Abstract
The intrinsic enhancer–promoter specificity and chromatin boundary/insulator function are two general mechanisms that govern enhancer trafficking in complex genetic loci. They have been shown to contribute to gene regulation in the homeotic gene complexes from fly to mouse. The regulatory region of the Scr gene in the Drosophila Antennapedia complex is interrupted by the neighboring ftz transcription unit, yet both genes are specifically activated by their respective enhancers from such juxtaposed positions. We identified a novel insulator, SF1, in the Scr–ftz intergenic region that restricts promoter selection by the ftz-distal enhancer in transgenic embryos. The enhancer-blocking activity of the full-length SF1, observed in both embryo and adult, is orientation- and enhancer-independent. The core region of the insulator, which contains a cluster of GAGA sites essential for its activity, is highly conserved among other Drosophila species. SF1 may be a member of a conserved family of chromatin boundaries/insulators in the HOM/Hox complexes and may facilitate the independent regulation of the neighboring Scr and ftz genes, by insulating the evolutionarily mobile ftz transcription unit.
Keywords: boundary/enhancer–promoter specificity/ftz/HOM/Hox/insulator/Scr
Introduction
The evolutionary conservation of the homeotic genes, both in function and organization, has been attributed to their important roles in animal body patterning, and to their coordinated regulation (Lewis, 1978; Harding et al., 1985; Kmita et al., 2000; Cai et al., 2001). Such coordination in the Drosophila homeotic complexes often involves extensive regulatory DNA, and control elements that function over long distances. In genomic intervals where neighboring genes are closely positioned, long-range enhancers present a challenge for independent gene control. Two complementary mechanisms are implicated in enhancer– promoter specification within complex genetic loci. The first mechanism, promoter competition, is the result of the preferential interaction between an enhancer and one promoter that reduces or excludes its interaction with other available promoters (Foley and Engel, 1992; Merli et al., 1996; Ohtsuki et al., 1998). An example is the AE1 enhancer of the fushi tarazu (ftz) gene in the Drosophila Antennapedia complex (ANT-C), which selectively activates the ftz promoter but not the neighboring Sex combs reduced (Scr) promoter (see diagram in Figure 1). This selectivity is due to the preference of AE1 for the TATA-containing ftz promoter, over the TATA-less Scr promoter, rather than its incompatibility with the Scr promoter (Ohtsuki et al., 1998).
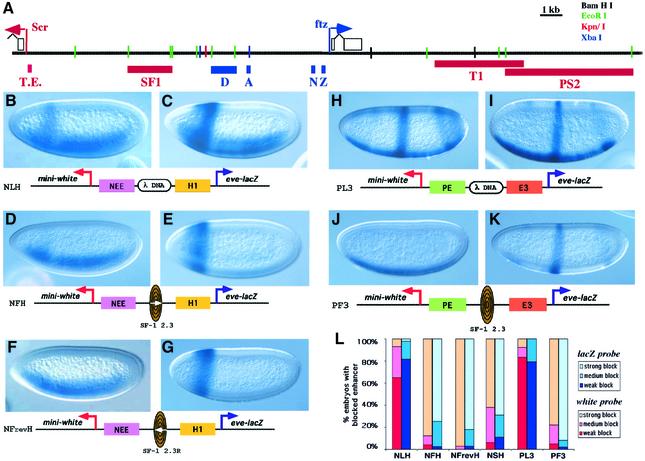
Fig. 1. Enhancer-blocking activity of SF1 in transgenic Drosophila embryos. (A) Schematic diagram of the ftz–Scr region. Arrows indicate the location and orientation of the Scr and ftz promoters. Open boxes represent exons and the thick lines represent introns and regulatory regions. Short vertical lines represent selected sites of several restriction enzymes, the names of which are indicated on the top right of the panel. Labeled boxes indicate selected enhancers and specialized DNA sequences: T.E., Scr Tethering Element (Calhoun et al., 2002); D, ftz-distal enhancer; A, ftz AE1 enhancer; T1, Scr enhancer for T1a; and PS2, Scr enhancer for C3p and T1a. The size and distance of the DNA elements are drawn to scale. (B–K) Reporter expression (white and eve/lacZ fusion gene) in blastoderm stage transgenic embryos visualized by whole-mount in situ hybridization (see Materials and methods). Embryos are shown anterior to the left and dorsal side up. Each test transgene is shown below the embryo image. (B and C) NLH embryos show a composite pattern consisting of comparable levels of NEE-directed ventral lateral expression and the anterior H1 stripe on the white (B) and lacZ (C) reporters. (D and E) NFH embryos show the reporter expression activated only by the proximal enhancers: NEE-directed ventrolateral stripes detected from white (D) and H1-specific expression from the lacZ reporter (E). (F and G) NFrevH embryos, which contain the SF1 element in reverse orientation, exhibit reporter expression by the proximal enhancers only: NEE from white (F), and H1 from lacZ (G). (H and I) PL3 embryos show a composite pattern consisting of PE-directed ventral expression and E3-directed mid-embryo stripe on the white (H) and lacZ (I) reporter genes. (J and K) PF3 embryos show that only the proximal enhancer can activate reporter expression: PE-directed ventral stain detected from white (J) and E3-specific expression from the lacZ reporter (K). (L) Quantitative assessment of the enhancer-blocking activity of each transgene. Thirty to 200 transgenic embryos from multiple lines were visually inspected for enhancer-blocking activity, which was categorized into weak, moderate or strong groups according to the level of reporter expression (see Materials and methods for details). The most frequently observed staining pattern is used in the figure.
