Abstract
Translesion DNA synthesis (TLS) and homologous DNA recombination (HR) are two major postreplicational repair (PRR) pathways. The REV3 gene of Saccharomyces cerevisiae encodes the catalytic subunit of DNA polymerase ζ, which is involved in mutagenic TLS. To investigate the role of REV3 in vertebrates, we disruped the gene in chicken DT40 cells. REV3–/– cells are sensitive to various DNA-damaging agents, including UV, methyl methanesulphonate (MMS), cisplatin and ionizing radiation (IR), consistent with its role in TLS. Interestingly, REV3–/– cells showed reduced gene targeting efficiencies and significant increase in the level of chromosomal breaks in the subsequent M phase after IR in the G2 phase, suggesting the involvement of Rev3 in HR-mediated double-strand break repair. REV3–/– cells showed significant increase in sister chromatid exchange events and chromosomal breaks even in the absence of exogenous genotoxic stress. Furthermore, double mutants of REV3 and RAD54, genes involved in HR, are synthetic lethal. In conclusion, Rev3 plays critical roles in PRR, which accounts for survival on naturally occurring endogenous as well as induced damages during replication.
Keywords: DNA polymerase ζ/genome instability/postreplication repair/REV3/translesion DNA synthesis
Introduction
Numerous and varied DNA lesions are generated continuously not only by environmental factors but also by endogenous damage. Unrepaired DNA damage can lead to replication fork arrest and the formation of gaps, and occasionally double-stranded DNA breaks (DSBs) in one of the daughter strands. To remove such DNA lesions arising during DNA replication, cells have evolved methods of postreplication DNA repair (PRR). PRR is carried out mainly by two pathways: translesion DNA synthesis (TLS) and homologous recombination (HR) repair (reviewed in Friedberg et al., 1995). TLS functions by filling gaps on a daughter strand using a number of specialized TLS polymerases, while HR repair promotes DNA synthesis from a damaged chromatid templated on the other intact sister chromatid. Compared with our understanding of HR in maintenance of chromosomal DNA (reviewed in Sonoda et al., 2001b), the role of TLS is only poorly characterized in vertebrate cells.
A number of TLS polymerases have been identified in Saccharomyces cerevisiae and mammals (reviewed in Ohmori et al., 2001; Wood et al., 2001). Three of TLS polymerases (Pol), Polη (McDonald et al., 1997; Johnson et al., 1999b; Masutani et al., 1999), Polζ (Morrison et al., 1989; Gibbs et al., 1998; Lin et al., 1999; Van Sloun et al., 1999; reviewed in Lawrence, 2002) and Rev1 (Nelson et al., 1996a; Gibbs et al., 2000; Simpson and Sale, 2003) are conserved between these species. Human Polη is mutated in a variant form of xeroderma pigmentosum (XP-V) (Johnson et al., 1999b; Masutani et al., 1999), which is characterized by predisposition to skin cancer and elevated UV sensitivity. Yeast and mammalian polζ is comprised of the Rev3 catalytic subunit and the Rev7 subunit (Nelson et al., 1996b; Murakumo et al., 2001). Yeast rev3 mutant strains exhibit a moderate sensitivity to UV as well as reduced levels of mutagenesis induced by a very broad range of mutagens (Lemontt, 1972; Lawrence and Christensen, 1976). Although a defect in Rev3 does not affect the viability of yeast, REV3–/– murine embryos died around midgestation, and rev3-deficient mammalian cell lines have not yet been established (Bemark et al., 2000; Esposito et al., 2000; Wittschieben et al., 2000). To analyze the essential function of Rev3 in higher eukaryotes, we generated Rev3-deficient cells from the chicken B lymphocyte line DT40 (Buerstedde and Takeda, 1991). Here, we report genetic evidence that Rev3 protein is involved in maintenance of chromosomal DNA as well as in tolerance of various DNA damages by TLS and HR.
Results
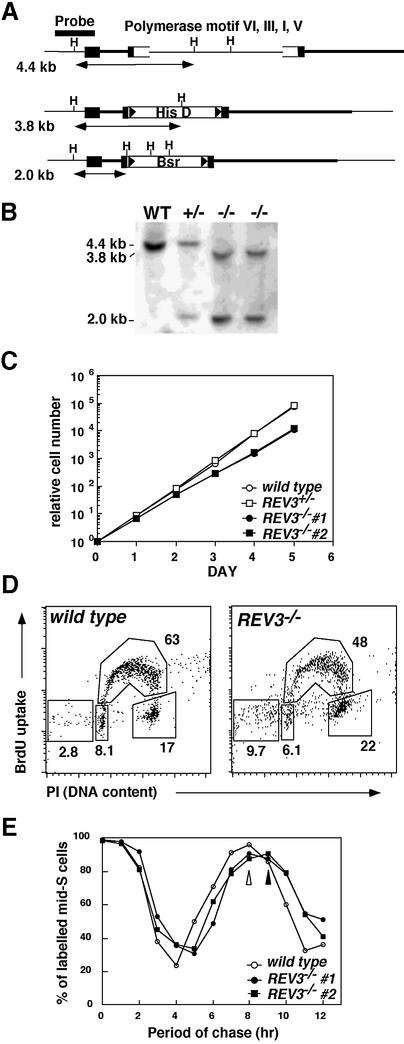
Slower growth kinetics of REV3-deficient cells
We isolated a chicken REV3 cDNA and determined its sequence. The six polymerase motifs show ∼95% identity, and the overall identity to the human Rev3 protein is ∼75%. Gene targeting constructs, which are expected to disrupt four polymerase motifs (VI, III, I, V; Figure 1A) were generated from the amplified genomic DNA. Gene targeting of the REV3 locus was confirmed by Southern blot analysis (Figure 1B). The proliferative properties of two independently isolated REV3–/– clones were monitored using growth curves and cell cycle analysis. REV3–/– cells proliferated in slower kinetics compared with wild-type cells, while REV3+/– cells proliferated with normal kinetics (Figure 1C). Pulse–chase bromodeoxyuridine (BrdU) labeling revealed that the length of cell cycle time was extended to 9 h (wild type, 8 h) in the absence of Rev3 (Figure 1E). Furthermore, a higher fraction of the REV3–/– cells accumulated in the G2 phase as well as in a sub-G1 fraction when compared with wild-type cells (Figure 1D). These observations indicate that the G2 phase is elongated in REV3–/– cells. Accumulation of cells in the G2 phase is also observed in HR-deficient cells, such as RAD51–/– DT40 cells (Sonoda et al., 1998), which show extensive chromosomal breaks probably leading to stimulation of the G2 damage checkpoint. Likewise, REV3–/– cells exhibited a significant increase in the level of spontaneous chromosomal breaks (Table I) as well as elevated level of dying sub-G1 cells (Figure 1D). These observations indicate that the slower proliferation rate of REV3–/– cells is explained by both an extended G2 phase and increased cell death, presumably caused by spontaneous chromosomal breaks.
Fig. 1. Gene targeting of the REV3 locus. (A) Schematic representation of part of the REV3 locus, the gene disruption constructs, and the configuration of the targeted alleles. Solid boxes indicate the position of exons. Only disrupted exons are indicated. Relevant HindIII sites and the position of the probe used in Southern blot analysis are indicated. (B) Southern blot analysis of HindIII-digested genomic DNA from cells with the indicated genotypes of the REV3 gene, using the probe shown in (A). The positions and sizes of hybridizing fragments of the wild-type and targeted loci are indicated. (C) Growth curves corresponding to the indicated cell cultures. Data shown are the average of the results from two separate clones for each genotype. Error bars show the standard deviation of the mean for three experiments. (D) Representative cell cycle distribution of the indicated cell cultures as measured by BrdU incorporation and DNA content in flow cytometry analysis. Cells were pulse–labeled for 10 min, and subsequently stained with FITC-anti-BrdU to detect BrdU incorporation (y-axis, log scale) and propidium iodide to detect total DNA (x-axis, linear scale). The upper gate identifies cells incorporating BrdU (∼S phase), the lower-left gate identifies G1 cells, and the lower-right gate displays G2/M cells. Sub-G1 cells reflect dead cells. Numbers show the percentages of cells falling in each gate. (E) The calculated relative percentages of cells in the indicated gate are plotted with time. The symbols for each sample are shown on the right. The progression of BrdU-labeled (i.e. S phase) cells out of S phase, and into first G2/M (4n DNA content), then G1 (2n DNA content), and back into S phase with time is indicated.
