Abstract
We have isolated the complete coding sequences for two Xenopus laevis isoforms of heterochromatin protein 1, corresponding to HP1α and HP1γ. The sequence of xHP1α shows considerable divergence from its mammalian homologues, whereas xHP1γ is highly conserved. Functionally, xHP1α behaves identically to human HP1α. We observe unexpected differences between the two HP1 variants in binding native soluble chromatin, which seem to correlate with their distinct nuclear distributions in vivo. A surprising finding is that the characteristic interaction of HP1 chromodomains with histone H3 at methylated lysine 9 is not detected in preformed chromatin due to its inaccessibility. Instead, we localize a strong chromatin-binding activity to the short hinge region between the chromodomain and the chromoshadow domain of xHP1α but not xHP1γ. This novel chromatin-binding activity has a non-specific DNA-binding component in addition to a linker histone-dependent preference for an altered chromatin structure with a likely heterochromatin organization.
Keywords: chromodomain/heterochromatin/HP1 hinge/linker histones
Introduction
Heterochromatin is a specialized chromatin structure that is distinguished from euchromatin by being condensed, late replicating, gene poor and generally incompatible with gene activity. When euchromatic genes are placed next to centromeric heterochromatin, they become prone to progressive silencing. This is thought to be caused by spreading of the condensed heterochromatic higher-order structure into adjacent euchromatin (Richards and Elgin, 2002). Silencing typically occurs in a proportion of cells and is heritable, leading to mosaic patterns of gene expression known as position-effect variegation (PEV). Genetic analysis in Drosophila has identified mutations that can enhance or suppress PEV. The Su(var)2-5 suppressor mutation locus encodes heterochromatin protein 1 (HP1), which was first identified biochemically and shown to localize to the chromocentre in polytene chromosomes (James et al., 1989; Eissenberg et al., 1990). Null mutations of HP1 cause mis-segregation of chromosomes and embryonic lethality in Drosophila. HP1 proteins are evolutionary conserved, with orthologues in yeast, plants and animals (Li et al., 2002). In mammals, three isoforms of HP1 (α, β and γ) have been identified, which distribute to different sub-nuclear locations. HP1α is found primarily in centromeric heterochromatin. HP1β and, to a much lesser extent, HP1γ are also detected at centromeric heterochromatin but are localized at many additional euchromatic sites dispersed within the nucleus (Nielsen et al., 2001).
HP1 has a tripartite structure of two chromodomains (CDs), separated by a hinge region (Richards and Elgin, 2002). A variety of studies has shown that both the amino CD and carboxyl chromoshadow domain (CSD) are versatile protein interaction modules implicated in HP1 function (Li et al., 2002). For example, the CD of mammalian HP1s and the Schizosaccharomyces pombe homologue Swi6 binds the methylated lysine 9 residue of histone H3 (Bannister et al., 2001; Lachner et al., 2001). In S.pombe, histone H3 tail methylation is required to maintain silencing at centromeres (Bannister et al., 2001). However, these analyses have not revealed any biochemical differences between the different HP1 isoforms that explain their different chromatin locations. CDs are also found in many non-HP1 proteins, including ATP-dependent chromatin modifiers and histone H3-specific methyltransferases (Jenuwein and Allis, 2001).
The hinge region of HP1 is less well characterized, but a few studies indicate that it is more than a linker connecting the CD modules and plays a direct role in HP1 function (Li et al., 2002). As regards a role in nuclear location, truncated forms of HP1 capable of targeting to heterochromatic regions require at least one CD in addition to a substantial portion of the hinge (Platero et al., 1995; Yamada et al., 1999; Muchardt et al., 2002). Recently, additional HP1 proteins have been identified in Drosophila, termed HP1b and HP1c, the localization of which is partially dependent on the sequence composition of their respective hinge regions (Smothers and Henikoff, 2001). Furthermore, studies in S.pombe describe a nuclear localization signal within the hinge that targets the HP1 homologue Swi6 to the nucleus (Wang et al., 2000).
In this paper, we characterize the HP1α and HP1γ isoforms of Xenopus laevis. Our biochemical analysis identifies a novel chromatin-binding activity in the hinge region of xHP1α that is not present in xHP1γ. This adds a new mode of interaction to a number of other contacts known to be possible between HP1 and chromatin components, suggesting that HP1 can make multiple contacts with chromatin or that each contact may be functionally different. We set out to determine which of these associations are predominant in the binding of HP1 to native oligonucleosomes isolated from chicken erythrocyte nuclei, a standard chromatin substrate not previously tested. A surprising finding is that the interaction between the HP1 CD and the methylated Lys9 residue of histone H3 cannot be detected when HP1 binds to this preformed chromatin. We present evidence to suggest that this contact is inaccessible in native oligonucleosomes, whereas HP1α can interact almost quantitatively and non-specifically via the HP1-hinge– DNA contact. Intriguingly, our results show that an additional interaction between the HP1α hinge region and linker histones allows for a preferential association with chromatin consistent with a heterochromatic organization. These functional differences between HP1 proteins may underlie their specialized roles in regulating chromatin structure and transcriptional competence at different loci. The implications of our findings for the model of HP1 involvement in setting up and spreading of the heterochomatin structure are discussed.
Results
Isolation of complete xHP1α and xHP1γ cDNAs
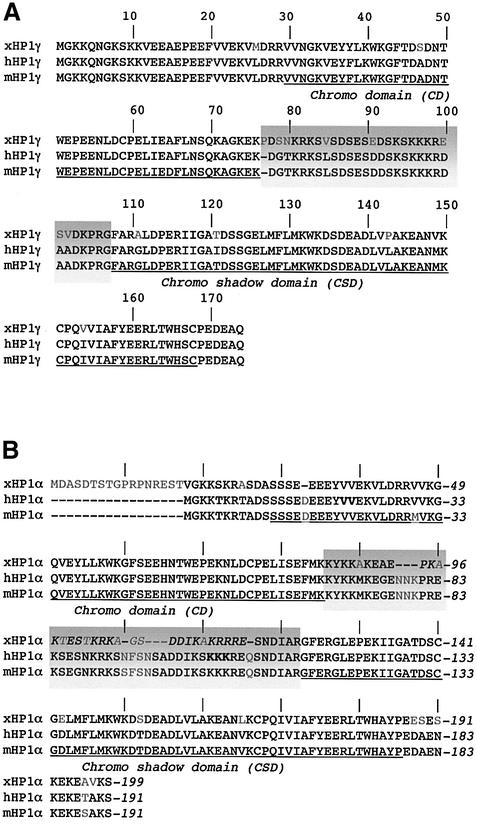
Partial sequences for xHP1α and xHP1γ truncated at the N-terminus have been isolated previously in a two-hybrid screen for proteins binding to xOrc1p (Pak et al., 1997). We have cloned full-length cDNAs for both HP1α and HP1γ from X.laevis oocytes by a combination of 5′ and 3′ RACE and deduced the complete ORF for each isoform [DDBJ/EMBL/GenBank accession Nos: AY168926 (xHP1γ) and AY168927 (xHP1α)]. The xHP1γ protein sequence is slightly longer (174 amino acids) than previously reported and different at two positions (G9S and A93S). We note that serine is present at both of these sites in mouse and human HP1γ (Figure 1A). Overall, xHP1γ shows 92% identity with its mammalian counterpart. A single amino acid change (A47S) occurs in the CD. There are five differences in the CSD between hHP1γ and xHP1γ, one of which (I121T) is also found in mHP1γ. The eight remaining amino acid changes are confined to the hinge region between the CD and CSD.
