Abstract
Activation of the oncogenic potential of the MEK kinase TPL-2 (Cot) requires deletion of its C terminus. This mutation also weakens the interaction of TPL-2 with NF-κB1 p105 in vitro, although it is unclear whether this is important for the activation of TPL-2 oncogenicity. It is demonstrated here that TPL-2 stability in vivo relies on its high-affinity, stoichiometric association with NF-κB1 p105. Formation of this complex occurs as a result of two distinct interactions. The TPL-2 C terminus binds to a region encompassing residues 497 to 534 of p105, whereas the TPL-2 kinase domain interacts with the p105 death domain. Binding to the p105 death domain inhibits TPL-2 MEK kinase activity in vitro, and this inhibition is significantly augmented by concomitant interaction of the TPL-2 C terminus with p105. In cotransfected cells, both interactions are required for inhibition of TPL-2 MEK kinase activity and, consequently, the catalytic activity of a C-terminally truncated oncogenic mutant of TPL-2 is not affected by p105. Thus, in addition to its role as a precursor for p50 and cytoplasmic inhibitor of NF-κB, p105 is a negative regulator of TPL-2. Insensitivity of C-terminally truncated TPL-2 to this regulatory mechanism is likely to contribute to its ability to transform cells.
The rat serine/threonine kinase TPL-2 was initially identified as a target for provirus integration in Moloney murine leukemia virus-induced T-cell lymphomas (25). The provirus integrates into the last intron of the TPL-2 gene, which results in enhanced expression of a truncated TPL-2 mRNA transcript encoding a protein that is altered at its C terminus (23). A C-terminally truncated form of the human homolog of TPL-2, known as Cot, was independently identified as a transforming gene for a human thyroid carcinoma cell line (5). Transgenic mice expressing a C-terminally deleted form of TPL-2 under the control of a T-cell-specific promoter develop T-cell lymphoblastic lymphomas, confirming its oncogenic potential (4). In contrast, transgenic expression of full-length TPL-2 has no transforming effect (4). More recently, this has been substantiated in two separate large-scale retroviral tagging screens for oncogenes in mice in which retroviral insertions in the TPL-2 gene were identified in multiple lymphoid and myeloid tumors (22, 24).
Studies in cell lines have demonstrated that overexpressed TPL-2 activates the ERK, JNK, and p38 mitogen-activated protein (MAP) kinase pathways (6, 26, 27). In vitro experiments indicate that this activity results from TPL-2 acting as a MAP 3-kinase, which directly phosphorylates and activates the respective MAP 2-kinases for each of the MAP kinase pathways (6, 27). However, the physiological relevance of the MAP 3-kinase activity of TPL-2 has only recently been established by analysis of TPL-2 knockout mice (10). Thus, lipopolysaccharide (LPS) activation of the MAP 2-kinases MEK-1 and -2, which phosphorylate and activate ERK-1 and -2 MAP kinases, is blocked in bone marrow-derived macrophages from TPL-2-deficient mice (10). In contrast, LPS activation of JNK and p38 in the knockout macrophages is normal, demonstrating that in these cells TPL-2 is physiologically a MAP 3-kinase only for the ERK MAP kinase cascade.
Our laboratory has previously demonstrated that TPL-2 binds to the C-terminal half of NF-κB1 p105 and also that overexpressed TPL-2 stimulates p105 proteolysis, thereby activating NF-κB (3). However, from analysis of TPL-2 knockout mice, it is unclear whether TPL-2 plays a physiological role in NF-κB activation (10). Interestingly, interaction with p105 requires the C terminus of TPL-2 in vitro, suggesting that oncogenic activation of TPL-2 may involve alteration of its association with p105. In the present study, the nature and consequences of TPL-2 association with p105 were examined in more detail. Evidence is presented that binding to p105 is required for TPL-2 metabolic stability and also inhibits TPL-2 MEK kinase activity. Significantly, C-terminally truncated oncogenic TPL-2 is insensitive to p105 negative regulation and consequently its kinase activity is dramatically increased compared to wild-type TPL-2 in p105-expressing cells.
MATERIALS AND METHODS
cDNA constructs, recombinant proteins, and antibodies.
All hemagglutinin (HA) epitope-tagged NF-κB1 p105 (HA-p105) cDNAs were cloned into the pcDNA3 vector (Invitrogen). Wild-type HA-p105, HA-p1051-801, HA-p105ΔDD (containing an internal deletion of the death domain [DD; residues 802 to 892]), and HA-p105L841A (containing a point mutation in the p105 DD) have been described previously (2, 28), as have Myc-tagged and untagged versions of TPL-2 and TPL-2ΔC (3). HA epitope-tagged NF-κB2 p100 (HA-p100), HA-p105Δ497-538, HA-p105Δ497-538; ΔDD, HA-p105Δ497-538; L841A, and the panel of N-terminal deletion mutants of Myc-TPL-2ΔC and Myc-TPL-2ΔC(D270A) were all generated by using PCR and verified by DNA sequencing. Myc-Raf1(CAAX) in the pEXV expression vector was kindly provided by Chris Marshall (Cancer Research UK, London, United Kingdom) (20).
To generate recombinant p105 protein, p105 cDNA sequences were cloned into pGEX-2T or pGEX-6P-1 (Amersham Pharmacia Biotech). Glutathione S-transferase (GST) fusion proteins were expressed at 30°C in Escherichia coli BL21(DE3) and purified by affinity chromatography on glutathione (GSH)-Sepharose 4B (Amersham Pharmacia Biotech). The p105 fragments were then cleaved from GST with either thrombin (Calbiochem) or PreScission Protease (Amersham Pharmacia Biotech) and further purified by gel filtration on a Superdex column (Amersham Pharmacia Biotech). The resulting proteins, which were >95% pure as judged by Coomassie brilliant blue (Novex) staining of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels, were concentrated by ultrafiltration and stored at −80°C. Expected masses of all proteins were confirmed by mass spectrometry.
Antibodies used for immunoprecipitation and Western blotting of HA and Myc epitope-tagged proteins have been described previously (2). TSP3 anti-TPL-2 antibody, which has been described previously (27), was used for Western blot detection of TPL-2. Anti-MEK-1/2 and anti-phospho-MEK-1/2 antibodies were purchased from Cell Signaling Technology. Tubulin and actin were used as loading controls for cell lysates and were detected on Western blots with TAT-1 anti-tubulin monoclonal antibody (MAb; kindly provided by Keith Gull, University of Manchester, Manchester, United Kingdom) and a commercial anti-actin MAb (Sigma), respectively.
Protein analyses.
293 cells (3 × 105 to 5 × 105 cells per 60-mm diameter Nunc dish) were transiently transfected by using Lipofectamine (Life Technologies, Inc.) and cultured for a total of 24 to 48 h, as described previously (2). Cell lysates were prepared by using buffer A (2) (which contains 1% NP-40). Immunoprecipitation and Western blotting of transfected proteins were carried out as described previously (17), with the indicated antibodies. For reconstitution of NF-κB1 knockout 3T3 fibroblasts with HA-p105, cells were cotransfected with HA-p105 vector and a reporter plasmid, pEGFP (Clontech), by using Lipofectamine. After 48 h of culture, transfected cells were sorted for expression of green fluorescent protein on a MoFlo cytometer (DakoCytomation) prior to cell lysis.
To determine the turnover of Myc-TPL-2, 293 cells were transfected as described above. After 24 h culture, cells were washed in phosphate-buffered saline and incubated for 1 h in methionine-cysteine-free minimal essential Eagle medium (Sigma). Cells were pulse-labeled with 2.65 MBq of [35S]methionine-[35S]cysteine (Pro-Mix; Amersham Pharmacia Biotech) for 30 min and chased for the times indicated in Dulbecco modified Eagle medium plus 2% fetal calf serum. Cells were lysed in buffer A supplemented with 0.5% deoxycholate and 0.1% SDS (radioimmunoprecipitation assay buffer). Myc-TPL-2 was isolated by immunoprecipitation with anti-Myc MAb and labeled bands revealed by autoradiography after SDS-PAGE on 10% acrylamide.
TPL-2 was synthesized in vitro and labeled with [35S]methionine-[35S]cysteine (Pro-Mix) by cell-free translation (Promega TNT-coupled rabbit reticulocyte system). Translated proteins were diluted by buffer (250 mM NaCl, 0.01% NP-40, 50 mM HEPES [pH 7.9], 50 mM EDTA) prior to incubation with GST fusion proteins and precipitated as described previously (27).
RNA isolation and RT-PCR analysis.
Total RNA from NF-κB1-deficient or wild-type 3T3 fibroblasts (29) was isolated by using Trizol reagent (Invitrogen). Semiquantitative reverse transcription-PCR (RT-PCR) was performed by utilizing the Qiagen OneStep RT-PCR kit. The primer pairs used were as follows: (5′) primer 5′-CATTGCTGATTCATCATGC-3′ and (3′) primer 5′-ACTGGGCTCATACACTGC-3′ for TPL-2 and 5′ primer 5′-GGCGGCTTGGTGACTCTAGATA-3′ and (3′) primer 5′-GCTCGGGCCTGCTTTGAACAC-3′ for 18SrRNA. The lengths of the TPL-2 and 18SrRNA amplicons were 1,169 and 560 bp, respectively. PCR products were visualized on 1.5% agarose gels, and TPL-2/18SrRNA ratios were determined by using a densitometric imager (Kodak ID 3.5).
