Abstract
Pseudomonas aeruginosa delivers the toxin ExoU to eukaryotic cells via a type III secretion system. Intoxication with ExoU is associated with lung injury, bacterial dissemination and sepsis in animal model and human infections. To search for ExoU targets in a genetically tractable system, we used controlled expression of the toxin in Saccharomyces cerevisiae. ExoU was cytotoxic for yeast and caused a vacuolar fragmentation phenotype. Inhibitors of human calcium-independent (iPLA2) and cytosolic phospholipase A2 (cPLA2) lipase activity reduce the cytotoxicity of ExoU. The catalytic domains of patatin, iPLA2 and cPLA2 align or are similar to ExoU sequences. Site-specific mutagenesis of predicted catalytic residues (ExoUS142A or ExoUD344A) eliminated toxicity. ExoU expression in yeast resulted in an accumulation of free palmitic acid, changes in the phospholipid profiles and reduction of radiolabeled neutral lipids. ExoUS142A and ExoUD344A expressed in yeast failed to release palmitic acid. Recombinant ExoU demonstrated lipase activity in vitro, but only in the presence of a yeast extract. From these data we conclude that ExoU is a lipase that requires activation or modification by eukaryotic factors.
Keywords: cytotoxicity/ExoU/lipase/Pseudomonas/type III secretion
Introduction
Pseudomonas aeruginosa is an opportunistic pathogen that infects individuals with compromised immune systems and patients with cystic fibrosis. Pseudomonas aeruginosa utilizes a specialized cellular intoxication mechanism, termed the type III secretion system (TTSS), to deliver toxins into eukaryotic cells and to eliminate or modify host innate responses (Frank, 1997; Kurahashi et al., 1999; Roy-Burman et al., 2001). At least four proteins are translocated by the P.aeruginosa TTSS (Frank, 1997). These proteins include two bi-functional enzymes ExoS and ExoT (reviewed in Barbieri, 2000), ExoY (Yahr et al., 1998) and ExoU (Finck-Barbançon et al., 1997; Finck-Barbançon and Frank, 2001).
ExoS, ExoT and ExoY have clearly defined enzymatic activities and in general, are responsible for altering cellular cytoskeletal components. ExoS and ExoT possess N-terminal domains that encode a GTPase activating protein activity (GAP) (Goehring et al., 1999). ExoY is an adenylyl cyclase produced by some strains of P.aeruginosa (Yahr et al., 1998). Intoxication with ExoS, ExoT and ExoY causes cell rounding and detachment, and may contribute to infection by inhibiting or preventing bacterial uptake and phagocytosis. The C-terminal domain of ExoS also encodes an ADP-ribosyltransferase (ADPRT) activity that covalently modifies several members of the Ras superfamily of G-proteins (Barbieri, 2000). ExoS ADPRT activity correlates with cellular cytotoxicity (Pederson and Barbieri, 1998).
Another protein delivered by the TTSS, ExoU, possesses a unique cytotoxic effect, which is rapid and potent. Pseudomonas aeruginosa strains producing ExoU are capable of destroying cellular monolayers during short infection periods (Finck-Barbançon et al., 1997). ExoU production is associated with accelerated lung injury in experimental animals and in patients, and plays a role in the development of septic shock (Finck-Barbançon et al., 1997; Kurahashi et al., 1999; Allewelt et al., 2000).
ExoU is expressed and secreted as a 74 kDa protein (687 amino acids) and predicted to be mainly hydrophilic (Finck-Barbançon et al., 1997). Transient transfection analyses suggest that the domain structure of the protein is complex (Finck-Barbançon and Frank, 2001). Plasmids encoding full-length but not truncated forms of ExoU inhibit reporter gene expression and result in cellular permeability and death. Toxicity required only with nanogram levels of plasmid DNA and biological effects could be detected within 3 h of transfection. Functional domains were localized to three different regions of the molecule (Finck-Barbançon and Frank, 2001).
Transfection studies in mammalian cells were useful for analyzing the domain structure of ExoU; however, the extreme toxicity limited biochemical and trafficking analyses. Other investigators have utilized expression of bacterial cytotoxins in Saccharomyces cerevisiae as a model system to overcome some of the issues related to transfection studies (Von Pawel-Rammingen et al., 2000; Lesser and Miller, 2001). Expression of Yersinia YopE in yeast linked the induction of cytotoxicity with a yeast growth inhibition phenotype (Von Pawel-Rammingen et al., 2000; Lesser and Miller, 2001). YopE was also shown to also block the polarization of the yeast cytoskeleton and cell cycle progression (Lesser and Miller, 2001).
In this manuscript we report the use of yeast hosts and controlled expression to characterize the mechanism of action of ExoU. Identification of a vacuolar fragmentation phenotype led to the hypothesis that ExoU encoded an enzymatic activity resulting in membrane disruption. Subsequent genetic and biochemical analyses demonstrate that ExoU possesses lipase activity and utilizes a serine-aspartate catalytic dyad similar to patatin, cPLA2 and iPLA2. ExoU represents the first lipase delivered by a type III system.
Results
ExoU expression correlates with a loss in yeast viability
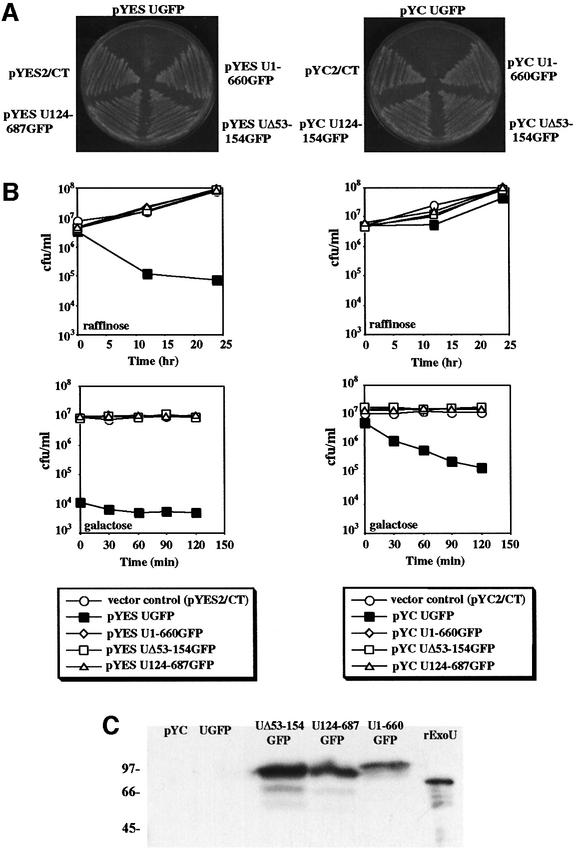
Under conditions of constitutive expression, full-length clones of ExoU could not be obtained in yeast. To determine if ExoU was toxic and to regulate ExoU expression, the gene was cloned into a high copy number (pYES2/CT, 2-µ origin) or a low copy number (pYC2/CT, CEN6) vector. In each case the GAL1 promoter controlled exoU transcription. Expression from the GAL1 promoter is repressed when transformants are grown in glucose, derepressed (basal transcript levels) when cells are grown in raffinose, and induced when transformants are grown in galactose. Transformants containing a full-length clone of exoU grew on glucose plates (not shown) but failed to grow on galactose plates (Figure 1A). Growth inhibition was observed when ExoU was expressed from both 2-µ and CEN6 plasmids, indicating that biological activity was detectable when the gene was present in a low copy number vector. ExoU deletion derivatives appeared to have no affect on yeast viability after growth on glucose (not shown) or galactose plates (Figure 1A).
