Abstract
The TOR protein is a phosphoinositide kinase-related kinase widely conserved among eukaryotes. Fission yeast tor1 encodes an ortholog of TOR, which is required for sexual development and growth under stressed conditions. We isolated gad8, which encodes a Ser/Thr kinase of the AGC family, as a high-copy suppressor of the sterility of a tor1 mutant. Disruption of gad8 caused phenotypes similar to those of tor1 disruption. Gad8p was less phosphorylated and its kinase activity was undetectable in tor1Δ cells. Three amino acid residues corresponding to conserved phosphorylation sites in the AGC family kinases, namely Thr387 in the activation loop, Ser527 in the turn motif and Ser546 in the hydrophobic motif, were important for the kinase activity of Gad8p. Tor1p was responsible for the phosphorylation of Ser527 and Ser546, whereas Ksg1p, a PDK1-like kinase, appeared to phosphorylate Thr387 directly. Altogether, Tor1p, Ksg1p and Gad8p appear to constitute a signaling module for sexual development and growth under stressed conditions in fission yeast, which resembles the mTOR–PDK1–S6K1 system in mammals and may represent a basic signaling module ubiquitous in eukaryotes.
Keywords: AGC family kinase/PDK1-like kinase/sexual development/stress response/TOR kinase
Introduction
Organisms employ various signal transduction pathways to respond to environmental changes as well as to factors for cell-to-cell communication. The TOR (target of rapamycin) protein is a widely conserved phosphoinositide kinase-related kinase, which is thought to participate in the signal transduction of nutritional availability. TOR proteins have been found in fungi, worms, flies, plants and mammals (Schmelzle and Hall, 2000; Gingras et al., 2001). The original members of the TOR gene family were Saccharomyces cerevisiae TOR1 and TOR2, which were identified in a screen for resistant mutants against rapamycin, an immunosuppressant drug causing growth arrest in the budding yeast (Heitman et al., 1991; Kunz et al., 1993). It is well characterized that TOR has a function relevant to regulation of translation. However, previous reports indicate that the TOR signaling is complex and may potentially also be involved in many biological processes (Schmelzle and Hall, 2000; Gingras et al., 2001). In S.cerevisiae, TOR1 and TOR2 have been shown to participate in the regulation of G1 progression, transcription, cytoskeletal organization, sporulation and autophagy (Schmelzle and Hall, 2000; Gingras et al., 2001).
PDK1 (phosphoinositide-dependent kinase-1) is another conserved Ser/Thr protein kinase in eukaryotes. PDK1 regulates signal transduction by activating members of the AGC family kinases [protein kinases (PK) A, G and C], which include isoforms of Akt/PKB, serum and glucocorticoid induced kinase (SGK), p90 ribosomal S6 kinase (RSK) and p70 ribosomal S6 kinase 1 (S6K1) (Toker and Newton, 2000; Alessi, 2001). PDK1 activates these kinases through direct phophorylation of the activation loop located in their catalytic domain. AGC family kinases also carry conserved phosphorylation sites that are C-terminal to the catalytic domain. One site is positioned in a region called the hydrophobic motif, which exists in most AGC family kinases except PKA, and phosphorylation of this site is essential for full activation of the kinase. Some members of the AGC kinase family, including PKC, S6K1, RSK and PKA, carry another phosphorylation site in a region called the turn motif, and its phosphorylation is important for kinase activity. However, in some cases, including PKB and SGK1, phosphorylation of the turn motif does not appear to affect the activity (Newton, 2001). Although it is widely accepted that PDK1 is responsible for the phosphorylation of the activation loop of the AGC family kinases, a variety of protein kinases appear to be involved in phosphorylation of the hydrophobic motif and the turn motif, and the significance of the phosphorylation of the turn motif requires further investigation. Interestingly, mTOR (mammalian ortholog of TOR) phosphorylates both the hydrophobic and turn motifs of mammalian S6K1, a downstream target of the mTOR signaling pathway (Burnett et al., 1998; Isotani et al., 1999; Saitoh et al., 2002).
Fission yeast arrests the mitotic cell cycle in G1 phase and enters sexual development upon nutrient starvation. Previous studies have shown that conserved signal transduction pathways participate in the onset of sexual development, such as the cAMP–PKA pathway, the Ras–MAPK cascade and the SAPK cascade (Yamamoto, 1996). Recently, Tor1p, which is an ortholog of TOR, and Ksg1p, which is a PDK1-like kinase, have been reported as positive regulators of sexual development in fission yeast (Niederberger and Schweingruber, 1999; Kawai et al., 2001; Weisman and Choder, 2001). However, no possible AGC family kinase potentially cooperating with Tor1p or Ksg1p has been identified.
To search for a Tor1p target, we screened for high-copy suppressors of the sterility of the tor1-g2 mutant, which carried a nonsense tor1 allele and was defective in G1 arrest under nitrogen starvation. We consequently identified a novel gene gad8, which encodes a Ser/Thr kinase of the AGC family. In this report we clarify the relationship of Gad8p with Tor1p and Ksg1p, and propose that these three protein kinases constitute a module for signaling, which may be conserved ubiquitously among eukaryotes.
Results
Identification of tor1-g2, a nonsense allele of tor1
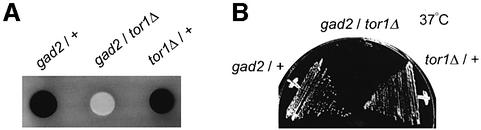
According to the previously described screening protocol (Kanoh et al., 1996), we isolated a fission yeast mutant, tentatively named gad2-1, that was defective in G1 arrest under nitrogen starvation and was sterile. This mutant was temperature sensitive for growth and grew slowly with an elongated cell shape even at the permissive temperature 30°C (data not shown). It has been reported that tor1 is required for sexual development and growth under stressed conditions, such as high osmolarity or high temperature (Kawai et al., 2001; Weisman and Choder, 2001). We noted that the gad2-1 mutant was phenotypically similar to the tor1Δ mutant, although the phenotype of gad2-1 was somewhat weaker. To examine the allelism, we produced a diploid strain by crossing a tor1Δ haploid with a gad2-1 haploid, as described in Materials and methods. This strain was sporulation-defective and exhibited temperature-sensitive growth (Figure 1), suggesting that gad2-1 might be allelic to tor1. We then analyzed the tor1 gene isolated from the gad2-1 mutant. The wild-type tor1 gene encodes a protein of 2335 amino acids (Kawai et al., 2001; Weisman and Choder, 2001). We found that the mutant gene carried a stop codon (TGA) in place of the arginine codon (CGA) at 1071. Thus, we concluded that gad2-1 is a new allele of tor1, which we call tor1-g2 hereafter.
Fig. 1. The gad2-1 (tor1-g2) mutation does not complement the tor1 disruption. (A) Sporulation assay. Diploid strain JW958 (denoted as gad2/tor1Δ), constructed by crossing a gad2-1 haploid and a tor1Δ haploid, was incubated on MEA medium together with control diploids heterozygous for either gad2-1 (JW956; denoted as gad2/+) or tor1Δ (JW957; denoted as tor1Δ/+). After four days, patches were exposed to iodine vapor, which stains sporulated cells dark brown. (B) Complementation for the growth at high temperature. The same diploid strains as in (A) were streaked on the YE medium and incubated for three days at 37°C.
The nonsense mutation found in tor1-g2, which was located upstream of the kinase domain, did not appear to be very tight, because the mutant phenotype was considerably weaker than the tor1 deletion. For instance, the tor1-g2 mutant could grow, though slowly, on medium containing 0.4 M KCl, whereas the tor1Δ mutant could not. We have evidence suggesting that the tor1-g2 mutation allows production of a small amount of the read-through gene product (data not shown).