Table I. Frequency of spontaneous chromosomal aberrations in wild-type and Rev3-deficient DT40 cells.
| Cells | No. of cells analyzed | Chromatid-type |
Chromosome-type |
Chromatid exchange | Total (per cell) | ||
|---|---|---|---|---|---|---|---|
| Gaps | Breaks | Gaps | Breaks | ||||
| Wild type | 100 | 3 | 2 | 1 | 0 | 0 | 6 (0.06) |
| REV3–/– | 100 | 2 | 3 | 7 | 4 | 0 | 18 (0.18) |
REV3–/– cells are sensitive to a variety of genotoxic stress
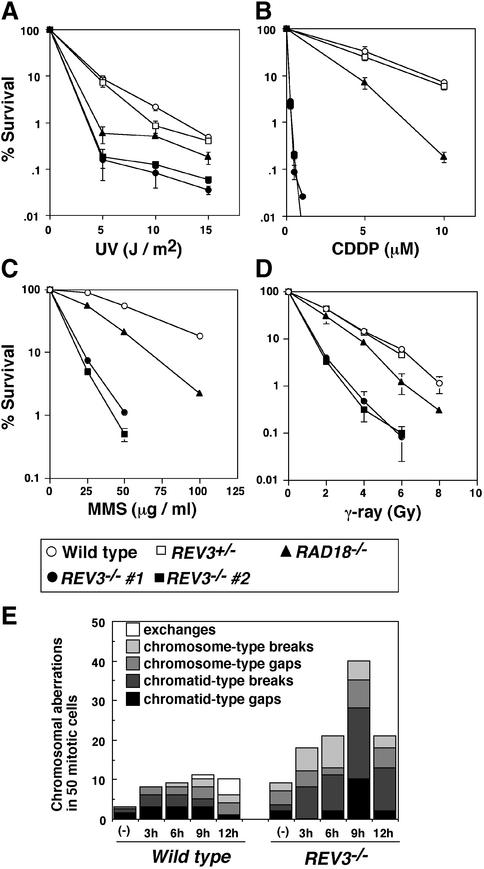
We examined viability of wild-type and REV3–/– cells after genotoxic treatment by colony survival assays. REV3–/– cells exhibited elevated sensitivity to UV, methyl methane sulphonate (MMS), cisplatin, and ironizing radiation (IR), when compared with wild-type and REV3+/– cells (Figure 2). Elevated sensitivity to a variety of DNA-damaging agents is reminiscent of the phenotype of yeast mutants of the Rad6/Rad18 pathway (McKee and Lawrence, 1980; Keszenman et al., 1992; Simon et al., 2000).
Fig. 2. Sensitivity of wild-type and REV3–/– cells to DNA-damaging agents, and UV-induced chromosomal aberrations. (A–D) The fractions of surviving colonies after the indicated treatment of cells compared with untreated controls of the same genotype are shown on the y-axis on a logarithmic scale. (A) UV; (B) cisplatin (CDDP); (C) MMS; (D) γ-rays. The dose of 137Cs γ-rays and UV, and concentrations of cisplatin and MMS are displayed on the x-axis on a linear scale in each graph. The data shown are the representative results from three separate experiments. (E) UV-induced chromosome aberrations of wild-type and REV3–/– cells. Cells were exposed to 5 J/m2 UV.
To investigate the cause of cell death following UV irradiation, we measured UV-induced chromosomal breaks, which reflect DSBs in the chromosomal DNA. It is known that DSBs are not induced by UV alone, but can be induced by DNA replication over UV-damaged template DNA (van Zeeland et al., 1980). Chromosome breaks were measured at 3, 6, 9 or 12 h after UV irradiation (Figure 2E). The data show that a defect in Rev3 dramatically increased UV-induced chromosomal breaks. In particular, cells that were exposed to UV when they were at early S phase showed a maximum amount of chromosome breaks (9 h after irradiation). This suggests that Rev3 may process DNA replication block caused by UV damage.
In yeast, all the components of the TLS pathways including Rev3 are epistatic to the Rad6–Rad18 molecules (reviewed in Lawrence, 1994; Broomfield et al., 2001). It is known that Rad6 is a ubiquitin-conjugating enzyme (E2), forming a tight complex with the Rad18 protein (Bailly et al., 1994, 1997). We have previously shown that RAD18–/– cells are sensitive to IR, UV and crosslinking agents (Yamashita et al., 2002). This phenotype is similar to that observed in yeast rad18 mutant (Lawrence and Christensen, 1976; Prakash, 1981; Fabre et al., 1989). Interestingly, REV3–/– cells exhibited significantly higher sensitivity to cisplatin, MMS and IR when compared with RAD18–/– cells (Figure 2). Thus, unlike in yeast, Rev3 may not be fully regulated by the Rad18 protein in vertebrate cells.
Defective DSB repair after completion of DNA replication in REV3–/– cells
REV3–/– as well as RAD18–/– cells exhibited elevated IR sensitivity when compared with wild-type cells (Figure 2D). This could be explained by a defect in any of the following four systems: a damage checkpoint, TLS, and two major DSB repair pathways (reviewed in Kanaar et al., 1998; van Gent et al., 2001)—nonhomologous end-joining (NHEJ) and HR. Two Gray γ-irradiation of wild-type and REV3–/– cells suppressed DNA replication and stimulated the G2 checkpoint (Supplementary figure 1, available at The EMBO Journal Online), indicating that Rev3 deficiency does not affect damage checkpoints. No difference between REV3–/– and wild-type cells was seen in recircularization of transfected linearized plasmid (Supplementary figure 2) indicating that NHEJ also appears to work normally (Verkaik et al., 2002).
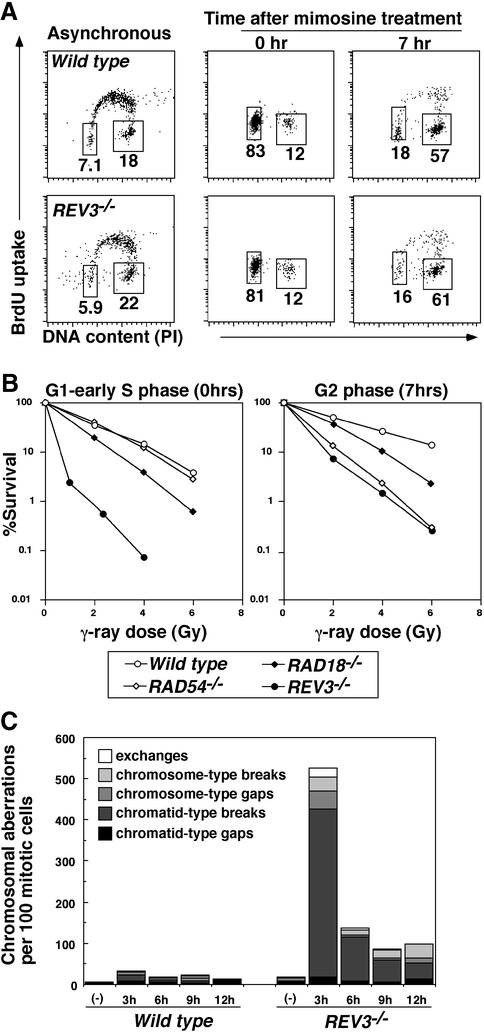
To dissect the contribution of the TLS and HR pathways to IR tolerance, we analyzed IR sensitivity of synchronised populations of REV3–/– and wild-type cells. Defective TLS would increase IR sensitivity particularly when the cells were exposed to IR prior to DNA replication. In contrast, defective HR would increase IR sensitivity in late S to G2 phases, because HR between sister chromatids, but not HR between homologous chromosomes, is responsible for DSB repair in higher eukaryotes (Takata et al., 1998). We synchronized cells using either nocodazole–mimosine treatments (Sonoda et al., 2001a; Figure 3A) or elutriation (Takata et al., 1998; data not shown), and obtained consistent data. As expected, REV3–/– cells exhibited elevated IR sensitivity in the G1 and early S phases more prominently when compared with late S to G2 phases (Figure 3B), suggesting that defective TLS accounts for their IR sensitivity at least partially as well as for the MMS sensitivity (Figure 2C).