Fig. 1. (A) Saccharomyces cerevisiae strain K699 containing high copy number pYES2/CT or low copy number pYC2/CT with ExoUGFP or derivatives of ExoUGFP (ExoU124-687GFP, ExoU1-660GFP or ExoUΔ53-154GFP). Both plates shown contain galactose. (B) Quantitation of the number of colony forming units after the induction of ExoU expression in pYES2/CT (left column) or pYC2/CT (right column). Open symbols represent strains containing vector controls or non-toxic expression constructs of ExoUGFP [as demonstrated in (A)] and filled squares represent strains expressing ExoUGFP under the control of the GAL1 promoter. (C) Western blot analysis of ExoU expression in the low copy number vector pYC2/CT. Proteins present in yeast lysates were separated by SDS-PAGE and transferred to nitrocellulose for western blot analysis with 5 ng of recombinant ExoU (rExoU) as a positive control.
To quantitate the effects of ExoU expression on yeast viability we measured colony forming units (c.f.u.)/ml during derepression (raffinose) and induction (galactose) from exoU cloned in low and high copy number plasmids. Biological effects on cell viability could be measured in both cases (Figure 1B). Differences in cell viability compared with vector control cultures were not detectable with ExoU deletion derivatives, regardless of vector copy number or GAL1 transcriptional level (derepression or induction). In contrast, ExoU expressed from the 2-µ plasmid correlated with a reduction in viability relative to vector control and deletion derivative transformants under conditions that only derepress GAL1 (raffinose). Cultures shifted from derepression to full induction failed to grow further. Cultures containing full-length exoU cloned into the lower copy number CEN6 plasmid exhibited a pattern in which the number of colony-forming units were minimally affected during growth in raffinose. When shifted from raffinose to galactose, however, there was a 100-fold reduction in c.f.u. over a 2 h period.
The expression of ExoU and derivatives was monitored by western blot analysis. ExoU was not detectable by western blot analysis when the gene was cloned into either low (Figure 1C) or high copy number vectors, even though cultures were harvested at a number of time points and after cultivation under various conditions of depression and induction (data not shown). In contrast, using similar growth conditions and harvesting methods, deletion derivatives of ExoU were detectable even when expressed in low copy number vectors. These results indicate that the amount of ExoU required for toxicity is below the detection limit of western blots. Our results indicate that ExoU is toxic to yeast and that toxicity requires intact N- and C-terminal domains. The fusion of green fluorescent protein (GFP) to the C-terminus of ExoU appears not to alter biological activity. A reduction in yeast cell viability is detectable within 30 min of ExoU induction from a low copy number plasmid, suggesting that the molecule is potent. When ExoU is expressed from a high copy number plasmid, basal levels of GAL1 transcription are sufficient for ExoU-mediated cytotoxicity.
Suppressor analysis
The toxicity of ExoU for yeast indicated that its expression would permit the selection of resistant mutants in yeast or in ExoU-coding sequences. Survivors of ExoU induction were isolated from galactose plates at similar frequencies (3 × 10–6 for pYC UGFP and 4 × 10–6 for pYES UGFP) using low and high copy number vectors. Plasmid DNA was isolated from these colonies and screened by restriction digestion or for protein expression in a coupled transcription/translation assay followed by western blot analysis using anti-ExoU and anti-GFP. All survivors of a galactose suppressor screen either rearranged or lost plasmid sequences, or were intragenic suppressors encoding C-terminal-truncated or -deleted forms of ExoU. The most frequent mutations in ExoU included a stop codon at amino acid 598, an in-frame deletion (Δ634–644) and an out-of-frame deletion (Δ580–592). Non-toxic ExoU derivatives appeared to be stable proteins that were detectable by western blot analysis (data not shown).
ExoU expression correlates with vacuole fragmentation
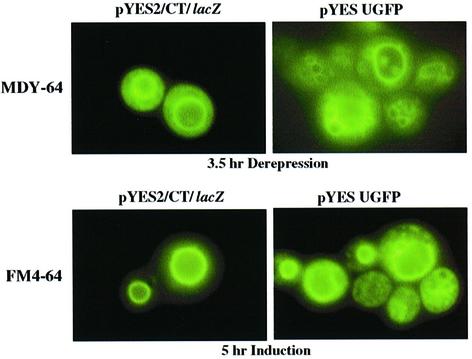
To determine if microscopic changes in yeast organelles occurred during ExoU expression, Nomarski differential interference contrast microscopy was used to monitor cell morphology after galactose induction. Yeast expressing ExoU accumulated numerous vesicles that appeared as dimples or pock marks (data not shown). Vector controls and strains expressing non-toxic forms of ExoU appeared smooth. To determine if specific cellular organelles were affected by ExoU expression, live cells were induced and examined with different fluorescent probes. Staining with Rhodamine B hexyl ester, rhodamine 123 or Mito Tracker Green FM indicated that there were no detectable changes in mitochondria after ExoU induction (data not shown). Similarly, staining with BODIPY TR ceramide for Golgi membranes showed no visible differences (data not shown). In contrast, staining with markers for the yeast vacuole (CellTracker Blue CMAC, CMAC-Arg, CMAC-Ala-Pro, Carboxy-DCFDA) and the yeast vacuolar membrane marker (MDY-64) demonstrated a fragmented phenotype in ExoU expression strains as compared with vector controls (Figure 2, upper panels: staining with MDY-64 as an example). This phenotype occurred when cells were grown under derepressed conditions, suggesting that even low amounts of ExoU induced vacuolar fragmentation.
Fig. 2. Fluorescence microscopy of yeast stained with a vacuolar membrane marker. Yeast cells with vector controls (left panels) or strains expressing an ExoUGFP fusion protein (right panels) were stained with the vacuole membrane marker MDY-64 after 3.5 h (top) of growth in raffinose, or stained with FM4-64 overnight and induced for 5 h (bottom). Yeast cells were not fixed during these experiments.
Staining with the lipophilic styryl dye FM 4-64 is specific for yeast vacuolar membranes, and allows assessment of the vacuolar morphology both at steady state and over several generations (Vida and Emr, 1995; Seeley et al., 2002). Fluorescence microscopy of induced cultures indicated that compared with vector controls, ExoU expression resulted in fragmentation of the vacuole and a weakening in the FM4-64 signal intensity (Figure 2, lower panels). Immunostaining of the vacuolar membrane with anti-v-ATPase (60 and 69 kDa subunits) also resulted in a weak pattern of staining in ExoU-expressing strains (data not shown). The loss of signal intensity from the v-ATPase marker and the fragmentation pattern seen with FM4-64 most closely resembles vacuolar morphology Class C mutants (Raymond et al., 1992; Seeley et al., 2002). Although changes were not detected in mitochondria or Golgi using vital staining, immunofluorescent signals of vacuole lumen, mitochondrial, late Golgi and endosomal membrane markers were more intense after ExoU induction than in vector controls (data not shown). These observations suggest that membrane epitopes are being exposed in yeast during ExoU induction.