Fig. 1. CLUSTAL alignment of xHP1α and xHP1γ with their mammalian homologues. (A) HP1γ. The CD and CSD of mHP1γ are underlined. The hinge region is shaded. Divergent amino acids are in grey. (B) HP1α. The DNA and chromatin-binding domain of xHP1α (amino acids 94–117) is in italics. Mutations in amino acids 21–22 and 104–106 of the constructs hHP1α-2V and hHP1α-3K, respectively (see text), are shown in bold. Note that xHP1α has an N-terminal extension but a shorter hinge region.
In contrast, the protein sequence of xHP1α shows considerably more divergence from its human and mouse counterparts (74% identity). There is a novel 17 amino acid N-terminal extension within the 58 amino acids of extra sequence that we identified. Overall, xHP1α is eight amino acids longer than its mammalian homologue (199 amino acids; Figure 1B). The hinge region of xHP1α is only 39 amino acids long, compared with 47 amino acids for mHP1α, and displays many sequence differences. However, it does contain the hinge sequence KRK that is invariant among many HP1 proteins and may have a role in nuclear import and localization (Smothers and Henikoff, 2001).
Previous work has shown that HP1 isoforms from different species can form homodimers and heterodimers in vitro (Nielsen et al., 2001; and references therein). In view of the sequence divergence of xHP1s, we used this distinctive feature as a biochemical validation of their HP1 homology. Using a number of different in vitro binding assays (see Supplementary data available at The EMBO Journal Online), we confirmed that purified recombinant forms of xHP1α and xHP1γ can self-associate as well as associate with each other. The results were consistent with an interaction dependent on the CSD region of xHP1α and xHP1γ, as is the case for HP1 from other organisms (Li et al., 2002).
xHP1α, but not xHP1γ, has DNA-binding activity
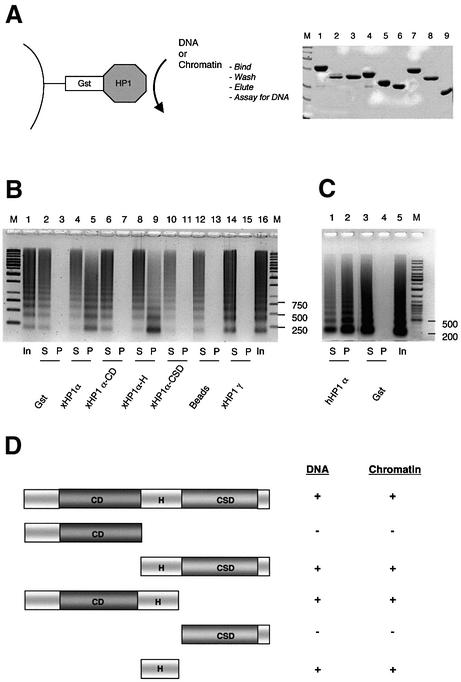
hHP1α was reported to have a DNA-binding activity localized to an internal 64 amino acid stretch comprising half the CD and most of the hinge region (Sugimoto et al., 1996). This region is less conserved between xHP1α and mammalian HP1α. We therefore determined whether xHP1α and xHP1γ had similar properties in this regard. We challenged GST–xHP1 fusion proteins immobilized on glutathione S–Sepharose beads with chicken oligonucleosomal DNA (Figure 2B and C). We found that xHP1α, like hHP1α, retained the DNA on the matrix, whereas xHP1γ or GST alone did not. Additional experiments using various truncated GST fusions (Figure 2A) were performed to define the minimum DNA-binding region of xHP1α. These narrowed down binding to within the hinge region itself, amino acids 94–117 (Figures 1B and 2B), and are summarized in Figure 2D. Thus, notwithstanding the sequence divergence in this segment, xHP1α, like hHP1α, has a non-specific DNA-binding activity, and this is localized to the hinge region. xHP1γ does not bind DNA, however. HP1 from Drosophila can also bind DNA, but in this case intact HP1 was required (Zhao et al., 2000).
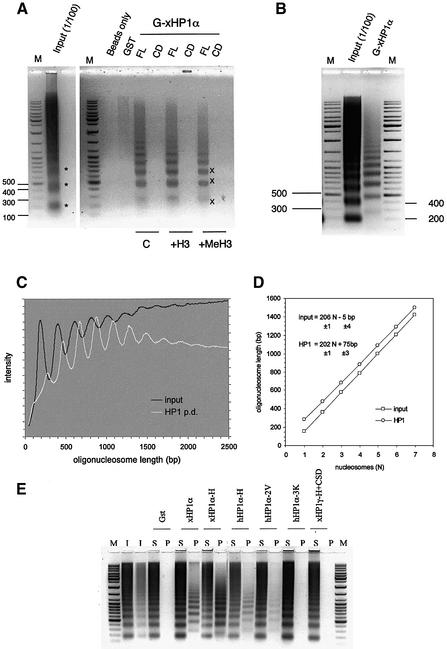
Fig. 2. xHP1α, but not xHP1γ, can bind DNA. (A) Scheme for analysing DNA or chromatin interaction with HP1. SDS–PAGE of purified GST proteins used in binding assays. M, protein size marker (Biorad); 1, xHP1α; 2, xHP1α-CD (chromodomain); 3, xHP1α-H+CSD (hinge plus chromoshadow domain); 4, xHP1α-CD+H (chromodomain plus hinge); 5, xHP1α-CSD (chromoshadow domain); 6, xHP1α-H (hinge); 7, xHP1γ; 8, xHP1γ-H-CSD; 9, GST. (B) Pull-down assay of chicken nucleosomal DNA (6 µg), incubated with 5 µg of GST or the indicated GST–xHP1 proteins coupled with glutathione–Sepharose. Bound (P, pellet) and unbound (S, supernatant) DNA was visualized on an ethidium bromide stained 1.5% agarose gel. xHP1α and xHP1α-H bound DNA but not the xHP1γ, xHP1α-CD or xHP1α-CSD GST fusions. M, DNA size marker; In, input DNA. (C) hHP1α (5 µg) also binds DNA in this assay. (D) Summary of the DNA and chromatin-binding properties of xHP1α (includes data not shown; –, no binding; +, binding). The hinge region of xHP1α can bind DNA and chromatin.
xHP1α, but not xHP1γ, has a chromatin-binding activity localized to the hinge region
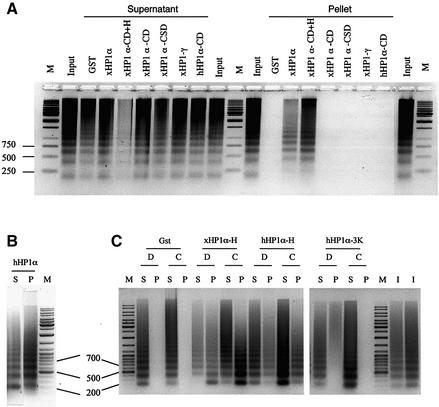
It has been shown that Drosophila and mammalian HP1 can associate with nucleosomes in vivo and in vitro (Zhao et al., 2000; Nielsen et al., 2001). To test whether xHP1 proteins can bind to chromatin directly, the pull-down assay described above was performed with native soluble oligonucleosomes from chicken nucleated erythrocytes (also found in Xenopus). Any resulting interaction was detected by resolving on a DNA agarose gel if a chromatin ladder was present in the pellet fraction. Chromatin was retained on the matrix by GST–xHP1α (but not by GST alone); in contrast, up to 20 µg of GST–xHP1γ did not pull down any chromatin in our assay (Figure 3A; data not shown). Although HP1γ does not show chromatin (or DNA) binding activity, it can, with the assistance of HP1α, associate indirectly with chromatin at least in vitro. This was verified by adding His6-tagged xHP1α to GST–xHP1γ coupled with glutathione beads and using the heterodimers in a pull-down assay (data not shown). We note that HP1α and HP1γ are mostly found in different sub-nuclear locations in vivo.
