Abstract
Many signal transduction pathways involve heterotrimeric G proteins. The accepted model for activation of heterotrimeric G proteins states that the protein dissociates to the free Gα (GTP)-bound subunit and free Gβγ dimer. On GTP hydrolysis, Gα (GDP) then reassociates with Gβγ [Gilman, A. G. (1987) Annu. Rev. Biochem. 56, 615–649]. We reexamined this hypothesis, by using the mating G protein of the yeast Saccharomyces cerevisiae encoded by the genes GPA1, STE4, and STE18. In the absence of mating pheromone, the Gα (Gpa1) subunit represses the mating pathway. On activation by binding of pheromone to a serpentine receptor, the Gβγ (Ste4, Ste18) dimer transmits the signal to a mitogen-activated protein kinase cascade, leading to gene activation, arrest in the G1 stage of the cell cycle, production of shmoos (mating projections), and cell fusion. We found that a Ste4-Gpa1 fusion protein transmitted the pheromone signal and activated the mating pathway as effectively as when Ste4 (Gβ) and Gpa1 (Gα) were coexpressed as separate proteins. Hence, dissociation of this G protein is not required for its activation. Rather, a conformational change in the heterotrimeric complex is likely to be involved in signal transduction.
Many environmental signals (such as light, odorants, and hormones) are transduced from cell surface receptors to intracellular signaling cascades by means of heterotrimeric G (GTP-binding) proteins (1). These proteins consist of three subunits: α, β, and γ. The Gα subunit contains the guanine nucleotide binding site and has intrinsic GTPase activity. On activation by agonist–receptor complexes, the GDP bound to the Gα subunit is exchanged by GTP. Hydrolysis of the GTP to GDP inactivates the G protein.
G proteins are commonly believed to operate by subunit dissociation (2): on G protein activation, the protein dissociates to the free Gα (GTP)-bound subunit and free Gβγ dimer. On GTP hydrolysis, Gα (GDP) then reassociates with Gβγ. Irreversible activation of G proteins in vitro by certain GTP analogs, such as GTPγS, has been seen to be accompanied by dissociation of Gα from Gβγ, particularly at high concentrations of Mg2+. The biochemical evidence is controversial, however, and we and others have reported that active G protein is not necessarily dissociated into the Gα and Gβγ moieties at physiological concentrations of Mg2+ (reviewed in refs. 3 and 4).
The yeast Saccharomyces cerevisiae mating cascade provided us with an ideal system in which to test the subunit dissociation hypothesis in vivo. The yeast mating G protein is encoded by the haploid-specific genes GPA1, STE4, and STE18. In the absence of mating pheromone, the Gα (Gpa1) subunit represses the mating pathway. On activation by binding of pheromone to a serpentine receptor, the Gβγ (Ste4, Ste18) dimer transmits the signal to a mitogen-activated protein (MAP) kinase cascade, leading to gene activation, arrest in the G1 stage of the cell cycle, production of shmoos (mating projections) and, finally, cell fusion to produce diploid cells (reviewed in refs. 5 and 6). Inactivation of GPA1 by mutation, or overexpression of STE4, leads to uncontrolled expression of the mating cascade and growth arrest (7–9).
In this study, we fused the STE4 and GPA1 genes. The nondissociable Ste4-Gpa1 fusion protein thus produced was fully active in transducing the pheromone signal and promoting mating. Thus, in this system, subunit dissociation is not required for signal transduction.
Materials and Methods
Yeast Media and Genetic Techniques.
S. cerevisiae were grown on SD (synthetic dextrose) medium (10) containing glucose (2%) or galactose (2%). All strains (except NKY102) were isogenic to S. cerevisiae βwt (MATa ura3–52 lys2 leu2 trp1 his3 met). SK1006 is βwt ste4∷LEU2, and SK1007 is βwt ste4∷LEU2 gpa1∷HIS3. Transformants were selected on SD-glucose plates lacking the relevant nutrients. Patch matings were performed by replicating the relevant strains onto a lawn of tester strain NKY102 (MATα ade8) on an SD-glucose or SD-galactose plate lacking uracil. After 5–18 h of incubation at 30°C, the mixture was replicated to an SD-glucose plate with no supplements, to select for diploids. For quantitative mating tests, 4 × 107 exponentially growing washed cells were incubated at 30°C with a large excess of washed tester cells in 200 μl of galactose medium. After washing, appropriate dilutions were plated on SD-glucose plates with no supplements (to score diploids), SD-glucose lacking lysine (to score tester), and SD-glucose lacking adenine (to score the plasmid-carrying strain). For the “halo” assay for sensitivity to α-factor, cells were pregrown for several hours in galactose medium, plated in a lawn of top agar on selective SD glucose or galactose plates, and 1 μg of α-factor (Sugen, South San Francisco, CA) was spotted on the solidified top agar, in 4-μl drops. The plates were incubated for 40 h at 30°C.