Fig. 3. Increased radiosensitivity in G2 as well as G1/S boundary in the absence of Rev3 (A) Cells of the indicated genotypes were synchronized at the G1/S boundary with nocodazole–mimosine treatment for 16 h, and released into culture at 0 h. The vast majority of the cells were in G1/S boundary at 0 h and in late S–G2 phases at 7 h after the release. Live cells were gated according to their FSC and SSC profiles. (B) IR sensitivity of these synchronized populations is shown as in Figure 2D. (C) IR-induced chromosome aberrations. Results from wild-type cells and REV3–/– are shown. Cells were exposed to 2 Gy γ-rays.
Interestingly, REV3–/– cells exhibited significant increase in IR sensitivity also in late S to G2 phases, as observed in HR deficient RAD54–/– cells (Bezzubova et al., 1997; Figure 3B). The increased IR sensitivity in the G2 phase in REV3–/– cells can be explained by the following two mechanisms. First, IR induces base damages, which interfere with DNA, leading to formation of DSBs. Secondly, IR-induced DSBs are not repaired efficiently in REV3–/– cells. To investigate the role of Rev3 in DSB repair, asynchronous populations of cells were exposed to IR, and chromosome breaks were measured at 3, 6, 9 or 12 h after IR (Figure 3C; Takata et al., 1998; Takao et al., 1999). The majority of the cells that entered mitosis between 0 and 3 h after 2 Gy IR should have been irradiated in the G2 phase, but not in the late S phase, for the following two reasons. First, the irradiated REV3–/– cells were partially arrested up to 4 h in the G2 phase (Supplementary figure 1). Secondly, no BrdU-labeled mitotic chromosomes were detected at 3 h after IR when the irradiated cells were exposed to both BrdU and colcemid between 0 and 3 h (Supplementary table 1). Strikingly, like RAD54–/– cells (Takata et al., 1998), REV3–/– cells exhibited marked increase in the levels of chromosomal breaks at 3 h after IR, while these levels decreased at later time points (Figure 3C). This pattern of IR-induced chromosomal breaks indicates that Rev3 plays a direct role in IR-induced DSB repair even after completion of S phase. Since the absence of Rev3 does not appear to affect NHEJ, we favor the possibility that Rev3 is involved in DSB repair by participating in the HR pathway. In conclusion, Rev3 deficiency may cause defects in both TLS- and HR-mediated DSB repair, resulting in hypersensitivity to IR.
To evaluate HR capability of REV3–/– cells, we measured the rate of the immunoglobulin (Ig) gene conversion and gene targeting efficiency. Chicken B lymphocyte precursors as well as DT40 cells diversify the variable region of the Ig genes through HR between V(D)J joints and their upstream pseudo V segments (reviewed in Reynaud et al., 1994). We examined the rate of gene conversion at the Ig λ locus in REV3–/– cells by measuring the gain and loss of surface Ig expression as described previously (Buerstedde et al., 1990; Sale et al., 2001). However, we were not able to detect any effect of the Rev3 deficiency on Ig gene conversion frequency (Supple mentary figure 3) or nontemplated base substitution (two mutations out of 18 and one mutation out of eight gain of surface Ig expression events in wild-type and REV3–/– cells, respectively). In contrast, the efficiency of gene targeting onto three loci was consistently reduced in the absence of Rev3 (Table II), indicating the involvement of Rev3 in some types of HR. In summary, Rev3 may play a direct role in HR, particularly in repair of IR induced DSBs.
Table II. Targeted integration frequenciesa.
| Genotype | Targeted locus |
||
|---|---|---|---|
| OVALBUMIN | Ig LAMBDA | RAD54 | |
| Wild type | 43/47 (91.5%) | 28/59 (47.4%) | 53/77 (68.8%) |
| REV3–/– | 32/48 (66.7%)b | 15/75 (20.0%)c | 6/34 (17.6%)d |
| RAD18–/– | 22/54 (40.7%) | ND | 3/38 (7.9%) |
| RAD54–/– | 0/44 (0%) | 3/48 (6.3%) | ND |
Wild-type and REV3–/– cells were transfected with targeting constructs of the indicated loci.
aThe data shown are the number of targeted clones at each locus divided by the number of drug-resistant clones analyzed. The percent frequency is in parentheses.
b–dSignificantly different from wild-type levels (bχ2 = 8.8037, P < 0.003; cχ2 = 11.4244, P < 0.0007; dχ2 = 24.8146, P < 0.0001). ND, not determined.
Postreplication repair in REV3–/– cells
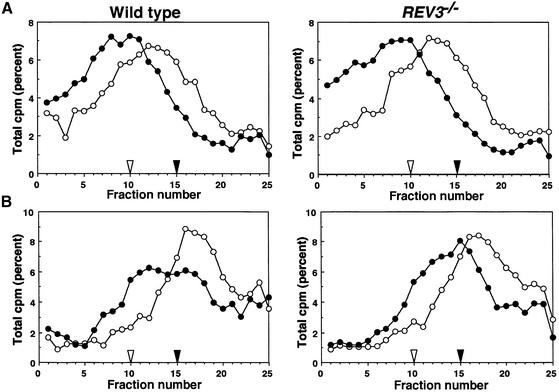
To assess the capability of PRR of REV3–/– cells, we measured the size of newly replicated DNA before and after UV-irradiation by velocity sedimentation analyses in alkaline sucrose gradients (Prakash, 1981; Tateishi et al., 2000; Yamashita et al., 2002). There was no obvious difference in the strand growth after UV irradiation between wild-type and REV3–/– cells (Figure 4, bottom panels), in agreement with the yeast rev3 mutation, which has no effect on PRR of UV-damaged DNA (Prakash, 1981).
Fig. 4. Postreplication repair of Rev3-deficient DT40. Alkaline sucrose-gradient analysis of DNA from wild-type and REV3–/– cells. (A) Wild-type (left) and Rev3-deficient (right) DT40 cells were pulse-labeled with [3H]thymidine (0.93 MBq/ml) for 15 min without UV irradiation (open circles). In a pulse–chase experiment, the pulse-labeled cells were further incubated for 30 min (closed circles) in fresh medium containing 10 µM unlabeled thymidine and uridine. Samples were sedimented on 5–20% alkaline sucrose gradients from right to left. (B) Wild-type DT40 cells and Rev3-deficient DT40 cells were irradiated with UV (8 J/m2), incubated for 10 min, and then pulse-labeled with [3H]thymidine (0.93 MBq/ml) for 15 min (open circles). In a pulse–chase experiment, the pulse-labeled cells were further incubated for 90 min (closed circles) in the chase medium containing 10 µM unlabeled thymidine and uridine. The samples were analyzed by the same method as (A). Closed and open arrowheads indicate the positions of bacteriophage λ DNA (42 kb) and T4GT7 DNA (165.6 kb), respectively.
