Abstract
Cellular metabolic needs are fulfilled by transport of amino acids across the plasma membrane by means of specialized transporter proteins. Although many of the classical amino acid transporters have been characterized functionally, less than half of these proteins have been cloned. In this report, we identify and characterize a cDNA encoding a plasma membrane amino acid transporter. The deduced amino acid sequence is 505 residues and is highly hydrophobic with the likely predicted structure of 9 transmembrane domains, which putatively place the amino terminus in the cytoplasm and the carboxy terminus on the cell surface. Expression of the cRNA in Xenopus laevis oocytes revealed strong transport activities specific for histidine and glutamine. This protein is a Na+- and pH-dependent transporter and tolerates substitution of Na+ by Li+. Furthermore, this transporter is not an obligatory exchanger because efflux occurs in the absence of influx. This transporter is expressed predominantly in the liver, although it is also present in the kidney, brain, and heart. In the liver, it is located in the plasma membrane of hepatocytes, and the strongest expression was detected in those adjacent to the central vein, gradually decreasing towards the portal tract. Because this protein displays functional similarities to the N-system amino acid transport, we have termed it mNAT, for murine N-system amino acid transporter. This is the first transporter gene identified within the N-system, one of the major amino acid transport systems in the body. The expression pattern displayed by mNAT suggests a potential role in hepatocyte physiology.
Amino acid transport across the plasma membrane introduces essential nutrients into cells, providing critical substrates for protein biosynthesis, hormone metabolism, regulation of cell growth, and production of metabolic energy, to name a few (1). Defective amino acid transport systems and the candidate transporter genes are known to be involved in several diseases (2). For example, the dicarboxylic amino aciduria developed by the null knockout EAAT3 mice revealed the putative role of an anionic amino acid transporter in this inherited disease (3). The pioneer work on amino acid transport was documented approximately three decades ago; however, more recently based on substrate specificity and sodium dependence, a classification of several systems was reached (4). In 1990, the first amino acid transporter gene for GABA was identified (5) and served as the starting point for the cloning of other mammalian amino acid transporter genes. The genes identified so far correspond to eight classical transport systems; whereas the genes encoding the remaining eight of the major amino acid transport systems are unknown (6).
Although the transporters identified thus far have multiple transmembrane domains and display some degree of substrate overlap, they share little sequence homology. The lack of high-affinity inhibitors, resistance to expressional cloning especially for nonneural amino acid transporters, as well as the fact that the transporters are expressed in low abundance in the plasma membrane complicates their structural identification and isolation. Thus, progress towards the molecular identification of amino acid transporters across the various systems has proceeded very slowly. In this report, we have identified a plasma membrane amino acid transporter gene with nine predicted transmembrane domains. This protein has no sequence homology with other mammalian amino acid transporter families expressed in the plasma membrane, but shares less than 20% homology with the vesicular γ-aminobutyric acid (GABA) transporter (6, 7). This protein preferentially transports histidine and glutamine, and possesses other functional features, which are similar to N-system amino acid transport. Thus we propose to name it mNAT (for N-system amino acid transporter). N-system amino acid transport is known to have an important role in glutamine uptake for the urea cycle (8) and export of newly synthesized glutamine for glutamine metabolism in the liver (9, 10). These transporting activities could be efficiently blocked by histidine (11). In addition, N-system transport is identified in the brain, where it appears to have a functional role in the blood–brain barrier (12, 13). Interestingly, mNAT is also abundantly expressed in liver and brain. In the liver, it is localized in the plasma membrane of hepatocytes surrounding the central vein and the levels of expression gradually decrease towards portal tract, suggesting a potential physiological function of this transporter in this organ.
Materials and Methods
Isolation of mNAT cDNA Clones.