The second mechanism involves the function of chromatin boundaries or insulators. These DNA elements can block transcriptional influences such as enhancer– promoter interactions and chromatin-mediated effects on gene expression (Gerasimova and Corces, 2001; West et al., 2002). Chromatin insulator function has been observed in HOM/Hox complexes of several species (Galloni et al., 1993; Hagstrom et al., 1996; Mihaly et al., 1998; Zhou et al., 1999; Kmita et al., 2000). In mouse, the functional range of the global hernia and digit enhancers flanking the Hox d10–d13 genes appears to be restricted by chromatin boundary element(s) positioned between the d13 and d11 genes (Kmita et al., 2000). In the Drosophila bithorax homeotic complex (BX-C), multiple boundary elements, including Mcp-1, Fab7 and Fab8, have been identified between the tissue-specific iab enhancers in the regulatory region of the Abdominal B (Abd-B) gene (Gyurkovics et al., 1990; Galloni et al., 1993). Although these boundaries have been implicated in modulating the iab–Abd-B interactions (Zhou et al., 1996), and maintaining the autonomy between neighboring iab enhancers (Mihaly et al., 1998), little is known about the mechanism of their function.
Here we report the presence of a novel insulator, SF1, in the Scr–ftz region in Drosophila, the first such activity identified in the ANT-C (see Figure 1 for map of the region). The SF1 activity persists throughout the animal life cycle, consistent with its potential role in regulating homeotic genes. As the Fab-7 insulator from the Drosophila BX-C, the highly conserved SF1 core sequence contains multiple GAGA sites that are essential for its activity. The intergenic position of SF1 and its ability to restrict promoter access by the promiscuous ftz-distal enhancer suggest that SF1 may direct enhancer trafficking in the Scr–ftz genomic interval.
Results
A novel enhancer-blocking activity in the Scr–ftz intergenic region
Although intrinsic properties of certain ftz enhancers, such as AE1, can account for their exclusive interaction with the cognate promoters, the same mechanism may not apply to all ftz enhancers in the region. Furthermore, the Scr-distal enhancers, separated from the Scr promoter by the entire ftz gene, would have to overcome the interference from a highly competitive ftz promoter. To test if insulator elements play a role in defining enhancer–promoter interactions in the Scr–ftz region, we examined DNA fragments from the Scr–ftz intergenic region for enhancer-blocking activity. Two tissue-specific enhancers were used in the enhancer-blocking assay, the hairy stripe 1 enhancer (H1) and the rhomboid neuroectoderm enhancer (NEE), which are active in a transverse anterior band and two ventral lateral stripes, respectively (Zhou et al., 1996; Cai and Levine, 1997). When a neutral DNA spacer from the λ phage is inserted between the two enhancers, both the lacZ and white reporters are expressed in a composite pattern directed by both H1 and NEE, as shown by whole-mount in situ hybridization (NLH, Figure 1B, C and L; Cai and Levine, 1997). Insertion of a 2.3 kb EcoRI fragment from the Scr–ftz intergenic region reduces the H1-directed white expression and NEE-directed lacZ expression but not the H1-directed lacZ or NEE-directed white expression, indicating a selective block of the distal enhancer activities (NFH, Figure 1D, E and L, see map in Figure 1). The enhancer-blocking activity of the element, named SF1 for the Scr–ftz boundary, appears comparable or even stronger than that of the Su(Hw) insulator from the gypsy retrotransposon (NSH, see Figure 1L). In contrast, other DNA fragments of comparable size from the 10 kb region surrounding SF1 exhibited little or no enhancer-blocking activity (data not shown). Importantly, the 15 kb intergenic region contains many closely spaced enhancers required for the tissue-specific regulation of Scr and ftz genes. The 2.3 kb SF1 region, however, appears to be devoid of any enhancer activities, as assayed in transgenic embryos with several promoters including those from the white, evenskipped (eve) and ftz genes (Figure 1; data not shown).
We also tested the ability of SF1 to block a different pair of embryonic enhancers, PE (twist proximal element) and E3 (eve stripe 3 enhancer; Cai and Levine, 1997). When the lambda spacer is inserted between the two enhancers, they direct the white and lacZ reporter expression in the ventral region and in the mid-embryo stripe, respectively (PL3, Figure 1H and I). Replacing the spacer with SF1 resulted in the block of E3-mediated expression of the white reporter and PE-mediated expression of the lacZ reporter (PF3, Figure 1J–L). Again, SF1 appears to block the distal enhancers more efficiently than the Su(Hw) insulator (data not shown). The insulator activity of SF1 is also orientation independent. When the 2.3 kb element is inserted in an inverted orientation between the NEE and H1 enhancers, it blocks the distal enhancers to a comparable level as in the forward orientation (NFrevH, see Figure 1F and G). In addition to the enhancer-blocking activity, the 2.3 kb SF1 element also contains a potent chromatin barrier activity as shown by its ability to protect the mini-white transgenes against chromosomal position effects (P.Majumder, unpublished data).