Proteolytic mapping.
0.8 nmol of p105497-968 protein was digested at room temperature for 60 min with trypsin (1/250 of total protein concentration; Roche Diagnostics GmbH) in the absence or presence of 1.6 nmol of a synthetic peptide corresponding to the C-terminal 70 amino acids of TPL-2 (purchased from W. Mawby, University of Bristol, Bristol, United Kingdom). Samples were then resolved by 10% Bis-Tris gel electrophoresis and Western blotted onto polyvinylidene difluoride membrane (Millipore Corp.). Bands were visualized by Coomassie brilliant blue (Novex) staining and the major differential cleavage products identified by Edman degradation (N-terminal 6 amino acids). For mass spectroscopy, p105497-968 protein was digested with trypsin with or without TPL-2 C-terminal peptide for 60 min. The protein molecular weights were determined by using a platform electrospray mass spectrometer (Micromass).
Surface plasmon resonance.
Interaction between the TPL-2 C terminus and recombinant p105 proteins was measured by determining the surface plasmon resonance by using the BIAcore system (BIAcore AB) (31). The C-terminal 70 amino acids of TPL-2 (TPL-2398-467) were synthesized as a biotinylated peptide (conjugated to the equivalent of Cys398 of full-length TPL-2) and immobilized on a streptavidin-coated sensor surface (Sensorchip SA; BIAcore AB). Recombinant p105 proteins were then injected at different concentrations, and resonance signals were monitored. Dissociation at 25°C was measured by flushing the TPL-2-p105 complexes with HBS-P buffer (10 mM HEPES [pH 7.4], 150 mM NaCl, 0.005% [vol/vol] polysorbate 20 [BIAcore AB]). Data were evaluated by using BIAevaluation software (version 3.0.2).
TPL-2 peptide pulldown assay.
Interaction between the TPL-2 C terminus and p105 was also determined in pulldown assays. For these experiments, 30 μg of biotinylated TPL-2398-467 peptide was incubated for 2 h with lysates of 293 cells transfected with plasmids encoding wild-type or mutant HA-p105 and then captured on streptavidin-agarose beads (Sigma). Isolated protein was resolved by 10%-acrylamide SDS-PAGE and Western blotted.
Isothermal titration calorimetry (ITC).
Calorimetric binding measurements were carried out by using a MicroCal Omega VP-ITC isothermal titration calorimeter (MicroCal, Inc.). For these experiments, recombinant p105 proteins were dialyzed against 20 mM K2HPO4 (pH 7.4) containing 2 mM dithiothreitol. Lyophilized TPL-2 peptide was directly dissolved in this dialysis buffer. TPL-2 peptide was titrated at concentrations of 150 to 200 μM into 15 to 40 μM solutions of the indicated recombinant p105 proteins. Heats of dilution were determined by injection of the TPL-2 peptide solutions into dialysis buffer. All titrations were carried out at 20°C. ITC data were evaluated by using the Microcal Origin programme (version 5.0).
Analytical equilibrium ultracentrifugation.
Sedimentation equilibrium studies of recombinant p105 proteins were carried out by using a Beckman Optima XLA analytical ultracentrifuge. Aliquots (110 μl) of protein solution, with optical densities at 280 nm of 1.0, 0.75, 0.5, or 0.25 were centrifuged into sedimentation equilibrium at two different rotor speeds (14,000 and 18,000 rpm; An-60ti Beckman rotor). After sedimentation equilibrium had been reached, radial scans of 280 nm absorbance were measured at 0.001-cm intervals and 20-fold averaged. All data sets were analyzed by using the Beckman Optima XLA/XL-1 data analysis software (version 4.1).
MEK kinase and GST-MEK pulldown assays.
For assays of TPL-2 MEK kinase activity, Myc-TPL-2 or Myc-TPL-2ΔC were isolated by immunoprecipitation from lysates of transiently transfected 293 cells, as described above. Immunoprecipitates were washed four times in buffer A, followed by two washes in kinase buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 5 mM β-glycerophosphate, 2 mM dithiothreitol, 0.1 mM sodium vanadate, 10 mM MgCl2, 1 mM EGTA, 0.01% Brij-35). Beads were then resuspended in 50 μl of kinase buffer plus the indicated amounts of recombinant p105 protein and incubated at 4°C for 30 min. A portion (1 μg) of kinase inactive GST-MEK1(K207A) protein (Upstate Biotechnology) was then added in 50 μl of kinase buffer plus 2 mM ATP and incubated a further 30 min at room temperature. The supernatant was removed, mixed with an equal volume of 2× sample buffer, Western blotted after SDS-PAGE in 8% acrylamide, and probed with anti-phospho-MEK-1/2 antibody. Immunoprecipitated Myc-TPL-2 or Myc-TPL-2ΔC was eluted with Laemmli sample buffer from the remaining anti-Myc MAb beads and then quantified by Western blotting.
To assay the interaction of TPL-2 with MEK, lysates from 293 cells (lysis in 0.5% NP-40 buffer A) cotransfected with a plasmid encoding Myc-TPL-2 with or without HA-p105 plasmid were incubated overnight with 2 μg of GST-MEK(K207A) protein. Recombinant p105 protein was incubated with lysates for 2 to 3 h prior to the addition of GST-MEK(K207A) protein. Then, 10 μl of GSH-Sepharose 4B (Amersham Pharmacia Biotech) was then added to capture the GST fusion protein (30-min incubation). Beads were extensively washed in 0.5% NP-40 buffer A and isolated TPL-2 assayed by Western blotting of 10% acrylamide SDS-PAGE gels.
RESULTS
TPL-2 kinase domain interacts with the p105 DD.
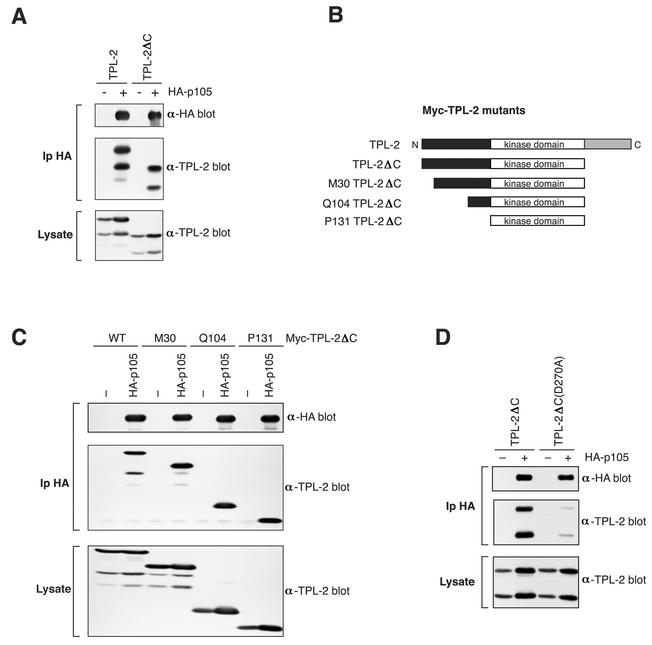
Transgenic mice expressing a C-terminally truncated TPL-2 under the control of a T-cell-specific promoter develop T-cell lymphoblastic lymphomas (4). Interestingly, previous binding experiments with in vitro-translated protein indicate that the C terminus of TPL-2 is required for efficient interaction with NF-κB1 p105 (3), which is expressed as an endogenous protein in T-lineage cells (11). Very low levels of C-terminally truncated TPL-2 (TPL-2ΔC) do in fact coimmunoprecipitate with p105 in vitro, which are revealed only after long exposure of autoradiographs (data not shown). To investigate the role of the TPL-2 C terminus in binding to p105 in more detail, it was important to determine whether TPL-2ΔC associated with p105 in vivo. To do this, 293 cells were transiently cotransfected with plasmids encoding HA-p105 and TPL-2ΔC, which lacks the C-terminal 70 amino acids (3). TPL-2ΔC was clearly detected in anti-HA immunoprecipitates, although at slightly lower levels than full-length TPL-2 (Fig. 1A). Consistent with previous data (3), neither TPL-2 nor TPL-2ΔC coimmunoprecipitated with HA-p50 (data not shown). Thus, TPL-2 can bind to p105 independently of its C terminus in vivo.
FIG. 1.
TPL-2 interacts with NF-κB1 p105 via its kinase domain. (A) 293 cells were cotransfected with vectors encoding TPL-2 or TPL-2ΔC and HA-p105. Anti-HA immunoprecipitates and cell lysates were Western blotted for HA-p105 and TPL-2. The amounts of vector encoding TPL-2 or TPL-2ΔC were adjusted to achieve equal protein expression in lysates with or without HA-p105 (B) Schematic diagram of Myc-TPL-2 deletion mutants. M30 corresponds to the natural internal initiation site (1). (C and D) 293 cells were cotransfected with vectors encoding the indicated Myc-TPL-2 or TPL-2 mutants and HA-p105 or EV(−). Anti-HA immunoprecipitates and cell lysates were sequentially immunoblotted with the indicated antibodies.
