Abstract
Excitatory neurotransmission mediated by the N-methyl-d-aspartate subtype of ionotropic glutamate receptors is fundamental to the development and function of the mammalian central nervous system. NMDA receptors require both glycine and glutamate for activation with NR1 and NR2 forming glycine and glutamate sites, respectively. Mechanisms to describe agonist and antagonist binding, and activation and desensitization of NMDA receptors have been hampered by the lack of high-resolution structures. Here, we describe the cocrystal structures of the NR1 S1S2 ligand-binding core with the agonists glycine and d-serine (DS), the partial agonist d-cycloserine (DCS) and the antagonist 5,7-dichlorokynurenic acid (DCKA). The cleft of the S1S2 ‘clamshell’ is open in the presence of the antagonist DCKA and closed in the glycine, DS and DCS complexes. In addition, the NR1 S1S2 structure reveals the fold and interactions of loop 1, a cysteine-rich region implicated in intersubunit allostery.
Keywords: ligand-gated ion channel/NMDA receptor/S1S2 ligand-binding core
Introduction
N-methyl-d-aspartate (NMDA) receptors occupy a unique position amongst ligand-gated ion channels because they require both glycine and glutamate for activation, and membrane depolarization to relieve block by magnesium (Cull-Candy et al., 2001). The prerequisite for simultaneous chemical and electrical stimuli, and the subsequent influx of calcium through the ion channel, distinguish NMDA receptors from (S)-2-amino-3-(3-hydroxy-5-methyl-4-isoxazole) propionic acid (AMPA) and kainate ionotropic glutamate receptors (iGluRs) (Dingledine et al., 1999). Under normal circumstances, NMDA receptors are involved in activity-dependent synaptic plasticity (Lisman and McIntyre, 2001) and in learning and memory (Nakazawa et al., 2002). NMDA receptors are also implicated in a number of disease and injury states, including schizophrenia and excitotoxicity (Mohn et al., 1999; Tsai and Coyle, 2002). In fact, NMDA receptor agonists such as d-cycloserine (DCS) show promise in the treatment of individuals with schizophrenia and Alzheimer’s disease (Kemp and McKernan, 2002).
Consonant with the requirement of glycine and glutamate for activation of NMDA receptors, the intact receptor is a complex of four or five subunits that typically include the NR1 subunit together with the NR2A-D or NR3A-B subunits (Cull-Candy et al., 2001). NR1 comprises the glycine-binding subunit and the NR2A-D subunits possess the glutamate-binding site (Hollmann, 1999). Interestingly, NR3A-B subunits can combine with NR1 upon expression in Xenopus oocytes to form receptors that are solely activated by glycine (Chatterton et al., 2002).
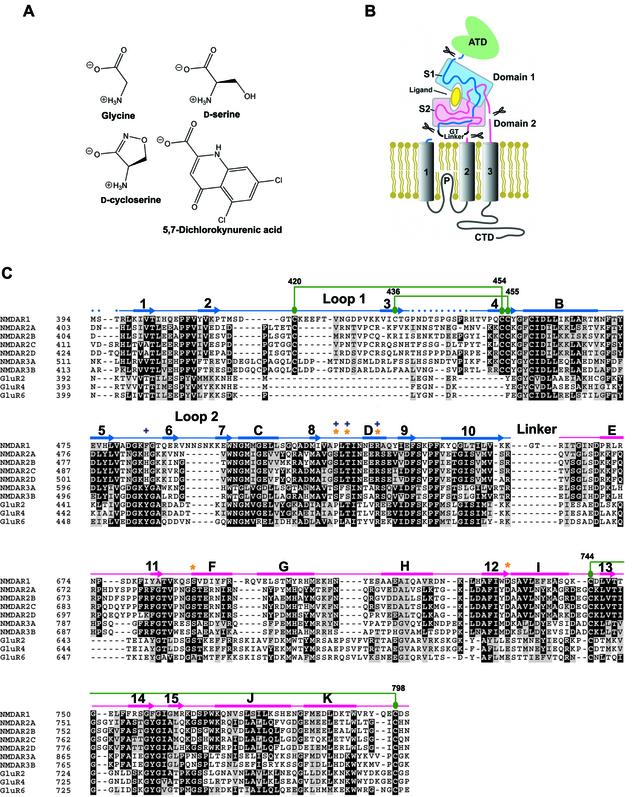
The architecture of NMDA receptors is similar to non-NMDA receptors: each NMDA receptor subunit has three transmembrane segments (M1, M2 and M3), a re-entrant membrane loop (P loop), an extracellular N-terminus and an intracellular C-terminus (Figure 1B). Like the other iGluR subtypes, the agonist-recognition region of an NMDA receptor subunit is defined by polypeptide segments S1 and S2 (Stern-Bach et al., 1994). Inspection of an alignment of the S1 and S2 regions of iGluRs, however, demonstrates that loop 1 of NMDA receptors is cysteine rich and ∼30 residues longer than the corresponding regions of AMPA and kainate receptors. The N-terminal domain (ATD) is defined by the first ∼400 amino acid residues and, while it is not directly involved in agonist binding, it is implicated in subunit assembly (Perez-Otano et al., 2001) and in receptor modulation by protons, polyamines, Zn2+ and ifenprodil (Zheng et al., 2001). The intracellular C-terminal domain (CTD) is a localization and regulatory module, and it interacts with numerous post-synaptic molecules.
Fig. 1. (A) Chemical structures of NMDA NR1 ligands. (B) Domain organization of a NR1 subunit showing the S1 and S2 segments in light blue and pink, respectively. The N-terminal domain (ATD), transmembrane segments and C-terminal domain (CTD) are not included within the S1S2 construct. (C) Multiple sequence alignment of S1 and S2 segments from rat NMDA, AMPA and kainate receptors. DDBJ/EMBL/GenBank accession Nos: X63255 (NR1), M91561 (NR2A), M91562 (NR2B), M91563 (NR2C), L31611 (NR2D), AF073379 (NR3A), AF440691 (NR3B), M85035 (GluR2), M85037 (GluR4) and Z11548 (GluR6). Drawn above the aligned sequences is the secondary structure determined from the NR1 S1S2 glycine structure where α-helices and β-strands are represented as rectangles and arrows, respectively. The color of the S1 and S2 segments is the same as that used in (B). Dots indicate the region where no electron density for the main chain is available. Cysteine residues participating in disulfide bond formation (green circles) are connected to their partners by green lines. Residues directly involved in agonist binding are marked with orange stars, whereas those specifically involved in antagonist binding are marked with blue + symbols. The sequences are numbered according to the predicted mature and immature polypeptides for non-NMDA and NMDA receptors, respectively.
Structural and functional studies of GluR2 S1S2 have yielded insights into the pharmacology and mechanism of desensitization (Armstrong and Gouaux, 2000; Jin et al., 2002; Sun et al., 2002). However, structural studies on NMDA receptors have lagged behind, to some extent, because of the difficulty in obtaining milligram quantities of pure functional protein. Nevertheless, several groups have performed molecular modeling studies of the NMDA receptor S1S2 (Laube et al., 1997; Tikhonova et al., 2002). While modeling studies can provide important insights, they are limited when conformational changes accompany ligand binding and when solvent molecules mediate key interactions. Moreover, the structure of loop 1, as well as subunit–subunit interaction surfaces and modes of association, are currently without precedent. Therefore, the structure of the NMDA receptor ligand-binding core, its modes of antagonist, partial agonist and full agonist binding, and its conformational states are unknown. Here we report the structures of the rat NR1 S1S2 ligand-binding core in complexes with two physiological full agonists, glycine and d-serine (DS), the partial agonist DCS and an antagonist, 5,7-dichlorokynurenic acid (DCKA).
