Abstract
The origin recognition complex (ORC) plays a central role in eukaryotic DNA replication. Here we describe a unique ORC-like complex in Tetrahymena thermophila, TIF4, which bound in an ATP-dependent manner to sequences required for cell cycle-controlled replication and gene amplification (ribosomal DNA [rDNA] type I elements). TIF4's mode of DNA recognition was distinct from that of other characterized ORCs, as it bound exclusively to single-stranded DNA. In contrast to yeast ORCs, TIF4 DNA binding activity was cell cycle regulated and peaked during S phase, coincident with the redistribution of the Orc2-related subunit, p69, from the cytoplasm to the macronucleus. Origin-binding activity and nuclear p69 immunoreactivity were further regulated during development, where they distinguished replicating from nonreplicating nuclei. Both activities were lost from germ line micronuclei following the programmed arrest of micronuclear replication. Replicating macronuclei stained with Orc2 antibodies throughout development in wild-type cells but failed to do so in the amplification-defective rmm11 mutant. Collectively, these findings indicate that the regulation of TIF4 is intimately tied to the cell cycle and developmentally programmed replication cycles. They further implicate TIF4 in rDNA gene amplification. As type I elements interact with other sequence-specific single-strand breaks (in vitro and in vivo), the dynamic interplay of Orc-like (TIF4) and non-ORC-like proteins with this replication determinant may provide a novel mechanism for regulation.
The initiation of eukaryotic DNA replication is precisely regulated. A key transactivator in the yeast Saccharomyces cerevisiae is the six-subunit origin recognition complex (ORC), which binds to the 11-bp autonomous replication sequence autonomous replication sequence element (5). Putative orthologs have been identified in many eukaryotes (17). In vitro replication assays have established a requirement for several nonyeast ORCs (7, 11); however, the underlying basis for origin specification is largely unknown, as replication initiates at random sites in vitro.
S. cerevisiae replicons typically span ≈100 bp and include discrete binding sites for sequence-specific double-stranded DNA proteins, including ORC (31). The ORC binding site is flanked by an A+T-rich DNA-unwinding element that may facilitate origin unwinding. Replication determinants are much more dispersed in other eukaryotes, encompassing hundreds to thousands of base pairs (17). The genetic organization of replicons is best understood in four model systems, S. cerevisiae, Schizosaccharomyces pombe, Tetrahymena thermophila, and Drosophila melanogaster, for which robust genetic assays have been developed.
An attractive feature of Tetrahymena spp. is the ability to study both cell cycle-controlled replication and locus-specific gene amplification (reviewed in reference 21). The T. thermophila macronucleus harbors a natural minichromosome encoding the 26S, 5.8S, and 16S rRNAs. This chromosome is generated by programmed excision and rearrangement of the germ line (micronuclear) ribosomal DNA (rDNA) locus into a 21-kb palindrome (Fig. 1). Macronuclear rDNA is amplified from 2 to 10,000 copies in a single S phase during development but is replicated on average once per cell cycle during vegetative growth. By comparison, non-rDNA chromosomes achieve a copy number of ≈45.
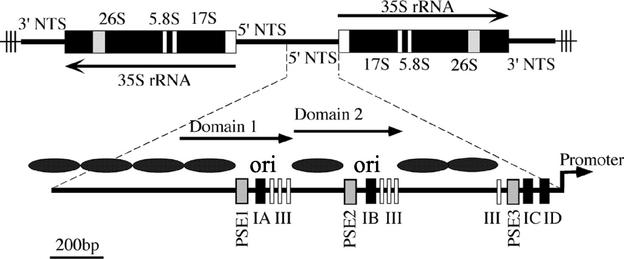
FIG. 1.
Organization of T. thermophila rDNA minichromosome. The Tetrahymena rDNA minichromosomes encode two inverted copies of the rRNA coding region (35S) and 5′ and 3′ nontranscribed spacers (NTS), bound by telomeres (thin lines with vertical bars). The 1.9-kb 5′ nontranscribed spacer contains positioned nucleosomes (black ovals) that flank two nucleosome-free regions that contain initiation sites for cell cycle-regulated DNA replication and gene amplification (ori). Dispersed, repeated type I and PSE elements are required for both replication initiation and the transient pausing of replication forks. Domains 1 and 2 are 430-bp tandemly duplicated segments that have undergone subsequent sequence divergence.
The 1.9-kb 5′ nontranscribed spacer is both necessary and sufficient for replication of artificial rDNA minichromosomes (6, 41, 43). Dispersed type I and pause site elements are required for rDNA replication (16, 25, 47). The type IA and IB elements colocalize with replication initiation sites, while promoter-proximal type I elements act at a distance to regulate origin firing (Fig. 1) (16, 43, 56). Type I elements are multifunctional; in addition to controlling replication initiation, they mediate replication fork pausing at pause site elements (30) and are required for rRNA transcription (42).
During development, amplifying rDNA molecules initiate replication from the same origins that mediate cell cycle-controlled vegetative replication (Fig. 1) (56). Understanding how the switch from gene amplification to cell cycle-controlled replication occurs will require the identification of both constitutive initiator proteins (presumably including ORC) and factors that modulate ORC activity. Efforts to identify ORC orthologs by degenerate PCR have proven difficult. Consequently, we have focused our efforts on biochemical approaches for identifying proteins that recognize genetically defined replication control elements.
Using electrophoretic mobility shift assays (EMSAs), we previously identified three distinct type I element-binding activities (type I factors 1 to 3 [TIF1 to TIF3]). In contrast to previously described replication initiation factors, these proteins bind exclusively to single-stranded DNA, specifically recognizing the A-rich strand of type I elements (33, 46, 52). Affinity-purified TIF1 also binds to the T-rich type I element strand in vitro. Using antisense ribosomes to inhibit TIF1 expression, we discovered that this protein exhibits a remarkable strand bias for origin- and promoter-proximal type I elements. In vivo, TIF1 interacts with just the A-rich strand at the rDNA origins and the T-rich strand at the rDNA promoter (47). This observation indicates that these regions are predominantly single-stranded in native chromosomes. Furthermore, it raised the possibility that T-rich strand-binding proteins might be selectively recruited to the rDNA replication origins.
Here we describe the purification and characterization of a T-rich strand-specific type I element binding complex, TIF4, that exhibits many of the biochemical properties of ORC but has a distinct mode of DNA recognition. In contrast to previously characterized ORCs, we provide evidence for cell cycle and developmental regulation of TIF4 DNA binding activity and the subcellular localization of its Orc2-related subunit. In conjunction with previous studies, these findings suggest that the interplay of an essential cis-acting replication determinant with Orc-like (TIF4) and non-ORC-like (TIF1 to TIF3) proteins may regulate origin activity.
MATERIALS AND METHODS
Culture, cell cycle synchronization, and developmental time courses.
Tetrahymena thermophila strains were cultured as previously described (39). Wild-type (CU428, SB1915, and SB1934) and amplification-defective mutant heterokaryons (FH210 and FH211) were studied (20, 54). Cell cycle synchronization was achieved by modifying a stationary-phase synchronization protocol (34) in which saturated cultures were placed in starvation medium for 8 h prior to dilution into growth medium. Following release from G0 arrest, cells were isolated at defined time intervals for Western blot, gel shift, and immunofluorescence analyses. To study development, synchronous matings were initiated between wild-type strains (SB1934 × SB1915) and amplification-defective rmm11 mutant heterokaryons (FH210 × FH211). Established morphological landmarks were used to stage development and assess the synchrony of mating cultures (reviewed in reference 38).
Fractionation of Tetrahymena cells.
Cytoplasmic and whole-cell nuclear extracts were prepared by lysing washed cells in ice-cold TMS buffer (10 mM Tris [pH 7.5], 10 mM MgCl2, 3 mM CaCl2, 0.25 M sucrose, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride) with the addition of NP-40 to a final concentration of 0.16%. After 30 min, solid sucrose (0.815 g/ml) was added, and nuclei were pelleted at 9,000 × g for 30 min. The supernatant was centrifuged for 1 h at 100,000 × g, and the resulting S100 supernatant was collected. Pelleted nuclei were resuspended in a low-salt buffer (20 mM HEPES [pH 7.9], 25% glycerol, 1.5 mM MgCl2, 20 mM KCl, 0.2 mM EDTA, 0.2 mM phenylmethylsulfonyl fluoride, 0.2 mM dithiothreitol), extracted with 800 mM KCl for 30 min, and centrifuged for 1 h at 100,000 × g.
Micro- and macronuclei were purified by a modification of a published protocol (1). Briefly, harvested cells were resuspended at 3.3 × 106 cells/ml in medium A (0.1 M sucrose, 4% gum arabic, 0.1% spermidine HCl, 2 mM MgCl2); 0.63% N-octyl alcohol was added, followed by mechanical disruption in a Waring blender (25 s at high speed, 15 s at low speed). Nuclei were pelleted at 34,000 × g for 40 min. Nuclear pellets were resuspended in medium A without N-octyl alcohol and pelleted through a 25% Percoll gradient by centrifugation at 1,500 × g for 20 min. Micro- and macronuclei were separated by three successive rounds of centrifugation at 1,500 × g for 8.5 min on a 50% Percoll gradient. Purity and recovery were assessed by light microscopy following staining with methyl green.
