Abstract
It is not clear how biological pathways evolve to mediate a certain physiological response and why they show a level of complexity that is generally above the minimum required to achieve such a response. One possibility is that pathway complexity increases due to the nature of evolutionary mechanisms. Here, we analyze this possibility by using mathematical models of biological pathways and evolutionary simulations. Starting with a population of small pathways of three proteins, we let the population evolve with mutations that affect pathway structure through duplication or deletion of existing proteins, deletion or creation of interactions among them, or addition of new proteins. Our simulations show that such mutational events, coupled with a selective pressure, leads to growth of pathways. These results indicate that pathways could be driven toward complexity via simple evolutionary mechanisms and that complexity can arise without any specific selective pressure for it. Furthermore, we find that the level of complexity that pathways evolve toward depends on the selection criteria. In general, we find that final pathway size tends to be lower when pathways evolve under stringent selection criteria. This leads to the counterintuitive conclusion that simple response requirements on a pathway would facilitate its evolution toward higher complexity.
Keywords: robustness, evolutionary simulations, mathematical models, protein pathways, signal transduction
A central question in evolutionary biology is the evolution of complex features. Research in this field has associated complexity at the organism level with complexity at the sequence level (1, 2), with transcriptional control (3, 4), or with cellular differentiation (5). Although these studies provided important insights into how complexity at organism level could have arisen, we still lack an explicit analysis of complexity in a specific system at molecular level. Here, we attempt such an analysis for biological signaling pathways. These pathways, i.e., systems of proteins that act in an orchestrated fashion, mediate the response of a cell toward internal and external signals. It is usually observed that pathways show high complexity, which manifests itself in various forms including number of components in a given pathway, interaction among different pathways, and compartmentalization (6). Although a sequential evolution as proposed by Darwin (7) could explain the existence of such complex pathways, it is not clear why pathways should be driven toward a level of complexity that is higher than required by their function (i.e., producing a specific response). Both experimental and theoretical studies indicate that certain biological responses such as oscillations and switches or phenomena such as pattern formation and chemotaxis could be achieved via pathways that are simpler than found in nature (5, 8–12). One possible explanation for pathway complexity (or biological complexity in general) is that selection for increased fitness necessitates an increase in complexity. Although there is support for such an explanation from studies in digital organisms (13), it is not clear why an increase in complexity should always be accompanied with an increase in fitness and vice versa (14, 15).
There is a much simpler explanation for the emergence of complexity in biological systems that has been first formulated by Saunders and Ho (15). In their words: “a system which optimizes its structure with respect to some criterion (which one may call fitness) will tend to permit the addition of components more readily than their removal.” This hypothesis does not assume that complexity should be related to increases in fitness or any other property that is under selection. However, complexity arising neutrally because of the aforementioned tendency in time could result in functional advantages and a fitness benefit.
Here, we formulate a similar hypothesis to explain the emergence of complexity in biological pathways: Pathways have an intrinsic tendency to become more complex that results from imbalanced effects of mutational events on pathway response. In other words, mutations resulting in the addition of new interactions or proteins to a given pathway have less deleterious effects on its response than mutations that result in loss of interactions or proteins. If such an imbalance exists, it would drive pathways to become larger and more complex in time. To test this hypothesis, we analyze evolution of pathways by using mathematical models and computer simulations. Starting with a population of small pathways, we let pathways replicate with various mutations affecting their structure. We find that such mutations, coupled with a selective pressure for function, lead to growth of pathways under various model assumptions. These results indicate that pathways have an intrinsic tendency to grow larger than required for proper functioning, arising simply from the nature of mutational events affecting pathway structure.
Results and Discussion
To test the possibility that pathways have an intrinsic tendency to become more complex because of mutational mechanisms affecting their structure, we perform evolutionary simulations and monitor pathway size as a proxy for complexity. These simulations (see Methods) start with a population of random pathways composed of only three proteins. Fig. 1 illustrates a sample pathway used in such simulations, its mathematical description, and response to a signal. At each successive generation of the simulation, pathways replicate based on their fitness value and are exposed to mutations with a certain probability during this replication.
Fig. 1.