Results
Preparation and ligand-binding activity of NR1 S1S2
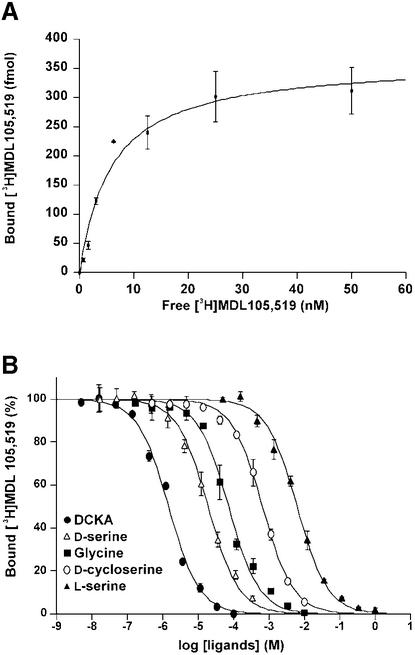
The boundaries of the NR1 S1S2 construct were based on GluR2 S1S2 (Figure 1). After removal of the His tag, the amino acid sequence begins with GM394S395T396…, where the native NR1 sequence starts at Met394 and ends with Ser800; a GT linker connects the S1 and S2 segments. In contrast to previous studies of NMDA receptor S1S2 constructs expressed in insect cells (Ivanovic et al., 1998; Miyazaki et al., 1999), the construct reported here is monomeric, as judged by size exclusion chromatography (SEC) and sedimentation equilibrium experiments at concentrations up to 1 mg/ml (data not shown). The recombinant NR1 S1S2 showed specific and saturable binding of the glycine-site antagonist [3H]MDL105,519 (Figure 2). The [3H]MDL105,519 binding was displaced by glycine, DS, DCS and DCKA (Figure 2B), and the measured Kd and Ki values were similar to those reported for an S1S2 construct or a full-length receptor expressed in insect cells (Ivanovic et al., 1998; Miyazaki et al., 1999).
Fig. 2. Ligand-binding properties of NR1 S1S2 as assessed by (A) saturation and (B) displacement experiments using the competitive antagonist of [3H]MDL105,519. The measured Kd value for [3H]MDL105,519 is 5.86 nM and the Ki values are 26.4 µM (glycine), 7.02 µM (DS), 241 µM (DCS), 0.54 µM (DCKA) and 2.30 mM (l-serine).
NR1 ligand-binding core structure and glycine-binding site
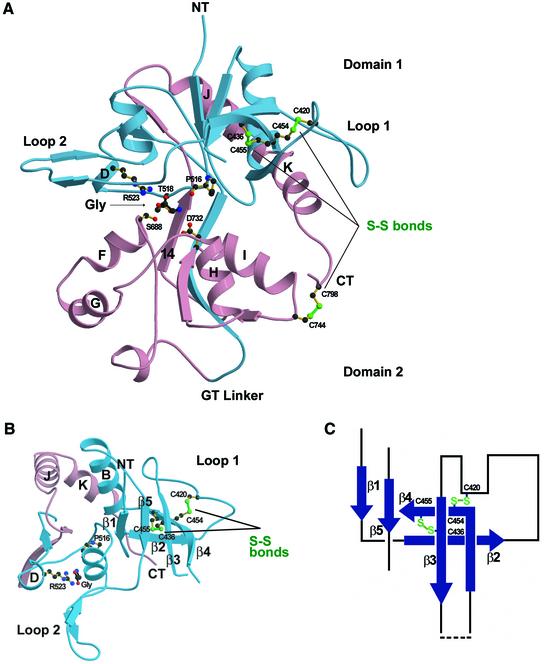
The glycine-bound NR1 S1S2 structure unambiguously reveals a bilobed or ‘clamshell’ structure consisting of domains 1 and 2 (Figure 3). Following model building and refinement, there was clear density for 281 out of 292 residues in the S1S2 construct. Three residues at the N-terminus of S1 and eight residues in loop 1 (Asp441–Arg448) were disordered. The fold of the NR1 S1S2 is similar to that of GluR2 S1S2 (Armstrong et al., 1998) even though the level of amino acid sequence identity is low (27%). Following superposition, the root-mean-square (r.m.s.) deviation on Cα positions between GluR2 S1S2 and NR1 S1S2 is 1.02 Å, where loops 1 and 2 and helix G of NR1 S1S2 and the corresponding regions of GluR2 S1S2 were excluded from the calculation.
Fig. 3. Structure of glycine-bound NR1 S1S2. (A) Ribbon representation of the glycine-bound state with S1 and S2 colored as in Figure 1B and viewed from the side. Glycine binds in the crevice between domains 1 and 2, and is surrounded by Pro516, Thr518, the N-terminal regions of helices D, F and H, and β-strand 14; residues from both domain 1 and domain 2 make contacts with the α substituents of glycine. The three disulfide bonds in NR1 S1S2 (Cys420–Cys454, Cys436–Cys455 and Cys744–Cys798) are drawn as green lines. The first two are in loop 1 and the last one is near the C-terminus (CT). (B) Ribbon representation of domain 1 viewed from the top of the N-terminus (NT). Protruding as far as 15 Å from domain 1 are loops 1 and 2. The disulfide bonds (Cys420–Cys454 and Cys436–Cys455) drawn as green lines are helping to knit together the β-strands and loop regions of loop 1. (C) Schematic representation of the loop 1 region. The dashed line indicates the region (Pro441–Arg448) where no electron density for the main chain is available.
The greatest structural difference between NR1 and GluR2 S1S2 is in loop 1, which contains a pair of antiparallel β-strands and intervening loops that are bound by two disulfide bridges. Interestingly, there is clear electron density for an alternate conformation for the side chain of Cys454 which would preclude formation of a disulfide bridge with Cys420. The length of loop 1, in conjunction with an extended loop 2, make domain 1 of NR1 ∼15 Å wider than GluR2 and GluR0 (Mayer et al., 2001). More specifically, strands 1–5 form a protein ‘wall’ projecting from domain 1. Loop 2 forms a pair of antiparallel β-strands projecting from domain 1, as in GluR2 S1S2. As with all eukaryotic iGluRs, there is a disulfide bond in NR1 located between Cys744 on helix I and Cys798 at the C-terminus of S2, linking the end of helix K on domain 1 with domain 2.
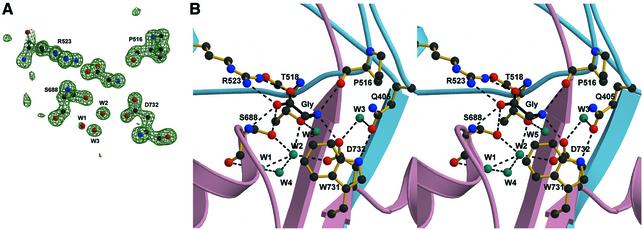
Glycine binds in the domain 1–domain 2 crevice and is surrounded by the N-terminus of helix D, helix F, helix H and β-strand 14. Residues from domains 1 and 2 make contacts with the α substituents of glycine through eight direct hydrogen bonds and electrostatic interactions (Figure 4B). In addition, water molecules W1, W2, W4 and W5 form interactions between glycine and NR1 S1S2. The α-carboxy group of glycine makes an essential interaction with the guanidinium group of Arg523, a residue conserved among all iGluRs. In GluR2 S1S2, the corresponding arginine residue interacts with the α-carboxy group of glutamate (Armstrong and Gouaux, 2000). The α-carboxy group of glycine also hydrogen bonds to the backbone amide groups of Thr518 and Ser688, and to the hydroxyl group of Ser688. The positively charged amino group of glycine interacts with the carbonyl oxygen of Pro516, the hydroxyl group of Thr518 and the carboxylate oxygen of Asp732, which is either an aspartate or a glutamate residue in iGluRs. Gln405, which is near the bound glycine, interacts with two residues in domain 2: a direct hydrogen bond to the indole nitrogen of Trp731 and, via W3, a water-mediated hydrogen bond to Asp732.
Fig. 4. The mechanism of glycine binding. (A) An Fo – Fc ‘omit’ electron density map using data to 1.35 Å resolution where the atoms corresponding to glycine, selected ligand-binding residues and waters W1, W2 and W3 were omitted from the Fc calculation. The contour level is 4.2 σ. (B) Stereo view of glycine (black bonds) and the interacting residues (yellow bonds). Dashed lines indicate hydrogen bonds and ionic interactions (interatom distance <3.2 Å). Water molecules (cyan) make important contributions to the hydrogen bond network and stabilize the binding of glycine.