Inhibitors of ExoU-mediated toxicity
Two possible mechanisms could account for biological effects caused by ExoU. Either ExoU interferes with the normal biogenesis of the vacuole or ExoU disrupts membranes, perhaps through aggregation or pore formation. As ExoU is a generally hydrophilic and monomeric molecule (Finck-Barbançon and Frank, 2001), we focused on testing inhibitors that might alter the vacuolar environment for their effects on yeast cell viability upon ExoU induction. Exposure to ouabain (0.9 mM; an inhibitor of Na+-K+ ATPases), bafilomycin A1 (1 μM; an inhibitor of vacuolar H+-ATPases), 2-(3-phenylpropylamino) benzoic acid [(NPPB) 500 μM; a chloride channel blocker], an inhibitor of serine and cysteine proteases, and Mn(III) tetra kis (4-benzoic acid) porphyrin [(TBAP) 120 μM; a metallophorphyrin antioxidant] had no effect on ExoU-mediated toxicity (data not shown).
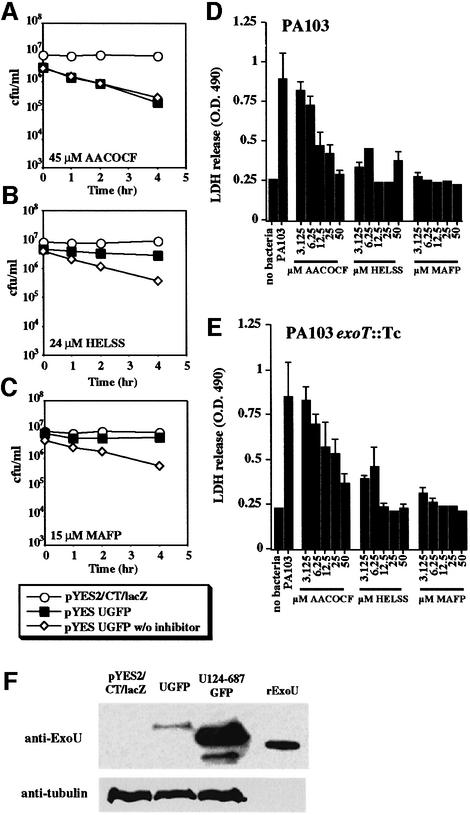
We then investigated whether a step in ExoU-mediated toxicity required the activity of intracellular lipases. Arachidonyltrifluoromethyl ketone [(AACOCF) 45 µM; a reversible inhibitor of iPLA2 and cPLA2], haloenol lactone suicide substrate [(HELSS) 24 µM; an irreversible inhibitor of iPLA2] and methyl arachidonyl fluorophosphonate [(MAFP) 15 µM; an irreversible inhibitor of cPLA2] (Cummings et al., 2000) were tested in the yeast viability assay. Each inhibitor was used at a level corres ponding to ∼3-fold higher than its inhibitory concentration (IC50) determined in mammalian cells (Figure 3A–C) (Ackermann et al., 1995; Balsinde and Dennis, 1996). In yeast, AACOCF had no effect on ExoU-mediated cell death, HELSS reduced ExoU-mediated toxicity, and MAFP significantly inhibited ExoU-mediated toxicity.
Fig. 3. Inhibitors of human cytosolic (cPLA2) and Ca2+-independent (iPLA2) lipases reduce the cytotoxic effects of ExoU expression in yeast (A–C) and during infection of BEAS-2B human bronchial cells with P.aeruginosa (D and E). Yeast transformants or BEAS-2B cells were treated with inhibitors at the indicated concentrations. In (A–C), yeast viability was measured over time after a shift to induce ExoU (by growth in galactose) in the presence or absence of the inhibitor. In (D), BEAS-2B cells were infected with a strain of P.aeruginosa producing both ExoU and ExoT, and the level of cytotoxicity was measured using the release of lactate dehydrogenase. In (E), BEAS-2B cells were co-cultivated with a P.aeruginosa strain delivering only ExoU. (F) Western blot of yeast extracts derived from cultures grown with 15 µM MAFP. pYES2/CT/lacZ is a vector control lane and UGFP contains an extract from yeast cells induced to express a toxic fusion protein (ExoUGFP). U124-687GFP, an extract from a yeast strain producing a non-toxic N-terminal truncation of ExoU, and 5 ng of His-tagged rExoU were used as markers in this experiment. Reactivity with an antibody to yeast tubulin was used as a loading control (anti-tubulin).
Since the IC50 for each inhibitor (15 µM AACOCF, 8 µM HELSS and 5 µM MAFP) had been determined in mammalian cell systems, we examined the relationship between inhibitor dose and ExoU-mediated cytotoxicity during cellular infections with P.aeruginosa. Two strains of P.aeruginosa were tested in BEAS-2B cell infections. Strain PA103 synthesizes and delivers both ExoU and ExoT into the cytoplasm of mammalian cells by the TTSS. An isogenic derivative of PA103, PA103exoT::Tc, was used in the infection analysis to eliminate any possible cytotoxic contributions of ExoT, a protein with Rho-GAP activity. Infections were carried out in the presence of the inhibitor for 4 h and the release of lactate dehydrogenase (LDH) was measured (Figure 3D and E).
During infection with either ExoU-producing strain there was a significant release of LDH relative to control, uninfected cells. Treatment of cells with AACOCF caused a dose-dependent reduction in LDH release. Both HELSS and MAPF significantly inhibited cytotoxicity at the lowest dose tested. This pattern was unaltered during infection with a strain that produces only ExoU and not ExoT, suggesting that ExoT does not contribute directly to cellular permeability.
Detection of toxic forms of ExoU was not possible in either mammalian cell transfection (Finck-Barbançon and Frank, 2001) or in yeast expression analysis (Figure 1C). Data presented in Figure 3 indicate that ExoU toxicity can be inhibited effectively with MAFP. To determine if the failure to detect ExoU was due to its toxicity, we induced ExoU production in yeast in the presence of MAFP (Figure 3F) and performed western blot analysis. Minute amounts of wild-type ExoU appear to be detectable when cytotoxicity is inhibited by MAFP. From the inhibitor analysis we concluded that ExoU either activates cellular lipases or that ExoU itself possesses lipase activity.
ExoU has a patatin-like phospholipase domain
We identified a patatin-like phospholipase domain when ExoU sequences were used to search the Conserved Domain Database (Altschul et al., 1997). Alignments of ExoU, plant patatins, mammalian iPLA2 and cPLA2 illustrate three highly conserved regions (Figure 4). A glycine-rich nucleotide binding motif, G-X-G-X-X-G, is located at amino acids 111–116 of ExoU. This motif in cPLA2 is responsible for polarizing the sn-2 ester and stabilizing the transition state of the tetrahedral intermediate (Dessen et al., 1999). A serine hydrolase motif, G-X-S-X-G, spans amino acids 140–144 of ExoU. Finally, an active site aspartate residue is conserved in the motif D-X-G/A (ExoU amino acids 344–346). The serine–aspartate catalytic dyad is required for the activity of cPLA2 and its conservation in ExoU suggested that cytotoxic activity could be related to expression of a lipase activity.
Fig. 4. The alignment of the primary sequences of P.aeruginosa ExoU (amino acids 107–154 and 316–352), and patatin-like phospholipase A2 domains of human iPLA2 (A and B), human cPLA2 (α, β and γ) and plant patatins using the NCBI Conserved Domain Database. A color index for conserved amino acid residues among listed proteins was based on the PAM250 substitution score matrix (Dayhoff et al., 1978).