A high-copy suppressor of tor1-g2 encodes a novel Ser/Thr kinase Gad8p
We set out to identify possible downstream targets of Tor1p by screening for high-copy suppressors of the sterility of the tor1-g2 mutant. We transformed homothallic haploid tor1-g2 cells with fission yeast genomic and cDNA libraries, and picked up transformants that could mate and sporulate on SSA medium. Among 370 000 transformants examined, thirteen were judged to be positive. Sequence analysis indicated that they were attributed to five genes, namely rst2, which encodes a transcription factor crucial for sexual development (Kunitomo et al., 2000; Higuchi et al., 2002), N-terminally truncated byr2/ste8, which encodes a MAPKKK in the pheromone-signaling pathway (Wang et al., 1991) and three novel genes. In this study we characterized one of these novel genes, which we named gad8, and is annotated as SPCC24B10.07 in the fission yeast genomic database (Wood et al., 2002).
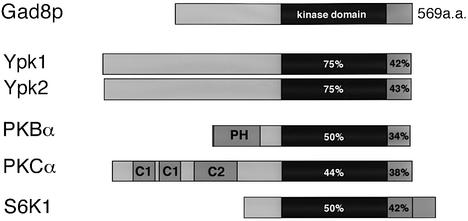
The deduced gad8 gene product was a Ser/Thr kinase. Gad8p showed high similarity to AGC family kinases in the catalytic domain (Figure 2), especially to budding yeast Ypk1 and Ypk2/Ykr2 (Maurer, 1988; Chen et al., 1993) (75% amino acid identity in each case). Gad8p also showed similarity to AGC family kinases in the C-terminal region (Figure 2).
Fig. 2. The deduced gad8 gene product, Gad8p, is a Ser/Thr kinase of the AGC family. Gad8p is aligned with some members of the AGC family, including budding yeast Ypk1 and Ypk2/Ykr2 and human PKBα, PKCα and S6K1. The kinase domain is represented by a black box and the C-terminal region conserved in AGC family kinases, by a shaded box. The percentage values indicates the amino acid identity of the respective region compared with the corresponding region in Gad8p. PH, pleckstrin homology domain; C1, diacylglycerol/phorbol ester binding domain; C2, Ca2+/TPA binding domain.
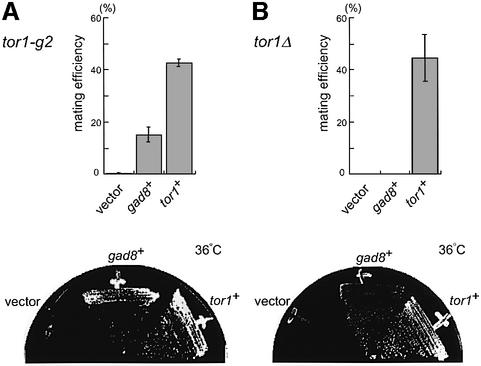
Overexpression of gad8 suppressed both sterility and temperature-sensitive growth of the tor1-g2 mutant (Figure 3A, top and bottom). However, overexpression of gad8 failed to suppress the sterility of the tor1Δ mutant (Figure 3A, top), although it could suppress the temperature sensitivity and high-osmolarity sensitivity of this strain (Figure 3B, bottom; data not shown). This may indicate that a higher activity of Tor1p and/or Gad8p is required for sexual development than for growth under stressed conditions.
Fig. 3. Suppression of tor1-g2 and tor1Δ by overexpression of gad8. (A) Suppression of the tor1-g2 mutant. A homothallic haploid tor1-g2 strain (JW948) was transformed with either pREP41-gad8, or pREP41-tor1 or the vector pREP41. Transformants were grown on SSA medium at 30°C for four days. The mating efficiency of each transformant was scored under the microscope (top panel). Each transformant was also incubated on MM medium at 36°C for four days (bottom panel). (B) Suppression of the tor1Δ strain. A homothallic haploid tor1Δ strain (JW951) was examined as in (A), except that the incubation time on MM medium was three days.
Gad8p and Tor1p are likely to be on the same regulatory pathway for sexual development and growth
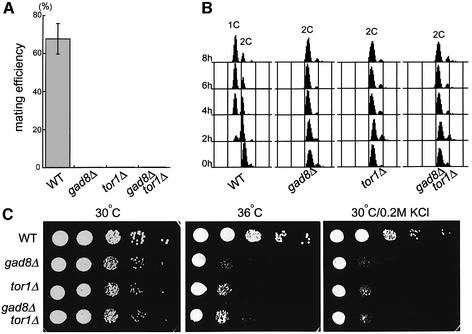
We disrupted the gad8 gene, as described in Materials and methods. Disruption of gad8 was not lethal. However, the haploid gad8 disruptant failed to mate on sporulation medium (Figure 4A), and apparently could not arrest the cell cycle at G1 under nitrogen starvation (Figure 4B). Furthermore, gad8Δ cells could not grow under stressed conditions, such as at 37°C or in the presence of 1 M KCl or 1.2 M sorbitol (data not shown; see below). As these gad8Δ phenotypes appeared very similar to those of tor1Δ, we constructed a gad8Δ tor1Δ double disruptant and compared it with the parental strains.
Fig. 4. Phenotypic similarities between gad8Δ and tor1Δ. (A) A comparison of the mating efficiencies of the wild-type (JY476), gad8Δ (JW945), tor1Δ (JW951) and gad8Δ tor1Δ (JW959) strains. Cells were incubated on SSA medium for four days and the mating efficiency was scored under the microscope. (B) Flow-cytometric analysis of the four strains subjected to nitrogen starvation. Cells were grown to logarithmic phase in MM liquid medium at 30°C, harvested, washed once and transferred to MM without a nitrogen source. Their DNA content was measured by FACS analysis at 2 h intervals. (C) Temperature-sensitivity and high-osmolarity sensitivity of each strain. Cells of each strain, grown to logarithmic phase at 30°C, were spotted on YE medium with sequential ten-fold dilution and incubated at either 30°C (left panel) or 36°C (middle panel) for three days. Cells were also spotted similarly on YE medium containing 0.2 M KCl and incubated at 30°C for three days (right panel).
All of the three strains, namely gad8Δ, tor1Δ and gad8Δ tor1Δ, were completely mating-defective (Figure 4A), and behaved almost identically in the failure of G1 arrest under nitrogen starvation (Figure 4B), in defective growth at 36°C (Figure 4C, middle), and in defective growth in the presence of 0.2 M KCl (Figure 4C, right). In addition, all of them grew slowly showing elongated cell shape. The doubling time in YE liquid medium was 2.8 ± 0.1 h for gad8Δ, 2.7 ± 0.1 h for tor1Δ and 2.8 ± 0.1 h for gad8Δ tor1Δ, while that of the wild type was 2.3 ± 0.1 h. The average length of septated cells was 17.1 ± 1.8 µm for gad8Δ, 17.2 ± 1.3 µm for tor1Δ and 17.3 ± 1.7 µm for gad8Δ tor1Δ, while that of the wild type was 14.2 ± 1.3 µm. These data strongly suggested that Tor1p and Gad8p should function in the same pathway. Because overexpression of gad8 could suppress the stress sensitivity of the tor1Δ strain (Figure 3) but overexpression of tor1 failed to suppress gad8Δ (data not shown), Gad8p was believed to be a downstream factor of Tor1p.
Tor1p affects the kinase activity and the phosphorylation state of Gad8p
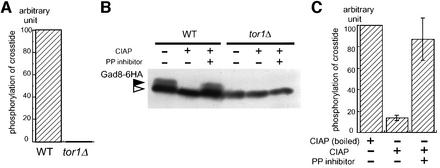
To measure the kinase activity of Gad8p, we immunoprecipitated 6HA (six times tandem-repeated hemagglutinin)-tagged Gad8p from the extracts of tor1+ and tor1Δ cells. We performed in vitro phosphorylation assay using ‘crosstide’ as a substrate, which is a peptide known to be phosphorylated by AGC family kinases such as PKB (Cross et al., 1995). Gad8-6HAp recovered from the tor1Δ background was inactive in phosphorylating crosstide, whereas Gad8-6HAp from tor1+ was fairly active (Figure 5A). We also found that the mobility of Gad8-6HAp varied depending on the genetic background of the host from which it was derived. Gad8-6HAp from the wild type showed a small fraction that migrated slower than the majority, but this fraction was missing in the preparation from the tor1Δ strain (Figure 5B). Treatment of Gad8-6HAp from the wild type with calf intestine alkaline phosphatase (CIAP) diminished the slow-migrating fraction, indicating that this fraction was generated by phosphorylation (Figure 5B). Moreover, CIAP treatment of Gad8-6HAp from the wild type abolished its ability to phosphorylate crosstide, indicating that phosphorylation is crucial for the kinase activity of Gad8p (Figure 5C). These data reinforce the suggestion that Tor1p regulates the kinase activity of Gad8p via phosphorylation.