Structural distinction between NR1 S1S2 and GluR2 S1S2
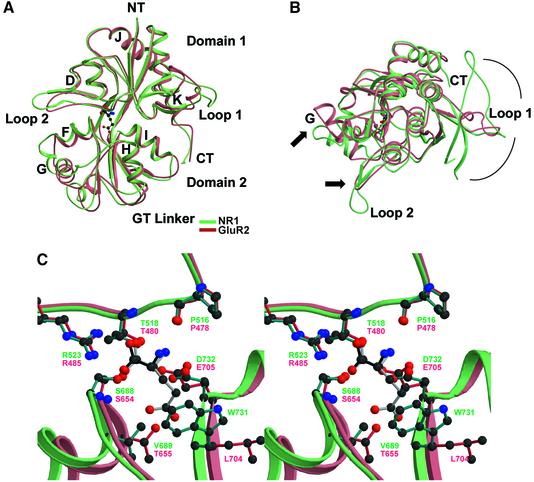
There are significant differences between the NR1 S1S2 and GluR2 S1S2 structures, even though their overall folds are similar (Figure 5). First, when comparing NR1 S1S2 and GluR2 S1S2 in their respective complexes with full agonists, NR1 S1S2 adopts a more closed conformation. Secondly, loop 1 in NR1 has a more substantial and complex structure in comparison with loop 1 in GluR2. Thirdly, loop 2 in NR1 S1S2 is longer and protrudes farther from domain 1 in comparison with loop 2 of GluR2. Lastly, the orientation of helix G, relative to the remainder of domain 2, is different in NR1 S1S2 (Figure 5).
Fig. 5. Superposition of the glycine-bound NR1 S1S2 (light cyan) and the l-glutamate-bound GluR2 S1S2 (light coral) structures using only Cα atoms viewed from (A) the side and (B) the top of the N-terminus. (C) Stereo view of the same superposition structures at the ligand-binding pocket with glycine (black) and l-glutamate (gray) interacting with the residues from NR1 S1S2 (cyan) and GluR2 S1S2 (crimson), respectively. The specificity of NR1 for glycine can be explained by (i) the hydrophobic environment created by Val689 and Trp731 and (ii) a steric constraint caused by the positioning of Trp731, which in GluR2 is Leu704 pointing away from the binding pocket, which disallow the γ-carboxyl group of l-glutamate to reside.
The essential functional difference between NR1 and other iGluR subtypes is that NR1 has a high affinity for glycine and an unmeasurably low affinity for l-glutamate (Miyazaki et al., 1999). To understand the agonist selectivity of NR1, we have superimposed NR1 S1S2 and GluR2 S1S2 (Figure 5C). Strikingly, the binding site residues of NR1 and GluR2 S1S2 superpose well and most of the agonist-contacting residues are identical or are conservative substitutions (NR1/GluR2 = Pro516/478, Thr518/480, Arg523/485, Ser688/654 and Asp732/Glu705) and share similar orientations. However, there are two critical differences between the NR1- and GluR2-binding sites. Residue 655 in GluR2 is a threonine and the hydroxyl group makes a hydrogen bond to a γ-carboxylate oxygen of glutamate; in NR1, the equivalent residue is Val689, which cannot form a similar interaction. The second key difference is residue Trp731 in NR1, which in GluR2 is Leu704. The indole ring in NR1 faces the agonist-binding pocket and, in the context of GluR2, would clash with the γ-carboxylate of glutamate. In GluR2, the leucine not only takes up less volume, but it is also oriented differently, thus allowing for the binding of the γ-carboxylate of glutamate. In NR2 subunits, the equivalent residue is a tyrosine. We suggest that the local hydrophobic environment created by Val689 and Trp731, as well as the steric constraint caused by the orientation of Trp731, prevents the binding of the γ-carboxyl group of l-glutamate.
d-serine and d-cycloserine complexes
The conformation of NR1 S1S2 in the DS and DCS complexes is similar to the S1S2 conformation in the glycine-bound form and the three complexes have essentially the same degree of domain closure. Indeed, the glycine, DS and DCS cocrystals are all isomorphous. Superpositions of α–carbon atoms of the DS and DCS S1S2 structures with the glycine structure yield r.m.s. deviations of 0.10 and 0.20 Å, respectively.
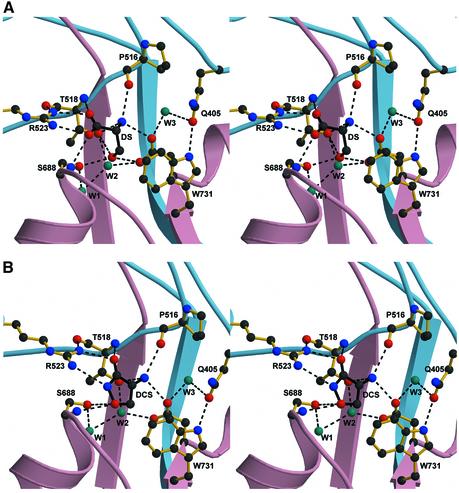
At the agonist-binding pocket, however, there are similarities and differences between the three agonists that are clearly defined by their respective electron densities. In the case of DS, the important differences are localized to the hydroxyl group as the α substituents are bound similarly to the α groups of glycine, with only one exception (Figure 6A). There are 10 salt-link and hydrogen-bonding interactions between DS and NR1 S1S2, along with interactions with two water molecules that form a hydrogen-bond network with residues Thr518, Ser688 and Asp732. At the α-carboxylate of DS, the hydroxyl of Ser688 is pointed away from the binding pocket and cannot make a hydrogen bond to an α-carboxylate oxygen, as it does in the glycine complex. Nevertheless, the DS hydroxyl group forms hydrogen bonds with the hydroxyl groups of Thr518 and Ser688 and the carboxyl group of Asp732.
Fig. 6. The binding mechanisms of DS and DCS. Stereo view of (A) DS and (B) DCS and interacting residues. In both cases, dashed lines indicate the potential hydrogen bonds and ionic interactions (interatom distance <3.2 Å). Water molecules (cyan), located in the binding pocket, are also forming a critical hydrogen-bond network to stabilize the binding of both DS and DCS.
DCS binds similarly to DS, even though the cyclic agonist contains a unique functional group. In the DCS complex, the exocyclic oxygen and the nitrogen mimic the α-carboxylates of glycine and DS, interacting with the guanidinium group of Arg523 (Figure 6B). The isoxazolidinone ring oxygen, instead of an α-carboxylate oxygen in glycine, hydrogen bonds with Ser688. The α-amino group of DCS follows the same pattern of interactions as the α-amino groups of glycine and DS.
An open-cleft conformation is stabilized by antagonist
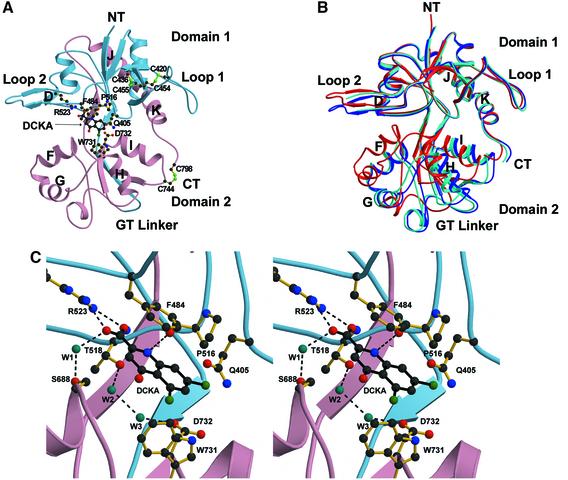
The NR1–DCKA cocrystals contain two molecules in each asymmetric unit with an expanded cleft between domain 1 and domain 2, relative to the complexes with full and partial agonists (Figure 7). The two molecules in the asymmetric unit are not identical in terms of domain closure, however, and they differ by ∼6° with molecule A being the most open. Besides the degree of domain separation, there are no large differences between the structures of molecules A and B including the orientation of the residues and water molecules surrounding the DCKA-binding pocket. The mechanism of DCKA binding is the same for both molecules. Relative to the NR1 S1S2 glycine structure, molecule A of the DCKA complex is 24° more open, i.e. there is 24° of domain closure in going from molecule A of the DCKA structure to the glycine complex.
