Abstract
The cryptochrome blue light photoreceptors mediate various photomorphogenic responses in plants, including hypocotyl elongation, cotyledon expansion, and control of flowering time. The molecular mechanism of cryptochrome function in Arabidopsis is becoming increasingly clear, with recent studies showing that both CRY1 and CRY2 are localized in the nucleus and that CRY2 is regulated by blue light–dependent phosphorylation. Despite these advances, no positive cryptochrome signaling component has been identified to date. Here, we demonstrate that a novel Ser/Thr protein phosphatase (AtPP7) with high sequence similarity to the Drosophila retinal degeneration C protein phosphatase acts as an intermediate in blue light signaling. Transgenic Arabidopsis seedlings with reduced AtPP7 expression levels exhibit loss of hypocotyl growth inhibition and display limited cotyledon expansion in response to blue light irradiation. These effects are as striking as those seen in hy4 mutant seedlings, which are deficient in CRY1. We further demonstrate that AtPP7 transcript levels are not rate limiting and that AtPP7 probably acts downstream of cryptochrome in the nucleus, ensuring signal flux through the pathway. Based on our findings and recent data regarding cryptochrome action, we propose that AtPP7 acts as a positive regulator of cryptochrome signaling in Arabidopsis.
INTRODUCTION
Because of their sessile nature, plants need to be extremely adaptable to light. Not only does light act as an energy source for photosynthesis, but plants have to monitor the light quality and quantity input to execute appropriate physiological and developmental responses. To enable them to do this, plants have evolved a set of photoreceptors, including the blue/UV A light–absorbing cryptochromes and the red/far-red light–absorbing phytochromes, of which the latter are the best understood (Furuya, 1993; Quail et al., 1995; Neff et al., 2000). Although it has been demonstrated clearly that phytochromes and cryptochromes act together and that there is crosstalk between these two pathways (Ahmad et al., 1998; Jarillo et al., 2001), little is known about the cryptochrome signaling pathway(s).
The first molecular genetic breakthrough regarding blue light signaling in plants came when Ahmad and Cashmore (1993) isolated the disrupted locus in hy4, an Arabidopsis mutant showing loss of hypocotyl growth inhibition in response to blue light, and showed that HY4 encodes a blue light receptor protein, CRY1. CRY1 contains 681 amino acids and has ∼30% sequence similarity to DNA photolyases (Ahmad and Cashmore, 1993). Although CRY1 is able to bind the chromophore typically found in photolyases, it has no photolyase activity (Lin et al., 1995). The notion that additional blue light photoreceptors exist came from findings demonstrating that hy4 seedlings are impaired only in certain responses (Koornneef et al., 1980; Ahmad and Cashmore, 1993). A second gene whose product shows ∼55% similarity to CRY1 at the amino acid level was identified and was named CRY2 (Hoffman et al., 1996; Lin et al., 1996). Like CRY1, CRY2 has no photolyase activity (Hoffman et al., 1996). The late-flowering Arabidopsis mutant fha1 has a genetic lesion in CRY2 (Guo et al., 1998), demonstrating that CRY2 is involved in sensing daylength.
Recently, it was demonstrated that both CRY1 and CRY2 can be detected in the nucleus (Cashmore et al., 1999; Guo et al., 1999; Kleiner et al., 1999). It also has been reported that blue light induces the phosphorylation of Arabidopsis CRY2, which in turn triggers photomorphogenic responses and protein degradation. Conversely, in darkness, CRY2 remains unphosphorylated in a stable but inactive form (Shalitin et al., 2002). These results suggest that potential light-dependent protein kinases may be involved in the blue light–dependent activation of CRY2.
Although our understanding of cryptochrome action is increasing, insight into how the perceived blue light signals are transduced, leading to morphological responses and altered gene expression patterns, remains sparse. To date, only one signaling intermediate, SUB1, has been identified. SUB1 encodes an Arabidopsis Ca2+ binding protein (Guo et al., 2001), and SUB1 deficiency results in hypersensitivity toward blue light irradiation, indicating that the protein acts as a negative regulator of the blue light signaling cascade. In addition, SUB1 deficiency results in hypersensitivity toward far-red light, and genetic studies have shown that SUB1 acts as an integral component of blue light signaling and as a modulator of phytochrome signaling. Therefore, SUB1 defines a point of crosstalk between the two pathways.
To date, no positive signaling component has been identified in plants involved in cryptochrome action. The novel Ser/Thr protein phosphatase PP7 (AtPP7) isolated from Arabidopsis shows structural features similar to those of the Drosophila retinal degeneration C (rdgC) subfamily of protein phosphatases (Andreeva et al., 1998). Subsequent biochemical studies have demonstrated that AtPP7 is in fact a Ser/Thr protein phosphatase able to bind calmodulin in a Ca2+-dependent manner (Kutuzov et al., 2001). Moreover, AtPP7 has been shown to be a nuclear protein (Andreeva and Kutuzov, 2001).
Because protein phosphorylation plays an important role in cryptochrome activation in response to blue light irradiation (Shalitin et al., 2002), it is reasonable to assume that protein dephosphorylation has equally important regulatory functions (Luan, 1998). Indeed, it has been shown that the human blue light photoreceptor hCRY2 interacts with Ser/Thr protein phosphatase 5 and that hCRY2 modulates the phosphatase activity (Zhao and Sancar, 1997). Here, we demonstrate that the Ser/Thr protein phosphatase AtPP7 represents a positive regulator and signaling component of cryptochrome signal transduction. Our results suggest that this nucleus-localized Ser/Thr phosphatase probably acts downstream but in concert with cryptochrome in the nucleus to activate blue light signaling. These findings represent an example of a positive regulator of cryptochrome signal transduction.