Fig. 5. Tor1p is essential for the kinase activity of Gad8p and affects its phosphorylation state. (A) Assay of the kinase activity of Gad8p recovered from either the wild-type (JW960) or the tor1Δ (JW965) background. Gad8-6HAp expressed in each strain was immunopurified from the cell extract and assayed for its ability to phosphorylate crosstide, as described in the Materials and methods. (B) Detection of Gad8-6HAp in wild-type and tor1Δ cells. Extracts of JW960 and JW965, expressing Gad8-6HAp, were separated by SDS–PAGE, and the protein was detected by western blotting using anti-HA antibody. Extracts were also treated with CIAP, with or without the addition of the phosphatase inhibitor mixture, to examine possible phosphorylation of Gad8p. The white arrowhead indicates the major band of Gad8p, and the black arrowhead indicates the minor band. (C) Effects of phosphatase treatment on the kinase activity of Gad8p. Gad8-6HAp immunopurified as in (A) was treated with CIAP, either native or boiled, in the presence or absence of the phosphatase inhibitor mixture. The kinase assay was performed as in (A).
Three phosphorylation sites conserved among AGC family kinases are important for the function of Gad8p
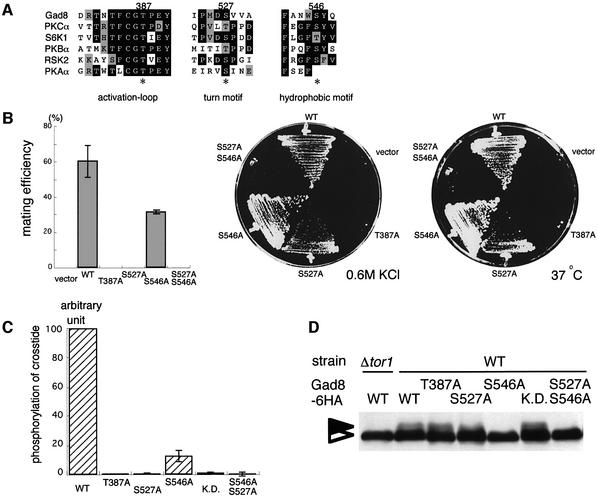
Members of the AGC kinase family undergo phosphorylation at conserved sites, which is crucial for their activity (Figure 6A). One phosphorylation site is located in the activation loop, another in the turn motif and a third in the hydrophobic motif. Gad8p apparently carried these three sites, namely Thr387 in the activation loop, Ser527 in the turn motif and Ser546 in the hydrophobic motif. To examine whether phosphorylation of these sites was necessary for Gad8p function, we mutated each of them to alanine. The three types of mutant gad8 alleles carried on the pR3C vector were transformed into gad8Δ cells, and the transformants were examined for their phenotypes (Figure 6B). The gad8-T387A transformant was sterile and did not grow under stressed conditions, like the gad8Δ host strain transformed with the vector. The gad8-S527A transformant was also sterile, but could grow weakly under stressed conditions. The gad8-S546A transformant was moderately fertile and showed no obvious growth defect at high temperature or under high osmolarity. These results suggested that the gad8-S527A and gad8-S546A alleles were partially active, the former being less active than the latter, whereas the gad8-T387A allele was completely inactive. We then constructed a gad8 allele carrying both S527A and S546A mutations. This allele, denoted gad8-S527A/S546A, was clearly less active than gad8-S527A and did not detectably complement the gad8Δ strain (Figure 6B).
Fig. 6. Effects of mutations in the conserved phosphorylation sites on Gad8p. (A) Alignment of the sequences around the three possible phosphorylation sites on Gad8p, in the activation loop, in the turn motif and in the hydrophobic motif, with those of human PKCα, S6K1, PKBα, PKAα and mouse RSK2. The predicted phosphorylation sites on Gad8p, namely Thr387, Ser527 and Ser546, are indicated by asterisks. (B) Unphosphorylatable forms of Gad8p are not functional. A homothallic haploid gad8Δ strain (JW945) was transformed with pR3C-gad8, pR3C-gad8-T387A, pR3C-gad8-S527A, pR3C-gad8-S546A, pR3C-gad8-S527A S546A or the vector pREP41. Transformants were examined for mating efficiency (left panel), growth at high osmolarity (middle panel) and growth at high temperature (right panel). Their mating efficiency was scored after incubation on SSA medium at 30°C for three days. Growth was examined on MM medium containing 0.6 M KCl at 30°C for six days, or on MM medium at 37°C for six days. (C) The kinase activity of the unphosphorylatable forms of Gad8p. Gad8-6HAp immunopurified from each strain, namely the gad8-T387A (JW962), gad8-S527A (JV107), gad8-S546A (JW963) and gad8-S527A S546A (JV108) mutants was subjected to the in vitro kinase assay, together with the control wild-type (JW960) and the gad8-K259R mutant (JW961) supposed to generate a kinase-dead product. (D) Detection of mutant forms of Gad8-6HAp in SDS–PAGE followed by western blot analysis.
We assayed the kinase activities of the mutant forms of Gad8p in vitro. Gad8-T387Ap, Gad8-S527Ap and Gad8-S527A/S546Ap hardly phosphorylated crosstide, like the kinase-dead form Gad8-K259Rp used as a negative control (Figure 6C). In contrast, Gad8-S546Ap revealed a modest but significant kinase activity, which was about one-seventh of the wild type. These results indicate that the observed phenotypes of the mutant gad8 alleles correlate with the kinase activity of their gene products, and that the phosphorylation sites in the activation loop, in the turn motif and in the hydrophobic motif of Gad8p are important for its full activity.
Phosphorylation of Ser527 and Ser546 depends on Tor1p
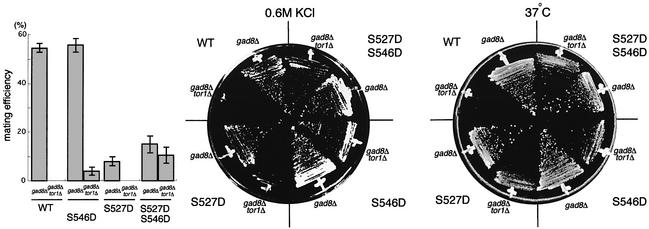
We investigated the mobility of Gad8-T387Ap, Gad8-S527Ap and Gad8-S546Ap, each tagged with 6HA, in gel electrophoresis (Figure 6D). The mobility of Gad8-T387Ap was indistinguishable from that of wild-type Gad8p, showing a slow-migrating fraction. In contrast, the slow-migrating fraction slightly shifted its position in Gad8-S527Ap, and was completely missing in Gad8-S546Ap. As wild-type Gad8p prepared from tor1Δ cells lacked the slow-migrating band, we suspected that Tor1p might be involved in phosphorylation of Ser527 and Ser546. To examine this hypothesis, we constructed gad8-S527D, gad8-S546D, and gad8-S527D/S546D alleles. The products of these mutant alleles, in which Ser527 and/or Ser546 was replaced by aspartate, were expected to mimic wild-type Gad8p phosphorylated at the respective serine residue. When we transformed the gad8Δ strain with the gad8-S546D allele carried on a pR3C vector, all the phenotypes of gad8Δ, including mating deficiency and defects in growth at high temperature (37°C) or under high osmolarity (0.6 M KCl), were suppressed as efficiently as by the wild-type gad8 allele (Figure 7), suggesting that the S546D mutation indeed mimicked phosphorylated Ser546. Interestingly, gad8-S546D could suppress the tor1Δ gad8Δ double mutant weakly, while wild-type gad8 could not. This observation was consistent with the idea that phosphorylation of Ser546 was regulated by Tor1p, but at the same time implied the possibility that Ser546 was not the sole target of Tor1p.