SF1 is active in late Drosophila development
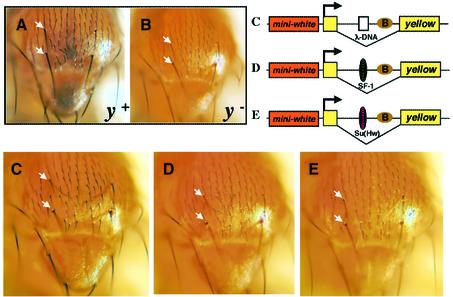
Activity of the homeotic selector genes such as Scr is required to maintain body segment identity throughout the animal life cycle. If SF1 is involved in regulating Scr and ftz genes, its boundary activity would be expected to persist to later stages of development. To test this, we examined the enhancer-blocking activity of SF1 in adult tissues with a transgenic yellow gene. The wild-type activity of yellow is required for the pigmentation of cuticle structures in larval and adult Drosophila (Figure 2A and B, arrows indicate macrochete bristles; Geyer and Corces, 1987). The yellow expression is activated in the adult bristles by the bristle-specific enhancer (B, see construct diagrams C–E, Figure 2) located in the first intron of the gene. A transgenic mini-yellow gene including the 400 bp upstream sequences and the first intron can produce the dark pigmentation in the bristles in a yellow null background (pYW, data not shown, see construct diagrams in Figure 2). Similar dark bristles are observed in flies carrying a transgene with the lambda spacer DNA inserted between the bristle enhancer and the mini-yellow gene promoter (pYW-λ, Figure 2C and construct C). When the full-length SF1 is inserted in place of the spacer DNA, it efficiently blocks the B enhancer, reducing the bristle pigmentation to that of the yellow1 mutant background (pYW-SF1, Figure 2D and construct D). Again, the enhancer-blocking activity of SF1 appears slightly stronger than that of the Su(Hw) insulator in a similar assay [pYW-Su(Hw), Figure 2E and construct E]. Thus the activity of SF1 is present in post-embryonic tissues, consistent with its potential role in regulating the homeotic gene Scr.
Fig. 2. SF1 boundary activity in adult Drosophila. (A) The notum of a Canton-S adult female. Arrows indicate the macrochete bristles on the notum cuticle, both of which exhibit dark pigmentation. (B) The notum of a yellow1 adult female. Note the yellow-colored cuticle and bristles (arrows). (C) The notum of a transgenic adult female carrying the pYW-λ in a yellow mutant background (construct diagram shown on the top-right of the figure), showing the restored pigmentation in the bristles due to the activity of the B enhancer. (D) The notum of a transgenic adult carrying the pYW-SF1 transgene. The bristles are yellow, indicating the lack of yellow expression due to the blockage of the B enhancer by SF1. (E) The notum of a transgenic adult containing the pYW-Su(Hw) transgene. Similar yellow bristles are seen, indicating the Su(Hw)-mediated block of the B enhancer.
The core insulator sequence of SF1 is highly conserved
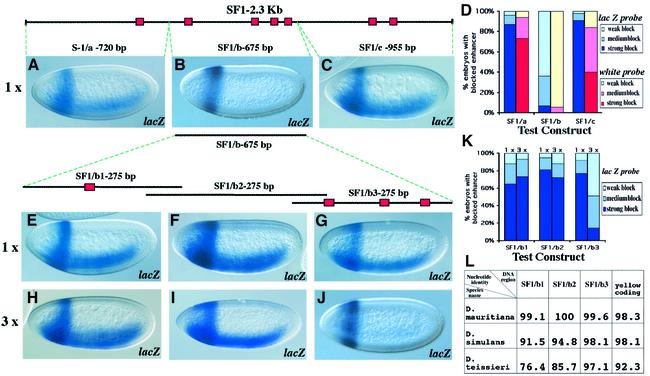
In order to understand the enhancer-blocking mechanism of SF1 and identify its protein components, we sought to define the minimal sequences required for its insulator activity. The 2.3 kb SF1 was divided into three fragments of comparable size (SF1/a–c, see Figure 3 diagram on top) which were then individually tested for enhancer-blocking activity (see Figure 3, transgene diagrams). The lacZ and white reporter expression show that the 720 bp Fragment a and the 955 bp Fragment c contain little or no enhancer-blocking activity (Figure 3A, C and D). In contrast, Fragment b strongly blocks the distal enhancers from the downstream reporter genes (Figure 3B and D). Compared with the full-length SF1, Fragment b is only slightly weaker in blocking the NEE and H1 enhancers (Figures 3D and 1L). Further truncation of Fragment b produced three sub-fragments (SF1/b1–b3) that exhibit little enhancer-blocking activity when tested as monomers between NEE and H1 (Figure 3E–G and K). However, when tested as tandem trimers, these elements showed striking differences in enhancer-blocking activity. The SF1/b1 and b2 fragments did not block distal enhancers (Figure 3H, I and K), whereas the SF1/b3 fragment exhibited substantial enhancer-blocking activity (∼40% activity of the full-length SF1, Figure 3J and K).
Fig. 3. Evolutionarily conserved core sequence of SF1 contains a cluster of GAGA sites. (A–J) Enhancer-blocking assay (only eve–lacZ reporter activity is shown) using the pNH vector was carried out (see Figure 1C) with SF1 sub-fragments inserted between the NEE and the H1 enhancers. (A–C) First round of enhancer-blocking tests using SF1/a (A), SF1/b (B) and SF1/c (C) fragments. The diagram above the embryos indicates the position and the size of the SF1/a, b and c fragments within the context of the full-length insulator (the orientation of the insulator is the same as in Figure 1A). Red boxes indicate GAGA sites. (D) Quantitative assessment of enhancer-blocking activity in the above transgenes with both eve–lacZ and the white reporter activity (see Figure 1 and methods for details). (E–G) Second round of enhancer-blocking tests using a single copy of SF1/b1 (E), SF1/b2 (F) and SF1/b3 (G) fragments. The diagram above the embryos indicates the position (orientation same as above) and the size of the SF1/b1, b2 and b3 fragments within the context of SF1/b. Note the terminal overlap between neighboring fragments. Red boxes indicate GAGA sites. (H–J) The enhancer-blocking activity of the trimerized fragments SF1/b1 (H), SF1/b2 (I) and SF1/b3 (J). (K) Quantitative assessment of the enhancer-blocking activity of the above transgenes using the eve–lacZ reporter activity (see Figure 1 and Materials and methods for details). (L) Sequence comparison of the SF1/b active fragment between four Drosophila species. Numbers represent the percent nucleotide identity between SF1/b1, SF1/b2 and SF1/b3 in the three species indicated and respective sequences in D.melanogaster. The conservation of the yellow gene coding region is given as an indication of evolutionary distance between the four fly species.
