Abstract
Fast anterograde transport of membrane-bound organelles delivers molecules synthesized in the neuronal cell body outward to distant synapses. Identification of the molecular “zipcodes” on organelles that mediate attachment and activation of microtubule-based motors for this directed transport is a major area of inquiry. Here we identify a short peptide sequence (15 aa) from the cytoplasmic C terminus of amyloid precursor protein (APP-C) sufficient to mediate the anterograde transport of peptide-conjugated beads in the squid giant axon. APP-C beads travel at fast axonal transport rates (0.53 μm/s average velocity, 0.9 μm/s maximal velocity) whereas beads coupled to other peptides coinjected into the same axon remain stationary at the injection site. This transport appears physiologic, because it mimics behavior of endogenous squid organelles and of beads conjugated to C99, a polypeptide containing the full-length cytoplasmic domain of amyloid precursor protein (APP). Beads conjugated to APP lacking the APP-C domain are not transported. Coinjection of APP-C peptide reduces C99 bead motility by 75% and abolishes APP-C bead motility, suggesting that the soluble peptide competes with protein-conjugated beads for axoplasmic motor(s). The APP-C domain is conserved (13/15 aa) from squid to human, and peptides from either squid or human APP behave similarly. Thus, we have identified a conserved peptide zipcode sufficient to direct anterograde transport of exogenous cargo and suggest that one of APP's roles may be to recruit and activate axonal machinery for endogenous cargo transport.
Keywords: fast axonal transport, herpes simplex virus, kinesin anterograde transport, squid giant axon
Membrane-bound vesicles are primarily transported in neurons by fast axonal transport apparently on cytoskeletal tracks. Intracellular pathogens, such as herpes simplex virus (HSV), appear to coopt this cellular transport machinery (1–5) and thus may serve as tools to uncover the mechanisms governing cargo–motor interactions (5, 6). HSV, a neurotropic virus that causes the recurrent cold sore, travels back and forth within neuronal processes at different stages in its life cycle (7). After the initial infection of the mucus membrane, HSV travels within the sensory nerve process to the trigeminal ganglion, where it enters latency. Upon reactivation, newly synthesized viral particles are packaged in the cell body and then travel out to the periphery by fast axonal transport within neuronal processes (1–3, 8–10). Recently we reconstituted HSV transport in the giant axon of the squid in both directions [retrograde, as detergent-stripped particles (11), and anterograde (6)] by injecting GFP-labeled virus into the giant axon of the squid. Viral particles transported in the anterograde direction were associated with a high copy number (≈3,000 molecules per virion) of amyloid precursor protein (APP) (6). Because APP has been implicated as a motor receptor for transport in other systems (12–14), we became interested in whether APP might play a role in anterograde transport of virus. In this study we focus on APP and its ability to drive transport by substituting polystyrene beads for virus and coating the beads with APP.
If APP were sufficient to serve as a receptor for transport machinery, then APP should be able to mediate transport of exogenous cargo, such as polystyrene beads, in the giant axon. Transport of APP-conjugated beads would thus determine whether APP could recruit the axonal motor machinery independent of any other motor receptor that might be present in viral or cellular membranes. Here we mimic viral particles and endogenous transport vesicles with fluorescent beads of comparable size, conjugate the beads to peptides and fusion proteins derived from APP, and determine whether any are sufficient to render the beads capable of fast axonal transport when introduced into the axon.
Results
The C-Terminal Domain of APP Is Sufficient to Mediate Anterograde Transport.
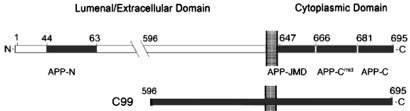
Initial experiments focused on whether a peptide containing the conserved GYENPTY motif, implicated in a variety of protein–protein interactions (15), might be sufficient to mediate transport of exogenous cargo within the squid giant axon. This peptide, termed APP-C, represents the final 15 aa of the 47-aa C terminus of APP, which extends into the cytoplasm, where it has access to cytoplasmic motors. A second peptide, APP-N, corresponding to a domain in the extracellular N terminus, was used as a negative control because this region of APP is internal to the organelle and thus should have no ability to facilitate transport (Table 1).
Table 1.
Diagram of APP showing location of peptide sequences and C99 construct used in this study
| Peptide | Position | Amino acid sequence |
|---|---|---|
| APP-N | 44–63 | HMNCQNGKWDSDPSGTKTCI |
| hJMD | 647–665 | KKKQYTSIHHGVVEVDA |
| sJMD | 565–582 | KRRTQRQRVTHGFVEVDP |
| hCmid | 666–681 | AVTPEERHLSKMQQN |
| sCmid | 583–597 | AASPEERHVANMQMS |
| hAPP-C | 681–695 | GYENPTYKFFEQMQN |
| sAPP-C | 598–612 | GYENPTYKYFE-MQN |
| hAPP-Cj | 681–695 | GEPYFEMNYNTKFQQ |
APP-N and APP-C peptides were each conjugated to a different color of fluorescent bead, and the behaviors of the two types of beads were compared when coinjected into the same axon (Fig. 1). The stability of a coinjected oil droplet was monitored as a sign of axonal integrity. In all experiments two colors of beads, each conjugated to a different peptide, were coinjected. A peptide was determined to lack transport capability only when tested in axons where directed transport of other particles could be observed.
