Abstract
The LKB1 gene encodes a serine/threonine kinase mutated in Peutz–Jeghers cancer syndrome. Despite several proposed models for LKB1 function in development and in tumour suppression, the detailed molecular action of LKB1 remains undefined. Here, we report the identification and characterization of an LKB1-specific adaptor protein and substrate, STRAD (STe20 Related ADaptor). STRAD consists of a STE20- like kinase domain, but lacks several residues that are indispensable for intrinsic catalytic activity. Endo genous LKB1 and STRAD form a complex in which STRAD activates LKB1, resulting in phosphorylation of both partners. STRAD determines the subcellular localization of wild-type, but not mutant LKB1, translocating it from nucleus to cytoplasm. One LKB1 mutation previously identified in a Peutz–Jeghers family that does not compromise its kinase activity is shown here to interfere with LKB1 binding to STRAD, and hence with STRAD-dependent regulation. Removal of endogenous STRAD by siRNA abrogates the LKB1-induced G1 arrest. Our results imply that STRAD plays a key role in regulating the tumour suppressor activities of LKB1.
Keywords: adaptor/LKB1/pseudokinase/STE20-like kinase/STRAD
Introduction
The tumour suppressor protein LKB1 is a serine/threonine kinase that has been causally linked to Peutz–Jeghers syndrome (PJS) (Hemminki et al., 1998; Jenne et al., 1998). Inactivating mutations in the LKB1 gene are found in ∼80% of PJS patients. Such patients are clinically characterized by mucocutaneous pigmentation, the development of hamartomas in the gastrointestinal tract and a predisposition to rare types of cancer (Peutz, 1921; Jeghers et al., 1949; Giardiello et al., 2000). Lkb1+/– mice develop gastrointestinal polyps and hepatocellular carcinomas, confirming its role as a tumour suppressor (Bardeesy et al., 2002; Jishage et al., 2002; Miyoshi et al., 2002; Nakau et al., 2002; Rossi et al., 2002). LKB1 also plays an essential role in development, since Lkb1–/– mice die at embryonic day 9 due to a variety of developmental abnormalities, affecting the yolk sac and placenta (Ylikorkala et al., 2001; Jishage et al., 2002). Genetic studies in Caenorhabditis elegans and Drosophila have indicated an essential function of LKB1 homologues in establishing cell polarity (Watts et al., 2000; Martin and St Johnston, 2003). This may suggest a model in which LKB1 loss causes hamartoma formation through disruption of epithelial polarity.
Forced expression of LKB1 in cells lacking this enzyme induces a G1 cell cycle arrest (Tiainen et al., 1999). Phosphorylation of LKB1 by cyclic AMP-dependent kinase or by p90 ribosomal S6 kinase may be required for this growth arrest (Collins et al., 2000; Sapkota et al., 2001), as is the chromatin modifier BRG-1 (Marignani et al., 2001). The subcellular localization of LKB1 seems to be crucial for its role in mediating G1 cell cycle arrest. This was demonstrated by a study with an LKB1 mutant lacking the nuclear localization signal (Tiainen et al., 2002). This solely cytoplasmic LKB1 protein was still able to induce growth arrest, suggesting that this portion of LKB1 is responsible for its growth regulating activities. Recently, LKB1-interacting protein 1 (LIP1) was identified and shown to translocate LKB1 from the nucleus to the cytoplasm upon forced expression of both proteins (Smith et al., 2001). Due to an observed interaction between LIP1 and SMAD4, LIP1 may link LKB1 to the transforming growth factor β pathway. LKB1 has also been suggested to play a central role in p53-mediated apoptosis (Karuman et al., 2001). Despite several proposed models for LKB1 function, the molecular activities of LKB1 in development and in preventing cellular transformation remain poorly understood. This is mainly due to the fact that LKB1 exhibits weak catalytic activity and that no in vivo LKB1 substrates have been identified to date.
In the present study, we identified a novel STE20-related kinase, which we termed STRAD (for STE20 Related ADaptor), as an LKB1-specific adaptor and substrate. Members of the STE20-like kinase family are implicated in the stimulation of mitogen-activated protein kinase (MAPK) pathways, by direct activation of MAPKKK. It has been suggested that this activity of STE20-like kinases is independent of their catalytic capacities (Dan et al., 2001). Interestingly, in contrast to previously described STE20-like kinases, STRAD encodes a pseudokinase due to the lack of several residues that are indispensable for intrinsic catalytic activity. We provide compelling evidence that STRAD acts as an upstream activator of the LKB1 kinase. Moreover, we show that STRAD is the first LKB1 substrate in vivo, and that STRAD directs the subcellular localization of LKB1 by anchoring it in the cytoplasm.
Results
STRAD is a pseudokinase which specifically binds to LKB1
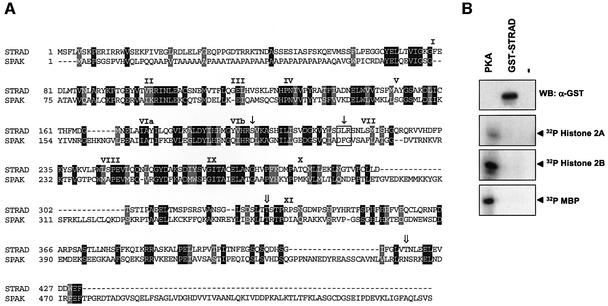
To gain more insight into the activities of LKB1, we performed a yeast two-hybrid screen to identify potential binding partners. We screened a fetal brain library using kinase-dead (KD) LKB1 (D176Y) as bait, and identified a single clone that interacted specifically with LKB1 and not with control baits. This clone encoded a protein consisting almost entirely of a kinase-like domain with strong homology to STE20-like kinases. We named this protein, which was previously submitted as NY-BR-96 (DDBJ/EMBL/GenBank accession No. AF308302), STRAD. STRAD most closely resembles the recently described STE20 homologues SPAK (Johnston et al., 2000) and ILPIP (Sanna et al., 2002) or PAP kinase (Nishigaki et al., 2003). Sequence analysis revealed that STRAD lacked several residues that are indispensable for intrinsic kinase activity compared with SPAK (Figure 1A) (Huse and Kuriyan, 2002). Specifically, the DFG motif of the potential magnesium interacting site of the ATP-binding cleft was absent. Furthermore, the essential aspartic acid residue, crucial for mediating proton transfer in the catalytic site, was replaced by serine. This replacement was found to be conserved in mouse and fly orthologues of STRAD (data not shown). The STRAD gene localizes to human chromosome 17, and its 13 exons produce a 2.1 kb cDNA, encoding a putative protein of ∼50 kDa.
Fig. 1. (A) Deduced amino acid sequence of human STRAD aligned with SPAK. Identical residues are represented in black and similar residues in grey. The subdomains of the pseudokinase domain are indicated with Roman numerals. The serine replacement of the essential aspartic acid residue in the catalytic site and the absence of the DFG motif in STRAD compared with SPAK are designated with closed arrows. The STRAD phosphorylation sites Thr329 and Thr419 are designated with open arrows. (B) STRAD does not phosphorylate the exogenous substrates myelin basic protein (MBP), histone 2A, histone 2B and CRE binding protein (CREB). Exogenous proteins (10 µg) were subjected to an in vitro kinase (IVK) assay with either 1 U of cAMP-dependent kinase (PKA) or 1 µg of GST–STRAD. Proteins were separated by SDS–PAGE and immunoblotted (WB) with α-GST or visualized by autoradiography.