Fig. 7. The binding mechanisms of DCKA. (A) Ribbon representation of the DKCA-bound NR1 S1S2 structure (DCKA molecule A) with S1 and S2 colored as in Figure 1B. (B) Superposition of the two DCKA-bound NR1 S1S2 molecules in an asymmetric unit (DCKA molecules A and B in blue and light blue, respectively) and the glycine-bound molecule (red). The superposition was calculated using Cα atoms. The r.m.s. deviation for DCKA molecules A and B is 0.74 Å. The glycine-bound form has a bilobed structure closed by 23.8° and 18.2° compared with the DCKA molecules A and B, respectively. (C) Stereo view of DCKA and interacting residues.
DCKA binds primarily to the ‘upper’ side of the binding pocket and makes the largest number of direct interactions with residues from domain 1. Like agonists, the carb oxylate of DCKA forms a salt link with Arg523 and a hydrogen bond with the amino group of Thr518, while the amino group of DCKA forms a hydrogen bond with the main-chain carbonyl oxygen of Pro516. The quinoline ring of DCKA is ∼3.5 Å below the aromatic ring of Phe484, participating in a π-stacking interaction (Figure 7C) as predicted by modeling studies (Tikhonova et al., 2002). The chlorine atoms at the 5 and 7 position of DCKA are in van der Waals contact with the aromatic rings of Phe408 and Trp731, respectively. The carbonyl oxygen of DCKA, while not directly interacting with the protein, does form a hydrogen bond with a water molecule at the base of helix F. DCKA acts like a wedge between Gln405 and Trp731/Asp732 by occupying the space between the residues. Hydrogen bonds between the amide oxygen of Gln405 and the indole nitrogen of Trp731, together with a water-mediated (W3) contact between Gln405 and Asp732, are maintained in the full and partial agonist-bound states. However, these interdomain contacts are disrupted by DCKA.
Discussion
Native NMDA receptors are composed of subunits with distinct ligand-recognition properties and are modulated by a large number of effector molecules and various pre- and post-translational modifications. Thus, elucidating mechanisms of NMDA receptor function based on high-resolution structural information is a particularly important and as yet unexplored area of investigation. In order to understand how the two different types of subunits recognize glycine and glutamate, and to probe possible modes of subunit–subunit interaction, we have undertaken a crystallographic investigation of the NR1 S1S2 ligand-binding core.
Ligand-binding specificity and affinity
The crystal structures described here demonstrate that the mode of agonist binding to NR1 S1S2 involves two electrostatic interactions with residues on domain 1 and a series of hydrogen bonds with amino acids on domain 2: the α-carboxylate binds to the guanidinium group of Arg523, the α-amino moiety interacts with the carboxy group of Asp732, and the α-carboxylate and/or R-group forms hydrogen bonds to residues at the N-terminus of helix F on domain 2. Disruption of Arg523 abolishes the agonist response (Hirai et al., 1996) and the mutation of Asp732 to Asn or Glu shifts the glycine EC50 ∼14 500- and 4200-fold, respectively (Williams et al., 1996). These studies reinforce the importance of correctly positioned positive and negative charges in the binding pocket (Lampinen et al., 1998). Agonist binding is critically dependent upon a series of hydrogen bonds to side-chain and main-chain atoms, as well as to water molecules. Indeed, we suggest that DS binds more tightly to the receptor in comparison with glycine because it makes three additional hydrogen bonds and displaces a water molecule (W2).
The selectivity of glycine over l-glutamate, or DS over l-serine, is very stringent; the affinity of l-glutamate to the NR1 subunit is so weak as to be unmeasurable (Miyazaki et al., 1999) and l-serine binds 300-fold less tightly than DS. Our NR1 S1S2 crystal structure, together with the GluR2 S1S2 glutamate structure (Armstrong and Gouaux, 2000), clearly explain the selectivity of glycine over glutamate and of DS over l-serine. The glycine selectivity is defined by the chemical environment and steric restraint created by primarily two amino acid side chains: Val689 and Trp731 preclude the binding of the l-glutamate γ-carboxyl group by removing a hydrogen bond donor and installing a steric barrier. d-isomer specificity is the consequence of steric constraints imposed by Phe484 and the hydrophobic environment created by Phe484 and Trp731. If one models l-serine into the binding pocket using the α-substituents of DS as a guide, then the hydroxyl group of l-serine unfavorably interacts with the phenyl ring of Phe484. By contrast, the hydroxyl group of DS is correctly positioned to form hydrogen bonds with Thr518, Asp732 and Ser688.
The binding of the partial agonist DCS is similar to the binding of glycine and DS, and involves electrostatic interactions with the side chains of Arg523 and Asp732. However, the affinity of NR1 S1S2 for DCS is 34-fold lower than that for DS. To some extent, this difference in affinity can be explained by the fact that the pKa values of the key ionizable groups on DCS are closer to neutrality in comparison with the pKa values for the corresponding groups of DS. Indeed, the pKa values for the exocyclic oxygen and amino group of DCS are 4.5 and 7.5, respectively, whereas the values for the α-carboxy group and amino group of DS are 2.2 and 9.1, respectively (McBain et al., 1989). Therefore, the concentration of the doubly ionized and presumably tightly binding form of DCS is relatively lower in comparison with the corresponding form of DS. An additional difference is that DCS does not contain a hydroxyl group that can form a hydrogen bond with the carboxylate group of Asp732.
The binding of DCKA is distinct from the binding of agonists in that DCKA is involved in π-stacking interactions with the aromatic ring of Phe484. Mutation of Phe484 severely disrupts the binding of 7-chlorokynurenic acid, an antagonist closely related to DCKA, to the NR1 receptor (Kuryatov et al., 1994). In GluR2 S1S2, the residue located at the same relative position as Phe484 is Tyr450 and in the GluR2 S1S2 5,6-dinitroquinoxalinedione (DNQX) cocrystal structure the quinoxaline ring of DNQX participates in π-stacking interactions with Tyr450 (Armstrong and Gouaux, 2000). π-stacking interactions are probably a common feature in the binding of aromatic antagonists to iGluRs. Kynurenic acid is less potent than the halogen-substituted antagonists, such as 7-chloro- and 5,7-dichlorokynurenic acid. In our DCKA structure, the 5- and 7-chloro substituents are ∼3.4 and 3.2 Å from the edges of the aromatic residues Trp731 and Phe408, respectively, and thus participate in weak but favorable hydrogen bonding and van der Waals interactions. The chloro substituents also lower the pKa of the aromatic ring substituents, which may enhance DCKA binding because the hydroxyl/carbonyl group is near the N-terminus of helix F, a known site for anion binding in both AMPA and NMDA receptors.
Relationship between domain closure and receptor activation
The crystallographic analysis of GluR2 S1S2 has revealed that the bilobed structure is ‘closed’ in the presence of agonist and ‘open’ in the apo state. A GluR2 antagonist, DNQX, stabilizes the ‘open’ apo-like conformation (Armstrong and Gouaux, 2000). The molecular movement resulting from the agonist-induced domain closure, in the context of the non-desensitized dimer, leads to separation of the regions proximal to the ion-channel gate and to activation or opening of the ion channel (Sun et al., 2002). Consistent with GluR2 S1S2, agonists and the antagonist DCKA stabilize NR1 S1S2 in ‘closed’ and ‘open’ conformations, respectively. The extent of domain closure in going from the antagonist- to agonist-bound states is greater in the case of NR1 S1S2 (∼21°) than in GluR2 S1S2 (∼16°). In terms of domain separation, the DCKA-bound NR1 S1S2 is more open than the DNQX-bound form of GluR2 S1S2 and is more closed than the apo state of GluR2 S1S2. We predict that the apo state of NR1 S1S2 will have a conformation that is similar to the DCKA-bound state. Furthermore, we suggest that the mechanism of NMDA receptor gating involves agonist-induced domain closure followed by the opening of the ion channel.