To identify which additional region(s) of TPL-2 is involved in interaction with p105, 293 cells were cotransfected with plasmids encoding HA-p105 and a panel of N-terminal deletion mutants of TPL-2ΔC (see Fig. 1B). Immunoprecipitation and Western blotting revealed that the N terminus of TPL-2 was not required for binding to HA-p105 in 293 cells (Fig. 1C). Consistent with these results, a GST fusion protein encoding the TPL-2 N terminus (residues 1 to 131) did not detectably interact with p105 (data not shown). Furthermore, a kinase domain activation loop mutant of TPL-2ΔC, Myc-TPL-2ΔC(D270A), which is catalytically inactive, interacted with HA-p105 at much reduced levels compared to wild-type TPL-2ΔC (Fig. 1D). Taken together with previous in vitro experiments (3), this suggests that both the TPL-2 kinase domain and C terminus are involved in interaction with p105.
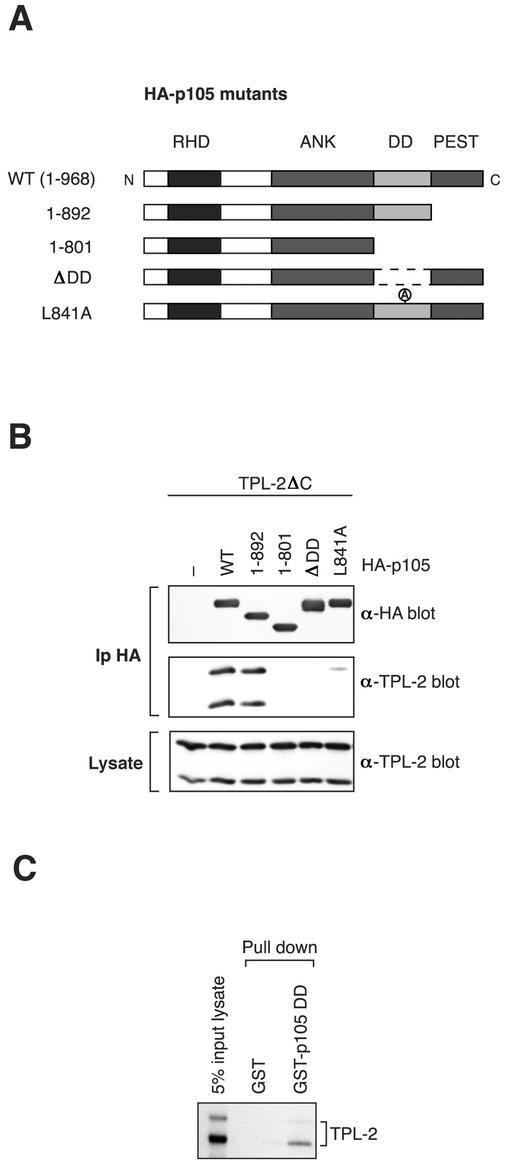
To characterize the binding site for TPL-2ΔC on p105, a panel of C-terminal deletion mutants of HA-p105 were individually coexpressed with TPL-2ΔC in 293 cells (Fig. 2A) (2) and association determined by immunoprecipitation and Western blotting. TPL-2ΔC interacted with HA-p1051-892, which lacks the PEST region, to a similar degree to full-length HA-p105 (Fig. 2B). However, TPL-2ΔC did not interact with three mutants HA-p1051-801, HA-p105ΔDD or HA-p105L841A, which lack a functional p105 DD. Thus, the p105 DD appears to be essential for interaction with TPL-2ΔC. A GST-p105 DD (p105 residues 808 to 892) fusion protein also associated with in vitro-translated TPL-2 (Fig. 2C), suggesting a direct interaction between TPL-2 and the p105 DD. Consistent with this notion, a GAL4-binding domain-TPL-2 fusion protein associates with the isolated p105 DD fused to the GAL4 activation domain in a yeast two-hybrid assay (data not shown). These data suggest that the p105 DD is the binding site for the TPL-2 kinase domain.
FIG. 2.
The p105 DD is a binding site for TPL-2. (A) Schematic diagram of HA-p105 mutants. (B) 293 cells were cotransfected with TPL-2ΔC and the indicated HA-p105 mutants. Anti-HA MAb immunoprecipitates and cell lysates were Western blotted and probed with the indicated antibodies. The amounts of vector encoding TPL-2ΔC were adjusted to achieve equal protein expression in lysates with or without HA-p105. (C) TPL-2 was synthesized and labeled with [35S]methionine-[35S]cysteine by in vitro cell-free translation. Pulldowns were then performed with GST-p105808-892 or GST control. Isolated protein was resolved by SDS-PAGE and revealed by fluorography.
Identification of the binding site on p105 for the C terminus of TPL-2.
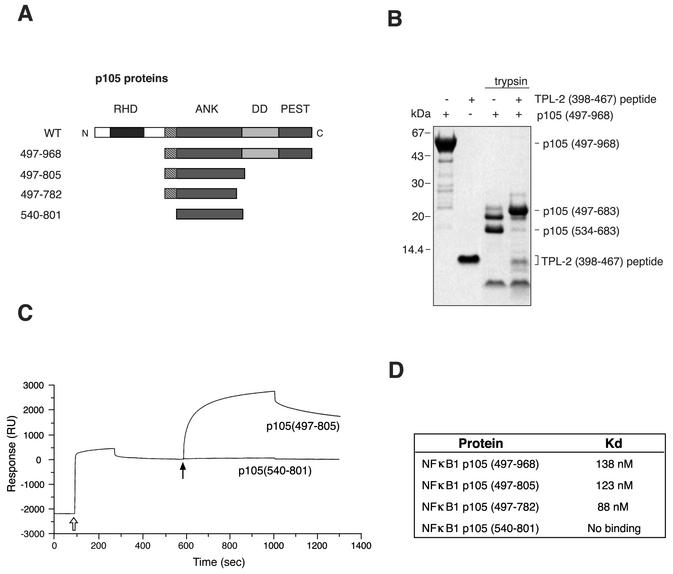
In our earlier studies, it was demonstrated that the C-terminal 70 amino acids of TPL-2 are required for efficient interaction with the C-terminal half of p105 in vitro (3). To identify the actual binding site on p105 for the TPL-2 C terminus, recombinant protein encoding the C terminus of p105 (residues 497 to 968) (Fig. 3A) was digested with trypsin in the presence or absence of a synthetic peptide corresponding to the C terminus of TPL-2 (residues 398 to 467). In the absence of TPL-2 peptide, a 16-kDa fragment accumulated in the digest (Fig. 3B), which was identified by N-terminal sequencing and mass spectrometry to correspond to amino acids 534 to 683 of p105. A larger fragment, with an apparent molecular mass of 20 kDa, was generated in the presence of TPL-2 peptide, which was identified as p105 amino acids 497 to 683 (Fig. 3B). Thus, the binding of TPL-2 peptide prevents access of the protease to target sites within the region from residues 497 to 534 of p105 and suggests that this region constitutes an important component of the binding site for the C terminus of TPL-2.
FIG. 3.
Characterization of the interaction of the TPL-2 C terminus with p105. (A) Schematic diagram of recombinant p105 proteins. (B) Recombinant p105497-968 protein was digested with trypsin with or without TPL-2398-497 peptide for 60 min, resolved by 10% Bis-Tris gel electrophoresis, and Western blotted. Protein fragments were visualized by Coomassie brilliant blue staining. The mobilities of p105497-968 and the fragments corresponding to p105497-683 and p105534-683 are indicated. (C) Surface plasmon resonance analysis of the interaction of p105497-805 and p105540-801 protein with biotinylated TPL-2398-497 peptide immobilized on a streptavidin-coated sensor surface. An open arrow denotes the injection of biotinylated TPL-2 C-terminal peptide, closed arrow denotes the injection of indicated p105 protein. (D) Binding affinities of TPL-2398-497 peptide for the indicated recombinant p105 proteins, as determined by surface plasmon resonance.
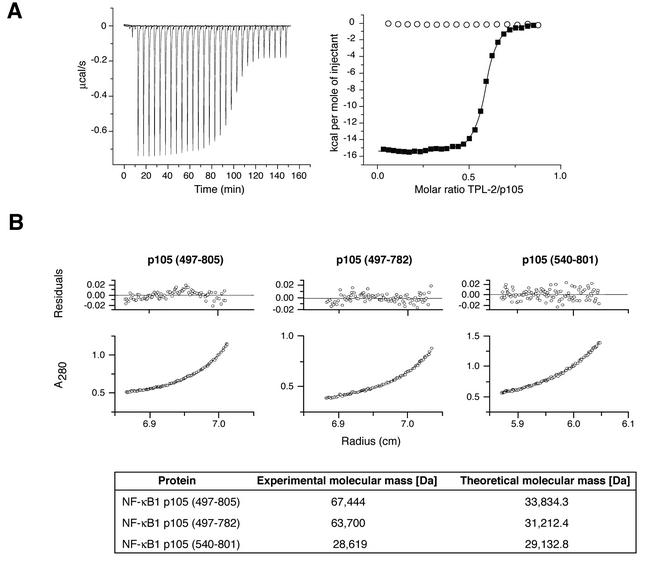
In order to obtain corroborating evidence for the TPL-2 C terminus binding site on p105, surface plasmon resonance experiments were carried out by using the panel of recombinant p105 proteins shown in Fig. 3A. These data revealed that p105540-801 did not detectably bind to the TPL-2 peptide (Fig. 3C), whereas recombinant p105 proteins that included the segment 497 to 539 bound to the TPL-2 peptide with nanomolar affinity constants (Fig. 3C and D). ITC experiments were also carried out to determine the affinity of interaction between the TPL-2 C terminus and p105 more accurately. The TPL-2 C-terminal peptide was found to bind to p105497-805 (Fig. 4A) and p105497-757 (data not shown) with Kd values of 58 and 64 nM, respectively. In contrast, p105540-801 did not detectably bind to the TPL-2 C-terminal peptide (Fig. 4A). Taken together, the proteolytic mapping data and binding experiments strongly suggest that residues 497 to 534 constitute at least part of a high-affinity binding site for the C terminus of TPL-2.