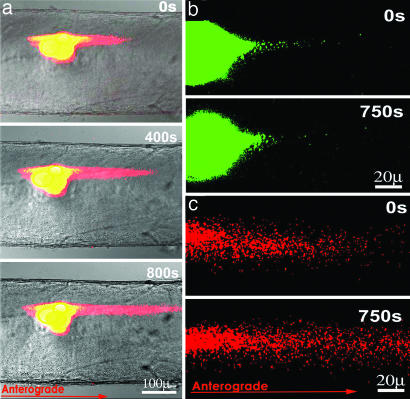
Fig. 1.
Fast anterograde transport of APP-C beads in the giant axon. (a) Select frames from a time sequence (83 frames at 10-s intervals) of APP-C (red) and APP-N (green) beads coinjected into the squid giant axon begun ≈6.5 min after injection. Overlap of red and green fluorescence appears yellow at the injection site (see Movie 1). (b and c) Sequence-specific transport of APP-C beads. Shown are two sequences (100 frames, 8-s intervals each) from the same axon coinjected with green beads conjugated to the jumbled peptide APP-CJ (b) and red beads conjugated to APP-C (c). The sequence with the APP-CJ green beads was captured first (see Movies 2 and 3).
Beads conjugated to APP-C began transport immediately upon injection and already formed a plume heading toward the synapse by the time (1–3 min) the axon was transferred to the confocal microscope (Fig. 1a, red beads, and Movie 1, which is published as supporting information on the PNAS web site). The rate of advance of the plume was calculated by measuring the increase in intensity along a line of pixels drawn from the injection site to the edge of the movie frame. The leading edge of plumes advanced over a 1-mm distance at fast axonal transport rates from 0.33 μm/s to 0.4 μm/s (n = 5). Individual beads conjugated to APP-C displayed fast transport capability (0.41 ± 07 μm/s instantaneous velocity, and 0.9 maximum velocity; n = 227). These transport rates are similar to those of endogenous organelles and exogenous particles transported by active transport in squid and other invertebrate axons (Table 2
Table 2.
Comparisons of velocities
| Cargo type | Average velocity, μm/s (n) | Maximum velocity, μm/s |
|---|---|---|
| Beads conjugated to | ||
| APP-C (human/rat) | 0.41 ± 0.07 (227) | 0.9 |
| APP-C (squid) | 0.53 ± 0.09 (54) | 0.9 |
| C99 | 0.66 ± 0.08 (194) | 1.46 |
| Beads, negatively charged | ||
| in squid axons (17) | 0.078 (70) | 0.4 |
| in crab axons* | 0.3 (22) | 0.6 |
| Mitochondria in squid axons (6) | 0.2 ± 0.08 (45) | 0.36 |
| Endogenous organelles | 0.4–2.0 | 5.0 |
| in squid axoplasm (20, 21) | ||
| HSV in squid axons | ||
| Anterograde (6) | 0.9 ± 0.3 (73) | 1.23 |
| Retrograde (11) | 2.2 ± 0.26 (76) | 2.7 |
*Calculated from data presented in ref. 16.
As expected, APP-N beads remained stationary at the injection site for >1 h of observation (Fig. 1a, green beads) whereas coinjected APP-C beads were rapidly transported past them (Fig. 1a, red beads, and Movie 1). Unconjugated carboxylated beads undergo transport when injected into invertebrate axons (16, 17). Thus, conjugation of the APP-N peptide to beads masks the carboxylic residue and inhibits this transport.
To test whether electrostatic charge or sequence of APP-C was responsible for this transport, we synthesized a peptide with the same amino acid composition as APP-C but in jumbled order, APP-Cj. This peptide had the same charge but not the same sequence as APP-C. APP-Cj conjugated to beads with the same efficiency as APP-C. Coinjection of APP-Cj with APP-C beads revealed that APP-Cj beads remained near the injection site whereas adjacent APP-C beads were rapidly transported (Fig. 1 b, green APP-Cj, and c, red APP-C, and Movies 2 and 3, which are published as supporting information on the PNAS web site). Rare individual APP-Cj beads found beyond the injection site were not seen to move during imaging sessions. Thus, the transport of APP-C was sequence-specific and not a consequence of electrostatic charge.
Next we tested whether other domains from the cytoplasmic tail of APP were also capable of mediating transport. Synthetic peptides designed to span the cytoplasmic domain of APP (Table 2) were coupled to beads and coinjected with APP-C beads into axons. Beads conjugated to equivalent amounts of peptides from other regions of the cytoplasmic domain were never seen to move, even though adjacent APP-C beads were rapidly transported. The difference between APP-C and any other peptide was robust and reproducible, occurring in >40 different axons independent of injection order or bead color.
A significant number of APP-C beads were transported. When 106 APP-C beads were injected, ≈0.5–1.3 × 103 left the injection site per minute. In comparison, only ≈0.001 × 103 APP-N beads were observed to leave the injection site per minute. None of the beads conjugated to the jumbled peptide were observed to leave the injection site during 10 min of recording time in three axons in which adjacent APP-C beads were transported. Similar low rates were observed for beads conjugated to all peptides except APP-C. At this rate, it would take up to 2,000 min for all 106 APP-C beads to leave the injection site. To determine whether all APP-C beads could leave the injection site, we injected the smallest number of beads that could be imaged successfully, 0.5 × 103. Under these conditions, individual beads left the injection site in single file, with approximately half of the beads transported out within 20 min of injection, for a rate of ≈12.5 beads per minute.