Fig. 7. Effects of the tor1 disruption were alleviated by gad8-S527D S546D. Homothallic haploid gad8Δ (JW945) or gad8Δ tor1Δ (JW959) strains were transformed with pR3C-gad8, pR3C-gad8-S527D, pR3C-gad8-S546D and pR3C- gad8-S527D S546D. Each transformant was tested for mating efficiency (left panel), growth at high osmolarity (middle panel) and growth at high temperature (right panel). The mating efficiency was scored after incubation on SSA medium at 30°C for five days. Growth was examined on MM medium containing 0.6 M KCl at 30°C for six days, or on MM medium at 37°C for five days.
In the case of the S527D mutation, Gad8p function appeared to be compromised by this substitution, although the mutant allele could suppress the mating deficiency of the gad8Δ strain to some extent and was obviously more active than gad8-S527A (Figure 7, also see Figure 6B). The gad8-S527D allele weakly suppressed growth defects of the tor1Δ gad8Δ double mutant at high temperature or under high osmolarity, but apparently failed to suppress its mating deficiency (Figure 7). Notably, however, the gad8-S527D/S546D allele could suppress the mating deficiency of the tor1Δ gad8Δ strain at considerably high efficiency, clearly more effectively than either the gad8-S527D or the gad8-S546D allele (Figure 7). Furthermore, the gad8-S527D/S546D allele suppressed the tor1Δ gad8Δ strain and the gad8Δ strain at nearly the same efficiency, while the wild-type and single mutant alleles of gad8 suppressed tor1Δ gad8Δ much less efficiently than gad8Δ (Figure 7). A simple interpretation of these observations is that the S527D mutation mimics the phosphorylated state of Ser527, though only to a limited extent. Also, the major targets of Tor1p on Gad8p appear to be Ser527 and Ser546, because replacement of both of them with aspartate suppresses gad8Δ effectively even in the absence of Tor1p. This is consistent with the aforementioned observation that the gad8-S527A/S546A double mutant allele cannot detectably rescue the gad8Δ strain, leaving all the phenotypes similar to those of the tor1Δ strain as they are.
The kinase activity of Gad8p is also regulated by Ksg1p, a PDK1-like kinase
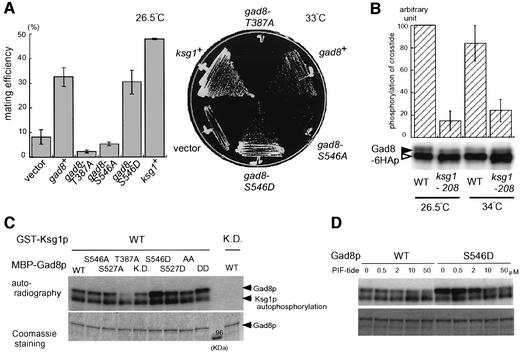
The activation loop of most AGC family kinases undergoes phosphorylation by PDK1 or PDK1-like kinase in mammals and budding yeast (Casamayor et al., 1999; Inagaki et al., 1999; Toker and Newton, 2000). It has been previously shown that fission yeast ksg1 encodes a PDK1-like kinase that is required for both vegetative growth and sexual development (Niederberger and Schweingruber, 1999). The ksg1-208 mutant is temperature sensitive for cell growth, and is impaired in mating and sporulation at the temperature permissive for growth (Niederberger and Schweingruber, 1999). We found that overexpression of gad8 could suppress these defective phenotypes of ksg1-208 (Figure 8A), suggesting that Gad8p might function downstream of Ksg1p. This possibility was examined using an in vitro phosphorylation assay. Gad8p was prepared from either the wild-type strain or the temperature-sensitive ksg1-208 mutant and its kinase activity was measured. As shown in Figure 8B, the kinase activity of Gad8p recovered from the ksg1-208 mutant was considerably low, whether the cells were cultured at the permissive temperature (26.5°C) or the restrictve temperature (34°C). These results indicate that Ksg1p is a determinant of the kinase activity of Gad8p.
Fig. 8. Functional relationship of Gad8p with Ksg1p. (A) Suppression of ksg1-208 by overexpression of gad8. The ksg1-208 mutant (JW237) was transformed with pREP41-gad8, pREP41-gad8-T387A, pREP41-gad8-S546A, pREP41-gad8-S546D, pREP41-ksg1 and the vector pREP41. Each transformant was examined for mating efficiency (left panel) and growth at the restrictive temperature (right panel). Transformants were cultured on SSA at 26.5°C for six days before scoring mating efficiency, and on MM medium at 33°C for three days to estimate temperature sensitivity. (B) The ksg1-208 mutation affects the kinase activity of Gad8p. Cells expressing Gad8-6HAp in either the wild-type (JW960) or the ksg1-208 (JW967) background were grown to logarithmic phase in YE liquid medium at 26.5°C and shifted to 34°C. Cells were harvested before and 4.5 h after the shift, and Gad8-6HAp was immunopurified from each sample. The kinase activity of Gad8-6HAp from each sample was assayed in vitro using crosstide. Immunoprecipitated Gad8-6HAp, detected by western blotting, is also shown. (C) Ksg1p phosphorylates Gad8p in vitro. Wild-type and mutant Gad8p fused to MBP, expressed bacterially and immunopurified, was incubated with GST-tagged Ksg1p in the presence of [γ-32P]ATP. Phosphorylation of MBP–Gad8p was detected by autoradiography. Ksg1-K128Rp, a kinase-dead form of Ksg1, was also examined as a negative control. An autoradiograph is shown in the upper panel and a Coomassie staining pattern in shown in the lower panel. (D) Inhibition of the Ksg1p kinase activity by PIF-tide. Phosphorylation reaction of Gad8p and Gad8-S546Dp was conducted as described in (C), in the presence of PIF-tide at the concentration indicated.
Overexpression of the gad8-T387A allele did not suppress the ksg1-208 mutant (Figure 8A). Although this observation was consistent with Thr387 being a target of Ksg1p kinase, it was not conclusive, because overexpressed gad8-S546A, or gad8-S527A or gad8-S527D could not suppress ksg1-208, while gad8-S546D could, suggesting that overexpression of any weak gad8 allele generally fails to suppress ksg1-208 (Figure 8A; data not shown). We also examined mutation of Thr387 into aspartate or glutamate, which might mimic the state of Gad8p activated by Ksg1p, but these mutants failed to complement gad8Δ, suggesting that neither gad8-T387D nor gad8-T387E was functional. As we could not establish genetic evidence for phosphorylation of Thr387 by the Ksg1p kinase, we carried out a direct phosphorylation assay as described below.
Ksg1p phosphorylates Gad8p on Thr387
To see whether Ksg1p might directly phosphorylate Gad8p, we prepared and purified Ksg1p tagged with glutathione S-transferase (GST) and Gad8p tagged with maltose binding protein (MBP) from Escherichia coli. Using these proteins we performed an in vitro phosphorylation assay. Ksg1p was found to phosphorylate Gad8p and Ksg1p itself, but Ksg1-K128Rp, a kinase-dead form, was not (Figure 8C). Gad8-S527Ap, Gad8-S546Ap or Gad8-S527A/S546Ap were phosphorylated by Ksg1p, but phosphorylation of Gad8-T387Ap was very weak, if any, indicating that Ksg1p was likely to phosphorylate Gad8p on Thr387. These data indicate that Ksg1p activates Gad8p via direct phosphorylation of Thr387, and activated Gad8p in turn controls cell growth and sexual development.