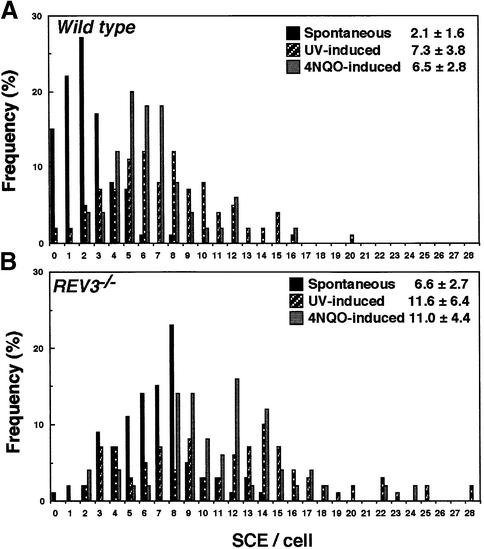
Since TLS and HR are thought to constitute major PRR pathways in yeast (Prakash, 1981; Broomfield et al., 2001) as well as in Escherichia coli (Friedberg et al., 1995; Berdichevsky et al., 2002), we wanted to know which pathway is mainly affected in REV3–/– cells. To evaluate HR-mediated PRR events, we measured the level of microscopically visible sister chromatid exchange (SCE) events (Sonoda et al., 1999; Dronkert et al., 2000; Wang et al., 2000). REV3–/– cells showed a few-fold higher spontaneous SCE level than wild-type cells (Figure 5A). This result suggests that Rev3 plays an important role during DNA replication even in the absence of exogenous genotoxic stress. We next measured the level of SCE following exposure of cells to UV and 4-nitroquinoline 1-oxide (4NQO), which damages base residues in a manner similar to UV irradiation (reviewed in Friedberg et al., 1995). The levels of UV- and 4NQO-induced SCE were 5.2 and 4.1 SCEs/cell, respectively, for wild-type cells, and 5.0 and 4.4 SCEs/cell, respectively, for REV3–/– cells (Figure 5). Thus, a defect in Rev3 elevates the level of spontaneous SCE but not UV- or 4NQO-induced SCE. It is surprising that a defect in Rev3 does not significantly affect DNA ζ synthesis on UV-damaged templates (Figure 4B), although the same defect significantly reduced colony survival (Figure 2A) and increased induced chromosomal breaks (Figure 2E) in response to UV. Presumably, a subtle defect in TLS past each UV damage may not be detectable by measuring DNA synthesis on UV-damaged templates for the following reason. Thirty seven per cent survival dose of UV irradiation induces as many as 40 000 UV damages per human cell (BEIRV, 1990), and a half-life of cyclobutane pyrimidine dimer (CPD), the major UV damage, is nearly one day (Setlow, 1985). Thus, even a slight decrease in TLS past each UV damage may substantially increase the number of gaps in each cell, leading to significant increase in the level of chromosomal breaks and reduction of colony survival.
Fig. 5. Spontaneous sister chromatid exchange is elevated in REV3–/– cells. Levels of SCE per cell are shown in wild-type and REV3–/– cells. Mean ± standard error is shown at the top of each panel. Solid box, distribution of SCE/cell without treatment; shaded and dotted box, distribution of SCE/cell with 0.25 J/m2 UV light and 0.2 ng/ml 4NQO treatment, respectively.
Double mutants of REV3 and RAD54 are not able to proliferate
Increased SCE in REV3–/– cells led us to investigate the functional overlap between Rev3-dependent PRR and HR in processing gaps caused by replication block. To this end, we generated cells deficient in both Rev3 and Rad54. We showed previously that Rad54-deficient DT40 cells exhibit hypersensitivity to IR, but are able to proliferate with nearly normal kinetics (Bezzubova et al., 1997; Takata et al., 1998). Since we have failed to generate cells deficient in both Rev3 and Rad54, we generated conditional REV3–/–RAD54–/– double mutant cells using the tamoxifen-inducible Cre–loxP system (Zhang et al., 1998; Fujimori et al., 2001). Upon the addition of tamoxifen to the culture media, virtually all cells lost their RAD54 transgene (designated as loxP-RAD54 hereafter) through Cre-recombinase (the transgene designated as CRE hereafter) action within 24 h (data not shown), generating mutant cells that had completely lost functional REV3 and RAD54 genes.
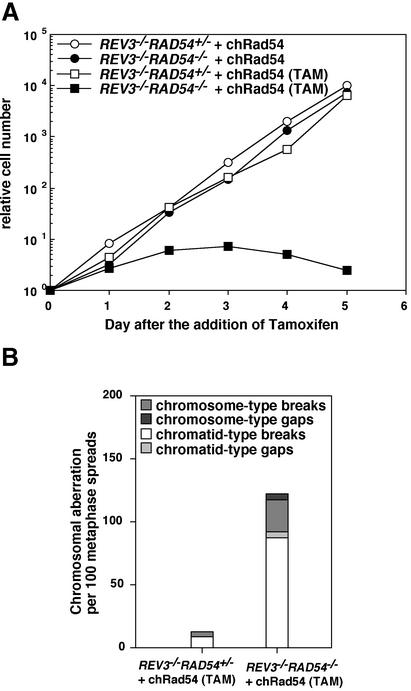
Continuous exposure of RAD54–/– or REV3–/– cells to tamoxifen did not affect their exponential proliferation (Figure 6A). In marked contrast, the REV3–/–RAD54–/– (w/loxP-RAD54, CRE) cells exhibited dramatic reduction in growth rate in the presence of tamoxifen and eventually died. To investigate the cause of cell death, we analyzed chromosomal breaks in REV3–/–RAD54–/–cells. The REV3–/–RAD54+/– cells showed only few chromosomal aberrations even after tamoxifen treatment (Figure 6B). In contrast with these control populations, the REV3–/–RAD54–/– cells had 1.25 aberrations/cell at day 3 after adding tamoxifen (Figure 6B). We showed previously that the level of spontaneous chromosomal breaks of various HR-deficient DT40 clones is closely correlated with the rate of cell death during the cell cycle (reviewed in Morrison and Takeda, 2000; Sonoda et al., 2001b). These observations indicate that the increased chromosomal breaks may account for the massive cell death of REV3–/–RAD54–/– cells.
Fig. 6. REV3–/–RAD54–/– double mutant cells are not viable. (A) Growth curves of the indicated cell cultures in the absence and presence of tamoxifen (TAM). (B) Chromosomal breaks are accumulated in dying REV3–/–RAD54–/– double mutant cells. Cells were exposed to tamoxifen for 3 days. One hundred mitotic cells were analyzed in each case.
Discussion
Requirement of Rev3 for PRR in DT40 cells
The present study reveals multiple roles of chicken Rev3 in PRR. First, the data show that a defect in Rev3 caused increase in sensitivities to several different types of DNA lesions. Secondly, we present genetic evidence that Rev3 is involved in targeted integration and HR-mediated DSB repair particularly following IR. Thirdly, we show that REV3–/– cells exhibited increased levels of spontaneous SCE events and chromosomal breaks. These observations as well as synthetic lethality of the rev3 and rad54 mutations suggests that Rev3 plays an important role in maintaining chromosomal DNA during the cell cycle. Fourthly, REV3–/– cells show a more prominent phenotype compared with RAD18–/– cells in terms of the level of spontaneous chromosomal breaks as well as sensitivity to genotoxic treatments. Thus, Rad18 does not fully regulate TLS DNA polymerases in higher eukaryotes, which is in marked contrast with yeast Rad18. Like Rad18-deficient DT40 cells, the REV3–/– DT40 cells exhibited high sensitivity to UV, IR, MMS and cisplatin, which is consistent with their role in PRR. These observations support the notion that chicken Rev3 is involved in TLS, as has been demonstrated for yeast Rev3. However, the phenotype of the DT40 rev3 mutant is not necessarily the same as that of the yeast mutant. The yeast rev3 mutant shows only mild sensitivity to UV and marginal sensitivity to IR and MMS (Lemontt, 1972; McKee and Lawrence, 1979; Johnson et al., 1998; Xiao et al., 1999). Thus, yeast Rev3 appears to account for only a small component of PRR.
The present study highlights Rev3 as a key factor for determining cellular response to cisplatin. There are a few explanations for the extremely high sensitivity of the rev3 mutant to cisplatin. First, after other TLS polymerases insert deoxynucleotides opposite cisplatin damages, such as intrastrand crosslinks, Rev3 may be essential for extending from such deoxynucleotides. Another explanation is synergistic effect of defective TLS and HR on processing of cisplatin damage in the rev3 mutant. The present study suggests that analysis of the expression of polζ would be useful for predicting cisplatin sensitivity of tumor cells.