Squid APP Is Expressed in Axons and Is Also Sufficient for Cargo Transport.
Squid APP was cloned from the stellate ganglion that contains the cell bodies that give rise to the giant axon. The predicted amino acid sequence is 612 aa long, with greater identities to human APP (38% identity) than to the other human homologues, APLP1 (32%) and APLP2 (34%) (GenBank accession no. DQ913735). The cytoplasmic domain is 64% identical to human and rat, which are 100% identical to each other (Fig. 2b). The squid APP-C domain was nearly identical to rat/human (13/15 aa identity). Antipeptide antibodies against the conserved C terminus that recognize rat brain APP also detected the squid protein, which migrates at slightly lower molecular weight in SDS/PAGE (Fig. 2a).
Fig. 2.
Squid APP is expressed in the giant axon and is homologous to human/rat APP. (a) Western blot of rat brain extract (RB) and squid axoplasm (SA) probed in parallel for APP. Molecular masses are indicated on left: 150, 100, 55, 45, and 25 kDa. (b) Amino acid sequence comparison of the cytoplasmic domain of squid APP with human and rat. Identities with squid are highlighted in yellow. The antigenic epitope used to generate the antibody is indicated by a red bar above the sequence (amino acids 681–695).
Peptides spanning the 47-aa cytoplasmic domain of squid APP were synthesized (Table 1). These peptides behaved similarly to those derived from the human sequence. Only the APP-C peptide was uniquely capable of driving fast transport of beads (0.53 ± 0.09 instantaneous velocity, 0.9 μm/s maximal velocity; n = 54) (Table 2). No motility was observed in 37 axons for other squid peptides.
Analysis of APP-C Bead Transport.
APP-C beads traveled uniquely in the anterograde direction with pauses of varying intervals but no reversals, as determined by analyzing time-lapse sequences at high magnification with intervals of 4–6 s (Fig. 3a and b, and Movie 4, which is published as supporting information on the PNAS web site). This transport was sustained for >1 h at distances >2 mm from the injection site. Tracings of multiple individual bead tracks passing through the same microscopic field over time revealed that APP-C beads traveled on interweaving parallel tracks (Fig. 3 b and c). In some cases several beads traced the same track, suggesting that beads follow each other on the same or neighboring microtubule–actin bundles (18). Some beads appeared or disappeared during the sequence, as if they moved on tracks tangential to the optical section.
Fig. 3.
Analysis of individual APP-C bead movements. (a) During 240 s of this example (60 frames at 4-s intervals), nine beads move and six remain stationary. Arrowheads mark two representative beads that travel in the anterograde direction past a group of stationary beads (see Movie 4). (b) Tracings of a few bead tracks from a demonstrate linearity of movement. Bead tracks are indicated in a different color so that overlapping trajectories of individual beads can be followed during the time sequence of the movie; stationary beads are black. (c) Tracings of a larger number of APP-C beads in another axon. APP-C beads travel exclusively in the anterograde direction with no reversals (100 frames at 4-s intervals). Bead tracks are colored as in b.
C99, a Polypeptide Containing the Full-Length Cytoplasmic Domain of APP, Mediates Bead Transport.
C99 contains the C-terminal 99 aa of APP, spanning from a short segment of the extracellular domain, the transmembrane domain, and the complete cytoplasmic domain from which we derived our peptides (Table 1). Because each HSV particle with anterograde motility has on average 3,000 copies of APP (6), we conjugated beads with equivalent amounts of C99 (3,000 copies per 100-nm bead), thereby mimicking the physiological conditions of viral transport in terms of size and APP display.
In axons coinjected with C99 beads and APP-C beads, both types of beads were transported (Fig. 4a and Movie 5, which is published as supporting information on the PNAS web site). Beads conjugated to either protein could be observed to follow each other along the same apparent track at similar velocities (Fig. 4b). Distance per time plots of two representative beads moving on superimposable tracks shows equivalent transport rates and pauses (Fig. 4c). Calculations of instantaneous velocities of C99 and APP-C obtained from multiple bead runs further confirms similarity in movement (Fig. 4d and Table 2). Despite slightly decreased average instantaneous velocities compared with C99, APP-C beads accomplish similar distances because of longer run lengths and fewer pauses (Table 4, which is published as supporting information on the PNAS web site).
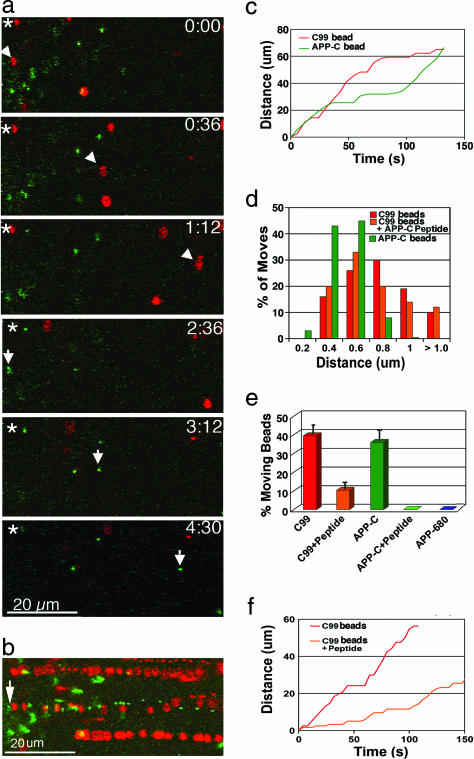
Fig. 4.
