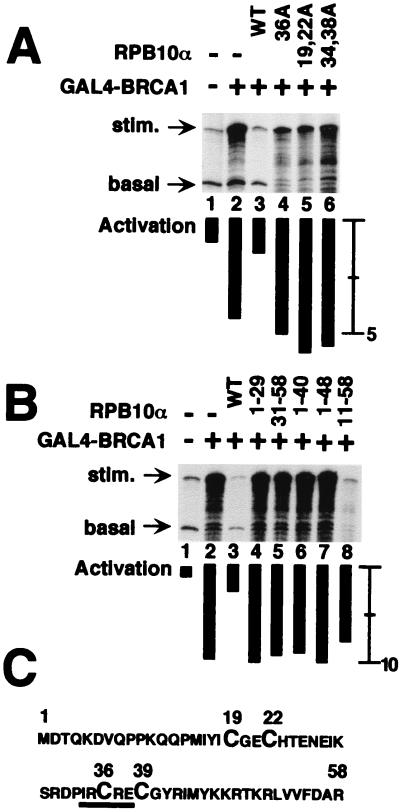
To test the specificity of hRPB10α function in activated transcription, we generated site-specific and truncation mutants of the 58 amino acid hRPB10α. Mutant hRPB10α did not inhibit GAL4-BRCA1-dependent transcription, suggesting that regulation of transcriptional activation is a function of the wild-type protein (Fig. 3). For example, cysteine-to-alanine substitutions in a zinc finger motif (Fig. 3A, lanes 4 and 5) or disruption of a putative Rb binding motif (Ile-X-Cys-X-Glu; Fig. 3A, lane 6) had no effect on activated transcription. Because there was no Rb protein in the reaction, it is likely that the altered residues of the Rb-binding motif disrupt the native conformation of the polypeptide. Truncation mutants of hRPB10α similarly did not inhibit transcriptional activation (Fig. 3B). One truncation, hRPB10α(11–58), which did not modify any obvious structural motifs (Fig. 3C), nonspecifically inhibited both activated and basal transcription (Fig. 3B). The loss of function by hRPB10α mutants in this assay system supports a specific role for this subunit in activated transcription.
Figure 3.
Mutant forms of hRPB10α do not mediate BRCA1-dependent activation. All reactions contained the general transcription factors, PC4, and Pol II in the absence or presence of GAL4-BRCA1. (A) Inclusion in reactions of site-specific mutants of hRPB10α that disrupt the zinc finger motif (lanes 4 and 5) or an Rb-binding motif (lane 6) do not inhibit BRCA1-dependent activated transcription. (B) Inclusion in reactions of hRPB10α truncation mutants that disrupt the zinc finger motif (lanes 4 and 5) or a highly basic region (lanes 6 and 7) do not inhibit BRCA1-dependent activated transcription. The hRPB10α(11–58) truncation mutant does not delete an obvious structural motif, but cannot specifically inhibit BRCA1-dependent activated transcription (lane 8). (C) Amino acid sequence of hRPB10α, with the zinc finger (Cys-X3-Cys-X13-Cys-X3-Cys) motif enlarged and the Rb-binding motif underlined.
The 140-kDa hRPB2 polypeptide was divided into four fragments and was assayed for inhibition of BRCA1-dependent activated transcription (Fig. 2C). Similar to the hRPB10α subunit, hRPB2(315–606) specifically inhibited activated transcription (Fig. 2C). Full inhibition of activated transcription was observed by using 1,200 ng of GST-hRPB2(315–606), a 250-fold molar excess relative to Pol II. We suggest that excess subunit, or subunit fragment, is necessary because the local concentration of individual GST-fused Pol II subunits at the promoter is expected to be lower than that of DNA-tethered transcription factors and intact core Pol II. Neither hRPB10α nor hRPB2(315–606) inhibited the interaction of the GAL4 DNA binding domain with its binding site in electrophoretic mobility shift assays (data not shown). This indicates that the specificity of inhibiting the GAL4-stimulated transcription is not explained by blocking the DNA binding of the activator.
Staged Initiation of Transcription.