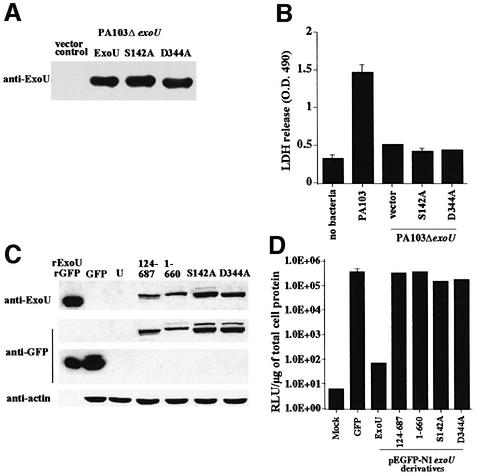
To test this hypothesis, alanine residues were used to replace the predicted ExoU catalytic residues. Site-specific mutants were fully sequenced and tested in two systems of ExoU-mediated toxicity; infection with P.aeruginosa and transfection of Chinese hamster ovary (CHO) cells. Infection studies measure several molecular characteristics that include synthesis and secretion of the toxin by the bacterium and translocation of the toxin by the type III system. Each construct was used to complement a strain of P.aeruginosa in which exoU had been deleted from the chromosome (PA103ΔexoU). After induction of the TTSS, effector proteins accumulate in the supernatant. Complementation of PA103ΔexoU with plasmids containing either wild-type or site-specific mutations demonstrated that each form of ExoU could be synthesized and exported by the type III apparatus (Figure 5A). Each strain was used to infect BEAS-2B cells and LDH release was monitored. As in previous assays, the parental strain caused release of LDH while the deletion strain (PA103ΔexoU vector control) was not cytotoxic. Alanine substitution for either of the catalytic residues eliminated LDH release mediated by ExoU (Figure 5B).
Fig. 5. Expression of point mutants during infection (A and B) and transfection (C and D). (A) Point mutations in the predicted catalytic dyad were constructed in exoU cloned in vectors compatible for replication in P.aeruginosa strain PA103ΔexoU. Vector control strains and transformants with plasmids encoding wild-type ExoU or point mutations in ExoU (S142A or D344A) were induced for type III secretion. Supernatant material was concentrated 20-fold. Each lane was loaded with an equivalent quantity of supernatant proteins based on the optical density of the culture at the time of harvest. Monoclonal antibody was used to detect ExoU after transfer to nitrocellulose. (B) Release of LDH after infection of BEAS-2B cells with strains of PA103ΔexoU (1 × 108 c.f.u./ml) expressing ExoUS142A or D344A. No bacteria or vector containing strains served as the negative controls and strain PA103 served as a positive ExoU-expressing strain. (C) Expression of ExoU during transfection analysis of CHO cells. Cells were transfected as described and analyzed for protein expression using western blots. The amount of sample loaded per lane was normalized to actin content in the cellular extract. Recombinant ExoU (rExoU) or GFP (rGFP) were loaded as positive controls. Cells transfected with GFP expression vectors, non-toxic ExoUGFP derivatives (amino acids 124–687GFP and 1–660GFP) and luciferase were processed as negative controls for ExoU-mediated cytotoxicity. (D) Cytotoxicity of ExoU or ExoU derivatives as measured by luciferase activity. CHO cells were co-transfected with a luciferase expression plasmid and constructs producing either GFP or ExoU, or derivatives of ExoU. All constructs with ExoU were made in pEGFP-N1 as GFP fusions. Cells in the ‘Mock’ lane were exposed to transfection reagents alone (no DNA). Luciferase activity is measured as relative light units (RLU) and normalized to micrograms of total cellular protein.
To confirm our results during infection, co-transfection studies (luciferase and ExoU) were performed. Expression of ExoU kills cells and causes a severe reduction in luciferase activity (Finck-Barbançon and Frank, 2001). To monitor intracellular protein expression, western blot analysis was performed on cellular extracts (Figure 5C). Lanes were normalized to actin and the position of recombinant ExoU or recombinant GFP is indicated. As in our yeast experiments, ExoU was not detectable in transfected CHO cells not exposed to a lipase inhibitor; however, non-toxic deletion derivatives and both of the alanine substitution clones produced proteins of the expected size that were reactive with anti-ExoU and anti-GFP antibodies. As shown in previous studies, ExoU was toxic and prevented the expression of luciferase (Figure 5D). Luciferase activity, however, was not affected by cells co-transfected with luciferase and GFP, an N- or C-terminal deletion of ExoU, or either of the two alanine substitutions (S142A or D344A). These data confirm that each of the point mutations is expressed as a full-length molecule in vivo and that the toxicity is probably due to ExoU-lipase activity, which is eliminated by alanine substitution of predicted catalytic residues.
Analysis of the lipolytic activity of ExoU in yeast
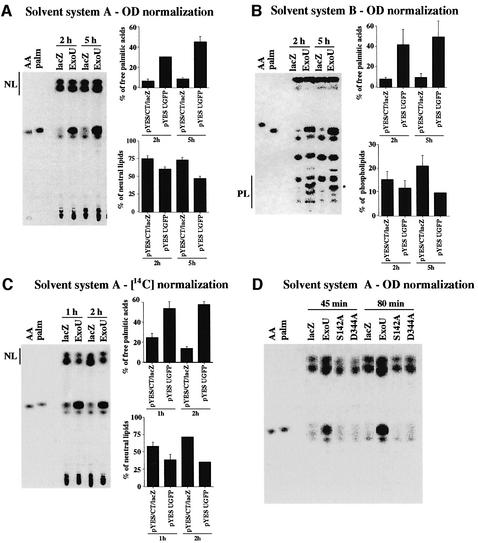
Patatin is a storage protein that possesses acyl transferase, lipid acyl hydrolase and broad esterase activity. The protein is activated by transit to the cytosol in response to environmental stress or pathogenic infection (Hirschberg et al., 2001). To investigate the potential lipolytic activity of ExoU, including phospholipase activity, total lipids were labeled with [U-14C]palmitic acid. In yeast cells, exogenous palmitic acids are incorporated into the sn-2 position of phosphatidylcholine and phosphatidylethanolamine (Wagner and Paltauf, 1994). After the incorporation of labeled palmitic acid, cells were normalized with OD600 and ExoU was induced. The change of lipid composition in yeast cells was analyzed by thin-layer chromatography (TLC) using two different solvent systems. Using solvent system A and normalizing to the culture optical density, yeast vector controls released only 6.4 to 8.5% of the total radioactivity as free palmitate (Figure 6A). In contrast, free palmitic acid accounted for 30.6% of the total radioactivity after a 2-h induction of ExoU, which increased to 45.4% after 5 h of induction. Radiolabeled neutral lipids were 75.1 and 73.2% in vector control strains. A 2 h induction of ExoU resulted in a decrease in radiolabeled neutral lipids to 60.5%, and to 47.1% after 5 h of induction.