To prepare whole-cell lysates for Western blot analysis, log-phase vegetative cells were washed and lysed by boiling in Laemmli sodium dodecyl sulfate (SDS) sample buffer. Fractionation into soluble and chromatin-bound fractions was achieved by lysing cells in 10 mM HEPES-10 mM KCl-1.5 mM MgCl2-1% Triton X-100-10% glycerol-340 mM sucrose-1 mM dithiothreitol-5 μg of aprotinin per ml-5 μg of leupeptin per ml-0.5 μg of pepstatin A per ml-100 mM phenylmethylsulfonyl fluoride. Following incubation on ice for 15 min, the soluble and insoluble protein fractions were separated by centrifugation at 16,000 × g for 15 min at 4°C. The nuclear pellet was resuspended in a volume equal to that of the soluble supernatant.
Electrophoretic mobility shift assays.
Gel-purified oligonucleotides were either labeled on their 5′ end with [γ-32P]dATP or used as unlabeled competitors. For standard binding reactions, oligonucleotide competition and antibody supershift studies, subsaturating amounts of crude cell lysates or purified protein were incubated with 0.1 pmol of radiolabeled DNA for 15 min on ice in 0.6× TGE (50 mM Tris, 220 mM glycine, 6 mM EDTA) supplemented with 12.5% glycerol. For experiments involving antibodies, crude rabbit serum, or protein A-Sepharose, purified antibodies were added to the binding reaction. Electrophoresis was typically carried out on 5% polyacrylamide-0.6× TGE gels. Where indicated, gel shift complexes from native EMSA gels were electroeluted for 1 h at 120 V in 1× SDS running buffer and concentrated on a Centricon 10 membrane (Amicon).
Oligonucleotide substrates included the C3 type IB element A-rich (ssA53) and T-rich (ssT51) strands, the B rDNA allele of the type IB element [T-rich strand only; ssT51(B)] (25), complementary oligonucleotides from the rDNA pause site element 2 region (30), and two sequences from the coding region of the TIF1 gene (47). The C3 type IB element confers a ≈5% replication advantage over B rDNA in the macronucleus of heterozygous cells (24).The following primers were used: ssT51 (T-rich strand, C3 rDNA type IB allele, 50-mer, 74% A+T; 5′-CTCAAAAGTTGCAAAAGTTCGGAAGGTTTACTATTTTTGTTTTTTTTTTT); ssA53 (A-rich strand, C3 rDNA type IB allele, 53-mer, 70% A+T; 5′-GGCAAAAAAAAAAACAAAAATAGTAAACCTTCCGAACTTTTGCAACTTTTGAG); ssT51(B) (T-rich strand, B rDNA type IB allele, 50-mer, 82% A+T; 5′-TTTCATTTTTCAAAAATTTTTCTAAGTTTACTATTTTGTTTTTTTTTTTGCC); TRC1 (TIF1 gene coding region, 69-mer, 67% A+T; 5′-GGATTAGTAACAAAGAAGACAGAGTTGTTCTTTGAACCCAATAATGAGTTCAGTGAAATTTGGAAAAAG); TRC2 (TIF1 gene coding region, 73-mer, 66% A+T; 5′-CCTGAAGATACCCATTTAAATATAGAAATCAAAGAAAGCAACTTATTACCTGATGAGAATAAATGGGCTCACC); PSE1 (upper strand, rDNA pause site 2 element, 74-mer, 68% A+T; 5′-CGCGGCCGCTATCAATTTCATTTATTCATTTTAGTTAAATTTTTCATTCACAAAAAACTTTTTTTTGGTGGGCC); and PSE2 (lower strand, rDNA pause site 2 element, 75-mer, 74% A+T; 5′-GTGACCGCGGCCGCTATCAATTTCATTTATTCATTTTAGTTAAATTTTACATTCAAAATAAATTTTTTTTGATTG).
Western blotting.
Xenopus Orc2 polyclonal rabbit antiserum (9) was either provided by David Gilbert or raised in our laboratory by using bacterially expressed Xenopus laevis Orc2 protein (plasmid pC187-1B, kindly provided by Phil Carpenter and William Dunphy). For Western blotting, protein samples were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on either 7.5 or 10% gels and transferred to polyvinylidene difluoride membranes (Millipore). Membranes were blocked in 10% nonfat dry milk and incubated with antibodies in TBST (Tris-buffered saline with 0.15% Tween) at the following dilutions (anti-Xenopus Orc2 or preimmune serum, 1:3,000; anti-human Orc2 serum, 1:500; anti-hv1 serum, 1:1,000; anti-mic α-peptide, 1:1,000; and anti-histone H3 serum, 1:1,000). Immunoreactivity was detected by enhanced chemiluminescence with horseradish peroxidase-conjugated anti-rabbit immunoglobulin secondary antibody according to the manufacturer's instructions (Jackson ImmunoResearch Laboratories); 105 cell equivalents were loaded per lane.
Purification of TIF4.
All purification steps were performed at 4°C and monitored by gel shift analysis of the radiolabeled substrate ssT51. Four liters of vegetative log-phase T. thermophila strain CU428 (2.5 × 105 cells/ml) was harvested by centrifugation at 3,000 × g for 5 min, washed twice in 10 mM Tris (pH 7.5), resuspended in 240 ml of TMS buffer (10 mM Tris [pH 7.5], 10 mM MgCl2, 3 mM CaCl2, 0.25 M sucrose, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride), and lysed by addition of NP-40 to a final concentration of 0.2%. After incubation for 20 min, solid sucrose (0.815 g/ml) was added, followed by centrifugation for 1 h at 9,000 × g to generate the TMS-100 extract (300 ml). Then 150 ml of TMS-100 was applied to a 25-ml SP-Sepharose column (Pharmacia) equilibrated with TIF4 buffer (TMS buffer containing 1.6 M sucrose). The column was washed with 50 ml of TIF4-0.2 M NaCl and eluted with a 100-ml linear (0.2 to 0.8 M) NaCl gradient.
TIF4-containing fractions (0.32 to 0.54 M NaCl pool) were diluted 1:3 in TIF4 buffer and loaded on a 25-ml Q-Sepharose column (Pharmacia) equilibrated with TIF4 buffer. The column was washed with 50 ml of TIF4-0.1 M NaCl and eluted with a 100-ml linear 0.1 to 0.8 M NaCl gradient. Active fractions (0.3 to 0.5 M NaCl) were dialyzed against oligonucleotide affinity buffer (OAB; 20 mM HEPES [pH 7.9], 10 mM MgCl2, 40 mM KCl, 1 mM CaCl2, 12.5% glycerol, 0.4 M sucrose) and loaded onto a 1-ml A-strand type I element (ssA53) oligonucleotide affinity column equilibrated with OAB buffer. The flowthrough fraction was adjusted to 4 mM ATP and loaded onto a 1-ml T-rich-strand type I element (ssT51) oligonucleotide affinity column. This column was washed with 5 ml of 0.1 M NaCl-4 mM ATP in OAB buffer, and bound proteins were eluted with a stepwise gradient of NaCl (0.2 to 1.0 M) in OAB buffer with 4 mM ATP (two 1-ml step elutions per NaCl concentration tested). Final column fractions were subjected to SDS-PAGE for silver staining and Western blot analysis.
Immunocytochemistry.
The protocol for processing cells was adapted from previously described methods (48, 49). Briefly, growing or conjugating (mating) cells were harvested by centrifugation, rinsed in PHEM buffer (60 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2, pH 6.9), and incubated in PHEM plus 0.5% Triton X-100 for 3 to 5 min. Cells were recentrifuged and fixed in PHEM plus 3% paraformaldehyde for 2 to 4 h at 4°C, followed by three washes with Tris-buffered saline (TBS). Fixed cells were placed in PBT blocking buffer (PHEM with 3% bovine serum albumin and 0.1% Tween) for >1 h prior to incubation with primary antiserum in PBT for 1 h at room temperature. Following three washes with PBT, cells were incubated for 1 h at room temperature with rhodamine-conjugated secondary antibody (Jackson ImmunoResearch Laboratories; 1:100 dilution in PBT-bovine serum albumin). Cells were washed once with PBT and suspended in 10 nM Sytox Green nucleic acid stain (Molecular Probes) or 0.1 μg of 4′,6′-diamidino-2-phenylindole (DAPI) (Sigma) for 10 min and washed two to three times with TBS. Cells were mounted onto slides in glycerol-TBS (4:1) and examined by epifluorescence microscopy.
RESULTS
Identification of a Tetrahymena replication origin-binding activity that is biochemically similar to ORC.
Type I elements are required for cell cycle-controlled replication and amplification of rDNA minichromosomes. Several different biochemical activities compete for binding to these sequences in vitro and in vivo (33, 47), suggesting that the contribution of this determinant to the regulation of origin activity may be complex. One such protein, TIF1, binds to either the A-rich or T-rich strand of the type I element in vitro. However, TIF1 interacts differently with origin- and promoter-proximal type I elements in vivo, selectively binding to just the A-rich strand at the origin. This finding led us to search for T-rich strand-specific proteins that might be preferentially targeted to replication initiation sites.
The type IB element T-rich strand oligonucleotide ssT51 formed several abundant ATP-independent gel shift complexes upon incubation with crude S100 cell extracts (Fig. 2A, left panel). A less abundant, slower migrating complex appeared upon the addition of ATP and MgCl2. This activity, here designated TIF4, was highly enriched in nuclear extracts (Fig. 2A, right panel). Similar to S. cerevisiae and Drosophila melanogaster ORC (3, 5), DNA binding was enhanced with the poorly hydrolyzable analog ATPγS (Fig. 2B). No protein-DNA complex was observed when GTP, UTP, or CTP was substituted for ATP (data not shown). In marked contrast to TIF1 to TIF3 (33), TIF4 failed to recognize the A-rich strand (Fig. 2B). In addition to strong ATP-dependent T-rich-strand binding, very weak duplex DNA binding was occasionally detected on long exposures.