Construction of the STE4-GPA1 Fusions.
The STE4-GPA1 fusions were constructed by using plasmid pGT-STE4–1, in which the STE4 gene is transcribed from the GAL1 promoter (8). The XhoI-MfeI fragment encompassing the last 78 bp of the STE4 coding region and an additional 0.6 kb downstream was replaced with a PCR fragment in which the termination codon (TAG) was converted to TTG, generating an MfeI restriction site and deleting the downstream 0.6 kb (generating plasmid pSK15). A synthetic double-stranded (ds) oligonucleotide encoding [(Gly)4Ser] 2 (Gly) 4Phe and the first several amino acids of Gpa1 (but excluding the first Met, which is normally removed by myristoylation), with a 5′ EcoRI compatible overhang was ligated in frame with GPA1 at the BsrGI site. The entire EcoRI fragment, encompassing the linker and GPA1, was inserted into the MfeI site in pSK15, obliterating that MfeI site. A plasmid with the insert in the desired orientation, i.e., with an in-frame (STE4-linker-GPA1) fusion, was designated pSTE4-GPA1-a (Fig. 1a).
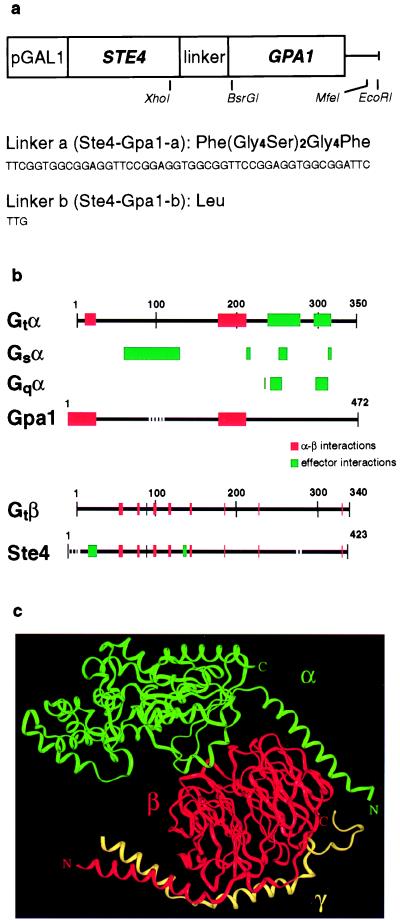
Figure 1.
(a) The STE4-GPA1 fusion constructs, showing relevant restriction sites, and the sequences of the linkers inserted. (b) The known sites of α-β interactions and of α-effector (Upper) or β-effector interactions (Lower) do not overlap. The diagram shows the homology between mammalian G proteins and yeast Gpa1 and Ste4. Mammalian Gα interaction domains are drawn as in ref. 4. Additional data are from refs. 14 and 15 and references cited within. (c) Crystallographic structure of the G protein heterotrimer αi1(GDP)β1γ2, showing the proximity of the carboxy terminus of the β1 subunit to the amino terminus of the αi1 subunit. Coordinates were from file 1GP2.pdb, and the figure was prepared with insightii biosym.
To generate pSTE4-GPA1-b, with a single codon between the fused genes, a small ds oligonucleotide with XhoI and BsrGI overhangs was ligated to the 5′ end of GPA1. A partial MfeI digest generated a 1-kb fragment encompassing the GPA1 gene, which was inserted into pSK15, and plasmids with the insert in the desired orientation (STE4-TTG (Leu)-GPA1) were identified (Fig. 1a). The MfeI site at the STE4-TTG junction was retained.