We generated an expression vector which produces a glutathione S-transferase (GST)-tagged STRAD fusion protein. GST–STRAD was isolated from HEK-293T cells and subjected to several in vitro kinase assays in the presence of [γ-32P]ATP. As predicted from the sequence analysis, GST–STRAD was not able to autophosphorylate (data not shown) nor to phosphorylate histone 2A, histone 2B, myelin basic protein (MBP) and CRE binding protein (CREB). These exogenous substrates were readily phosphorylated by cAMP-dependent kinase (PKA) (Figure 1B). We therefore tentatively concluded that STRAD is a pseudokinase.
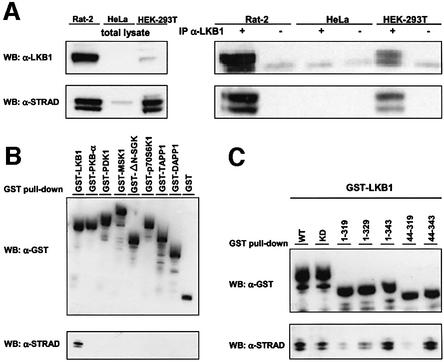
To establish whether endogenous LKB1 and STRAD interact, we generated a monoclonal antibody against STRAD. This anti-STRAD antibody recognized two specific bands on immunoblots at 45–48 kDa (Figure 2A). A direct interaction could be readily visualized in Rat-2 and HEK-293T cells, as determined by endogenous STRAD being co-immunoprecipitated with endogenous LKB1 (Figure 2A). HeLa cells, which do not express LKB1, were included as a negative control. A GST–LKB1 fusion protein, transfected into HEK-293T cells, associated with endogenous STRAD (Figure 2B). Unrelated GST fusion proteins, including several kinases, did not bind to STRAD, confirming the specificity of the interaction between LKB1 and STRAD. Deletion of either N- or C-terminal amino acids from the LKB1 kinase domain abolished STRAD binding completely (data not shown). Because GST–LKB1[1–319] and GST–LKB1[44–319] failed to bind strongly to STRAD, whereas GST– LKB1[1–343] and GST–LKB1[44–343] did bind (Figure 2C), we concluded that the entire kinase domain as well as the C-terminal amino acids 319–343 of LKB1 were necessary for binding STRAD.
Fig. 2. (A) Endogenous LKB1 (55 kDa) interacts with STRAD (45–48 kDa) in Rat-2 and HEK-293T cells. Co-immunoprecipitations (IP) were performed on 1 mg of indicated cell lysates with either α-LKB1 polyclonal antibody (sheep) or an irrelevant sheep polyclonal antibody. Protein complexes (right panel) and 20 µg of total lysates (left panel) were immunoblotted (WB) with α-LKB1 or α-STRAD. (B) STRAD specifically interacts with LKB1. GST–LKB1 and several unrelated GST fusion proteins were isolated from HEK-293T cells using GST-affinity chromatography. Protein complexes were immunoblotted (WB) with α-GST or α-STRAD. (C) Amino acids 319–343 of LKB1 are necessary for STRAD binding. Indicated GST–LKB1 deletion constructs were isolated from HEK-293T using GST-affinity chromatography. Protein complexes were immunoblotted (WB) with α-GST or α-STRAD.
LKB1 phosphorylates STRAD in vitro and in vivo
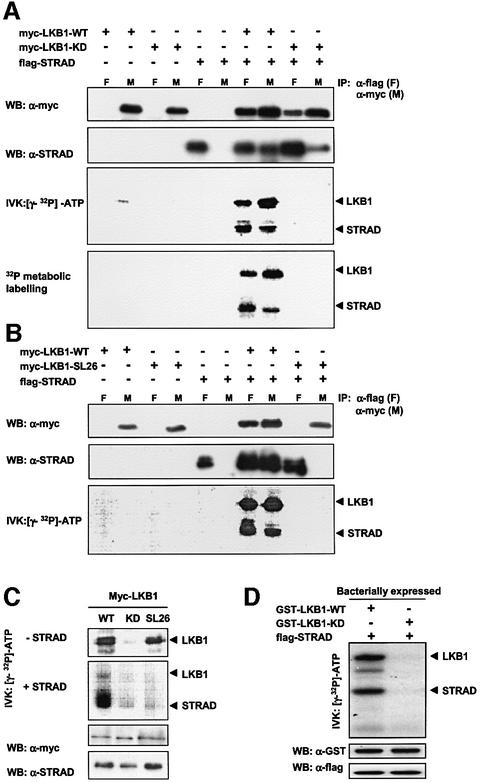
After confirming the LKB1–STRAD interaction in mammalian cells, we were interested in the molecular consequences of STRAD binding to LKB1. For this purpose, we used myc-tagged LKB1-WT (wild type) and LKB1-KD (kinase dead; D176Y) constructs, and a myc-tagged LKB1 mutant found in a PJS family: SL26. The SL26 mutation consists of a small in-frame deletion of 9 bp at the C-terminus of the kinase domain (Hemminki et al., 1998), and is the only LKB1 mutant found in PJS patients that retains its intrinsic kinase activity (Nezu et al., 1999). Flag-tagged STRAD and myc-tagged LKB1 could be co-immunoprecipitated following co-transfection of HEK- 293T cells (Figure 3A). Remarkably, when the isolated complexes were analysed in an in vitro kinase assay in the presence of [γ-32P]ATP, STRAD became phosphorylated and LKB1 autophosphorylation was strongly enhanced (Figure 3A and B). Moreover, by means of metabolic phosphate labelling of the transfected cells, we found that these processes also occurred in vivo (Figure 3A, bottom panel). The phosphorylation events were dependent on the kinase activity of LKB1, since no phosphorylation was observed with LKB1-KD (Figure 3A, last two lanes).
Fig. 3. LKB1 phosphorylates STRAD directly in vitro and in vivo. (A) LKB1-WT (wild type), but not LKB1-KD (kinase dead; D176Y), mediates phosphorylation of STRAD and shows enhanced autophosphorylation upon forced STRAD expression. Myc-tagged LKB1-WT, LKB1-KD and flag-STRAD were transfected into HEK-293T cells as indicated. Protein complexes were co-immunoprecipitated (IP) using either myc-Ab (M) or flag-Ab (F), and immunoblotted (WB) with α-myc or α-STRAD. In addition, the protein complexes were subjected to an in vitro kinase (IVK) assay. Cells transfected with the indicated constructs were metabolically labelled with [32P]phosphate for 3 h prior to lysing and co-immunoprecipitation. Protein complexes were separated by SDS–PAGE and visualized by autoradiography. (B) The LKB1-SL26 mutant does not bind to STRAD, which prevents STRAD phosphorylation and STRAD-mediated enhanced autophosphorylation of LKB1. Myc-tagged LKB1-WT, LKB1-SL26 and flag-STRAD were transfected into HEK-293T cells as indicated. Protein complexes were co-immunoprecipitated (IP) using either myc-Ab (M) or flag-Ab (F). Subsequently, they were immunoblotted (WB) with α-myc or α-STRAD. In addition, the protein complexes were subjected to an in vitro kinase assay (IVK), separated by SDS–PAGE and visualized by autoradiography. (C) LKB1–STRAD interaction is required for STRAD phosphorylation by LKB1. Anti-myc immunoprecipitated LKB1-WT, LKB1-KD and LKB1-SL26 were subjected to an in vitro kinase assay (IVK) alone, or after mixing with α-flag immunoprecipitated STRAD. Protein complexes were immunoblotted (WB) with α-myc or α-STRAD, and visualized by autoradiography. (D) LKB1 phosphorylates STRAD directly. Bacterially produced GST–LKB1- WT, GST–LKB1-KD and flag-STRAD were subjected to an in vitro kinase assay as indicated. Proteins were separated by SDS–PAGE, and immunoblotted (WB) with α-GST or α-flag. Phosphorylated proteins were visualized by autoradiography.