Fig. 3. HP1α, but not HP1γ, can bind chromatin. (A) In vitro pull-down assay of native chicken chromatin (12 µg), incubated with 5 µg of GST or the indicated Xenopus or human GST–HP1 proteins coupled with glutathione–Sepharose. DNA extracted from the bound chromatin (P, pellet) and unbound chromatin (S, supernatant) was visualized as in Figure 2B. Only GST–xHP1α and xHP1α-CD+H (chromodomain plus hinge) interacted with chromatin. (B) Full-length hHP1α binds chromatin. (C) Same assay using 12 µg of native chicken chromatin (C) or the DNA derived from it (D) with 5 µg of GST–hinge proteins xHP1α-H (94–117), hHP1α-H (67–119) or full-length hHP1α-3K carrying mutations K104–6A in the hinge region.
The interaction of mammalian HP1 with chromatin has not previously been reported as being isoform specific. Using the same series of truncated GST fusions employed to identify the DNA-binding domain (Figure 2A and D), we found that the chromatin-binding domain of xHP1α was also localized to the hinge region (Figure 3A, compare xHP1α-CD+H with xHP1α-CD). Neither the CD nor the CSD of xHP1α can bind chromatin in our assay (Figure 3A). The CD region of hHP1α was also unable to bind chromatin (Figure 3A, hHP1α-CD), whereas full-length hHP1α could (Figure 3B). The GST–hinge fusion containing amino acids 94–117 of xHP1α (xHP1α-H) was sufficient and possibly more efficient in binding chromatin than DNA (Figure 3C). Binding of chromatin and DNA was also observed with the human GST–hinge (67–119) construct hHP1α-H. Interestingly, within the context of the full-length hHP1α sequence, a triple mutation K104–6A in the hinge region (hHP1α-3K) was sufficient to abolish the ability of the protein to bind chromatin and also severely compromised its DNA-binding ability (Figure 3C). This mutation is known to prevent RNA binding (Muchardt et al., 2002). These results prove that the short hinge region sequence enables hHP1α and xHP1α to bind chromatin and DNA in vitro. Drosophila HP1 binding to nucleosomes was reported to require intact native HP1 (Zhao et al., 2000). Surprisingly, our results do not support an HP1 association with native chromatin mediated by the CD region as seen with nucleosome core particles (Bannister et al., 2001).
Both xHP1α and xHP1γ can bind histone H3 tails in vitro when methylated at Lys9
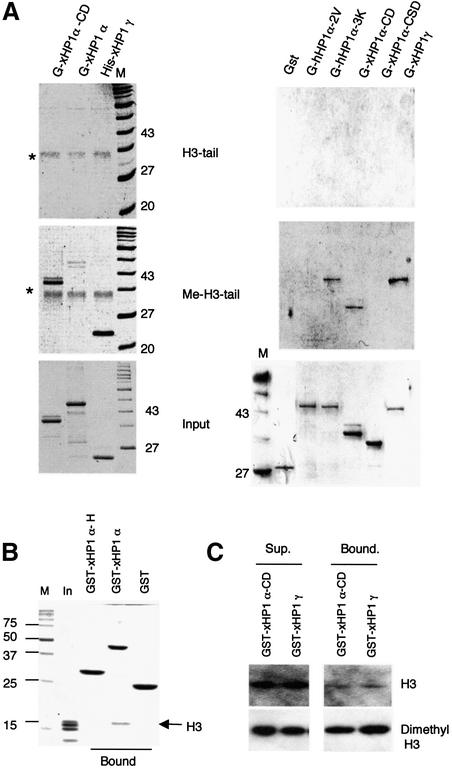
Two reports (Bannister et al., 2001; Lachner et al., 2001) showed that all three mammalian HP1 variants can bind to histone H3 tails methylated at Lys9 via their CDs. Since we failed to observe this binding activity with native chromatin, we used antibodies that specifically detect histone H3 (Lys9) dimethylation or trimethylation to confirm that a proportion of histone H3 tails in chicken chromatin is methylated at this residue (see Supple mentary data). Unlike the globular domains of the core histones responsible for most of the histone–histone and histone–DNA interactions within the nucleosome, the N-terminal regions (tails) of the histones are more mobile (Cary et al., 1978). Therefore, we investigated the contribution of histone H3 tail binding by challenging recombinant HP1 proteins with amino acids 1–21 of histone H3 attached to an affinity matrix, either in an unmodified form or dimethylated at Lys9.
In accordance with the results of Lachner et al. (2001) for hHP1α (data not shown), we found that xHP1γ (whether a GST or His fusion) as well as xHP1α can also bind to methylated histone H3 peptide (Me-H3-tail, Figure 4A) but not its unmodified counterpart (H3-tail, Figure 4A). The affinity of xHP1α for the methylated H3 tail is reproducibly lower. This is possibly due to structural hindrance, as the CD of xHP1α in isolation had the same preference for Lys9-dimethylated histone H3 peptide as xHP1γ. As expected, xHP1α-CSD did not bind the peptides. The hHP1α-3K hinge mutation did not prevent full-length HP1 from binding methylated peptide (although it abolished binding to chromatin; Figure 3C). A V21M-V22A double mutation in the CD of full-length hHP1α-2V effectively abolished binding to methylated peptide (but not to chromatin; see Figure 5E). These results illustrate a disconnection between the two modes of binding. Far-western analysis of native core histones confirmed the specificity of the xHP1α interaction with histone H3 (see Supplementary data). In addition, we also detected an interaction between linker histones and xHP1α. An interaction with histone H1 was also observed for mHP1α and localized to its hinge region (Nielsen et al., 2001). Far-western analysis showed that the hinge region of xHP1α could bind histone H1 and also histone H5 when used as a probe but that H5 was easily displaced by H1 (see Supplementary data).
Fig. 4. The chromodomains of xHP1α and xHP1γ can bind methylated Lys9 histone H3 tails. (A) Pull-down assays of the indicated GST and His-tagged proteins with affinity matrices presenting either unmodifed (H3-tail) or dimethylated Lys9 (Me-H3-tail) biotinylated histone H3 tail peptides. Lower, SDS–PAGE gels, input HP1 proteins. Middle, proteins bound to the dimethylated histone H3 tail. Top, no proteins bound the unmodified histone H3 tail (*, avidin matrix). Note that hHP1α-2V carrying mutations V21A-V22M in the chromodomain does not bind dimethylated Lys9, but hHP1α-3K mutated in the hinge does. (B) Pull-down experiment with free native chicken core histones separated on 15% SDS–PAGE. GST–xHP1α, but not GST–xHP1α-H, can interact with histone H3 (arrow). (C) Western blot analysis with antibodies specific for dimethylated Lys9 of histone H3 or total H3 tail shows that the GST–xHP1α-CD and full-length GST–xHP1γ can also pull down histone H3 (Bound) and that this interaction is preferentially for the methylated form of histone H3.