RESULTS
Generation of Transgenic Plants Deficient for AtPP7 and Initial Characterization
To dissect the functional role of AtPP7 in plants, we generated transgenic Arabidopsis plants containing an antisense AtPP7 transgene. To this end, we initially amplified a full-length Arabidopsis AtPP7 cDNA followed by further amplification of the 311-bp 5′ end of the AtPP7 cDNA fragment. This smaller fragment (nucleotides 116 to 426) was used to construct a binary vector containing the AtPP7 5′ fragment in the antisense orientation transcribed from the 35S promoter of Cauliflower mosaic virus (Figure 1A). We selected the 5′ region of AtPP7 because of the relatively low sequence similarity in this region. In addition, the use of a short antisense transcript enables the simultaneous detection of both the endogenous AtPP7 transcript and the AtPP7 antisense transcript by RNA gel blot analysis. The resulting construct was transformed into Arabidopsis ecotype Columbia (Col), and T1 seedlings were selected for further analysis. During the T1 selection process, we observed a high proportion (∼20%) of Basta-resistant seedlings exhibiting loss of hypocotyl growth inhibition under white light conditions. Transgenic seedlings exhibiting both elongated hypocotyls and normal wild-type hypocotyl length were analyzed further in the T2 generation, revealing elongated hypocotyls as observed in the T1 generation.
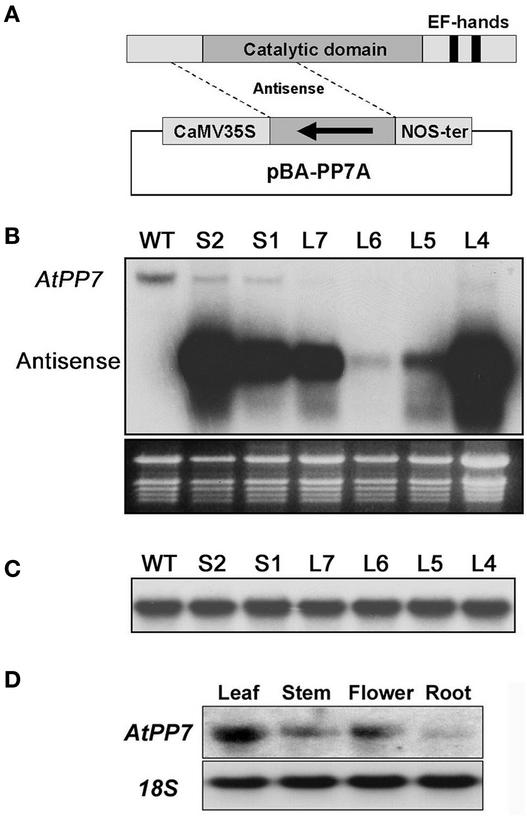
Figure 1.
Molecular Analysis of Arabidopsis AtPP7 Antisense Plants and Tissue-Specific Expression Profiles.
(A) Scheme of the AtPP7 antisense fragment used to generate AtPP7 antisense transgenic Arabidopsis plants. CaMV, Cauliflower mosaic virus; NOS, nopaline synthase.
(B) The top gel shows the analysis of AtPP7 transcript levels. The RNA gel blot was probed with the AtPP7 antisense cDNA fragment. Note the presence of the endogenous AtPP7 transcript in wild-type Columbia (WT) plants and in two wild-type-length hypocotyl transgenic seedlings (S1 and S2) and the lack of endogenous AtPP7 transcript in four independent long-hypocotyl transgenic Arabidopsis seedlings (L7, L6, L5, and L4) harboring the construct shown in (A). The presence of the antisense AtPP7 transcript is indicated. The bottom gel shows the stained RNA gel with rRNA bands as loading controls.
(C) RNA gel blot analysis using the membrane shown in (A) probed with a genomic region homologous with AtPP7 demonstrating no changes in transcript level between wild-type Columbia (WT) and transgenic seedlings.
(D) Tissue-specific expression of AtPP7. The RNA gel blot was probed with the full-length AtPP7 cDNA. Note that AtPP7 is highly expressed in leaves.
To ensure that the observed phenotype correlated with a reduction of endogenous AtPP7 transcript, total RNA was harvested from leaf tissue from four transgenic lines (L4, L5, L6, and L7) showing elongated hypocotyls and from two transgenic lines (S1 and S2) showing wild-type-length hypocotyls. Leaf tissue was used because of the higher AtPP7 transcript levels in this part of wild-type plants (Figure 1D). After RNA gel blot analysis using the antisense AtPP7 cDNA as a probe, we observed a clear reduction in endogenous AtPP7 transcript levels in all four long-hypocotyl transgenic lines tested compared with the wild type (Figure 1B), with virtually undetectable endogenous AtPP7 transcript. By contrast, transgenic seedlings with wild-type-length hypocotyls (S1 and S2) showed a much less severe reduction in endogenous AtPP7 transcript levels (Figure 1B).
To ensure that the observed antisense effect was the result of a true deficiency of AtPP7 transcript and not of spurious hybridization events to other RNA species, we scanned the Arabidopsis genome for regions homologous with the AtPP7 antisense fragment. From this analysis, we found two short genomic regions (region 1, 29 bp, chromosome I; region 2, 108 bp, chromosome V) with similarity (95 and 84%, respectively) to the AtPP7 antisense fragment. We amplified these two regions in addition to some flanking sequence using PCR and used the amplified products as probes on the same RNA gel blot shown in Figure 1B. This analysis revealed that only one of the homologous regions was part of an open reading frame. Moreover, the transcript level of this open reading frame did not vary between the wild-type and the AtPP7 antisense transgenic lines (Figure 1C). From this analysis, it was clear that the reduction of endogenous AtPP7 transcript levels correlates with the loss of hypocotyl growth inhibition in response to white light irradiation.