The full and partial agonist structures of NR1 S1S2 clearly indicate that agonists stabilize hydrogen-bonding interactions between residues on domain 1 (Gln405) and domain 2 (Trp731 and Asp732) in the vicinity of the ligand-binding pocket. By contrast, the antagonist DCKA disrupts the interdomain hydrogen bonds and stabilizes an ‘open’-cleft conformation of the NR1 S1S2. Consistent with the above observation, the non-conservative substitution of Gln405 by Lys increases the glycine EC50 14 230-fold (Kuryatov et al., 1994). In GluR2 S1S2, there is an analogous interdomain interaction between Glu402 and Thr686 that stabilizes the activated closed-cleft conformation of the receptor (Armstrong et al., 1998; Armstrong and Gouaux, 2000). In the NR1 S1S2 structure, Ala714 occupies a position that is equivalent to Thr686 in GluR2 S1S2, and when Ala714 is mutated to leucine, the resulting receptor has an apparent reduced affinity for glycine but DCKA inhibition is unaffected (Wood et al., 1999). In the agonist/partial agonist-bound NR1 S1S2 structures, Ala714 is located at the N-terminus of helix I and is only 3.8 Å away from side chain of Gln405. We suggest that the Ala714 to leucine mutation destabilizes the glycine-bound closed-cleft conformation of NR1 S1S2, and therefore the effect of the mutation is greatest on full and partial agonists.
The finding that the degree of domain closure for partial agonists acting on GluR2 S1S2 is correlated to the extent of receptor activation has led to the assertion that partial agonists stabilize different, and partially closed, conformations of the ligand-binding core (Armstrong and Gouaux, 2000). In contrast to the studies on GluR2 S1S2, we see no substantial difference in the degree of domain closure of NR1 S1S2 when it is bound to the full agonists glycine and DS, and to the partial agonist DCS. While it is perhaps premature to draw firm conclusions from only one partial agonist structure, we nevertheless suggest that DCS may simply not bind as tightly to the closed-cleft state of the NR1 S1S2, relative to glycine and DS, and thus DCS does not lead to as great a stabilization of the closed-cleft activated state of the ion channel as glycine and DS. Therefore, DCS may act like a classic partial agonist (Li et al., 1997). Alternatively, DCS may stabilize conformations of the NR1 S1S2 that have intermediate degrees of domain closure and we may have simply sampled only one conformation that happens to be fully closed. Interestingly, in the series of cyclic ligands that range from 1-aminocyclopropane-1-carboxylate and 1-amino-cyclobutane-1-carboxylate to cycloleucine, the efficacy of the compounds correspondingly ranges from that of a partial agonist to an antagonist (Watson and Lanthorn, 1990). In these cases, as the ring expands, we suggest that the ability of NR1 S1S2 to adopt full glycine-like domain closure diminishes, and the efficacy of the ligands shifts toward the antagonist end of the efficacy spectrum.
Insights into subunit interactions in the NMDA receptor
A growing number of studies suggest that AMPA receptors are tetrameric complexes composed of a dimer-of-dimers (Madden, 2002). Recent biophysical and crystallographic studies demonstrate that the GluR2 S1S2, in particular, forms a 2-fold symmetric dimer (Sun et al., 2002). In AMPA receptors, the rearrangement of S1S2 subunit–subunit interactions in the dimer is coupled to receptor desensitization or inactivation, and contained within the dimer interface are a number of conserved hydrophobic residues that form key contacts between the two S1S2 subunits (Sun et al., 2002).
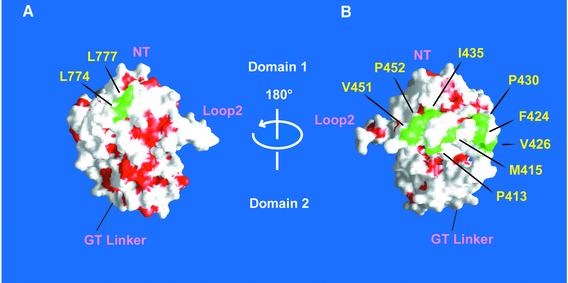
In NMDA receptors, there is not only a lack of consensus on the stoichiometry of the intact receptors, but there is also an absence of information on the nature of contacts between S1S2 subunits. While the crystal structures reported here do not directly address the NR1 S1S2 contacts in the intact receptor, inspection of the NR1 S1S2 structure and amino acid sequence alignments of iGluRs provide helpful insight. For example, in the GluR2 S1S2 dimer, Leu748 and Leu751 on helix J form an exposed hydrophobic surface that defines an important portion of the dimer interface (Armstrong and Gouaux, 2000). Strikingly, the corresponding residues in helix J of NR1 and NR2A–D subunits are also leucines (Figures 1A and 8). This observation strongly suggests that helix J in NMDA receptors participates in subunit–subunit contacts. However, there are also differences between the residues in the dimer interface of GluR2 and the corresponding residues of NMDA receptors, and thus the mode of subunit–subunit association in NMDA receptors may still diverge from that of AMPA receptors.
Fig. 8. Solvent-accessible surface of the glycine-bound NR1 S1S2 protein facing (A) helices J and K or (B) loop 1. Hydrophobic residues (Ala, Val, Leu, Ile, Met, Pro, Phe, Tyr and Trp) are in red, whereas the exposed hydrophobic residues in helices J and K and loop 1 are in green.
In the NR1 S1S2 structures, there is no contact similar to those observed in the crystallographic studies of GluR2 S1S2. In other words, even at the high protein concentrations required to form a crystal lattice, the NR1 S1S2 subunits do not display a propensity to form GluR2 S1S2-like dimers. Buttressing this observation is the fact that the Escherichia coli expressed NR1 S1S2 protein exists exclusively as a monomer in solution, as assessed by SEC and sedimentation equilibrium experiments. To ensure that glycosylation or other post-translational modifications were not necessary for assembly, we expressed the NR1 S1S2 construct in insect cells and showed that it also behaves as a monomer in solution. Our results demonstrating that the S1S2 portion of NR1 is not sufficient for oligomerization are consistent with a previous report that the first 380 residues of the ATD are important for NR1 homodimerization as well as for NR1 association with NR2A (Meddows et al., 2001). However, there are data showing that a somewhat longer NR1 S1S2 construct than the one described here, when expressed in insect cells, forms a dimer (Ivanovic et al., 1998). At the present time, it is unclear why the NR1 S1S2 species studied by Ivanovic and colleagues appears to be a dimer, but it is likely due to the fact that it is a longer construct.
A recent study has shown that loop 1 of NR2A allosterically couples the binding of glutamate on an NR2A to the binding of glycine on an NR1 subunit (Regalado et al., 2001). Even though the level of sequence identity between loop 1 of the NR1 and NR2 subunits is not high, there are a number of conserved amino acids, including the four cysteines that form two disulfide bonds in the NR1 S1S2 structure. Indeed, one of the key residues in loop 1 of NR2A that confers negative cooperativity between glutamate binding on NR2A and glycine binding on NR1 is Glu427. This residue is proximal to Cys429 (NR2A), which in turn is located at the same relative position as Cys420 in the NR1 S1S2 structure, and we have shown that Cys420 forms a disulfide bond with Cys454. It is plausible that loop 1 of the NR2 subunits will resemble loop 1 of NR1 S1S2 and that the pattern of disulfide bonds will be preserved. If NR1 and NR2 subunits assembled into a heterodimer similar to the GluR2 S1S2 dimer, then it is unlikely that loop 1 will interact with a pseudo 2-fold related subunit (Sun et al., 2002). We would suggest, instead, that loop 1 interacts with a subunit on an adjacent dimer, perhaps via exposed hydrophobic residues such as Phe424, Val426, Ile435, Val451 or Pro452, shown in Figure 8.