It was noted that Ksg1p phosphorylated Gad8p more intensively when Ser546 was mutated to aspartate (Figure 8C). The phosphorylated threonine or serine in the hydrophobic motif of S6K1, SGK, PKB or RSK2 is known to be captured by the PIF (PDK1-interacting fragment)-binding pocket of PDK1, which stimulates phosphorylation of these AGC family kinases in the activation loop (Frodin et al., 2000; Biondi et al., 2001; Frodin et al., 2002). To examine whether the observed hyperphosphorylation of Gad8-S546Dp by Ksg1p was due to a similar mechanism, we used PIF-tide, which is known to bind to the PIF-binding pocket on PDK1 and inhibit its ability to phosphorylate substrates (Biondi et al., 2001). As shown in Figure 8D, the addition of PIF-tide reduced phosphorylation of both Gad8p and Gad8-S546Dp by Ksg1, showing more prominent reduction with Gad8-S546Dp. Thus, it appears likely that Tor1p-dependent phosphorylation of Gad8p on Ser546 facilitates the interaction of Ksg1p with Gad8p via the PIF-binding pocket and enhances phosphorylation of Gad8p by Ksg1p on Thr387.
Discussion
The fission yeast gad8 gene encodes a novel Ser/Thr kinase of the AGC family. Disruption of gad8 results in the same phenotypes as disruption of tor1, both strains being defective in sexual development, in proper G1 arrest under nitrogen starvation, and in growth at high temperature or high osmolarity. Importantly, both the kinase activity of Gad8p and its phosphorylation state change depending on the function of tor1, with strong suggestions that Tor1p is responsible for the phosphorylation of Gad8p on Ser527 and Ser546. Replacement of both Ser527 and Ser546 with alanine abolishes Gad8p function almost completely, highlighting the importance of the dual phosphorylation in the turn and hydrophobic motifs for Gad8p activity. Although it is probable that Tor1p might have a third target site on Gad8p in addition to Ser527 and Ser546, the extensive similarity between the phenotypes of the gad8Δ and the gad8Δ tor1Δ strains each transformed with gad8-S527D/S546D indicates that Ser527 and Ser546 are the major target sites for Tor1p. We have seen that Tor1p prepared from fission yeast cells can phosphorylate Gad8p in vitro (our unpublished results), but this phosphorylation is very weak and it is as yet inconclusive as to whether the observed phosphorylation is physiologically meaningful or merely artefactual. Comparison of the Tor1p/Gad8p system to the mammalian system in which mTOR has been shown to phosphorylate S6K1 on Ser371 in the turn motif and on Thr389 in the hydrophobic motif in vitro (Burnett et al., 1998; Isotani et al., 1999; Saitoh et al., 2002), may suggest that Tor1p is likely to phosphorylate the two residues directly. However, it should be noted that the experiments on mTOR have not completely excluded the possibility that phosphorylation of S6K1 is due to an intermediate kinase co-immunoprecipitated with mTOR. Thus, it remains inconclusive whether Tor1p directly phosphorylates Gad8p on Ser527 and Ser546.
Our analysis has demonstrated that Ksg1p is responsible for the phosphorylation of Gad8p on Thr387. From the results of the in vitro study using bacterially expressed Ksg1p (Figure 8A), it appears plausible that Ksg1p phosphorylates Thr387 directly. Furthermore, it is also evident that Ksg1p has a target protein(s) other than Gad8p, which is essential for vegetative growth, as ksg1Δ is lethal and gad8Δ is not. Consistently, while overexpression of gad8 can recover the growth of ksg1-208 at high temperature (Figure 8), it cannot rescue the lethality of ksg1Δ (our unpublished results).
Gad8p recovered from the ksg1-208 mutant is partially active, irrespective of the culture temperature (Figure 8). This suggests that the product of the ksg1-208 allele is unlikely to be a thermo-labile protein but likely to have reduced Ksg1p activity at all temperatures, which is insufficient to support cell growth at a high temperature. The phenotypes of the ksg1-208 mutant, namely sterility and temperature-sensitive growth, may be largely attributed to limited Gad8p activity in this strain due to the reduced Ksg1p kinase activity, as overproduction of Gad8p suppresses ksg1-208 fairly well (Figure 8A and B).
Altogether, phosphorylation of Gad8p by Tor1p and Ksg1p appears to play a crucial role in the regulation of its kinase activity (schematically illustrated in Figure 9). The three phosphorylation sites on Gad8p, namely Thr387 in the activation loop, Ser527 in the turn motif and Ser546 in the hydrophobic motif, correspond to the conserved residues in AGC family kinases that are important for their activity (Toker and Newton, 2000; Newton, 2001). There are great similarities between the Tor1p–Ksg1p– Gad8p system we have identified in fission yeast and the mTOR–PDK1–S6K1 system found previously in mammals and Drosophila. S6K1, an AGC family kinase in mammalian cells, has been identified as a downstream effector of mTOR (Chung et al., 1992; Price et al., 1992). Rapamycin, which disrupts the function of mTOR, diminishes the kinase activity and the phosphorylation of S6K1. Thr229 in the activation loop of S6K1 is phosphorylated by PDK1 (Alessi et al., 1998; Pullen et al., 1998), whereas Ser371 in the turn motif and Thr389 in the hydrophobic motif are targeted by mTOR (Burnett et al., 1998; Isotani et al., 1999; Saitoh et al., 2002). Substitution of each phosphorylation site with alanine results in a reduction in the S6K1 kinase activity (Pearson et al., 1995; Moser et al., 1997). Drosophila has a homologous signaling system, although less characterized than the one in mammals. The Drosophila ortholog of S6K1; named dS6K, is regulated by dTOR via phosphorylation of the hydrophobic motif (Oldham et al., 2000; Zhang et al., 2000; Radimerski et al., 2002). It is also regulated by dPDK1 (Rintelen et al., 2001; Radimerski et al., 2002), however, at an unknown site and phosphorylation in the turn motif is unclear. In summary, although the upstream signals and downstream effectors may not be entirely the same, the system composed of the three kinases is apparently conserved in mammals, fly and fission yeast. We suspect that this three kinase system, involving TOR, PDK1-like and AGC family kinases, may be a basic module for signal transduction, which might compare with the ubiquitous MAP kinase cascade.
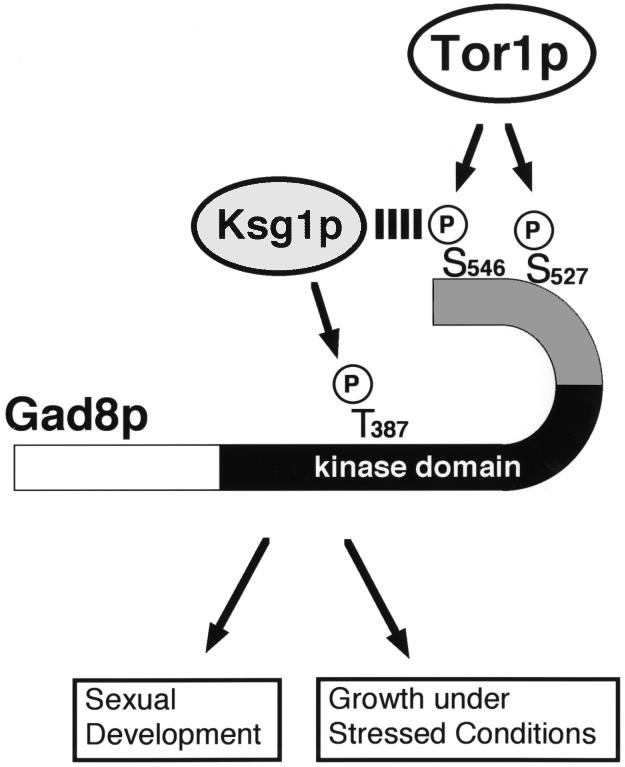
Fig. 9. A model for the activation of Gad8p by Tor1p and Ksg1p. Gad8p is an essential factor required for sexual development and growth under stressed conditions. Tor1p is responsible for phosphorylation of Ser546 in the hydrophobic motif and Ser527 in the turn motif. Ksg1p phosphorylates Thr387 in the activation loop. Phosphorylation of these three residues is essential for the full activation of Gad8p. Phosphorylation of Ser546 by Tor1p may facilitate the access of Ksg1p to Gad8p and hence enhance the phosphorylation of Thr387.