AtPP7 Deficiency Results in Loss of Hypocotyl Growth Inhibition and Cotyledon Opening in Response to Blue Light Irradiation
To further examine the effect of AtPP7 deficiency on hypocotyl elongation, we analyzed the wavelength specificity of the observed long-hypocotyl phenotype. We subjected two independent transgenic Arabidopsis lines with reduced AtPP7 expression (L5 and L7) to darkness and high-fluence blue light, red light, far-red light, and white light irradiation for 4 to 5 days. As a control, we used transgenic seedlings containing the empty binary vector (pBA002), which in our analysis behaved like wild-type seedlings. Our results showed no significant difference in hypocotyl elongation between vector control seedlings and L5 and L7 AtPP7-deficient seedlings in darkness or when treated with high-fluence far-red or red light (Figure 2A). As described above, we observed a twofold increase in hypocotyl elongation in response to white light irradiation in L5 and L7 seedlings compared with vector control seedlings (Figure 2). Interestingly, in response to blue light irradiation (12 μmol·m−2·s−1), L5 and L7 seedlings showed a striking loss of hypocotyl growth inhibition, with an approximately threefold increase in hypocotyl elongation compared with vector control seedlings (Figure 2).
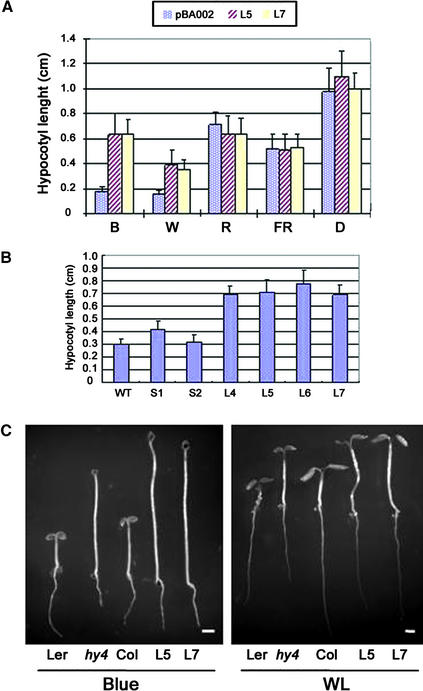
Figure 2.
Phenotypic Analysis of AtPP7-Deficient Arabidopsis Seedlings in Response to Different Light Treatments.
(A) Hypocotyl lengths of vector control seedlings (pBA002) and two AtPP7 antisense transgenic Arabidopsis seedlings (L5 and L7) in darkness (D) and in response to blue (B; 12 μmol·m−2·s−1), white (W; 32 μmol·m−2·s−1), red (R; 25 μmol·m−2·s−1), and far-red (FR; 4.4 μmol·m−2·s−1) light irradiation. n = 30, and standard error bars are shown.
(B) Hypocotyl lengths of wild-type (WT) and six AtPP7 antisense transgenic Arabidopsis seedlings (S1, S2, L4, L5, L6, and L7) in response to blue light irradiation. The fluence rate of blue light is the same as in (A). Col was used as a wild-type control.
(C) Hypocotyl lengths of AtPP7-deficient seedlings and CRY1-deficient (hy4) seedlings in response to white light (WL) and blue light irradiation. Fluence rates of blue and white light are the same as in (A). Col was used as a wild-type control for AtPP7-deficient seedlings, and Landsberg erecta (Ler) was used as the wild-type control for hy4. Bars = 1 mm.
To expand these findings, we tested two additional long-hypocotyl transgenic lines (L4 and L6) and two wild-type-length hypocotyl transgenic lines (S1 and S2) in response to blue light treatment. This analysis showed that the severity of the observed hypocotyl phenotype (Figure 2B) correlated with the level of endogenous AtPP7 transcript (Figure 1B). The observed blue light hypocotyl phenotype prompted us to compare the response of CRY1-deficient hy4-1 seedlings and AtPP7-deficient seedlings grown under blue light and white light. In this analysis, AtPP7-deficient seedlings showed similar hypocotyl length to hy4-1 seedlings under white light conditions and in response to blue light irradiation (Figure 2C). To ensure that our seed stocks were not contaminated with hy4-1 seeds or that the transgenic plants had a CRY1 mutation, we sequenced the CRY1 gene in our transgenic plants and found that the CRY1 nucleotide sequence was identical to that in wild-type Arabidopsis.
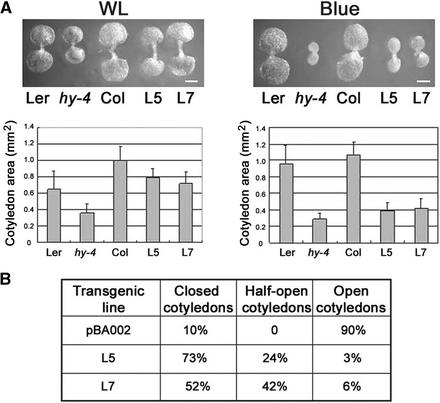
We also analyzed the effect of AtPP7 deficiency on cotyledon opening, because impaired blue light signaling in CRY-deficient seedlings has a marked effect on cotyledon size. As expected, similar to hy4-1 seedlings, transgenic seedlings deficient for AtPP7 expression had smaller cotyledons than wild-type seedlings in response to blue light irradiation (Figure 3A). The difference in cotyledon size between wild-type seedlings and CRY1- or AtPP7-deficient seedlings was less striking in white light (Figure 3A). We also analyzed the effect of blue light irradiation on cotyledon expansion and found that AtPP7-deficient seedlings had between 50 and 70% closed cotyledons compared with vector control seedlings, which had ∼90% open cotyledons (Figure 3B). Thus, it is clear that AtPP7 deficiency in Arabidopsis results in the impairment of blue light signaling events, leading to loss of hypocotyl growth inhibition and cotyledon expansion in response to blue light irradiation. This finding suggests that AtPP7 is a positive regulator of blue light signaling in Arabidopsis.
Figure 3.