Cartoon and mathematical representation of a sample three-protein pathway and its response (solid line) to a specific signal (dashed line). The mathematical pathway representation gives coefficients for interactions among proteins in a table format, listing actions of other proteins on a given protein at each row. The absolute values of negative and positive numbers stand for activating and deactivating interactions, respectively. The shown signal is used throughout all evolutionary simulations to assess pathway fitness.
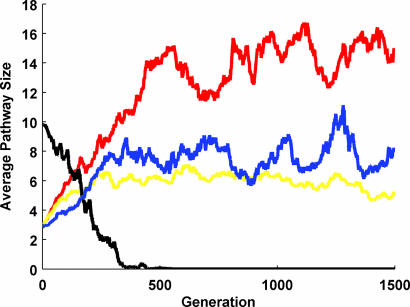
To decouple fitness and complexity, we use a step-like fitness function that is adjusted only by the cost of having multiple proteins. In other words, pathways are either viable [fitness = 1 − (number of proteins) × (cost per protein)] or not (fitness = 0) and the only determinants of fitness are pathway size and the ability to fulfill a chosen selection criteria (see Methods). Furthermore, in the presented simulations, we assume that mutations leading to an increase in pathway complexity (i.e., mutations adding a protein or an interaction) and mutations leading to a decrease (i.e., mutations deleting a protein or an interaction) have equal chances to occur, ensuring that we do not artificially introduce complexity (see Methods). Despite such an assumption, this null-model biases toward decreasing pathway size because interaction-deleting mutations can occasionally result in loss of proteins, whereas interaction-adding mutations cannot have a balancing effect. Hence, we find that in absence of any selection (i.e., all pathways are viable) pathways shrink to a size of zero (see Fig. 2).
Fig. 2.
The average pathway size for populations evolving under different selection schemes: black (no selection), red (selection for any type of response to a given signal), blue (selection for existence), and yellow (selection for derivative-type of response to a given signal). The latter simulation starts with a homogenous population of three-protein pathways (shown in Fig. 1) that are capable of a derivative-like response, whereas all other simulations start with a random population. See Methods section for details.
This situation reverses in presence of selection. Fig. 2 shows that when a population of randomly created three-protein pathways are under selection for existence (i.e., having at least two proteins) or for giving any type of response to a signal (see Methods), the average pathway size increases until it settles to stochastic fluctuations around a certain size. Given the nature of the used fitness function and the fact that even small pathways can achieve high fitness under these selection criteria, it is unlikely that the observed increase in pathway size is due to increasing fitness. To make this point more clear, we run a simulation starting with a homogenous population of three-protein pathways that are capable of producing a response that loosely tracks the derivative of the signal (see Fig. 1). Hence, under selection for this type of response, all members of the population have the same fitness value and there is no possible fitness gain from increased complexity but even a small cost associated with having more proteins in the pathway. Still, we find that the average pathway size increases in this population as evolution progresses (see Fig. 2). It is important to note that the observed increase in average pathway size is not due to few pathways dominating the population but rather due to an increase in the frequency of larger pathways (see Fig. 5, which is published as supporting information on the PNAS web site).
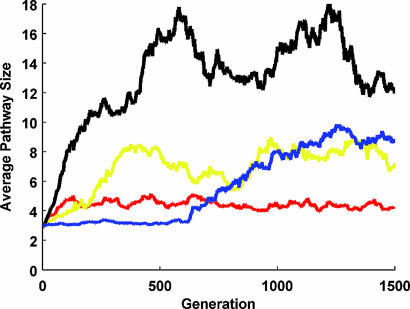
To see whether these results are extendable to various biologically relevant responses, we have run additional simulations with the following selection schemes: (i) “transient/base,” a transient response with equal pre- and postsignal levels of active effector protein concentration; (ii) “transient/magnitude,” a transient response with a magnitude that is larger than the presignal level of active effector protein concentration; (iii) “transient/base and magnitude,” a transient response that fulfills both criteria (i) and (ii); and (iv) “switch,” a “switch-like” response where the postsignal level of the active effector protein concentration is higher than the presignal level by at least 0.01. Starting with a random population of three-protein pathways, we find that under such selection criteria pathways grow and reach a certain size level that is different for different selection schemes but always above the minimum possible (see Fig. 3). In Fig. 6, which is published as supporting information on the PNAS web site, we show sample pathways from the last generation of some of these evolutionary simulations.