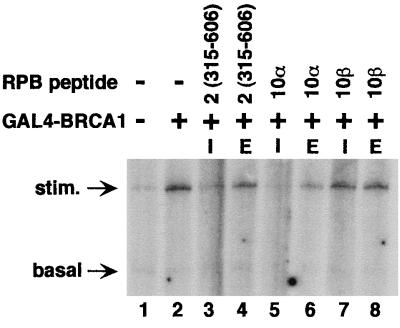
hRPB10α had no effect on the accumulation of the 380-nt transcript in the absence of added activator (data not shown), suggesting that this inhibition was not attributable to a selective inhibition of transcriptional elongation. To test whether the inhibition of transcriptional activation by hRPB10α or hRPB2 functioned at a particular stage of the transcription cycle, an experiment was designed to separate the transcription reaction into initiation and elongation phases. By excluding the nucleotide triphosphate UTP, only the first seven nucleotides would be incorporated into the nascent transcript (see Materials and Methods). Subsequent addition of UTP permits elongation of previously initiated transcripts. Addition of excess unlabeled CTP at this elongation step prevents visualization of newly initiated transcripts. When GST-hRPB2(315–606) and GST-hRPB10α are included in the reactions before initiation of RNA synthesis, BRCA1-dependent activation is blocked (Fig. 4, lanes 3, and 5), consistent with a role for both subunits in initiation of transcription. GST-hRPB10β was included as a negative control (Fig. 4, lanes 7 and 8). When GST-hRPB2(315–606) and GST-hRPB10α are added after initiation has already occurred, these subunits have minimal effect on transcriptional elongation. (Fig. 4, lanes 4 and 6).
Figure 4.
Pol II subunits hRPB2 and hRPB10α mediate BRCA1-dependent transcriptional activation before initiation. All reactions were performed as in Fig. 2 except that UTP was excluded, stopping synthesis of the nascent transcript after incorporation of seven nucleotides. Individual Pol II subunits in concentrations identical to Fig. 2 were added concurrent with initiation of the reaction (I), or later upon resumption of transcript elongation by the addition of UTP (E).
VP16- and Sp1-Dependent Transcriptional Activation.
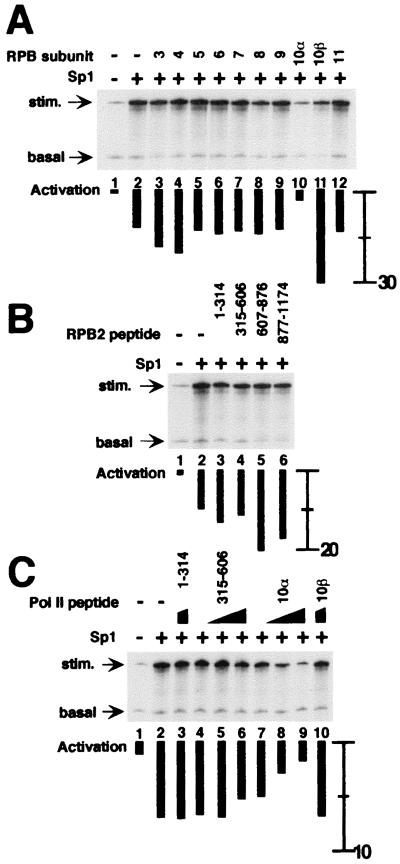
To test whether inhibition of transcriptional activation by hRPB10α or hRPB2 was specific to BRCA1, GAL4-VP16 and Sp1 were tested for the effects of hRPB subunits. Activation of transcription by GAL4-VP16 was inhibited by both hRPB10α and hRPB2 (315–606) (Fig. 5). No other Pol II subunits inhibited VP16-dependent activation in this assay. Sp1 did not activate transcription from the appropriate GC-box containing template under the same conditions as used for GAL4-BRCA1 and GAL4-VP16. Instead, a relatively crude fraction containing the USA activity (5) and that contains the Sp1-specific coactivator CRSP (43) was included in transcription reactions. Sp1-dependent activated transcription was inhibited by GST-hRPB10α, and no new interactions with other Pol II subunits were revealed in this the assay (Fig. 6A). In contrast to the case with BRCA1, none of the hRPB2 peptides inhibited the Sp1-activated transcription (Fig. 6B). Titration of either hRPB2(315–606) or hRPB10α into Sp1-dependent activation reactions confirms that only the latter inhibited the Sp1-activated RNA synthesis with little effect on the accumulation of the basal control transcript (Fig. 6C).