Critical roles of Rev3 in repair of IR-induced DNA damage
We have shown that a defect in Rev3 as well as Rad18 causes an elevation of IR sensitivity. This phenotype appears to be explained by defects in both TLS and probably another repair pathway, which accounts for the increased levels of chromosomal aberrations observed in cells that are irradiated in the G2 phase of the cell cycle. The obvious first candidate repair pathways are the two major DSB repair pathways NHEJ and HR (Kanaar et al., 1998; van Gent et al., 2001). We presume that the major DSB repair pathway in G2 cells is HR, as we showed previously (Takata et al., 1998). Furthermore, involvement of the Rev3 polymerase in the NHEJ pathway has not been shown in yeast genetic studies, while Rev3 is implicated in mutagenesis associated with homology-dependent DSB repair (Holbeck and Strathern, 1997). Indeed, we have failed to detect a defect in NHEJ in REV3–/– cells, using an assay to measure the relative usage of accurate recircularization of transfected linear plasmid (Tauchi et al., 2002; Verkaik et al., 2002; Supplementary figure 2). However, we did find indications of reduced HR, most notably a reduced efficiency of gene targeting in three different loci in the absence of Rev3 (Table II). The reduced HR capability can be explained by the following two explanations. First, since the HR pathway in REV3–/– cells has to repair a large number of endogenous DNA damage caused by defective TLS in the S phase, it could not efficiently repair additional IR-induced DSBs. This explanation is, however, unlikely, because cells that were exposed to γ-rays in the S phase (3–6 and 6–9 h in Figure 3C) exhibited a significant reduction in the levels of induced chromosomal breaks when compared with cells irradiated in the G2 phase (0–3 in Figure 3C). Secondly, Rev3 may play a critical role in HR-mediated DSB repair of IR-induced DSBs, because IR-induced DSBs may be associated with chemical modification, which may make the 3′ ends unsuitable as primers for most DNA polymerases. Rev3, however, is relatively indifferent to structural distortions in the primer and template strands, including a mismatch near the 3′ end of a primer (Johnson et al., 2000; Lawrence et al., 2000; reviewed in Lawrence and Maher, 2001; Woodgate, 2001). Therefore, Rev3, but not other DNA polymerases, may be critical for initiation of DNA synthesis from the 3′ end of IR-induced DSBs in HR-mediated repair.
Although the data discussed above suggest that Rev3 is involved in HR, it should be noted that Rev3 does not play an important role in some types of HR, such as the Ig gene conversion. Likewise, a defect in Rev3 appears to result in only mild impairment of HR-mediated PRR of spontaneously arising DNA damage, since the level of spontaneous SCE was rather increased in the absence of Rev3. On the other hand, a defect in Rev3 appears to impair HR as well as TLS in repairing gaps at UV damage. Indeed, it is surprising that a defect in Rev3 did not cause an increase in the level of UV-induced SCE, although the number of UV-induced gaps should be dramatically increased in the absence of Rev3. Presumably, Rev3-dependent HR might also play a role in repairing gaps at UV damage. Alternatively, since 37% survival dose of UV irradiation induces as many as 40 000 UV damages per human cell (BEIRV, 1990), the capability of HR-mediated PRR might be saturated particularly in REV3–/– cells. Taken together, we favor the hypothesis that HR is directly affected by the Rev3 mutation.
Rev3-dependent PRR contributes to the maintenance of chromosomal DNA
Accumulating evidence has suggested that Rev3 plays an important role in maintaining chromosomal integrity in cycling cells. First, loss of Rev3 leads to embryonic lethality in the mouse (Bemark et al., 2000; Esposito et al., 2000; Wittschieben et al., 2000). Furthermore, a significant increase in DSBs as well as chromatid and chromosome aberrations was observed in primary cells from REV3–/– murine embryos (Van Sloun et al., 2002).
In this study, we show that REV3–/– DT40 cells exhibited increase in spontaneous chromosomal breaks (Table I). These observations indicate frequent employment of Rev3 in PRR during the normal cell cycle. We hypothesize that the main DNA lesions that trigger PRR in vertebrate cells may be abasic sites, as they are among the most frequently formed DNA lesions, which block the replicative DNA machinery. Since biochemical studies have shown that polζ efficiently bypass AP sites in combination with other polymerases such as polδ and Rev1 (Nelson et al., 1996a, 2000; Haracska et al., 2001), polζ may make a significant contribution to the tolerance to AP sites in vivo. Indeed, even if yeast cells simultaneously lack Ntg1, Ntg2 and Apn1, all of which are responsible for the processing of AP sites following base damages, they can survive because polζ readily bypasses the AP sites (Swanson et al., 1999). Since yeast polζ is implicated in spontaneous mutation (Quah et al., 1980; Roche et al., 1994; Harfe and Jinks-Robertson, 2000), vertebrate polζ also may have a significant impact on accumulation of base substitution and tumorigenesis by participating in PRR during physiological cell cycle.
We also found that REV3–/– cells exhibited a significant increase in the level of spontaneous SCE (Figure 5). We interpret this increase as more frequent usage of HR-mediated PRR pathway due to a defect in the TLS-mediated PRR pathway. Alternatively, it is possible that the HR pathway is hyperactivated in REV3–/– cells. This is unlikely because the frequency of targeted integration is reduced in REV3–/– cells compared with wild-type cells (Table II). Thus, we conclude that a defect in TLS in REV3–/– cells increases the number of gaps, leading to more frequent usage of HR-mediated PRR in the mutant cells than in wild-type cells. This conclusion supports the notion that a substantial fraction of the DNA lesions that occur during DNA replication can be processed by both HR and Rev3-dependent PRR pathways. To investigate functional redundancy between these two pathways, we generated REV3–/– cells that were conditionally deficient for Rad54. The Rad54 appears to have a minor role in HR when compared with Rad51 in vertebrates, as manifested by normal embryogenesis in Rad54-deficient mice (Essers et al., 1997), which is in marked contrast with lethality of Rad51 depletion to the cells (Sonoda et al., 1998). We previously found that rad54 and rad18 mutations are synthetic lethal (Yamashita et al., 2002). Likewise, rad54 and rev3 mutations were synthetic lethal to the cells exhibiting extensive chromosomal breaks, although either type of the single mutants is able to proliferate. Presumably, since a large numbers of gaps, some of which could then be processed into DSBs, are generated in the absence of Rev3, even a minor defect in HR, i.e. a defect in RAD54, may be lethal to the cells. In conclusion, synthetic lethality of rev3 and rad54 and that of rad18 and rad54 suggest that two major PRR pathways, TLS and HR, cooperatively contribute to the maintenance of chromosomal DNA in higher eukaryotes.
Materials and methods
Plasmid construction
Two REV3 disruption constructs, REV3-hisD and REV3-bsr, were generated from genomic PCR products combined with hisD- and bsr-selection marker cassettes. Genomic DNA sequences were amplified using the primers 5′-TCCCGCTTCTTCAGCAACGCTGG-3′and 5′-TCGTGAAATACTGTGAGATGGTGCC-3′ (for the left arm of the KO construct); and 5′-ACAGAGGTAGCTCTGCTTACAAGCC-3′ and 5′-ACGGGATCTGTCGGTCAAAGCAGCC-3′ (for the right arm of the KO construct). Amplified PCR products were cloned into pCRII-TOPO vecotor (Invitrogen). The 1.1 kb EcoRV fragment from the left arm were cloned into XhoI (blunt ended)–EcoRV site of pCRII containing the 4 kb right-arm sequence. The EcoRV site was used to clone marker gene cassettes. The 0.8 kb HindIII fragment from the genomic DNA amplified using the primers 5′-ATTACGTTAGCCGGGTCCATGGG-3′ and 5′-AGAACAGCGTTGCTGTAGAAGCGGG-3′was used as a probe for Southern blot analysis. RAD54-puro and RAD54-bsr disruption constructs were described previously (Takata et al., 1998). We constructed an expression vector pCR3-loxP-RAD54/IRES-EGFP-loxP (named loxP-RAD54 in short), in which RAD54 and green fluorescent protein (EGFP) genes are flanked by the loxP sequences, by inserting a RAD54 BamHI fragment into the BamHI site of pCR3-loxP-MCS-loxP (Fujimori et al., 2001; Yamashita et al., 2002).