We confirmed that LKB1-SL26 retains its kinase activity, as demonstrated by its ability to autophosphorylate in vitro (Figure 3C, upper panel, overexposure). However, the 9 bp deletion abolished the interaction with STRAD (Figure 3B, last two lanes). As predicted, this prevented LKB1-SL26 from phosphorylating STRAD and from increasing its autophosphorylation (Figure 3B, bottom panel). We postulate that LKB1-SL26 is a loss-of-function mutant causing PJS due to its inability to interact with STRAD.
To investigate whether the interaction of LKB1 and STRAD is necessary for STRAD phosphorylation by LKB1, we mixed immunoprecipitated LKB1-WT, -KD or -SL26 with immunoprecipitated STRAD (Figure 3C). While LKB1-WT was still able to phosphorylate STRAD under these conditions, LKB1-KD and LKB1-SL26 were not. This means that both LKB1 kinase activity and LKB1 ability to bind to STRAD were required for LKB1-mediated STRAD phosphorylation.
In order to prove that STRAD is directly phosphorylated by LKB1, and not by putative kinases that could be bound to the LKB1–STRAD complex, we isolated bacterially expressed GST–LKB1-WT, GST–LKB1-KD and GST– flag-STRAD. The GST tag of flag-STRAD was subsequently removed. An in vitro kinase analysis of the purified proteins showed that LKB1-WT directly phosphorylated STRAD, whereas LKB1-KD did not (Figure 3D). Thus, our results indicate that the pseudokinase STRAD is the first direct in vivo LKB1 substrate to be identified.
STRAD activates the catalytic activity of LKB1
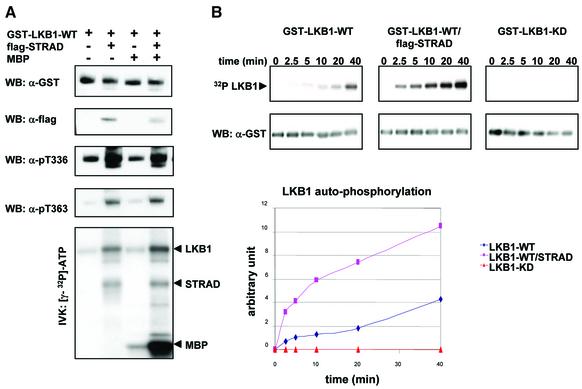
The previous observations led us to hypothesize that STRAD could activate the autophosphorylation, and hence the catalytic activity of LKB1. However, upon transfection of STRAD, LKB1 accumulates in cells (data not shown), which makes it difficult to distinguish between enhanced activation and stabilization of LKB1. To examine the activation of LKB1 in more detail, we therefore isolated GST–LKB1 and the GST–LKB1/flag-STRAD complex from transfected HEK-293T cells, since these purified complexes were easily quantified. Equal amounts of the protein complexes were subjected to an in vitro kinase assay in the presence of [γ-32P]ATP. Immunoblots were performed with phospho-specific antibodies against the major in vitro autophosphorylation sites of LKB1, namely Thr336 and Thr363 (Thr366 in mouse Lkb1) (Sapkota et al., 2002). Upon transfection of STRAD, we found a strong increase in LKB1 autophosphorylation at these sites (Figure 4A). Alone, LKB1 is a weakly active kinase with minimal potential to phosphorylate exogenous substrates (D.R.Alessi, unpublished data). We investigated whether STRAD could enhance phosphorylation of the exogenous substrate MBP by LKB1. The phosphorylation of MBP by the GST–LKB1/flag-STRAD complex was strongly enhanced compared with its phosphorylation by GST–LKB1 alone (Figure 4A, bottom panel).
Fig. 4. (A) STRAD activates LKB1 as visualized by LKB1s enhanced autophosphorylation and its increased ability to phosphorylate MBP. GST–LKB1-WT and GST–LKB1-WT/flag-STRAD protein complexes were isolated from transfected HEK-293T cells using GST-affinity chromatography. Protein complexes were subjected to an in vitro kinase assay (IVK) in the presence or absence of 10 µg of MBP. Protein complexes were immunoblotted (WB) with α-GST or α-flag to determine equal loading. Autophosphorylation of LKB1 was determined by immunoblotting (WB) with phospho-specific antibodies against the LKB1 autophosphorylation sites Thr336 and Thr363. Phosphorylated proteins were separated by SDS–PAGE and visualized by autoradiography. (B) LKB1 autophosphorylation is enhanced 3- to 4-fold upon forced expression of STRAD. A time-course experiment was performed using GST–LKB1-WT, GST–LKB1-WT/flag-STRAD and GST–LKB1-KD isolated from transfected HEK-293T cells. The in vitro kinase assay (IVK) was initiated by adding [γ-32P]ATP and kinase buffer to the GST proteins, and stopped at the indicated time points by the addition of SDS sample buffer. The protein complexes were separated by SDS–PAGE and immunoblotted (WB) with α-GST. Phosphorylated proteins were visualized by autoradiography. Levels of phosphorylation were quantified and are represented graphically.
To study the kinetics of STRAD-mediated activation of LKB1 autophosphorylation, we performed a time-course experiment (Figure 4B). GST–LKB1-WT, GST–LKB1-KD and the GST–LKB1-WT/flag-STRAD complex were isolated from transfected HEK-293T cells, and subjected to an in vitro kinase assay in the presence of [γ-32P]ATP for time periods ranging from 0 to 40 min. LKB1 autophosphorylation increased 3- to 4-fold upon forced STRAD expression, and this enhancement was already visible after 2.5 min (Figure 4B, bottom panel).
STRAD activates LKB1, but does not alter its substrate specificity
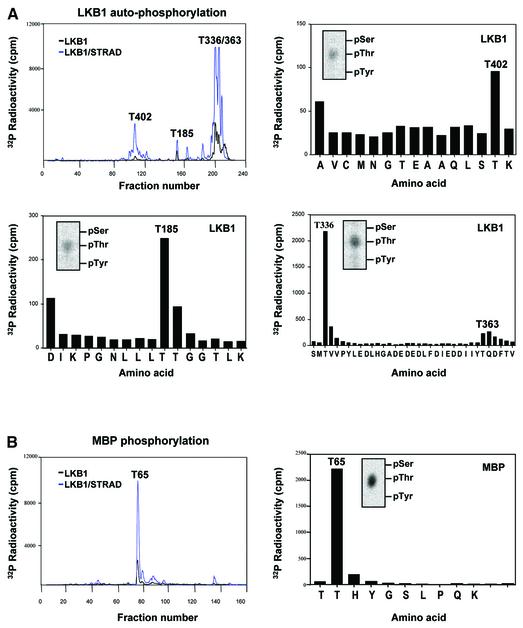
To address the specificity of STRAD-mediated LKB1 activation, we investigated the phosphorylated sites on LKB1 and MBP. GST–LKB1 and the GST–LKB1/flag-STRAD complex were isolated from transfected HEK-293T cells and subjected to an in vitro kinase assay in the presence of [γ-32P]ATP. HPLC fractionation of trypsin-digested LKB1 and STRAD-activated LKB1 generated comparable peak patterns (Figure 5A, upper left), confirming that the increased phosphorylation of LKB1 upon transfection of STRAD was caused by intrinsic activation of LKB1. The stoichiometry of LKB1 autophosphorylation in the presence of STRAD was dramatically increased. The 32P fractions were analysed by MALDI-TOF mass spectrometry, followed by phospho-amino acid analysis and solid-phase Edman degradation. In addition to the previously identified Thr336 and Thr363 sites (Figure 5A, bottom right; Table I), two novel autophosphorylation sites, namely Thr185 and Thr402, were characterized (Figure 5A, upper right and bottom left; Table I).