Cotyledon Expansion and Size of AtPP7-Deficient Seedlings in Response to White Light and Blue Light Irradiation.
(A) White light (WL; 32 μmol·m−2·s−1) or blue light (12 μmol·m−2·s−1) was given for 5 days. Col was used as a wild-type control for AtPP7-deficient seedlings, and Landsberg erecta (Ler) was used as the wild-type control for hy4. n = 20. Bars = 0.1 mm.
(B) Cotyledon opening of empty vector–transformed and AtPP7-deficient Arabidopsis seedlings after 5 days of blue light (12 μmol·m−2·s−1) irradiation. Thirty to 40 seedlings were measured for each data point.
AtPP7 Levels Are Not Rate Limiting in Response to Blue Light Irradiation
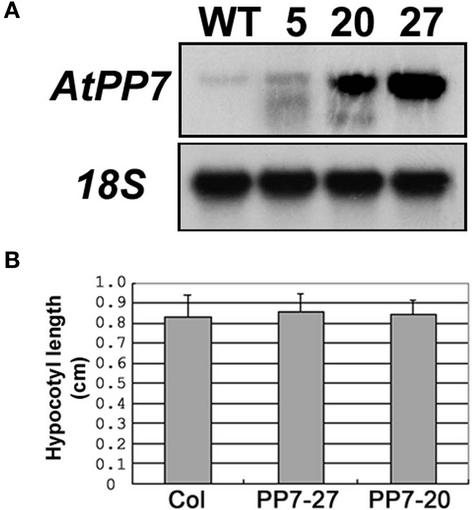
Because AtPP7 deficiency results in insensitivity to blue light irradiation, we analyzed whether increased levels of AtPP7 results in hypersensitivity. We generated a binary vector containing the full-length AtPP7 cDNA under the control of the 35S promoter of Cauliflower mosaic virus. After transformation into Arabidopsis ecotype Col, we selected several T1 transgenic seedlings and performed RNA gel blot analysis using the AtPP7 cDNA as a probe. We identified five transgenic lines showing increased AtPP7 transcript levels, and two of these overexpressing lines (PP7-20 and PP7-27; Figure 4) were analyzed for hypersensitivity in the subsequent T2 generation. We found that increased levels of AtPP7 had no effect on hypocotyl length in response to low-fluence (0.6 μmol·m−2·s−1) blue light irradiation (Figure 4B). However, under low fluences of blue light, the signal flux through the CRY1 pathway may not be sufficient to evoke hypersensitivity. Thus, we also analyzed hypocotyl lengths in response to higher blue light fluences (12 μmol·m−2·s−1), which also had no effect on hypocotyl length (data not shown). These findings indicate that in wild-type plants AtPP7 is present at sufficiently high levels so that it is not rate limiting for blue light signaling.
Figure 4.
Overexpression of AtPP7 in Transgenic Arabidopsis Plants.
(A) RNA gel blot probed with the full-length AtPP7 cDNA showing two transgenic Arabidopsis lines (PP7-20 and PP7-27) exhibiting AtPP7 overexpression. WT, wild type.
(B) Hypocotyl lengths of wild-type (Col) and AtPP7-overexpressing Arabidopsis lines PP7-20 and PP7-27 in response to low-fluence blue light irradiation (0.6 μmol·m−2·s−1). n = 30 and standard error bars are shown.
AtPP7 Deficiency Results in the Attenuation of Light-Regulated Gene Expression
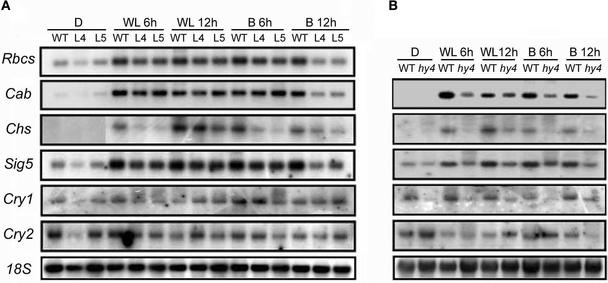
It has been demonstrated that cryptochromes mediate the general light regulation of gene expression (Lin, 2002). In view of this finding, we analyzed the effect of AtPP7 deficiency on expression levels of several light-regulated transcripts in response to white light and blue light irradiation. Wild-type and AtPP7-deficient seedlings (L4 and L5) were exposed to white light and blue light for 6 and 12 h followed by RNA gel blot analysis. In CRY1-deficient seedlings (hy4), we found that AtPP7 deficiency resulted in a clear attenuation of chalcone synthase (Chs) expression in response to 6 and 12 h of both white light and blue light irradiation compared with wild-type seedlings (Figure 5B). In L4 and L5 seedlings, the attenuation in blue light was more severe than that in white light conditions, in which only limited reduction was found in response to 12 h of white light treatment (Figure 5A), similar to the effect seen in hy4 seedlings (Figure 5B). In addition, we observed a clear reduction of ribulose-1,5-bisphosphate carboxylase small subunit (Rbcs) and chlorophyll a/b binding protein (Cab) expression after 12 h of blue light treatment compared with the wild type (Figure 5A). However, in response to white light treatment, no attenuation was observed (Figure 5A), a finding different from that seen in hy4 seedlings, which showed a reduction in Cab expression under white light conditions (Figure 5B).
Figure 5.
RNA Gel Blot Analysis of Light-Regulated Genes in Response to White Light and Blue Light Irradiation.
(A) RNA samples were collected from wild-type Col seedlings (WT) and from two transgenic Arabidopsis lines deficient for AtPP7 (L4 and L5) in darkness (D), in response to 6 and 12 h of blue light irradiation (25 μmol·m−2·s−1) (B 6h and B12h), and in response to 6 and 12 h of white light irradiation (32 μmol·m−2·s−1) (WL 6h and WL 12h). The same membrane was probed with Rbcs, Cab, Chs, Sig5, CRY1, and CRY2. 18S rRNA was used as a loading control.