Cell culture and DNA transfection
Cells were cultured in RPMI1640 supplemented with 10–5 M β-mercaptethanol, 10% fetal calf serum, and 1% chicken serum (Sigma, St Louis, MO) at 39.5°C. Methods of DNA transfection and genotoxic treatments are as described previously (Takata et al., 1998). Cell synchronization at the G1/S phase transition was achieved as described previously, either by sequential nocodazole and mimosine blocks (Sonoda et al., 2001a) or by elutriation (Takata et al., 1998).
Generation of REV3–/– and REV3–/–RAD54–/– (w/loxP-RAD54, CRE) cells
The wild-type DT40 cells were sequentially transfected with REV3-bsr- and REV3-hisD-targeting constructs to obtain REV3–/– cells. Then, the plasmid containing CreER chimeric recombinase (pANMerCreMer; Zhang et al., 1998) was transfected to delete the drug-resistant cassettes. The REV3–/– w/CRE cells without drug-resistant cassettes were transfected with RAD54-puro-targeting construct. The resulting REV3–/–RAD54+/– clone was transfected with loxP-RAD54, which can be selected by neo-selection marker (Fujimori et al., 2001). REV3–/–RAD54+/– (w/loxP-RAD54) cells were subsequently transfected with RAD54-bsr-targeting construct to obtain REV3–/–RAD54–/– (w/loxP-RAD54) cells. We also transfected the plasmid containing the CreER chimeric recombinase into RAD54–/– cells, and obtained Rad54–/– w/CRE cells. These cells were treated by 100 nM tamoxifen as described previously (Yamashita et al., 2002).
Measurement of the length of cell cycle time using pulse–chase labeling of cells
The method of flowcytometric analysis of cell cycle is as described previously (Takata et al., 1998).
Colony formation assay following genotoxic treatments
Colony formation assay was performed as described previously (Okada et al., 2002).
Measurement of the size of newly synthesized DNA strands following UV irradiation
Postreplication repair was analyzed by the sedimentation velocity method as described previously (Tateishi et al., 2000; Yamashita et al., 2002).
Analysis of irradiation-induced damage checkpoints
To monitor the S phase checkpoint, cells were either untreated or irradiated with 4 Gy γ-rays, then incubated for 1 h. [3H]thymidine (20 µCi/ml) was pulsed for the last 15 min. Inhibition of DNA synthesis, which indicates the activation of S phase checkpoint, was monitored by the [3H]thymidine incorporation. To monitor the G2/M checkpoint, cells were either untreated or irradiated with 2 Gy γ-rays, then incubated with colcemid and aliquots of the culture were taken every 1 h. Cells in mitosis were identified by co-staining with PI and antibody to anti-phospho-histone H3 (Upstate, New York).
Chromosome aberration analysis
Karyotype analysis was performed as described previously (Sonoda et al., 1998). For the morphological analysis of chromosome aberrations, cells were treated with colcemid for 3 h to enrich mitotic cells.
Measurement of SCE levels
Measurement of SCE levels was performed as described previously (Okada et al., 2002; Yamashita et al., 2002).
Measurement of targeted integration frequencies.
To analyze targeted integration events at the Ovalbumin Igλ (Buerstedde and Takeda, 1991) and RAD54 (Bezzubova et al., 1997) loci, disruption construct DNAs were transfected into cells, and Southern blot analysis was performed following selection of clones resistant to appropriate antibiotics.
Analysis of Ig gene conversion
CL18 is a surface IgM– (sIgM–) subclone of DT40 (Buerstedde et al., 1990) and is the parental clone for the Rev3 mutants described here. We confirmed Rev3-deficient clones retained the same frameshift mutation in the Vλ sequence as do wild-type CL18 cells. As described previously (Buerstedde et al., 1990; Sale et al., 2001), we assessed the Ig gene conversion by measuring the gain and loss of sIgM expression during a 3 week period.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We would like to thank Y.Sato, M.Nagao and C.Tateishi for their technical assistance. We also thank to Drs C.W.Lawrence (University of Rochester School of Medicine and Dentistry, MD), J.E.Sale (Medical Research Council Laboratory of Molecular Biology, UK) and Y.M.Yamashita (Stanford University, CA) for critical reading and discussion. Financial support was provided in part by CREST.JST. (Saitama, Japan) and the center of excellence (COE) grant for Scientific Research from the Ministry of Education, Culture, Sports and Technology and by grants from The Uehara Memorial Foundation, and The Naito Foundation. This work was funded in part by grants of the Virtual Research Institute of Aging of Nippon Boehringer Ingelheim.
References
- Bailly V., Lamb,J., Sung,P., Prakash,S. and Prakash,L. (1994) Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev., 8, 811–820. [DOI] [PubMed] [Google Scholar]
- Bailly V., Lauder,S., Prakash,S. and Prakash,L. (1997) Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J. Biol. Chem., 272, 23360–23365. [DOI] [PubMed] [Google Scholar]
- BEIRV (1990) Health Effects of Exposure to Low Levels of Ionizing Radiation. BEIR Report V. National Academy of Science, National Research Council, Washington, DC, p. 15.
- Bemark M., Khamlichi,A.A., Davies,S.L. and Neuberger,M.S. (2000) Disruption of mouse polymerase ζ (Rev3) leads to embryonic lethality and impairs blastocyst development in vitro. Curr. Biol., 10, 1213–1216. [DOI] [PubMed] [Google Scholar]
- Berdichevsky A., Izhar,L. and Livneh,Z. (2002) Error-free recombinational repair predominates over mutagenic translesion replication in E. coli. Mol. Cell, 10, 917–924. [DOI] [PubMed] [Google Scholar]
- Bezzubova O., Silbergleit,A., Yamaguchi-Iwai,Y., Takeda,S. and Buerstedde,J.M. (1997) Reduced X-ray resistance and homologous recombination frequencies in a RAD54–/– mutant of the chicken DT40 cell line. Cell, 89, 185–193. [DOI] [PubMed] [Google Scholar]
- Broomfield S., Hryciw,T. and Xiao,W. (2001) DNA postreplication repair and mutagenesis in Saccharomyces cerevisiae. Mutat. Res., 486, 167–184. [DOI] [PubMed] [Google Scholar]
- Buerstedde J.M. and Takeda,S. (1991) Increased ratio of targeted to random integration after transfection of chicken B cell lines. Cell, 67, 179–188. [DOI] [PubMed] [Google Scholar]
- Buerstedde J.M., Reynaud,C.A., Humphries,E.H., Olson,W., Ewert,D.L. and Weill,J.C. (1990) Light chain gene conversion continues at high rate in an ALV-induced cell line. EMBO J., 9, 921–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dronkert M.L., Beverloo,H.B., Johnson,R.D., Hoeijmakers,J.H., Jasin,M. and Kanaar,R. (2000) Mouse RAD54 affects DNA double-strand break repair and sister chromatid exchange. Mol. Cell. Biol., 20, 3147–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito G., Godindagger,I., Klein,U., Yaspo,M.L., Cumano,A. and Rajewsky,K. (2000) Disruption of the Rev3l-encoded catalytic subunit of polymerase ζ in mice results in early embryonic lethality. Curr. Biol., 10, 1221–1224. [DOI] [PubMed] [Google Scholar]
- Essers J., Hendriks,R.W., Swagemakers,S.M., Troelstra,C., de Wit,J., Bootsma,D., Hoeijmakers,J.H. and Kanaar,R. (1997) Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell, 89, 195–204. [DOI] [PubMed] [Google Scholar]
- Fabre F., Magana-Schwencke,N. and Chanet,R. (1989) Isolation of the RAD18 gene of Saccharomyces cerevisiae and construction of rad18 deletion mutants. Mol. Gen. Genet., 215, 425–430. [DOI] [PubMed] [Google Scholar]
- Faili A., Aoufouchi,S., Flatter,E., Gueranger,Q., Reynaud,C.A. and Weill,J.C. (2002) Induction of somatic hypermutation in immunoglobulin genes is dependent on DNA polymerase iota. Nature, 419, 944–947. [DOI] [PubMed] [Google Scholar]
- Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. ASM Press, Washington, DC.