Conservation in DNA sequences is often an indication of important biological function. To test if SF1 is evolutionarily conserved, we cloned and sequenced the Fragment b homologs from three species closely related to Drosophila melanogaster, namely, D.mauritiana, D.simulans and D.teissieri (see Materials and methods). As shown in Figure 3L, the extent of sequence identity within the b1 and b2 regions reduces significantly with the increase in phylogenetic distance despite the close relationship among the species. However, the b3 fragment that contains the strongest insulator activity remains >97% conserved across all four species separated by 2–5 million years. The level of conservation is higher even than that of the coding region of the yellow gene, which is expected to be more conserved than non-coding regions in general. Indeed, the yellow sequence is more conserved than either the b1 or the b2 fragment. The enhancer-blocking activity of the SF1 sub-fragments correlated well with the level of evolutionary conservation of the DNA. The high degree of conservation of the SF1 element in the absence of any detectable accompanying enhancer activity indicates that the insulator may play an important role in gene regulation.
GAGA sites are essential for the enhancer-blocking activity of the SF1/b3 minimal insulator
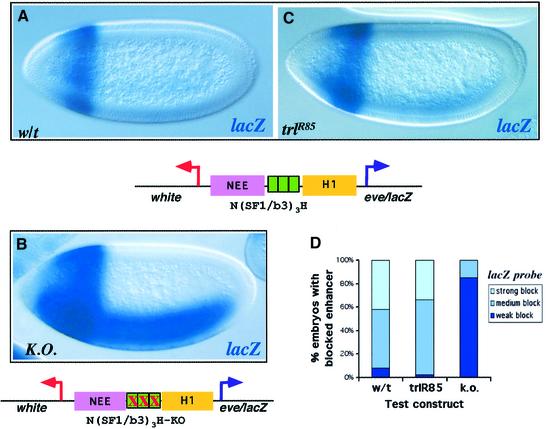
Sequence analysis of both SF1/b and SF1/b3 fragments revealed multiple binding sites for the GAGA factor, encoded by the Drosophila Trithorax-like (Trl) gene (see Figure 3 diagram on top; Farkas et al., 1994). GAGA sites are found frequently in regulatory sequences of Drosophila homeotic genes and at the heat shock loci (Poux et al., 2001; Leibovitch et al., 2002). Recently, they were also implicated in insulator/boundary function. It was reported that binding of the GAGA factor to a single GAGAG site in the Drosophila eve promoter was essential for insulator activity (Ohtsuki and Levine, 1998). GAGA sites are also required for the insulator function of the Mcp-1 and Fab-7 boundaries from the BX-C (Paul Schedl, personal communication; Busturia et al., 2001). We tested the functional significance of GAGA sites in the minimal insulator SF1/b3, using site-directed mutagenesis. Replacement of all three GAGA sites in SF1/b3 with unrelated sequences abolished its enhancer-blocking activity in the pN(SF1/b3ko)3H transgene (Figure 4A, C and D). This result indicates that the GAGA sites are essential for the enhancer-blocking activity of SF1. The presence of a common protein component in several insulators from the Drosophila homeotic complexes suggests that they may belong to a conserved family of boundary elements important in regulating homeotic genes.
Fig. 4. The enhancer-blocking activity of the core SF1 depends on the GAGA factor. (A) LacZ expression in the wild-type embryo carrying the transgene shown diagrammatically below the figure. The reporter is expressed only in the domain of H1 enhancer, indicating a strong insulator activity of the SF1/b3 trimer. (B) The pattern of lacZ expression in the embryo carrying the knock-out transgene in which all nine GAGA sites present in the SF1/b3 trimer have been changed to the unrelated sequences. The reporter is expressed in both H1- and NEE-specific domains, demonstrating the lack of enhancer-blocking activity in the mutant insulator. (C) The homozygous males carrying the N(SF1/b3)3H transgene were mated with the hetero zygous TrlR85 females. The reporter expression in the F1 embryos was visualized by the whole-mount in situ hybridization. Compared to the wild type, a larger fraction of the mutant embryos exhibit NEE-induced lacZ expression, suggesting that the enhancer-blocking activity of SF1/b3 depends on the concentration of GAGA factor. (D) Quantitative assessment of the enhancer-blocking activity of the above transgenes using the eve–lacZ reporter activity (see Figure 1 and Materials and methods for details).
We further tested if the GAGA factor is required for the insulator activity of SF1. Since the GAGA protein level in early embryos is heavily influenced by maternal contribution we examined the SF1 activity in embryos collected from the TrlR85 heterozygous females mated with wild-type males carrying the pN(Fb3)3H transgene. The SF1/b3 mediated enhancer-blocking activity showed a small but consistent decrease in the TrlR85 mutant, which may be due to the reduced GAGA protein level in these embryos. The reduction of the SF1/b3 insulator activity appears to be less pronounced than that observed for the GAGA insulator in the eve promoter (Ohtsuki and Levine, 1998). This difference may be due to the clustering of GAGA sites in SF1/b3 trimer, which could buffer the insulator from the reduced GAGA protein level by facilitating cooperative binding of the protein (van Steensel et al., 2003). Alternatively, different flanking sequences in the GAGA binding region of the two insulators and the resulting differences in their binding affinity may account for the less sensitive response to changes in the GAGA protein level. Finally, it is possible that GAGA binding proteins other than the GAGA factor are at least partly responsible for the SF1 activity.