Originally, we attempted to use a mouse genomic library to screen for unknown gap junction proteins, connexins (Cx). The mouse genomic library was constructed by using the GenomeWalker Kit (CLONTECH; K1805-1). The genomic DNA was isolated from a mouse cell line, MC3T3-E1. Two degenerate primers were designed based on the known connexin coding sequences (Cx43, Cx45, and Cx46), sense primer: GGT/C TGT/C, A/GAC/A/G AAC/T GTC/T TGC/T TAC/T GAC/T; antisense primer: CTC C/AGT C/GGG G/A/CCTG T/GGA A/G/CAG/T/C GT/AA C/G/ACA. After PCR amplification of over 30 cycles with annealing temperatures of 40°C, we obtained several fragments. In addition to fragments encoding connexins, a fragment of 0.6 kb was obtained. To determine the full-length cDNA of this gene, a preliminary Northern blot analysis was performed using this fragment as a probe: a 2.4-kb transcript in mouse kidney, liver, brain, and heart. To obtain the cDNA sequence, polyadenylated RNA was isolated from mouse kidney using a mRNA Separator Kit (CLONTECH). mRNA was converted to double-stranded cDNA using the Two-Hybrid cDNA Library Construction Kit (CLONTECH). The cDNA fraction between 2.0 and 2.5 kb was isolated using a 1.5% agarose gel, purified with a Gel Extraction kit (Qiagen, Santa Clarita, CA) and then subcloned into vector pGAD10 (CLONTECH). PCR-based screening was performed using the primer pairs: sense 5′-AATTCGCGGCCGCGTCGAC (linker primer), and antisense 5′-GCTGTAAGCAGGAACAGGAAAAGGATGAT (corresponding to nucleotides 426–398 of mNAT). To obtain the full-length transcript, 5′-rapid amplification of cDNA ends (RACE) was performed with the Marathon cDNA amplification Kit (CLONTECH) according to the instructions from the manufacturer. Primers 5′-GCTAGGGGCAGAATGATGGTGACAG (nucleotides 725–702) and 5′-CATGATGGCGTTGCTGAGAT (nucleotides 352–333) were used as gene-specific primers for the first and nested PCR of 5′-RACE, respectively. Each DNA fragment was sequenced automatically on both strands with specific oligonucleotide primers (sequenced in the UTHSCSA DNA Sequencing Facility). To ensure that the sequence of mNAT obtained from kidney was exactly the same as that expressed in other tissues, reverse transcription (RT)-PCR and sequencing were performed using RNA isolated from brain and liver (1 μg) as previous described (14).
Northern Blot Analysis.
Northern blots were performed as described (14). Briefly, total RNA was isolated from mouse tissues (liver, brain, kidney, lung, heart, small intestine, muscle, and testis) using TRI REAGENT (Molecular Research Center, Cincinnati). Equal amounts of RNA were loaded and separated on a 1% agarose gel in the presence of formaldehyde, and transferred to a nylon membrane (Oncogene). The membrane was hybridized at 45°C in 50% formamide with a [32P]dCTP-labeled 602 bp of PCR fragment corresponding to nucleotides 1,135–1,737 of mNAT. The probed membrane was washed at 65°C in 0.1× SSC and 0.1% SDS.
Expression of mNAT and Human g17 in Xenopus laevis Oocytes.
To prepare mRNA for oocyte injection, cDNAs were synthesized by PCR and subcloned between the 5′- and 3′-flanking sequences of Xenopus β globin gene previously described (15). The primer pairs used to amplify the full ORF from mNAT and human g17 were designed with a restriction site BamHI at 5′ end and HindIII at 3′ end. Primer pairs for mNAT consist of a sense 5′-AATGGATCCATGGAGATACCCCGACAGACA and an antisense primer 5′-ATCGAAGCTTAGGTCATCCTAATGGTTTC. Primer pairs for human g17 included a sense 5′-AATGGATCCATGGAGGCGCCTTTGCAGACA and an antisense primer 5′-ATCGAAGCTTAGGTCATCCTAGTGGTTTC. PCR products were gel-purified with the Gel Extraction kit (Qiagen) and digested with BamHI and HindIII, before subcloning into the Xenopus expression vector. The constructs were verified by sequencing.
The plasmids were linearized with NotI, and in vitro transcription reactions by T7 RNA polymerase were performed with mMESSAGE mMACHINE (Ambion, Austin, TX). Capped cRNAs were extracted with phenol/CHCl3, precipitated with ethanol as described previously (15), resuspended in DEPC-H2O at a concentration of 1.5–2.0 μg/μl and stored in aliquots of 3 μl at −80°C before use.
Antibody Preparation.
Rabbit anti-mNAT IgG antibody was produced using a glutathione S-transferase (GST) tagged fusion protein as described previously (14). A DNA fragment encoding amino acids 1–79 was produced by PCR using a full-length mNAT as a template (sense primer: 5′-AATGGATCCATGGAGATACCCCGACAGACA, anti-sense primer: 5′-AGGAATTCATGATGGCGTTGCTGAGAT) and inserted into the expression pGEX-2T vector (21). The recombinant fusion protein was expressed in Escherichia coli induced by isopropylthio-β-d-galactoside and isolated with GST-beads (Sigma). The purified protein was used to raise a rabbit polyclonal antiserum (Pocono Rabbit Farm, Canadensis, PA). The antisera generated were affinity-purified by passage through two Sepharose CL-4B columns, GST-conjugated and GST-mNAT fusion protein conjugated, respectively.
Membrane Protein Preparation, Western Blot, Glycosylation, and Phosphorylation Analyses.