Fig. 5. STRAD-mediated activation of LKB1 does not affect its substrate specificity. (A) Clockwise, starting upper left. Tryptic peptide map of GST–LKB1 and GST–LKB1 activated by STRAD, and the corresponding Edman degradation diagrams and phospho-amino acid (PAA) analysis (inserts). The increased stoichiometry of LKB1 autophosphorylation upon STRAD activation enabled the identification of two novel autophosphorylation sites: Thr185 and Thr402. (B) Tryptic peptide map of MBP phosphorylated by GST–LKB1 and GST–LKB1/flag-STRAD (left panel), and the corresponding Edman degradation diagram (right panel) and PAA analysis (insert). The LKB1 phosphorylation site on MBP was determined as Thr65.
Table I. Identification of tryptic peptides isolated from the in vitro kinase reactions.
| Residues | Phosphorylationsite | Modification | Mass | |
|---|---|---|---|---|
| |
|
|
Measured |
Theoretical |
| 389–403 LKB1 | T402 | P04 + Met-Ox | 1724.697 | 1724.776 |
| 176–191 LKBI | T185 | P04 | 1720.915 | 1720.959 |
| 334–383 LKB1 | T336 + T363 | 2× P04 | 5843.049 | 5842.028 |
| 64–73 MBP | T65 | P04 | 1211.533 | 1211.546 |
| 317–346 STRAD | T329 | P04 | 3204.390 | 3204.470 |
| 386–431 STRAD | T419 | P04 | 5213.557 | 5212.619 |
The site phosphorylated in MBP by GST–LKB1 alone and by the GST–LKB1/flag-STRAD complex was similarly determined to be Thr65 (Figure 5B; Table I). The comparable peak patterns of the HPLC traces of trypsin-digested MBP in this experiment (Figure 5B, left panel) indicated that STRAD enhanced the kinase activity of LKB1, but did not alter its substrate specificity.
LKB1 phosphorylates STRAD at Thr329 and Thr419
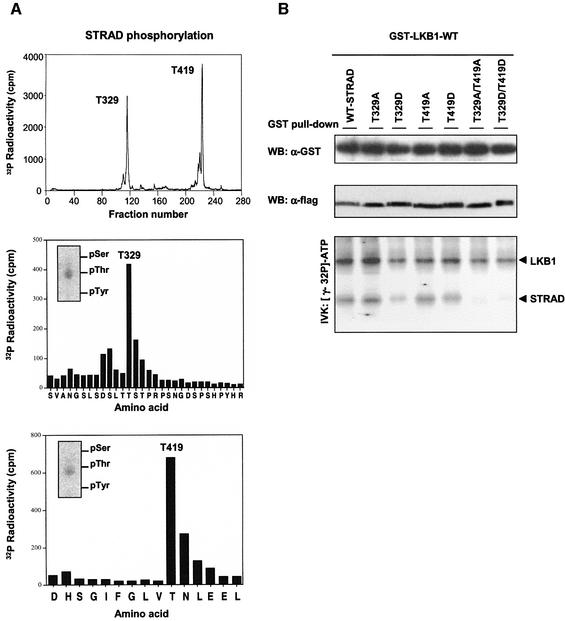
Since STRAD is the first physiological substrate of LKB1 identified so far, we were interested in mapping the phosphorylated sites on STRAD. For this purpose, a GST–STRAD/myc–LKB1-WT complex was expressed in HEK-293T cells and subsequently isolated. After the complex was subjected to an in vitro kinase assay in the presence of [γ-32P]ATP, the sites were mapped as described above and were found to be Thr329 and Thr419 (Figure 6A; Table I). Mutation of these sites to either alanine or aspartic acid residues abolished LKB1-mediated STRAD phosphorylation, as is demonstrated by an in vitro kinase assay with the corresponding mutants (Figure 6B). However, these mutations did not alter the ability of STRAD to bind or activate LKB1.
Fig. 6. (A) LKB1 phosphorylates STRAD at Thr329 and Thr419. Tryptic map of GST–STRAD phosphorylated by myc–LKB1-WT (upper panel), and the corresponding Edman degradation diagrams (bottom two panels) and phospho-amino acid (PAA) analysis (inserts). (B) Mutation of Thr329 and Thr419 abolishes STRAD phosphorylation by LKB1, but does not interfere with STRAD’s ability to bind or activate LKB1. Thr329 and/or Thr419 of flag-STRAD were mutated to Ala (T329A, T419A) or Asp (T329D, T419D). The indicated protein complexes were isolated from transfected HEK-293T cells using GST-affinity chromatography and immunoblotted (WB) with α-GST or α-STRAD. In addition, protein complexes were subjected to an in vitro kinase assay (IVK), separated by SDS–PAGE and visualized by autoradiography.
STRAD determines the subcellular localization of LKB1
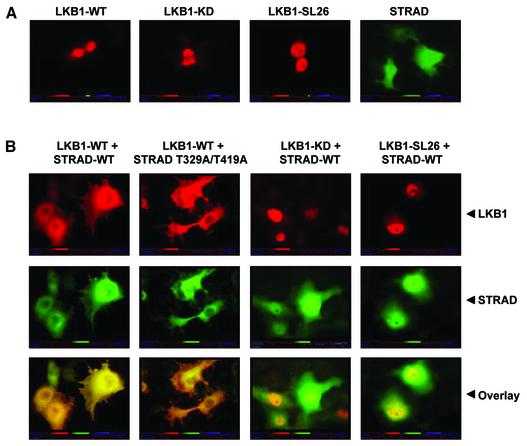
Cytoplasmic localization of LKB1 is reportedly important for its role in mediating G1 cell cycle arrest (Tiainen et al., 2002). Previously, it was shown that overexpression of an LKB1-interacting protein, LIP1, induces translocation of LKB1 from the nucleus to the cytoplasm (Smith et al., 2001). To study whether STRAD affects the subcellular distribution of LKB1, we carried out localization studies in COS cells. Myc-tagged LKB1 constructs and flag-tagged STRAD constructs were transfected into COS cells, and the localization of the corresponding proteins was determined by immunofluorescence. LKB1-WT and mutants localized mainly to the nucleus, while STRAD was diffusely expressed in the cell (Figure 7A). Upon transfection of STRAD, LKB1-WT re-localized to the cytoplasm together with STRAD (Figure 7B). The STRAD mutant in which the Thr329 and Thr419 LKB1 phosphorylation sites are altered to alanine was still able to translocate LKB1 to the cytoplasm, indicating that phosphorylation of STRAD by LKB1 is not essential for this process. The nuclear localization of LKB1-KD and of the SL26 mutant did not alter upon STRAD transfection. This implied that the cytoplasmic re-localization of LKB1 was dependent both on LKB1 kinase activity and on its ability to associate with STRAD.
Fig. 7. STRAD determines the cytoplasmic subcellular localization of LKB1, which is dependent on the kinase activity of LKB1 and its ability to associate with STRAD. Phosphorylation of STRAD by LKB1 is not essential for this process. (A) Myc-tagged LKB1-WT, -KD, -SL26 and flag-STRAD were transfected into COS cells. The localization of proteins was determined by incubation with α-myc/TRITC–goat α-mouse or α-STRAD–biotin/FITC–streptavidin and visualized by fluorescence microscopy. (B) Co-transfection of STRAD with LKB1-WT results in translocation of the LKB1-WT–STRAD complex to the cytoplasm. The STRAD mutant T329A/T419A is also able to re-localize LKB1-WT. LKB1-KD and LKB1-SL26 are not able to translocate to the cytoplasm upon forced expression of STRAD. Detection was performed as above.