Western Blot Analysis.
Cultures were grown to 1 × 107 cells/ml in selective glucose medium, washed twice, and transferred to selective galactose medium. Yeast lysates were prepared from trichloroacetic acid-treated cells, as described (11). Proteins were separated on 8–12% SDS-polyacrylamide gradient gels, blotted onto Sartorius nitrocellulose membranes, reacted with rabbit polyclonal antibody to Ste4 (12) or to Gpa1 (13), and visualized by enhanced chemiluminescence.
Results
Design of the STE4-GPA1 Fusions.
The yeast and mammalian G protein subunits are highly homologous. Therefore, we designed our fusion construct with the published crystal structures of mammalian G protein in mind (14–16). Specifically, the crystal structures of the mammalian heterotrimer show that the C terminus of the Gβ subunit is close to the N terminus of the Gα subunit (Fig. 1c). Accordingly, we arranged the genes in the order STE4-GPA1. Because the Gβ and Gγ subunits are known to associate strongly (12, 17), we expected that the γ subunit would be able to associate with our fusion gene product, as long as the fusion did not seriously distort the conformation of the Gβ subunit.
We actually constructed two versions of the fusion (Fig. 1a): Ste4-Gpa1-a (encoded on plasmid pSTE4-GPA1-a), with a linker of 16 amino acid residues between the Gβ and Gα subunits, to allow the two subunits to assume as natural a conformation as possible, and Ste4-Gpa1-b (encoded on plasmid pSTE4-GPA1-b), with only a single amino acid residue (Leu) linking the two elements. As it turned out, both constructs were equally active (see below). Because overexpression of Ste4 without Gpa1 is lethal to the cell (7–9), we expressed the fusion under the control of the inducible GAL1 promoter. Thus, we could easily verify that both the Ste4 and Gpa1 elements of the fusions were functional: Ste4 by its ability to complement a ste4 deletion and promote mating, and Gpa1 by its ability to rescue the cell from lethality because of overproduction of Ste4.
Mating and Growth of Strains Carrying the STE4-GPA1 Fusions.
The fusion plasmids were introduced into the haploid S. cerevisiae strain SK1006, which has a null mutation in ste4. Control plasmids were also introduced, as follows: the vector (pGT5[URA3]; ref. 18); the same vector carrying GPA1 under control of the GAL1 promoter (pG1501; ref. 18); the same vector carrying STE4 under control of the GAL1 promoter (pGT-STE4–1; ref. 8); and GPA1 and STE4 under control of the GAL1 promoter on separate plasmids (pG1501 and pGT-STE4–2 [TRP1]; ref. 8).
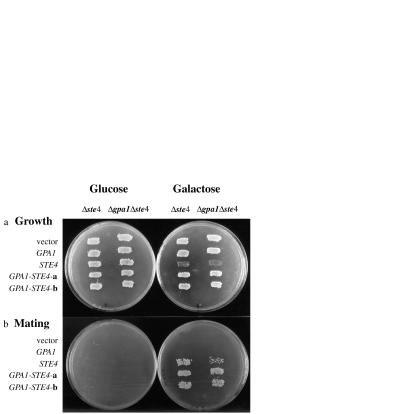
To verify that the STE4 gene on the fusion plasmids was functional, we looked for complementation of the mating defect of the ste4 mutation. As shown in Fig. 2b, strain SK1006 (ste4) carrying plasmid pGT-STE4–1, or either of the STE4-GPA1 fusion plasmids, pSTE4-GPA1-a or pSTE4-GPA1-b, mated with a MATα tester strain (NKY102) efficiently, on galactose plates. Controls carrying only the vector (pGT5) or the GPA1-carrying plasmid (pG1501) could not mate. None of the plasmids promoted mating on glucose plates, on which the GAL1 promoter was repressed.
Figure 2.