Fig. 5. xHP1α preferentially binds a subfraction of soluble chicken chromatin. (A) Agarose gel showing pull-down fractions from a high concentration of native chromatin (300 µg) with GST, and full-length (FL) or the chromodomain (CD) of xHP1α, in the absence or presence of 2 µM unmodified (+H3) and dimethylated Lys9 histone H3 tail peptides (+MeH3). Note that the CD did not bind chromatin and that the histone H3 tails did not compete with xHP1α binding. The monomer, dimer and trimer of the input chromatin (asterisks) and shifted pull-down pattern (X) are marked. (B) The input and xHP1α pull-down from (A) side by side, showing the distinct nucleosomal ladder patterns. (C) Scan of the same lanes from (B) converted to a linear base-pair scale. (D) Oligonucleosome length was plotted versus nucleosome number to determine the nucleosomal repeat lengths of the two chromatin samples, which were similar. The main size differences observed are due to an extra length of DNA (of 75 ± 3 bp) in the xHP1α pull-down nucleosome arrays. (E) The hinge regions of xHP1α or hHP1α can pull down the altered chromatin pattern as in (A). Mutations (hHP1α-2V) in the CD region do not prevent the interaction with this chromatin, whereas mutations in the hinge region (hHP1α-3K) do. In addition, an xHP1γ-H+CSD (77–174) truncation mutant cannot interact with chromatin under these conditions.
An important point remains that the same constructs xHP1γ, the CD of xHP1α, and hHP1α-3K that can bind exposed methylated histone H3 tails are unable to interact with native chicken chromatin under the same conditions. These data support the notion that it is the hinge region of xHP1α (and of hHP1α) that has the predominant intrinsic chromatin-binding activity. However, a surprising implication is that histone H3 (Lys9) methylated tails may not be accessible to HP1 in native soluble chromatin. To verify this possibility, we isolated the core histones from the soluble chromatin by salt elution and used these as input in a pull-down assay. Figure 4B shows that xHP1α, but not its hinge, was indeed able to interact with histone H3 when the core histones were presented outside of their chromatin context. Western blotting confirmed that H3 histones pulled down by the CD of xHP1α or xHP1γ were significantly enriched for Lys9 dimethylation (Figure 4C).
The hinge of HP1α interacts preferentially with chromatin that has an altered nucleosomal pattern
Bannister et al. (2001) used 200 µg of core particles in their pull-down assays and found ∼1% of the input associated with mouse GST–HP1β, a binding attributed to the CD. In our 20 mM salt condition, neither the CSD nor the CD can bind oligonucleosomes, and we attribute the almost quantitative binding by full-length HP1α to the hinge region. To resolve this discrepancy, we repeated our pull-down experiment with 300 µg of input chromatin instead of 5–10 µg, but we found that the CD of xHP1α still showed no detectable binding (Figure 5A). Under the same conditions, full-length xHP1α was very efficient in interacting with this substrate, whereas xHP1γ was unable to bind chromatin (data not shown, but see Figure 6A).
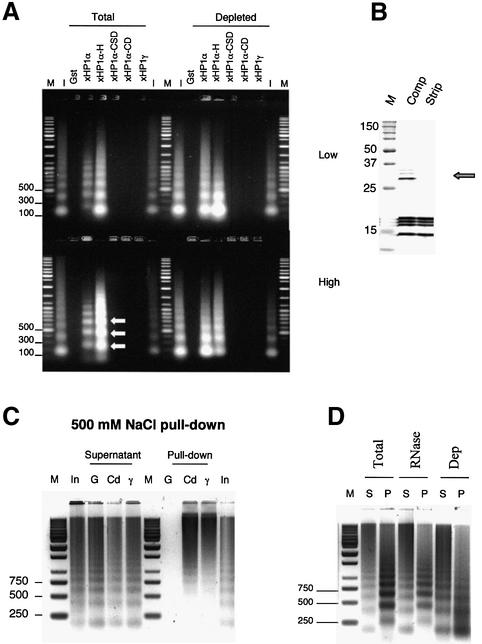
Fig. 6. Linker histones are required to detect the alternate chromatin pattern pulled down by xHP1α. (A) Pull-down assay using 5 and 50 µg of total or linker histone-depleted soluble chicken chromatin with GST and various GST fusions of xHP1α and xHP1γ coupled with glutathione–Sepharose. Input (1 and 0.5 µg) and bound fractions are shown. The alternative chromatin pattern (arrows) is observed with xHP1α and its hinge region only at a high input of complete chromatin, but interaction also occurs with linker histone-depleted chromatin. Note that xHP1γ and xHP1α-CD cannot bind chromatin, whether depleted of linker histones or not. (B) A Coomassie-stained 15% SDS–PAGE showing the protein composition of complete and stripped chromatin (no linker histones H5, H1a and H1b, arrow). (C) Pull-down assay performed in 500 mM NaCl using poly-lysine preincubated Sepharose beads to prevent non-specific binding. In this situation the chromodomain (Cd) of xHP1α and full-length xHP1γ can pull down chromatin, but GST (G) cannot. The resulting pattern contains a less digested fraction of the input nucleosomal ladder (In). (D) GST–xHP1α pull-down experiment with 200 µg of soluble chromatin (Total) or chromatin pre-treated with RNase A (RNase) or depleted of linker histones (Dep). RNase A treatment does not prevent the preferential interaction with the altered chromatin, whereas loss of linker histones does.
A striking observation is that the nucleosomal pattern of chromatin pulled down by xHP1α was altered (Figure 5A, B and C) when input native chromatin was in large excess. Quantitative analysis indicates that, in these conditions, xHP1α pulled down a specific population of chromatin with a similar nucleosome repeat length of 202 ± 1 bp to the input chromatin of 206 ± 1 bp. However, this pull-down population appeared to have additional DNA extending from the ends of the nucleosomal arrays (or, alternatively, internal gaps), of a total length of 75 ± 3 bp compared with none in the input (Figure 5D). This difference accounts for the upward shift of the nucleosome ladder on agarose gels whenever xHP1α was presented with excess input chromatin (Figure 5B). The pull-down fraction may represent chromatin of a specific sequence or, alternatively, an altered configuration within the input soluble chromatin. One possibility was that this fraction contained methylated histone H3 and that this interacted with xHP1α. However, in a competition experiment, we found that the addition of unmodified or dimethylated histone H3 tails did not affect xHP1α chromatin binding (Figure 5A).
Figure 5E shows that the hinge region xHP1α-H is responsible for the shifted nucleosomal ladder pulled down by xHP1α. This altered pattern was also selected by hHP1α-H (and full-length hHP1α; data not shown) and by full-length hHP1α-2V carrying the CD mutations (Muchardt et al., 2002), but not by hHP1α-3K carrying the triple hinge mutation. The xHP1γ hinge presented as an xHP1γ-H+CSD truncation was unable to bind excess input chromatin any better than in its full-length context, despite the similar lysine-rich character of the γ-hinge sequence.
Detection of the alternative chromatin fraction depends on the presence of linker histones
Since HP1α can bind linker histones, it is possible that this mediates the interaction with soluble chromatin in our assay. We therefore tested the ability of xHP1α to bind chromatin that had been stripped of linker histones (Figure 6B). Figure 6A shows that both full-length xHP1α and the hinge region are able to pull down with low (5 µg) and high (50 µg) amounts of either linker histone-depleted or complete chromatin, indicating a linker histone-independent association. At low input, the pattern of the pull-down fractions is similar (Figure 6A). However, at high input, we observe an upward shift in the pattern, but only with complete chromatin (arrows in Figure 6A) and not with stripped chromatin. This suggests that the interaction of xHP1α with the alternative chromatin fraction is dependent on the presence of linker histones. The association is not reliant on ionic conditions favouring the formation of the linker histone-containing higher-order chromatin structure (data not shown) but may involve direct binding to linker histones.