Analysis and comparisons of movements of C99 beads with various other bead conjugates and the effect of APP-C peptide preinjection. (a) Fast anterograde transport of C99 (red) and APP-C (green) beads in a coinjected axon (77 frames at 6-s intervals). A green APP-C bead (arrow, bottom three panels) follows the track of a red C99 bead (arrowhead, top three panels) (see Movie 5). The asterisks in the upper left corners indicate stationary beads. (b) Track traces. Same movie sequence as in a with the 77 frames superimposed showing the positions of beads as they traverse the field. An arrow indicates the starting point for the two beads that follow each other. (c) Plot of distance moved per time comparing representative C99 (red) and APP-C (green) beads. Bead movements were measured in a 100-frame, 6-s time-lapse movie of a coinjected axon. (d) Histogram of instantaneous velocities for C99 and APP-C with and without soluble peptide. In all cases, the distance moved per frame was measured in 4-s time-lapse movies for C99 (194 moves, four different beads), APP-C (227 moves, six beads), and C99 plus peptide (49 moves, three beads). No moves were observed for APP-C with coinjected soluble peptide (see Movie 6). (e) Comparison of the percentage of beads that move for different bead conjugates with and without peptide. Comparable amounts of beads were injected for each experiment (see Fig. 5, which is published as supporting information on the PNAS web site). An imaging field was selected at random within 500 μm of the injection site. Areas with large masses of beads were discarded. All beads appearing in each 100-frame movie were counted. Moving beads were any that translocated more than three consecutive frames. The percentage of moving beads was calculated for three different 100-frame movie sequences and then averaged. C99, 39.5 ± 4.5%; C99 plus peptide, 10.3 ± 4.4%; APP-C, 36 ± 6.2%; APP-C plus peptide, 0%; APP-680, 0%. Fewer C99 beads and no APP-C beads were found outside the injection site after peptide preinjection. APP-680 beads were rarely found beyond the injection site, and none moved (see Movies 5 and 6). (f) Plot of distance moved per time comparing C99 beads with and without preinjection of APP-C peptide. Shown are plots of two representative C99 beads moving in two different axons, one not injected with peptide (C99) and the other (C99 plus peptide) preinjected with 500 pl of APP-C peptide at 2.5 mg/ml (0.66 picomoles). Each movie sequence was 100 frames at 4-s intervals.
To test whether motility was due to an interaction of the APP-C domain with axoplasmic motors, we preinjected axons with soluble APP-C peptide followed by coinjection of C99 and APP-C beads. Soluble peptide would be expected to compete with protein-conjugated beads for axoplasmic motors. Peptide preinjection resulted in a 4-fold reduction of moving C99 beads and no motility of APP-C beads (Fig. 4e and Movie 6, which is published as supporting information on the PNAS web site). Movement of a few C99 beads in peptide-injected axons confirmed that transport was still possible, although diminished. When C99 beads do move in the presence of peptide, the instantaneous velocity is similar to beads without preinjected peptide (Fig. 4d and Table 2), as would be expected if the peptide competed for the motor. In addition to reducing the proportion of beads that moved, peptide preinjection decreased the average run length by half, thus doubling the occurrence of pauses (Fig. 4f and Table 2).
Beads conjugated to a construct similar to C99 but truncated at amino acid 680, and thus lacking the APP-C domain (APP-680), were immotile in axons where other coinjected beads were transported out from the injection site.
The APP-C Domain Is Highly Conserved.
The 15-aa APP-C domain is identical in APP of vertebrates (Table 3), and the GYENPTY within that sequence is identical also with invertebrates: Drosophila melanogaster, Caenorhabditis elegans, and Loligo pealei (squid). The squid sequence reported here is more similar to human than is either worm or fly, having only two differences: one amino acid substitution and one omission. The entire cytoplasmic domain of APP is almost as highly conserved as APP-C, with 100% identity among mammals and >90% identity between mammals and other vertebrates. These identities are higher than those for the full-length APP sequence because of the more divergent extracellular N terminus.
Table 3.