(a) Growth of transformants on glucose (Left) and galactose (Right) medium. On each plate, the SK1006 (ste4) derivatives are on the left and the SK1007 (ste4 gpa1) derivatives are on the right. Plasmids are, from top to bottom: pGT5 (vector), pG1501 (GPA1), pGT-STE4–1 (STE4), pSTE4-GPA1-a (STE4-GPA1-a), pSTE4-GPA1-b (STE4-GPA1-b). (b) Mating of transformants on glucose (Left) and galactose (Right) medium. Cells were patched onto a lawn of tester strain NKY102 (MATα ade8) on an SD-glucose or SD-galactose plate lacking uracil (10), and incubated at 30°C. The mixture was replicated to an SD-glucose plate with no supplements and incubated at 30°C, to select for diploids.
We also tested whether the GPA1 moiety of the fusion construct could compensate for the overexpression of the STE4 moiety. There were no obvious effects of any of the plasmids on growth when glucose was the sole carbon source (Fig. 2a). When the various transformants were replicated to galactose plates, strain SK1006 (ste4) carrying only plasmid pGT-STE4–1 (Fig. 2a) or pGT-STE4–2 (data not shown) grew very poorly, because of overexpression of STE4. Simultaneous overexpression of GPA1 and STE4 (pG1501 + pGT-STE4–2) compensated for the overexpression of STE4, as expected (data not shown; refs. 7–9). Transformants with the STE4-GPA1 fusion plasmids, pSTE4-GPA1-a or pSTE4-GPA1-b, grew well on galactose (Fig.2a). Furthermore, we dissected tetrads from a GPA1/gpa1∷HIS3 diploid strain, and the His+ segregants carrying the STE4-GPA1 fusion were viable. Hence, the GPA1 moiety on the fusions was able to compensate for overexpression of STE4. Myristoylation of Gpa1 is necessary for repression of the pheromone pathway (13, 19), and is believed to be required for association of Gα and Gβγ (20). The STE4-GPA1 fusion constructs clearly cannot be myristoylated, but the covalent association of the Ste4 and Gpa1 subunits apparently overrides the need for myristoylation.
We next introduced the various plasmids into S. cerevisiae strain SK1007, which is isogenic to strain SK1006, but has null mutations in both ste4 and gpa1. A STE4-plasmid alone [pGT-STE4–1 (Fig. 2b) or pGT-STE4–2 (data not shown)] allowed only inefficient mating. This was expected, because in the absence of Gpa1, there is no way to couple the pheromone receptor to activation of the mating pathway. The cells then shmoo in random directions, impairing the mating process. In contrast, the STE4-GPA1 fusion plasmids (pSTE4-GPA1-a and pSTE4-GPA1-b) promoted efficient mating with the MATα tester (NKY102) on galactose plates (Fig. 2b). These strains also grew well on galactose medium (Fig. 2a). These results confirmed that both the STE4 and GPA1 moieties on the fusion plasmids were functional. Coexpression of STE4 and GPA1 from plasmids pGT-STE4–2 and pG1501 also promoted efficient mating and growth on galactose medium (data not shown).
Response to α-Factor.
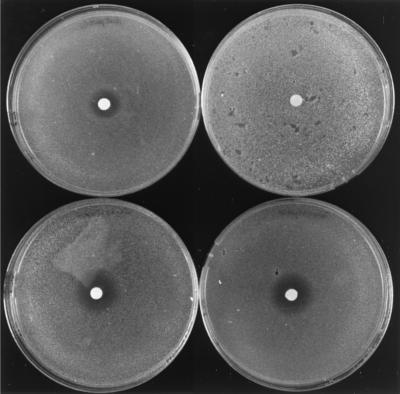
We examined whether the fusion strains could respond to α-factor. When cultures carrying the fusion plasmids were pregrown overnight with galactose before addition of α-factor, shmoos were detectable 2 h after addition of α-factor. No shmoos were seen in the control strain carrying the vector pGT5. Pheromone-induced arrest was also shown by the “halo assay.” Strains carrying pGT5 or pG1501 (GPA1) were insensitive to α-factor on both galactose plates (Fig. 3) and glucose plates (data not shown). Strains carrying either of the STE4-GPA1 fusion plasmids were sensitive to α-factor on galactose plates (Fig. 3), but not on glucose (data not shown). A wild-type control strain (βwt carrying pGT5) was sensitive to α-factor on both glucose and galactose plates (Fig. 3 and data not shown).
Figure 3.