(B) RNA samples were collected from wild-type Landsberg erecta (WT) and hy4 seedlings in darkness and in response to 6 and 12 h of both white light and blue light irradiation (abbreviations as in [A]). The same membrane was probed with Chs, Sig5, CRY1, and CRY2. 18S rRNA was used as a loading control.
These results suggest a disruption in the cryptochrome signaling cascade slightly different from those observed in cryptochrome-deficient mutants. Cryptochromes have been shown to regulate the expression of chloroplast genes, such as psbD (Thum et al., 2001). This regulation is mediated indirectly through the regulation of nucleus-encoded σ factors, such as Sig5, which exhibits blue light–specific transcript induction (Tsunoyama et al., 2002). To determine whether AtPP7 deficiency affects chloroplast gene expression, we analyzed Sig5 transcript levels. As excepted, we found that in response to blue light treatment, Sig5 was clearly induced in wild-type seedlings, whereas in AtPP7-deficient seedlings, Sig5 levels were reduced after 12 h of blue light treatment (Figure 5A). In response to white light treatment, virtually no effect on Sig5 expression was observed in AtPP7-deficient seedlings (Figure 5A). We also analyzed CRY1 and CRY2 transcript levels in response to white light and blue light and found no significant differences between wild-type, L5, and L7 seedlings (Figure 5), demonstrating that AtPP7 probably resides downstream of both CRY1 and CRY2.
Together, our data demonstrate that AtPP7 acts as a component of blue light signaling, ensuring the appropriate regulation of gene expression.
DISCUSSION
The blue/UV A light–perceiving cryptochromes and the red/far-red light–perceiving phytochromes are the main photoreceptors in plants that mediate light-regulated growth and development (Kendrick and Kronenberg, 1994; Quail et al., 1995). Although the molecular and cell biological mechanisms of phytochrome signaling are being elucidated (Møller et al., 2002), signal transduction downstream of plant cryptochrome still remains largely unclear. To our knowledge, no positive component of cryptochrome signaling in plants has been identified to date. Our studies reveal the involvement of a novel Ser/Thr protein phosphatase as a positive regulator of blue light signaling in Arabidopsis.
Arabidopsis Ser/Thr Protein Phosphatase Acts as a Positive Regulator of Blue Light Signaling
AtPP7 was identified initially as a novel Ser/Thr protein phosphatase from Arabidopsis showing limited amino acid similarity (∼30%) to other phosphatases (Andreeva et al., 1998). Interestingly, AtPP7 displays remarkable structural similarity to the protein phosphatase identified from the Drosophila rdgC mutant (Steele et al., 1992), showing a unique structure among PPP family members. Fruit fly PP7, human PP7 (Huang and Honkanen, 1998), and AtPP7 contain two putative Ca2+ binding EF hands downstream of the phosphatase catalytic domain. This is different from the PP2B family of phosphatases, which exhibits Ca2+ binding through the B regulatory subunit and calmodulin. Despite these differences, there is evidence that AtPP7 can bind calmodulin, albeit weakly, in a Ca2+-dependent manner (Kutuzov et al., 2001). However, Kutuzov et al. (1998) reported that the phosphatase activity of recombinant AtPP7 is stimulated by Mn2+ and not by Ca2+, an observation confirmed by us (data not shown). It is clear from these studies that some information exists concerning the biochemical properties of this novel AtPP7 phosphatase, although the role of this protein in plants remains unknown.
In light of this fact and to dissect the functional role of AtPP7 in Arabidopsis, we generated transgenic plants harboring an AtPP7 antisense construct. Molecular analysis identified several transgenic lines with reduced AtPP7 transcript levels (Figure 1B). We demonstrated that all of these plants exhibited loss of hypocotyl growth inhibition, primarily in response to blue light irradiation but also in response to white light conditions, and that the severity of the observed insensitivity correlated with the slight fluctuations of endogenous AtPP7 transcript levels (Figure 2B). This insensitivity towards blue light irradiation was similar to that in CRY1-deficient hy4 seedlings (Figure 2C), indicating a severe impairment of blue light signaling. Although one concern could be possible hy4 seed contamination in our transgenic seed stocks or a mutation in the CRY1 gene itself, we have several lines of evidence against these possibilities. First, only a fraction of Basta-resistant T1 seedlings showed the long-hypocotyl phenotype. Second, the long-hypocotyl phenotype segregated at the T2 generation. Third, the phenotype varied in severity between independent transgenic lines. Fourth, the CRY1 nucleotide sequence in our transgenic plants was identical to that in wild-type Arabidopsis seedlings. Confocal microscopy further verified that the loss of hypocotyl growth inhibition was attributable to hypocotyl cell expansion, as seen in hy4 (data not shown). In addition, we also observed a large proportion of AtPP7-deficient seedlings with closed cotyledons in response to blue light irradiation, another phenotype similar to that observed in hy4 seedlings (Figure 3). Our results indicate that AtPP7 acts as a positive regulator of blue light signaling in Arabidopsis, because a reduction in AtPP7 expression levels resulted in decreased signal flux.
Although no positive regulator of blue light signaling has been isolated to date, the Arabidopsis SUB1 protein has been shown to act as a negative regulator downstream of CRY1 and CRY2 (Guo et al., 2001). Interestingly, SUB1 is a Ca2+ binding protein that has two EF hands at the C-terminal region, similar to AtPP7. SUB1 deficiency in Arabidopsis results in hypersensitivity toward both blue and far-red light, suggesting that SUB1 acts as a signaling component downstream of cryptochrome but as a modulator of phytochrome A signaling (Guo et al., 2001). These results suggest that SUB1 defines a point of crosstalk between cryptochrome and phytochrome. By contrast, our findings here demonstrate that AtPP7 deficiency has no effect on hypocotyl elongation or cotyledon expansion in response to monochromatic red or far-red light (Figure 2). Therefore, AtPP7 represents a true blue light–specific signaling component.