Conservation of APP-C and APP
| Species | GenBank accession no. | APP-C amino acid sequence lineup | % identity (C terminus/full-length) |
|---|---|---|---|
| Homo sapiens | P05067 | GYENPTYKFFEQMQN | 100/100 |
| Macaca fascicularis | A49795 | GYENPTYKFFEQMQN | 100/99 |
| Rattus rattus | S00550 | GYENPTYKFFEQMQN | 100/97 |
| Mus musculus | A27485 | GYENPTYKFFEQMQN | 100/97 |
| Xenopus laevis | JH0773 | GYENPTYKFFEQMQN | 96/87 |
| Danio rerio | Q90W28 | GYENPTYKFFEQMQN | 92/74 |
| Takifugu radiatus | O93279 | GYENPTYKFFEQMQN | 92/71 |
| Loligo p.(squid) | GYENPTYKYFE-MQN | 64/38 | |
| D. melanogaster | A32758 | GYENPTYKYFE-VKE | 53/30 |
| C. elegans | T15795 | GYENPTYSFFD-SKA | 48/36 |
| H. sapiensAPLP1 | AAH12889 | GYENPTYRFLEERP | 53/41 |
| H. sapiensAPLP2 | AAH12889 | GYENPTYKYLEQMQI | 68/52 |
Discussion
Here we show that APP contains a 15-aa domain sufficient to drive the transport of beads in the giant axon of the squid. A squid APP homologue is expressed in the giant axon and encodes an identical domain equally capable of mediating transport of beads. In contrast, no other peptide from the cytoplasmic domain of either human or squid APP had this activity, nor did a peptide with the same amino acids as APP-C but in jumbled order. The activity of the APP-C peptide paralleled the activity of C99, a polypeptide containing the complete cytoplasmic domain. C99 also mediated transport of beads when conjugated to beads at molar amounts equivalent to that associated with motile HSV (3,000 copies of APP per particle) (6). Truncation of the terminal 15 aa removes transport capability. Preinjection of soluble APP-C peptide abolishes APP-C bead transport and decreases the number of C99 beads moving by 75% while doubling pause length and frequency. Thus, C99 may have a higher affinity for the axoplasmic motor than APP-C. The APP-C sequence is nearly identical across species, from human to squid, worm, and fly, arguing that these results uncover a highly conserved physiological interaction between APP and anterograde motor machinery. Thus, APP would be sufficient to mediate the anterograde transport of HSV during egress and likely plays a role in endogenous membrane trafficking.
Transport of APP Beads Resembles Physiological Fast Axonal Transport.
A test for sufficiency of any motor receptor is difficult to design inside cells, because cellular organelles contain many different proteins and may display more than one type of motor receptor. Injection of beads coated with only one type of protein into the axon allowed us to test directly whether any protein on its own was sufficient for transport independent of other organelle motor receptors. These studies do not address whether APP is necessary for cargo transport, which is unlikely because there may be many ways for cargo to recruit motors, including binding to lipids (19).
The behavior of APP-C and C99 beads in axons resembled that of endogenous organelles, suggesting that APP-C beads moved by normal physiologic mechanisms (see Supporting Materials and Methods and Table 4). Of particular interest to us was the similarity in direction and speed of C99 beads (+0.66 ± 0.08 μm/s) to those of APP-associated HSV particles (+0.9 ± 0.3 μm/s) in the giant axon (6). Thus, APP is sufficient to mediate transport of 100-nm beads at concentrations similar to those found on motile virus. This finding supports our hypothesis that APP serves to mediate the transport of HSV during egress.
APP Bead Transport in Context.
The average velocity and maximal velocity of moving APP-C and C99 beads are consistent with kinesin-1-mediated fast axonal transport: in vitro rates of 0.5 μm/s for soluble kinesin passively adherent to carboxylated beads (20) and 0.4–2.0 μm/s for axoplasmic organelles (21, 22). Variation in transport rates of cargo carried by kinesin is attributed in part to regulation of the motor head by a cargo-binding domain in the tail (23–25) and by phosphorylations (26, 27).
The GYENPTY domain in the active peptide APP-C is also found in APLP1 and APLP2 (Table 3), and thus these homologues may also mediate transport. It has been shown that kinesin light chain 1 binds APP in vitro (13) as well as APLP1 and APLP2 (28). These proteins may compensate for APP in knockouts where transport remains normal despite APP loss (13, 28, 29). Whether this kinesin light chain 1–APP interaction is specific has been questioned, and whether this interaction activates motor activity has not been tested. APP–kinesin interactions could occur by other mechanisms, either direct binding to heavy chains and/or indirect binding via scaffolding proteins such as JIP1/2 that bind both kinesin and APP (30, 31). In one report immunoprecipitation with anti-APP antibodies directed against the APP-C domain failed to detect an association with kinesin (28). Such antibodies could compete with kinesin light chain 1 for binding.
No single peptide sequence, no “universal code,” common to all receptors that bind kinesin has been found, consistent with kinesin's ability to mediate transport of many different cargos in both the fast and slow compartments, including neurofilament (32–34). Kinesin-1 binding partners, such as GRIP1 (35), kinectin (36), JIP1/2, and JIP3 (31, 37), do not contain the APP-C sequence. Thus, other sequences may also encode the capacity to recruit kinesin and mediate transport. Kinesin-binding domains in JIP1 (37) and GRIP1 (35) have been identified, and these sequences await testing in motility assays such as we report here.
The domain of APP695 that drives transport, amino acids 681–695, has been implicated in other cellular activities, including endocytosis and transcriptional regulation (38–41). This domain contains the NPXY motif involved in many protein–protein interactions (39, 42). Regulation of which protein interacts with the NPXY domain of APP could occur by phosphorylations or changes in conformation. Indeed, phosphorylation of the C-terminal fragment of APP apparently affects its association with kinesin light chain 1 (43) and localization to axonal processes (44, 45). The slight differences in transport behavior of C99 compared with APP-C beads could be secondary to posttranslational events on epitopes outside the APP-C domain, such as phosphorylations of Thr-668, that affect localization.