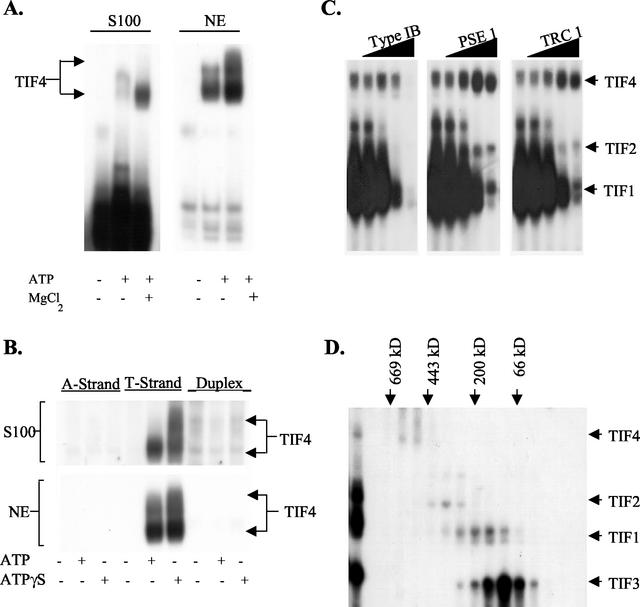
FIG. 2.
Identification and characterization of TIF4. (A) ATP-dependent binding of TIF4 to the T-rich strand of the type IB element. Electrophoretic mobility shift assays were performed with TMS S100 whole-cell or nuclear extracts (NE) and 0.1 pmol of radiolabeled ssT51; 4 mM ATP and 5 mM MgCl2 were added as indicated. (B) Type I element substrate specificity. Radiolabeled C3 type IB element oligonucleotides (A-rich strand, ssA53; T-rich strand, ssT51; duplex, ssA53::ssT51, labeled on the A-rich strand) were incubated with TMS-S100 or nuclear extracts in the presence or absence of 4 mM ATP or ATPγS and subjected to EMSA. (C) Sequence specificity of TIF4 DNA binding. S100 whole-cell extracts were incubated with 0.1 pmol of radiolabeled ssT51 in the presence of increasing concentrations of unlabeled single-stranded DNA competitors (0, 0.01, 0.1, 1, and 10 pmol). See Materials and Methods for details on competitor DNAs. (D) Size fractionation of T-rich-strand binding complexes. S100 whole-cell extracts were fractionated on a 6-ml Superose 6 fast protein liquid chromatography (FPLC) column (Amersham), and 0.3-ml fractions were assayed for binding to ssT51. The migration of protein molecular size markers is indicated with arrows.
Competition assays revealed that TIF4 exhibited a high degree of sequence specificity (Fig. 2C, and data not shown). Unlike the other type I element-binding proteins examined, TIF4 binding to the type I element T-rich strand was not competed away by unrelated single-stranded oligonucleotides of similar length and A+T content (Fig. 2C and data not shown; TIF1 gene coding regions TRC1 and TRC2, and rDNA pause site elements PSE1 and PSE2). Enhanced binding to the type I element T-rich strand was observed in the presence of oligonucleotides corresponding to PSE elements, which map 50 to 100 bp upstream of type I elements (Fig. 1). The molecular mass of TIF4 was estimated by Superose 6 gel filtration (Fig. 2D). TIF4 DNA binding activity peaked at 550 kDa, similar to yeast and metazoan ORCs (5, 18, 53). The remaining gel shift activities had apparent molecular masses of ≈300, 100, and 70 kDa, similar to the previously reported A-rich strand-binding complexes TIF1 to TIF3 (33).
TIF4 contains a chromatin-associated Orc2-related subunit.
Immunodepletion studies previously revealed that the Xenopus laevis Orc2-related protein is essential for DNA replication in cell-free egg extracts (9). Preliminary Western blot analysis revealed that rabbit anti-X. laevis Orc2 antiserum (kindly provided by David Gilbert) crosequence-specific reacted with recombinant D. melanogaster and S. cerevisiae Orc2 proteins (Fig. 3A, left panel). This antibody recognized a single polypeptide of ≈70 kDa in whole-cell lysates prepared from S. cerevisiae and T. thermophila (Fig. 3A, middle panel). Human Orc2 antiserum (kindly provided by Thomas Kelly) also crosequence-specific reacted with the 69-kDa Tetrahymena protein (Fig. 3A, right panel). As expected, Tetrahymena p69 was recognized by X. laevis Orc2 rabbit antiserum raised in our laboratory (Fig. 3B, whole-cell lysate, left two panels). The subcellular localization of p69 was investigated by partitioning whole-cell lysates into soluble and chromatin-associated fractions (8, 28). p69 was exclusively associated with the insoluble chromatin-enriched pellet (Fig. 3B). Similar to histone H3, p69 was released into the soluble fraction with increasing salt concentrations (Fig. 3C). DNase I digestion also solubilized p69 (Fig. 3D); however, the ratio of released p69 to H3 was lower than in salt-extracted nuclei, suggesting that it may also associate with a nonchromatin nuclear compartment.
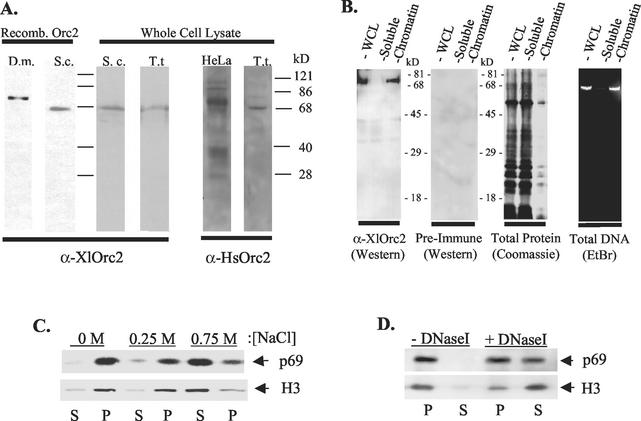
FIG. 3.
Chromatin association of an Orc2-related Tetrahymena protein. (A) Western blot analysis with rabbit anti-Xenopus and anti-human Orc2 antisera and recombinant D. melanogaster (D.m.) and S. cerevisiae (S.c.) Orc2 proteins or S. cerevisiae and T. thermophila (T.t.) whole-cell lysates. (B) Vegetative whole-cell lysates (WCL) and soluble and chromatin-associated cell fractions were resolved by SDS-PAGE and subjected to Western blot analysis with X. laevis Orc2 (XlOrc2) or preimmune serum (left panels). Right panels: protein (Coomassie blue stain) and DNA (ethidium bromide stain) profiles of fractioned nuclei. (C) Salt-sensitive chromatin association of p69. Chromatin-enriched fractions were treated with increasing concentrations of sodium chloride and incubated on ice for 10 min prior to recentrifugation. Pellet (P) and supernatant (S) fractions were analyzed by Western blotting with X. laevis Orc2 or histone H3 antibodies. (D) Association of p69 with DNA. Chromatin-enriched fractions were treated with DNase I, separated into soluble supernatant (S) and insoluble pellet (P) fractions, and analyzed by Western blotting as above.
We next sought to determine whether p69 was a component of the TIF4 origin-binding complex. Gel shift complexes formed with crude S100 extracts were examined by excising complexes from native gels and fractionating eluted proteins by SDS-PAGE. Seven prominent and several minor proteins were detected in the region containing the TIF4-DNA complex (Fig. 4A, TIF4). A limited number of proteins were detected in the region forming ATP-independent T-rich-strand complexes (Fig. 4A, TIF2+). Western blot analysis indicated that p69 was present in the region encompassing the TIF4-DNA complex and absent in the region spanning the other known type I element-binding activities (Fig. 4A).
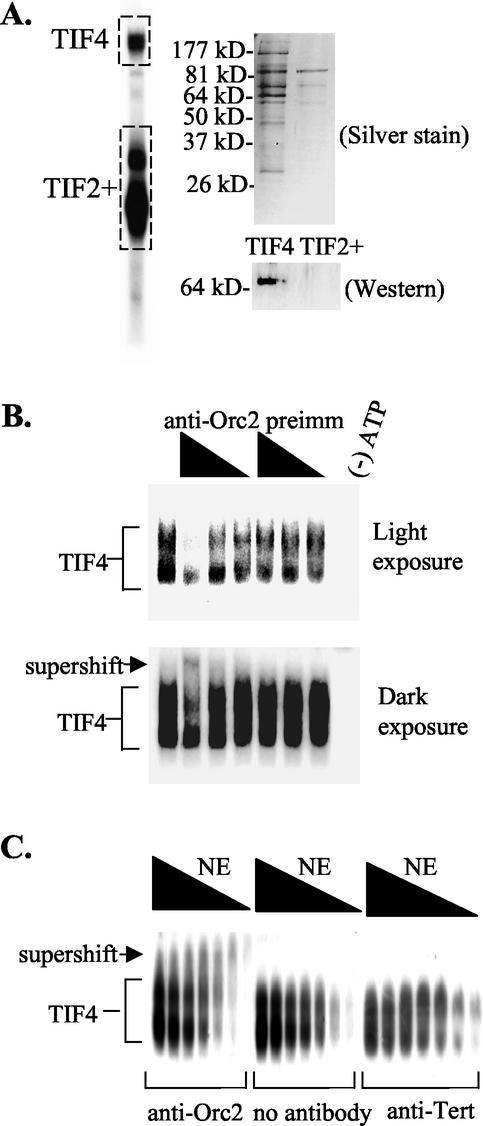
FIG. 4.