We also tested the ability of xHP1γ or the isolated CD and CSD domains of xHP1α to bind stripped chromatin, as it was possible that the presence of linker histones prevented access to docking sites for these proteins. However, no interaction was observed under the conditions tested (Figure 6A). Finally, we performed a chromatin pull-down assay in non-physiological 0.5 M NaCl buffer. In the range of 0.3–0.6 M NaCl, ionic interactions are severely reduced and the histone tails are mobilized, although the chromatin structure remains intact (Cary et al., 1978). Furthermore, pull-down experiments are often conducted at 300 mM NaCl (Bannister et al., 2001). Interestingly, in high salt, the CD of xHP1α and xHP1γ could pull down chromatin, and it consisted of a longer and thus relatively nuclease resistant fraction of the input chromatin (Figure 6C).
HP1 has recently been shown to bind RNA (Muchardt et al., 2002). We therefore tested the possibility that an RNA fraction naturally associated with the native chromatin might mediate the interaction with the alternative chromatin structure. However, RNase treatment of the input chromatin did not affect the observed upward shift in the pull-down pattern (Figure 6D).
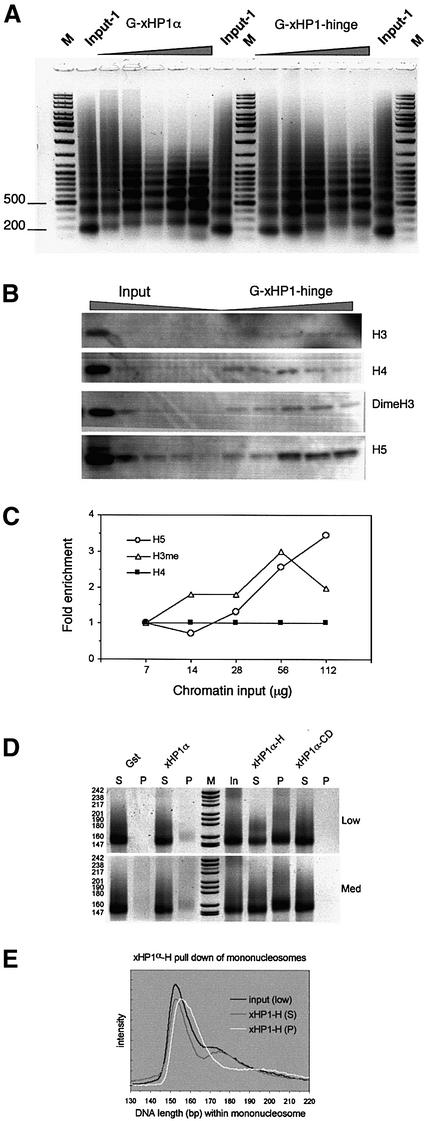
To determine whether there was a transition point at which the alternate chromatin ladder appeared, we titrated fixed amounts of xHP1α or the hinge region with increasing amounts of soluble chromatin. Figure 7A shows that the change in pattern is gradual, with the pull-down fraction exhibiting an increasingly shifted ladder in the range 7–112 µg of input chromatin. The inverse result was observed when a fixed amount of chromatin (50 µg) was titrated with increasing amounts of xHP1α or the hinge region (see Supplementary data), confirming preferential binding of this alternative chromatin. Western blotting of the chromatin pulled down from an increasing input range with a fixed amount of xHP1α or the hinge region (Figure 7B; data not shown) showed a sharp gradual increase of the level of histone H5 (data plotted in Figure 7C; we note histone H1 could not be visualized at these low levels with a number of antibodies tested). Interestingly, an increase was also observed for the level of histone H3 (Lys9) dimethylation (Figure 7B and C).
Fig. 7. The alternative chromatin pattern appears as the chromatin-to-xHP1α ratio increases. (A) Pull-down assay of increasing amounts (7, 14, 28, 56 and 112 µg) of soluble chicken chromatin incubated with GST, GST–xHP1α or GST–xHP1-hinge coupled with glutathione– Sepharose. Note the gradual upward shift relative to the Input in the bound nucleosomal ladders shown. GST pull-downs were negative (data not shown). (B) Proteins in the GST–xHP1-hinge pull-down samples were separated by 15% SDS–PAGE, western blotted and probed with antibodies against histone H3, histone H4, dimethylated H3 and histone H5. A dilution series of input chromatin is also shown (10, 1, 0.5, 0.25 and 0.125 µg). (C) The signal in (B) was quantitated, normalized against the histone H4 signal and plotted as fold enrichment relative to the data point with the lowest chromatin input. The alternative chromatin structure is enriched for histone H5 but also for dimethylated H3 (Lys9). (D) Pull-down assay of native chicken mononucleosomes (Low, 4 µg; Med, 16 µg) with GST or the indicated GST–xHP1α constructs. DNA from the input (I), pellet (P) and supernatant (S) fractions was visualized on a 12% polyacrylamide gel. Note that the chromodomain does not bind mononucleosomes, including core particles. (E) Scan showing slightly different DNA length distributions (on a linear base-pair scale) of the input, supernatant and pull-down lanes of xHP1α-H from the 4 µg assay. There is also a slight upward shift in the xHP1α pull-down pattern of (D).
We also investigated the binding preferences of full-length xHP1α, hinge and CD region using a native soluble mononucleosome substrate containing core particles as well as longer mononucleosomes. xHP1α was less efficient in binding mononucleosomes and seemed to prefer particles with some linker DNA over core particles, as indicated by the upward shift compared with the input (Figure 7D). The hinge region bound mononucleosomes very efficiently. At limiting amounts of input mononucleosomes, the residual supernatant and pull-down patterns were again suggestive of different DNA length distributions (Figure 7E). These shifts were modest in spite of the availability of longer linker-containing nucleosomes, however. Interestingly, the xHP1α-CD was not able to pull down even core particles (Figure 7D), leaving high salt as the only condition in which it could bind chromatin (Figure 6C).
The alternative chromatin pattern represents a nuclease resistant fraction
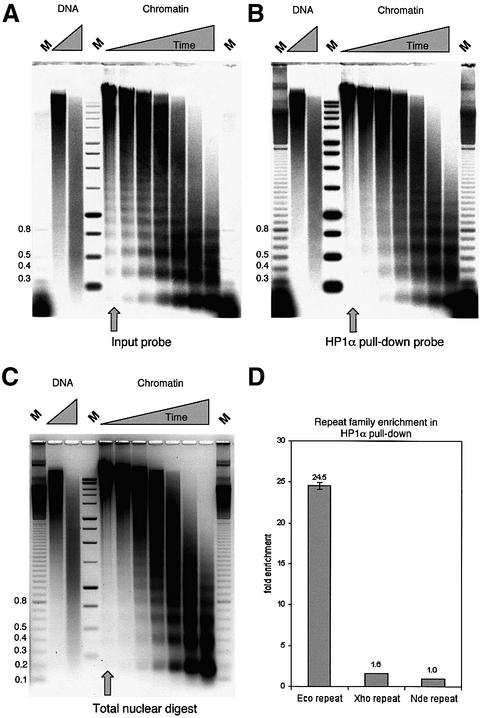
To determine the nuclear origin of the alternative chromatin species, isolated trimer–tetramer DNA from input and pull-down nucleosomal ladders (Figure 5B) was used as a probe against Southern blots of a time-course digestion of chicken erythrocyte nuclei with micrococcal nuclease. Figure 8C shows the ethidium bromide pattern of total digested nuclei, in which a nucleosomal ladder appears at later time points. The probe derived from the input trinucleosome/tetranucleosome fraction detects a nucleosomal ladder from the initial time point onwards (Figure 8A). This short soluble chromatin is normally enriched for active chromatin, which is known to be nuclease sensitive (Weintraub et al., 1978). In contrast, the xHP1α pull-down trimer–tetramer probe detects a pattern that is distinct from the input probe (Figure 8B) and more similar to the total nuclear ethidium bromide pattern. This suggests that the shifted pull-down ladder represents a less nuclease sensitive fraction within the input soluble chicken chromatin. The soluble input probe detects a nucleosomal ladder with a shorter repeat length compared with the pull-down probe (visible in Figure 8A and B). However, the pull-down probe does not light up a shifted ladder in the total nuclear chromatin pattern, suggesting that this shift is not linked to particular DNA sequences.