AtPP7 Is a Nuclear Protein That Acts Downstream of Cryptochrome to Mediate Blue Light Signaling Events
Because cryptochromes are largely localized to the nucleus in darkness and translocated to the cytosol in response to blue light (Guo et al., 1999; Kleiner et al., 1999), early signaling events may occur in the nucleus. We found that AtPP7 is localized to the nucleus in darkness and in response to blue light irradiation (data not shown), confirming and extending the findings of Andreeva and Kutuzov (2001). The finding that AtPP7 is a constitutively nuclear Ser/Thr protein phosphatase suggested that AtPP7 might function early in the signaling pathway during the dark phase. To test this possibility, we used yeast two-hybrid assays to determine whether AtPP7 interacts physically with cryptochrome. Our results showed that AtPP7 did not interact with CRY1 (data not shown), implying that AtPP7 probably acts physically independently of cryptochrome in the nucleus.
Recently, Shalitin et al. (2002) demonstrated that in response to blue light, CRY2 becomes phosphorylated, leading to activation and photomorphogenic responses. The blue light–induced phosphorylation also marks CRY2 for degradation, which may be involved in photoreceptor desensitization. Our data suggest that AtPP7 ensures signal flux through the pathway by dephosphorylation in response to blue light irradiation. It is possible that AtPP7 dephosphorylates a nuclear signaling intermediate and by doing so activates this intermediate to sustain and amplify blue light signals. Based on the hypocotyl growth inhibition studies (Figure 2) and the gene expression analysis of light-regulated genes (Figure 5), AtPP7 is probably not involved in the primary response to blue light. AtPP7 may not act only as a simple positive signaling component but also may act as an important signaling checkpoint to fine-tune the signal flux downstream of cryptochrome. One way of addressing this issue is to identify AtPP7-interacting proteins; these studies are under way at present.
Conclusions
In conclusion, we have identified a positive regulator of cryptochrome signaling in plants. We have shown that AtPP7, a Ser/Thr protein phosphatase, acts as a nucleus-localized signaling component that probably acts downstream of but in concert with cryptochrome in response to blue light irradiation. Our results suggest that AtPP7 ensures signal flux in response to blue light irradiation by possible dephosphorylation of a nuclear signaling intermediate. The nature of this dephosphorylation event and the nature of the target intermediate remain to be elucidated, representing exciting future challenges.
METHODS
Plant Material and Plant Transformation
Arabidopsis thaliana ecotypes Columbia (Col) and Landsberg erecta were used for all experiments. Both wild-type and transgenic plants were propagated in an identical manner. Surface-sterilized seeds were sown on Murashige and Skoog (1962) (MS) medium and placed at 4°C in darkness for 2 to 4 days followed by germination under continuous white light at 22°C. Seedlings then were transferred to 8-h-light/16-h-dark cycles at 22°C or transferred to soil and grown under standard greenhouse conditions. Transgenic Arabidopsis plants were generated using the Agrobacterium tumefaciens–mediated floral-dip method (Clough and Bent, 1998), and primary transformants were selected on 15 μg/mL dl-phosphinotricin (Basta; Research Products International, Mount Prospect, IL) and transplanted onto fresh MS medium or soil.
Cloning of AtPP7 and Generation of AtPP7 Antisense and Overexpression Plants
A full-length AtPP7 cDNA (1242 bp) was isolated by reverse transcriptase–mediated PCR (ProStar; Stratagene) from total RNA (RNeasy Plant Mini Kit; Qiagen, Valencia, CA) and Pwo DNA polymerase (Roche Diagnostics, Indianapolis, IN) using the primer pair PP7/5 (5′-TACTCGAGATGGAAACTGTTCCACCATCTCCC-3′) and PP7/3 (5′-ATACTAGTTCAGCTATTTGGTTGTTCGTTATTGG-3′) (incorporated XhoI and SpeI restriction sites are underlined). The PCR product was cloned into pCRScript (Stratagene) and subjected to DNA sequencing. For the antisense construct, a 311-bp AtPP7 5′ fragment was amplified by PCR (nucleotides 116 to 426) from the full-length cDNA using primers PP7A/5 (5′-TAACTAGTCCCAGTTACCTTCACTATTGCC-3′) and PP7A/3 (5′-ATCTCGAGGAGTAGGTAGACTCTGTCC-3′) (incorporated XhoI and SpeI restriction sites are underlined) followed by cloning into pBA002 (Kost et al., 1998) in the antisense orientation under the control of the 35S promoter of Cauliflower mosaic virus (CaMV). For the overexpression construct, the full-length AtPP7 cDNA was subcloned into pBA002 (Kost et al., 1998). The resulting constructs were transformed into Arabidopsis ecotype Col as described above.
RNA Gel Blot Analysis
Total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen), and all RNA gel blots contained 10 μg of total RNA. For analysis of AtPP7 antisense plants, RNA gel blots were hybridized to the randomly primed 32P antisense cDNA fragment. For analysis of antisense specificity, the same RNA gel blot described above was hybridized to two randomly primed 32P probes represented by nucleotides 75,322 to 75,430 on P1 clone MSJI (chromosome I) and nucleotides 29,782 to 29,852 on BAC T7A14 (chromosome V). For analysis of AtPP7-overexpressing plants and for tissue-specific expression in wild-type plants, RNA gel blots were hybridized to the randomly primed 32P full-length AtPP7 cDNA. RNA gel blots containing 5 μg of total RNA from vector-transformed and AtPP7 antisense seedlings were hybridized to 32P-labeled cDNA probes encoding Chs, Cab, Rbcs, Sig5, and CRY1. DNA probes of Chs, Cab, and Rbcs were as described elsewhere (Genoud et al., 1998; Møller et al., 2001). The Sig5 probe represented a 459-bp fragment described by Tsunoyama et al. (2002). The CRY1 probe represented a 578-bp fragment of the CRY1 3′ end that was not homologous with CRY2. The CRY2 probe represented a 384-bp fragment of the 3′ end of CRY2 that was not homologous with CRY1.