SF1 may direct enhancer trafficking and maintain independence of the Scr and ftz gene regulation
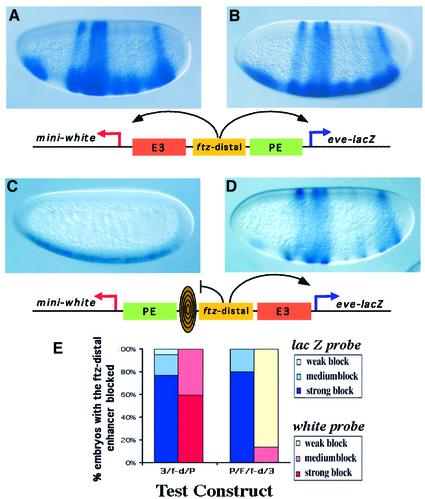
The location of SF1 raises the possibility of its role in maintaining the regulatory independence of the Scr and ftz genes. Although the promoter specificity of the ftz AE1 enhancer depends on competition from the TATA-containing ftz promoter, chromatin boundary function may be necessary to prevent Scr enhancers from interfering with the ftz expression, or to prevent other ftz enhancers from influencing Scr. We probed this possibility by testing the promoter preference of the most outlying ftz enhancer, the ftz-distal enhancer (Pick et al., 1990). The 1.2 kb ftz-distal enhancer was placed between the E3 and the PE enhancers between divergently transcribed white and lacZ reporters. In situ hybridization of transgenic embryos showed that the ftz-distal enhancer indeed strongly activates both the TATA-less white promoter and the TATA-containing eve promoter (Figure 5A, B and E). We tested the ability of SF1 to block the interaction of ftz-distal with the TATA-less white promoter by inserting the SF1/b element between them. The white expression directed by the ftz-distal enhancer is greatly attenuated (Figure 5C and E). In addition to the ftz enhancer, the distal E3 enhancer is also blocked, indicating that SF1 can simultaneously block multiple enhancers. Our findings show that different ftz enhancers exhibit distinct promoter preferences and may use alternative mechanisms to select their target promoters. The position of SF1 and its ability to prevent the ftz-distal enhancer from activating a TATA-less promoter suggest that it may be essential in maintaining independent gene regulation in the region.
Fig. 5. Evidence for the potential role of SF1 in defining the range of the ftz-distal enhancer. (A and B) The expression of the white (A) and lacZ (B) reporters was visualized by whole-mount in situ hybridization of the embryos carrying the transgene shown below the figure. In the 2–4 h old embryos the ftz-distal enhancer activates the expression of the white and lacZ reporters at comparable levels. The other two enhancers present in this construct, E3 and PE, are also active, resulting in the composite expression pattern. (C and D) The insertion of SF1/b between PE and the ftz-distal enhancer redirects the enhancer trafficking in the test transgene. In the majority of embryos the white reporter (C) is activated by PE, whereas the lacZ reporter (D) is activated by the ftz-distal enhancer and E3. (E) Quantitative assessment of the enhancer-blocking activity in the above transgenes using the white and eve–lacZ reporter activity (see Figure 1 and Materials and methods for details).
Discussion
Homeotic gene complexes emerge as an excellent model for studying genetic programming of development, and mechanisms of transcriptional regulation. The independent, yet coordinated control of multiple genes by multiple regulatory elements provides a unique opportunity to probe the diverse mechanisms governing the interplay between gene organization and gene regulation. In the Scr–ftz region in ANT-C, at least three distinct types of cis-acting elements define the promoter specificity for no less than ten different enhancers. Enhancers such as AE1 distinguish the available promoters based on the core promoter sequence and selectively interact with the TATA-containing ftz promoter (Ohtsuki et al., 1998). The Scr-distal T1 enhancer appears to depend on a newly identified ‘promoter tethering element’ located near the Scr gene for specific interaction (Calhoun et al., 2002).
Here we present evidence that a third type of regulatory DNA, the SF1 boundary/insulator, may be responsible for target promoter specification by the ftz-distal enhancer. We show that the ftz-distal enhancer does not share the same promoter preferences as AE1 and can equally activate TATA or TATA-less promoters. The intergenic position of the SF1 chromatin boundary at the junction of the ftz transcriptional unit and the neighboring Scr gene, and its ability to block the ftz-distal enhancer from a TATA-less, Scr-like promoter suggest that SF1 may be essential for maintaining independent gene regulation in the region. Consistent with this proposed role in regulating the Scr homeotic gene, the boundary activity of SF1 persists through the later stages of development. Another indication of the functional role of the SF1 insulator in the genomic interval is the conservation of the insulator DNA during evolution. While the flanking region has diverged significantly (76% identity) in D.teissieri, the core insulator sequence remains highly conserved (>97% identity) in this species.
However, it is unclear how SF1, an insulator positioned within the Scr regulatory region, is circumvented by the Scr-distal enhancers located downstream of ftz. Similar questions exist for the Mcp-1, Fab7 and Fab8 boundaries between the Abd-B promoter and the distal iab enhancers in BX-C. A specialized DNA element named promoter targeting sequence (PTS) near the Abd-B promoter may facilitate the enhancers in overcoming the intervening Fab boundaries (Zhou and Levine, 1999). An alternative mechanism is based on the recent finding that the Su(Hw) enhancer-blocking activity is abolished by the tandem arrangement of insulators (Cai and Shen, 2001; Muravyova et al., 2001). SF1 or other specialized DNA elements such as the Scr tethering element may interact with similar elements positioned downstream of ftz, thereby ‘looping out’ the intervening ftz domain and facilitating the Scr enhancer–promoter interactions.