Fig. 6. TLC of lipid populations after ExoU induction. Yeast cells were labeled with 1 µCi [14C]palmitic acid. The change of lipid composition after ExoU induction was analyzed by TLC. (A) Samples were loaded onto silica gel TLC plates, based on cell number (OD600), and resolved using solvent system A. Percentages of free palmitic acids and neutral lipids of the total radioactivity were analyzed after induction times of 2 and 5 h. Arachidonic acid (AA) and palmitate (palm) were used as markers. The migration of neutral lipids is marked as NL. (B) TLC with OD600 normalization using solvent system B to resolve phospholipids. The migration of phospholipids is marked as PL. A unique radiolabeled phospholipid product (*) appears only in cultures after induction of ExoU. The radioactivity in this spot was not included in the calculation of the percentage of radioactivity in the phospholipid fraction. (C) Samples were normalized using 14C instead of cell numbers. Percentages of free palmitic acids and neutral lipids of the total radioactivity were analyzed after 1 and 2 h induction using solvent system A. (D) TLC with optical density normalization using solvent system A. TLC was performed on vector controls pYES/CT/lacZ (lacZ), strains expressing ExoUGFP (ExoU) and strains containing an alanine substitution for either of the catalytic residues (S142A and D344A) after 45 and 80 min of induction.
Solvent system B was used to resolve phospholipids (Figure 6B). We observed approximately 7.7 to 9.6% of total radioactivity from the vector control as free palmitic acid. The amount of free palmitic acid in the ExoU strain increased to 41.5 and 49.4% of total radioactivity after 2 and 5 h of induction, respectively. The amount of radiolabel in phospholipid species in control strains was between 15.3 and 21.0%. The percentage of radiolabeled phospholipids decreased to 11.8 and 9.6% after 2 and 5 h of induction, respectively. One species of phospholipid was only observed after ExoU induction (Figure 6B). This unique spot (*) is probably the product of ExoU enzymatic activity.
The total amount of 14C in each lane was always higher in the ExoU expression strain than vector controls when samples were normalized to culture optical density. This result could be explained by the activity of ExoU, which might weaken cellular membranes and may make lipid extractions more efficient. Alternatively, phospholipid turnover may be affected. To eliminate this potential artifact, samples were normalized to 14C content prior to lipid analysis by TLC. An increase in free palmitate was also observed in the ExoU-expressing yeast strain with 14C normalization after 1 and 2 h of induction (Figure 6C).
ExoUS142A and D344A were transformed into yeast hosts and analyzed for lipid profiles after induction (Figure 6D). An increase in free palmitic acid was observed with wild-type ExoU after 45 and 80 min of induction. The substitution of an alanine residue at either active site abolished the release of free palmitate by ExoU. These data support the notion that ExoU is a lipase and suggest that ExoU has broad substrate specificity as it appears to hydrolyze neutral and phospholipids.
Lipase activity of rExoU
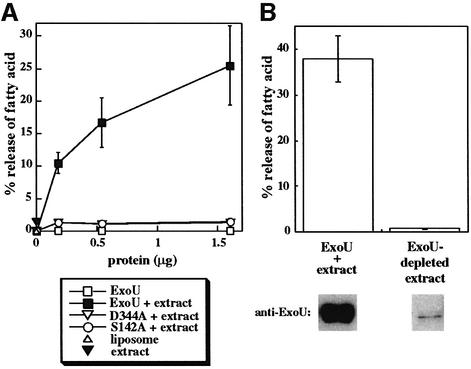
To determine whether recombinant ExoU (rExoU) possesses lipase activity, we purified histidine-tagged derivatives of wild-type ExoU, S142A and D344A. Proteins were mixed with 14C-labeled liposomes and fatty acid release was monitored by TLC. Fatty acid released was not detectable with rExoU protein preparations, although the positive control, bee venom PLA2, showed activity (Figure 7A; data not shown). We reasoned that like ExoS and ExoY, ExoU might require a eukaryotic cofactor for enzymatic activity. To test this hypothesis, rExoU, S144A and D344A were preincubated with a yeast extract before addition to the radioactive liposomes. As shown in Figure 7A, rExoU exposed to the yeast extract possesses lipase activity. Lipase activity increases with the amount of rExoU added to the reaction. Site-specific mutants predicted to be defective for catalysis, liposomes, yeast extract alone or untreated ExoU have no activity. rExoU did not activate an endogenous yeast lipase, as no activity was detected in an ExoU-depleted yeast extract (Figure 7B). Our data confirm that ExoU is a lipase and demonstrate further that ExoU requires either a eukaryotic cofactor or modification to express its lipase activity.
Fig. 7. (A) Lipase activity of rExoU. Purified His-tagged proteins at the indicated concentrations were added to an in vitro assay with 14C-labeled liposomes. In some cases purified proteins were preincubated with a yeast extract (+ extract) before addition to the labeled liposomes. The release of free fatty acids was measured using TLC. (B) The activity of an rExoU-replete (ExoU + extract) and an rExoU-depleted yeast extract was quantified by fatty acid release. Removal of rExoU from the treated yeast extract was accomplished using cobalt beads (97.6% reduction in protein as assessed by western blot analysis). Lipase activity was reduced by 98.1% upon removal of His-tagged rExoU.
Discussion
Lipolytic enzymes are indispensable to cellular metabolism as they govern the turnover of lipids and the biogenesis of membranes (Six and Dennis, 2000). These enzymes are involved in digestion and the transfer of lipids from one organism to another, the deposition and utilization of fat stores and the metabolism of intracellular lipids, which is critical to membrane function. Notably, the biological activity of these enzymes as venoms or toxins in reptiles, invertebrates and microbes appears to have a protective role for the organism. In this manuscript we report the lipolytic activity of the bacterial cytotoxin ExoU. The synthesis and TTSS-mediated delivery of ExoU to the cytoplasm of mammalian cells is correlated with cell death, lung injury, bacterial dissemination and death in both experimental animals and in patients (Finck-Barbançon et al., 1997; Kurahashi et al., 1999; Allewelt et al., 2000). Our results describing ExoU as a lipase are consistent with the observed loss of cytoplasmic contents and membrane permeability in mammalian cells, and the vacuolar fragmentation phenotype in yeast. Mutations that are predicted to abolish the lipase activity of ExoU eliminate cytotoxicity when the protein is expressed during transfection, delivered by TTSS during infection, or expressed in a non-mammalian eukaryotic host post-transformation and -induction. These data, combined with our in vitro data demonstrating the lipase activity of rExoU, argue strongly that ExoU alone may be responsible for extensive membrane damage leading to cell death.
The selection of yeast as a host to investigate the mechanism of ExoU activity allowed tight control of ExoU expression using both vector copy number and repressible promoters. We observed that only basal levels of transcription were required to elicit biological responses. The presence of the cell wall in yeast appears to prevent immediate lysis and allowed observation of morphological changes in the vacuole and differences in immunostaining patterns with other organelle markers.
The use of yeast hosts offered a genetically tractable system to identify potential eukaryotic target molecules for an ExoU-mediated activity. Screening for yeast survivors of ExoU induction resulted in the isolation of only intragenic suppressor strains, suggesting that a critical target was being modified. Biochemical approaches were subsequently used to identify ExoU–protein interactants from yeast and mammalian cellular extracts, but we were unable to find such inter actants (V.Finck-Barbançon and D.W.Frank, unpublished data). Yeast two-hybrid screens with HeLa cell expression banks identified many potential ExoU interactants; however in subsequent tests, stable associations were undetectable (V.Finck-Barbançon and D.W.Frank, unpublished data). Thus, we were unable to identify protein targets or protein interactants using several approaches. Combined with the biological potency of ExoU, these data suggested that ExoU may be transiently associated with a target and support the model of ExoU as an enzyme.