STRAD is an essential co-factor for LKB1-mediated G1 cell cycle arrest
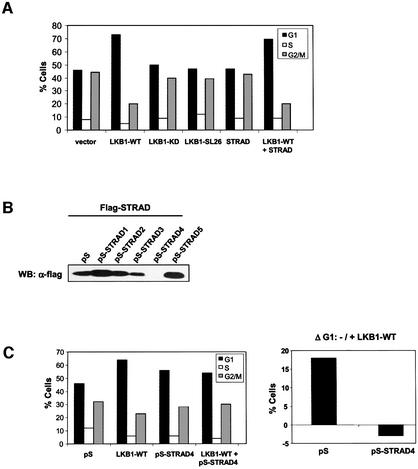
Previously, forced expression of LKB1-WT was shown to induce a G1 cell cycle arrest in LKB1-deficient G361 melanoma cells (Tiainen et al., 1999). Since our results predict that STRAD is important for LKB1 function, we investigated its effect on this LKB1-induced growth arrest. Expression of LKB1-WT induced a very potent G1 arrest which could not be further enhanced by STRAD (Figure 8A). As predicted, transient transfection of STRAD did not affect the cell cycle, since no endogenous LKB1 is present in G361 cells. Interestingly, both kinase-inactive LKB1 and the SL26 mutant failed to induce G1 arrest. This suggested not only that LKB1 kinase activity was required for its growth-suppressive activities, but also that LKB1 had to be able to associate with its adaptor STRAD. Western blot analysis confirmed the presence of endogenous STRAD in G361 cells (data not shown). We therefore hypothesized that endogenous STRAD cooperated with exogenous LKB1 in mediating G1 cell cycle arrest. To examine this in detail, we generated STRAD-specific small interfering RNA by taking advantage of the pSUPER system (Brummelkamp et al., 2002). Several siRNA sequences were tested by co-transfection of flag-STRAD and the indicated pSUPER (pS) constructs in HEK-293T cells. pSUPER-STRAD4 completely eliminated the expression of co-transfected STRAD (Figure 8B). We analysed the ability of LKB1-WT to induce G1 cell cycle arrest upon co-transfection with pSUPER-STRAD4, and found that removal of STRAD by RNA interference abolished the LKB1-directed growth arrest (Figure 8C). This indicated that STRAD is essential for the growth-suppressive activity of LKB1, most probably by activating LKB1 and anchoring it in the cytoplasm, and underscored the importance of STRAD as an LKB1-specific co-factor in a biological setting.
Fig. 8. STRAD is an essential co-factor for LKB1-mediated G1 cell cycle arrest in G361 cells. (A) Cell cycle profiles of G361 cells, transfected with the indicated plasmids and blocked with nocadozole. Both LKB1 kinase activity and its ability to associate with STRAD are required for its G1 arrest-inducing potential, as is demonstrated by the failure of LKB1-KD and LKB1-SL26 to induce cell cycle arrest. (B) pS-STRAD4 efficiently knocks-down STRAD expression. The indicated pS-STRAD constructs were co-transfected with flag-STRAD and Renilla luciferase. Cell lysates were immunoblotted with α-flag to analyse STRAD expression. Transfection efficiencies were determined to be equal by measuring Renilla luciferase units (data not shown). (C) Cell cycle profiles of G361 cells, transfected with pS or pS-STRAD4 in the absence or presence of LKB1-WT (left panel). The amount of cells in G1 was determined and ΔG1 is represented in a graph (right panel).
Discussion
In this study we provide evidence that the STE20-like pseudokinase STRAD acts as an upstream activator of LKB1. This has several implications. First, the discovery of a STE20-like molecule in the yet to be defined LKB1 pathway potentially links LKB1 to a pathway activated similarly to MAPK signalling. MAPKs are activated by a variety of stimuli, in which STE20-like kinases mediate the activation of MAPKKKs (Dan et al., 2001).
Secondly, our observations with a naturally kinase-dead STE20 homologue demonstrate that activation by STE20-like molecules can be independent of their intrinsic catalytic activity. Mutagenesis studies of the STE20-like kinases NIK/HGK, TNIK and MINK have similarly suggested activation functions for these proteins unrelated to their kinase activity (Su et al., 1997; Fu et al., 1999; Dan et al., 2000).
Thirdly, our results suggest that a pseudokinase can regulate the catalytic activity of the kinase to which it binds. Interestingly, Janus kinases (JAKs) contain both a C-terminal kinase domain (JH1) and an adjacent pseudokinase domain (JH2) (Wilks et al., 1991). Several studies have implied an essential regulatory role for the JH2 pseudokinase domain. Substitutions and in-frame deletions in the JH2 domain of JAK3 cause severe combined immunodeficiency (Macchi et al., 1995; Candotti et al., 1997). A more detailed study showed that the JH2 domain of JAK3 binds to the JH1 kinase domain, and is essential for its intrinsic catalytic activity (Chen et al., 2000). Similarly, mutagenesis of the JH2 domain of TYK-2 JAK kinase impaired its catalytic function (Yeh et al., 2000). These findings imply a positive regulatory role for pseudokinase domains, most likely through intramolecular interactions with the adjacent kinase domain. However, the JH2 domain may also inhibit JAK-2 kinase activity in other settings. Mutation of the JH2 domain of the Drosophila JAK orthologue Hopscotch yielded a gain-of-function phenotype (Luo et al., 1997). Accordingly, the corresponding JAK-2 mutant was a hyperactive kinase, while deletion of the entire JH2 domain of JAK-2 results in increased kinase activity, as measured by activation of downstream Stat transcription factors (Saharinen et al., 2000). A recent report proposes a switch function for the JH2 domain, which may unify the opposing results regarding the JAK pseudokinase domain (Saharinen and Silvennoinen, 2002). STRAD may activate LKB1 by an intermolecular interaction between the pseudokinase domain of STRAD and the kinase domain of LKB1. Since this activation is very rapid, it is likely to be a significant physiological process. How LKB1 activation by STRAD is achieved remains to be investigated. X-ray crystallography of the individual proteins and of the complex will provide more insight into this process. The enhanced LKB1 activity will be of great importance for future investigations into other substrates and for the rigorous analysis of LKB1 function.
Recently, it was shown that Thr336 and Thr366 are the major autophosphorylation sites of mouse Lkb1 (Sapkota et al., 2002). We confirmed these data on the human orthologues Thr336 and Thr363. Moreover, the enhanced stoichiometry of LKB1 autophosphorylation by STRAD enabled us to identify two novel sites: Thr185 and Thr402. We show that increased LKB1 autophosphorylation of all sites correlates with the activation of its catalytic activity. The role of STRAD phosphorylation by LKB1 remains to be established, but it did not affect the STRAD regulatory activities described in this study. However, LKB1-mediated phosphorylation of STRAD at Thr329 and Thr419 may be of importance for signalling downstream of LKB1 and STRAD.
We show that STRAD coordinates the cytoplasmic subcellular localization of LKB1, which has been reported to be critical for its role in mediating cell cycle arrest (Tiainen et al., 2002). Previously, LIP1 was shown to translocate LKB1 from the nucleus to the cytoplasm (Smith et al., 2001). However, the effects of the two proteins on LKB1 localization appear to be different. LIP1-forced expression re-localizes LKB1 to distinct cytoplasmic foci, while STRAD-directed translocation results in diffuse cytoplasmic LKB1 localization. Furthermore, LKB1 cytoplasmic re-localization by LIP1 was shown to occur in 30% of LIP1/LKB1 co-transfected cells, while STRAD-directed LKB1 translocation was consistently observed. Removal of endogenous STRAD by RNA interference abolishes LKB1-directed G1 cell cycle arrest, indicating that STRAD is an essential co-factor for LKB1 growth-suppressive activities.