FIG. 4.
p105 amino acids 497 to 539 are required for dimerization of the C-terminal half of p105. (A) Binding of TPL2398-497 peptide to p105497-805 protein was determined by ITC. The right panel shows integrated heat changes (▪), corrected for the heat of dilution, which were fitted by using a single binding site model (Kd = 58 nM; stoichiometry, n = 0.57). Under the same conditions, p105540-801 protein did not bind TPL2398-497 peptide (○). The left panel shows the raw data of the p105497-805 titration in which the heat change of this endothermic binding reaction was measured in microcalories/second. (B) Molecular masses of recombinant p105 proteins were determined by sedimentation equilibrium ultracentrifugation. The lower panels show the protein distribution at equilibrium. The upper panels give the residuals in relation to the radial position. Experimental and theoretical molecular masses of the indicated recombinant p105 proteins are shown.
In previous immunoprecipitation experiments with in vitro-cotranslated and [35S]methionine-labeled protein, the stoichiometery of the TPL-2-p105 complex was deduced to be 1:1 (3). However, the ITC titrations demonstrate that the binding stoichiometry between the TPL-2 C terminus and p105 is in fact 1:2 (Fig. 4A), suggesting that TPL-2 may bind to p105 dimers. To better understand the stoichiometry of this interaction, a series of sedimentation equilibrium ultracentrifugation experiments were undertaken with recombinant p105 proteins. These revealed that both p105487-805 and p105497-782 exist as dimers in the absence of TPL-2 peptide (Fig. 4B). In contrast, p105540-801 was found to be monomeric. Thus, p105 homodimerizes through its C-terminal moiety, in addition to its known ability to dimerize with Rel subunits via its N-terminal Rel homology domain and ankyrin repeat region (30). Amino acids 497 to 539 of p105, which are required for binding to the C terminus of TPL-2, are also necessary for dimerization of the p105 C-terminal half.
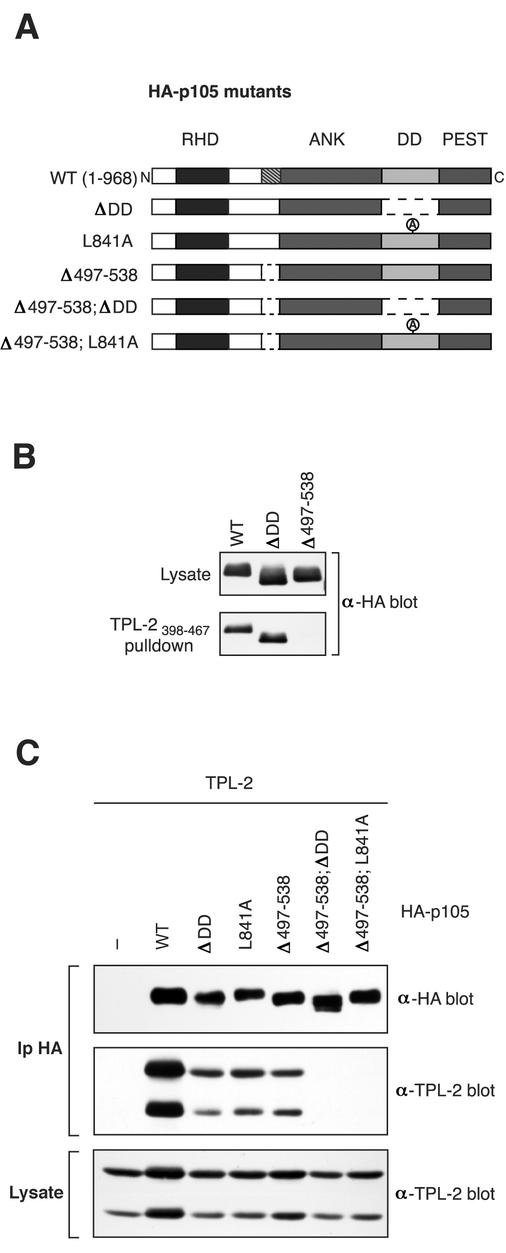
The importance of the region encompassing residues 497 to 534 for binding to the TPL-2 C terminus was also determined in full-length p105. To do this, residues 497 to 538, just prior to the start of the ankyrin repeats, were deleted in HA-p105 to generate HA-p105Δ497-538 (Fig. 5A). Biotinylated TPL-2398-467 peptide coupled to streptavidin-agarose beads clearly interacted with both wild-type HA-p105 or HA-p105ΔDD in pulldown assays (Fig. 5B). However, HA-p105Δ497-538 failed to detectably interact in this assay (Fig. 5B), a finding consistent with binding experiments with recombinant p105 fragments (Fig. 3 and 4).
FIG. 5.
Both TPL-2 binding sites on p105 are required for optimal association with TPL-2. (A) Schematic diagram of HA-p105 mutants. (B) 293 cells were transfected with vectors encoding wild-type or deletion mutant forms of HA-p105. Biotinylated TPL-2398-467 peptide, coupled to streptavidin-agarose beads, was then used to affinity purify HA-p105 from cell lysates. Isolated protein was detected by Western blotting. (C) 293 cells were cotransfected with vectors encoding TPL-2 and the indicated HA-p105 mutants or EV. In this experiment, to assay the interaction between TPL-2 and HA-p105 under more stringent conditions, cells were lysed and immunoprecipitated in radioimmunoprecipitation assay buffer. Anti-HA immunoprecipitates and cell lysates were Western blotted and probed sequentially with the indicated antibodies. The amount of TPL-2 epression vector was adjusted so that similar steady-state levels of protein expression in cell lysates were obtained with or without HA-p105.
Both p105 binding sites are required for optimal TPL-2 interaction.
To determine the relative contributions of the two TPL-2 binding sites on p105, additional HA-p105 constructs were generated with residues 497 to 538 deleted in combination with deletion (HA-p105Δ497-538; ΔDD) or point mutation (HA-p105Δ497-538; L841A) of the p105 DD (see Fig. 5A). These HA-p105 constructs and those in which each binding site alone was mutated (HA-p105Δ497-538, HA-p105ΔDD, and HA-p105L841A; see Fig. 2A) were individually coexpressed with wild-type TPL-2 in 293 cells and immunoprecipitated from cell lysates with anti-HA antibody. The extent of coimmunoprecipitation of TPL-2 was reduced by deletion or point mutation of the DD compared to wild-type HA-p105 (Fig. 5B). Deletion of residues 497 to 538 of p105 reduced association with TPL-2 to a similar extent. However, no association of TPL-2 was detected with either of the double binding site mutants. Control experiments demonstrated that all of the HA-p105 mutants were cytoplasmic, similar to wild type, indicating that altered subcellular localization did not contribute to their differential association with TPL-2 (data not shown). Thus, each of the identified binding sites makes a substantial contribution to TPL-2 association with p105 in vivo, and both are required for optimal complex formation. This implies that the binding affinity of TPL-2 kinase domain for the p105 DD is of an order similar to that observed between the TPL-2 C terminus and its binding site on p105 (nanomolar range). The overall affinity of TPL-2 for p105, therefore, might be very high (perhaps picomolar), which is consistent with the finding that all of the cellular pool of TPL-2 is complexed with p105 in HeLa cells (3).
Interaction with both binding sites on p105 regulates the metabolic stability of TPL-2.
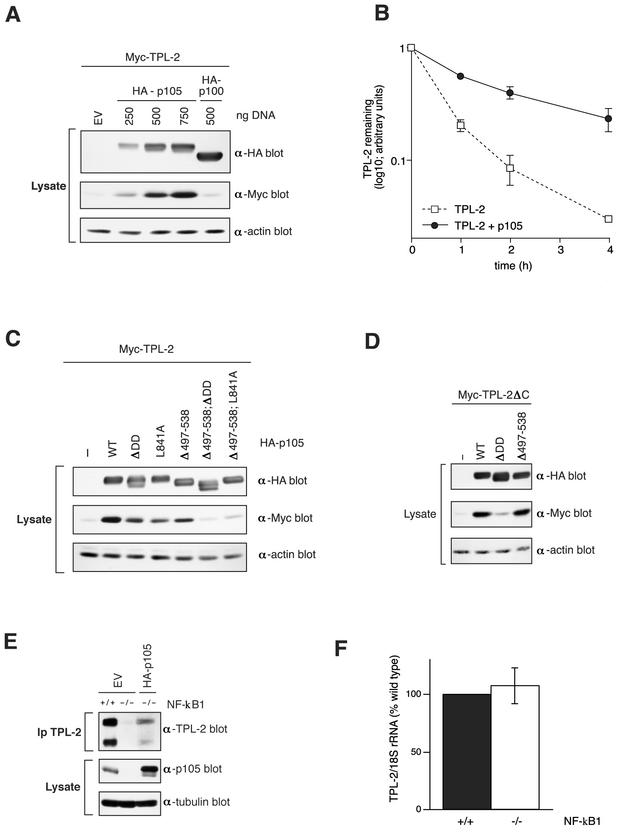
Our earlier observation that endogenous TPL-2 does not exist in p105-free form in cells (3) suggests that p105 might be important for the stability of TPL-2. To investigate this possibility, 293 cells were transiently cotransfected with a fixed quantity of vector encoding TPL-2 and increasing amounts of HA-p105 vector. Expression of HA-p105 resulted in a concentration-dependent increase in the amount of TPL-2 detected in cell extracts compared to empty vector (EV) cotransfected cells (Fig. 6A). In contrast, coexpression with HA epitope-tagged NF-κB2 p100 (HA-p100), a structurally related homolog of p105 that does not associate with TPL-2 (18; data not shown), had little effect on TPL-2 protein levels (Fig. 6A). Semiquantitative PCR demonstrated that coexpression of HA-p105 did not alter the level of Myc-TPL-2 mRNA (data not shown). Thus, the effect of HA-p105 on steady-state levels of TPL-2 protein was not due to increased transcription from the TPL-2 expression vector but rather suggests that HA-p105 association stabilizes TPL-2 protein. This was confirmed by pulse-chase metabolic labeling, which demonstrated that HA-p105 coexpression increased the half-life of TPL-2 (Fig. 6B).