The vacuolar fragmentation phenotype in intoxicated yeast hinted that ExoU might inhibit vesicle/protein trafficking involved in vacuolar biogenesis. To determine whether ExoU caused the breakdown of an assembled vacuole or interfered with vacuolar biogenesis, we used immunofluorescence microscopy to localize vacuolar membrane and luminal markers after ExoU induction (data not shown). Our results indicated that vacuolar membrane markers tended to lose signal relative to vector controls. In contrast, vacuolar lumenal markers stained with strong intensity throughout the cell. Quinacrine staining of acidic compartments suggested that vesicular remnants of the vacuole retained their acidic characteristics (data not shown). Our model of cytotoxicity was refined to postulate that ExoU enzymatically disrupted the yeast vacuole to cause an irreversible event leading to cell death. Since protein interaction experiments were inconclusive, we entertained the notion that ExoU targeted a non-protein cellular component.
Homology data from Basic Logic Alignment Search Tool (BLAST) searches were generally not useful in identifying the potential function of ExoU. Domain searches were more informative and identified a region of ExoU with a phospholipase-like domain, which was homologous to patatin, a soluble storage protein in potato tubers. Patatin, in turn, is homologous to cPLA2, an enzyme that possesses a serine-aspartate active site dyad (Nalefski et al., 1994; Pickard et al., 1996; Dessen et al., 1999). Inhibitors that block nucleophilic serine residues by covalent attachment of a methyl-octylphosphonate group severely reduce patatin activity (Hirschberg et al., 2001). Conserved residues in ExoU align with the patatin family of proteins as well as human iPLA2 and cPLA2 enzymes (Six and Dennis, 2000). Mutagenesis of the predicted active site residues (S142 and D344) inhibits ExoU-mediated cytotoxicity and reduces the release of free palmitic acid in vivo. Recombinant forms of the protein do not possess enzymatic activity using labeled liposomes. Although it could be argued that the tertiary structure of the molecule might be affected by point mutations, in transfection studies the fused GFP molecule is visible in CHO cells, suggesting that a proportion of the expressed protein is folded correctly (Waldo et al., 1999). The effect of inhibitors of iPLA2 and cPLA2 on the cytotoxic response, the absence of cytotoxicity of ExoU point mutants expressed in CHO cell and in yeast systems, and the in vitro activity of rExoU support the conclusion that ExoU is a lipase.
ExoU lipase activity appears to have broad substrate specificity that includes the hydrolysis of neutral lipids, presumably diacylglycerols and triacylglycerols as well as phospholipids. Patatin is also known to have a broad range of substrates and hydrolyzes phospholipids, monoacylglycerols and p-nitrophenyl esters, but not diacylglycerols and triacylglycerols (Andrews et al., 1988; Hirschberg et al., 2001). Patatin is an inactive protein and localizes mainly to plant vacuoles (Hirschberg et al., 2001). Wounding or invasion by pathogens triggers the translocation of patatin to the cytosol, where the enzyme is activated by a more basic environment. The TTSS system of P.aeruginosa delivers ExoU directly to the cytosol of eukaryotic cells, where the broad substrate specificity of the enzyme would allow damage to internal and plasma membrane components, leading to cell lysis and necrotic death. These properties may explain observations from several investigators indicating that ExoU is cytotoxic for many cell types, including social amoeba (Finck-Barbançon et al., 1997; Kurahashi et al., 1999; Allewelt et al., 2000; Pukatzki et al., 2002). Moreover, the major lipid lung surfactant is dipalmitoyl phosphatidylcholine, which is essential for pulmonary function. Lipolytic activity may contribute to the ability of ExoU-expressing bacterial strains to disseminate rapidly from lung tissue into the bloodstream (Allewelt et al., 2000).
This study sheds some light on the structural requirements for ExoU cytotoxicity and enzymatic activity. In previous work, ExoU cytotoxicity was dependent on the expression of three regions of the molecule: an N-terminal domain consisting of residues 52–155, a middle region that appeared to encompass amino acids 300–400, and a C-terminal region from residue 580 to 687 (Finck-Barbançon and Frank, 2001). This mapping analysis is consistent with the catalytic dyad residing at S142 and D344, and the patatin homology spanning ExoU amino acids 107–357. Interestingly, all ExoU derivatives isolated from suppressor screens occurred within the C-terminal domain, well outside of the predicted catalytic domain. Protein folding or degradation could be cited as potential confounding issues. On the other hand, all non-toxic C-terminal disruptions of ExoU were detected in western blot analysis with little or no degradation. In addition, they were visible in fluorescent microscopy experiments as GFP fusions during CHO cell transfection, suggesting that some of the protein is properly folded (Waldo et al., 1999). These observations and prior domain mapping data suggest that the ExoU C-terminus may play a critical role in the activation state of the molecule or its interaction with lipid substrates. In addition, our observation that lipase activity depends upon the exposure of ExoU to a eukaryotic extract further implicates the requirement of a cofactor or a post-translational modification. Based on the cPLA2 structure, C-terminal residues of ExoU may need to be phosphorylated for ExoU to be active (Nalefski et al., 1994; Pickard et al., 1996; Dessen et al., 1999; Gijon et al., 1999). Alternatively, other processing events or modifications may need to occur to express full lipase activity. Modifications of ExoU, a requirement for a cofactor, or the recognition of specific eukaryotic lipid substrates could explain the apparent lack of toxicity for P.aeruginosa.
In summary, the mechanism of ExoU acute cytotoxicity is related to its ability to destroy eukaryotic membranes by using an iPLA2/patatin-like phospholipase activity. This is the first lipase reported that is secreted by a type III system. The direct delivery to the cytoplasm and the broad substrate specificity of ExoU appears to contribute to necrotic death, cell permeability and the rapid dissemination of ExoU-producing strains of P.aeruginosa. The use of yeast hosts led to insights into the structure and function of ExoU through genetic, biochemical and microscopic approaches. The development of an in vitro assay system demonstrates further that, similar to ExoS and ExoY of P.aeruginosa, ExoU requires exposure to an eukaryotic environment to express enzymatic activity.
Materials and methods
Yeast strains and growth conditions
Yeast cells (strain K699; Jansen et al., 1996) were grown in minimal medium [0.17% yeast nitrogen base without amino acids (Difco Laboratories)] supplemented with appropriate amino acids and with 2% glucose, raffinose or galactose (Sigma) as a carbon source. Transformation of yeast with recombinant plasmids was achieved using standard methods (Ito et al., 1983). Yeast vector DNAs (pYES2/CT, pYES2CT/lacZ, pYC2/CT, pYC2CT/lacZ) were obtained commercially (Invitrogen).
Colony forming unit assays
Yeast cells were grown in medium with 2% glucose overnight, transferred into medium with 2% raffinose for derepression for 24 h, and induced in medium with 2% galactose. During the transition to different growth or GAL1-inducing growth conditions, cells were washed with supplemented minimal medium without glucose, raffinose or galactose. Diluted cultures of derepressed or induced cells were plated on medium agar with 2% glucose and incubated at 30°C for 2 days. Viability graphs are the average of duplicate experiments.