Fig. 8. The xHP1α pull-down fraction detects more nuclease-resistant chromatin. DNA agarose gel and Southern blot of chicken erythrocyte whole nuclei and DNA digested with micrococcal nuclease (DNA, 1 and 2 min; nuclei, 0.5, 1, 2, 4, 8, 16 and 32 min). Hybridization was with isolated trimer–tetramer DNA from (A) the input soluble chromatin or (B) the xHP1α pull-down fraction of Figure 5B. (C) The ethidium bromide stained gel. The xHP1α pull-down fraction does not contain the nuclease-sensitive, presumed active chromatin material for which the soluble input chromatin is enriched. (D) Spot blot analysis of 20, 15, 10 and 5 ng of xHP1α pull-down and input probe DNA from (A) and (B), spotted on nylon. The same filter was consecutively probed with the EcoRI (twice), XhoI and NheI chicken repeat family sequences. The hybridized signal per nanogram was determined by phosphoimaging and linear regression analysis. Pull-down over input ratios for the EcoRI and XhoI repeat were normalized against the NheI repeat to give their enrichment within the xHP1α pull-down fraction.
EcoRI and XhoI repeats are localized to W-hetero chromatin in chicken fibroblast cells, and this is also where the chicken heterochromatin protein CHCB1 is found (Yamaguchi et al., 1998). Dot blotting demonstrates that the pull-down fraction is enriched for the chicken EcoRI but not the closely related XhoI repeat or the unrelated Z-terminal NheI repeat (Figure 8D). The enrichment is proportional to the amount of excess input chromatin and is also seen with hHP1α (data not shown). We note that our results are unlikely to be influenced by endogenous chicken HP1, since the chromatin used in these experiments does not contain significant amounts of nuclear protein other than histones (Figure 6B). In addition, no HP1 proteins are detected in chicken erythrocyte nuclear extracts (J.Allan, personal communication).
Discussion
The significant sequence variation between amphibian and vertebrate HP1α prompted us to compare the biochemical properties of xHP1α and xHP1γ in vitro. Both of these proteins can form homodimers and heterodimers, consistent with the CSD dimerization typical of HP1 homologues. Both exhibit the characteristic CD interaction with methylated histone H3 tails. Yet clear differences emerged between xHP1α and xHP1γ in DNA and native chromatin pull-down assays. Only xHP1α, but not xHP1γ, can bind DNA (Figure 2) or chromatin (Figures 3 and 6A). In both cases, binding activity was localized to the hinge region. The binding region consists of 24 amino acids with only 50% consensus to mammalian counterparts (Figure 1B). Despite this sequence divergence, xHP1α was not found to behave differently from hHP1α in our binding assays.
The interaction of the HP1α hinge with chromatin seems primarily mediated by DNA. Firstly, the hinge region binds naked DNA, but it does not bind to any of the core histones (Figure 4B). Secondly, although we observe an interaction with linker histones, their presence is not required because the hinge also binds depleted chromatin very well (Figure 6A). An important conclusion from our work is that the interaction involving the HP1α hinge is the predominant association responsible for the high affinity of full-length xHP1α and hHP1α for native oligonucleosomes. Truncated constructs and point mutations demonstrate that the HP1α-CD and HP1α-CSD domains do not contribute any individual contacts to this association (Figures 3, 5E and 6A).
The lack of any affinity of the CD for native oligonucleosomes was very surprising. Its known association with the methylated Lys9 residue of histone H3 (Bannister et al., 2001; Lachner et al., 2001) was proposed to be responsible for targeting HP1 to heterochromatin-forming regions. A demonstration of the importance of this methylated mark in cells is that removal of the histone H3 (Lys9) methylase Suv39h disrupts the formation of pericentromeric heterochromatin in vivo (Lachner et al., 2001). Yet, our evidence points to the inaccessibility of the methylated Lys9 residue to the HP1 CD in preformed chromatin isolated from nuclei. Both components of this interaction are existent and functional in our system. First of all, methylated Lys9 is present in histone H3 of the chicken oligonucleosomes we use. Our recombinant HP1 CD constructs are able to interact specifically with synthetic methylated histone H3 tail peptides, although point mutations in full-length hHP1α indicate that this interaction is independent of the ability to bind native chromatin. Finally, the HP1-CD-containing constructs that fail to bind the intact oligonucleosome substrate do associate with histone H3 specifically when presented with free core histones isolated from this chromatin. These bound histone H3 molecules are enriched for Lys9 methylation (Figure 4).
So what explains the absence of an interaction of the HP1 CD with methylated Lys9 of histone H3 of native chromatin fragments? The linker histones are not responsible for shielding this possible contact, as their removal has no effect (Figure 6A). However, the presence of linker DNA between consecutive nucleosomes is likely to be significant. Recent cross-linking experiments have shown that, in oligonucleosomes, at ionic strengths ranging from 10 to 120 mM NaCl, the majority of core histone tails are bound to the linker DNA (Angelov et al., 2001). In particular, the N-terminal tail of histone H3 exhibited increased cross-linking in linker-containing oligonucleosomes compared with core particles. The combined electrostatic interactions between the N-terminal arginines plus lysines and DNA are expected to produce a very high affinity of the histone H3 tail for linker DNA at low ionic strength [for comparison, the KD of the binding of histone H4 (1–23) peptide to free DNA was determined to be 2 × 10–12 M (Hong et al., 1993)]. Only at ionic strengths of 0.3–0.6 M NaCl do the histone tails dissociate and become fully mobile (Cary et al., 1978).
Significantly, the KD of the binding of methylated N-terminal histone H3 peptides into the fitted site of the HP1 CD was determined to be only ∼5 × 10–6 M (Jacobs and Khorasanizadeh, 2002; Nielsen et al., 2002). The likelihood that the affinity of histone H3 tails for HP1 is a few orders of magnitude lower than for DNA means that this interaction would not be competitive in the range of low to physiological ionic strengths. Thus, the histone H3 tails need not be inaccessible inside the chromatin structure in order to preclude their interaction with the HP1 CD; besides, they are known to be accessible to trypsin or antibodies. Instead, the likely explanation is that, in native oligonucleosomes, the histone H3 tails are stoichiometrically engaged in electrostatic interactions with linker DNA, with which the weaker HP1 binding can neither coexist nor compete. This hypothesis was confirmed by performing a chromatin-binding assay in non-physiological 0.5 M NaCl buffer, so as to abolish electrostatic interactions with DNA (of histone H3 tails as well as HP1 hinge). Under these conditions, both HP1α and HP1γ (which did not interact with chromatin before) were now able to bind chromatin via the CD. The pull-down pattern corresponded to a nuclease-resistant fraction of the input chromatin of plausible heterochromatic origin in the nucleus (Figure 6C). In spite of a somewhat weaker association of histone tails in core particles, no HP1α-CD binding occurred in low ionic conditions, suggesting that the high salt in the original experiments contributed to the observed interaction.