In budding yeast, Ypk1 and Ypk2/Ykr2 (Maurer, 1988; Chen et al., 1993) are highly similar to Gad8p in the kinase domain and the C-terminal region. They are activated by PDK1-like kinases Pkh1 and Pkh2 in the sphingolipid-mediated signaling pathway, and regulate endocytosis and cell wall integrity (Sun et al., 2000; Friant et al., 2001; deHart et al., 2002; Roelants et al., 2002), with Pkh1 phosphorylating the activation loop of Ypk1 (Casamayor et al., 1999). Some observations have implied that Ypk1 and Ypk2/Ykr2 are relevant to the TOR pathway (Gelperin et al., 2002; Schmelzle et al., 2002), presumably through Tor1 and Tor2 regulating Ypk1 and Ypk2/Ykr2 by phosphorylation of the apparently conserved turn and hydrophobic motifs.
Finally, the significance of the three kinase module in fission yeast physiology is considered. TOR proteins are thought to positively regulate translation in response to nutrient availability (Schmelzle and Hall, 2000; Gingras et al., 2001). As shown in this study, fission yeast tor1Δ and gad8Δ cells grow slowly in rich medium. A previous study reported that tor1Δ cells grow as fast as wild-type cells (Weisman and Choder, 2001); we suspect that this discrepancy may have stemmed from the fact that tor1Δ as well as gad8Δ cells readily produce fast-growing pseudo-revertants (our unpublished results). Thus, it appears that tor1 and gad8 function positively for growth, as ksg1 does. The three kinase module of fission yeast may have a role in harmonizing the balance of nutrients in the environment with cell growth and sexual development. A decrease in the Gad8p kinase activity apparently affects sexual development more severely than growth under stressed conditions. It is possible that cells may keep a certain level of Gad8p kinase activity under nutrient starvation in order to complete mating and meiosis, the energy-consuming processes that should proceed in the absence of fuel supply. The three kinase module may monitor the level of environmental nutrition or intracellular ATP and assure completion of sexual development.
Materials and methods
Yeast strains, media and genetic methods
Schizosaccharomyces pombe strains used in this study are listed in Table I. Yeast media YEA, SD, MM, SSA and MEA were used for routine culture of S.pombe strains (Sherman et al., 1986; Moreno et al., 1990). General genetic methods for S.pombe were described previously (Gutz et al., 1974). Specific mutations were introduced into the gad8 and ksg1 genes according to a standard method (Kunkel, 1985). When a cross of a sterile haploid strain was desired, the strain was made fertile by introducing a plasmid carrying the respective wild-type gene, and the plasmid was removed after the cross.
Table I. Schizosaccharomyces pombe strains used in this study.
| Strain | Genotype |
|---|---|
| JV107 | h90 ade6-M210 leu1 ura4-D18 gad8S527A-6HA<< kanr gad8::ura4+ |
| JV108 | h90 ade6-M210 leu1 ura4-D18 gad8S527A/S546A-6HA<< kanr gad8::ura4+ |
| JW237 | h90 ade6-M216 leu1 ksg1-208 |
| JW945 | h90 ade6-M210 leu1 ura4-D18 gad8::ura4+ |
| JW948 | h90 ade6-M216 leu1 gad2-1 |
| JW951 | h90 ade6-M210 leu1 ura4-D18 tor1::ura4+ |
| JW956 | h90/h90 ade6-M210/ade6M216 leu1/leu1 gad2-1/+ |
| JW957 | h90/h90 ade6-M210/ade6M216 leu1/leu1 tor1::ura4+/+ ura4-D18/+ |
| JW958 | h90/h90 ade6-M210/ade6M216 leu1/leu1 tor1::ura4+/gad2-1 ura4-D18/+ |
| JW959 | h90 ade6-M210 leu1 ura4-D18 gad8::ura4+ tor1::ura4+ |
| JW960 | h90 ade6-M210 leu1 ura4-D18 gad8-6HA<<kanr gad8::ura4+ |
| JW961 | h90 ade6-M210 leu1 ura4-D18 gad8K259R-6HA<< kanr gad8::ura4+ |
| JW962 | h90 ade6-M210 leu1 ura4-D18 gad8T387A-6HA<< kanr gad8::ura4+ |
| JW963 | h90 ade6-M210 leu1 ura4-D18 gad8S546A-6HA<< kanr gad8::ura4+ |
| JW965 | h90 ade6-M210 leu1 ura4-D18 tor1::ura4gad8-6HA<< kanr gad8::ura4+ |
| JW967 | h90 ade6-M210 leu1 ura4-D18 ksg1-208gad8-6HA<< kanr gad8::ura4+ |
| JY476 | h90 ade6-M210 leu1 |
| JY878 | h90 ade6-M216 leu1 ura4-D18 |
| JZ489 | h90/h90 ade6-M210/ade6M216 leu1/leu1 ura4-D18/ura4-D18 |
Gene disruption
One-step gene disruption of gad8 was carried out as follows. A 0.4 kb HincII–HincII fragment within the gad8 open reading frame (ORF) corresponding to the kinase domain was replaced by a ura4+ cassette. A diploid strain JZ489 (ura4-D18/ura4-D18) was transformed with a 3.4 kb EcoRI–PstI fragment carrying this gad8::ura4+ construct, and cells were spread on SD medium without uracil. Stable Ura4+ transformants were selected and proper disruption of gad8 was confirmed in some of the transformants by PCR. To disrupt tor1, we employed the direct chromosomal integration method described previously (Bahler et al., 1998), with some modifications. We first constructed a template vector for PCR, named pKS(FAura4), which was designed for the disruption of a target gene with ura4+ instead of kanr employed in the original protocol. We then used the following primers to generate a fragment for gene replacement. Forward: CATTGTGATGAATGCCTAAGTGGAAGAAT TGAACACCGCGACTATTAGAAAGTCTATCGTTTCACTCGCTCT CTTTGATTCCGGATCCCCGGGTTAATTAA. Reverse: CATTTTAA AAAAAGGTAAAAGAGAAGTCTCTTTGAAATTTTTGATGAGTA TGAGAAATAAAATAGTCATCCAGGAAAAGAGTTTAAACGAG CTCGAATTC.
The PCR-generated fragment, in which the entire tor1 ORF was substituted by ura4+, was transformed into JY878 (ura4-D18). We selected and confirmed tor1 disruptants in the same way as gad8 disruptants.
Construction of strains producing wild-type or mutant Gad8p tagged with 6HA
We followed a standard integration method using the integration vector int8, which was derived from int2 (Hirota et al., 2001) by replacing the green fluorescent protein (GFP) ORF with 6HA. To connect the gad8+ or each mutant gad8 ORF, which was cloned in the vector pR3C (Matsuyama et al., 2000), to the 6HA ORF in frame, a fragment flanked by the SalI site on the vector and the BglII site created at the C-terminus of each gad8 ORF was inserted between the SalI and BamHI sites of the int8 vector. The resultant plasmid was cleaved at the KpnI site in the N-terminal region of the gad8 ORF to boost integration efficiency and transformed into JW945 (gad8::ura4+). The correct integration of the fusion gene was confirmed by PCR.
Detection of Gad8p by western blotting
Harvested cells were boiled, and then disrupted with glass beads in buffer A [50 mM Tris–Cl pH 7.6, 150 mM KCl, 5 mM EDTA, 1 mM dithiothreitol (DTT), 10% glycerol]. Each suspension containing 6 µg of protein was separated by SDS–PAGE. We used 8% separating acrylamide gel with the mono/bis ratio of 29.8:0.2 to detect Gad8p. Mouse anti-HA antibody 12CA5 (Sigma) was used as the primary antibody, and sheep anti-mouse IgG conjugated with horseradish peroxidase (Amersham-Pharmacia) was used as the secondary antibody. Enhanced chemiluminescence (Amersham-Pharmacia) was used for immunodetection on the membrane. For phosphatase treatment, cells were disrupted in buffer A containing 1% SDS. The supernatant containing 15 µg of protein was incubated with 20 U of CIAP (Takara Biomedicals) at 37°C for 30 min, either with or without the inhibitor mixture [10 mM EGTA, 0.1 mM Na3VO4, 20 mM β-glycerophosphate, 15 mM p-nitrophenyl phosphate (PNPP)]. Immunodetection of Gad8p was performed similarly to that described above.