Fig. 3.
The average pathway size for populations evolving under different selection schemes: black (selection for transient base), yellow (selection for transient magnitude), blue (selection for transient base and magnitude), and red (selection for switch). All simulations start with a random population of three-protein pathways. See Methods section for simulation details.
These results clearly indicate that the effects of the modeled mutational events on pathway response are distributed in an asymmetric manner. Deletion of interactions and proteins render pathways nonfunctional (e.g., not able to produce the required response) more often than additions do, pushing pathways to become larger and more complex. However, the fact that populations reach “equilibrium” with respect to size indicates that the functional effects of these two types of mutations depend on the pathway size itself. As the strength of these effects change, so does the imbalance among them, and at some point, the average pathway size settles to stochastic fluctuations around a certain value. This point is most clearly explained for the simplest selection scheme we use: existence. Under such a selection scheme, there is a strong imbalance between the effects of size-increasing and -decreasing mutations on the simplest pathways (namely those with two and three proteins). Whereas size-increasing mutations have only a slight effect on fitness (due to fitness cost of proteins), size-decreasing mutations are almost certain to turn such pathways inviable. As pathways grow, this deleterious effect of size-decreasing mutations weakens allowing pathways to survive both types of mutations with equal chance, and the average pathway size of the population reaches equilibrium.
To show that such an explanation for increasing complexity can be extended to more complex selection criteria, we analyze different types of mutations that occurred during the replication process and that did not render a pathway inviable (i.e., nonlethal mutations) for the simulations where we applied selection for any type of response and for derivative-like response (see Fig. 4). Whereas the percentage of nonlethal, interaction-adding and -deleting mutations were the same on average, there were more nonlethal, protein-adding mutations than protein-deleting mutations, especially at the beginning of the simulations where most pathways were small. Hence, at the early generations the average pathway size of the population quickly increases (see Figs. 2 and 3). Then, as the evolutionary simulation progresses the percentage of nonlethal, protein-adding and -deleting mutations approach each other, resulting in the average pathway size of the population to settle at an equilibrium level. Note that such equilibrium can be maintained even though nonlethal, protein-adding mutations continue occurring more frequently than protein-deleting mutations, because of above-discussed effects of interaction-deleting mutations. Another interesting observation is that in the homogenous population capable of derivative-like response and under selection for such response, protein duplications were more tolerated than addition of new proteins to the pathway. This was not the case for random pathways that were under selection for any type of response (see Fig. 4 A Inset and B Inset). This suggests that for pathways that are already functional, protein duplications are the dominant evolutionary mechanism, consistent with observations of transcription control pathways (16, 17).
Fig. 4.
Analysis of mutations that did not render a pathway inviable (i.e., nonlethal mutations). A and B show the results for populations under selection for any type of response and for derivative-like response respectively. (Upper) Summary of interaction-adding (red) and -deleting (blue) mutations as a percentage of all nonlethal mutations. (Lower) Percentages for protein-adding (red) and –deleting (blue) mutations. (Insets) The results for different protein-adding mutations: duplication of existing proteins (green) and addition of new proteins (black).
These simulations suggest that for pathways that achieve a certain response with the simplest possible configuration, the effects of size-decreasing mutations typically will be more severe than size-increasing mutations. This is the main reason why pathways grow large in the presented simulations as soon as there is some sort of selection applied. How long such growth continues before pathways reach an equilibrium size depends on the function that they are selected for and on the distribution of functional pathways in the pathway structure (i.e., topology) space. If for a given function and pathway size there are more functional neighbors in the topology space that are larger than the current pathway size, this manifests itself as size-increasing mutations being less deleterious than size-decreasing ones. This leads to the aforementioned imbalance, which then leads to pathway growth. The growth trend continues until pathways reach a region in the topology space where they have comparable numbers of functional neighboring topologies that are larger and smaller than the current size.