Our studies with the LKB1 mutant SL26 suggest a very significant role for STRAD in LKB1-mediated tumour suppression. The SL26 mutation, which was found in a PJS family (Hemminki et al., 1998), is unique in that it has retained its basal catalytic activity (Nezu et al., 1999). However, this mutation abolishes STRAD interaction, and hence STRAD-mediated activation and re-localization of LKB1. Interestingly, the SL26 LKB1 mutant is unable to mediate LKB1-directed G1 cell cycle arrest. We postulate that patients harbouring the SL26 mutation develop PJS due to SL26’s inability to be regulated by STRAD.
In conclusion, our results imply that the regulation of LKB1 by STRAD is very likely to be central to the physiological activities of LKB1. Therefore, this study will open up new avenues of research to define the biological context in which LKB1 functions.
Materials and methods
Reagents
HEK-293T, HeLa, Rat-2, COS and G361 cells were cultured in Dulbecco’s modified Eagle’s medium with standard supplements. HEK-293T and COS cells were transfected using FUGENE 6 reagent (Roche, Meylan, France) according to the manufacturer’s instructions. G361 cells were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). The following antibodies were used: 9E10 (mouse) for Myc-tagged proteins; anti-flag monoclonal antibody M2 (mouse) (Sigma, Poole, Dorset, UK); anti-GST monoclonal antibody (mouse) (Sigma); anti-LKB1 antibodies (sheep and rabbit) were described previously (Sapkota et al., 2001, 2002); anti-STRAD monoclonal antibody (5A11, mouse) was generated as described below. STRAD cDNA was cloned from a yeast two-hybrid screen of a fetal brain library (Clontech, Basingstoke, UK) and subsequently subcloned into pCDNA3 (Flag tagged), pGEX-1N and pEBG-2T (both GST tagged) according to standard cloning procedures. MBP was obtained from Invitrogen. PKA, histone 2A and histone 2B were kind gifts of Dr Boudewijn Burgering (UMC, The Netherlands). pSUPER was kindly provided by Dr Reuven Agami (NKI, The Netherlands). Myc-tagged LKB1 mutants and flag-tagged STRAD mutants were generated by QuikChange site-directed mutagenesis (Stratagene, La Jolla, CA).
Generation of STRAD monoclonal antibody
Six-week-old BALB/c mice were immunized by intraperitoneal injection of 100 µg of bacterially expressed GST–STRAD fusion protein in Freund’s complete adjuvant (Difco, Detroit, MI), with a second injection in Freund’s incomplete adjuvant (Difco) 14 days later. Three additional injections were performed using 100 µg of GST–STRAD in phosphate-buffered saline (PBS) at weekly intervals. A mouse with an anti-STRAD titre of 1/1000 was killed, the spleen isolated and splenocytes fused to SP2/0 myeloma cells using a standard polyethylene glycol protocol as described previously (Castrop et al., 1995). The fused cell population was resuspended in hypoxanthine aminopterin thymidine selection medium (Life Technologies, Gaithersburg, MD) and plated into 96-well flat-bottom culture plates. Selection was allowed to occur over a 2 week period and hybridoma supernatants were screened for anti-STRAD antibodies (see below). Positive hybridomas were repeatedly subcloned to generate clonal hybridomas secreting STRAD antibodies. Anti-STRAD was purified over a protein A column (Bio-Rad, Hercules, CA), and dialysed extensively against PBS. Purified STRAD antibody was biotinylated by incubation of 1 mg of antibody with 100 µg of EZ-Link Sulfo-NHS-LC-LC-Biotin (Pierce, Chester, UK) for 1 h at room temperature. Biotinylated antibody was dialysed extensively against PBS.
Hybridoma screening assay
Approximately 10 × 106 COS cells were transiently transfected with 10 µg of pCDNA3 vector expressing flag-tagged STRAD. Cells were subsequently plated into 96-well culture plates at a concentration of 100 per well. The cells were cultured for 48 h and fixed with 100% methanol for 2 h at –20°C. Screening the hybridomas for anti-STRAD antibodies was performed by incubating 100 µl of hybridoma supernatant for 1.5 h at room temperature. Detection was carried out with a rabbit anti-mouse horseradish peroxidase-coupled antibody (Dako) and 0.02% amino ethyl carbonate/0.1% hydrogen peroxide in 0.3 M sodium acetate pH 4.8 as a colour substrate. Individual wells were examined for staining using an inverted microscope.
Immunoprecipitation, affinity chromatography and immunoblotting
Cells were lysed in 1% Triton X-100 lysis buffer (50 mM Tris–HCl pH 7.5, 1 mM EGTA, 1 mM EDTA, 1% Triton X-100, 1 mM orthovanadate, 50 mM sodium fluoride, 5 mM sodium pyrophosphate, 0.27 M sucrose) with complete Mini protease inhibitor cocktail tablets (Roche). Protein complexes were either immunoprecipitated with the indicated antibodies or affinity chromatographed using glutathione– Sepharose (Amersham, Little Chalfont, UK) according to standard procedures. GST-tagged proteins were eluted from the resin in 20 mM glutathione. Isolated protein complexes were separated by SDS–PAGE and transferred to PVDF membranes (Hybond-P, Amersham). Blots were probed with the indicated antibodies and bound antibodies were visualized using horseradish peroxidase-conjugated antibodies and ECL (Amersham) according to standard procedures.
Expression of GST-tagged fusion proteins in Escherichia coli
The pGEX constructs (expressing GST–LKB1, GST–STRAD, GST–flag-STRAD and GST–CREB) were transformed into E.coli BL21 cells, and a 0.5 l culture was grown at 37°C until an OD600 of 0.7. Isopropyl-β-d-galactosidase was added to 250 µM to induce protein expression, and the cells were cultured for another 16 h at 26°C. Cells were harvested by centrifugation, and lysed by freeze–thawing in 1% Triton X-100 lysis buffer and sonication. GST-tagged proteins were purified from the lysates using glutathione–Sepharose and eluted from the resin in 20 mM glutathione. For GST–flag-STRAD, the flag-STRAD protein was removed from the resin by incubation with PreScission protease, which cleaved the N-terminal GST tag.
In vitro kinase assay
Immunoprecipitated and GST–protein complexes were incubated in a kinase buffer containing 50 mM Tris–HCl pH 7.5, 10 mM manganese chloride and 100 µM [γ-32P]ATP (500 c.p.m./pmol). After incubation for 60 min at 30°C, the reactions were terminated by addition of SDS sample buffer, the proteins separated by SDS–PAGE and analysed by autoradiography.
32P-labelling of transfected HEK-293T cells
Twenty-four hours post-transfection, cells were washed with phosphate-free Dulbecco’s modified Eagle’s medium (DMEM), incubated for 3 h with [32P]phosphate (1 mCi/ml) and subsequently lysed as described before.