Halo assay for sensitivity to α-factor on galactose plates. Cells were pregrown for several hours in galactose medium, plated in a lawn of top agar on selective SD galactose plates, and 1 μg of α-factor (Sugen) was spotted on the solidified top agar, in 4-μl drops. The plates were incubated for 40 h at 30°C. Strains are (clockwise from top left): βwt carrying vector pGT5 (wild-type), and SK1007 (ste4Δ gpa1Δ) with pGT5, pSTE4-GPA1-a (STE4-GPA1-a), or pSTE4-GPA1-b (STE4-GPA1-b).
Western Blot Analysis of the Fusion Product.
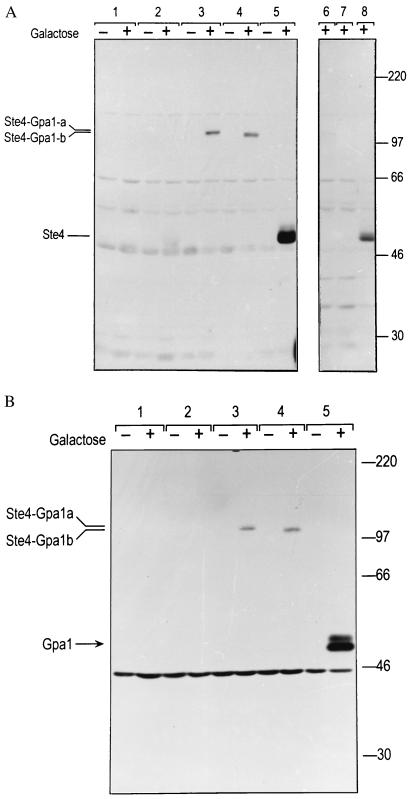
To verify that the STE4-GPA1 gene fusion resulted in a chimeric protein of the expected size (100 kDa), we performed Western blot analysis by using rabbit polyclonal antibody against Ste4 (12) (Fig. 4a). Lysates were prepared from strain SK1007 (ste4 gpa1) carrying the relevant plasmids, after growth with glucose and after induction for 9 h with galactose. The cultures were tested in parallel to confirm mating capabilities (data not shown). No specific bands were detected in any of the lysates from glucose-grown cells, or in the lysate from galactose-grown cells carrying the vector (Fig. 4a, 1). A band of the expected size for Ste4 protein (47 kDa) was detected from galactose-grown cells carrying pGT-STE4–1 alone (Fig. 4a, 2) or pG1501(GPA1) + pGT-STE4–2 (Fig. 4a, 5). In the strain with pGT-STE4–1 alone, this band was faint, perhaps because overexpression of STE4 on its own is lethal. Bands consistent with the expected sizes for the Ste4-Gpa1 fusion proteins were detected from galactose-grown cells carrying the fusion plasmids, pSTE4-GPA1-a (Fig. 4a, 3; Ste4-Gpa1-a; predicted molecular mass = 101.2 kDa) or pSTE4-GPA1-b (Fig. 4a, 4; Ste4-Gpa1-b; predicted molecular mass = 100.1 kDa). To confirm that the designated bands indeed represented Ste4 protein, recombinant Ste4 protein was produced in Escherichia coli BL21(DES) from plasmid pBH18 (12) and used to block the anti-Ste4 antiserum (Fig. 4a, 6–8). The fusion proteins were also detected with rabbit anti-Gpa1 antibody (13), as shown in Fig. 4b. We note that there was no evidence of proteolysis in the lysates from the strains with the fusions, nor could we detect any anti-Ste4 reactive proteins or fragments other than the ≈100-kDa fusion products. Furthermore, no proteolysis of the fusion protein was detected after exposure to α-factor (data not shown).
Figure 4.