- Fujimori A. et al. (2001) Rad52 partially substitutes for the Rad51 paralog XRCC3 in maintaining chromosomal integrity in vertebrate cells. EMBO J., 20, 5513–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima T. et al. (2001) Genetic analysis of the DNA-dependent protein kinase reveals an inhibitory role of Ku in late S–G2 phase DNA double-strand break repair. J. Biol. Chem., 276, 44413–44418. [DOI] [PubMed] [Google Scholar]
- Gibbs P.E., McGregor,W.G., Maher,V.M., Nisson,P. and Lawrence, C.W. (1998) A human homolog of the Saccharomyces cerevisiae REV3 gene, which encodes the catalytic subunit of DNA polymerase ζ. Proc. Natl Acad. Sci. USA, 95, 6876–6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs P.E., Wang,X.D., Li,Z., McManus,T.P., McGregor,W.G., Lawrence,C.W. and Maher,V.M. (2000) The function of the human homolog of Saccharomyces cerevisiae REV1 is required for mutagenesis induced by UV light. Proc. Natl Acad. Sci. USA, 97, 4186–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L., Unk,I., Johnson,R.E., Johansson,E., Burgers,P.M., Prakash,S. and Prakash,L. (2001) Roles of yeast DNA polymerases δ and ζ and of Rev1 in the bypass of abasic sites. Genes Dev., 15, 945–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe B.D. and Jinks-Robertson,S. (2000) DNA polymerase ζ introduces multiple mutations when bypassing spontaneous DNA damage in Saccharomyces cerevisiae. Mol. Cell, 6, 1491–1499. [DOI] [PubMed] [Google Scholar]
- Holbeck S.L. and Strathern,J.N. (1997) A role for REV3 in mutagenesis during double-strand break repair in Saccharomyces cerevisiae. Genetics, 147, 1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.E., Torres-Ramos,C.A., Izumi,T., Mitra,S., Prakash,S. and Prakash,L. (1998) Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites. Genes Dev., 12, 3137–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.E., Liu,N. and Jasin,M. (1999a) Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature, 401, 397–399. [DOI] [PubMed] [Google Scholar]
- Johnson R.E., Kondratick,C.M., Prakash,S. and Prakash,L. (1999b) hRAD30 mutations in the variant form of xeroderma pigmentosum. Science, 285, 263–265. [DOI] [PubMed] [Google Scholar]
- Johnson R.E., Washington,M.T., Haracska,L., Prakash,S. and Prakash,L. (2000) Eukaryotic polymerases iota and ζ act sequentially to bypass DNA lesions. Nature, 406, 1015–1019. [DOI] [PubMed] [Google Scholar]
- Kanaar R., Hoeijmakers,J.H. and van Gent,D.C. (1998) Molecular Mechanisms of DNA double strand break repair. Trends Cell Biol., 8, 483–489. [DOI] [PubMed] [Google Scholar]
- Keszenman D.J., Salvo,V.A. and Nunes,E. (1992) Effects of bleomycin on growth kinetics and survival of Saccharomyces cerevisiae: a model of repair pathways. J. Bacteriol., 174, 3125–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C. (1994) The RAD6 DNA repair pathway in Saccharomyces cerevisiae: what does it do, and how does it do it? BioEssays, 16, 253–258. [DOI] [PubMed] [Google Scholar]
- Lawrence C.W. (2002) Cellular roles of DNA polymerase ζ and Rev1 protein. DNA Repair, 1, 425–435. [DOI] [PubMed] [Google Scholar]
- Lawrence C.W. and Christensen,R. (1976) UV mutagenesis in radiation-sensitive strains of yeast. Genetics, 82, 207–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C.W. and Maher,V.M. (2001) Mutagenesis in eukaryotes dependent on DNA polymerase ζ and Rev1p. Philos. Trans. R. Soc. Lond. B. Biol. Sci., 356, 41–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C.W. et al. (2000) Roles of DNA polymerase ζ and Rev1 protein in eukaryotic mutagenesis and translesion replication. In Proceedings of the Cold Spring Harbor Symposia on Quantitative Biology. Vol. 65. CSLH Press, Cold Spring Harbor, NY, pp. 61–69. [DOI] [PubMed]
- Lemontt J.F. (1972) Induction of forward mutations in mutationally defective yeast. Mol. Gen. Genet., 119, 27–42. [DOI] [PubMed] [Google Scholar]
- Lin W., Wu,X. and Wang,Z. (1999) A full-length cDNA of hREV3 is predicted to encode DNA polymerase ζ for damage-induced mutagenesis in humans. Mutat. Res., 433, 89–98. [DOI] [PubMed] [Google Scholar]
- Masutani C. et al. (1999) The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature, 399, 700–704. [DOI] [PubMed] [Google Scholar]
- McDonald J.P., Levine,A.S. and Woodgate,R. (1997) The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics, 147, 1557–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee R.H. and Lawrence,C.W. (1979) Genetic analysis of γ-ray mutagenesis in yeast. I. Reversion in radiation-sensitive strains. Genetics, 93, 361–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee R.H. and Lawrence,C.W. (1980) Genetic analysis of γ-ray mutagenesis in yeast. III. Double-mutant strains. Mutat. Res., 70, 37–48. [DOI] [PubMed] [Google Scholar]
- Morrison A., Christensen,R.B., Alley,J., Beck,A.K., Bernstine,E.G., Lemontt,J.F. and Lawrence,C.W. (1989) REV3, a Saccharomyces cerevisiae gene whose function is required for induced mutagenesis, is predicted to encode a nonessential DNA polymerase. J. Bacteriol., 171, 5659–5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison C. and Takeda,S. (2000) Genetic analysis of homologous DNA recombination in vertebrate somatic cells. Int. J. Biochem. Cell Biol., 32, 817–831. [DOI] [PubMed] [Google Scholar]
- Moynahan M.E., Pierce,A.J. and Jasin,M. (2001) BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell, 7, 263–272. [DOI] [PubMed] [Google Scholar]
- Murakumo Y., Ogura,Y., Ishii,H., Numata,S., Ichihara,M., Croce,C.M., Fishel,R. and Takahashi,M. (2001) Interactions in the error-prone postreplication repair proteins hREV1, hREV3, and hREV7. J. Biol. Chem., 276, 35644–35651. [DOI] [PubMed] [Google Scholar]
- Nelson J.R., Lawrence,C.W. and Hinkle,D.C. (1996a) Deoxycytidyl transferase activity of yeast REV1 protein. Nature, 382, 729–731. [DOI] [PubMed] [Google Scholar]
- Nelson J.R., Lawrence,C.W. and Hinkle,D.C. (1996b) Thymine-thymine dimer bypass by yeast DNA polymerase ζ. Science, 272, 1646–1649. [DOI] [PubMed] [Google Scholar]
- Nelson J.R., Gibbs,P.E., Nowicka,A.M., Hinkle,D.C. and Lawrence,C.W. (2000) Evidence for a second function for Saccharomyces cerevisiae Rev1p. Mol. Microbiol., 37, 549–554. [DOI] [PubMed] [Google Scholar]
- Ohmori H. et al. (2001) The Y-family of DNA polymerases. Mol. Cell, 8, 7–8. [DOI] [PubMed] [Google Scholar]
- Okada T., Sonoda,E., Yamashita,Y.M., Koyoshi,S., Tateishi,S., Yamaizumi,M., Takata,M., Ogawa,O. and Takeda,S. (2002) Involvement of vertebrate Polkappa in Rad18-independent post- replication repair of UV damage. J. Biol. Chem., 277, 48690–48695. [DOI] [PubMed] [Google Scholar]
- Prakash L. (1981) Characterization of postreplication repair in Saccharomyces cerevisiae and effects of rad6, rad18, rev3 and rad52 mutations. Mol. Gen. Genet., 184, 471–478. [DOI] [PubMed] [Google Scholar]