SDS–PAGE and immunoblot analysis
In general, 10% polyacrylamide SDS gels were used to resolve proteins, and western blot analysis was carried out using standard techniques. For detection of ExoU expression in yeast, induced strains were normalized to optical density and prepared as outlined in the Supplementary data (available at The EMBO Journal Online). β-galactosidase (not shown) or tubulin was used as a loading control in yeast expression experiments. Supernatants of P.aeruginosa expressing ExoU were concentrated and subjected to western blot analysis as described previously (Finck-Barbançon et al., 1997). Each sample was normalized to the optical density of the bacterial culture at the time of harvest. Transfected CHO cell lysates were normalized for western blotting to actin content. Proteins were transferred to nitrocellulose membranes and probed with mouse anti-ExoU monoclonal antibody U29F8 (1:20 000), rat anti-α-tubulin IgG [1:15 000 dilution (Serotec)], mouse anti-GFP [1:5000 dilution (Covance)] or rabbit anti-actin (1:5000 dilution; Lei et al., 1996). Horseradish peroxidase conjugates appropriate for each IgG were used to detect primary antibody (Roche). SuperSignal® West Pico Chemilu minescent Substrate (Pierce) was used for detection of bound peroxidase.
In vitro cytotoxicity assay with BEAS-2B cells
A human bronchial epithelial cell line, immortalized by SV40-adeno hybrid virus (BEAS-2B, ATCC CRL9609) was cultured in Dulbecco’s modified Eagle medium (DME) with 10% heat-inactivated fetal calf serum, penicillin and streptomycin. Cells were kept in an incubator with 5% CO2. When the cells reached confluence, 2 × 104 cells were transferred to 96-well tissue culture plates and incubated for 18 h. After washing three times with phosphate-buffered saline, 1 ml of fresh DME medium with 10 mM HEPES was added, per well. In the experiments using PLA2 inhibitors, the agent was added 1 h before the addition of bacteria. Pseudomonas aeruginosa strains (∼3 × 107 c.f.u./ml was used in experiments depicted in Figure 3D and E, and 1 × 108 c.f.u./ml was used for experimental data shown in Figure 5B) were mixed with medium and applied to the cells for 4 h. Cytotoxicity was quantified by the production of LDH by using a cytotoxicity assay kit (Cytotox 96; Promega, Madison, WI). Reported values are the mean and standard deviation in triplicate assays.
Live cell staining
Yeast cells were stained with the yeast vacuolar membrane marker MDY-64, as recommended by the distributor (Molecular Probes, Inc., Eugene, OR) after derepression for 3.5 h. For staining with N-(3-triethylammoniumpropyl)-4-(p-diethyl-amino phenyl hexatrienyl) pyridinium dibromide (FM 4-64; Molecular Probes, Inc.), yeast cells were grown overnight in medium with 1% glucose and 3 µM FM 4-64, and then induced in medium with 2% galactose and 3 µM FM 4-64 for 5 h. Rhodamine B hexyl ester, rhodamine 123, or Mito Tracker Green FM, BODIPY TR ceramide, CellTracker Blue CMAC, CMAC-Arg, CMAC-Ala-Pro and Carboxy-DCFDA were used according to the manufacturer’s instructions (Molecular Probes, Inc.; data not shown).
Lipid extraction and TLC
Yeast cells were incubated in medium with 1% glucose (Sigma) and 1 µCi [U-14C] palmitic acid (Amersham Bioscience) at 30°C overnight. The culture was washed and diluted into the medium with 2% galactose (Sigma) for induction. Lipids were extracted as described previously (Hayashi et al., 1976). Total lipids were separated by TLC (LK6D silica gel 60 Å; Whatman) with either solvent system A (hexane/ether/acetone/acetic acid (50:40:10:1)] or solvent system B [chloroform/methanol/ammonium hydroxide/water (65:35:5:1)]. Solvent system A was used to separate total lipids, mainly free fatty acids and neutral lipids. Solvent system B was more acidic and used to separate glycerides with a charged head group, such as phospholipids. Radioactivity on the TLC plate was quantified using InstantImager Electronic Autoradiography (Packard). Reported values are the mean and standard deviation of assays performed in triplicate.
Lipase activity of rExoU
His-tagged recombinant ExoU, D344A and S142A proteins were purified by cobalt chromatography as described by the manufacturer (Clontech). Liposomes consisted of a mixture of 2.5 µM phosphatidylcholine, 2.5 µM phosphatidylserine (Avanti Polar lipids) and 0.027 µM phosphatidylcholine (1-palmitoyl-2-[1-14C]oleoyl; Amersham Biosciences). The mixture was dried, hydrated with 20 mM MOPS and extruded through a 0.1 µm polycarbonate filter. Yeast extract was prepared from the pYES/CT/lacZ strain. Briefly, yeast cells were suspended in 1.2 ml (per 250 OD600 unit) of 50 mM sodium phosphate, 1 mM EDTA and 5% glycerol (Sigma) with protease inhibitors, as outlined in Supplementary data (protein extraction from yeast). Cells were lysed by passage through a French pressure cell (four passages), and unbroken cells and large debris were removed from the extract by centrifugation at 14 000 g for 30 min. Approximately 100 µg of the extract was incubated with various amounts of wild-type or catalytic mutants of ExoU for 1 h at 30°C. The preincubation step was followed by incubation with 8 µl of the liposome mixture for 3 h at 37°C. TLC was used to measure the release of free fatty acids. Reported values are the mean and standard deviations of assays performed in triplicate.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
Expert technical assistance was provided by M.Casey and S.Kalivendi. This work was supported by funds from the National Institutes of Health (AI49577 to D.W.F. and M.L., HL59239 to J.W.K. and D.W.F., AI44101 to J.W.K. and HL67600 to T.S.), the American Lung Association of Wisconsin Award (RG009L to V.F.B.) and the American Lung Association (RG004N to T.S.).
References
- Ackermann E.J., Conde-Frieboes,K. and Dennis,E.A. (1995) Inhibition of macrophage Ca2+-independent phospholipase A2 by bromoenol lactone and trifluoromethyl ketones. J. Biol. Chem., 270, 445–450. [DOI] [PubMed] [Google Scholar]
- Allewelt M., Coleman,F.T., Grout,M., Priebe,G.P. and Pier,G.B. (2000) Acquisition of expression of the Pseudomonas aeruginosa ExoU cytotoxin leads to increased bacterial virulence in a murine model of acute pneumonia and systemic spread. Infect. Immun., 68, 3998–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Madden,T.L., Schåffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews D.L., Beames,B., Summers,M.D. and Park,W.D. (1988) Characterization of the lipid acyl hydrolase activity of the major potato (Solanum tuberosum) tuber protein, patatin, by cloning and abundant expression in a baculovirus vector. Biochem. J., 252, 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsinde J. and Dennis,E.A. (1996) Distinct roles in signal transduction for each of the phospholipase A2 enzymes present in P388D1 macrophages. J. Biol. Chem., 271, 6758–6765. [DOI] [PubMed] [Google Scholar]
- Barbieri J.T. (2000) Pseudomonas aeruginosa exoenzyme S, a bifunctional type-III secreted cytotoxin. Intern. J. Med. Microbiol., 290, 381–387. [DOI] [PubMed] [Google Scholar]
- Cummings B.S., Mchowat,J. and Schnellmann,R.G. (2000) Phospholipase A2s in cell injury and death. J. Pharmacol. Exp. Ther., 294, 793–799. [PubMed] [Google Scholar]
- Dayhoff M.O., Schwartz,R.M. and Orcutt,B.C. (1978) A model of evolutionary change in proteins. In Dayhoff,M.O. (ed.), Atlas of Protein Sequence and Structure. National Biomedical Research Foundation, Washington, DC, pp. 345–352.