FIG. 6.
TPL-2 metabolic stability is regulated by association with NF-κB1 p105. (A) 293 cells were transiently transfected with 0.25 μg of Myc-TPL-2 plasmid and the indicated amounts of HA-p105 plasmid or HA-p100 (NF-κB2) plasmid. Total plasmid DNA was adjusted to 1 μg with EV. Expression of TPL-2, HA-p105, and HA-p100 was determined by Western blotting of cell lysates. (B) 293 cells were cotransfected with expression vectors encoding Myc-TPL-2 and HA-p105 or EV. After 24 h, cells were metabolically pulse-labeled with [35S]methionine-[35S]cysteine (30 min) and then chased for the times indicated. Anti-Myc immunoprecipitates were resolved by 8% acrylamide SDS-PAGE and visualized by fluorography. Amounts of immunoprecipitated Myc-TPL-2 were quantified by laser densitometry (n = 2). (C and D) Cell lysates of 293 cells cotransfected with fixed ratios of the expression vectors encoding TPL-2 (0.25 μg of DNA) and the indicated HA-p105 mutants (0.75 μg of DNA; see Fig. 2A and 4A). Cell lysates were Western blotted and probed sequentially with the indicated antibodies. (E) NF-κB1-deficient or wild-type 3T3 fibroblasts were transfected with vector encoding HA-p105 or EV, as indicated. Transfected cell lysates were immunoprecipitated with anti-TPL-2 antibody. Isolated TPL-2 and p105 in cell lysates were revealed by Western blotting. (F) TPL-2 mRNA levels in total RNA isolated from NF-κB1-deficient and wild-type 3T3 fibroblasts were assayed by semiquantitative PCR. 18SrRNA amplicon was used as an internal control. Scanned data from three experiments are presented.
The effect of each of the individual binding sites on p105 for stability of TPL-2 protein in 293 cells was investigated. Coexpression of HA-p105 containing deletion or point mutation of the DD increased the steady-state level of TPL-2 to a lesser extent than coexpression with wild-type HA-p105 (Fig. 6C). Similarly, HA-p105 lacking residues 497 to 538 resulted in lower levels of TPL-2. Moreover, HA-p105 mutants lacking both binding sites had no effect on the levels of cotransfected TPL-2 protein. Pulse-chase metabolic labeling experiments confirmed that HA-p105Δ497-538; ΔDD did not alter the half-life of coexpressed TPL-2 (data not shown). Thus, the optimal effect of p105 on TPL-2 protein stabilization requires association via both binding sites.
Myc-TPL-2ΔC was also stabilized by coexpression with HA-p105 or HA-p105Δ497-538 (Fig. 6D), which both associate with TPL-2ΔC (Fig. 2B, data not shown). However, coexpression with HA-p105ΔDD, which cannot bind TPL-2ΔC (Fig. 2B), had a minimal effect on Myc-TPL-2ΔC levels (Fig. 6D).
To determine whether p105 regulates the steady-state levels of endogenous TPL-2 protein, cell lysates were prepared from 3T3 fibroblasts derived from wild-type and NF-κB1-deficient mice. TPL-2 was readily detected by Western blotting of anti-TPL-2 immunoprecipitates from wild-type fibroblasts (Fig. 6E) and was associated with endogenous p105 (data not shown). In contrast, TPL-2 protein was barely detectable in the NF-κB1 knockout fibroblasts, although semiquantitative RT-PCR demonstrated that both cell types expressed similar levels of TPL-2 mRNA (Fig. 6E and F). Expression of HA-p105 in NF-κB1 knockout cells by transfection significantly increased the level of detectable endogenous TPL-2 protein (Fig. 6E). Thus, NF-κB1 expression is required for the maintenance of steady-state levels of endogenous TPL-2 protein, a finding consistent with the results of the 293 pulse-chase experiments (Fig. 6B).
Interaction with the p105 DD inhibits TPL-2 MEK kinase activity.
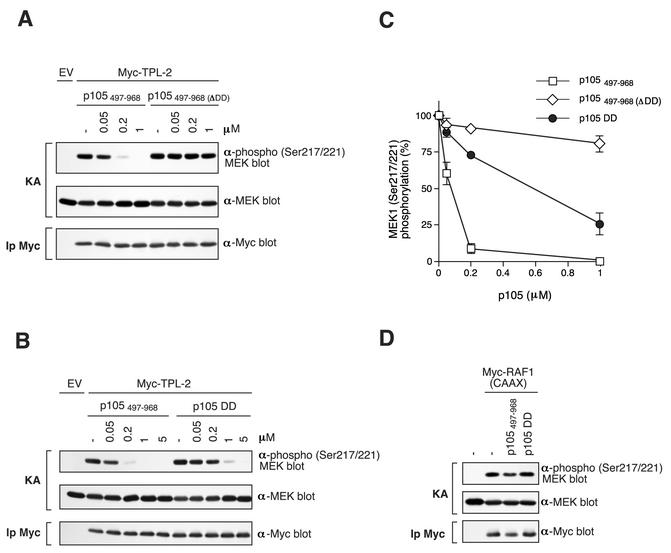
The interaction of the kinase domain of TPL-2 with the p105 DD raised the interesting possibility that TPL-2 kinase activity might be regulated by its association with p105. To investigate this, Myc-TPL-2 was purified by immunoprecipitation from transiently transfected 293 cells and its kinase activity assayed in vitro, with GST-MEK1(K207A) protein as a substrate. Only a small fraction of overexpressed Myc-TPL-2 is associated with p105 (data not shown), presumably due to titration of endogenous p105 protein. The effect of p105 binding on Myc-TPL-2 MEK kinase activity was investigated by incubation of TPL-2 immunoprecipitates with recombinant p105497-968 protein prior to in vitro kinase assay. MEK phosphorylation was assayed by Western blotting with anti-phospho-MEK1/2 antibody. Control experiments confirmed that p105497-968 protein bound to TPL-2 in vitro (data not shown). Addition of p105497-968 dramatically inhibited TPL-2 MEK kinase activity in a dose-dependent fashion (Fig. 7A and C). A 50% inhibititory concentration (IC50) was achieved at 0.09 μM p105497-968, which represents a molar p105497-968/GST-MEK ratio of 0.6:1. The inhibitory effect of p105497-968 was specific since the MEK kinase activity of activated Raf1 [Myc-Raf1(CAAX) (20)] was not affected by the addition of p105497-968 protein (Fig. 7D).
FIG. 7.
TPL-2 MEK kinase activity is inhibited by p105 in vitro. (A and B) Myc-TPL-2 was isolated by immunoprecipitation from lysates of transfected 293 cells and then preincubated with the different amounts of the indicated recombinant p105 proteins or control buffer (−). In vitro kinase assays (KAs) were performed with GST-MEK1(K207A) as a substrate and phosphorylation determined by Western blotting of reaction mixtures and probing with an anti-phospho-MEK1/2 Ser217/Ser221 antibody. Equal loading of GST-MEK1(K207A) protein was confirmed by reprobing blots with anti-MEK1/2 antibody. Western blotting of anti-Myc immunoprecipitates (Ip) demonstrated that similar amounts of TPL-2 were assayed in each reaction (lower panel). (C) MEK1 phosphorylation in replicates of the experiments shown in A and B was quantified by laser densitometry (n = 3). Data are presented as percentages of control MEK kinase activity. (D) Myc-Raf1(CAAX) was immunoprecipitated from lysates of transfected 293 cells and MEK kinase activity assayed as in panel A. Recombinant p105 protein was added to a final concentration of 5 μM.
Addition of p105497-968(ΔDD) protein, which lacks the p105 DD, did not affect Myc-TPL-2 MEK kinase activity at the equivalent molar concentrations to p105497-968 (Fig. 7A and C), suggesting that interaction of the p105 DD with the kinase domain of Myc-TPL-2 inhibited its kinase activity. This hypothesis was tested directly by assaying the effect of the addition of recombinant p105808-892 protein, which consists solely of the p105 DD. The p105 DD clearly inhibited Myc-TPL-2 MEK kinase activity (Fig. 7B and C), although less efficiently than p105497-968 protein (TPL-2 IC50 [p105 DD] = 0.55 μM). As expected, the p105 DD had no effect on Myc-Raf1(CAAX) MEK kinase activity (Fig. 7D). Thus, TPL-2 MEK kinase activity is negatively regulated by binding to the p105 DD.