The robust motility of C99 beads in the intact axon argues for a physiologic role of APP in recruitment of anterograde transport machinery inside cells. The ability of the short APP-C peptide to tag exogenous cargo for fast axonal transport has implications beyond demonstrating a role for APP in transport. This peptide sequence could be used to send exogenous molecules expressed within the neuron to the synapse. The identification of a minimal amino acid sequence that is capable of delivering exogenous cargo to the presynaptic terminal offers exciting possibilities, such as tagging tract tracers to delineate neuronal pathways or tagging exogenous molecules for delivery to presynaptic termini for therapeutic or engineering purposes.
Methods
Dissection and Microinjection of the Giant Axon of the Squid.
The giant axon was obtained from squid (L. pealei) at the Marine Biological Laboratory (46). Red and green beads, each conjugated to a different peptide (1.5 nl at 1012 to 109 beads per milliliter), were loaded into a mercury micropipette either as a mixture or in equal volumes separated by oil (11, 17, 47) (Fig. 5). For each pair of beads the order of injection was alternated such that each color would have an opportunity to recruit motors first.
Peptide Conjugation to Fluorescent Microspheres.
Carboxylated microspheres (beads, 0.1-μm diameter) with red (580/605 nm) and green (505/515 nm) fluorescence (Invitrogen, Carlsbad, CA) were washed through a Low Binding Durapore filter (100-nm cutoff; Millipore, Billerica, MA). Uncoated beads were diluted in motility buffer (21) and injected without further treatment.
Custom synthesized peptides (Aves Labs, Tigard, OR) were cross-linked to beads via their N termini to allow presentation of the carboxyl end of the peptide to the cytoplasm, mimicking the physiological orientation of APP. APP constructs (C99 and APP-680) were generated by PCR from human APP–yellow fluorescent protein in pShuttleCMV (E.-M. Mandelkow and Jacek Biernat, Max Planck Institute, Hamburg, Germany) (48) (Supporting Materials and Methods, which is published as supporting information on the PNAS web site). APP–yellow fluorescent protein is actively transported in neurons (48). For C99 and APP-680 the yellow fluorescent protein moiety was replaced with monomer red fluorescent protein (49) and GFP, respectively. Each construct was cross-linked to a bead of similar spectrum as the fluorescent protein, and conjugated beads were individually checked for monochromic emission by confocal microscopy. C99 beads displayed only red and no green fluorescence, and APP-680 beads were only green with no red. Accuracy of all sequences was confirmed by sequencing. Recombinant protein was generated in PC12 cells and purified by antifluorescent protein affinity columns (Vector Laboratories, Burlingame, CA). Protein concentrations were determined by protein assay (Bio-Rad, Hercules, CA), and composition and purity were determined by SDS/PAGE.
Peptides or recombinant proteins (20 μl of a 2 mg/ml stock) were mixed in 300 μl of 50 mM MES buffer with 10 μl of 2% beads for 15 min. Cross-linker [1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride; Molecular Probes, Eugene, OR] was added to a concentration of 10 μg/ml, and the solution was adjusted to pH 6.5 with 1 M NaOH. After 5 h the reaction was quenched by adding 2 M glycine for a final concentration of 0.1 M. Conjugation was confirmed by analyzing peptide concentrations before and after by protein assay. The protein:bead ratio was adjusted so that equivalent molar amounts of each protein would be conjugated to the beads. Efficiency of conjugation was relatively similar for all peptides. Binding sites on the beads (106) were saturated by using 300- to 1,000-fold excess peptide. For C99 and APP-680, protein concentration was adjusted so that 3,000 copies per bead were conjugated to the surface and unoccupied sites were blocked with glycine.
Imaging Axonal Transport by Confocal Microscopy.
Detection parameters were set to ensure that each color was uniquely detected in its appropriate channel by imaging beads on coverslips with the LSM510 Laser Scanning Confocal Microscope (Zeiss, Thornwood, NY). Fluorescent bead movements in the axon were collected with ×10 Plan Neofluor 0.3 N.A. air and ×40 Achroplan 0.8 N.A. water correctible objectives. Green and red fluorescence and phase images were collected simultaneously by using LSM510 multitracking (Zeiss).
Analysis of Transport.
Rates and trajectories of APP-C beads were measured by stepping through the frames in either the LSM Image Browser (Zeiss) or NIH ImageJ (http://rsb.info.nih.gov/nih-image). Only particles moving into and out of a frame were included. Rates of moving particles were statistically analyzed and graphed by using Excel (Microsoft, Redmond, WA).
Cloning and Sequencing of Squid APP.
Squid APP was cloned as part of a new project to acquire a database of genes expressed in the squid nervous system (J.A.D., unpublished data). Extracted mRNA from squid stellate ganglia was reverse-transcribed by oligo(dT) priming. Double-stranded cDNA was directionally ligated into pCMVSport6 vector (Invitrogen). Resultant clones were sequenced by using universal primers from the 5′ end. A Blastp search of predicted amino acid sequences from these clones identified one homologous to human APP.
Supplementary Material
Acknowledgments
We thank Thomas S. Reese for his support crucial to this work. We also thank Marine Biological Laboratory colleagues James Galbraith, Laurinda Jaffe, Mark Terasaki, Louie Kerr, and Rudi Rottenfusser, as well as ChengBiao Wu, Janice Valetta in Bill Mobley's laboratory at Stanford University (Stanford, CA), Michael Liebling in Scott Fraser's laboratory at California Institute of Technology (Pasadena, CA) for computational imaging assistance, and other members of the E.L.B. laboratory: Paulette Ferland, Bryan Kinney, Kelly Cleveland-Donovan, and Octavian Biris for technical assistance and/or advice. This work was supported by National Institute of General Medical Sciences Grant GM-47562 and National Institute of Neurological Disorders and Stroke Grant NS046810 (to E.L.B.); Dart Neurosciences (E.L.B.); a Molecular Biology, Cell Biology, and Biochemistry/National Institutes of Health Graduate Program Training grant (P.S.-K.); and intramural support from the National Institute of Neurological Disorders and Stroke (to J.A.D.).