This point is clearly visible in the presented run by using the selection scheme transient/base and magnitude (see Fig. 3). Here, the average pathway size of the population seemingly reaches equilibrium at three to four proteins. Then, as pathways accumulate mutations, they reach a region of the topology space where most of functional neighbors are larger than the current size. At this point, the imbalance between size-increasing and -decreasing mutations is regenerated and pathway growth continues. We also observed that in rare cases, it is possible for the population to get stuck in a certain region of the topology space that would not allow growth (see Fig. 7B Bottom, which is published as supporting information on the PNAS web site). In this case, the population is dominated by a specific pathway topology (see Fig. 6A Bottom), which has very rare or hardly accessible larger functional neighbors that could not overtake the population in the presented simulations. Under natural circumstances where fitness function would not have a step-like shape as presented here, it would be more difficult for the population to get stuck in such topologies.
Based on the above explanation for pathway growth, results presented in Figs. 2 and 3 are in line with earlier studies on similar models. These studies indicate that, at least for small pathways, the transient responses occur with increasing frequency as pathway size increases and that “switch-” and “derivative-like” responses occur rarely compared with transient responses (12).
To summarize, the key finding of the presented study is that there is an imbalance between the effects of size-decreasing and -increasing mutations on pathway response, the amount of which is related to pathway size and the selection criterion. It is important to note that the ability of such imbalance to produce complexity will depend on the cost of maintaining additional proteins in the pathway and the assumptions regarding the frequency of size-increasing and -decreasing mutations (see Methods). The presented findings do not contradict the results showing that systems of interacting units become unstable with increasing size and connectivity (18). Although we find pathway size to increase in the presented simulations, connectivity (the ratio of actual interactions to all possible interactions) decreases (results not shown). This is because the number of interactions should increase exponentially, whereas pathway size increases linearly, in order for connectivity not to decrease (i.e., newly added proteins should connect to all existing proteins for connectivity to remain constant), a scenario that is not observed in our simulations and may be unlikely in nature.
Conclusion
The intuitive view for the emergence of complexity is that it is due to increasing fitness, a view that has found support from studies on digital organisms (13). However, this study suggests that an imbalance in the effects of size-decreasing and -increasing mutations on function could lead to increase in complexity, supporting a mechanistic or neutral explanation (15). Hence, as long as there is selection acting on a system, even neutral processes that do not cause any immediate fitness benefit would force the system toward higher complexity.
We find that the amount of the aforementioned imbalance is related to complexity itself. It is highest for the simplest possible pathways that could achieve the task for which there is selection operating and decreases with increasing complexity. Hence, as evolution progresses, pathways achieve a certain size or level of complexity that is always well above the minimum required for their function. Further, we find that although simulations with different selection criteria start with pathways of the same size, for pathways that are under selection for functions that are attainable by many different topologies (i.e., that are simpler to achieve), the final level of complexity achieved tends to be higher.
These findings have implications for our understanding of the evolution of new features. For example, in simulating the evolution of a random population of three-protein pathways under selection for responding to a given signal, we reach a structurally diverse population with an average pathway size of ≈14 proteins. Given the exponential relation between pathway size and available pathway topologies, it is plausible to assume that such growth would facilitate the emergence of pathways with various response dynamics that would not have been possible with three proteins only. If such pathways achieve new functions or high-fitness solutions under the current selection criterion, they would be strongly selected. Hence, even though complexity can emerge neutrally, once it results in evolutionary favorable pathways, it may be maintained subsequently by selection.
Finally, we note that the presented results have an interesting connection to robustness. In general, robustness in biological pathways is associated with the ability of such pathways to withstand knockout mutations or disturbances in interaction parameters. Based on this description, many natural pathways have been found to have high robustness (19–24). This observation becomes obvious in light of the presented results. Our simulations suggest that in general, simplest-solution pathways are nonrobust toward deletions. Thus, populations predominantly consisting of such pathways drift toward larger, more complex solutions, and, in this sense, selection for function leads to emergence of robustness. A similar scenario is given for the evolution of robustness in general (25); through evolutionary mechanisms populations move toward wide plateaus on an imaginary fitness landscape, where they would be protected against small perturbations (i.e., they have high robustness). The idea of an imbalance between the functional effects of various mutations having different effects could be seen as the driving force behind such move, leading both to high robustness and high complexity. This statement overlaps with the theories that see complexity and robustness tying closely together (26).