Disulfide bonds and redox modulation
There is a rich and complex history of the modulation of NMDA receptor activity by reducing and oxidizing reagents (Lipton et al., 2002), beginning with the observation that reducing agents, such as dithiothreitol, potentiate NMDA responses (Aizenman et al., 1989). Perhaps the best studied cysteine residues are Cys744 and Cys798 in the NR1 S1S2 structure. When Cys744 and Cys798 are changed to alanines, there is a 6-fold decrease in the NMDA EC50 (Sullivan et al., 1994) and a decrease in the voltage-independent Zn2+ inhibition of NR1/NR2A receptors (Traynelis et al., 1998). In both NR1 S1S2 and GluR2 S1S2, Cys744 (NR1) and Cys798 (NR1) and their equivalents in GluR2 form a disulfide bond. On the basis of the NR1 S1S2 complexes with glycine and with DCKA, we see that domain closure results in a movement of the end of helix K, which is adjacent to the Cys744–Cys798 disulfide bond, toward the binding cleft. We suggest that removal of the disulfide bond releases a constraint on this rearrangement, facilitating domain closure and agonist binding.
The cysteine residues in loop 1 have also been the subject of site-directed mutagenesis and functional analysis, albeit with somewhat contradictory results. Laube et al. (1993) found that when the cysteine residues equivalent to Cys420 and Cys436 were mutated to alanines in the NR1 subunit, the relative extent of potentiation by glutamate and glycine for NR1/NR2B receptors was reduced ∼15-fold. However, mutation of cysteine residues corresponding to Cys454 and Cys455 had only a minimal effect on receptor activation by glutamate and glycine (Laube et al., 1993). In a separate study, NR1 Cys420 or Cys436 was mutated to alanine with no noted effect on receptor activity (Köhr et al., 1994). Consistent with Laube et al. (1993), Köhr et al. (1994) reported that substitution of the vicinal cysteines (Cys454, Cys455) with alanines was also without effect. Replacement of the equivalent vicinal cysteines in NR2A did cause a loss of channel activity, however (Köhr et al., 1994). We cannot resolve the discrepancies in these previous experiments, or point directly to how substitution of the cysteine residues leads to a specific effect on receptor activity. Nevertheless, from the NR1 S1S2 structure, we see that loop 1 adopts a defined fold and that the disulfide bonds in loop 1 mediate specific interactions between elements of secondary structure. Disruption of the disulfides, therefore, may perturb the structure of loop 1 and result in alteration of receptor expression and function.
Conclusions
Our high-resolution crystallographic study addresses important elements of NMDA receptor structure and function relationships such as the fold of loop 1, the molecular basis of glycine and d-isomer selectivity, and the conformational changes accompanying antagonist, partial agonist and full agonist binding. The structures provide deeper insights into the mechanisms of receptor function and subunit–subunit association. In addition, this new structural information should enhance efforts to design subtype-selective agonists and antagonists that may have beneficial therapeutic properties.
Materials and methods
Protein purification and crystallization of NR1 S1S2
The DNA sequences encoding S1 (Met394 to Lys544) and S2 (Arg663 to Ser800) connected by Gly-Thr linker were expressed in Origami B (DE3) cells (Novagen) using a T7 expression vector pET22b(+). After growing the cells to an OD600 of 0.8, expression was induced with 0.5 mM isopropyl-β-d-thiogalactoside followed by incubation at 20°C for 24 h. The protein was purified using Ni-NTA (Pharmacia), followed by the removal of polyhistidine tag by thrombin digestion. The digest was further purified by SP Sepharose column (Pharmacia) and concentrated to 5 mg/ml (ε280 = 1.1). Ligands were added to 10 mM for glycine, DS and DCS, and to 1 mM for DCKA. The glycine-, DS- or DCS-bound NR1 S1S2 crystals were grown at 4°C in hanging drops containing 1:2 or 1:1 (v/v) ratio of protein to reservoir solution composed of 100 mM sodium cacodylate pH 6.0, 100 mM Li2SO4 and 20% PEG 1000. The DCKA-bound NR1 S1S2 crystals were grown at 4°C in hanging drops containing 1:2 (v/v) ratio of protein to reservoir solution containing 100 mM HEPES pH 7.0 and 20–25% PEG 2000. Clusters of thin plates appeared within 2 days and larger crystals were grown by microseeding. The crystals were soaked in the crystal buffer supplemented with 12–15% glycerol and the appropriate ligand, and flash frozen in liquid nitrogen. For the multiple-wavelength anomalous diffraction (MAD) experiment (Hendrickson, 1991), the glycine crystals were soaked in a cryobuffer supplemented with 0.5 M NaBr (Dauter and Dauter, 1999).
Structural determination
All diffraction data sets were collected at the X4A beamline at the National Synchrotron Light Source and were processed using the HKL suite of programs (Otwinowsky and Minor, 1997). A three-wavelength MAD dataset was collected on the glycine-bound crystal soaked in NaBr at 13 474.4 eV (edge), 13 479.0 eV (peak) and 13 573.8 eV (remote) using inverse-beam geometry. The bromide sites were located using SOLVE (Terwilliger, 1997), and the resulting phases were improved using the companion program RESOLVE. The resulting phases yielded a readily interpretable map, and the structure was built using the program O (Jones and Kjeldgaard, 1997). After one round of Powell minimization and B-factor refinement using CNS (Brunger et al., 1998), Rwork and Rfree were 34.0 and 38.1%, respectively. The structure was then refined against a native dataset measured to 1.35 Å resolution. After convergence of the R values (Rwork = 0.285 and Rfree = 0.295), glycine and water molecules were built into |Fo| – |Fc| omit maps. Refinement using Powell minimization and individual B-factor refinement was continued until the R values converged.
The DS and DCS complexes were isomorphous to the glycine cocrystal and thus the glycine structure, minus glycine and solvent molecules, was the starting point for the refinement of DS and DCS structures. These refinements were also carried out using CNS in a manner similar to the refinement protocol for the glycine structure. The density for the agonists was unambiguous.
The DCKA structure, in which there are two molecules in the asymmetric unit, was determined by molecular replacement (MR) using domain 1 of the glycine structure (Leu398 to Gln536 and Phe758 to Ser800) as the search probe (Navaza, 1994). The two domain 1 molecules were subjected to rigid body refinement followed by a slow-cool simulated-annealing run that started at 5000 K. Structure factor and phases were then calculated and weighted using the programs SFALL and SIGMAA, respectively (CCP4, 1994). The resulting phases were extended to 1.9 Å and were improved by averaging, solvent flattening and histogram matching using the program DM, yielding an interpretable electron density for most of domain 2. A new structure for both molecules was then automatically built using the program ARP_WARP. Two rounds of Powell minimization and individual B-factor refinement were carried out without applying NCS restraints. The two molecules in the asymmetric unit were manually rebuilt between each refinement cycle. When Rfree reached 0.30, the DCKA molecules were built into an |Fo| – |Fc| map. Addition of solvent molecules and further refinement reduced the crystallographic Rwork and Rfree values to 0.217 and 0.258, respectively. Analysis of the structure using PROCHECK (Laskowski et al., 1993) demonstrated that all residues in all of the structures are in the most favored or additionally favored regions of the Ramachandran plot. Statistics on the diffraction data sets and the refinements are listed in Tables I and II. Solvent-accessible surface areas were calculated using the program GRASP (Nicholls et al., 1991). The Cα superpositions and the r.m.s. deviations were calculated by LSQMAN (Kleywegt, 1996).