The question remains whether the non-physiological circumstances of this binding of histone H3 tails in preformed chromatin might occur in vivo. Chromatin remodelling complexes, chaperones, HP1 binding partners, RNA, as well as histone tail or HP1 hinge modifications reducing their positive charge, could all plausibly change local ionic conditions at the chromatin so as to make the histone H3 tails available for HP1 binding. However, these changes would not improve upon the binding parameters of the methylated Lys9 segment for HP1 and would therefore need to be permanent for this interaction to survive. xHP1γ has no other mode of chromatin binding, although its localization may depend on association with other proteins. Alternatively, nearby exposed methylated histone H3 tails may bind dimers of HP1 cooperatively, enhancing the overall affinity. On the other hand, a transitory weak interaction with the methylated Lys9 residue of histone H3 might be sufficient to target xHP1α to heterochromatin-forming regions of the nucleus, where it could be stabilized by subsequent strong binding through the hinge.
The targeting of HP1 to methylated Lys9 of histone H3 is not absolute, as the mere presence of this modification is not sufficient for HP1 binding to chromosomes (Cowell et al., 2002). The alternative, straightforward interpretation of our data is that, under physiological conditions, HP1α does not bind preformed nuclear chromatin through interaction of the HP1 CD with histone H3 methylated Lys9, but rather through the very strong HP1-hinge–DNA interactions that we have identified. In view of the undeniable importance of the methylated mark for the nuclear organization of pericentromeric heterochromatin in vivo, we propose that this interaction might occur instead at replicating chromatin, prior to the association of histone H3 tails with the linker DNA. In this view, the heterochromatic condensed chromatin structure would be assembled de novo rather than being imposed on a pre-existing template. This may explain some of the features of heterochromatin, such as its very regular nucleosome array spacing (Richards and Elgin, 2002). The observed association of HP1 with ORC proteins (Pak et al., 1997) could signify a link with silencing as well as with replication.
The other conclusion derived from our results is that, apart from the non-specific DNA-mediated chromatin-binding activity, the HP1 hinge can also impose specificity mediated by linker histones. Linker histones can be contacted directly by the xHP1α hinge, as has been observed for murine HP1α (Nielsen et al., 2001). The striking upward shift in the nucleosome ladder suggests that this is accompanied by an additional preference for up to 75 bp extra length of DNA (Figure 5). Drosophila HP1 was shown to have an affinity for oligonucleotides that increased with DNA length (Zhao et al., 2000). This length requirement may become apparent if less free linker DNA is available when linker histones are in place. The shift in the pull-down pattern may, for example, be caused by enrichment for linker histone–DNA end complexes. The mononucleosome pull-down assay shows a limited preference for longer linker lengths compared with core particles.
The oligonucleosome pull-down titration does not reveal a bias for binding the most undigested material in the input. Given a large excess of chromatin, full-length HP1α and hinge nevertheless first titrate chromatin from a relatively digestion-resistant fraction of the nucleus, which is strongly enriched in linker histones and EcoRI repeat sequences (Figures 7 and 8) normally localized to W-heterochromatin in chicken embryonic fibroblast cells (Yamaguchi et al., 1998). Linker histones are an essential component of nucleosomal arrays forming the highly folded transcriptionally repressive 30 nm diameter chromatin fibre (van Holde, 1988), but many details about this structure are not known. Our observation of linker histone enrichment may tie in with proposals that chromatin condensation levels depend on linker histone load or subtype composition (Bates and Thomas, 1981; Coles et al., 1987). The chromatin pulled down by the HP1α hinge is also enriched for histone H3 Lys9 dimethylation (Figure 7C and D). This is interesting, because the hinge region itself has no affinity for histone H3. Thus, this enrichment seems to be a corollary of the preference of the HP1α hinge for heterochromatic regions and implies that HP1 can target correctly to such regions independently of the CD.
This study of xHP1α and xHP1γ provides some clarification as to why HP1 subtypes have different distributions in nuclei. Although all isoforms are capable of the CD–H3 methylated Lys9 interaction, it appears that this weak binding is strongly regulated by the availability of histone tails and does not occur in preformed chromatin. In contrast, isoforms vary greatly in the affinity of their hinge region for chromatin via DNA and linker histones, ranging from no binding at all (xHP1γ) to a very strong binding with a predisposition for likely heterochromatic regions (xHP1α). This mimics their respectively diffuse and heterochromatic localizations in vivo. In agreement with our in vitro analysis, sequence variations within the hinge account for HP1 targeting distinctions in Drosophila (Smothers and Henikoff, 2001). The similar behaviour we observe for xHP1α and hHP1α suggests that hinge regions may be functionally preserved in spite of their poor sequence conservation.
HP1 is an exceptionally versatile protein in its interactions with numerous binding partners, mostly through the CD and CSD (Li et al., 2002). It is likely to be involved in different associations during the formation, spreading and heritable transmission of pericentromeric heterochromatin, as well as at its many non-heterochromatic sites. This study using preformed native chromatin substrates separates a plausible role for the xHP1α hinge region in targeting and spreading of heterochromatin from the role of the CD, which seems to necessitate a remodelling of chromatin that could be linked to heterochromatin formation.
Materials and methods
Recombinant proteins
xHP1α and xHP1γ cDNAs were subcloned in-frame into vectors pET6H and pGEX-2T. PCR primer sequences used to produce the various His-tagged and GST-tagged constructs are available upon request. Fusion proteins were expressed in Escherichia coli and purified by affinity chromatography according to Pharmacia Biotech protocols. hHP1 constructs are described in Cowell and Austin (1997), Muchardt et al. (2002) and Nielsen et al. (2001).
Pull-down assays for chromatin, DNA and histone proteins
GST proteins were coupled with 20 µl of glutathione–Sepharose 4B matrix (Pharmacia Biotech) in 20 mM NaCl, 1× HEPES buffer (10 mM HEPES, 0.5 mM EGTA, 1 mM DTT, 10% glycerol, proteinase inhibitor cocktail). Chicken erythrocyte chromatin, chromatin-derived DNA or purified histones were added in 200 µl of the same buffer. The mix was incubated at 20°C for 1 h, followed by three buffer washes. DNA was recovered from the resuspended pellet and supernatant fractions by proteinase K digestion, phenol/chloroform extraction and ethanol precipitation and analysed on 1.5% agarose TAE gels. For concomitant protein analysis of the chromatin pull-down fractions, samples were resuspended in 1.5% SDS, and proteins were recovered by acetone precipitation of the phenol/chloroform phase after DNA extraction. Core histones interaction assays used Sepharose beads preincubated with poly-lysine (100 µg/ml) to prevent non-specific binding. GST proteins (10 µg) were coupled with 25 µl of beads and mixed with 100 µg of purified chicken histones in 500 µl of 500 mM NaCl, 1× HEPES buffer. Recovered proteins were resuspended in loading buffer and analysed by 15% SDS–PAGE and western blotting.
Histone H3 tail pull-downs
HP1 binding reactions were as above but with 2 µg of biotinylated amino acids 1–21 of histones H3, either unmodified or dimethylated at Lys9 (Upstate Biotech), coupled with 25 µl of tetrameric avidin resin (Promega).
Western blotting
SDS–PAGE separated histone proteins were transferred onto PVDF. After blocking, membranes were incubated with primary antibodies against histone H4, histone H3, dimethylated H3 (Lys9) (Upstate Biotech) or histone H5. Positive signals were detected using secondary HRP-linked antibody and ECL reagent (Pharmacia Biotech).
Chromatin preparation
Chicken erythrocyte nuclei (2.25 DNA mg/ml) were digested with 60 units/ml micrococcal nuclease (Worthington) for 8–10 min at 37°C in nuclease digestion buffer (250 mM sucrose, 10 mM Tris–HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1 mM PMSF), adding 1 mM CaCl2. Digestion was stopped on ice by adding 5 mM EDTA. After gently spinning down, nuclei were lysed by vortexing in TE buffer (10 mM Tris–HCl pH 7.4, 10 mM NaCl, 0.2 mM EDTA, 0.1 mM PMSF). Soluble chromatin released in the supernatant after pelleting the nuclear debris was kept on ice and used within 10 days.