Abbreviations
- APP
amyloid precursor protein
- HSV
herpes simplex virus.
Footnotes
This work was presented in preliminary form at the Annual Meeting of the American Society for Investigative Pathology, San Diego, CA, April 11–15, 2003; and at the Marine Biological Laboratories General Scientific meeting, Woods Hole, MA, August 9–11, 2004.
The authors declare no conflict of interest.
Data deposition: The sequence for squid APP reported in this paper has been deposited in the GenBank database (accession no. DQ913735).
References
- 1.Enquist LW, Tomishima MJ, Gross S, Smith GA. Vet Microbiol. 2002;86:5–16. doi: 10.1016/s0378-1135(01)00486-2. [DOI] [PubMed] [Google Scholar]
- 2.Holland DJ, Miranda-Saksena M, Boadle RA, Armati P, Cunningham AL. J Virol. 1999;73:8503–8511. doi: 10.1128/jvi.73.10.8503-8511.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miranda-Saksena M, Armati P, Boadle RA, Holland DJ, Cunningham AL. J Virol. 2000;74:1827–1839. doi: 10.1128/jvi.74.4.1827-1839.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dohner K, Nagel CH, Sodeik B. Trends Microbiol. 2005;13:320–327. doi: 10.1016/j.tim.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 5.Bearer EL, Satpute-Krishnan P. Current Drug Targets Infect Disord. 2002;2:247–264. doi: 10.2174/1568005023342407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Satpute-Krishnan P, DeGiorgis JA, Bearer EL. Aging Cell. 2003;2:305–318. doi: 10.1046/j.1474-9728.2003.00069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roizman B, Knipe DM. In: Fields Virology. Knipe DM, Howley PM, editors. Vol. 2. Philadelphia: Lippincott Williams; 2001. pp. 2399–2459. [Google Scholar]
- 8.Ohara PT, Chin MS, LaVail JH. J Virol. 2000;74:4776–4786. doi: 10.1128/jvi.74.10.4776-4786.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lycke E, Kristensson K, Svennerholm B, Vahlne A, Ziegler R. J Gen Virol. 1984;65:55–64. doi: 10.1099/0022-1317-65-1-55. [DOI] [PubMed] [Google Scholar]
- 10.Kristensson K, Lycke E, Roytta M, Svennerholm B, Vahlne A. J Gen Virol. 1986;67:2023–2028. doi: 10.1099/0022-1317-67-9-2023. [DOI] [PubMed] [Google Scholar]
- 11.Bearer EL, Breakefield XO, Schuback D, Reese TS, LaVail JH. Proc Natl Acad Sci USA. 2000;97:8146–8150. doi: 10.1073/pnas.97.14.8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gunawardena S, Goldstein LS. Neuron. 2001;32:389–401. doi: 10.1016/s0896-6273(01)00496-2. [DOI] [PubMed] [Google Scholar]
- 13.Kamal A, Stokin GB, Yang Z, Xia CH, Goldstein LS. Neuron. 2000;28:449–459. doi: 10.1016/s0896-6273(00)00124-0. [DOI] [PubMed] [Google Scholar]
- 14.Kamal A, Almenar-Queralt A, LeBlanc JF, Roberts EA, Goldstein LS. Nature. 2001;414:643–648. doi: 10.1038/414643a. [DOI] [PubMed] [Google Scholar]
- 15.De Strooper B, Annaert W. J Cell Sci. 2000;113:1857–1870. doi: 10.1242/jcs.113.11.1857. [DOI] [PubMed] [Google Scholar]
- 16.Adams RJ, Bray D. Nature. 1983;303:718–720. doi: 10.1038/303718a0. [DOI] [PubMed] [Google Scholar]
- 17.Terasaki M, Schmidek A, Galbraith JA, Gallant PE, Reese TS. Proc Natl Acad Sci USA. 1995;92:11500–11503. doi: 10.1073/pnas.92.25.11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bearer EL, Reese TS. J Neurocytol. 1999;28:85–98. doi: 10.1023/a:1007025421849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klopfenstein DR, Tomishige M, Stuurman N, Vale RD. Cell. 2002;109:347–358. doi: 10.1016/s0092-8674(02)00708-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vale RD, Schnapp BJ, Reese TS, Sheetz MP. Cell. 1985;40:559–569. doi: 10.1016/0092-8674(85)90204-1. [DOI] [PubMed] [Google Scholar]
- 21.Brady ST, Lasek RJ, Allen RD. Science. 1982;218:1129–1131. doi: 10.1126/science.6183745. [DOI] [PubMed] [Google Scholar]
- 22.Vale RD, Schnapp BJ, Reese TS, Sheetz MP. Cell. 1985;40:449–454. doi: 10.1016/0092-8674(85)90159-x. [DOI] [PubMed] [Google Scholar]
- 23.Jiang MY, Sheetz MP. Biophys J. 1995;68:283S–284S. [PMC free article] [PubMed] [Google Scholar]
- 24.Friedman DS, Vale RD. Nat Cell Biol. 1999;1:293–297. doi: 10.1038/13008. [DOI] [PubMed] [Google Scholar]