- Dessen A., Tang,J., Schmidt,H., Stahl,M., Clark,J.D., Seehra,J. and Somers,W.S. (1999) Crystal structure of human cytosolic phospholipase A2 reveals a novel topology and catalytic mechanism. Cell, 97, 349–360. [DOI] [PubMed] [Google Scholar]
- Finck-Barbançon V. and Frank,D.W. (2001) Multiple domains are required for the toxic activity of Pseudomonas aeruginosa ExoU. J. Bacteriol., 183, 4330–4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finck-Barbançon V., Goranson,J., Zhu,L., Sawa,T., Wiener-Kronish,J.P., Fleiszig,S.M.J., Wu,C., Mende-Mueller,L. and Frank,D.W. (1997) ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol. Microbiol., 25, 547–557. [DOI] [PubMed] [Google Scholar]
- Frank D.W. (1997) The exoenzyme S regulon of Pseudomonas aeruginosa. Mol. Microbiol., 26, 621–629. [DOI] [PubMed] [Google Scholar]
- Gijon M.A., Spencer,D.M., Kaiser,A.L. and Leslie,C.C. (1999) Role of phosphorylation sites and the C2 domain in regulation of cytosolic phospholipase A2. J. Cell Biol., 145, 1219–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehring U.M., Schmidt,G., Pederson,K.J., Aktories,K. and Barbieri,J.T. (1999) The N-terminal domain of Pseudomonas aeruginosa exoenzyme S is a GTPase-activating protein for Rho GTPases. J. Biol. Chem., 274, 36369–36372. [DOI] [PubMed] [Google Scholar]
- Hayashi E., Hasegawa,R. and Tomita,T. (1976) Accumulation of neutral lipids in Saccharomyces carlsbergensis by myo-inositol deficiency and its mechanism. J. Biol. Chem., 251, 5759–5769. [PubMed] [Google Scholar]
- Hirschberg H.J.H.B., Simons,J.-W.F.A., Dekker,N. and Egmond,M.R. (2001) Cloning, expression, purification and characterization of patatin, a novel phospholipase A. Eur. J. Biochem., 268, 5037–5044. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukudua,Y., Murata,K. and Kimura,A. (1983) Transformation of intact yeast cells treated with alkali cations. J. Bacteriol., 153, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R.-P., Dowzer,C., Michaelis,C., Galova,M. and Nasmyth,K. (1996) Mother cell-specific HO expression in budding yeast depends on the unconventional myosin Myo4p and other cytoplasmic proteins. Cell, 84, 687–697. [DOI] [PubMed] [Google Scholar]
- Kurahashi K., Kajikawa,O., Sawa,T., Ohara,M., Gropper,M.A., Frank, D.W., Martin,T.R. and Wiener-Kronish,J.P. (1999) Pathogenesis of septic shock in Pseudomonas aeruginosa pneumonia. J. Clin. Invest., 104, 743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M., Kawasaki,Y. and Tye,B.K.(1996) Physical interactions among Mcm proteins and effects of Mcm dosage on DNA replication in Saccharomyces cerevisiae. Mol. Cell Biol., 16, 5081–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser C.F. and Miller,S.I. (2001) Expression of microbial virulence proteins in Saccharomyces cerevisiae models mammalian infection. EMBO J., 20, 1840–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalefski E.A., Sultzman,L.A., Martin,D.M., Kriz,R.W., Towler,P.S., Knopf,J.L. and Clark,J.D. (1994) Delineation of two functionally distinct domains of cytosolic phospholipase A2, a regulatory Ca2+-dependent lipid-binding domain and a Ca2+-independent catalytic domain. J. Biol. Chem., 269, 18239–18249. [PubMed] [Google Scholar]
- Pederson K.J. and Barbieri,J.T. (1998) Intracellular expression of the ADP-ribosyltransferase domain of Pseudomonas exoenzyme S is cytotoxic to eukaryotic cells. Mol. Microbiol., 30, 751–759. [DOI] [PubMed] [Google Scholar]
- Pickard R.T. et al. (1996) Identification of essential residues for the catalytic function of 85-kDa cytosolic phospholipase A2. J. Biol. Chem., 271, 19225–19231. [DOI] [PubMed] [Google Scholar]
- Pukatzki S., Kessin,R.H. and Mekalanos,J.J. (2002) The human pathogen Pseudomonas aeruginosa utilizes conserved virulence pathways to infect the social amoeba Dictyostelium discoideum. Proc. Natl Acad. Sci. USA, 99, 3159–3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond C.K., Howald-Stevenson,I., Vater,C.A. and Stevens,T.H. (1992) Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell, 3, 1389–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy-Burman A., Savel,R.H., Racine,S., Swanson,B.L., Revadigar,N.S., Fujimoto,J., Sawa,T., Frank,D.W. and Wiener-Kronish,J.P. (2001) Type III protein secretion is associated with death in lower respiratory and systemic Pseudomonas aeruginosa infections. J. Infect. Dis., 183, 1767–1774. [DOI] [PubMed] [Google Scholar]
- Seeley E.S., Kato,M., Margolis,N., Wickner,W. and Eitzen,G. (2002) Genomic analysis of homotypic vacuole fusion. Mol. Biol. Cell, 13, 782–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Six D.A. and Dennis,E.A. (2000) The expanding superfamily of phospholipase A2 enzymes: classification and characterization. Biochim. Biophys Acta, 1488, 1–19. [DOI] [PubMed] [Google Scholar]
- Vida T.A. and Emr,S.D. (1995) A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol., 128, 779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Pawel-Rammingen U., Telepnev,M.V., Schmidt,G., Aktories,K., Wolf-Watz,H. and Rosqvist,R. (2000) GAP activity of the Yersinia YopE cytotoxin specifically targets the Rho pathway: a mechanism for disruption of actin microfilament structure. Mol. Microbiol., 36, 737–748. [DOI] [PubMed] [Google Scholar]
- Wagner S. and Paltauf,F. (1994) Generation of glycerophospholipid molecular species in the yeast Saccharomyces cerevisiae. Fatty acid pattern of phospholipid classes and selective acyl turnover at sn-1 and sn-2 positions. Yeast, 10, 1429–1437. [DOI] [PubMed] [Google Scholar]
- Waldo G.S., Standish,B.M., Berendzen,J. and Terwilliger,T.C. (1999) Rapid protein-folding assay using green fluorescent protein. Nature Biotechnol., 17, 691–695. [DOI] [PubMed] [Google Scholar]
- Yahr T.L., Vallis,A.J., Hancock,M.K., Barbieri,J.T. and Frank,D.W. (1998) ExoY, an adenylate cyclase secreted by the Pseudomonas aeruginosa type III system. Proc. Natl Acad. Sci. USA, 95, 13899–13904. [DOI] [PMC free article] [PubMed] [Google Scholar]