Chromatin boundary function has been shown to be important for gene regulation in the Hox clusters from fly to mouse. However, the protein components involved in the Hox boundary activity, as well as the mechanism of the boundary function are unknown. We have identified multiple GAGA binding sites that are essential for the enhancer-blocking activity of the SF1 core insulator. We have also shown that the Drosophila GAGA factor may be involved in the SF1 boundary function. Similar findings that GAGA sites are critical for the function of Mcp1 and Fab7 boundary elements from the BX-C have been reported recently (Paul Schedl, personal communication; Busturia et al., 2001). These observations suggest that the chromatin insulators from the ANT-C and the BX-C may share common components and mechanism, and belong to a family of conserved boundary elements that regulate enhancer–promoter interactions in the Hox complexes.
It is interesting that the GAGA factor is implicated in the boundary activity in the Drosophila Hox clusters. The GAGA factor has been known to regulate transcription by recruiting chromatin remodeling and transcription initiation complexes (Mishra et al., 2001; Leibovitch et al., 2002). However, its role in boundary/insulator activity may not be attributed to its ability to activate transcription but rather to the ability of this protein to forge links among distant DNA elements through its BTB domain (Ohtsuki and Levine, 1998; Mahmoudi et al., 2002). This property of the GAGA factor is consistent with the looping models proposed for the insulator/boundary mechanism (Cai and Shen, 2001).
The existence of an independent ftz transcription domain flanked by boundary elements is also consistent with the observed mobility of ftz during evolution. ftz is an ‘accessory’ gene unique to the invertebrate homeotic complex. Although it has been found in all major arthropod groups, the protein sequence and function of ftz have diverged from the neighboring homeotic genes (Akam et al., 1994; Telford, 2000; Lohr et al., 2001). Nonetheless, the internal organization of the ftz transcription unit including regulatory sequences is highly conserved, possibly due to its important role in segmentation and neural development (Maier et al., 1993; Dawes et al., 1994; Ferrier and Akam, 1996; Mouchel-Vielh et al., 1998). The shift in ftz function appears to coincide with an increased mobility of the transcription unit as a whole, as the 16 kb genomic region is found inverted in certain Drosophila subgenera or missing entirely from the complex in certain insect species (Maier et al., 1990). The presence of the SF1 boundary element at the junction of such an evolutionary mobile unit is consistent with its role in maintaining gene independence during evolution.
Materials and methods
P-element transformation, whole-mount in situ hybridization and visual assessment of reporter gene expression
The y1w67c23 and w1118 Drosophila strains were used to generate all transgenic lines reported. P-element-mediated germ line transformation was carried out as described previously (Rubin and Spradling, 1982). Three or more independent transgenic lines were obtained and characterized for each test construct. Transgenic embryos were collected and fixed as described previously (Cai et al., 2001). Reporter gene expression in blastoderm stage embryos was detected using whole-mount in situ hybridization with the digoxigenin–UTP labeled antisense RNA probes. Expression patterns were visualized by colorimetric reaction following incubation with anti-digoxigenin antibody conjugated to alkaline phosphatase (Genius Kit, Boehringer; Tautz and Pfeifle, 1989; Cai et al., 2001). All in situ stains were carried out under the same conditions and using the same amount of reporter probes. Thirty to 200 blastoderm transgenic embryos from multiple lines were visually inspected. To ensure objectivity, the label of each slide was covered and scored in a double blind fashion with a large group of slides that contained samples from the control transgenes. The extent of enhancer block was judged by the expression level directed by the distal enhancers compared with that of the proximal enhancers. In most cases, the H1-directed staining was used as a reference for the NEE-directed expression. Both numbers of stained cells and intensity of stain were considered during the visual inspection. Each embryo was assigned to one of three groups: weak (<30% block: NEE/H1<70%), moderate (30–70% block: NEE/H1<30–70%), and strong (>70% block: NEE/H1<30%). All quantification procedures were repeated by at least two different authors and the average was used in the report. The most frequently observed staining patterns were used to produce the image in the figures.
Construction of transgenes and test of enhancer-blocking activity in mutant strains
All P-element constructs used in the embryo enhancer-blocking assays were derivatives of pCaSPeR. The lacZ coding region was fused in frame with the eve promoter (–42 to +200) and the eve–lacZ reporter was inserted into pCaSPeR generating pEb vector (Small et al., 1992; Cai and Levine, 1997). Construction of the NLH, PL3, NSH and PS3 was as described previously (Cai and Levine, 1997). The 2.3 kb SF1 DNA was sub-cloned from a λ phage genomic clone that hybridized to probes from the Scr region (H.N.Cai, unpublished data). Sequences of the PCR primers used to subclone the full-length SF1 element and sub-fragments of SF1 are as follows (all primers contain a NotI site at the end): SF1-1: 5′-ATTGCGGCCGCGAATTCGGTTTTCGAAGCC-3′, SF1-2: 5′-ATTG CGGCCGCAACTATGGTAGCGCAGAGC-3′, SF1-3: 5′-ATTGCGGC CGCAGTGTTGCTGTAAGGACCG-3′, SF1-4: 5′-ATTGCGGCCGCA TTCTGAGCAGCGGAGTCG-3′, SF1-5: 5′-ATTGCGGCCGCTCCGC TGCTCAGAATTAGG-3′, SF1-6: 5′-ATTGCGGCCGCGGATTCCCC ATCCTATACC-3′. The sub-fragments of SF1 were generated by PCR and cloned into pCRII/TOPO vector (Invitrogen). These sequences were subsequently inserted into the NotI site between the NEE and H1 enhancers in pEbNH, PE and E3 enhancers in pEbP3 vectors (Cai and Levine, 1997). SF1-3 and SF1-4 primers were used to clone by PCR the SF1/b related sequences from D.mauritiana, D.simulans and D.teissieii with an annealing temperature of 53°C. The cloned fragment was sequenced by the MGIF sequencing facility at the University of Georgia and analyzed with conventional DNA analysis software. The DDBJ/EMBL/GenBank accession numbers for the SF1/b related sequences in D.mauritiana, D.simulans and D.teissieii are AY256571, AY256573 and AY256572, respectively. Site-directed mutagenesis of the GAGA sites in the SF1/b3 element was performed using the single-stranded DNA method as described previously (Ip et al., 1992). The base substitution of the three GAGA sites in the SF1/b3 element was done using the following oligonucleotides: 5′-GCTGAAAACAAGCTTCATTGACATT-3′, 5′-GT TTCAAGGCATCGATTGTTTTGTG-3′ and 5′-ATTTCACTGGCTGC AGTTGCACATGT-3′. The 1.2 kb ftz-distal enhancer was provided by L.Pick (Pick et al., 1990). The mini-yellow gene in pYW constructs was made using the yellow genomic region (from –400 to 400 bp downstream of the polyA site) provided by J.Zhou (personal communication). An 1181 bp ClaI fragment in the first intron was deleted and the EcoRV site at +778 was converted to a NotI site. The test DNA sequences, such as the λ-spacer, SF1 or Su(Hw) were inserted into the NotI site. The position and orientation of enhancers and insulators were determined by restriction digestions, PCR analyses using P-element specific primers, and in some cases by DNA sequencing. To test the enhancer-blocking activity of SF1 in the GAGA mutant background the homozygous males carrying the test transgene were mated with the heterozygous TrlR85 females. The reporter expression in the embryos was determined by in situ hybridization.