Association of p69 with the TIF4 origin binding complex. (A) Two-dimensional gel analysis of TIF4. EMSA was performed on whole-cell S100 extracts with ssT51. The boxed regions (TIF4 complex and TIF2+ complex) were excised from the EMSA gel. Proteins were electroeluted and separated by SDS-PAGE for silver staining and Western blot analysis with X. laevis Orc2 antibodies. Preimmune serum showed no detectable signal (data not shown; see Fig. 2A), and X. laevis Orc2 antiserum recognized a single 69-kDa polypeptide. (B and C) X. laevis Orc2 antibodies form a ternary complex with TIF4 and the type I element T-rich strand. (B) EMSA analysis with a fixed amount of Q-Sepharose-purified TIF4 and a twofold dilution series of X. laevis Orc2 or preimmune antiserum. Radiolabeled oligonucleotide, ssT51.(C) EMSA analysis with a twofold dilution series of nuclear extracts in the presence and absence of a fixed amount of protein A-Sepharose-purified X. laevis Orc2 or Euplotes crassus telomerase reverse transcriptase (Tert) antibody.
To establish a more direct connection between p69 and the TIF4 DNA-binding complex, mobility shift experiments were performed in the presence of X. laevis Orc2 or control antibodies. Initially, a partially purified TIF4 preparation was incubated with serially diluted rabbit preimmune or X. laevis Orc2 antiserum (S100 extract Q-Sepharose peak fraction; Fig. 5). A reproducible decrease in the abundance of the normally migrating TIF4 complexes was observed with the p69-specific antiserum (Fig. 4B, light exposure), along with the formation of a slower-migrating (supershifted) complex (Fig. 4B, dark exposure). As expected, exogenous ATP was required to generate both the normal and antibody-dependent supershift complexes (data not shown). The antibody-dependent supershift was more evident when fixed amounts of purified (protein A-Sepharose) antibodies were incubated with serially diluted nuclear extract. At low protein-to-antibody ratios, virtually all of the TIF4 DNA-protein complex was retarded by the X. laevis Orc2 antibody (Fig. 4C, left panel, supershift). As a control, rabbit antibodies specific for the Euplotes crassus telomerase reverse transcriptase subunit were added to gel shift reactions; they had no effect on the migration of the TIF4 DNA-protein complex (Fig. 4C, right panel). The collective data suggest that the Orc2-related protein p69 is a component of the TIF4 origin-binding complex.
FIG. 5.
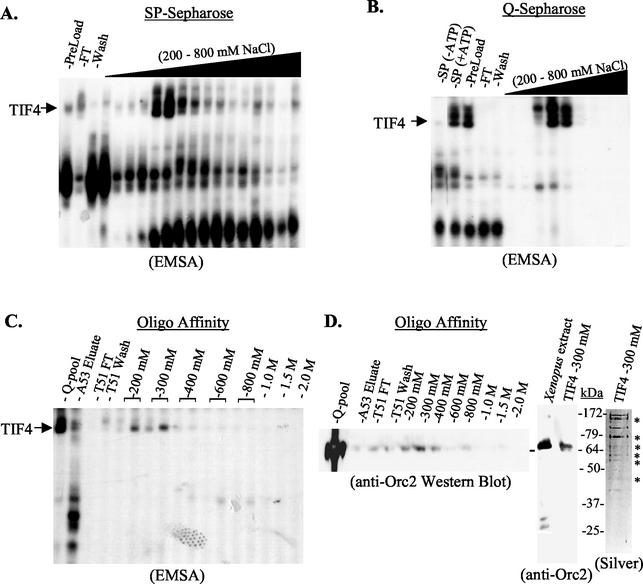
Copurification of p69 with TIF4 DNA-binding complexes. EMSA analysis of column fractions following sequential purification on (A) SP-Sepharose, (B) Q-Sepharose, and (C) oligonucleotide affinity chromatography (ssA53 Sepharose [−ATP], followed by ssT51 Sepharose [+ATP]). Unless otherwise indicated, gel shift reaction mixes contained 4 mM ATP. FT, flowthrough fraction. (D) Polypeptide analysis of ssT51 affinity chromatography fractions. Stepwise-eluted fractions were subjected to SDS-PAGE and probed with X. laevis Orc2 antiserum (left panel, column fractions; middle panel, peak DNA-binding fraction and X. laevis extract control). The peak ssT51 DNA-binding fraction (300 mM NaCl) was also silver stained to determine its protein composition (right panel).
p69 copurifies with TIF4.
The association of p69 with the TIF4 complex was further substantiated by purifying TIF4 to near homogeneity by conventional and oligonucleotide affinity chromatography. The initial S100 lysate (SP-Sepharose preload) generated a single TIF4 gel shift complex. However, two slower-migrating complexes were subsequently resolved following SP-Sepharose chromatography (Fig. 5A). All three complexes were ATP dependent (Fig. 5B, lanes 1 and 2) and coeluted from SP-Sepharose at between 320 and 540 mM NaCl. They further cofractionated on Q-Sepharose column (Fig. 5B). Sequential purification on ssA53- and ssT51-Sepharose removed A-rich strand and nonspecific DNA-binding proteins. TIF4 DNA-binding activity eluted in the 200 and 300 mM NaCl ssT51 fractions, generating just the lower (original) gel shift complex (Fig. 5C). Ten major protein bands were detected in the peak TIF4 fraction, six of which (Fig. 5D, silver stain, asterisks) displayed the same mobility as prominent polypeptides eluted from TIF4 EMSA complexes, including a protein of ≈70 kDa. Western blot analysis revealed that this protein crosequence-specific reacted with the Xenopus Orc2 antiserum, peaking in fractions containing maximal TIF4 DNA-binding activity (Fig. 5D, left panel, anti-Orc2). As expected, this antiserum recognized a single TIF4 polypeptide which comigrated with Xenopus Orc2 protein (Fig. 5D, right panel). Consistent with the antibody supershift studies, these data support the conclusion that p69 is a component of the TIF4 origin binding complex.
TIF4 and p69 are present in both micro- and macronuclei.
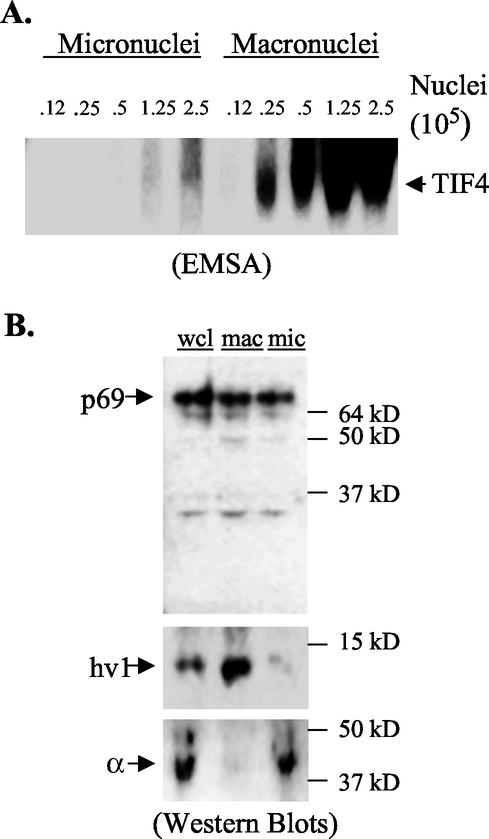
To determine whether TIF4 and its chromatin-associated p69 subunit are rDNA specific or have more global cellular roles, S100 extracts were prepared from purified vegetative micro- and macronuclei. While there are just two rDNA copies per diploid micronucleus, the rDNA is amplified to 10,000 copies in the macronucleus. Gel shift assays revealed that TIF4 was present in both nuclear compartments (Fig. 6A). Nuclear extract titrations indicated that a 20-fold excess of input micronuclei was required to produce a signal intensity comparable to that in macronuclear extracts. When normalized to total nuclear DNA content (macronucleus, 45N; micronucleus, 2N), the TIF4-DNA ratio was found to be roughly equivalent in micro- and macronuclei, even though the normalized macronuclear to micronuclear rDNA ratio is 5,000:1.
FIG. 6.
Distribution of p69 and TIF4 in vegetative growing cells. (A) EMSA analysis of extracts prepared from purified micro- and macronuclei. Extracts were prepared from equal numbers of purified micro- and macronuclei. The nuclear input for each binding reaction is indicated. (B) Western blot analysis of whole-cell lysates (wcl) and lysates from purified micronuclei (mic) and macronuclei (mac). Extracts derived from equivalent numbers of micro- and macronuclei were loaded. Upper panel, p69 Western blot with X. laevis Orc2 antibodies. Middle and lower panels, Western blot analysis with antibodies specific for the macronuclear-limited histone H2 hv1 variant and micronuclear-limited histone H1-like α-peptide subunit.
Despite differences in the abundance of TIF4 DNA-binding activity in micro- and macronuclear extracts, p69 protein levels were indistinguishable when an equivalent number of input micro- and macronuclei were examined (Fig. 6B, upper panel). The purity of each preparation was verified with antibodies specific for micro- or macronuclear-limited histone variants (Fig. 6B, middle and lower panels) (55). When differences in nuclear DNA content were considered, the p69-DNA ratio was predicted to be >10-fold higher in the micronucleus.
Cell cycle regulation of TIF4 and p69.