Table I. Data collection statistics.
| Ligand | Space group | Unit cell dimensions (Å) | No. per a.u.a | λ | dmin (Å)b | Total hkl | Unique hkl | Rmerge (%)c,d | Completeness (%)d | I/σI |
|---|---|---|---|---|---|---|---|---|---|---|
| Glycine | P212121 | a = 41.7 | 1 | 0.9202 | 1.35 (1.4) | 383 080 | 57 637 | 6.5 (28.0) | 87.7 (41.1) | 34.3 |
| b = 73.0 | ||||||||||
| c = 96.9 | ||||||||||
| Glycine–NaBr | P212121 | a = 41.6 | 1 | |||||||
| b = 72.7 | ||||||||||
| c = 96.7 | ||||||||||
| (e1-edge) | 0.9202 | 1.90 (1.97) | 150 212 | 22 891 | 5.3 (11.2) | 95.4 (73.1) | 36.6 | |||
| (e2-peak) | 0.9198 | 1.90 (1.97) | 149 431 | 22 865 | 5.7 (10.8) | 95.4 (73.3) | 35.6 | |||
| (e3-remote) | 0.9134 | 1.90 (1.97) | 151 935 | 23 095 | 5.7 (15.3) | 96.1 (75.4) | 32.2 | |||
| DCKA | C2 | a = 85.7 | 2 | 0.9202 | 1.90 (1.97) | 323 126 | 47 403 | 3.9 (15.4) | 88.0 (47.7) | 31.1 |
| b = 65.3 | ||||||||||
| c = 124.3 | ||||||||||
| β = 98.1° | ||||||||||
| d-serine | P212121 | a = 41.5 | 1 | 0.9795 | 1.45 (1.50) | 414 093 | 51 309 | 6.6 (17.2) | 97.0 (85.2) | 28.9 |
| b = 73.1 | ||||||||||
| c = 96.9 | ||||||||||
| d-cycloserine | P212121 | a = 41.6 | 1 | 0.9795 | 1.60 (1.66) | 293 632 | 38 432 | 8.5 (21.4) | 96.8 (78.3) | 26.0 |
| b = 72.9 | ||||||||||
| c = 96.7 |
aNumber of molecules per asymmetric unit (a.u.).
bValues in parentheses define the low-resolution limits for the highest-resolution shell of data.
cRmerge = (Σ|Ii – <Ii>|)/Σi|Ii|, where <Ii> is the mean Ii over symmetry-equivalent reflections.
dValues in parentheses are for the highest-resolution shell of data.
Table II. Refinement statistics.
| Ligand | Resolution (Å) | Rworka (%) | Rfreeb (%) | No. of protein atoms | No. of ligand atoms | No. of H2O | Average B value |
R.m.s. deviations |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Overall | Main | Ligand | Bonds (Å) | Angles (°) |
B values |
||||||||
| Bonds | Angles | ||||||||||||
| Glycine | 10–1.35 | 19.5 | 21.6 | 2151 | 5 | 331 | 17.42 | 14.37 | 8.26 | 0.006 | 1.28 | 1.64 | 2.33 |
| DCKA | 10–1.90 | 21.8 | 25.9 | 4072 | 32 | 511 | 28.00 | 26.32 | 15.06 | 0.006 | 1.18 | 1.37 | 2.13 |
| d-serine | 10–1.45 | 20.4 | 22.6 | 2171 | 7 | 353 | 17.83 | 15.13 | 10.56 | 0.005 | 1.17 | 1.06 | 1.65 |
| d-cycloserine | 10–1.60 | 20.4 | 22.8 | 2141 | 7 | 288 | 19.96 | 18.05 | 14.00 | 0.005 | 1.17 | 1.12 | 1.74 |
aRwork = (Σ||Fo| – |Fc||)/Σ|Fo|, where Fo and Fc denote observed and calculated structure factors, respectively.
bTen per cent of the reflections were set aside for calculation of the Rfree value.
Activity assay
NR1 S1S2 protein was dialyzed exhaustively against ligand-binding buffer (LBB) (20 mM HEPES pH 7.0, 150 mM NaCl, 1mM EDTA and 10% glycerol) following ion exchange chromatography. In the saturation experiments, NR1 S1S2 was incubated with 0, 0.78, 1.56, 3.13, 6.25, 12.5, 25.0 and 50.0 nM [3H]MDL105,519 (Perkin–Elmer) in LBB for 1 h on ice. In the competition binding experiments, NR1 S1S2 was incubated with 10 nM [3H]MDL105,519 in the presence of various concentrations of glycine, DS, l-serine, DCS and DCKA. Non-specific binding was measured for each reaction condition by including 100 mM glycine. Ligand-bound NR1 S1S2 protein was trapped on GSWP 02500 membranes by vacuum filtration on a 12-place manifold (Millipore), the filters were washed three times with 2 ml of ice-cold LBB and placed in vials containing 6 ml of scintillation fluid, and radioactivity was measured using a scintillation counter. Kaleidagraph was employed for calculations involving ligand binding.
Coordinates
Coordinates for the NR1 S1S2 ligand binding core in complex with glycine, DS, DCS and DCKA have been deposited in the Protein Data Bank with accession codes 1PB7, 1PB8, 1PB9 and 1PBQ.
Acknowledgments
Acknowledgements
The NR1 cDNA was a generous gift from Dr S.Heinemann. Rich Olson is gratefully acknowledged for performing the analytical ultracentrifugation experiments. We thank Joe Lidestri for expert maintenance of the X-ray facility at Columbia, and Craig Ogata and Randy Abramowitz for assistance with collection of X-ray diffraction data at beamline X4A at the National Synchrotron Light Source. M.Mayer and S.K.Singh are thanked for critical reading of this manuscript. This research was supported by the National Alliance for Research on Schizophrenia and Depression, and the NIH. E.G. is an assistant investigator in the Howard Hughes Medical Institute.
References
- Aizenman E., Lipton,S.A. and Loring,R.H. (1989) Selective modulation of NMDA responses by reduction and oxidation. Neuron, 2, 1257–1263. [DOI] [PubMed] [Google Scholar]
- Armstrong N. and Gouaux,E. (2000) Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron, 28, 165–181. [DOI] [PubMed] [Google Scholar]
- Armstrong N., Sun,Y., Chen,G.-Q. and Gouaux,E. (1998) Structure of a glutamate receptor ligand binding core in complex with kainate. Nature, 395, 913–917. [DOI] [PubMed] [Google Scholar]
- Brunger A.T. et al. (1998) Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr.D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Chatterton J.E. et al. (2002) Excitatory glycine receptors containing the NR3 family of NMDA receptor subunits. Nature, 415, 793–798. [DOI] [PubMed] [Google Scholar]
- Cull-Candy S., Brickley,S. and Farrant,M. (2001) NMDA receptor subunits: diversity, development and disease. Curr. Opin. Neurobiol., 11, 327–335. [DOI] [PubMed] [Google Scholar]
- Dauter Z. and Dauter,M. (1999) Anomalous signal of solvent bromides used for phasing of lysozyme. J. Mol. Biol., 289, 93–101. [DOI] [PubMed] [Google Scholar]
- Dingledine R., Borges,K., Bowie,D. and Traynelis,S.F. (1999) The glutamate receptor ion channels. Pharmacol. Rev., 51, 7–61. [PubMed] [Google Scholar]
- Hendrickson W.A. (1991) Determination of macromolecular structures from anomalous diffraction of synchrotron radiation. Science, 254, 51–58. [DOI] [PubMed] [Google Scholar]
- Hirai H., Kirsch,J., Laube,B., Betz,H. and Kuhse,J. (1996) The glycine binding site of the N-methyl-d-aspartate receptor subunit NR1: identification of novel determinants of co-agonist potentiation in the extracellular M3–M4 loop region. Proc. Natl Acad. Sci. USA, 93, 6031–6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollmann M. (ed.) (1999) Structure of Ionotropic Glutamate Receptors. Springer, Berlin, Germany.