Acknowledgments
Acknowledgements
We thank Dimple Bosu and Alyssa Ingmundson for technical assistance, Susan Schweinsberg, Paul Schedl, Krishna Bhat, John McDonald and Wyatt Anderson for fly stocks, Mike Levine and Paul Schedl for sharing unpublished results. This work is supported by the NIH.
References
- Akam M., Averof,M., Castelli-Gair,J., Dawes,R., Falciani,F. and Ferrier,D. (1994) The evolving role of Hox genes in arthropods. Dev. Suppl., 209–215. [PubMed] [Google Scholar]
- Busturia A., Lloyd,A., Bejarano,F., Zavortink,M., Xin,H. and Sakonju,S. (2001) The MCP silencer of the Drosophila Abd-B gene requires both Pleiohomeotic and GAGA factor for the maintenance of repression. Development, 128, 2163–2173. [DOI] [PubMed] [Google Scholar]
- Cai H.N. and Levine,M. (1997) The gypsy insulator can function as a promoter-specific silencer in the Drosophila embryo. EMBO J., 16, 1732–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H.N. and Shen,P. (2001) Effects of cis arrangement of chromatin insulators on enhancer-blocking activity. Science, 291, 493–495. [DOI] [PubMed] [Google Scholar]
- Cai H.N., Zhang,Z., Adams,J.R. and Shen,P. (2001) Genomic context modulates insulator activity through promoter competition. Development, 128, 4339–4347. [DOI] [PubMed] [Google Scholar]
- Calhoun V.C., Stathopoulos,A. and Levine,M. (2002) Promoter-proximal tethering elements regulate enhancer–promoter specificity in the Drosophila Antennapedia complex. Proc. Natl Acad. Sci. USA, 99, 9243–9247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawes R., Dawson,I., Falciani,F., Tear,G. and Akam,M. (1994) Dax, a locust Hox gene related to fushi-tarazu but showing no pair-rule expression. Development, 120, 1561–1572. [DOI] [PubMed] [Google Scholar]
- Farkas G., Gausz,J., Galloni,M., Reuter,G., Gyurkovics,H. and Karch,F. (1994) The Trithorax-like gene encodes the Drosophila GAGA factor. Nature, 371, 806–808. [DOI] [PubMed] [Google Scholar]
- Ferrier D.E. and Akam,M. (1996) Organization of the Hox gene cluster in the grasshopper, Schistocerca gregaria. Proc. Natl Acad. Sci. USA, 93, 13024–13029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley K.P. and Engel,J.D. (1992) Individual stage selector element mutations lead to reciprocal changes in beta- vs epsilon-globin gene transcription: genetic confirmation of promoter competition during globin gene switching. Genes Dev., 6, 730–744. [DOI] [PubMed] [Google Scholar]
- Galloni M., Gyurkovics,H., Schedl,P. and Karch,F. (1993) The bluetail transposon: evidence for independent cis-regulatory domains and domain boundaries in the bithorax complex. EMBO J., 12, 1087–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimova T.I. and Corces,V.G. (2001) Chromatin insulators and boundaries: effects on transcription and nuclear organization. Annu. Rev. Genet., 35, 193–208. [DOI] [PubMed] [Google Scholar]
- Geyer P.K. and Corces,V.G. (1987) Separate regulatory elements are responsible for the complex pattern of tissue-specific and developmental transcription of the yellow locus in Drosophila melanogaster. Genes Dev., 1, 996–1004. [DOI] [PubMed] [Google Scholar]
- Gyurkovics H., Gausz,J., Kummer,J. and Karch,F. (1990) A new homeotic mutation in the Drosophila bithorax complex removes a boundary separating two domains of regulation. EMBO J., 9, 2579–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom K., Muller,M. and Schedl,P. (1996) Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev., 10, 3202–3215. [DOI] [PubMed] [Google Scholar]
- Harding K., Wedeen,C., McGinnis,W. and Levine,M. (1985) Spatially regulated expression of homeotic genes in Drosophila. Science, 229, 1236–1242. [DOI] [PubMed] [Google Scholar]
- Ip Y.T., Park,R.E., Kosman,D., Bier,E. and Levine,M. (1992) The dorsal gradient morphogen regulates stripes of rhomboid expression in the presumptive neuroectoderm of the Drosophila embryo. Genes Dev., 6, 1728–1739. [DOI] [PubMed] [Google Scholar]
- Kmita M., Kondo,T. and Duboule,D. (2000) Targeted inversion of a polar silencer within the HoxD complex re-allocates domains of enhancer sharing. Nat. Genet., 26, 451–454. [DOI] [PubMed] [Google Scholar]
- Leibovitch B.A., Lu,Q., Benjamin,L.R., Liu,Y., Gilmour,D.S. and Elgin,S.C. (2002) GAGA factor and the TFIID complex collaborate in generating an open chromatin structure at the Drosophila melanogaster hsp26 promoter. Mol. Cell. Biol., 22, 6148–6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis E.B. (1978) A gene complex controlling segmentation in Drosophila. Nature, 276, 565–570. [DOI] [PubMed] [Google Scholar]