Figure 5.
Pol II subunits hRPB10α and hRPB2 mediate VP16-dependent transcriptional activation. All reactions were performed as in Fig. 2 except that GAL4-VP16 was used as an activator. (A) Effect on VP16-dependent activated transcription by hRPB3 to hRPB11. For comparison, the activation in lane 2 (absence of added Pol II subunit) is 12-fold. (B) Effect on VP16-dependent activated transcription by hRPB2 peptides.
Figure 6.
hRPB10α, but not hRPB2, mediates Sp1-dependent activation. All reactions contained the general transcription factors, USA fraction, and Pol II in the absence or presence of Sp1. (A) Effect on Sp1-dependent activated transcription by hRPB3 to hRPB11. For comparison, the activation in lane 2 (absence of added Pol II subunit) is 10-fold. (B) Effect Sp1-dependent transcription by hRPB2 peptides. (C) Titration, as in Fig. 2, of hRPB10α (lanes 7–9), but not hRPB2(315–606) (lanes 4–6), results in an incremental decrease in Sp1-dependent activated transcription.
Discussion
In these experiments we find that individual Pol II subunits mediate template-specific activation of transcription. There is a direct interaction between the activation domain of BRCA1 and core mammalian Pol II. Analysis of the individual subunits in binding assays similar to that in Fig. 1 was complicated by nonspecific binding of many subunits (data not shown). Instead, roles for individual subunits can be demonstrated in a functional assay in which an excess of a single subunit inhibits the activated portion of a transcription reaction with minimal effect on basal transcription. The repression with these Pol II subunits likely reflects positive interactions between the Pol II subunit and activator that are inhibited by squelching. Polymerase subunit hRPB10α, when provided in excess, can specifically inhibit BRCA1-, VP16-, and Sp1-dependent activated transcription when assayed in vitro, with minimal effect on basal levels of transcription. It is possible that inhibition is caused by interactions of buried surfaces of Pol II subunits that do not normally occur in the context of the native transcription complex. However, hRPB10α mutants do not block activated transcription in this assay, suggesting that wild-type hRPB10α in the core Pol II complex mediates the activation of transcription by these activators. To our knowledge, this is the first identified function for hRPB10α. In contrast to the results with the hRPB10α subunit, the GST-hRPB2(315–606) peptide inhibits only BRCA1- and VP16-dependent activation, implicating hRPB2 in factor-specific activation of transcription.
The finding that hRPB10α mediates template-specific transcription in all three activators tested suggests that this subunit may in fact function in regulated transcription for many activators. Further experiments with many more activators should determine how generally this subunit functions in regulated transcription. This small subunit, common to all three polymerases, is well conserved and essential (44). The identified role for hRPB10α in activated transcription may explain why it is essential for viability in yeast.
The human RPB2(315–606) region is highly conserved across eukaryotic RPB2 subunits. RPB2 mutations clustered within this domain suppress δ-element insertions in HIS4 and LYS2 promoters in yeast, suggesting a role for RPB2 in start site selection (25). It is unclear how this phenotype correlates with the observed role of hRPB2 in activated transcription. hRPB2(315–606) did not inhibit Sp1-dependent activation in this assay, but it did inhibit BRCA1- and VP16-dependent activation. This survey of different activators is too limited to determine whether many activators will function via hRPB2. VP16 is a powerful viral activator for which a number of mechanisms have been described (45), and it may usurp activation pathways specific for endogenous transcription factors, such as BRCA1. Alternatively, finer mapping of the hRPB2(315–606) domain may reveal a divergence in the domains regulating BRCA1- and VP16-dependent transcriptional activation.