Assay of the kinase activity of Gad8p
Schizosaccharomyces pombe cells producing 6HA-tagged Gad8p were disrupted with glass beads in buffer B [50 mM Tris–Cl pH 7.6, 150 mM KCl, 5 mM EDTA, 1 mM DTT, 10% glycerol, 0.2% NP-40, 20 mM β-glycerophosphate, 0.1 mM Na3VO4, 15 mM PNPP, 1 mM phenylmethylsulfonyl fluoride and the protease inhibitor cocktail (Roche)]. Gad8p was immunoprecipitated with monoclonal anti-HA antibody 16B12 (Berkeley Antibody Co.) and protein G–Sepharose (Amersham-Pharmacia) from each crude lysate containing 750 µg protein. The Sepharose beads were washed three times with buffer B and then twice with buffer C (20 mM HEPES–KOH pH 7.5, 10 mM MgCl2, 1 mM DTT, 20 mM β-glycerophosphate, 0.1 mM Na3VO4, 15 mM PNPP). The reaction mix, adjusted to 25 µl with buffer C, contained 25 µM cold ATP, 5 µCi of [γ-32P]ATP (Amersham-Pharmacia) and 100 µM crosstide (GRPR TSSFAEG) (Upstate Biotechnology). After a 30 min incubation at 32°C, the reaction was stopped by boiling. Ten microliter of the mixture was spotted onto phosphocellulose paper (Whatman P81), which was washed five times by 75 mM phosphate buffer for 1 min, followed by an acetone wash for 5 min. The radioactivity was measured in a Scintillation Counter (LSC-5100; Aloka) and normalized by the quantum of Gad8p after subtracting radioactivity in the control specimen.
To evaluate the effect of phosphatase treatment on the kinase activity of Gad8p, Gad8p was immuprecipitated as described above. The Sepharose beads were washed three times with buffer B containing 500 mM NaCl instead of 150 mM KCl, and then washed three times by alkaline phosphatase buffer (Takara Biomedicals). The beads were incubated with 20 U of either natural or boiled CIAP (Takara Biomedicals) at 32°C for 30 min, with or without the inhibitor mixture. After the reaction, the beads were washed five times with buffer C, and then the activity of Gad8p was measured as described above.
Phosphorylation assay of Ksg1p in vitro
GST-fused Ksg1p and MBP-fused Gad8p were produced in E.coli BL21 by using expression vector pGEX 4T-3 (Pharmacia Biotech) and pMAL-cRI (New England Biolabs), respectively, together with the mutant variants of these fusion proteins. Affinity purification of the fusion proteins was carried out according to the protocol described previously (Guan and Dixon, 1991) or provided by the manufacturer, except that buffer D (50 mM Tris–Cl pH 7.6, 0.1 mM EGTA, 10 mM β-mercaptoethanol) was used as the solvent for elution. MBP–Gad8p (0.5 µg) was incubated with 0.05 µg of GST–Ksg1p in buffer D containing 4 µCi of [γ-32P]ATP, 100 µM cold ATP and 10 mM MgCl2 for 30 min at 30°C. In some experiments, PIF-tide (PILTPPREPRI LSEEEQEMFRDFAYL) (Biondi et al., 2001), supplied by a custom peptide synthesis service (BEX Corporation), was added to the reaction mix. The reaction was stopped by boiling the mix with SDS–PAGE sample buffer, and proteins were separated by 10% SDS–PAGE, followed by Coomassie Brilliant Blue staining and autoradiography.
Acknowledgments
Acknowledgements
We thank Dr M.Ernst Schweingruber for the generous gift of the S.pombe ksg1 strain and plasmid, and Dr Yuichi Iino for his help in the early stage of the project. This work was supported by a Grant-in-Aid for Specially Promoted Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- Alessi D.R. (2001) Discovery of PDK1, one of the missing links in insulin signal transduction. Colworth Medal Lecture. Biochem. Soc. Trans, 29, 1–14. [DOI] [PubMed] [Google Scholar]
- Alessi D.R., Kozlowski,M.T., Weng,Q.P., Morrice,N. and Avruch,J. (1998) 3-Phosphoinositide-dependent protein kinase 1 (PDK1) phosphorylates and activates the p70 S6 kinase in vivo and in vitro. Curr. Biol., 8, 69–81. [DOI] [PubMed] [Google Scholar]
- Bahler J., Wu,J.Q., Longtine,M.S., Shah,N.G., McKenzie,A.,III, Steever,A.B., Wach,A., Philippsen,P. and Pringle,J.R. (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast, 14, 943–951. [DOI] [PubMed] [Google Scholar]
- Biondi R.M., Kieloch,A., Currie,R.A., Deak,M. and Alessi,D.R. (2001) The PIF-binding pocket in PDK1 is essential for activation of S6K and SGK, but not PKB. EMBO J., 20, 4380–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett P.E., Barrow,R.K., Cohen,N.A., Snyder,S.H. and Sabatini,D.M. (1998) RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc. Natl Acad. Sci. USA, 95, 1432–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casamayor A., Torrance,P.D., Kobayashi,T., Thorner,J. and Alessi,D.R. (1999) Functional counterparts of mammalian protein kinases PDK1 and SGK in budding yeast. Curr. Biol., 9, 186–197. [DOI] [PubMed] [Google Scholar]
- Chen P., Lee,K.S. and Levin,D.E. (1993) A pair of putative protein kinase genes (YPK1 and YPK2) is required for cell growth in Saccharomyces cerevisiae. Mol. Gen. Genet., 236, 443–447. [DOI] [PubMed] [Google Scholar]
- Chung J., Kuo,C.J., Crabtree,G.R. and Blenis,J. (1992) Rapamycin–FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 protein kinases. Cell, 69, 1227–1236. [DOI] [PubMed] [Google Scholar]
- Cross D.A., Alessi,D.R., Cohen,P., Andjelkovich,M. and Hemmings,B.A. (1995) Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature, 378, 785–789. [DOI] [PubMed] [Google Scholar]
- deHart A.K., Schnell,J.D., Allen,D.A. and Hicke,L. (2002) The conserved Pkh–Ypk kinase cascade is required for endocytosis in yeast. J. Cell Biol., 156, 241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friant S., Lombardi,R., Schmelzle,T., Hall,M.N. and Riezman,H. (2001) Sphingoid base signaling via Pkh kinases is required for endocytosis in yeast. EMBO J., 20, 6783–6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frodin M., Jensen,C.J., Merienne,K. and Gammeltoft,S. (2000) A phosphoserine-regulated docking site in the protein kinase RSK2 that recruits and activates PDK1. EMBO J., 19, 2924–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frodin M., Antal,T.L., Dummler,B.A., Jensen,C.J., Deak,M., Gammeltoft,S. and Biondi,R.M. (2002) A phosphoserine/threonine-binding pocket in AGC kinases and PDK1 mediates activation by hydrophobic motif phosphorylation. EMBO J., 21, 5396–5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelperin D., Horton,L., DeChant,A., Hensold,J. and Lemmon,S.K. (2002) Loss of ypk1 function causes rapamycin sensitivity, inhibition of translation initiation and synthetic lethality in 14-3-3-deficient yeast. Genetics, 161, 1453–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A.-C., Raught,B. and Sonenberg,N. (2001) Regulation of translation initiation by FRAP/mTOR. Genes Dev., 15, 807–826. [DOI] [PubMed] [Google Scholar]
- Guan K.L. and Dixon,J.E. (1991) Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem., 192, 262–267. [DOI] [PubMed] [Google Scholar]
- Gutz H., Heslot,H., Leupold,U. and Loprieno,N. (1974) Schizosaccharomyces pombe. In Handbook of Genetics, Vol. 1. Plenum Publishing Corporation, New York, NY, pp. 395–446.