The presented discussion approximates the behavior of biological pathways with a simple mathematical model and makes several assumptions regarding the nature and rate of mutations affecting their structure. Although the true nature of these pathways clearly is more complicated, we believe that the qualitative conclusions of this theoretical study would remain under biologically relevant parameter values (see Methods). These conclusions could also be extendable to any system that is under selection and is subject to mutations affecting its structure. Under such conditions, the idea of complexity arising neutrally could be more general.
Methods
Pathway Model.
We use a generic mathematical model to capture the basic properties of biological pathways. The model has been explained in detail previously (11, 12). In brief, it considers a pathway as a collection of interacting proteins, each of which can exist either in an active (P*i) or inactive (Pi) state. Proteins in the active state can interact with other proteins and influence their activation. There are many different biochemical mechanisms that make such influence possible including phosphorylation, methylation, and physical interaction. Here, we do not distinguish among these different interaction types and model the influence of an active protein on others simply through an interaction coefficient. We assume that the energy required for these activation and deactivation reactions are provided from outside sources such as high-energy molecules. Each active protein is free to interact with any other protein in the pathway, resulting in the following chemical equilibrium
 |
where [P*j] represents the concentration of active form of protein j and kij and lij represent the strength of the interaction between protein i and j. We assume that proteins do not interact with themselves (i.e., kii = lii = 0) and their influence on other proteins can be only activating or deactivating (i.e., kij·lji = 0), resulting in n2 − n parameters for a pathway of n proteins. We do not consider processes such as autophosphorylation or intrinsic phosphotase activity in this model.
To allow the pathway to respond to a signal, we arbitrarily define two proteins in the system as the receptor and effector. We couple the equilibrium state of the receptor to the ligand concentration (i.e., signal) and take the concentration of the effector active form as the response produced by the pathway. In a natural pathway, this response can have various forms, depending on the effector, and can range from transcriptional control to enzymatic activity. To summarize, this model defines a biological pathway by the number of proteins and a set of interaction coefficients. Pathway response to a changing ligand concentration thus can be obtained by solving the set of differential equations resulting from the collection of reactions as shown in Eq. 1:
 |
where δi1 = 1 for i = 1 and δi1 = 0 for i ≠ 1. Note, that the total concentration of each protein [Pitot] is constant and set to 1 (i.e., [P*i] = 1 − [Pi]). Also, we set the maximum value that an interaction coefficient can attain to 1 for computational ease.
To assess the response of a pathway, we initiate the model with equal amounts of active and inactive proteins (i.e., [P*i] = [Pi] = 0.5). After equilibrating the system by integrating the set of differential equations for 1,000 time steps or until steady state is reached, we introduce a signal in the form of a changing ligand concentration (a Gaussian shape with a variance of 6, as shown in Fig. 1). The integration then is continued in presence of this signal for 1,000 time steps or until the system reaches the steady state. At the end of the integration, we validate that we reached a steady state with an eigenvalue analysis. The pathway response then is deduced from the active effector concentration recorded during the second integration period, in the presence of the signal. We classify pathway responses based on the number of sign changes in the derivative of the time course of active effector concentration. To avoid any effects of numerical artifacts on this classification, we consider only responses >10−7 and changes in the response derivative >1%.
Evolutionary Simulations.
We start evolutionary simulations with a population of 1,000 random pathways composed of three proteins. For each generation, we assess the response of each pathway as explained above and calculate its fitness. The calculation of pathway fitness directly defines the selection pressure we impose. Here, we use a step-like fitness function adjusted by the cost of having multiple proteins in the system:
where F stands for pathway fitness, α for a Boolean parameter defined by the selection criteria (i.e., α = 1 if the pathway satisfies the selection criterion, and α = 0 otherwise), n is the number of proteins in the pathway, and c is the fitness cost of each protein (for the reported results, c was 0.001). The use of such a fitness function allows decoupling of fitness and complexity that, in turn, allows us to study the mechanistic effects of mutational events on emergence of complexity. In other words, the fitness function does not allow a continuous increase in fitness with changing performance, which could lead to complexity. In contrast, it assigns essentially the same fitness value to all functional pathways, adjusted by a weak dependence on size.