Depleted (H1/H5) chromatin and core histone preparation
Soluble chicken chromatin was bound onto hydroxylapatite (HAP; Calbiochem) in 10 mM potassium phosphate pH 6.8. Linker histones were extracted with 600 mM NaCl, 10 mM potassium phosphate. The ‘H1/H5-depleted chromatin’ was eluted from HAP in 150 mM potassium phosphate pH 6.8, dialysed to TE buffer at 4°C and used fresh in chromatin pull-down experiments. Alternatively, core histones were prepared from H1/H5-depleted chromatin bound to HAP by extraction with 2 M NaCl, 10 mM potassium phosphate.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Jim Allan for H5 antibody; Bill Earnshaw, Pierre Chambon, Christian Muchardt and Caroline Austin for hHP1 constructs; and Shigeki Mizuno for chicken repeat family clones. We thank D.Pearson (Grampian Chickens) for chicken erythrocytes. We thank Robin Allshire, Janet Partridge and members of the chromatin laboratories for discussions. This work carried out in the R.M. laboratory and S.P. laboratory was funded by grants from the Wellcome Trust. C.-F.K. acknowledges financial support from Mr and Mrs Kao during his PhD study. S.P. is a Wellcome Trust Senior Research Fellow in Basic Biomedical Sciences.
References
- Angelov D., Vitolo,J.M., Mutskov,V., Dimitrov,S. and Hayes,J.J. (2001) Preferential interaction of the core histone tail domains with linker DNA. Proc. Natl Acad. Sci. USA, 98, 6599–6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister A.J., Zegerman,P., Partridge,J.F., Miska,E.A., Thomas,J.O., Allshire,R.C. and Kouzarides,T. (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature, 410, 120–124. [DOI] [PubMed] [Google Scholar]
- Bates D.L. and Thomas,J.O. (1981) Histones H1 and H5: one or two molecules per nucleosome? Nucleic Acids Res., 9, 5883–5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary P.D., Moss,T. and Bradbury,E.M. (1978) High-resolution proton-magnetic-resonance studies of chromatin core particles. Eur. J. Biochem., 89, 475–482. [DOI] [PubMed] [Google Scholar]
- Coles L.S., Robins,A.J., Madley,L.K. and Wells,J.R. (1987) Characterization of the chicken histone H1 gene complement. Generation of a complete set of vertebrate H1 protein sequences. J. Biol. Chem., 262, 9656–9663. [PubMed] [Google Scholar]
- Cowell I.G. and Austin,C.A. (1997) Self-association of chromo domain peptides. Biochim. Biophys. Acta, 1337, 198–206. [DOI] [PubMed] [Google Scholar]
- Cowell I.G. et al. (2002) Heterochromatin, HP1 and methylation at lysine 9 of histone H3 in animals. Chromosoma, 111, 22–36. [DOI] [PubMed] [Google Scholar]
- Eissenberg J.C., James,T.C., Foster-Hartnett,D.M., Hartnett,T., Ngan,V. and Elgin,S.C. (1990) Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc. Natl Acad. Sci. USA, 87, 9923–9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong L., Schroth,G.P., Matthews,H.R., Yau,P. and Bradbury,E.M. (1993) Studies of the DNA binding properties of histone H4 amino terminus. Thermal denaturation studies reveal that acetylation markedly reduces the binding constant of the H4 ‘tail’ to DNA. J. Biol. Chem., 268, 305–314. [PubMed] [Google Scholar]
- Jacobs S.A. and Khorasanizadeh,S. (2002) Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science, 295, 2080–2083. [DOI] [PubMed] [Google Scholar]
- James T.C., Eissenberg,J.C., Craig,C., Dietrich,V., Hobson,A. and Elgin,S.C. (1989) Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur. J. Cell Biol., 50, 170–180. [PubMed] [Google Scholar]
- Jenuwein T. and Allis,C.D. (2001) Translating the histone code. Science, 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- Lachner M., O’Carroll,D., Rea,S., Mechtler,K. and Jenuwein,T. (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature, 410, 116–120. [DOI] [PubMed] [Google Scholar]
- Li Y., Kirschmann,D.A. and Walrath,L.L. (2002) Does heterochromatin protein 1 always follow code? Proc. Natl Acad. Sci. USA, 99, 16462–16469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C., Guilleme,M., Seeler,J.S., Trouche,D., Dejean,A. and Yaniv,M. (2002) Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1α. EMBO Rep., 3, 975–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen A.L., Oulad-Abdelghani,M., Ortiz,J.A., Remboutsika,E., Chambon,P. and Losson,R. (2001) Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol. Cell, 7, 729–739. [DOI] [PubMed] [Google Scholar]
- Nielsen P.R., Nietlispach,D., Mott,H.R., Callaghan,J., Bannister,A., Kouzarides,T., Murzin,A.G., Murzina,N.V. and Laue,E.D. (2002) Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature, 416, 103–107. [DOI] [PubMed] [Google Scholar]
- Pak D.T., Pflumm,M., Chesnokov,I., Huang,D.W., Kellum,R., Marr,J., Romanowski,P. and Botchan,M.R. (1997) Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell, 91, 311–323. [DOI] [PubMed] [Google Scholar]
- Platero J.S., Hartnett,T. and Eissenberg,J.C. (1995) Functional analysis of the chromo domain of HP1. EMBO J., 14, 3977–3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards E.J. and Elgin,S.C. (2002) Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell, 108, 489–500. [DOI] [PubMed] [Google Scholar]
- Smothers J.F. and Henikoff,S. (2001) The hinge and chromo shadow domain impart distinct targeting of HP1-like proteins. Mol. Cell. Biol., 21, 2555–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K., Yamada,T., Muro,Y. and Himeno,M. (1996) Human homolog of Drosophila heterochromatin-associated protein 1 (HP1) is a DNA-binding protein which possesses a DNA-binding motif with weak similarity to that of human centromere protein C (CENP-C). J. Biochem. (Tokyo), 120, 153–159. [DOI] [PubMed] [Google Scholar]
- van Holde K.E. (1988) Chromatin. Springer, New York, NY.
- Wang G., Ma,A., Chow,C.M., Horsley,D., Brown,N.R., Cowell,I.G. and Singh,P.B. (2000) Conservation of heterochromatin protein 1 function. Mol. Cell. Biol., 20, 6970–6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Flint,S.J., Leffak,I.M., Groudine,M. and Grainger,R.M. (1978) The generation and propagation of variegated chromosome structures. Cold Spring Harb. Symp. Quant. Biol., 42, 401–407. [DOI] [PubMed] [Google Scholar]
- Yamada T., Fukuda,R., Himeno,M. and Sugimoto,K. (1999) Functional domain structure of human heterochromatin protein HP1(Hsα): involvement of internal DNA-binding and C-terminal self-association domains in the formation of discrete dots in interphase nuclei. J. Biochem. (Tokyo), 125, 832–837. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K., Hidema,S. and Mizuno,S. (1998) Chicken chromobox proteins: cDNA cloning of CHCB1, -2, -3 and their relation to W-heterochromatin. Exp. Cell Res., 242, 303–314. [DOI] [PubMed] [Google Scholar]
- Zhao T., Heyduk,T., Allis,C.D. and Eissenberg,J.C. (2000) Hetero chromatin protein 1 binds to nucleosomes and DNA in vitro. J. Biol. Chem., 275, 28332–28338. [DOI] [PubMed] [Google Scholar]