Phosphopeptide sequence analysis
In order to map the phosphorylation sites in LKB1, MBP and STRAD, 10 µg of the indicated proteins were phosphorylated in a 200 µl reaction as described above except that the specific activity of [γ-32P]ATP was 5000 c.p.m./pmol. The reactions were terminated by adding SDS (1% by mass final) and DTT (10 mM final), then alkylated with 4-vinylpyridine. The proteins were separated by SDS–PAGE, visualized by autoradiography, excised from the gel and digested with trypsin. The tryptic peptides were analysed by C18 chromatography on a Vydac 218TP54 C18 column (Separations Group, Hesperia, CA) equilibrated in 0.1% (w/v) trifluoroacetic acid in water. The column was developed with a linear acetonitrile gradient at a flow rate of 0.8 ml/min, and fractions of 0.4 ml were collected. At least 80% of the radioactivity applied to the column was recovered in the fractions for each of the runs shown in Figures 5 and 6. Each of the major eluting 32P-labelled peptides was analysed by MALDI-TOF mass spectrometry on a PerSeptive Biosystems Elite-STR mass spectrometer using α-cyanocinnamic acid as the matrix. Spectra were obtained in both the linear and reflector mode. The site of phosphorylation was determined by solid-phase Edman degradation of the peptides coupled to Sequelon-AA membrane (Milligen) on an Applied Biosystems 476A sequenator as described previously (Campbell and Morrice, 2002). The STRAD peptide 386–431 was subdigested in Asp-N prior to its coupling to Sequelon-AA membrane. Phospho-amino acid analysis was carried out by digestion of dried 32P-labelled peptides in HCl for 90 min at 100°C. 32P-labelled amino acids were separated by thin-layer chromatography and visualized by autoradiography.
Immunofluorescence
Twenty-four hours post-transfection, COS cells were fixed in 3.7% paraformaldehyde, followed by permeabilization in 0.1% Triton. To study the localization of LKB1, cells were stained with anti-myc antibody, followed by incubation with TRITC-labelled goat anti-mouse IgG (Southern Biotechnology Associates). Subsequently, unoccupied binding sites were blocked with normal mouse serum. Next, STRAD localization was determined by staining with biotinylated STRAD MoAb, followed by incubation with FITC-labelled streptavidin (Zymed). Cells were mounted using Vectashield with DAPI (Vector Laboratories, Burlingame, CA), and visualized by fluorescence microscopy.
G361 cell cycle analysis
G361 cells were co-transfected with GFP–spectrin and the indicated plasmids as described above. Thirty hours post-transfection, the cells were treated with nocodazole (70 ng/ml; Sigma), and grown for an additional 18 h to induce G2/M block. Subsequently, the cells were harvested, fixed in 70% ethanol, extensively washed in PBS, treated with RNase (100 µg/ml; Qiagen, Hilden, Germany), and stained with propidium iodide (50 µg/ml). The cell cycle profiles of the GFP-positive cells were determined by FACS analysis.
Generation of STRAD small interference RNAs
Several STRAD-specific sequences were targeted for RNA inter ference by cloning compatible oligos into pSUPER (pS): pS-STRAD1, 5′-AGGATTTGAGGACCTGATG-3′; pS-STRAD2, 5′-TATCGTGCC ATATCGAGCC-3′; pS-STRAD3, 5′-TGAGCTGTGGGTTGTCACA-3′; pS-STRAD4, 5′-GGTCTACCTGTCTGGTTTG-3′; pS-STRAD5, 5′-TT TTGAGGGCAGCCAGTCT-3′.
To test their ability to downregulate STRAD expression, the pS-STRAD constructs were co-transfected in duplicates with flag-STRAD and Renilla luciferase in HEK-293T cells. After 36 h, one set of transfectants was lysed in passive lysis buffer, and Renilla luciferase units were measured using Dual Luciferase Reporter Assay System (Promega, Madison, WI) in order to determine transfection efficiencies. The other set of transfectants was lysed in Triton X-100 buffer and analysed by immunoblotting with α-flag.
Acknowledgments
Acknowledgements
We thank Dr D.G.Campell, Dr M.Deak and A.Kieloch for technical assistance, Drs M.van de Wetering and J.van Es for helpful advice, and Dr R.H.Giles for critical reading of the manuscript. This work was supported by a grant from the Centre for Biomedical Genetics, Utrecht, The Netherlands.
References
- Bardeesy N., Sinha,M., Hezel,A.F., Signoretti,S., Hathaway,N.A., Sharpless,N.E., Loda,M., Carrasco,D.R. and DePinho,R.A. (2002) Loss of the Lkb1 tumour suppressor provokes intestinal polyposis but resistance to transformation. Nature, 419, 162–167. [DOI] [PubMed] [Google Scholar]
- Brummelkamp T.R., Bernards,R. and Agami,R. (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science, 296, 550–552. [DOI] [PubMed] [Google Scholar]
- Campbell D.G. and Morrice,N.A. (2002) Identification of protein phosphorylation sites by a combination of mass spectrometry and solid phase Edman sequencing. J. Biomol. Tech., 13, 121–132. [PMC free article] [PubMed] [Google Scholar]
- Candotti F. et al. (1997) Structural and functional basis for JAK3-deficient severe combined immunodeficiency. Blood, 90, 3996–4003. [PubMed] [Google Scholar]
- Castrop J., van Wichen,D., Koomans-Bitter,M., Van de,W.M., de Weger,R., van Dongen,J. and Clevers,H. (1995) The human TCF-1 gene encodes a nuclear DNA-binding protein uniquely expressed in normal and neoplastic T-lineage lymphocytes. Blood, 86, 3050–3059. [PubMed] [Google Scholar]
- Chen M., Cheng,A., Candotti,F., Zhou,Y.J., Hymel,A., Fasth,A., Notarangelo,L.D. and O’Shea,J.J. (2000) Complex effects of naturally occurring mutations in the JAK3 pseudokinase domain: evidence for interactions between the kinase and pseudokinase domains. Mol. Cell. Biol., 20, 947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins S.P., Reoma,J.L., Gamm,D.M. and Uhler,M.D. (2000) LKB1, a novel serine/threonine protein kinase and potential tumour suppressor, is phosphorylated by cAMP-dependent protein kinase (PKA) and prenylated in vivo. Biochem. J., 345, 673–680. [PMC free article] [PubMed] [Google Scholar]
- Dan I. et al. (2000) Molecular cloning of MINK, a novel member of mammalian GCK family kinases, which is up-regulated during postnatal mouse cerebral development. FEBS Lett., 469, 19–23. [DOI] [PubMed] [Google Scholar]
- Dan I., Watanabe,N.M. and Kusumi,A. (2001) The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol., 11, 220–230. [DOI] [PubMed] [Google Scholar]
- Fu C.A., Shen,M., Huang,B.C., Lasaga,J., Payan,D.G. and Luo,Y. (1999) TNIK, a novel member of the germinal center kinase family that activates the c-Jun N-terminal kinase pathway and regulates the cytoskeleton. J. Biol. Chem., 274, 30729–30737. [DOI] [PubMed] [Google Scholar]
- Giardiello F.M., Brensinger,J.D., Tersmette,A.C., Goodman,S.N., Petersen,G.M., Booker,S.V., Cruz-Correa,M. and Offerhaus,J.A. (2000) Very high risk of cancer in familial Peutz–Jeghers syndrome. Gastroenterology, 119, 1447–1453. [DOI] [PubMed] [Google Scholar]
- Hemminki A. et al. (1998) A serine/threonine kinase gene defective in Peutz–Jeghers syndrome. Nature, 391, 184–187. [DOI] [PubMed] [Google Scholar]