Crude membrane extracts including plasma and intracellular membrane proteins were prepared from Xenopus oocytes and mouse tissues as previous described (14, 16). Briefly, tissues and cells were homogenized in lysis-buffer (Tris 20 mM, NaCl 100 mM, PMSF 20 μM, and leupeptin 10 μg/ml). The homogenate was centrifuged at 10,000 × g at 4°C for 10 min to discard yolk and cellular debris. The supernatant was centrifuged at 100,000 × g at 4°C for 30 min. The membrane pellet was resuspended in lysis buffer containing 1% SDS and 1% β-mercaptoethanol. The protein concentration was determined using a microBCA assay (Pierce). One microgram of protein was loaded on a 10% SDS/PAGE and transferred to a nitrocellulose membrane. The membrane was probed with a 1:500 dilution of affinity-purified preimmune and anti-mNAT IgG. The primary antibody was detected using peroxidase-conjugated secondary anti-rabbit antiserum and followed by chemiluminlunance reagent kit (ECL) (Amersham Pharmacia) according to the manufacturer's instruction. The membranes were exposed to X-Omat AR films (Eastman Kodak, Rochester, NY) and detected by fluorography. To analyze the specificity of mNAT antibody toward the mNAT antigen, the mNAT fusion protein (0.2 μg/ml, 2 μg/ml, and 20 μg/ml) was preincubated with mNAT antibody (1:400) or Cx43 antibody (1:400), and Western blots were performed with the preabsorbed antibody.
The deglycosylation experiments were performed using a Glycoprotein Deglycosylation Kit (Calbiochem) strictly following the manufacturer's instructions. The deglycosylation enzymes contained in the kit include N-glycosidase F, Endo-α-N-acetylgalactosaminidase, α2,3,6,8-neuraminidase, β1,4-galactosidase and β-N-acetylglucosaminidase. The dephosphorylation assay was performed as previously described (14).
Transport Assays.
Stage V-VI oocytes from Xenopus laevis were dissected and injected with 50 nl of the synthetic mRNA or DEPC-H2O as control. After 3 days of incubation at 18°C, functional analyses were performed in groups of 5–10 oocytes per assay as described previously (17–19). Oocytes were rinsed briefly in uptake buffer (KCl, 2 mM; MgCl2, 1 mM; CaCl2, 1 mM; Hepes, 10 mM; and Tris, 50 mM) in the presence of 100 mM NaCl (Na+-buffer), 100 mM choline chloride (choline+-buffer), or 100 mM LiCl (Li+-buffer). These oocytes were transferred into a 24-well culture dish containing 2 ml uptake buffer and were incubated for 2–60 min at room temperature. Amino acid transport activities were measured by incubating oocytes in 0.5 ml of uptake buffer in the presence of 50 μM l-amino acids plus corresponding [3H]-labeled l-amino acids (New Life Science, Boston, MA) as tracers for 15 or 30 min. The oocytes were washed four times in uptake buffer. These oocytes were lysed in 100 μl of 2% SDS, and the radioactivity accumulated by each oocyte was measured with a scintillation counter (Beckman Coulter) in 5 ml of scintillation solution. The specificity of mNAT-mediated amino acid uptake was examined using an amino acid competition assay. l-histidine uptake (50 μM) was measured in the presence of 5 mM nonradioactive amino acid. The Na+-dependence and Li+-tolerance of histidine transport by mNAT were investigated using choline+-buffer or Li+-buffer instead of Na+-buffer. The effects of extracellular pH on l-histidine and l-glutamine uptake mediated by mNAT was investigated at pH 6–8.5 by adjusting Na+-buffer with Tris-base or hydrochloric acid as described (18, 19). l-histidine efflux was measured as previously described (20). Briefly, injected oocytes were incubated for 30 min in Na+-buffer containing 50 μM [3H] l-histidine. After washing in Na+-buffer containing 50 μM nonradioactive l-histidine, oocytes were transferred to 300 μl Na+-buffer or Na+-buffer containing 1 mM nonradioactive l-histidine. Samples (2 μl) were taken at the designated times to determine the levels of radioactivity. All experiments were repeated at least three times, and the data collected were presented as SEMs.
Immunofluorescence and Confocal Laser Microscopy.
The immunofluorescence detection of mNAT was performed as described previously (22). Briefly, oocytes and tissue samples were fixed in 2% paraformaldehyde for 2 h at 4°C, washed three times with PBS, incubated in 30% sucrose in PBS overnight at 4°C, embedded in OCT (Miles Scientific), and then frozen in liquid nitrogen. Frozen tissue sections (5–10 μm) were incubated with blocking solution (2% normal goat serum, 2% fish skin gelatin, and 1% BSA in PBS) for 30 min and then with affinity-purified anti-mNAT (1:500 dilution in blocking solution) at room temperature for 2 h. The primary antibodies were detected by fluorescein (FITC)-conjugated goat anti-rabbit Ig for 2 h at room temperature. The specimens were analyzed with either an upright fluorescence microscope (Olympus Optical, Tokyo, Japan) or a confocal laser scanning microscopy (Model: Fluoview, Olympus). FITC fluorescence was excited by a 488-nm argon laser light.