RPB10α and RPB2 may now be added to a growing list of Pol II subunits that are involved in template-specific transcriptional regulation. RPB7 interacts with the oncogenic EWS-Fli1 fusion protein to increase transcriptional activation in vivo (29). The viral transactivating protein HBx interacts with RPB5 (46, 47). Successive truncation of the C-terminal domain of RPB1 in yeast results in the decreased transcriptional induction from multiple genes and decreased response to specific activators (26, 27), probably because of effects on the Pol II holoenzyme. An RPB5 point mutant similarly has decreased transcriptional induction of multiple genes, and RPB5 and C-terminal domain truncation double mutants are synthetically lethal, suggesting an overlap in function for the two subunits (28). RPB5 did not inhibit activated transcription in our assay. This may reflect species differences because hRPB5 does not complement the yeast gene (48), or RPB5 mutants may disrupt the Pol II holoenzyme, whereas the assay in this study utilizes core Pol II.
Our finding that individual Pol II subunits mediate BRCA1-dependent activation strengthens the evidence that BRCA1 functions as a transcription factor. A growing number of proteins involved in transcription interact with BRCA1, including RHA, p53, BARD1, CtIP, and now Pol II (41, 49–54). The in vitro transcription assay only recapitulates a part of the process of gene expression in the cell. It is likely that the in vitro assay is limited by events that occur at initiation whereas in the cell Pol II must elongate over a chromatin template and the RNA must be properly processed. Truncated RHA protein blocks GAL4-BRCA1-dependent transcription in cultured mammalian cells (41) but not in the cell free assays used in this study (data not shown). We demonstrate that hRPB2 and hRPB10α function in the initiation of transcription, and it is possible that RHA functions in postinitiation events. In the context of gene activation by the Pol II holoenzyme, multiple contacts by BRCA1 to different holoenzyme components may reflect allosteric changes in the complex during the transcription process.
Acknowledgments
We thank E. Golemis for the gift of hRPB4 and hRPB7 cDNAs, M. Meisterernst for the expression vector for PC4, R. G. Roeder and J. DeJong for the expression vectors for TFIIA, and C. Chiang for the FLAG-tagged TFIID cell line. This work was supported by the National Institutes of Health Grants GM53504 (to J.D.P.) and HL52666 and AI34270 (to J.A.A.L.) and a research project grant from the American Cancer Society (to J.D.P.). B.P.S. was supported by a grant from the Susan G. Komen Breast Cancer Foundation. J.A.A.L. is an Established Investigator of the American Heart Association.
Abbreviations
- Pol II
RNA polymerase II
- GST
glutathione S-transferase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.070452397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.070452397
References
- 1.Buratowski S, Hahn S, Guarente L, Sharp P A. Cell. 1989;56:549–561. doi: 10.1016/0092-8674(89)90578-3. [DOI] [PubMed] [Google Scholar]
- 2.Maldonado E, Ha I, Cortes P, Weis L, Reinberg D. Mol Cell Biol. 1990;10:6335–6347. doi: 10.1128/mcb.10.12.6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orphanides G, Lagrange T, Reinberg D. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 4.Parvin J D, Young R A. Curr Opin Genet Dev. 1998;8:565–570. doi: 10.1016/s0959-437x(98)80012-9. [DOI] [PubMed] [Google Scholar]