- Heitman J., Movva,N.R. and Hall,M.N. (1991) Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science, 253, 905–909. [DOI] [PubMed] [Google Scholar]
- Higuchi T., Watanabe,Y. and Yamamoto,M. (2002) Protein kinase A regulates sexual development and gluconeogenesis through phosphorylation of the Zn finger transcriptional activator Rst2p in fission yeast. Mol. Cell. Biol., 22, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K., Tanaka,K., Watanabe,Y. and Yamamoto,M. (2001) Functional analysis of the C-terminal cytoplasmic region of the M-factor receptor in fission yeast. Genes Cells, 6, 201–214. [DOI] [PubMed] [Google Scholar]
- Inagaki M., Schmelzle,T., Yamaguchi,K., Irie,K., Hall,M.N. and Matsumoto,K. (1999) PDK1 homologs activate the Pkc1–mitogen-activated protein kinase pathway in yeast. Mol. Cell. Biol., 19, 8344–8352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isotani S., Hara,K., Tokunaga,C., Inoue,H., Avruch,J. and Yonezawa,K. (1999) Immunopurified mammalian target of rapamycin phosphorylates and activates p70 S6 kinase alpha in vitro. J. Biol. Chem., 274, 34493–34498. [DOI] [PubMed] [Google Scholar]
- Kanoh J., Watanabe,Y., Ohsugi,M., Iino,Y. and Yamamoto,M. (1996) Schizosaccharomyces pombe gad7+ encodes a phosphoprotein with a bZIP domain, which is required for proper G1 arrest and gene expression under nitrogen starvation. Genes Cells, 1, 391–408. [DOI] [PubMed] [Google Scholar]
- Kawai M., Nakashima,A., Ueno,M., Ushimaru,T., Aiba,K., Doi,H. and Uritani,M. (2001) Fission yeast tor1 functions in response to various stresses including nitrogen starvation, high osmolarity and high temperature. Curr. Genet., 39, 166–174. [DOI] [PubMed] [Google Scholar]
- Kunitomo H., Higuchi,T., Iino,Y. and Yamamoto,M. (2000) A zinc-finger protein, Rst2p, regulates transcription of the fission yeast ste11(+) gene, which encodes a pivotal transcription factor for sexual development. Mol. Biol. Cell, 11, 3205–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T.A. (1985) Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl Acad. Sci. USA, 82, 488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz J., Henriquez,R., Schneider,U., Deuter-Reinhard,M., Movva,N.R. and Hall,M.N. (1993) Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell, 73, 585–596. [DOI] [PubMed] [Google Scholar]
- Matsuyama A., Yabana,N., Watanabe,Y. and Yamamoto,M. (2000) Schizosaccharomyces pombe Ste7p is required for both promotion and withholding of the entry to meiosis. Genetics, 155, 539–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer R.A. (1988) Isolation of a yeast protein kinase gene by screening with a mammalian protein kinase cDNA. DNA, 7, 469–474. [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar,A. and Nurse,P. (1990) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol., 194, 795–826. [DOI] [PubMed] [Google Scholar]
- Moser B.A., Dennis,P.B., Pullen,N., Pearson,R.B., Williamson,N.A., Wettenhall,R.E., Kozma,S.C. and Thomas,G. (1997) Dual requirement for a newly identified phosphorylation site in p70s6k. Mol. Cell. Biol., 17, 5648–5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton A.C. (2001) Protein kinase C: structural and spatial regulation by phosphorylation, cofactors and macromolecular interactions. Chem. Rev., 101, 2353–2364. [DOI] [PubMed] [Google Scholar]
- Niederberger C. and Schweingruber,M.E. (1999) A Schizosaccharomyces pombe gene, ksg1, that shows structural homology to the human phosphoinositide-dependent protein kinase PDK1, is essential for growth, mating and sporulation. Mol. Gen. Genet., 261, 177–183. [DOI] [PubMed] [Google Scholar]
- Oldham S., Montagne,J., Radimerski,T., Thomas,G. and Hafen,E. (2000) Genetic and biochemical characterization of dTOR, the Drosophila homolog of the target of rapamycin. Genes Dev., 14, 2689–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson R.B., Dennis,P.B., Han,J.W., Williamson,N.A., Kozma,S.C., Wettenhall,R.E. and Thomas,G. (1995) The principal target of rapamycin-induced p70s6k inactivation is a novel phosphorylation site within a conserved hydrophobic domain. EMBO J., 14, 5279–5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price D.J., Grove,J.R., Calvo,V., Avruch,J. and Bierer,B.E. (1992) Rapamycin-induced inhibition of the 70-kilodalton S6 protein kinase. Science, 257, 973–977. [DOI] [PubMed] [Google Scholar]
- Pullen N., Dennis,P.B., Andjelkovic,M., Dufner,A., Kozma,S.C., Hemmings,B.A. and Thomas,G. (1998) Phosphorylation and activation of p70s6k by PDK1. Science, 279, 707–710. [DOI] [PubMed] [Google Scholar]
- Radimerski T., Montagne,J., Rintelen,F., Stocker,H., van der Kaay,J., Downes,C.P., Hafen,E. and Thomas,G. (2002) dS6K-regulated cell growth is dPKB/dPI(3)K-independent, but requires dPDK1. Nat. Cell. Biol., 4, 251–255. [DOI] [PubMed] [Google Scholar]
- Rintelen F., Stocker,H., Thomas,G. and Hafen,E. (2001) PDK1 regulates growth through Akt and S6K in Drosophila. Proc. Natl Acad. Sci. USA, 98, 15020–15025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelants F.M., Torrance,P.D., Bezman,N. and Thorner,J. (2002) Pkh1 and pkh2 differentially phosphorylate and activate ypk1 and ykr2 and define protein kinase modules required for maintenance of cell wall integrity. Mol. Biol. Cell, 13, 3005–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh M., Pullen,N., Brennan,P., Cantrell,D., Dennis,P.B. and Thomas,G. (2002) Regulation of an activated S6 kinase 1 variant reveals a novel mammalian target of rapamycin phosphorylation site. J. Biol. Chem., 277, 20104–20112. [DOI] [PubMed] [Google Scholar]
- Schmelzle T. and Hall,M.N. (2000) TOR, a central controller of cell growth. Cell, 103, 253–262. [DOI] [PubMed] [Google Scholar]
- Schmelzle T., Helliwell,S.B. and Hall,M.N. (2002) Yeast protein kinases and the RHO1 exchange factor TUS1 are novel components of the cell integrity pathway in yeast. Mol. Cell. Biol., 22, 1329–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Fink,G. and Hicks,J. (1986) Methods in Yeast Genetics: Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, NY.
- Sun Y., Taniguchi,R., Tanoue,D., Yamaji,T., Takematsu,H., Mori,K., Fujita,T., Kawasaki,T. and Kozutsumi,Y. (2000) Sli2 (Ypk1), a homologue of mammalian protein kinase SGK, is a downstream kinase in the sphingolipid-mediated signaling pathway of yeast. Mol. Cell. Biol., 20, 4411–4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toker A. and Newton,A.C. (2000) Cellular signaling: pivoting around PDK-1. Cell, 103, 185–188. [DOI] [PubMed] [Google Scholar]
- Wang Y., Xu,H.P., Riggs,M., Rodgers,L. and Wigler,M. (1991) byr2, a Schizosaccharomyces pombe gene encoding a protein kinase capable of partial suppression of the ras1 mutant phenotype. Mol. Cell. Biol., 11, 3554–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisman R. and Choder,M. (2001) The fission yeast TOR homolog, tor1+, is required for the response to starvation and other stresses via a conserved serine. J. Biol. Chem., 276, 7027–7032. [DOI] [PubMed] [Google Scholar]
- Wood V. et al. (2002) The genome sequence of Schizosaccharomyces pombe. Nature, 415, 871–880. [DOI] [PubMed] [Google Scholar]
- Yamamoto M. (1996) The molecular control mechanisms of meiosis in fission yeast. Trends Biochem. Sci., 21, 18–22. [PubMed] [Google Scholar]
- Zhang H., Stallock,J.P., Ng,J.C., Reinhard,C. and Neufeld,T.P. (2000) Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev., 14, 2712–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]