- Huse M. and Kuriyan,J. (2002) The conformational plasticity of protein kinases. Cell, 109, 275–282. [DOI] [PubMed] [Google Scholar]
- Jeghers H., McKusick,V.A. and Karz,K.H. (1949) Generalised intestinal polyposis and melanin spots of oral mucosa, lips and digits: a syndrome of diagnostic significance. N. Engl. J. Med., 241, 992–1005. [DOI] [PubMed] [Google Scholar]
- Jenne D.E., Reimann,H., Nezu,J., Friedel,W., Loff,S., Jeschke,R., Muller,O., Back,W. and Zimmer,M. (1998) Peutz–Jeghers syndrome is caused by mutations in a novel serine threonine kinase. Nat. Genet., 18, 38–43. [DOI] [PubMed] [Google Scholar]
- Jishage K. et al. (2002) Role of Lkb1, the causative gene of Peutz–Jegher’s syndrome, in embryogenesis and polyposis. Proc. Natl Acad. Sci. USA, 99, 8903–8908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston A.M., Naselli,G., Gonez,L.J., Martin,R.M., Harrison,L.C. and DeAizpurua,H.J. (2000) SPAK, a STE20/SPS1-related kinase that activates the p38 pathway. Oncogene, 19, 4290–4297. [DOI] [PubMed] [Google Scholar]
- Karuman P. et al. (2001) The Peutz–Jegher gene product LKB1 is a mediator of p53-dependent cell death. Mol. Cell, 7, 1307–1319. [DOI] [PubMed] [Google Scholar]
- Luo H., Rose,P., Barber,D., Hanratty,W.P., Lee,S., Roberts,T.M., D’Andrea,A.D. and Dearolf,C.R. (1997) Mutation in the Jak kinase JH2 domain hyperactivates Drosophila and mammalian Jak-Stat pathways. Mol. Cell. Biol., 17, 1562–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macchi P. et al. (1995) Mutations of Jak-3 gene in patients with autosomal severe combined immune deficiency (SCID). Nature, 377, 65–68. [DOI] [PubMed] [Google Scholar]
- Marignani P.A., Kanai,F. and Carpenter,C.L. (2001) LKB1 associates with Brg1 and is necessary for Brg1-induced growth arrest. J. Biol. Chem., 276, 32415–32418. [DOI] [PubMed] [Google Scholar]
- Martin S.G. and St Johnston,D. (2003) A role for Drosophila LKB1 in anterior-posterior axis formation and epithelial polarity. Nature, 421, 379–384. [DOI] [PubMed] [Google Scholar]
- Miyoshi H., Nakau,M., Ishikawa,T.O., Seldin,M.F., Oshima,M. and Taketo,M.M. (2002) Gastrointestinal hamartomatous polyposis in Lkb1 heterozygous knockout mice. Cancer Res., 62, 2261–2266. [PubMed] [Google Scholar]
- Nakau M., Miyoshi,H., Seldin,M.F., Imamura,M., Oshima,M. and Taketo,M.M. (2002) Hepatocellular carcinoma caused by loss of heterozygosity in lkb1 gene knockout mice. Cancer Res., 62, 4549–4553. [PubMed] [Google Scholar]
- Nezu J., Oku,A. and Shimane,M. (1999) Loss of cytoplasmic retention ability of mutant LKB1 found in Peutz–Jeghers syndrome patients. Biochem. Biophys. Res. Commun., 261, 750–755. [DOI] [PubMed] [Google Scholar]
- Nishigaki K., Thompson D., Yugawa T., Rulli K., Hanson C., Cmarik J., Gutkind J.S., Teramoto H. and Ruscetti S. (2003) Identification and characterization of a novel Ste-20/GCK-related kinase, PAP kinase (PAPK). J. Biol. Chem., 278, 13520–13530. [DOI] [PubMed] [Google Scholar]
- Peutz J.L. (1921) On a very remarkable case of familial polyposis of the mucous membrane of the intestinal tract and nasopharynx accompanied by peculiar pigmentation of the skin and mucous membranes. Ned. Tijdschr. Geneeskd., 10, 134–146. [Google Scholar]
- Rossi D.J. et al. (2002) Induction of cyclooxygenase-2 in a mouse model of Peutz–Jeghers polyposis. Proc. Natl Acad. Sci. USA, 99, 12327–12332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saharinen P. and Silvennoinen,O. (2002) The pseudokinase domain is required for suppression of basal activity of Jak2 and Jak3 tyrosine kinases and for cytokine-inducible activation of signal transduction. J. Biol. Chem., 277, 47954–47963. [DOI] [PubMed] [Google Scholar]
- Saharinen P., Takaluoma,K. and Silvennoinen,O. (2000) Regulation of the Jak2 tyrosine kinase by its pseudokinase domain. Mol. Cell. Biol., 20, 3387–3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanna M.G., Da Silva,C.J., Luo,Y., Chuang,B., Paulson,L.M., Nguyen,B., Deveraux,Q.L. and Ulevitch,R.J. (2002) ILPIP, a novel anti-apoptotic protein that enhances XIAP-mediated activation of JNK1 and protection against apoptosis. J. Biol. Chem., 277, 30454–30462. [DOI] [PubMed] [Google Scholar]
- Sapkota G.P., Kieloch,A., Lizcano,J.M., Lain,S., Arthur,J.S., Williams,M.R., Morrice,N., Deak,M. and Alessi,D.R. (2001) Phosphorylation of the protein kinase mutated in Peutz–Jeghers cancer syndrome, LKB1/STK11, at Ser431 by p90RSK and cAMP-dependent protein kinase, but not its farnesylation at Cys433, is essential for LKB1 to suppress cell growth. J. Biol. Chem., 276, 19469–19482. [DOI] [PubMed] [Google Scholar]
- Sapkota G.P., Boudeau,J., Deak,M., Kieloch,A., Morrice,N. and Alessi,D.R. (2002) Identification and characterization of four novel phosphorylation sites (Ser31, Ser325, Thr336 and Thr366) on LKB1/STK11, the protein kinase mutated in Peutz–Jeghers cancer syndrome. Biochem. J., 362, 481–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D.P., Rayter,S.I., Niederlander,C., Spicer,J., Jones,C.M. and Ashworth,A. (2001) LIP1, a cytoplasmic protein functionally linked to the Peutz–Jeghers syndrome kinase LKB1. Hum. Mol. Genet., 10, 2869–2877. [DOI] [PubMed] [Google Scholar]
- Su Y.C., Han,J., Xu,S., Cobb,M. and Skolnik,E.Y. (1997) NIK is a new Ste20-related kinase that binds NCK and MEKK1 and activates the SAPK/JNK cascade via a conserved regulatory domain. EMBO J., 16, 1279–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiainen M., Ylikorkala,A. and Makela,T.P. (1999) Growth suppression by Lkb1 is mediated by a G1 cell cycle arrest. Proc. Natl Acad. Sci. USA, 96, 9248–9251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiainen M., Vaahtomeri,K., Ylikorkala,A. and Makela,T.P. (2002) Growth arrest by the LKB1 tumor suppressor: induction of p21WAF1/CIP1. Hum. Mol. Genet., 11, 1497–1504. [DOI] [PubMed] [Google Scholar]
- Watts J.L., Morotn,D.G., Bestman,J. and Kemphues,J. (2000) The C.elegans par-4 gene encodes a putative serine-threonine kinase required for establishing embryonic asymmetry. Development, 127, 1467–1475. [DOI] [PubMed] [Google Scholar]
- Wilks A.F., Harpur,A.G., Kurban,R.R., Ralph,S.J., Zurcher,G. and Ziemiecki,A. (1991) Two novel protein-tyrosine kinases, each with a second phosphotransferase-related catalytic domain, define a new class of protein kinase. Mol. Cell. Biol., 11, 2057–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh T.C., Dondi,E., Uze,G. and Pellegrini,S. (2000) A dual role for the kinase-like domain of the tyrosine kinase Tyk2 in interferon-α signaling. Proc. Natl Acad. Sci. USA, 97, 8991–8996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylikorkala A., Rossi,D.J., Korsisaari,N., Luukko,K., Alitalo,K., Henkemeyer,M. and Makela,T.P. (2001) Vascular abnormalities and deregulation of VEGF in Lkb1-deficient mice. Science, 293, 1323–1326. [DOI] [PubMed] [Google Scholar]