- 5.Meisterernst M, Roy A L, Lieu H M, Roeder R G. Cell. 1991;66:981–993. doi: 10.1016/0092-8674(91)90443-3. [DOI] [PubMed] [Google Scholar]
- 6.Shykind B M, Kim J, Sharp P A. Genes Dev. 1995;9:1354–1365. doi: 10.1101/gad.9.11.1354. [DOI] [PubMed] [Google Scholar]
- 7.McAllister W T. Cell Mol Biol Res. 1993;39:385–391. [PubMed] [Google Scholar]
- 8.Hochschild A, Dove S L. Cell. 1998;92:597–600. doi: 10.1016/s0092-8674(00)81126-5. [DOI] [PubMed] [Google Scholar]
- 9.Dove S L, Joung J K, Hochschild A. Nature (London) 1997;386:627–630. doi: 10.1038/386627a0. [DOI] [PubMed] [Google Scholar]
- 10.Ishihama A. Mol Microbiol. 1992;6:3283–3288. doi: 10.1111/j.1365-2958.1992.tb02196.x. [DOI] [PubMed] [Google Scholar]
- 11.Lee J H, Hoover T R. Proc Natl Acad Sci USA. 1995;92:9702–9706. doi: 10.1073/pnas.92.21.9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller A, Wood D, Ebright R H, Rothman-Denes L B. Science. 1997;275:1655–1657. doi: 10.1126/science.275.5306.1655. [DOI] [PubMed] [Google Scholar]
- 13.Young R A. Annu Rev Biochem. 1991;60:689–715. doi: 10.1146/annurev.bi.60.070191.003353. [DOI] [PubMed] [Google Scholar]
- 14.Acker J, de Graaff M, Cheynel I, Khazak V, Kedinger C, Vigneron M. J Biol Chem. 1997;272:16815–16821. doi: 10.1074/jbc.272.27.16815. [DOI] [PubMed] [Google Scholar]
- 15.Sweetser D, Nonet M, Young R A. Proc Natl Acad Sci USA. 1987;84:1192–1196. doi: 10.1073/pnas.84.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolodziej P A, Young R A. Mol Cell Biol. 1991;11:4669–4678. doi: 10.1128/mcb.11.9.4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyao T, Yasui K, Sakurai H, Yamagishi M, Ishihama A. Genes Cells. 1996;1:843–854. doi: 10.1046/j.1365-2443.1996.730274.x. [DOI] [PubMed] [Google Scholar]
- 18.Kimura M, Ishiguro A, Ishihama A. J Biol Chem. 1997;272:25851–25855. doi: 10.1074/jbc.272.41.25851. [DOI] [PubMed] [Google Scholar]
- 19.Ishiguro A, Kimura M, Yasui K, Iwata A, Ueda S, Ishihama A. J Mol Biol. 1998;279:703–712. doi: 10.1006/jmbi.1998.1823. [DOI] [PubMed] [Google Scholar]
- 20.Yasui K, Ishiguro A, Ishihama A. Biochemistry. 1998;37:5542–5548. doi: 10.1021/bi972939b. [DOI] [PubMed] [Google Scholar]
- 21.Archambault J, Lacroute F, Ruet A, Friesen J D. Mol Cell Biol. 1992;12:4142–4152. doi: 10.1128/mcb.12.9.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Powell W, Reines D. J Biol Chem. 1996;271:6866–6873. doi: 10.1074/jbc.271.12.6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arndt K T, Styles C A, Fink G R. Cell. 1989;56:527–537. doi: 10.1016/0092-8674(89)90576-x. [DOI] [PubMed] [Google Scholar]
- 24.Edwards A M, Kane C M, Young R A, Kornberg R D. J Biol Chem. 1991;266:71–75. [PubMed] [Google Scholar]
- 25.Hekmatpanah D S, Young R A. Mol Cell Biol. 1991;11:5781–5791. doi: 10.1128/mcb.11.11.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scafe C, Chao D, Lopes J, Hirsch J P, Henry S, Young R A. Nature (London) 1990;347:491–494. doi: 10.1038/347491a0. [DOI] [PubMed] [Google Scholar]
- 27.Liao S M, Taylor I C, Kingston R E, Young R A. Genes Dev. 1991;5:2431–2440. doi: 10.1101/gad.5.12b.2431. [DOI] [PubMed] [Google Scholar]
- 28.Miyao T, Woychik N A. Proc Natl Acad Sci USA. 1998;95:15281–15286. doi: 10.1073/pnas.95.26.15281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petermann R, Mossier B M, Aryee D N, Khazak V, Golemis E A, Kovar H. Oncogene. 1998;17:603–610. doi: 10.1038/sj.onc.1201964. [DOI] [PubMed] [Google Scholar]