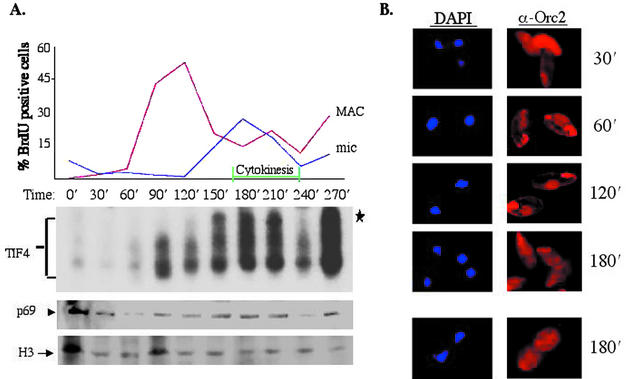
Recent studies indicate that the association of mammalian Orc1 and Orc2 subunits with the holocomplex is cell cycle regulated (15, 23, 28, 44). To assess whether TIF4 and it Orc2-related p69 subunit are similarly regulated, vegetative cultures were examined at defined intervals following cell cycle synchronization. Cells were synchronized in G0 by maintaining cultures at saturating cell densities, followed by a brief starvation period in Tris-containing buffer. Pulse-labeling with 5-bromodeoxyuridine (BrdU) revealed that refed cells entered S phase with a high degree of synchrony. Distinct but overlapping periods for macronuclear (90 to 150 min) and micronuclear (120 to 240 min) S phases were evident by BrdU immunocytological analysis (Fig. 7A, upper panel graph). Following the formation of a visible cleavage furrow (Fig. 7A, cytokinesis) and cell doubling, a new round of macronuclear BrdU labeling was observed almost immediately (270 min). Consequently, the second macronuclear S phase was preceded by a brief G1 period. Although the time of entrance into the first macronuclear S phase varied between experiments, the duration and timing of macronuclear S and subsequent cell cycle events were very consistent, including the brief gap between the first micronuclear and second macronuclear S phase.
FIG. 7.
Cell cycle regulation of TIF4 and p69. (A) BrdU, EMSA, and p69 Western blot analysis of vegetative cells synchronized by stationary-phase release. G0-arrested cultures were examined at 30-min intervals after refeeding. Cells were pulse-labeled for 5 min with BrdU and harvested for BrdU immunofluorescence and light microscopy to monitor micro- and macronuclear DNA replication, cytokinesis (upper panel graph), and cell doubling (data not shown). S100 extracts were also prepared and subjected to EMSA (lower panel) or Western blot analysis with X. laevis Orc2 or control (histone H3) antibodies. The ratio of p69 to H3 was determined, normalizing the zero time point ratio to 1 (upper panel graph). Histone H3 signals were consistent at time points following the release from G0 arrest and thus served not only as a normalization control for p69 protein levels, but also as a loading control for EMSA analysis. (B) p69 immunofluorescence in synchronized vegetative cells. Nuclear (DAPI, blue) and X. laevis Orc2 (red) staining profiles. The two fields at 180 min correspond to cells that have (lower panel) or have not (upper panel) initiated cytokinesis, which takes 3 to 5 min to complete. Staining of the oral apparatus is nonspecific, as it was observed with preimmune antiserum as well (data not shown).
EMSA analysis revealed that TIF4 DNA-binding activity was barely detectable in G0-arrested cells. A dramatic increase in TIF4 DNA-binding activity occurred at the onset of macronuclear S phase (90 min), peaking at the height of micronuclear S (180 min), and dropping precipitously as cells exited micronuclear S (240 min) (Fig. 7A, middle panel). Interestingly, the slowest-migrating TIF4 complex (asterisk) appeared as cells were exiting macronuclear S phase and entering micronuclear S (120 to 150 min). Control experiments verified that the three gel shift complexes were TIF4 derived. Not only did these complexes comigrate with species generated with TIF4 prepared from asynchronous vegetative cultures, DNA binding was ATP dependent and T-rich-strand specific (data not shown).
Western blot analysis was performed on gel shift extracts with p69 and histone H3 antibodies, the latter of which served as a normalization control. Less pronounced but reproducible evidence for p69 cell cycle regulation was observed (Fig. 7A, lower panel). p69 levels peaked during the overlapping macro- and micronuclear S phases. Furthermore, a consistent dip in p69 signal intensity was observed immediately after the first cycle of nuclear BrdU labeling (T = 240 min in the experiment shown; n = 3).
Indirect immunofluorescence with Xenopus Orc2 antibodies was used to examine the subcellular localization of p69. Intense cytoplasmic p69 staining was observed in starved cells (data not shown) and at the earliest refeeding time point (Fig. 7B, 30 min). Clear macronuclear staining became evident immediately prior to the onset of macronuclear S phase along with a concurrent decrease in cytoplasmic p69 staining (60 min). Nonspecific staining of the oral apparatus was observed with both preimmune (data not shown) and Xenopus Orc2 antiserum. Macronuclear immunoreactivity was strongest when macronuclear BrdU labeling was maximal (120 min and data not shown). This cyclic redistribution of p69 became apparent when cells exited macronuclear S phase (180-min time point); macronuclear immunofluorescence disappeared, while strong cytoplasmic staining pattern reappeared at this time. Definitive information on vegetative micronuclei could not be obtained due to their close association with macronuclei throughout most of the cell cycle (Fig. 7B, DAPI). However, they do not physically associate during development, when the relationship between micronuclear p69 staining and replication status was subsequently established (see below). The correlation between p69 nuclear localization and increased TIF4 DNA-binding activity during macronuclear S phase suggests that the interaction between TIF4 and the rDNA replication origin is cell cycle regulated.
Developmental regulation of p69 and TIF4 in wild-type cells.
Indirect immunofluorescence with X. laevis Orc2 antibodies was next used to examine p69 during development. After an overnight starvation period, strains of opposite mating types were mixed to initiate mating. Prior to nuclear exchange, the micronucleus is replicated during pre- and postmeiotic S phases to generate two genetically identical pronuclei (2 to 4 h after mixing the strains). Following reciprocal pronuclear exchange and fusion, the diploid zygotic nucleus undergoes two rounds of replication and mitotic nuclear division (4 to 7 h), producing cells that harbor four diploid micronuclei and the old polyploid parental macronucleus. Further replication is restricted to two progeny nuclei that differentiate into transcriptionally active macronuclei (anlagen). During the early stage of anlagen development (7 to 10 h), rDNA and non-rDNA chromosomes are replicated; however, non-rDNA chromosomes arrest with a four- to eight-copy DNA content unless the exconjugants are refed (2). In starved exconjugants, replication in the later stage macronuclear anlagen is restricted to the rDNA locus, which is amplified extensively (20). Well-established cytological criteria were used to stage cells throughout development (2; reviewed in reference 38).
Consistent with experiments performed on vegetative cells, p69 immunoreactivity was a reliable marker for the replication status in newly developing nuclei. Initial experiments were performed on matings between two wild-type strains (Fig. 8A, upper panels). Robust p69 staining was detected in the old parental macronucleus and pronuclei during the pre- and postmeiotic S phases (data not shown). A similar pattern was observed during the postzygotic nuclear divisions that follow pronuclear exchange and precede macronuclear anlagen formation (Fig. 8A, left panel, postzygotic S). Orc2 antibody staining was detected in the developing macronuclear anlagen throughout the remainder of development. However, staining was completely absent in micronuclei during the macronuclear anlagen formation stage, when micronuclear replication has ceased (Fig. 8A, middle and right panels, early and late macronuclear development). The loss of micronuclear Orc2 immunoreactivity cannot be ascribed to antibody inaccessibility, as anti-BrdU antibodies stained pulse-labeled micronuclei throughout development (data not shown). Although only three developmental time points are shown, cells were examined at 3-h intervals during the 24-h conjugal period. Samples taken at all time points exhibited a strong correlation between p69 staining and developmental landmarks for nucleus-specific replication.
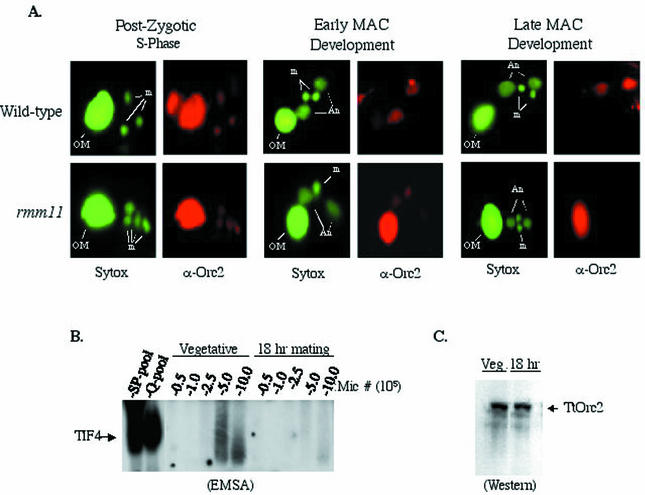
FIG. 8.
Developmental regulation of p69 and TIF4 DNA-binding activity. (A) Sytox DNA staining (green) and p69 immunofluorescence (red) of nonrefed conjugating wild-type cells and homozygous rmm11 mutant heterokaryons defective for rDNA gene amplification. A representative cell from a mating pair is shown for the developmental stages indicated (OM, old parental macronucleus; m, progeny micronuclei; An, developing progeny macronuclei). (B) EMSA analysis of micronuclear extracts from isolated log-phase vegetative cells or mating cells (18 h, late macronuclear development). (C) Western blot analysis of micronuclear lysates with X. laevis Orc2 antiserum.