- 25.Seiler S, Kirchner J, Horn C, Kallipolitou A, Woehlke G, Schliwa M. Nat Cell Biol. 2000;2:333–338. doi: 10.1038/35014022. [DOI] [PubMed] [Google Scholar]
- 26.Morfini G, Szebenyi G, Brown H, Pant HC, Pigino G, DeBoer S, Beffert U, Brady ST. EMBO J. 2004;23:2235–2245. doi: 10.1038/sj.emboj.7600237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morfini G, Szebenyi G, Elluru R, Ratner N, Brady ST. EMBO J. 2002;21:281–293. doi: 10.1093/emboj/21.3.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lazarov O, Morfini GA, Lee EB, Farah MH, Szodorai A, DeBoer SR, Koliatsos VE, Kins S, Lee VM, Wong PC, et al. J Neurosci. 2005;25:2386–2395. doi: 10.1523/JNEUROSCI.3089-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng H, Jiang M, Trumbauer ME, Sirinathsinghji DJ, Hopkins R, Smith DW, Heavens RP, Dawson GR, Boyce S, Conner MW, et al. Cell. 1995;81:525–531. doi: 10.1016/0092-8674(95)90073-x. [DOI] [PubMed] [Google Scholar]
- 30.Taru H, Iijima K, Hase M, Kirino Y, Yagi Y, Suzuki T. J Biol Chem. 2002;277:20070–20078. doi: 10.1074/jbc.M108372200. [DOI] [PubMed] [Google Scholar]
- 31.Verhey KJ, Meyer D, Deehan R, Blenis J, Schnapp BJ, Rapoport TA, Margolis B. J Cell Biol. 2001;152:959–970. doi: 10.1083/jcb.152.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prahlad V, Helfand BT, Langford GM, Vale RD, Goldman RD. J Cell Sci. 2000;113:3939–3946. doi: 10.1242/jcs.113.22.3939. [DOI] [PubMed] [Google Scholar]
- 33.Terada S, Hirokawa N. Curr Opin Neurobiol. 2000;10:566–573. doi: 10.1016/s0959-4388(00)00129-x. [DOI] [PubMed] [Google Scholar]
- 34.Shah JV, Cleveland DW. Curr Opin Cell Biol. 2002;14:58–62. doi: 10.1016/s0955-0674(01)00294-0. [DOI] [PubMed] [Google Scholar]
- 35.Setou M, Seog DH, Tanaka Y, Kanai Y, Takei Y, Kawagishi M, Hirokawa N. Nature. 2002;417:83–87. doi: 10.1038/nature743. [DOI] [PubMed] [Google Scholar]
- 36.Toyoshima I, Yu H, Steuer ER, Sheetz MP. J Cell Biol. 1992;118:1121–1131. doi: 10.1083/jcb.118.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horiuchi D, Barkus RV, Pilling AD, Gassman A, Saxton WM. Curr Biol. 2005;15:2137–2141. doi: 10.1016/j.cub.2005.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cescato R, Dumermuth E, Spiess M, Paganetti PA. J Neurochem. 2000;74:1131–1139. doi: 10.1046/j.1471-4159.2000.741131.x. [DOI] [PubMed] [Google Scholar]
- 39.Homayouni R, Rice DS, Sheldon M, Curran T. J Neurosci. 1999;19:7507–7515. doi: 10.1523/JNEUROSCI.19-17-07507.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okamoto M, Nakajima Y, Matsuyama T, Sugita M. Neuroscience. 2001;104:653–665. doi: 10.1016/s0306-4522(01)00124-5. [DOI] [PubMed] [Google Scholar]
- 41.Trommsdorff M, Borg JP, Margolis B, Herz J. J Biol Chem. 1998;273:33556–33560. doi: 10.1074/jbc.273.50.33556. [DOI] [PubMed] [Google Scholar]
- 42.Rodriguez-Boulan E, Musch A. Biochim Biophys Acta. 2005;1744:455–464. doi: 10.1016/j.bbamcr.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 43.Inomata H, Nakamura Y, Hayakawa A, Takata H, Suzuki T, Miyazawa K, Kitamura N. J Biol Chem. 2003;278:22946–22955. doi: 10.1074/jbc.M212160200. [DOI] [PubMed] [Google Scholar]
- 44.Muresan Z, Muresan V. J Cell Biol. 2005;171:615–625. doi: 10.1083/jcb.200502043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muresan Z, Muresan V. J Neurosci. 2005;25:3741–3751. doi: 10.1523/JNEUROSCI.0152-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bearer EL, DeGiorgis JA, Bodner RA, Kao AW, Reese TS. Proc Natl Acad Sci USA. 1993;90:11252–11256. doi: 10.1073/pnas.90.23.11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jaffe LA, Terasaki M. Methods Cell Biol. 2004;74:219–242. doi: 10.1016/s0091-679x(04)74010-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stamer K, Vogel R, Thies E, Mandelkow E, Mandelkow EM. J Cell Biol. 2002;156:1051–1063. doi: 10.1083/jcb.200108057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. Proc Natl Acad Sci USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.