Analysis of AtPP7 Localization
The full-length AtPP7 cDNA was amplified by PCR, removing the stop codon and incorporating XhoI and KpnI restriction sites (underlined), using the primer combination PP7/5 (5′-TACTCGAGATGGAAACTGTTCCACCATCTCCC-3′) and PP7/GFP3 (5′-TAGGTACCGCTATTTGGTTGTTCGTTATTGG-3′). The amplified fragments were subjected to DNA sequencing and cloned into pGFP2 (Kost et al., 1998) under the control of the CaMV 35S promoter as a translational fusion to the N terminus of green fluorescent protein (GFP), generating the construct pGFP2/AtPP7. The resulting construct was introduced into onion epidermal cells by particle bombardment (Kost et al., 1998) and analyzed for GFP fluorescence using a Zeiss Axioskop microscope (Jena, Germany). For Arabidopsis transformations, the entire CaMV 35S/AtPP7/GFP cassette was subcloned into a promoterless version of the binary vector pBA002 (Kost et al., 1998) and transformed into wild-type Arabidopsis (Col) plants as described above. Positive transgenic T1 and T2 seedlings were irradiated with blue light for 48 h (11 μmol·m−2·s−1) or kept in darkness and analyzed for GFP fluorescence as described above.
Phenotypic Analysis
Wild-type, vector-transformed, AtPP7 antisense, and hy4-1 seeds were surface-sterilized and sown on MS plates without sucrose. Seeds were incubated at 4°C in darkness for 4 days, followed by irradiation with continuous white light for 1 h (for experiments in blue light, red light, and darkness) or 1 day (for the far-red light experiment) to promote germination. Seedlings were grown for 4 to 5 days at 22°C in darkness and under far-red, red, blue, and white light followed by hypocotyl length and cotyledon expansion measurements. Wide-spectrum fluorescent bulbs (model GRO-LUX F96T12/GRO; Sylvania, Danvers, MA) were used for continuous light treatment. The usual white light fluence rate was 32 μmol·m−2·s−1. Continuous red light was provided by red light bulbs (model 660 F48T12/2364/VHO; Sylvania) filtered through one layer of Plexiglas (No. 2793; Atohaas North America, Philadelphia, PA), with a fluence rate of 25 μmol·m−2·s−1. Continuous far-red light (output of light bulbs; model F48T12/232/VHO; Sylvania) was filtered through one layer of Plexiglas (No. 067894; West Lake Plastics, Lenni, PA) and had a fluence rate of 4.4 μmol·m−2·s−1. Continuous blue light was provided by F20T12-B fluorescent bulbs (General Electric, Fairfield, CT) and filtered through one layer of Plexiglas (No. 2424; Dayton Plastics, Columbus, OH); the fluence rate under the filter was 12 μmol·m−2·s−1 or 0.6 μmol·m−2·s−1. Cotyledon sizes were measured using a confocal microscope (Carl Zeiss laser scanning microscope) and Carl Zeiss LSM software. Each sample contained 30 to 40 seedlings.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Numbers
Accession numbers for the sequences mentioned in this article are AJ000057 (AtPP7), AC005322.2 (region 1, chromosome I), and AB008268.1 (region 2, chromosome V).
Acknowledgments
We thank Thierry Genoud for discussion. S.G.M. was supported by a North Atlantic Treaty Organization Science Fellowship, and Y.-S.K. was supported by the Postdoctoral Fellowship Program of the Korea Science and Engineering Foundation. This work was supported by National Institutes of Health Grant GM 44640 to N.-H.C.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.008649.
References
- Ahmad, M., and Cashmore, A.R. (1993). HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366, 162–166. [DOI] [PubMed] [Google Scholar]
- Ahmad, M., Jarillo, J.A., Smirnova, O., and Cashmore, A.R. (1998). The CRY1 blue light photoreceptor of Arabidopsis interacts with phytochrome A in vitro. Mol. Cell 1, 939–948. [DOI] [PubMed] [Google Scholar]
- Andreeva, A.V., Evans, D.E., Hawes, C.R., Bennett, N., and Kutuzov, M.A. (1998). PP7, a plant phosphatase representing a novel evolutionary branch of eukaryotic protein Ser/Thr phosphatases. Biochem. Mol. Biol. Int. 44, 703–715. [DOI] [PubMed] [Google Scholar]
- Andreeva, A.V., and Kutuzov, M.A. (2001). Nuclear localization of the plant protein Ser/Thr phosphatase PP7. Mol. Cell. Biol. Res. Commun. 4, 345–352. [DOI] [PubMed] [Google Scholar]
- Cashmore, A.R., Jarillo, J.A., Wu, Y.J., and Liu, D. (1999). Cryptochromes: Blue light receptors for plants and animals. Science 284, 760–765. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Furuya, M. (1993). Phytochromes: Their molecular species, gene families, and functions. Annu. Rev. Plant Physiol. Plant Mol. Biol. 44, 617–645. [Google Scholar]
- Genoud, T., Miller, A.J., Nishizawa, N., Kay, S.A., Schafer, E., Nagatani, A., and Chua, N.H. (1998). An Arabidopsis mutant hypersensitive to red and far-red light signals. Plant Cell 10, 889–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H., Duong, H., Ma, N., and Lin, C. (1999). The Arabidopsis blue light receptor cryptochrome 2 is a nuclear protein regulated by a blue light-dependent post-transcriptional mechanism. Plant J. 19, 279–287. [DOI] [PubMed] [Google Scholar]
- Guo, H., Mockler, T., Duong, H., and Lin, C. (2001). SUB1, an Arabidopsis Ca2+-binding protein involved in cryptochrome and phytochrome coaction. Science 291, 487–490. [DOI] [PubMed] [Google Scholar]
- Guo, H., Yang, H., Mockler, T.C., and Lin, C. (1998). Regulation of flowering time by Arabidopsis photoreceptors. Science 279, 1360–1363. [DOI] [PubMed] [Google Scholar]
- Hoffman, P.D., Batschauer, A., and Hays, J.B. (1996). PHH1, a novel gene from Arabidopsis thaliana that encodes a protein similar to plant blue-light photoreceptors and microbial photolyases. Mol. Gen. Genet. 253, 259–265. [DOI] [PubMed] [Google Scholar]
- Huang, X., and Honkanen, R.E. (1998). Molecular cloning, expression, and characterization of a novel human serine/threonine protein phosphatase, PP7, that is homologous to Drosophila retinal degeneration C gene product (rdgC). J. Biol. Chem. 273, 1462–1468. [DOI] [PubMed] [Google Scholar]
- Jarillo, J.A., Capel, J., Tang, R.H., Yang, H.Q., Alonso, J.M., Ecker, J.R., and Cashmore, A.R. (2001). An Arabidopsis circadian clock component interacts with both CRY1 and phyB. Nature 410, 487–490. [DOI] [PubMed] [Google Scholar]
- Kendrick, R.E., and Kronenberg, G.H.M. (1994). Photomorphogenesis in Plants, 2nd ed. (Dordrecht, The Netherlands: Kluwer Academic Publishers).