(a) Western blot showing detection by anti-Ste4 antibody of Ste4-Gpa1 fusions from cells grown in glucose (−) or galactose (+) medium. 40 μg total protein were loaded per lane. Transformants of strain SK1007 (ste4 gpa1), carry the following plasmids: 1, pGT5 (vector); 2, pGT-STE4–1 (STE4); 3, pSTE4-GPA1-a (STE4-GPA1-a); 4, pSTE4-GPA1-b (STE4-GPA1-b); 5, pG1501 (GPA1) and pGT-STE4–2 (STE4). Lanes 6–8 show specific blocking of the anti-Ste4 reactive bands with recombinant Ste4 protein. Lane 6, pSTE4-GPA1-a (STE4-GPA1-a). Lane 7, pSTE4-GPA1-b (STE4-GPA1-b). Lane 8, pG1501 (GPA1) and pGT-STE4–2 (STE4). A number of nonspecific bands are evident, the most prominent migrating at approximately 29, 47, 56, 65, and 110 kDa. The band at 47 kDa seems to be nonspecific, as it appears in the empty vector control, it appears in the absence of galactose, its strength actually decreases in samples grown in galactose, and it is blocked by recombinant Ste4 protein to a much lesser extent (<2-fold) than the bona fide anti-Ste4 reactive products (blocked >10-fold). (b) Western blot by using anti-Gpa1 antibody. Strains as in a. Both myristoylated (lower band) and nonmyristoylated (upper band) forms of Gpa1 are evident. The expected molecular weight of nonmyristoylated Gpa1 is 54 kDa. The band at 45 kDa is nonspecific.
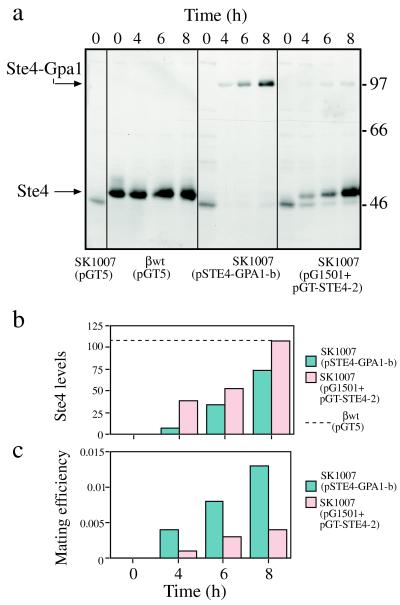
Figure 5.
(a) Induction of anti-Ste4 reactive protein on incubation in galactose. Cells were grown in selective glucose medium and transferred at time 0 h to galactose medium. Aliquots were removed at the indicated times and lysates prepared. Then, 20 μg total protein were loaded per lane. Nonspecific bands are as in Fig. 4a. Strain backgrounds: βwt (wild-type) or SK1007 (ste4 gpa1). Plasmids: pGT5 (vector), pSTE4-GPA1-b (STE4-GPA1-b), pG1501 (GPA1), pGT-STE4–2 (STE4). (b) The amounts of anti-Ste4 reactive protein were quantitated by using the National Institutes of Health image program. Panels were scanned from the same exposure. (c) Induction of mating ability on incubation in galactose. A total of 4 × 107 exponentially growing washed cells were incubated at 30°C for the times indicated with a large excess of washed tester NKY102 (MATα ade8) cells. After washing, appropriate dilutions were plated on selective plates to score diploids, tester, and plasmid-carrying strain. Mating efficiencies represent the number of diploid cells per SK1007(pSTE4-GPA1-b) or SK1007 (pG1501 + pGT-STE4–2) cell.
Mating Efficiencies.
We compared the amount of the Ste4-Gpa1 fusion protein expressed in strain SK1007(pSTE4-GPA1-b) to the amounts of the Ste4 and Gpa1 proteins coexpressed from separate plasmids in strain SK1007(pG1501 + pGT-STE4–2), as well as to the amounts of the Ste4 and Gpa1 proteins in a wild-type strain carrying the empty vector [βwt(pGT5)] during incubation in galactose medium (Fig. 5 a and b). At the start of galactose induction, aliquots of the same cultures were mixed with the mating tester. As can be seen in Fig. 5c, the strain with the fusion plasmid mated at least as efficiently as the strain with the separate plasmids, if not more efficiently, and the amounts of anti-Ste4 reactive protein were comparable, for at least 8 h. In the wild-type strain, Ste4 levels were easily detectable; it is therefore unreasonable to suppose that the biological activity of the fusion protein derived from tiny, undetectable amounts of cleaved subunits. Furthermore, after preincubation overnight in galactose, followed by incubation with the mating tester (NKY102) for 4 h, the fusion-carrying strain promoted mating as well as the wild-type strain. The amounts of anti-Ste4 reactive protein were similar at this point in the two strains (data not shown).