Results and Discussion
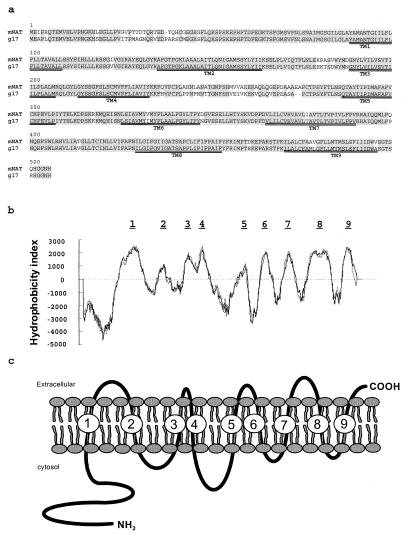
Screening a mouse genomic DNA library resulted in the isolation of a 0.6-kb DNA fragment, of which, 110 bp shared partial homology with members of transporter families present in databases [blast, National Center for Biotechnology Information (NCBI) file servers] (23), which include human transporter protein (g17) mRNA (U49082), amino acid transporter in arabidopsis (U397820), amino acid permease 1 in Nicotiana sylvestris (U31932), and transposase in Pseudomonas stutzeri (AF039534). Northern blot analysis using this fragment as a probe revealed a ≈2.4-kb transcript in mouse kidney. A PCR-based cloning strategy using a mouse kidney cDNA library yielded a 2,412-bp cDNA encoding a 505-amino acid protein with a predicted molecular mass of 56 kDa. The ORF is flanked by 115 nucleotides of 5′-untranslated region (UTR), 842 nucleotides of 3′-UTR, a long poly(A) tail, and a variant polyadenylation site (TATAAA) between nucleotides 2,394 and 2,400. Comparison of this sequence at the nucleotide and amino acid levels with known databases (blastn, blastp, and blastx, NCBI file servers) led to the identification of a single sequence, which was 89% homologous at the amino acid and 84% homologous at the nucleotide coding region with human g17 (a putative transporter, GDB g:1840044). Although this protein has no sequence homology with any identified mammalian plasma membrane amino acid transporters, homologies of less than 25% were found with 51 other transmembrane proteins. Twenty-one of them are related to amino acid transporters from plants, yeast, and worms, and two are vesicular GABA transporters from rat (AF030253) and mouse (AJ001598). The alignment of amino acid sequences of mNAT and human g17 is shown in Fig. 1a, in which most of the predicted transmembrane domains are conserved (TM1–TM9). Hydrophobicity studies predict that mNAT is a plasma membrane protein (certainty = 0.60, psort version 6, Nakai file server) with a higher probability of having 9 transmembrane domains (Swiss, embnet; psort and bcm Search Launcher) as opposed to having 10 (Swiss, embnet). The N-terminal segment is predicted to be in the cytoplasm and the C-terminal tail facing the extracellular side (higher probability score, psort version 6, Nakai file server) (Fig. 1 b and c). The predicted molecular structure of mNAT with 9 transmembrane domains is different from other known amino acid transporters, which are composed of 10–14 transmembrane domains (6, 24–26). Together, these results suggest that this protein belongs to a new family of amino acid transporters in mammalian cells.
Figure 1.
Amino acid sequence and molecular structure of mNAT. (a) The deduced amino acid sequence of mNAT in one-letter code is aligned to the human g17 using the blast 2 software (33). Identical residues are shaded in gray. Putative membrane-spanning domains are numbered and doubly underlined. (b) Transmembrane topology of mNAT as predicted from hydrophobicity plots determined with programs obtained from embnet, psort, and bcm Search Launcher servers. (c) Hypothetical membrane topology model of mNAT is based on its hydropathy profile.
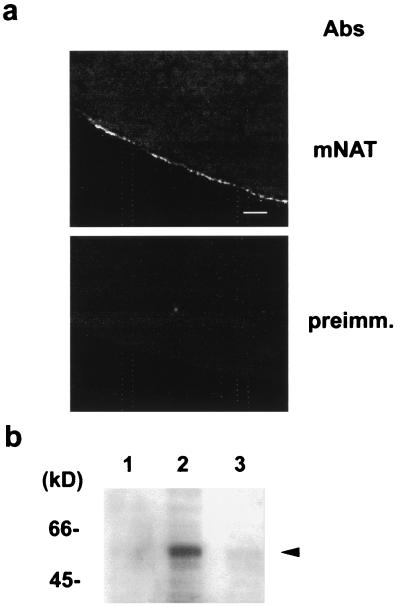
To examine whether mNAT is a novel amino acid transporter, the expression and function of mNAT was primarily investigated in Xenopus oocytes (17, 18). Expression of mNAT was detected by Western blot analysis and confocal immunofluorescence as early as 24 h following cRNA injection. mNAT expression was localized to the oocyte plasma membrane (at the limit of resolution imparted by fluorescence microscopy) as detected by an mNAT-specific antibody, whereas fluorescence levels were undetectable with preimmune serum (Fig. 2a). A protein band, ≈54 kDa was detected by Western blotting with a mNAT specific antibody, which was undetectable in control oocytes (Fig. 2b). Differences in the pattern of migration of mNAT were not altered by deglycosylation or dephosphorylation treatments (data not shown), indicating that the protein might not be glycosylated nor may support phosphorylation.