- Lohr U., Yussa,M. and Pick,L. (2001) Drosophila fushi tarazu: a gene on the border of homeotic function. Curr. Biol., 11, 1403–1412. [DOI] [PubMed] [Google Scholar]
- Mahmoudi T., Katsani,K.R. and Verrijzer,C.P. (2002) GAGA can mediate enhancer function in trans by linking two separate DNA molecules. EMBO J., 21, 1775–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier D., Preiss,A. and Powell,J.R. (1990) Regulation of the segmentation gene fushi tarazu has been functionally conserved in Drosophila. EMBO J., 9, 3957–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier D., Sperlich,D. and Powell,J.R. (1993) Conservation and change of the developmentally crucial fushi tarazu gene in Drosophila. J. Mol. Evol., 36, 315–326. [DOI] [PubMed] [Google Scholar]
- Merli C., Bergstrom,D.E., Cygan,J.A. and Blackman,R.K. (1996) Promoter specificity mediates the independent regulation of neighboring genes. Genes Dev., 10, 1260–1270. [DOI] [PubMed] [Google Scholar]
- Mihaly J. et al. (1998) Chromatin domain boundaries in the Bithorax complex. Cell. Mol. Life Sci., 54, 60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra R.K., Mihaly,J., Barges,S., Spierer,A., Karch,F., Hagstrom,K., Schweinsberg,S.E. and Schedl,P. (2001) The iab-7 polycomb response element maps to a nucleosome-free region of chromatin and requires both GAGA and pleiohomeotic for silencing activity. Mol. Cell. Biol., 21, 1311–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchel-Vielh E., Rigolot,C., Gibert,J.M. and Deutsch,J.S. (1998) Molecules and the body plan: the Hox genes of Cirripedes (Crustacea). Mol. Phylogenet. Evol., 9, 382–389. [DOI] [PubMed] [Google Scholar]
- Muravyova E., Golovnin,A., Gracheva,E., Parshikov,A., Belenkaya,T., Pirrotta,V. and Georgiev,P. (2001) Loss of insulator activity by paired Su(Hw) chromatin insulators. Science, 291, 495–498. [DOI] [PubMed] [Google Scholar]
- Ohtsuki S. and Levine,M. (1998) GAGA mediates the enhancer blocking activity of the eve promoter in the Drosophila embryo. Genes Dev., 12, 3325–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuki S., Levine,M. and Cai,H.N. (1998) Different core promoters possess distinct regulatory activities in the Drosophila embryo. Genes Dev., 12, 547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pick L., Schier,A., Affolter,M., Schmidt-Glenewinkel,T. and Gehring,W.J. (1990) Analysis of the ftz upstream element: germ layer-specific enhancers are independently autoregulated. Genes Dev., 4, 1224–1239. [DOI] [PubMed] [Google Scholar]
- Poux S., Melfi,R. and Pirrotta,V. (2001) Establishment of Polycomb silencing requires a transient interaction between PC and ESC. Genes Dev., 15, 2509–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G.M. and Spradling,A.C. (1982) Genetic transformation of Drosophila with transposable element vectors. Science, 218, 348–353. [DOI] [PubMed] [Google Scholar]
- Small S., Blair,A. and Levine,M. (1992) Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J., 11, 4047–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D. and Pfeifle,C. (1989) A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma, 98, 81–85. [DOI] [PubMed] [Google Scholar]
- Telford M.J. (2000) Evidence for the derivation of the Drosophila fushi tarazu gene from a Hox gene orthologous to lophotrochozoan Lox5. Curr. Biol., 10, 349–352. [DOI] [PubMed] [Google Scholar]
- van Steensel B., Delrow,J. and Bussemaker,H.J. (2003) Genomewide analysis of Drosophila GAGA factor target genes reveals context-dependent DNA binding. Proc. Natl Acad. Sci. USA, 100, 2580–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West A.G., Gaszner,M. and Felsenfeld,G. (2002) Insulators: many functions, many mechanisms. Genes Dev., 16, 271–288. [DOI] [PubMed] [Google Scholar]
- Zhou J. and Levine,M. (1999) A novel cis-regulatory element, the PTS, mediates an anti-insulator activity in the Drosophila embryo. Cell, 99, 567–575. [DOI] [PubMed] [Google Scholar]
- Zhou J., Barolo,S., Szymanski,P. and Levine,M. (1996) The Fab-7 element of the bithorax complex attenuates enhancer-promoter interactions in the Drosophila embryo. Genes Dev., 10, 3195–3201. [DOI] [PubMed] [Google Scholar]
- Zhou J., Ashe,H., Burks,C. and Levine,M. (1999) Characterization of the transvection mediating region of the abdominal-B locus in Drosophila. Development, 126, 3057–3065. [DOI] [PubMed] [Google Scholar]