Discussion
We have shown that a nondissociable fusion protein comprising the α and β subunits of the S. cerevisiae pheromone pathway G protein can function in transducing the pheromone signal and promoting growth arrest and mating. It is likely that conformational changes in the heterotrimeric complex are induced on receptor activation. These changes may reveal binding interfaces that were previously buried, allowing them to interact with effectors. Our data imply that in the wild-type G protein, even if the Gα and Gβγ moieties do detach from one another, they remain in close proximity to one another. When yeast Gpa1 is fused to the pheromone receptor, the mating signal can be transduced (21). Moreover, the mating G protein has been shown to recruit the Ste5-MAPK scaffold to the cell membrane (22). Thus, most of the molecules involved in signal transduction (receptor, G protein, effector) are very likely closely juxtaposed.
A comparison of the expected Gpa1-Ste4 contact sites, based on their homology with bovine Gtαβ, and the residues implicated in interactions between Ste4 and its effector Ste20 (23, 24), supports our contention that Gpa1-Ste4 association need not interfere with the interaction between Ste4 and Ste20; Fig. 1 b and c). An alternative interpretation of our results would be the occurrence of interactions between fusion protein molecules, allowing the β moiety of one chimera to interact with, and then dissociate from, the α moiety of another. Our data show that there is no decrease in mating efficiency supported by the chimera, even when the amounts of Ste4 are limiting (Fig. 5), suggesting that this interpretation is unlikely. Moreover, there is no evidence that heterotrimeric G proteins can form higher order complexes.
One of the attractions of the subunit dissociation hypothesis is that it allows for shuffling of the G protein subunits, which could reassociate in different combinations, to allow creation of a large number of G proteins of different specificities, from a small number of subunits. Indeed, several different combinations of Gβγ subunits have been shown to interact with the same Gα in vitro (25). The sequence of the S. cerevisiae genome revealed only two Gα homologs (GPA1 and GPA2), but eight candidate β genes and three candidate γ genes (26). None of these candidate β and γ genes has been shown to be functional, but it is possible that different combinations of these subunits are active in vivo, perhaps subject to compartmentalization in space (e.g., ref. 27) and/or time. We have noticed that our Ste4-Gpa1 fusion proteins apparently hypersensitize the cells to α-factor (Fig. 3 and unpublished data). This appears to be an artifact resulting from overexpression of the protein (our unpublished data).
It would be interesting to crystallize the fusion protein, as well as the native heterotrimer, in the presence of the nonhydrolyzable GTP analog, GppNHp. Comparing these crystal structures with the structure of the heterotrimer-GDP complex should shed light on the nature of the conformational changes that occur on activation and deactivation of the complex. It may also be possible to build similar constructs for mammalian G proteins, in which the Gα and Gβ subunits are covalently joined.
Previous biochemical experiments have shown that Gs dissociation is not required to activate adenylyl cyclase (28); in this system, it is Gα that interacts with the effector. In light of the strong homology between various G proteins, it is possible that dissociation of Gα from Gβγ does not have to take place in other pathways employing G proteins.
Acknowledgments
We thank J. Hirschman, D. Jenness, and D. Stone for gifts of antibodies and strains. Many thanks to all members of our laboratory and to D. Zenvirth, I. Marbach, and D. Engelberg for discussions and practical assistance.