Figure 2.
Expression of mNAT in Xenopus oocytes. (a) Frozen sections of Xenopus laevis oocytes 3 days after injection with cRNA of mNAT were labeled with anti-mNAT antibody or preimmune serum and were examined by confocal microscopy at a scanning interval section of 0.5 μm. (Bar, 30 μm.) (b) Western blot analysis demonstrates overexpression of mNAT (arrrowhead) using a specific antibody in extracts from oocytes injected with water (lane 1), mNAT cRNA (lane 2), or uninjected oocytes (lane 3).
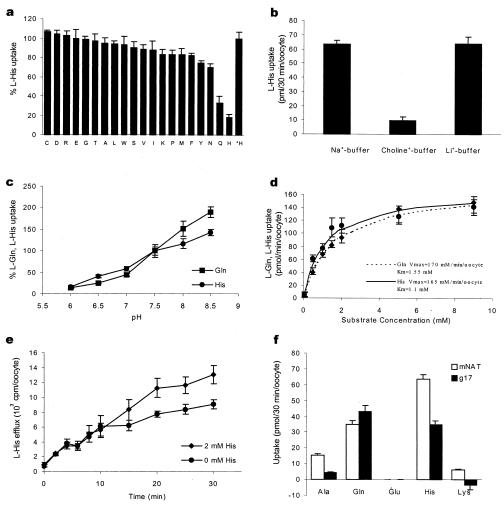
The uptake of [3H]-labeled l-alanine (zwitterionic), l-glutamate (anionic), and l-lysine (cationic) was measured in Xenopus oocytes injected with mNAT cRNA. Compared with controls, we found that mNAT expression induced a 10-fold increase in the uptake of l-alanine (17.2 ± 3.2 vs. 1.9 ± 0.48, measured in pmol/30 min per oocyte), 1.1-fold for lysine (69.9 ± 4.7 vs. 63.8 ± 3.0), whereas there was no significant transport of glutamate (0.55 ± 0.37 vs. 0.32 ± 0.30). These results suggest that mNAT is capable of transporting l-alanine across the plasma membrane. To determine the substrate selectivity of mNAT, a competition assay in the presence of all 20 amino acids was performed. In this assay, the rate of uptake of l-alanine was determined in oocytes injected with mNAT cRNA in the presence of nonradioactive-labeled amino acids. The uptake was significantly inhibited up to an efficiency of 90% by l-histidine. l-asparagine and l-glutamine showed a modest degree of inhibition of l-alanine uptake (data not shown). Therefore, l-histidine was selected as a model for amino acid competition assay to examine the transport specificity of mNAT (Fig. 3a). l-histidine uptake was significantly inhibited by l-histidine and l-glutamine, suggesting that mNAT is an amino acid transporter with greatest specificity for these two amino acids. To distinguish between the known classical amino acid transport systems, we examined the Na+-dependence and tolerance for substitution by Li+. In the presence of Na+ buffer, l-histidine uptake into oocytes injected with mNAT cRNA was significantly greater (23.8-fold higher) than that exhibited by control oocytes injected with water. Choline ion substitution for Na+ led to a significant decrease in l-histidine uptake, (6.8-fold lower) when compared with that in Na+-buffer. Replacement of Li+ for Na+ did not affect the rate of uptake of l-histidine (Fig 3b). These results demonstrate that l-histidine uptake mediated by mNAT is Na+-dependent and tolerant of replacement of Na+ by Li+. Together, these features are consistent with an N-system amino acid transporter as previously described (17, 27).
Figure 3.
Functional characterization of mNAT expressed in Xenopus laevis oocytes. (a) Inhibition of 50 μM l-histidine uptake in oocytes injected with cRNA of mNAT by 5 mM of 20 types of l-amino acids. The percentage values after subtracting controls injected with water are presented as 50 μM l-histidine uptake is defined as 100%. *Bar represents l-histidine uptake in the absence of nonlabeled amino acids. (b) Uptake of 50 μM [3H]histidine was measured using choline+-buffer or Li+-buffer to replace Na+-buffer. (c) pH dependence of l-histidine and l-glutamine uptake. Uptake of 50 μM [3H] l-histidine or l-glutamine was measured with Na+-buffer under indicated pH. (d) Concentration dependence of l-histidine uptake by mNAT. Net uptakes of l-histidine and glutamine were measured at different concentrations (0–10 mM) of l-histidine in Na+-buffer at pH 7.5. Km = 1.1 mM and Vmax = 170 pmol/min/oocyte for histidine, and Km = 1. 55 mM and Vmax= 165 pmol/min/oocyte for glutamine were determined. (e) l-histidine efflux was examined in Na+-buffer and in Na+-buffer containing 2 mM nonradioactive labeled-l-histidine. (f) Uptake rates for l-alanine, l-glutamine, l-glutamate, l-histidine, and l-lysine were determined for oocytes expressing mNAT and human g17.