Throughout the evolutionary simulation, the generation at a given time step is created from the previous one by randomly selecting individuals for replication with replacement. Randomly selected pathways replicate with a probability proportional to their fitness, and undergo mutations per protein with a certain probability (for the reported results this probability was 0.05). Such mutations can cause one of the following with the given probabilities: create a new interaction among existing proteins in the pathway (P = 0.3), delete an existing interaction (P = 0.3) or protein (P = 0.2) in the pathway, create a new protein (P = 0.1) or duplicate an existing one (P = 0.1), and attach this to the pathway. This scheme ensures that from the point of their frequency of occurrence, mutational events do not favor pathway growth or shrinkage. All these mechanisms are biologically plausible. For example, creation and loss of interactions can result from point mutations affecting the structure of proteins, whereas duplication and deletion of proteins can result both from point mutations and from larger genomic mutations.
The relevant frequency of these different mutational events and the fitness cost of each protein (parameter c in Eq. 3) are important parameters that determine whether complexity can arise neutrally under selection (see Results and Discussion). We believe a value of 0.001 for the latter parameter is biologically plausible as the fitness cost of producing one extra protein, compared with the overall fitness of an organism is likely very small, given the total number of proteins that any organism produces. In additional simulations with different c values, we find that increasing the cost for additional proteins decreases the equilibrium size of pathways (see Fig. 8, which is published as supporting information on the PNAS web site) and at c = 0.1 pathways stop growing altogether (results not shown). These results indicate that the quantitative conclusions presented here hold for cases with c as high as 0.01, i.e., where the cost of a single protein is 1% of the overall fitness of a pathway.
Similarly, increasing the frequency of size-decreasing mutations in relevance to size-increasing mutations slows pathway growth (see Fig. 9, which is published as supporting information on the PNAS web site). In this model, we consider as size-increasing and -decreasing mutations all types of mutations that lead to addition (deletion) of a protein to (from) the pathway. For example, one protein-adding mutation would be “to recruit” a nonparticipating, but already existing protein in the organism into the pathway. Given the frequently large number of closely related proteins in an organism, this might be a major mutational mechanism leading to pathway growth. The exact value of the processes that we consider as “protein deletion” and “protein addition” is difficult to determine, but given the large number of nonparticipating proteins, we regard the assumption that protein deletions and protein additions occur at comparable rates as a reasonable null model and as biologically justifiable.
Simulating interaction-deleting mutations involved randomly selecting an interaction in the pathway and setting the associated coefficient to zero, whereas interaction-adding mutations involved selecting two noninteracting proteins randomly and creating a connection among them. Protein-deleting mutations are simulated by randomly selecting a protein and setting coefficients for all interactions involving this protein to zero, whereas protein-adding mutations are simulated by connecting a new protein randomly to existing ones. In the latter case, we connect the newly created protein through so many interactions as a randomly selected existing protein has. This ensures that the effects of protein additions and deletions have similar effects on average. Coefficients for newly created interactions are chosen randomly from the interval [−1, 1]. If a selected interaction coefficient for proteins i and j was negative (positive) then lij (kij) is set to the absolute value of this number and kij (lij) is set to zero. Mutations leading to protein duplications result in addition of a new protein with exactly those interactions as the duplicated protein.
The evolutionary simulation continues until a certain generation and the average pathway properties and the distribution of pathway sizes are monitored. Each simulation is run three to five times to ascertain that the qualitative conclusions made here are robust to stochastic fluctuations inherent in these evolutionary simulations (see Fig. 7). The number of proteins and interactions in the pathway are determined by using the core part of the pathway related to response. In other words, for these analyses, we do not consider proteins that do not influence other proteins in the pathway. This leads to a conservative estimate of these measures and ensures that we do not detect an artificial pathway growth that does not affect pathway response. All simulations are written in C++, and the source code is available from the authors upon request.
Supplementary Material
Acknowledgments
We thank Marcel Salathé for insightful discussions. This work is funded by Gebert Ruef Stiftung Project Grant GRS-031/03.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission. S.S. is a guest editor invited by the Editorial Board.