To assess whether the programmed loss of micronuclear p69 staining during late macronuclear development coincides with decreased TIF4 origin binding activity or p69 protein levels, extracts from purified late stage micronuclei (18 h) were compared to micronuclei derived from an asynchronous vegetative culture (≈15% of which were in micronuclear S phase). TIF4 DNA-binding activity was significantly lower in the late stage developing micronuclei (Fig. 8B); however, p69 protein levels were indistinguishable (Fig. 8C). Similar to vegetative macronuclei, in vivo p69 immunoreactivity and TIF4 DNA-binding activity were markedly elevated in replication-competent (vegetative) versus nonreplicating (late developmental) micronuclei.
Macronuclear p69 immunoreactivity is lost in amplification-defective rmm11 mutant.
Previous fluorescent in situ hybridization studies revealed the formation of punctate perinuclear rDNA foci at late stages of macronuclear development (54). Punctate, perinuclear immunofluorescence was similarly detected in wild-type macronuclear anlagen with X. laevis Orc2 antibodies, along with strong disperse macronuclear staining (Fig. 8A and data not shown). To further explore the relationship between p69 and gene amplification, mutants defective in rDNA gene amplification were examined throughout development.
The rmm11 mutation resides in a chromosome breakage sequence element immediately downstream of the single-copy micronuclear rRNA gene. The rDNA fails to excise in 90% of progeny. Excision is sufficiently delayed in the remaining cells that rDNA gene amplification fails to occur (20). Heterokaryons that contain a wild-type macronucleus were used to propagate this recessive lethal mutation in the homozygous state in the germ line micronucleus (reviewed in reference 51).
Developmental time courses were performed on matings between homozygous germ line rmm11 heterokaryons. Prior to anlagen formation, the rmm11 p69 staining pattern was indistinguishable from that of the wild type (Fig. 8A, left panel, and data not shown). Diminished anlagen staining was detected during early macronuclear development (Fig. 8A, middle panel, and data not shown). The size of the macronuclear anlagen was comparable to that of wild-type progeny, suggesting that early macronuclear development was largely unperturbed. In marked contrast to wild-type matings, no Orc2 staining was detected in the mutant anlagen at later time points, when selective amplification of the rDNA normally occurs (Fig. 8A, right panel, late macronuclear development, and data not shown). Additionally, the old parental macronucleus retained Orc2 staining, indicating that the normal developmental program was arrested. Control antibodies that recognize lysine 4/lysine 9-diacetylated histone H3 stained the wild-type and mutant anlagen at all times, indicating that the p69-negative rmm11 anlagen are accessible to antibody (data not shown). The loss of p69 immunoreactivity in amplification-defective macronuclei suggests that this protein, and seemingly the intact TIF4 complex, is involved in locus-specific amplification of the rDNA.
DISCUSSION
Recognition of replicator sequences by TIF4.
Tetrahymena has served as a useful model system for studying many aspects of eukaryotic DNA replication. The modular rDNA replicon harbors several dispersed cis-acting regulatory determinants that function cooperatively and can act at a distance to control DNA replication (16, 43). Type I elements are required for cell cycle-regulated replication and developmentally programmed gene amplification and thus are candidate binding sites for replication initiation proteins. Since type I elements interact with several different proteins in vivo (47), competition between trans-acting factors may regulate origin activity. Thus far, cis-acting determinants in other eukaryotic replicons are known to interact with just one sequence-specific regulatory factor.
In this study we describe the identification and characterization of a novel type I element-binding activity, TIF4, that displays many features of eukaryotic ORCs. TIF4 exhibits three biochemical properties expected of an ORC: chromatin association, origin-specific DNA recognition, and organization into a multiprotein complex that contains at least one putative ORC subunit. What distinguishes TIF4 from yeast ORCs is the nature of its substrate recognition. Instead of interacting with duplex DNA, TIF4 binds exclusively to the T-rich strand of type I elements. Similar to the S. cerervisiae (5) and D. melanogaster (4) ORCs, TIF4 requires ATP for sequence-specific DNA binding. However, in contrast to the S. cerervisiae ORC (27), TIF4 does not form nonspecific DNA-protein complexes in the absence of ATP.
While origin-specific DNA binding has not yet been established for mammalian and amphibian ORCs (reference 53 and references therein), a recent study revealed that the Schizosaccharomyces pombe Orc4 subunit recognizes a preferred sequence within the degenerate ARS3001 replicon (23). In addition to binding to the duplex form of the δ3 element, S. pombe Orc4 binds specifically to the T-rich strand of this target sequence. Although S. pombe Orc4's preference for double-stranded DNA is ≈100-fold higher, its ability to recognize a single-stranded DNA target indicates that TIF4's substrate preference is within the spectrum possibilities for eukaryotic ORCs. In both organisms, the DNA-binding substrate is the T-rich strand of the replicator element (16, 25). By analogy, the vast majority of the contacts between the S. cerevisiae ORC and duplex DNA occur on one DNA strand, the A-rich strand (26).
The potential biological relevance of a single-stranded-DNA-binding activity such as TIF4 has been enhanced by experiments probing the in vivo configuration of the rDNA origin region. Potassium permanganate footprinting studies previously revealed that segments encompassing the rDNA origin and promoter are largely unwound in asynchronous cycling cells (47). Consequently, the binding of TIF4 and other sequence-specific single-stranded-DNA-binding proteins to their cognate target sequence is favored in native rDNA minichromosomes. Using antisense ribosomes to inhibit the expression of TIF1, we previously determined that this protein discriminates between origin- and promoter-proximal type I elements in vivo. Altered footprints were restricted to just one strand in these mutants, the A-rich strand at the rDNA origin and the T-rich strand at the rRNA promoter. The T-rich strand specificity that we describe here for TIF4 raises the possibility that TIF1 might facilitate the binding of this ORC-like complex to rDNA origins. Our recent discovery that TIF1 null mutants are delayed in S-phase progression further implicates TIF1 in replication-based processes (T. L. Morrison and G. M. Kapler, unpublished results).
Although relatively rare, eukaryotic sequence-specific single-strand breaks have been shown to regulate important chromosomal processes, such as transcription (13, 33, 37) and telomere maintenance (36). A human sequence-specific single-stranded binding protein that recognizes a specific target in the myc replicon has been identified. By virtue of its physical association with DNA polymerase α, MSSP was proposed to play a role in mammalian DNA replication (35). Whether the sequence-specific single-stranded binding protein TIF4 is indeed the Tetrahymena ORC awaits the sequencing and reverse genetic analysis of individual subunits. Whatever the outcome, the regulatory properties of TIF4 strongly suggest that it plays an important role in DNA replication, including programmed gene amplification.
Regulation of p69 and TIF4: potential mechanisms for DNA replication control.
Using polyclonal Xenopus Orc2 antiserum (2), which specifically recognizes Orc2 orthologs from diverse eukaryotes, we identified a 69-kDa chromatin-associated Tetrahymena protein that is present in both micro- and macronuclei. Three lines of experimentation argue that p69 is an integral component of the TIF4 origin-binding complex. (i) p69 is present in purified TIF4 gel shift complexes along with five similarly abundant proteins. (ii) p69-specific X. laevis Orc2 antibodies form a ternary complex with TIF4 and type I element DNA. (iii) The elution profiles of p69 and TIF4 DNA-binding activity are superimposable in affinity column fractions purified to near homogeneity. Ten prominent polypeptides were identified in these TIF4 preparations, six (including p69) of which correspond in size to abundant proteins in gel-purified TIF4 EMSA complexes. These data indicate that the multisubunit TIF4 complex has a complexity similar to that of eukaryotic ORCs.
We uncovered two strong positive correlations between TIF4 and replication in the micro- and macronucleus: (i) an association between nuclear replication and enhanced TIF4 DNA-binding activity, and (ii) the presence of robust p69 immunoreactivity in replicating nuclei. Because micro- and macronuclear S phases are offset during development and across the vegetative cell cycle, these nuclei probably respond to different cellular signals to regulate their replication. For example, while both macronuclear replication and karyokinesis precede cytokinesis, micronuclear division and, to a lesser extent, micronuclear S phase are not tightly coupled to cell division (Fig. 7 and data not shown). Similarly, micronuclear replication is selectively arrested during the period for macronuclear anlagen formation in developing progeny.
The cyclin-dependent kinase cdc2 regulates replication in all eukaryotes examined (reviewed in reference 14). Although cdc2 kinase activity is readily detected in isolated macronuclei, this activity is noticeably absent from the micronucleus (50). Consistent with these findings, cdc2 antibodies exclusively stain the macronucleus throughout the cell cycle and development (PSTAIRE monoclonal antibody, Santa Cruz Biotechnology; R. D. York and G. M. Kapler, unpublished results). Despite the apparent exclusion of cdc2 from the micronucleus, a dramatic increase in TIF4 DNA-binding activity is associated with micronuclear DNA replication. Hence, TIF4 (or the equivalent ORC complex) may be regulated differently in the two nuclear compartments. Interestingly, formation of the slowest-migrating TIF4 gel shift complex is temporally coupled to the exit from macronuclear S phase and entrance into micronuclear S (Fig. 7).
The most unanticipated finding of these studies is the consistent association between p69 in vivo immunoreactivity and nuclear replication status. This correlation has been established for developing micro- and macronuclei and for vegetative macronuclei. Although p69 is accessible to Orc2 antibodies in replicating postzygotic micronuclei, immunoreactivity is lost once these micronuclei exit S phase. In contrast, postzygotic nuclei that develop into macronuclear anlagen and undergo DNA replication retain their Orc2 reactivity. Furthermore, Orc2 staining is lost from the developing macronucleus in the amplification-defective rmm11 mutant. Finally, the localization of p69 to the vegetative macronucleus is cell cycle regulated, coinciding with the period for macronuclear BrdU labeling.