Since the N-system of amino acid transporters are known to have a pH dependence (19), we tested whether extracellular pH in the range between 6 and 8.5 had an effect on l-histidine and l-glutamine uptake mediated by mNAT. The results revealed that uptake of these two amino acids exhibited a similar pH-dependency increasing from low to high pH (Fig 3c), confirming that mNAT is a member of the N-system amino acid transporter family. Furthermore, we investigated the concentration dependence of l-histidine and l-glutamine uptake mediated by mNAT. At concentrations between 0 and 1 mM, the rate of uptake was an incremental function of concentration, whereas at higher amino acid concentrations (5–10 mM), uptake was saturated (Fig 3d). This observation indicated that l-histidine and l-glutamine uptake in oocytes expressing mNAT exhibited the characteristics of a carrier-mediated transport. Furthermore, to determine whether the substrate affects efflux of l-histidine, efflux of [3H] l-histidine was measured in mNAT overexpressing oocytes, which were prelabeled with [3H] l-histidine and incubated in Na+-buffer in the presence or absence of nonradioactive l-histidine. Interestingly, in the presence of external l-histidine, oocytes expressing mNAT showed significantly increased efflux of [3H] l-histidine at later time points, as compared with control oocytes incubated in the absence of external l-histidine (Fig 3e). The explanation of this phenomenon could be that the uptake of unlabeled histidine prevents depletion of intracellular substrate stores, prolonging the activity of the transporter.
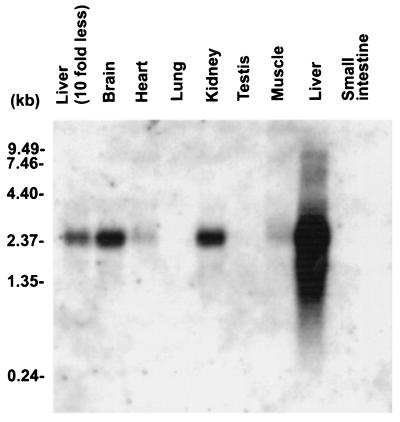
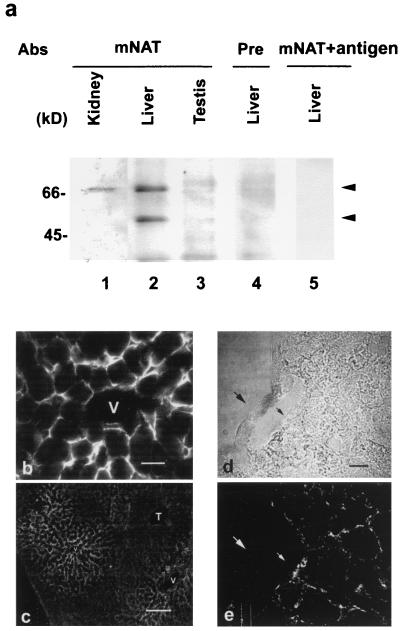
To elucidate the tissue-specific expression of mNAT, high-stringency Northern blots of equivalent amounts of RNA isolated from various tissues were probed with a mNAT DNA fragment labeled with [32P]dCTP. A full-length transcript of 2.4 kb was detected in abundance in mouse liver, moderately detected in kidney and brain, and barely detected in heart and muscle (Fig. 4). The transcript from liver was observed even when a 10-fold lower amount of RNA was loaded (see Fig. 4). This finding is consistent with the restrictive localization of the N-system amino acid transporters to the liver (6). Western blot analysis with an mNAT-specific antibody on crude membrane fractions isolated from liver, kidney, and testis revealed two immunoreactive protein bands of Mr 67 and 54 kDaa in the liver (Fig 5a, lane 2). The 67 kDa band, which is different from the predicted molecular mass (56 kDa) of mNAT was also detected in kidney (lane 1). There was no detectable mNAT expression in testis (lane 3). The specificity of the two protein bands was ascertained by the lack of detected immunoreactivity by probing with preimmune serum (lane 4) or by preincubating the mNAT antibody with the mNAT antigen (lane 5). Furthermore, an antibody for the nonrelated protein, connexin 43, was not competed off by the mNAT antigen (data not shown). The differences between the expected and observed protein bands could be caused by posttranslational modifications such as glycosylation or phosphorylation. However, deglycosylation or dephosphorylation treatment did not alter the mNAT migrating patterns (data not shown), suggesting that it is unlikely that both types of modifications can account for these differences. The discrepancy between predicted and measured molecular weight on SDS/PAGE and between the different migration patterns of mNAT expressed in the oocyte, and in liver/kidney, could be caused by the structural complexity of membrane proteins. The migration on SDS/PAGE is known to differ from predicted levels in other proteins with multiple transmembrane segments, such as gap junction proteins and connexin 56 (14, 28). The localization of mNAT appeared to be basolateral and not canalicular in the plasma membrane of hepatocytes surrounding the central vein (Fig. 5b). There is a graded distribution of mNAT expression from central vein to portal tract (Fig 5c). There was no detectable expression of mNAT in endothelial cells as revealed by confocal microscopy (Fig. 5 d and e). This specific distribution of mNAT in hepatocytes is consistent with a function of this protein in amino acid transport (10).