- Prakash L. and Taillon-Miller,P. (1981) Effects of the rad52 gene on sister chromatid recombination in Saccharomyces cerevisiae. Curr. Genet., 3, 247–250. [DOI] [PubMed] [Google Scholar]
- Quah S.K., von Borstel,R.C. and Hastings,P.J. (1980) The origin of spontaneous mutation in Saccharomyces cerevisiae. Genetics, 96, 819–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud C.A., Bertocci,B., Dahan,A. and Weill,J.C. (1994) Formation of the chicken B-cell repertoire: ontogenesis, regulation of Ig gene rearrangement, and diversification by gene conversion. Adv. Immunol., 57, 353–378. [DOI] [PubMed] [Google Scholar]
- Roche H., Gietz,R.D. and Kunz,B.A. (1994) Specificity of the yeast rev3 δ antimutator and REV3 dependency of the mutator resulting from a defect (rad1 δ) in nucleotide excision repair. Genetics, 137, 637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale J.E., Calandrini,D.M., Takata,M., Takeda,S. and Neuberger,M.S. (2001) Ablation of XRCC2/3 transforms immunoglobulin V gene conversion into somatic hypermutation. Nature, 412, 921–926. [DOI] [PubMed] [Google Scholar]
- Setlow R.B. (1985) DNA repair,aging and cancer. Natl Inst. Cancer Monogr., 60, 249–255. [PubMed] [Google Scholar]
- Simon J.A., Szankasi,P., Nguyen,D.K., Ludlow,C., Dunstan,H.M., Roberts,C.J., Jensen,E.L., Hartwell,L.H. and Friend,S.H. (2000) Differential toxicities of anticancer agents among DNA repair and checkpoint mutants of Saccharomyces cerevisiae. Cancer Res., 60, 328–333. [PubMed] [Google Scholar]
- Simpson L.J. and Sale,J.E. (2003) Rev1 is essential for DNA damage tolerance and non-templated immunoglobulin gene mutation in a vertebrate cell line.. EMBO J., 22, 1654–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E., Sasaki,M.S., Buerstedde,J.-M., Bezzubova,O., Shinohara,A., Ogawa,H., Takata,M., Yamaguchi-Iwai,Y. and Takeda,S. (1998) Rad51 deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J., 17, 598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E., Sasaki,M.S., Morrison,C., Yamaguchi-Iwai,Y., Takata,M. and Takeda,S. (1999) Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol. Cell. Biol., 19, 5166–5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E. et al. (2001a) Scc1/Rad21/Mcd1 is required for sister chromatid cohesion and kinetochore function in vertebrate cells. Dev. Cell, 1, 759–770. [DOI] [PubMed] [Google Scholar]
- Sonoda E., Takata,M., Yamashita,Y.M., Morrison,C. and Takeda,S. (2001b) Homologous DNA recombination in vertebrate cells. Proc. Natl Acad. Sci. USA, 98, 8388–8394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson R.L., Morey,N.J., Doetsch,P.W. and Jinks-Robertson,S. (1999) Overlapping specificities of base excision repair, nucleotide excision repair, recombination, and translesion synthesis pathways for DNA base damage in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 2929–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao N. et al. (1999) Disruption of ATM in p53-null cells causes multiple functional abnormalities in cellular response to ionizing radiation. Oncogene, 18, 7002–7009. [DOI] [PubMed] [Google Scholar]
- Takata M., Sasaki,M.S., Sonoda,E., Morrison,C., Hashimoto,M., Utsumi,H., Yamaguchi-Iwai,Y., Shinohara,A. and Takeda,S. (1998) Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J., 17, 5497–5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi S., Sakuraba,Y., Masuyama,S., Inoue,H. and Yamaizumi,M. (2000) Dysfunction of human Rad18 results in defective postreplication repair and hypersensitivity to multiple mutagens. Proc. Natl Acad. Sci. USA, 97, 7927–7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauchi H. et al. (2002) Nbs1 is essential for DNA repair by homologous recombination in higher vertebrate cells. Nature, 420, 93–98. [DOI] [PubMed] [Google Scholar]
- van Gent D.C., Hoeijmakers,J.H. and Kanaar,R. (2001) Chromosomal stability and the DNA double-stranded break connection. Nat. Rev. Genet., 2, 196–206. [DOI] [PubMed] [Google Scholar]
- Van Sloun P.P., Romeijn,R.J. and Eeken,J.C. (1999) Molecular cloning, expression and chromosomal localisation of the mouse Rev3l gene, encoding the catalytic subunit of polymerase ζ. Mutat. Res., 433, 109–116. [DOI] [PubMed] [Google Scholar]
- Van Sloun P.P., Varlet,I., Sonneveld,E., Boei,J.J., Romeijn,R.J., Eeken,J.C. and De Wind,N. (2002) Involvement of mouse Rev3 in tolerance of endogenous and exogenous DNA damage. Mol. Cell. Biol., 22, 2159–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zeeland A.A., Natarajan,A.T., Verdegaal-Immerzeel,E.A. and Filon,A.R. (1980) Photoreactivation of UV induced cell killing, chromosome aberrations, sister chromatid exchanges, mutations and pyrimidine dimers in Xenopus laevis fibroblasts. Mol. Gen. Genet., 180, 495–500. [DOI] [PubMed] [Google Scholar]
- Verkaik N.S., Esveldt-van Lange,R.E., van Heemst,D., Bruggenwirth, H.T., Hoeijmakers,J.H., Zdzienicka,M.Z. and van Gent,D.C. (2002) Different types of V(D)J recombination and end-joining defects in DNA double-strand break repair mutant mammalian cells. Eur. J. Immunol., 32, 701–709. [DOI] [PubMed] [Google Scholar]
- Wang W., Seki,M., Narita,Y., Sonoda,E., Takeda,S., Yamada,K., Masuko,T., Katada,T. and Enomoto,T. (2000) Possible association of BLM in decreasing DNA double strand breaks during DNA replication. EMBO J., 19, 3428–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittschieben J., Shivji,M.K., Lalani,E., Jacobs,M.A., Marini,F., Gearhart,P.J., Rosewell,I., Stamp,G. and Wood,R.D. (2000) Disruption of the developmentally regulated Rev3l gene causes embryonic lethality. Curr. Biol., 10, 1217–1220. [DOI] [PubMed] [Google Scholar]
- Wood R.D., Mitchell,M., Sgouros,J. and Lindahl,T. (2001) Human DNA repair genes. Science, 291, 1284–1289. [DOI] [PubMed] [Google Scholar]
- Woodgate R. (2001) Evolution of the two-step model for UV-mutagenesis. Mutat. Res., 485, 83–92. [DOI] [PubMed] [Google Scholar]
- Xiao W., Chow,B.L., Fontanie,T., Ma,L., Bacchetti,S., Hryciw,T. and Broomfield,S. (1999) Genetic interactions between error-prone and error-free postreplication repair pathways in Saccharomyces cerevisiae. Mutat. Res., 435, 1–11. [DOI] [PubMed] [Google Scholar]
- Yamashita Y.M., Okada,T., Matsusaka,T., Sonoda,E., Zhao,G.Y., Araki,K., Tateishi,S., Yamaizumi,M. and Takeda,S. (2002) RAD18 and RAD54 cooperatively contribute to maintenance of genomic stability in vertebrate cells. EMBO J., 21, 5558–5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wienands,J., Zurn,C. and Reth,M. (1998) Induction of the antigen receptor expression on B lymphocytes results in rapid competence for signaling of SLP-65 and Syk. EMBO J., 17, 7304–7310. [DOI] [PMC free article] [PubMed] [Google Scholar]