- Kleiner, O., Kircher, S., Harter, K., and Batschauer, A. (1999). Nuclear localization of the Arabidopsis blue light receptor cryptochrome 2. Plant J. 19, 289–296. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Rolf, E., and Spruit, C.J.P. (1980). Genetic control of the light-inhibited hypocotyl elongation in Arabidopsis thaliana. Heynh. Z. Pflanzenphysiol. 100S, 147–160. [Google Scholar]
- Kost, B., Spielhofer, P., and Chua, N.H. (1998). A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualises the actin cytoskeleton in growing pollen tubes. Plant J. 16, 393–401. [DOI] [PubMed] [Google Scholar]
- Kutuzov, M.A., Bennett, N., and Andreeva, A.V. (2001). Interaction of plant protein Ser/Thr phosphatase PP7 with calmodulin. Biochem. Biophys. Res. Commun. 289, 634–640. [DOI] [PubMed] [Google Scholar]
- Kutuzov, M.A., Evans, D.E., and Andreeva, A.V. (1998). Expression and characterization of PP7, a novel plant protein Ser/Thr phosphatase distantly related to RdgC/PPEF and PP5. FEBS Lett. 440, 147–152. [DOI] [PubMed] [Google Scholar]
- Lin, C. (2002). Blue light receptors and signal transduction. Plant Cell 14 (suppl.), S207.–S225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C., Ahmad, M., Chan, J., and Cashmore, A.R. (1996). CRY2: A second member of the Arabidopsis cryptochrome gene family (accession no. U43397) (PGR 96–001). Plant Physiol. 110, 1047.8819875 [Google Scholar]
- Lin, C., Robertson, D.E., Ahmad, M., Raibekas, A.A., Jorns, M.S., Dutton, P.L., and Cashmore, A.R. (1995). Association of flavin adenine dinucleotide with the Arabidopsis blue light receptor CRY1. Science 269, 968–970. [DOI] [PubMed] [Google Scholar]
- Luan, S. (1998). Protein phosphatases and signalling cascades in higher plants. Trends Plant Sci. 3, 271–275. [Google Scholar]
- Møller, S.G., Ingles, P.J., and Whitelam, G.C. (2002). The cell biology of phytochrome signalling. New Phytol. 154, 553–590. [DOI] [PubMed] [Google Scholar]
- Møller, S.G., Kunkel, T., and Chua, N.H. (2001). A plastidic ABC protein involved in intercompartmental communication of light signalling. Genes Dev. 15, 90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Neff, M.M., Fankhauser, C., and Chory, J. (2000). Light: An indicator of time and place. Genes Dev. 14, 257–271. [PubMed] [Google Scholar]
- Quail, P.H., Boylan, M.T., Parks, B.M., Short, T.W., Xu, Y., and Wagner, D. (1995). Phytochromes: Photosensory perception and signal transduction. Science 268, 675–680. [DOI] [PubMed] [Google Scholar]
- Shalitin, D., Yang, H., Mockler, T.C., Maymon, M., Guo, H., Whitelam, G.C., and Lin, C. (2002). Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature 417, 763–767. [DOI] [PubMed] [Google Scholar]
- Steele, F.R., Washburn, T., Rieger, R., and O'Tousa, J.E. (1992). Drosophila retinal degeneration C (rdgC) encodes a novel serine/threonine protein phosphatase. Cell 69, 669–676. [DOI] [PubMed] [Google Scholar]
- Thum, K.E., Kim, M., Christopher, D.A., and Mullet, J.E. (2001). Cryptochrome 1, cryptochrome 2, and phytochrome A co-activate the chloroplast psdD blue light–responsive promoter. Plant Cell 13, 2747–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunoyama, Y., Morikawa, K., Shiina, T., and Toyoshima, Y. (2002). Blue light specific and differential expression of a plastid σ factor, Sig5 in Arabidopsis thaliana. FEBS Lett. 516, 225–228. [DOI] [PubMed] [Google Scholar]
- Zhao, S., and Sancar, A. (1997). Human blue-light photoreceptor hCRY2 specifically interacts with protein serine/threonine phosphatase 5 and modulates its activity. Photochem. Photobiol. 66, 727–731. [DOI] [PubMed] [Google Scholar]