- 30.Neish A S, Anderson S F, Schlegel B P, Wei W, Parvin J D. Nucleic Acids Res. 1998;26:847–853. doi: 10.1093/nar/26.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson N E, Aronson D B, Burgess R R. J Biol Chem. 1990;265:7069–7077. [PubMed] [Google Scholar]
- 32.Hiremath C N, Ladias J A A. Protein Expression Purif. 1998;13:198–204. doi: 10.1006/prep.1998.0889. [DOI] [PubMed] [Google Scholar]
- 33.Haile D T, Parvin J D. J Biol Chem. 1999;274:2113–2117. doi: 10.1074/jbc.274.4.2113. [DOI] [PubMed] [Google Scholar]
- 34.DeJong J, Roeder R G. Genes Dev. 1993;7:2220–2234. doi: 10.1101/gad.7.11.2220. [DOI] [PubMed] [Google Scholar]
- 35.DeJong J, Bernstein R, Roeder R G. Proc Natl Acad Sci USA. 1995;92:3313–3317. doi: 10.1073/pnas.92.8.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiang C M, Roeder R G. Pept Res. 1993;6:62–64. [PubMed] [Google Scholar]
- 37.Kretzschmar M, Kaiser K, Lottspeich F, Meisterernst M. Cell. 1994;78:525–534. doi: 10.1016/0092-8674(94)90429-4. [DOI] [PubMed] [Google Scholar]
- 38.Ge H, Roeder R G. Cell. 1994;78:513–523. doi: 10.1016/0092-8674(94)90428-6. [DOI] [PubMed] [Google Scholar]
- 39.Chapman M S, Verma I M. Nature (London) 1996;382:678–679. doi: 10.1038/382678a0. [DOI] [PubMed] [Google Scholar]
- 40.Monteiro A N, August A, Hanafusa H. Proc Natl Acad Sci USA. 1996;93:13595–13599. doi: 10.1073/pnas.93.24.13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anderson S F, Schlegel B P, Nakajima T, Wolpin E S, Parvin J D. Nat Genet. 1998;19:254–256. doi: 10.1038/930. [DOI] [PubMed] [Google Scholar]
- 42.Scully R, Anderson S F, Chao D M, Wei W, Ye L, Young R A, Livingston D M, Parvin J D. Proc Natl Acad Sci USA. 1997;94:5605–5610. doi: 10.1073/pnas.94.11.5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ryu S, Tjian R. Proc Natl Acad Sci USA. 1999;96:7137–7142. doi: 10.1073/pnas.96.13.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Treich I, Carles C, Riva M, Sentenac A. Gene Expression. 1992;2:31–37. [PMC free article] [PubMed] [Google Scholar]
- 45.Flint J, Shenk T. Annu Rev Genet. 1997;31:177–212. doi: 10.1146/annurev.genet.31.1.177. [DOI] [PubMed] [Google Scholar]
- 46.Cheong J H, Yi M, Lin Y, Murakami S. EMBO J. 1995;14:143–150. doi: 10.1002/j.1460-2075.1995.tb06984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin Y, Nomura T, Cheong J, Dorjsuren D, Iida K, Murakami S. J Biol Chem. 1997;272:7132–7139. doi: 10.1074/jbc.272.11.7132. [DOI] [PubMed] [Google Scholar]
- 48.McKune K, Moore P A, Hull M W, Woychik N A. Mol Cell Biol. 1995;15:6895–6900. doi: 10.1128/mcb.15.12.6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ouchi T, Monteiro A N, August A, Aaronson S A, Hanafusa H. Proc Natl Acad Sci USA. 1998;95:2302–2306. doi: 10.1073/pnas.95.5.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang H, Somasundaram K, Peng Y, Tian H, Zhang H, Bi D, Weber B L, El-Deiry W S. Oncogene. 1998;16:1713–1721. doi: 10.1038/sj.onc.1201932. [DOI] [PubMed] [Google Scholar]
- 51.Wu L C, Wang Z W, Tsan J T, Spillman M A, Phung A, Xu X L, Yang M C, Hwang L Y, Bowcock A M, Baer R. Nat Genet. 1996;14:430–440. doi: 10.1038/ng1296-430. [DOI] [PubMed] [Google Scholar]
- 52.Kleiman F E, Manley J L. Science. 1999;285:1576–1579. doi: 10.1126/science.285.5433.1576. [DOI] [PubMed] [Google Scholar]
- 53.Yu X, Wu L C, Bowcock A M, Aronheim A, Baer R. J Biol Chem. 1998;273:25388–25392. doi: 10.1074/jbc.273.39.25388. [DOI] [PubMed] [Google Scholar]
- 54.Li S, Chen P L, Subramanian T, Chinnadurai G, Tomlinson G, Osborne C K, Sharp Z D, Lee W H. J Biol Chem. 1999;274:11334–11338. doi: 10.1074/jbc.274.16.11334. [DOI] [PubMed] [Google Scholar]