These findings collectively support a model in which programmed changes in p69 regulate origin activity. Several mechanisms could account for altered nuclear p69 immunoreactivity. p69 might be masked in nonreplicating stages due to a conformational change in TIF4 or the association of other regulatory proteins within the complex. Alternatively, p69 might dissociate from the TIF4 complex and relocalize to a nonchromatin nuclear compartment. The latter possibility is raised by our observation that less than half of the p69 present in asynchronous vegetative nuclei is released by DNase I. In addition to changes in nuclear p69 immunoreactivity during S phase, cell synchronization studies suggest that p69 relocalizes from the cytoplasm to the macronucleus at the onset of S phase.
Recent studies indicate that the human Orc1 and Orc2 subunits cycle between chromatin and nonchromatin nuclear compartments (15, 23). Cell cycle-regulated release of Orc1 from chromatin has also been reported in Chinese hamster ovary cells (15). Cyclin-mediated export of human Orc1 from the nucleus may represent yet another aspect of regulation (24). The possible shuttling of p69 between the cytoplasm and the nucleus may reflect mechanisms that have evolved to regulate nonyeast eukaryotic replicons.
Gene amplification: parallels between Tetrahymena rDNA and Drosophila chorion gene replicons.
The Tetrahymena rDNA and Drosophila chorion replicons have several similarities. (i) At the chromosomal level, discrete cis-acting regulatory determinants have been identified, some but not all of which colocalize with replication initiation sites (12, 19, 29). (ii) Drosophila ORC and TIF4 bind to genetically defined replication determinants in a sequence-specific manner (3; this work). (iii) Genome-wide endoreplication precedes gene amplification in both instances, indicating that these cells have exited their normal cell cycle. (iv) Locus-specific amplification is associated with the arrest of genome-wide replication. A major distinction between these systems is the fate of affected cells. Chorion gene amplification occurs in terminally differentiated cells that never reenter the cell cycle, while rDNA amplification precedes vegetative cell division in Tetrahymena cells. Consequently, replication competence must be maintained at non-rDNA Tetrahymena origins. Additionally, the Tetrahymena rDNA replicon is physically separated from neighboring DNA by site-specific fragmentation prior to gene amplification.
Since TIF4 and D. melanogaster ORC are not exclusively dedicated to the respective rDNA and chorion gene replicons, these proteins must play a more general role in replication initiation. Immunofluorescence studies link D. melanogaster ORC and TIF4 to the amplification process. In D. melanogaster follicle cells, diffusely distributed nuclear Orc2 is reorganized into discrete brightly staining foci concomitant with chorion gene amplification (9, 45). Although a similarly dramatic nuclear reorganization of p69 was not observed in wild-type T. thermophila, p69 staining is completely eliminated in the amplification-defective rmm11 macronucleus. By comparison, a Drosophila myeloblastosis oncoprotein ortholog was recently shown to bind a sequence in the chorion gene ACE3 element (4). Although this protein is required for chorion gene amplification, diffuse nuclear staining, similar to that in TIF4, was observed in amplifying follicle cells. We suggest that the diffuse macronuclear anlagen staining observed late in wild-type Tetrahymena development represents p69 molecules that have transiently dissociated from nonreplicating non-rDNA chromosomal origins. The absence of p69 staining in the rmm11 mutant macronuclear anlagen may reflect abortion of the macronuclear developmental program, a conclusion supported by the observation that p69 staining is retained in the old parental macronucleus of this mutant. Minimally, our studies of normal cell cycles and development indicate that p69 immunoreactivity is under the control of factors that regulate DNA replication in T. thermophila.
Perspective.
The work presented here, in conjunction with two-dimensional gel analysis of Tetrahymena rDNA replication, establishes T. thermophila as the first model organism for which the regulation of origin firing (56) and origin-binding activities (this work) have been examined during normal cell divisions and the amplification cell cycle. Previous work demonstrated that rDNA excision is required for amplification to occur (20, 54). Consequently, the “license to amplify” may result from the physical separation of rDNA origins from repressive flanking sequences or unfavorable chromatin environments. In the context of the work presented here, rDNA excision may establish persistent, productive interactions between TIF4 and type I elements during development. Temporal changes in the pattern of rDNA replication intermediates during development may reflect a chromosome “maturation” process that affects the binding of ORC and non-ORC regulatory proteins (56). It is tempting to speculate that rDNA chromatin is reorganized from a form that is permissive for gene amplification to one that is not (i.e., the vegetative rDNA form) (40). The progression of replication intermediates observed during development is consistent with this model. Germ line DNA transformation will allow us to examine the contribution of cis-acting determinants to rDNA gene amplification and determine the exact role of TIF4 in this process.
Acknowledgments
We are indebted to Anja Bielinsky for help with the initial Orc2 Western blot studies, advice, and encouragement. We acknowledge David Gilbert and Thomas Kelly for anti-Xenopus and human Orc2 antisera, Stephen Bell for Drosophila and S. cerevisiae Orc2 proteins, Marty Gorovsky and Dave Allis for antibodies against micro- and macronuclear-specific Tetrahymena histones, and Dorothy Shippen for Euplotes crassus TERT antibodies. We thank Phil Carpenter and William Dunphy for providing the X. laevis Orc2 plasmid, pC187-1B, for expression of recombinant X. laevis Orc2 protein. We are grateful to Suma Datta for advice on fluorescence microscopy and Dorothy Shippen and Sebastian Yakisich for careful reading of the manuscript.
This work was supported by NIH grant R01-GM53572.
The first two authors contributed equally to this work.
REFERENCES
- 1.Allen, S. L., T. C. White, J. P. Langmore, and M. A. Swancutt. 1983. Highly purified micro- and macronuclei from Tetrahymena thermophila isolated by Percoll gradients. J. Protozool. 30:21-30. [Google Scholar]
- 2.Allis, C. D., and D. K. Dennison. 1982. Identification and purification of young macronuclear anlagen from conjugating cells of Tetrahymena thermophila. Dev. Biol. 93:519-533. [DOI] [PubMed] [Google Scholar]
- 3.Austin, R. J., T. L. Orr-Weaver, and S. P. Bell. 1999. Drosophila ORC specifically binds ACE3, an origin of DNA replication control element. Genes Dev. 13:2639-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beall, E. L., J. R. Manak, S. Zhou, M. Bell, J. S. Lipsick, and M. R. Botchan. 2002. Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature 420:833-837. [DOI] [PubMed] [Google Scholar]
- 5.Bell, S. P., and B. Stillman. 1992. ATP-dependent recognition of eukaryotic origins of DNA replication by a multi-protein complex. Nature 357:128-134. [DOI] [PubMed] [Google Scholar]
- 6.Blomberg, P., C. Randolph, C.-H. Yao, and M.-C. Yao. 1997. Regulatory sequences for amplification and replication of the ribosomal DNA minichromosome in Tetrahymena thermophila. Mol. Cell. Biol. 17:7237-7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blow, J. J., and R. A. Laskey. 1986. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell 47:577-587. [DOI] [PubMed] [Google Scholar]
- 8.Burke, T., J. Cook, M. Asano, and J. Nevins. 2001. Replication factors MCM2 and ORC1 interact with the histone acetyltransferase HB01. J. Biol. Chem. 276:15397-15408. [DOI] [PubMed] [Google Scholar]
- 9.Calvi, B. R., M. A. Lilly, and A. C. Spradling. 1998. Cell cycle control of chorion gene amplification. Genes Dev. 12:734-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carpenter, P. B., P. R. Mueller, and W. G. Dunphy. 1996. Role of Xenopus Orc2-related protein in controlling DNA replication. Nature 379:357-360. [DOI] [PubMed] [Google Scholar]
- 11.Chesnokov, I., M. Gossen, D. Remus, and M. Botchan. 1999. Assembly of functionally active Drosophila origin recognition complex from recombinant proteins. Genes Dev. 13:1289-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delidakis, C., and F. C. Kafatos. 1989. Amplification enhancers and replication origins in the autosomal chorion gene cluster of Drosophila. EMBO J. 8:891-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desveaux, D., C. Despres, A. Joyeaux, R. Subramaniam, and N. Brisson. 2000. PBF-2 is a novel single-stranded DNA binding factor implicated in PR-10a gene activation in potato. Plant Cell 12:1477-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dutta, A., and S. P. Bell. 1997. Initiation of DNA replication in eukaryotic cells. Annu. Rev. Cell Dev. Biol. 13:293-332. [DOI] [PubMed] [Google Scholar]
- 15.Fugita, M., Y. Ishimi, H. Nakamura, T. Kiyono, and T. Tsurmi. 2002. Nuclear organization of DNA replication initiation proteins in mammalian cells. J. Biol. Chem. 277:10354-10361. [DOI] [PubMed] [Google Scholar]