Figure 4.
Northern blot analyses of tissue-specific expression patterns of mNAT. Northern blots with equal amount of total RNAs from different mouse tissues were hybridized with a DNA probe of mNAT under high-stringency conditions. The position of known size standards is shown on the left.
Figure 5.
Expression of mNAT in vivo. Immunoblot detection of mNAT expression in mouse liver, kidney, and testis (a). Crude membranes isolated from kidney (lane 1), liver (lanes 2, 4, and 5), and testis (lane 3) were treated with affinity-purified antibodies (1:400); anti-mNAT (lanes 1–3), preimmune (lane 4), and anti-mNAT pretreated with mNAT antigen (0.2 μg/ml) (lane 5). Immunofluorescence detection localized mNAT to the plasma membrane of hepatocytes that surround the central vein (V) (b). The expression diminishes gradually toward portal tract (T) (c). The confocal laser fluorescence of 0.5-μm interval section (d, phase; e, fluorescence) showed the expression of mNAT on hepatocyte membrane, but not in endothelial cells (arrow). [(b) Bar, 20 μm; (c) Bar, 200 μm; (d and e), Bar, 10 μm.]
Because human g17 shares more than 80% sequence homology with mNAT, the amino acid transporting activities of this gene were similarly analyzed and compared with those of mNAT. Oocytes were injected with g17 or mNAT cRNAs, and, after 3 days, uptake rates of l-alanine, l-glutamine, l-glutamate, l-histidine, and l-lysine were measured. Comparison between the two groups demonstrated similar transport patterns (Fig 3f). Together, the results suggest that g17 is an N-system amino acid transporter and, in fact, may be the human orthologue of mNAT.
In this report, we have identified mNAT as an N-system transporter expressed predominantly in the liver. The liver is known to contain four systems of transport for neutral amino acids, A, ASC, N, and L (1, 6). Of these four, only the proteins responsible for the ASC-transport system have been cloned and further characterized as being pH-insensitive (29). The l-system mainly transports bulky side chain-neutral amino acids and can be trans-stimulated by substrate. However, this system is known to be Na+-independent (4). Thus, the features of the above two systems distinguish them from mNAT. System A functions as a Na+-dependent transporter specific for small and N-methyl group amino acids (30, 31). Although the A-system shares similarities with mNAT in substrate selectivity, i.e., alanine, the even stronger transport efficiency of mNAT for histidine and glutamine matches the amino acid selectivity of the N-system. In addition, in contrast to the A-system which shows trans-inhibition, mNAT can be trans-stimulated by substrate (32). The Km value of mNAT for l-glutamine (1.55 mM) determined from the oocyte transport assay is similar to the value of 1.1 mM obtained for the N-system by Kilberg et al. (27), despite the fact that, in their study, the Km was derived from hepatocytes. The selectivity of the mNAT amino acid transporter and its differential expression in liver indicates that this molecule may be involved in urea and glutamine metabolism, a critical function of hepatocytes, especially in physiology and pathophysiology in which the gluconeogenic process is prominent. Taken together, our data suggest that mNAT is an active N-system amino acid transporter in the liver.
Acknowledgments
We thank Michael Lerman at National Cancer Institute-Frederick Cancer Research & Development Center for kindly providing cDNA clone of human g17. We thank D. Adan-Rice, L. M. John, L. Wang, and V. Frolick for technical assistance and M.S. Olson for critical reading of the manuscript. The work is supported by National Institutes of Health grants (to J.X.J. and P.C.).