References
- 1.Adami C. BioEssays. 2002;24:1085–1094. doi: 10.1002/bies.10192. [DOI] [PubMed] [Google Scholar]
- 2.Lynch M, Conery JS. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- 3.Zuckerkandl E. J Mol Evol. 1997;44:470. doi: 10.1007/pl00006169. [DOI] [PubMed] [Google Scholar]
- 4.Zuckerkandl E. J Mol Evol. 2001;53:539–554. doi: 10.1007/s002390010244. [DOI] [PubMed] [Google Scholar]
- 5.Furusawa C, Kaneko K. Phys Rev Lett. 2000;84:6130–6133. doi: 10.1103/PhysRevLett.84.6130. [DOI] [PubMed] [Google Scholar]
- 6.Weng G, Bhalla US, Iyengar R. Science. 1999;284:92–96. doi: 10.1126/science.284.5411.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darwin C. The Origin of Species. London: John Murray; 1859. [Google Scholar]
- 8.Atkinson MR, Savageau MA, Myers JT, Ninfa AJ. Cell. 2003;113:597–607. doi: 10.1016/s0092-8674(03)00346-5. [DOI] [PubMed] [Google Scholar]
- 9.Gardner TS, Cantor CR, Collins JJ. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 10.Rao CV, Kirby JR, Arkin AP. PLoS Biol. 2004;2:E49. doi: 10.1371/journal.pbio.0020049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soyer OS, Pfeiffer T, Bonhoeffer S. J Theor Biol. 2006;241:223–232. doi: 10.1016/j.jtbi.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 12.Soyer OS, Salathe M, Bonhoeffer S. J Theor Biol. 2006;238:416–425. doi: 10.1016/j.jtbi.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 13.Lenski RE, Ofria C, Pennock RT, Adami C. Nature. 2003;423:139–144. doi: 10.1038/nature01568. [DOI] [PubMed] [Google Scholar]
- 14.Maynard Smith J. In: Towards a Theoretical Biology. Waddington CH, editor. Vol 2. Edinburgh: Edinburg Univ Press; 1969. pp. 82–89. [Google Scholar]
- 15.Saunders PT, Ho MW. J Theor Biol. 1976;63:375–384. doi: 10.1016/0022-5193(76)90040-0. [DOI] [PubMed] [Google Scholar]
- 16.Babu MM, Teichmann SA. Nucleic Acids Res. 2003;31:1234–1244. doi: 10.1093/nar/gkg210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teichmann SA, Babu MM. Nat Genet. 2004;36:492–496. doi: 10.1038/ng1340. [DOI] [PubMed] [Google Scholar]
- 18.May RM. Nature. 1972;238:413–414. doi: 10.1038/238413a0. [DOI] [PubMed] [Google Scholar]
- 19.Alon U, Surette MG, Barkai N, Leibler S. Nature. 1999;397:168–171. doi: 10.1038/16483. [DOI] [PubMed] [Google Scholar]
- 20.Cheng P, Yang Y, Liu Y. Proc Natl Acad Sci USA. 2001;98:7408–7413. doi: 10.1073/pnas.121170298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edwards JS, Palsson BO. Biotechnol Prog. 2000;16:927–939. doi: 10.1021/bp0000712. [DOI] [PubMed] [Google Scholar]
- 22.Gonze D, Halloy J, Goldbeter A. Proc Natl Acad Sci USA. 2002;99:673–678. doi: 10.1073/pnas.022628299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.von Dassow G, Meir E, Munro EM, Odell GM. Nature. 2000;406:188–192. doi: 10.1038/35018085. [DOI] [PubMed] [Google Scholar]
- 24.Wagner A. Nat Genet. 2000;24:355–361. doi: 10.1038/74174. [DOI] [PubMed] [Google Scholar]
- 25.de Visser JA, Hermisson J, Wagner GP, Ancel Meyers L, Bagheri-Chaichian H, Blanchard JL, Chao L, Cheverud JM, Elena SF, Fontana W, et al. Evolution Int J Org Evolution. 2003;57:1959–1972. doi: 10.1111/j.0014-3820.2003.tb00377.x. [DOI] [PubMed] [Google Scholar]
- 26.Carlson JM, Doyle J. Proc Natl Acad Sci USA. 2002;99(Suppl 1):2538–2545. doi: 10.1073/pnas.012582499. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.