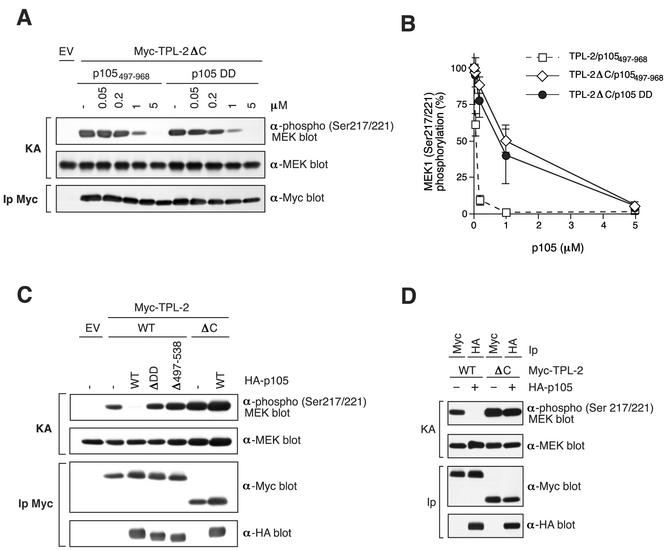
TPL-2ΔC MEK kinase activity is insensitive to p105 negative regulation.
The inhibition of TPL-2 MEK kinase activity by p105497-968 may be more efficient than the isolated p105 DD (Fig. 7C) since the former protein contains a binding site for the TPL-2 C terminus. It was, therefore, interesting to determine the sensitivity of ΤPL-2ΔC catalytic activity to inhibition by p105. The specific activity of Myc-TPL-2ΔC, isolated by immunoprecipitation from transfected 293 cell lysates, was modestly (ca. two- to threefold) increased relative to wild-type Myc-TPL-2 (Fig. 8C and D). However, Myc-TPL-2ΔC MEK kinase activity was significantly less sensitive to inhibition by p105497-968 protein (TPL-2ΔC IC50 [p105497-968] = 1 μM) than full-length Myc-TPL-2 (TPL-2 IC50 [p105497-968] = 0.09 μM) (Fig. 8A and B). The isolated p105 DD (p105802-892), that lacks the binding site for the TPL-2 C terminus, inhibited TPL-2ΔC MEK kinase activity with efficiency similar to that of the p105497-968 protein (Fig. 8A and B), in contrast to its decreased inhibitory effect on wild-type Myc-TPL-2 (Fig. 7B and C). These data suggest that efficient inhibition of TPL-2 MEK kinase activity by the p105 DD requires binding of the TPL-2 C terminus to p105.
FIG. 8.
TPL-2ΔC MEK kinase activity is less sensitive to inhibition by p105 than full-length TPL-2. (A) Myc-TPL-2 and Myc-TPL-2ΔC were isolated by immunoprecipitation from lysates of transfected 293 cells. MEK kinase assays were carried out as described in Fig. 6, in the presence of the indicated amounts of recombinant p105 proteins. (B) MEK phosphorylation in replicates of the experiment shown in panel A was quantified by laser densitometry (n = 3 for p105497-968 and n = 2 for p105 DD). Data are presented as percentages of control MEK kinase activity. For comparative purposes, the data from Fig. 7C showing the effect of p105497-968 protein on the MEK kinase activity of TPL-2 are included. (C and D) 293 cells were cotransfected with vectors encoding Myc-TPL-2 or Myc-TPL-2ΔC and the indicated HA-p105 proteins or EV. The amounts of TPL-2 and TPL-2ΔC vector were adjusted so that similar levels of protein expression were obtained. MEK kinase activity of Myc-TPL-2 and Myc-TPL-2ΔC, isolated by immunoprecipitation with anti-Myc or anti-HA MAb, was determined as in Fig. 6.
It was also determined whether binding to p105 affects TPL-2 kinase activity when Myc-TPL-2 was transiently expressed in 293 cells in the presence or absence of an excess of wild-type HA-p105 and isolated by immunoprecipitation with anti-Myc MAb. Myc-TPL-2 MEK kinase activity was dramatically inhibited by coexpression with HA-p105 (Fig. 8C). However, coexpression with HA-p105ΔDD or HA-p105Δ497-538 did not affect Myc-TPL-2 activity. Strikingly, although Myc-TPL-2ΔC clearly associates with HA-p105 (Fig. 8C, lower panel), its MEK kinase activity was completely unaffected by coexpression with HA-p105. Anti-HA MAb immunoprecipitates were also assayed for MEK kinase activity. Consistently, HA-p105-associated Myc-TPL-2 displayed minimal MEK kinase activity, whereas HA-p105-associated Myc-TPL-2ΔC was clearly very active (Fig. 8D).
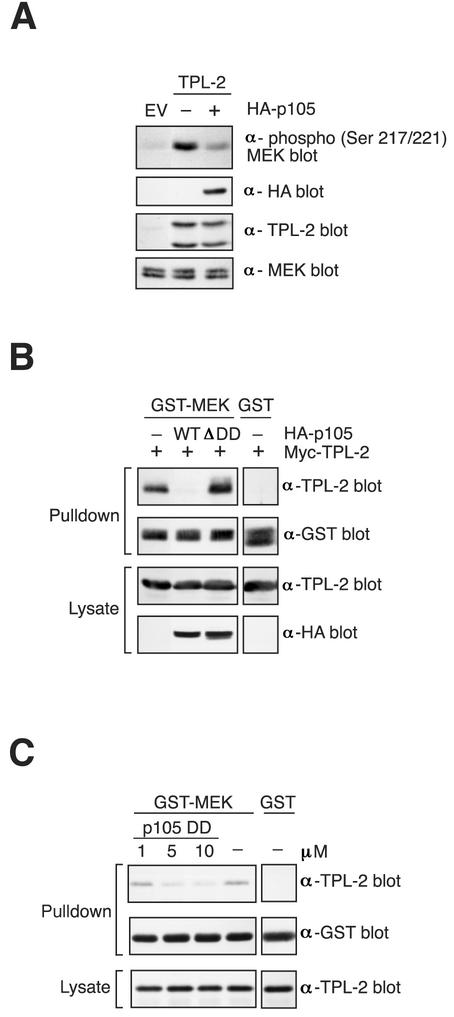
The experiments in this and the previous section clearly indicate that, in vitro, TPL-2 MEK kinase activity is negatively regulated by binding to p105. However, it was important to determine whether in vivo TPL-2 MEK kinase activity was similarly affected. To investigate this, 3T3 fibroblasts were transfected with TPL-2 plasmid on its own or together with HA-p105 plasmid and phosphorylation of endogenous MEK was then determined by Western blotting of cell lysates. TPL-2 induced clear phosphorylation of MEK compared to EV transfected cells (Fig. 9A). HA-p105 coexpression significantly reduced induction of MEK phosphorylation by TPL-2. Thus, p105 negatively regulates TPL-2 MEK kinase activity in vivo.
FIG. 9.
p105 blocks interaction between TPL-2 and MEK. (A) 3T3 fibroblasts were transfected with EV or TPL-2 vector or in combination with HA-p105 vector or EV. Cell lysates were Western blotted and probed sequentially with the indicated antibodies. (B) 293 cells were transfected with expression vectors encoding Myc-TPL-2 with or without wild-type or mutant version of HA-p105. GST-MEK1(K207A) protein, bound to GSH-Sepharose 4B beads, was used to affinity purify Myc-TPL-2 from cell lysates. Isolated protein and protein expression in lysates was assayed by Western blotting. (C) Lysates from 293 cells transfected with Myc-TPL-2 were incubated with the indicated concentrations of p105 DD protein prior to addition of GST-MEK1(K207A) affinity ligand. Pulldowns and lysates were Western blotted.
TPL-2 interaction with MEK is blocked by p105.
In vitro kinase assays demonstrate that TPL-2 does not phosphorylate the p105 DD, although the PEST region is clearly phosphorylated (data not shown). Thus, the p105 DD does not inhibit TPL-2 MEK kinase activity by acting as a competitive substrate. Raf kinase inhibitor protein (RKIP) is a physiological inhibitor of the Raf-1/MEK/ERK pathway (34). RKIP blocks Raf-1/MEK complex formation, acting as a competitive inhibitor of MEK phosphorylation by Raf-1 (33). It was interesting, therefore, to determine whether p105 might act in a similar fashion to regulate TPL-2 MEK kinase activity. As an initial step to investigate this possibility, TPL-2 was expressed in 293 cells on its own or together with HA-p105. The potential interaction of Myc-TPL-2 with MEK was then determined in a pulldown assay with GST-MEK1(K207A) as an affinity ligand. When expressed on its own, Myc-TPL-2 specifically associated with GST-MEK1(K207A) (Fig. 9B). However, this interaction was completely abrogated by coexpression with HA-p105 but not by coexpression with HA-p105ΔDD. Interaction between Myc-TPL-2 and GST-MEK1(K207A) was also blocked by addition of p105498-967 (data not shown) or the isolated p105 DD to lysates of Myc-TPL-2 transfected 293 cells (Fig. 9C). Thus, p105 negatively regulates TPL-2 MEK kinase activity by preventing access to its substrate.