Abbreviations
- MAP
mitogen-activated protein
- ds
double-stranded
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.050015797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.050015797
References
- 1.Hamm H E. J Biol Chem. 1998;273:669–672. doi: 10.1074/jbc.273.2.669. [DOI] [PubMed] [Google Scholar]
- 2.Gilman A G. Annu Rev Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- 3.Levitzki A. Physiol Rev. 1986;66:819–854. doi: 10.1152/physrev.1986.66.3.819. [DOI] [PubMed] [Google Scholar]
- 4.Rebois R V, Warner D R, Basi N S. Cell Signalling. 1997;9:141–151. doi: 10.1016/s0898-6568(96)00133-7. [DOI] [PubMed] [Google Scholar]
- 5.Herskowitz I. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- 6.Leberer E, Thomas D Y, Whiteway M. Curr Opin Genet Dev. 1997;7:59–66. doi: 10.1016/s0959-437x(97)80110-4. [DOI] [PubMed] [Google Scholar]
- 7.Cole G M, Stone D E, Reed S I. Mol Cell Biol. 1990;10:510–517. doi: 10.1128/mcb.10.2.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nomoto S, Nakayama N, Arai K, Matsumoto K. EMBO J. 1990;9:691–696. doi: 10.1002/j.1460-2075.1990.tb08161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whiteway M, Hougan L, Thomas D Y. Mol Cell Biol. 1990;10:217–222. doi: 10.1128/mcb.10.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sherman F. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 11.Foiani M, Marini F, Gamba D, Lucchini G, Plevani P. Mol Cell Biol. 1994;15:923–933. doi: 10.1128/mcb.14.2.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirschman J E, De Zutter G S, Simonds W F, Jenness D D. J Biol Chem. 1997;272:240–248. doi: 10.1074/jbc.272.1.240. [DOI] [PubMed] [Google Scholar]
- 13.Stone D E, Cole G M, de Barros Lopes M, Goebl M, Reed S I. Genes Dev. 1991;5:1996–1981. doi: 10.1101/gad.5.11.1969. [DOI] [PubMed] [Google Scholar]
- 14.Lambright D G, Sondek J, Bohm A, Skiba N P, Hamm H E, Sigler P B. Nature (London) 1996;379:311–319. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
- 15.Sondek J, Bohm A, Lambright D G, Hamm H E, Sigler P B. Nature (London) 1996;379:369–374. doi: 10.1038/379369a0. [DOI] [PubMed] [Google Scholar]
- 16.Wall M A, Coleman D E, Lee E, Iniguez-Liuhi J A, Posner B A, Gilman A G, Sprang S R. Cell. 1995;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt C J, Neer E J. J Biol Chem. 1991;266:4538–4544. [PubMed] [Google Scholar]
- 18.Miyajima I, Nakafuku M, Nakayama N, Brenner C, Miyajima A, Kaibuchi K, Arai K, Kaziro Y, Matsumoto K. Cell. 1987;50:1011–1019. doi: 10.1016/0092-8674(87)90167-x. [DOI] [PubMed] [Google Scholar]
- 19.Dohlman H G, Goldsmith P, Spegel A M, Thorner J. Proc Natl Acad Sci USA. 1993;90:9688–9692. doi: 10.1073/pnas.90.20.9688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boutin J A. Cell Signalling. 1997;9:15–35. doi: 10.1016/s0898-6568(96)00100-3. [DOI] [PubMed] [Google Scholar]
- 21.Medici R, Bianchi E, Di Segni G, Tocchini-Valentini G P. EMBO J. 1997;16:7241–7249. doi: 10.1093/emboj/16.24.7241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pryciak P M, Huntress F A. Genes Dev. 1998;12:2684–2697. doi: 10.1101/gad.12.17.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leberer E, Dignard D, Harcus D, Thomas D Y, Whiteway M. EMBO J. 1992;11:4815–4824. doi: 10.1002/j.1460-2075.1992.tb05587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leberer E, Dignard D, Hougan L, Thomas D Y, Whiteway M. EMBO J. 1992;11:4805–4813. doi: 10.1002/j.1460-2075.1992.tb05586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graf R, Mattera R, Codina J, Evans T, Ho Y-K, Estes M K, Birnbaumer L. Eur J Biochem. 1992;210:609–619. doi: 10.1111/j.1432-1033.1992.tb17461.x. [DOI] [PubMed] [Google Scholar]
- 26.Lorenz M C, Heitman J. EMBO J. 1997;16:7008–7018. doi: 10.1093/emboj/16.23.7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brunk I, Pahner I, Maier U, Jenner B, Veh R W, Nurnberg B, Ahnert-Hilger G. Eur J Cell Biol. 1999;78:311–322. doi: 10.1016/s0171-9335(99)80065-x. [DOI] [PubMed] [Google Scholar]
- 28.Bar-Sinai A, Marbach I, Shorr R G, Levitzki A. Eur J Biochem. 1992;207:703–708. doi: 10.1111/j.1432-1033.1992.tb17098.x. [DOI] [PubMed] [Google Scholar]