- 16.Gallagher, R. C., and E. H. Blackburn. 1998. A promoter mutation affecting replication of the Tetrahymena ribosomal DNA minichromosome. Mol. Cell. Biol. 18:3021-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gavin, K. A., M. Hidaka, and B. Stillman. 1995. Conserved initiator proteins in eukaryotes. Science 270:1667-1670. [DOI] [PubMed] [Google Scholar]
- 18.Gossen, M., D. Pak, S. Hansen, J. Acharya, and M. Botchan. 1995. A Drosophila homolog of the yeast origin recognition complex. Science 270:1674-1677. [DOI] [PubMed] [Google Scholar]
- 19.Heck, M. M. S., and A. C. Spradling. 1990. Multiple replication origins are used during Drosophila chorion gene amplification. J. Cell Biol. 110:903-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kapler, G. M., and E. H. Blackburn. 1994. A weak germline excision mutation blocks developmentally controlled amplification of the rDNA minichromosome of Tetrahymena thermophila. Genes Dev. 8:84-95. [DOI] [PubMed] [Google Scholar]
- 21.Kapler, G. M., D. L. Dobbs, and E. H. Blackburn. 1996. DNA replication in Tetrahymena, p. 915-932. In M. L. DePamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 22.Kong, D., and M. L. DePamphilis. 2002. Site-specific ORC binding, pre-replication complex assembly and DNA synthesis at Schizosaccharomyces pombe replication origins. EMBO J. 21:5567-5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kreitz, S., M. RItz, M. Baack, and R. Knippers. 2001. The human origin recognition compled protein 1 dissociates from chromatin during S phase in HeLa cells. J. Biol. Chem. 276:6337-6342. [DOI] [PubMed] [Google Scholar]
- 24.Laman, H., G. Peters, and N. Jones. 2001. Cyclin-mediated export of human Orc1. Exp. Cell Res. 271:230-237. [DOI] [PubMed] [Google Scholar]
- 25.Larson, D. D., E. H. Blackburn, P. C. Yaeger, and E. Orias. 1986. Control of rDNA replication in Tetrahymena involves a cis-acting upstream repeat of a promoter element. Cell 47:229-240. [DOI] [PubMed] [Google Scholar]
- 26.Lee, D. G., and S. P. Bell. 1997. Architecture of the yeast origin recognition complex bound to origins of DNA replication. Mol. Cell. Biol. 17:7159-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, D. G., A. M. Makhov, R. D. Klemm, J. D. Griffith, and S. M. Bell. 2000. Regulation of origin recognition complex conformation and ATPase activity: differential effects of single-stranded and double-stranded DNA binding. EMBO J. 19:4774-4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, C.-J., and M. DePamphilis. 2002. Mammalian Orc1 protein is selectively released from chromatin and ubiquitinated during the S-to-M transition in the cell division cycle. Mol. Cell. Biol. 22:105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu, L., H. Zhang, and J. Tower. 2001. Functionally distinct, sequence-specific replicator and origin elements are required for Drosophila chorion gene amplification. Genes Dev. 15:134-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacAlpine, D. M., Z. Zhang, and G. M. Kapler. 1997. Type I elements mediate replication fork pausing at conserved upstream sites in the Tetrahymena thermophila rDNA minichromosome. Mol. Cell. Biol. 17:4517-4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marahrens, Y., and B. Stillman. 1992. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science 255:817-823. [DOI] [PubMed] [Google Scholar]
- 32.Michelotti, G. A., E. F. Michelotti, A. Pullner, R. C. Duncan, D. Eick, and D. Levens. 1996. Multiple single-stranded cis-acting elements are associated with activated chromatin of the human c-myc gene in vivo. Mol. Cell. Biol. 16:2656-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohammad, M., S. Saha, and G. M. Kapler. 2000. Three different proteins recognize a multifunctional determinant that controls replication initiation, fork arrest and transcription in Tetrahymena. Nucleic Acids Res. 28:843-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murata-Hori, M., and M. Fujishima. 1996. Released substances from Tetrahymena thermophila arrest the cell cycle at G1 phase and removal of the substances induces highly synchronized cell division. Eur. J. Protistol. 32:481-489. [Google Scholar]
- 35.Niki, T., I. Galli, H. Ariga, and S. Iguchi-Ariga. 2000. MSSP. a protein binding to an origin of replication in the c-myc gene, interacts with a catalytic subuniit of DNA polymerase α and stimulates its polymerase activity. FEBS Lett. 475:209-212. [DOI] [PubMed] [Google Scholar]
- 36.Nugent, C., T. Hughes, N. Lue, and V. Lundblad. 1996. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274:249-252. [DOI] [PubMed] [Google Scholar]
- 37.Ohmori, M., M. Ohta, H. Shimura, Y. Shimurat, K. Suzuki, and L. D. Kohn. 1996. Cloning of the single strand DNA-binding protein important for maximal expression and thyrotropin (TSH)-induced negative regulation of the TSH receptor. Mol. Endocrinol. 10:1407-1424. [DOI] [PubMed] [Google Scholar]
- 38.Orias, E. 1986. Ciliate conjugation, p. 45-84. In J. G. Gall (ed.), The molecular biology of ciliated protozoa. Academic Press, Orlando, Fla.
- 39.Orias, E., and P. J. Bruns. 1975. Induction and isolation of mutants in Tetrahymena, p. 247-282. In D. M. Prescott (ed.), Methods in cell biology, vol. 13. Academic Press, New York, N.Y. [PubMed]
- 40.Palen, T. E., and T. R. Cech. 1984. Chromatin structure at the replication origins and transcription-initiation regions of the ribosomal RNA genes of Tetrahymena. Cell 36:933-942. [DOI] [PubMed] [Google Scholar]
- 41.Pan, W.-J., and E. H. Blackburn. 1995. Tandem repeats of the 5′ nontranscribed spacer of Tetrahymena rDNA function as high copy number autonomous replicons in the macronucleus, but do not prevent rRNA gene dosage regulation. Nucleic Acids Res. 23:1561-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan, W.-J., R. C. Gallagher, and E. H. Blackburn. 1995. Replication of an rDNA gene origin plasmid in the Tetrahymena thermophila macronucleus is prevented by transcription through the origin from an RNA polymerase I promoter. Mol. Cell. Biol. 15:3372-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reischmann, K. P., Z. Zhang, and G. M. Kapler. 1999. Long range cooperative interactions regulate the initiation of replication in the Tetrahymena thermophila rDNA minichromosome. Nucleic Acids Res. 26:4635-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Romanowski, P., M. A. Madine, A. Rowles, J. J. Blow, and R. A. Laskey. 1996. The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr. Biol. 6:1416-1425. [DOI] [PubMed] [Google Scholar]
- 45.Royzman, I., R. J. Austin, G. Bosco, S. P. Bell, and T. L. Orr-Weaver. 1999. ORC localization in Drosophila follicle cells and the effects of mutations in dE2F and dDp. Genes Dev. 13:827-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saha, S., and G. M. Kapler. 2000. Allele-specific protein-DNA interactions between the single-stranded DNA binding protein, ssA-TIBF, and DNA replication determinants in Tetrahymena. J. Mol. Biol. 295:423-439. [DOI] [PubMed] [Google Scholar]
- 47.Saha, S., A. Nicholson, and G. M. Kapler. 2001. Cloning and biochemical analysis of the Tetrahymena origin binding protein TIF1: in vitro and in vivo binding to critical rDNA replication determinants. J. Biol. Chem. 276:45417-45426. [DOI] [PubMed] [Google Scholar]
- 48.Smothers, J., C. Mizzen, M. Tubbert, R. Cook, and C. D. Allis. 1997. Pdd1p associates with germline-restricted chromatin and a second novel anlagen-enriched protein in developmentally programmed DNA elimination structures. Development 124:4537-4545. [DOI] [PubMed] [Google Scholar]
- 49.Stuart, K., and E. Cole. 2000. Nuclear and cytoskeletal fluorescence microscopy techniques. Methods Cell Biol. 62:291-311. [DOI] [PubMed] [Google Scholar]
- 50.Sweet, M., G. Carlson, R. Cook, D. Nelson, and C. D. Allis. 1997. Phosphorylation of linker histones by a protein kinase A-like activity in mitotic nuclei. J. Biol. Chem. 272:916-923. [DOI] [PubMed] [Google Scholar]
- 51.Turkewitz, A., E. Orias, and G. Kapler. 2002. Functional genomics: the coming of age for Tetrahymena thermophila. Trends Genet. 18:35-40. [DOI] [PubMed] [Google Scholar]
- 52.Umthun, A. R., Z. Hou, Z. A. Sibenaller, W.-L. Shiau, and D. L. Dobbs. 1994. Identification of DNA-binding proteins that recognize a conserved Type I repeat sequence in the replication origin region of Tetrahymena rDNA. Nucleic Acids Res. 22:4432-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vashee, S., P. Simancek, M. Challberg, and T. Kelly. 2001. Assembly of the human origin recognition complex. J. Biol. Chem. 276:26666-26673. [DOI] [PubMed] [Google Scholar]
- 54.Ward, J., P. Blomberg, N. Hoffman, and M.-C. Yao. 1997. The intranuclear organization of normal, hemizygous and excision-deficient rRNA genes during developmental amplification in Tetrahymena thermophila. Chromosoma 106:233-242. [DOI] [PubMed] [Google Scholar]
- 55.Wu, M., C. D. Allis, M. Sweet, R. Cook, T. Thatcher, and M. Gorovsky. 1994. Four distinct and unusual linker proteins in a mitotically dividing nucleus are derived from a 71-kilodalton polyprotein, lacks p32cdc2 sites and contains protein A kinase sites. Mol. Cell. Biol. 14:10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang, Z., D. M. MacAlpine, and G. M. Kapler. 1997. Developmental regulation of DNA replication: replication fork barriers and programmed gene amplification in Tetrahymena. Mol. Cell. Biol. 17:6147-6156. [DOI] [PMC free article] [PubMed] [Google Scholar]