- Ivanovic A., Reiländer,H., Laube,B. and Kuhse,J. (1998) Expression and initial characterization of a soluble glycine binding domain of the N-methyl-d-aspartate receptor NR1 subunit. J. Biol. Chem., 273, 19933–19937. [DOI] [PubMed] [Google Scholar]
- Jin R., Horning,M., Mayer,M.L. and Gouaux,E. (2002) Mechanism of activation and selectivity in a ligand-gated ion channel: structural and functional studies of GluR2 and quisqualate. Biochemistry, 41, 15635–15643. [DOI] [PubMed] [Google Scholar]
- Jones T.A. and Kjeldgaard,M. (1997) Electron-density map interpretation. Methods Enzymol., 277, 173–208. [DOI] [PubMed] [Google Scholar]
- Kemp J.A. and McKernan,R.M. (2002) NMDA receptor pathways as drug targets. Nat. Neurosci., 5, 1039–1042. [DOI] [PubMed] [Google Scholar]
- Kleywegt G.J. (1996) Use of non-crystallographic symmetry in protein structure refinement. Acta Crystallogr. D, 52, 842–857. [DOI] [PubMed] [Google Scholar]
- Köhr G., Eckardt,S., Luddens,H., Monyer,H. and Seeburg,P.H. (1994) NMDA receptor channels: subunit-specific potentiation by reducing agents. Neuron, 12, 1031–1040. [DOI] [PubMed] [Google Scholar]
- Kuryatov A., Laube,B., Betz,H. and Kuhse,J. (1994) Mutational analysis of the glycine-binding site of the NMDA receptor: structural similarity with bacterial amino acid-binding proteins. Neuron, 12, 1291–1300. [DOI] [PubMed] [Google Scholar]
- Lampinen M., Pentikäinen,O., Johnson,M.S. and Keinänen,K. (1998) AMPA receptors and bacterial periplasmic amino acid-binding proteins share the ionic mechanism of ligand recognition. EMBO J., 17, 4704–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Laube B., Kuryatov,A., Kuhse,J. and Betz,H. (1993) Glycine–glutamate interactions at the NMDA receptor: role of cysteine residues. FEBS Lett., 335, 331–334. [DOI] [PubMed] [Google Scholar]
- Laube B., Hirai,H., Sturgess,M., Betz,H. and Kuhse,J. (1997) Molecular determinants of agonist discrimination by NMDA receptor subunits: analysis of the glutamate binding site on the NR2B subunit. Neuron, 18, 493–503. [DOI] [PubMed] [Google Scholar]
- Li J., Zagotta,W.N. and Lester,H.A. (1997) Cyclic nucleotide-gated channels: structural basis of ligand efficacy and allosteric modulation. Q. Rev. Biophys., 30, 177–193. [DOI] [PubMed] [Google Scholar]
- Lipton S.A., Choi,Y.-B., Takahashi,H., Zhang,D., Li,W., Godzik,A. and Bankston,L.A. (2002) Cysteine regulation of protein function—as exemplified by NMDA-receptor modulation. Trends Neurosci., 25, 474–480. [DOI] [PubMed] [Google Scholar]
- Lisman J.E. and McIntyre,C.C. (2001) Synaptic plasticity: a molecular memory switch. Curr. Biol., 11, R788–R791. [DOI] [PubMed] [Google Scholar]
- Madden D.R. (2002) The structure and function of glutamate receptor ion channels. Nat. Rev. Neurosci., 3, 91–101. [DOI] [PubMed] [Google Scholar]
- Mayer M.L., Olson,R.A. and Gouaux,E. (2001) Mechanisms for ligand binding to GluR0 ion channels: crystal structures of the glutamate and serine complexes and a closed apo state. J. Mol. Biol., 311, 815–836. [DOI] [PubMed] [Google Scholar]
- McBain C.J., Kleckner,N.W., Wryick,S. and Dingledine,R. (1989) Structural requirements for activation of the glycine coagonist site of N-methyl-d-aspartate receptors expressed in Xenopus oocytes. Mol. Pharmacol., 36, 556–565. [PubMed] [Google Scholar]
- Meddows E., Le Bourdelles,B., Grimwood,S., Wafford,K., Sandhu,S., Whiting,P. and McIlhinney,R.A. (2001) Identification of molecular determinants that are important in the assembly of N-methyl-d-aspartate receptors. J. Biol. Chem., 276, 18795–18803. [DOI] [PubMed] [Google Scholar]
- Miyazaki J., Nakanishi,S. and Jingami,H. (1999) Expression and characterization of a glycine-binding fragment of the N-methyl-d-aspartate receptor subunit NR1. Biochem. J., 340, 687–692. [PMC free article] [PubMed] [Google Scholar]
- Mohn A.R., Gainetdinov,R.R., Caron,M.G. and Koller,B.H. (1999) Mice with reduced NMDA receptor expression display behaviors related to schizophrenia. Cell, 98, 427–436. [DOI] [PubMed] [Google Scholar]
- Nakazawa K. et al. (2002) Requirement for hippocampal CA3 NMDA receptors in associative memory recall. Science, 297, 211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaza J. (1994) AMoRe: An automated package for molecular replacement. Acta Crystallogr. A, 50, 157–163. [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: Insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Otwinowsky Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Perez-Otano I., Schulteis,C.T., Contractor,A., Lipton,S.A., Trimmer,J.S., Sucher,N.J. and Heinemann,S.F. (2001) Assembly with the NR1 subunit is required for surface expression of NR3A-containing NMDA receptors. J. Neurosci., 21, 1228–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regalado M.P., Villarroel,A. and Lerma,J. (2001) Intersubunit cooperativity in the NMDA receptor. Neuron, 32, 1085–1096. [DOI] [PubMed] [Google Scholar]
- Stern-Bach Y., Bettler,B., Hartley,M., Sheppard,P.O., O’Hara,P.J. and Heinemann,S.F. (1994) Agonist selectivity of glutamate receptors is specified by two domains structurally related to bacterial amino acid-binding proteins. Neuron, 13, 1345–1357. [DOI] [PubMed] [Google Scholar]
- Sullivan J.M., Traynelis,S.F., Chen,H.-S.V., Escobar,W., Heinemann,S.F. and Lipton,S.A. (1994) Identification of two cysteine residues that are required for redox modulation of the NMDA subtype of glutamate receptor. Neuron, 13, 929–936. [DOI] [PubMed] [Google Scholar]
- Sun Y., Olson,R.A., Horning,M., Armstrong,N., Mayer,M.L. and Gouaux,E. (2002) Mechanism of glutamate receptor desensitization. Nature, 417, 245–253. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C. (1997) Multiwavelength anomalous diffraction phasing of macromolecular structures: analysis of MAD data as single isomorphous replacement with anomalous scattering data using the MADMRG program. Methods Enzymol., 276, 530–537. [PubMed] [Google Scholar]
- Tikhonova I.G., Baskin,I.I., Palyulin,V.A. and Zefirov,N.S. (2002) A spatial model of the glycine site of the NR1 subunit of NMDA-receptor ligand docking. Dokl. Biochem. Biophys. 382, 67–70. [DOI] [PubMed] [Google Scholar]
- Traynelis S.F., Burgess,M.F., Zheng,F., Lyuboslavsky,P. and Powers,J.L. (1998) Control of voltage-independent zinc inhibition of NMDA receptors by the NR1 subunit. J. Neurosci., 18, 6163–6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai G. and Coyle,J.T. (2002) Glutamatergic mechanisms in schizophrenia. Annu. Rev. Pharmacol. Toxicol., 42, 165–179. [DOI] [PubMed] [Google Scholar]
- Watson G.B. and Lanthorn,T.H. (1990) Pharmacological characteristics of cyclic homologues of glycine at the N-methyl-d-aspartate receptor-associated glycine site. Neuropharmacology, 29, 727–730. [DOI] [PubMed] [Google Scholar]
- Williams K., Chao,J., Kashiwagi,K., Masuko,T. and Igarashi,K. (1996) Activation of N-methyl-d-aspartate receptors by glycine: role of an aspartate residue in the M3–M4 loop of the NR1 subunit. Mol. Pharmacol., 50, 701–708. [PubMed] [Google Scholar]
- Wood M.W., VanDongen,H.M.A. and VanDongen,A.M.J. (1999) A mutation in the glycine binding pocket of the N-methyl-d-aspartate receptor NR1 subunit alters agonist efficacy. Mol. Brain Res., 73, 189–192. [DOI] [PubMed] [Google Scholar]
- Zheng F., Errenger,K., Low,C.M., Banke,T.G., Lee,C.J., Conn,P.J. and Traynelis,S.F. (2001) Allosteric interaction between the amino terminal domain and the ligand-binding domain of NR2A. Nat. Neurosci., 4, 894–901. [DOI] [PubMed] [Google Scholar]