DISCUSSION
In the present study, TPL-2 stability in vivo is shown to require its high-affinity, stoichiometric association with NF-κB1 p105 via two distinct interactions. The TPL-2 C terminus binds to a region N-terminal to the p105 ankyrin repeats (residues 497 to 534), whereas the TPL-2 kinase domain binds to the p105 DD. Surprisingly, the MEK kinase activity of TPL-2 was found to be profoundly inhibited by its interaction with the p105 DD, which prevents access of TPL-2 to its substrate MEK. The simultaneous interaction of the C terminus of TPL-2 with p105 significantly augments this inhibitory effect of the p105 DD. These data suggest that p105 functions upstream of TPL-2 as a negative regulator.
C-terminal truncation of TPL-2 activates its oncogenic potential in T lineage cells (4), which express endogenous p105 (11). Our studies reveal a mechanism by which deletion of the C terminus could lead to aberrant TPL-2 activity. Although TPL-2ΔC binds to p105 when expressed in cells (Fig. 1A and 8C and D), its kinase activity is largely unaffected by this association, in contrast to the full-length protein, which is dramatically inhibited. The C terminus of TPL-2, therefore, appears to be essential for negative regulation of its kinase activity by the p105 DD. This may be important to facilitate appropriate interaction between the TPL-2 kinase domain and the p105 DD. Removal of the TPL-2 C terminus generates an oncogenic form that has much higher specific activity than wild type in cells that coexpress p105 (Fig. 8C and D). Persistent phosphorylation of MEK is likely to be important for cell transformation by TPL-2ΔC, since constitutively active MEK mutants will oncogenically transform tissue culture cells (7).
It has previously been suggested that TPL-2 kinase activity is regulated by an intramolecular interaction with its C terminus (4). This hypothesis was based on the observation that a GST fusion protein encoding the C terminus of TPL-2 binds to TPL-2ΔC when the two proteins are expressed in baculovirus-infected Sf9 cells and inhibits TPL-2ΔC kinase activity in vitro. In the present study, in which both kinases were isolated in an effectively p105-free form from lysates of transfected 293 cells, Myc-TPL-2ΔC was indeed found to have somewhat higher MEK kinase activity than Myc-TPL-2 (two- to threefold; see Fig. 8C and D). However, although TPL-2398-467 peptide bound to TPL-2ΔC, it was not found to affect its in vitro MEK kinase activity (data not shown). It is therefore unclear whether the stimulatory effect of C-terminal deletion on p105-free TPL-2 kinase activity is due to abrogation of an intramolecular interaction or simply the result a gross structural change in the kinase. More importantly, it is apparent from our work that the physiologically relevant forms of TPL-2 and TPL-2ΔC exist in a complex with p105. Under these conditions, it is apparent that there is a far more pronounced difference (>40-fold) in specific MEK kinase activity between TPL-2 and TPL-2ΔC because of the insensitivity of the latter protein to negative regulation by p105.
NF-κB1-deficient and wild-type 3T3 fibroblasts synthesize similar levels of TPL-2 mRNA (Fig. 6F). However, NF-κB1 knockout cells contain very low steady-state levels of TPL-2, which are significantly increased by expression of HA-p105 by transfection (Fig. 6E). Furthermore, cotransfection experiments in 293 cells reveal that binding to p105 via both identified binding sites substantially increases the half-life of TPL-2 protein. Thus, p105 is an obligate partner for TPL-2 in vivo, which is required to maintain its metabolic stability. Clearly, these results indicate that it will be important to reexamine the molecular phenotype of NF-κB1 p105/p50 and NF-κB1 p105 knockout mice (15, 29), which may arise in part because of defective ERK activation caused by TPL-2 deficiency. Consistent with this hypothesis, it was reported while this study was in review that LPS activation of MEK/ERK is substantially blocked in NF-κB1-deficient macrophages due to the absence of TPL-2 protein (32). The regulation of TPL-2 stability by p105 may have evolved to ensure that unstimulated cells do not contain p105-free, unregulated TPL-2 activity.
Amino acids 497 to 539 of p105, which constitute at least part of a high-affinity binding site for the TPL-2 C terminus, are also required for dimerization of the C-terminal half of p105 (Fig. 4). Significantly, there is no fragment with homology to p105497-539 in IκBα (data not shown), which is a monomer (14, 16). However, the sequence of p105497-539 is highly conserved between human, mouse, rat, and chicken p105, suggesting that all p105 orthologues may dimerize through this region. It has previously been concluded that p105 is a monomer based on glycerol gradient sedimentation experiments (13). However, the reported sedimentation at 6.2S (13) is actually consistent with either p105 existing as an extended monomer or a globular dimer, depending on the translational diffusion coefficient, which is not determined in this present study. Therefore, although the data in the present study clearly demonstrate that the C-terminal half of p105 in isolation dimerizes, it remains to be determined whether full-length p105 behaves in a similar fashion.
Previous studies from our laboratory, in which p105 was identified as a binding partner for TPL-2, demonstrated that overexpressed TPL-2 induces proteolysis of p105 (3). However, as TPL-2-deficient macrophages have no obvious defect in LPS activation of NF-κB (10), it is not clear whether this reflects a physiological function of TPL-2. TNF-α stimulation of p105 proteolysis is triggered by IKK complex phosphorylation of the destruction box in the p105 PEST region (19, 28). Since overexpressed TPL-2 can activate the IKK complex (21), it is likely that its effect on p105 proteolysis is mediated via the stimulation of the endogenous IKK complex. However, many MAP 3-kinases activate the IKK complex when overexpressed (18); presumably, this is due to the similarity of the activating phosphoacceptor sites on its two component kinase subunits, IKK1 and IKK2, to those in MAP 2-kinases (8). Thus, induction of p105 proteolysis by TPL-2 may simply arise as an artifact of overexpression. More extensive analysis of TPL-2 knockout mice will be necessary to determine whether TPL-2 does in fact play a physiological role in regulating p105 proteolysis.
A particularly important question that arises from the present study is the mechanism by which LPS stimulation triggers phosphorylation of MEK1 and MEK2 in macrophages via TPL-2 (10). Since, essentially all of TPL-2 is complexed with p105 in these cells (data not shown), TPL-2 would be expected to be subject to p105 negative regulation. Presumably, LPS stimulation activates TPL-2 to induce MEK phosphorylation. This could occur as a consequence of LPS stimulation inducing proteolysis of p105 (9), thereby releasing TPL-2 from its inhibitor. However, preliminary kinetic experiments with bone marrow-derived macrophages have indicated that LPS induction of MEK phosphorylation precedes p105 proteolysis. Indeed, LPS stimulation actually induces proteolysis of TPL-2 itself by the proteasome, which occurs coincidentally with p105 proteolysis (data not shown). TPL-2 proteolysis after LPS stimulation presumably constititutes a downregulatory mechanism. An alternative possible mechanism to activate TPL-2 is that LPS may induce the disruption of one or both of the interactions between TPL-2 and p105, thus relieving p105 negative regulation. This could occur as a consequence of either TPL-2 or p105 phosphorylation. Interestingly, it has been reported that the p105 DD is phosphorylated by IKK in vitro (12), and it will be important to determine whether this modification alters the inhibition of TPL-2 activity by p105. The mechanism by which ligand stimulation regulates TPL-2 kinase activity is currently being investigated.
In conclusion, the present study has revealed a novel and unexpected link between NF-κB and the regulation of MEK activity. Thus, interaction with the DD of NF-κB1 p105 profoundly inhibits the ability of TPL-2 to phosphorylate MEK when the TPL-2 C terminus simultaneously binds to p105. NF-κB1 p105 is therefore a negative regulator of TPL-2 MEK kinase, in addition to its functions as a precursor for p50 and an IκB protein (18). C-terminal truncation renders TPL-2 insensitive to p105 negative regulation in vivo, thus providing a rationale for why this mutation is oncogenic (4).
Acknowledgments
S.B. and J.D. contributed equally to this study.
We thank David Baltimore, Anne O'Garra, Alexander Hoffmann, Alain Israel, Mary Holman, Chris Marshall, Antony Symons, and Phillip Tsichlis for reagents used in this study. We are also indebted to Hamish Allen for helpful comments, Matoula Papoutsopoulou for carrying out semiquantitative PCR assay of TPL-2 mRNA, Christopher Atkins for cell sorting, and other members of the Ley laboratory for support during the course of this study.
This work was supported by the UK Medical Research Council, a Boehringer Ingelheim Fonds fellowship to S.B., a European Union Marie Curie fellowship to J.D., and the Arthritis Research Campaign (project grant L0536 to V.L.).
REFERENCES
- 1.Aoki, M., F. Hamada, T. Sugimoto, S. Sumida, T. Akiyama, and K. Toyoshima. 1993. The human cot proto-oncogene encodes two protein serine/threonine kinases with different transforming activities by alternative initiation of translation. J. Biol. Chem. 268:22723-22732. [PubMed] [Google Scholar]
- 2.Beinke, S., M. P. Belich, and S. C. Ley. 2002. The death domain of NF-κB1 p105 is essential for signal-induced p105 proteolysis. J. Biol. Chem. 277:24162-24168. [DOI] [PubMed] [Google Scholar]
- 3.Belich, M. P., A. Salmeron, L. H. Johnston, and S. C. Ley. 1999. TPL-2 kinase regulates the proteolysis of the NF-κB inhibitory protein NF-κB1 p105. Nature 397:363-368. [DOI] [PubMed] [Google Scholar]
- 4.Ceci, J. D., C. P. Patriotis, C. Tsatsanis, A. M. Makris, R. Kovatch, D. A. Swing, N. A. Jenkins, P. N. Tsichlis, and N. G. Copeland. 1997. TPL-2 is an oncogenic kinase that is activated by carboxy-terminal truncation. Gene Dev. 11:688-700. [DOI] [PubMed] [Google Scholar]
- 5.Chan, A. M.-L., M. Chedid, E. S. McGovern, N. C. Popescu, T. Miki, and S. A. Aaronson. 1993. Expression cDNA cloning of a serine kinase transforming gene. Oncogene 8:1329-1333. [PubMed] [Google Scholar]
- 6.Chiariello, M., M. J. Marinissen, and J. S. Gutkind. 2000. Multiple mitogen-activated protein kinase signaling pathways connect the Cot oncoprotein to the c-Jun promoter and to cellular transformation. Mol. Cell. Biol. 20:1747-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cowley, S., H. Paterson, P. Kemp, and C. J. Marshall. 1994. Activation of MAP kinase kinase is necessary and sufficient for PC12 differentiation and for transformation of NIH 3T3 cells. Cell 77:841-852. [DOI] [PubMed] [Google Scholar]
- 8.Delhase, M., M. Hayakawa, Y. Chen, and M. Karin. 1999. Positive and negative regulation of IκB kinase activity through IKK-β subunit phosphorylation. Science 284:309-313. [DOI] [PubMed] [Google Scholar]
- 9.Donald, R., D. W. Ballard, and J. Hawiger. 1995. Proteolytic processing of NF-κB/IκB in human monocytes. J. Biol. Chem. 270:9-12. [DOI] [PubMed] [Google Scholar]
- 10.Dumitru, C. D., J. D. Ceci, C. Tsatsanis, D. Kontoyiannis, K. Stamatakis, J.-H. Lin, C. Patriotis, N. A. Jenkins, N. G. Copeland, G. Kollias, and P. N. Tsichlis. 2000. TNFα induction by LPS is regulated posttranscriptionally via a TPL2/ERK-dependent pathway. Cell 103:1071-1083. [DOI] [PubMed] [Google Scholar]
- 11.Gerondakis, S., N. Morrice, I. B. Richardson, R. Wettenhall, J. Fecondo, and R. J. Grumont. 1993. The activity of a 70-kDa IκB molecule identical to the carboxyl terminus of the p105 NF-κB precursor is modulated by protein kinase A. Cell Growth Differ. 4:617-627. [PubMed] [Google Scholar]
- 12.Heissmeyer, V., D. Krappmann, F. G. Wulczyn, and C. Scheidereit. 1999. NF-κB p105 is a target on IκB kinases and controls signal induction of BCL-3-p50 complexes. EMBO J. 18:4766-4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henkel, T., U. Zabel, K. van Zee, J. M. Muller, E. Fanning, and P. A. Baeuerle. 1992. Intramolecular masking of the nuclear location signal and dimerization domain in the precursor for the p50 NF-κB subunit. Cell 68:1121-1133. [DOI] [PubMed] [Google Scholar]
- 14.Huxford, T., D. B. Huang, S. Malek, and G. Ghosh. 1998. The crystal structure of the IκBα/NF-κB complex reveals mechanisms of NF-κB inactivation. Cell 95:759-770. [DOI] [PubMed] [Google Scholar]
- 15.Ishikawa, H., E. Claudio, D. Dambach, C. Raventos-Suarez, C. Ryan, and R. Bravo. 1998. Chronic inflammation and susceptibility to bacterial infections in mice lacking the polypeptide (p) 105 precursor (NF-κB1) but expressing p50. J. Exp. Med. 187:985-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacobs, M. D., and S. C. Harrison. 1998. Structure of an IκBα/NF-κB complex. Cell 95:749-758. [DOI] [PubMed] [Google Scholar]
- 17.Kabouridis, P. S., A. I. Magee, and S. C. Ley. 1997. S-acylation of LCK protein tyrosine kinase is essential for its signalling function in T lymphocytes. EMBO J. 16:4983-4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karin, M., and Y. Ben-Neriah. 2000. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 18:621-663. [DOI] [PubMed] [Google Scholar]
- 19.Lang, V., J. Janzen, G. Z. Fischer, Y. Soneji, S. Beinke, A. Salmeron, H. Allen, R. T. Hay, Y. Ben-Neriah, and S. C. Ley. 2003. βTrCP-mediated proteolysis of NF-κB1 p105 requires phosphorylation of p105 serines 927 and 932. Mol. Cell. Biol. 23:402-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leevers, S. J., H. F. Paterson, and C. J. Marshall. 1994. Requirement for Ras in Raf activation is overcome by targetting Raf to the plasma membrane. Nature 369:411-414. [DOI] [PubMed] [Google Scholar]
- 21.Lin, X., E. T. Cunningham, Y. Mu, R. Geleziunas, and W. C. Greene. 1999. The proto-oncogene Cot kinase participates in CD3/CD28 induction of NF-κB acting through the NF-κB-inducing kinase and IκB kinases. Immunity 10:271-280. [DOI] [PubMed] [Google Scholar]
- 22.Lund, A. H., G. Turner, A. Trubetskoy, E. Verhoeven, E. Wientjens, D. Hulsman, R. Russell, R. A. DePinho, J. Lenz, and M. van Lohuizen. 2002. Genome-wide retroviral insertional tagging of genes involved in cancer in Cdkn2a-deficient mice. Nat. Gene 32:160-165. [DOI] [PubMed] [Google Scholar]
- 23.Makris, A., C. Patriotis, S. E. Bear, and P. N. Tsichlis. 1993. Genomic organization and expression of Tpl-2 in normal cells and moloney murine leukemia virus-induced rat T-cell lymphomas: activation by provirus insertion. J. Virol. 67:4283-4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mikkers, H., J. Allen, P. Knipscheer, L. Romeyn, A. Hart, E. Vink, and A. Berns. 2002. High throughput retroviral tagging to identify components of specific signaling pathways in cancer. Nat. Gene 32:153-159. [DOI] [PubMed] [Google Scholar]
- 25.Patriotis, C., A. Makris, S. E. Bear, and P. N. Tsichlis. 1993. Tumor progression locus 2 (Tpl-2) encodes a protein kinase involved in the progression of rodent T-cell lymphomas and in T-cell activation. Proc. Natl. Acad. Sci. USA 90:2251-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patriotis, C., A. Makris, J. Chernoff, and P. N. Tsichlis. 1994. Tpl-2 acts in concert with Ras and Raf-1 to activate mitogen-activated protein kinase. Proc. Natl. Acad. Sci. USA 91:9755-9759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salmeron, A., T. B. Ahmad, G. W. Carlile, D. Pappin, R. P. Narsimhan, and S. C. Ley. 1996. Activation of MEK-1 and SEK-1 by Tpl-2 proto-oncoprotein, a novel MAP kinase kinase kinase. EMBO J. 15:817-826. [PMC free article] [PubMed] [Google Scholar]
- 28.Salmeron, A., J. Janzen, Y. Soneji, N. Bump, J. Kamens, H. Allen, and S. C. Ley. 2001. Direct phosphorylation of NF-κB p105 by the IκB kinase complex on serine 927 is essential for signal-induced p105 proteolysis. J. Biol. Chem. 276:22215-22222. [DOI] [PubMed] [Google Scholar]
- 29.Sha, W. C., H.-C. Liou, E. I. Tuomanen, and D. Baltimore. 1995. Targeted disruption of the p50 subunit of NF-κB leads to multifocal defects in immune responses. Cell 80:321-330. [DOI] [PubMed] [Google Scholar]
- 30.Siebenlist, U., G. Franzoso, and K. Brown. 1994. Structure, regulation and function of NF-κB. Annu. Rev. Cell Biol. 10:405-455. [DOI] [PubMed] [Google Scholar]
- 31.Szabo, A., L. Stolz, and R. Granzow. 1995. Surface plasmon resonance and its use in biomolecular interaction analysis (BIA). Curr. Opin. Struct. Biol. 5:699-705. [DOI] [PubMed] [Google Scholar]
- 32.Waterfield, M. R., M. Zhang, L. P. Norman, and S.-C. Sun. 2003. NF-κB1/p105 regulates lipopolysaccharide-stimulated MAP kinase signaling by governing the stability and function of the TPL-2 kinase. Mol. Cell 11:685-694. [DOI] [PubMed] [Google Scholar]
- 33.Yeung, K. T., P. Janosch, B. McFerran, D. W. Rose, H. Mischak, J. M. Sedivy, and W. Kolch. 2000. Mechanism of suppression of the Raf/MEK/extracellular signal-regulated kinase pathway by the Raf kinase inhibitor protein. Mol. Cell. Biol. 20:3079-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yeung, K. T., T. Seitz, S. Li, P. Janosch, B. McFerran, C. Kaiser, F. Fee, K. D. Katsanakis, D. W. Rose, H. Mischak, J. M. Sedivy, and W. Kolch. 1999. Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature 401:173-177. [DOI] [PubMed] [Google Scholar]