Abbreviations
- mNAT
murine N-system amino acid transporter
- GABA
γ-aminobutyric acid
- RACE
rapid amplification of cDNA ends
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF159856).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.050318197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.050318197
References
- 1.Castagna M, Shayakul C, Trotti D, Sacchi V F, Harvey W R, Hediger M A. J Exp Biol. 1997;200:269–286. doi: 10.1242/jeb.200.2.269. [DOI] [PubMed] [Google Scholar]
- 2.Scriver C H, Beaudet A L, Sly W S, Valle D. The Metabolic and Molecular Bases of Inherited Diseases. New York: McGraw–Hill; 1995. [Google Scholar]
- 3.Peghini P, Janzen J, Stoffel W. EMBO J. 1997;16:3822–3832. doi: 10.1093/emboj/16.13.3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christensen H N. Physiol Rev. 1990;70:43–77. doi: 10.1152/physrev.1990.70.1.43. [DOI] [PubMed] [Google Scholar]
- 5.Guastella J N, Nelson H, Nelson L, Nelson L, Czyzyk S, Keynan M C, Miedel N, Davidson H A, Lester H A, Kanner B I. Science. 1990;249:1303–1306. doi: 10.1126/science.1975955. [DOI] [PubMed] [Google Scholar]
- 6.Palacin M, Estevez R, Bertran J, Zorzano A. Physiol Rev. 1998;78:969–1054. doi: 10.1152/physrev.1998.78.4.969. [DOI] [PubMed] [Google Scholar]
- 7.McIntire S, Reimer R J, Schuske K, Edwards R H, Jorgensen E M. Nature (London) 1997;389:870–876. doi: 10.1038/39908. [DOI] [PubMed] [Google Scholar]
- 8.Krebs H A. Biochem J. 1935;29:1951–1959. doi: 10.1042/bj0291951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haussinger D. Biochem J. 1990;267:281–290. doi: 10.1042/bj2670281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burger H-J, Gebhardt R, Mayer C, Mecke D. Hepatology. 1989;9:22–28. doi: 10.1002/hep.1840090105. [DOI] [PubMed] [Google Scholar]
- 11.Haussinger D, Soboll S, Meijer A J, Gerok W, Tager J M, Sies H. J Biochem. 1985;152:597–603. doi: 10.1111/j.1432-1033.1985.tb09237.x. [DOI] [PubMed] [Google Scholar]
- 12.Keep R F, Xiang J. J Neurochem. 1995;65:2571–2576. doi: 10.1046/j.1471-4159.1995.65062571.x. [DOI] [PubMed] [Google Scholar]
- 13.Ennis S R, Kawai N, Ren X-d, Abdelkarim G E, Keep R F. J Neurochem. 1998;71:2565–2573. doi: 10.1046/j.1471-4159.1998.71062565.x. [DOI] [PubMed] [Google Scholar]
- 14.Jiang J X, White T W, Goodenough D A, Paul D L. Mol Biol Cell. 1994;5:363–373. doi: 10.1091/mbc.5.3.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.John L M, Lechleiter J D, Camacho P. J Cell Biol. 1998;142:963–973. doi: 10.1083/jcb.142.4.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Camacho P, Lechleiter J D. Cell. 1995;82:765–771. doi: 10.1016/0092-8674(95)90473-5. [DOI] [PubMed] [Google Scholar]
- 17.Taylor P M, Hundal H S, Rennie M J. J Membr Biol. 1989;112:149–157. doi: 10.1007/BF01871276. [DOI] [PubMed] [Google Scholar]
- 18.Kim J W, Closs E I, Albritton L M, Cunningham J M. Nature (London) 1991;352:725–728. doi: 10.1038/352725a0. [DOI] [PubMed] [Google Scholar]
- 19.Taylor P M, Mackenzie B, Low S Y, Rennie M J. J Biol Chem. 1992;267:3873–3877. [PubMed] [Google Scholar]
- 20.Mastroberardino L, Spindler B, Pfeiffer R, Skelly P J, Loffing J, Shoemaker C B, Verrey F. Nature (London) 1998;395:288–291. doi: 10.1038/26246. [DOI] [PubMed] [Google Scholar]
- 21.Smith D B, Johnson K S. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 22.Jiang J X, White T W, Goodenough D A. Dev Biol. 1995;168:649–661. doi: 10.1006/dbio.1995.1109. [DOI] [PubMed] [Google Scholar]
- 23.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D L. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Closs E I, Albritton L M, Kim J W, Cunningham J M. J Biol Chem. 1993;268:7538–7544. [PubMed] [Google Scholar]
- 25.Olivares L, Aragon C, Gimenez C, Zafra F. J Biol Chem. 1997;272:1211–1217. doi: 10.1074/jbc.272.2.1211. [DOI] [PubMed] [Google Scholar]
- 26.Wahle S, Stoffel W. J Cell Biol. 1996;135:1867–1877. doi: 10.1083/jcb.135.6.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kilberg M S, Handlogten M E, Christensen H N. J Biol Chem. 1980;255:4011–4019. [PubMed] [Google Scholar]
- 28.Ebihara L, Xu X, Oberti C, Beyer E C, Berthoud V M. Biophys J. 1999;76:198–206. doi: 10.1016/S0006-3495(99)77189-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Utsunomiya-Tate N, Endou H, Kanai Y. J Biol Chem. 1996;271:14883–14890. doi: 10.1074/jbc.271.25.14883. [DOI] [PubMed] [Google Scholar]
- 30.Christensen H N, Liang M, Archer E G. J Biol Chem. 1967;242:5237–5242. [PubMed] [Google Scholar]
- 31.Christensen H N, Oxender D L, Liang M, Vatz K A. J Biol Chem. 1965;240:3609–3616. [PubMed] [Google Scholar]
- 32.Guidotti G G, Gazzola G C. In: Mammalian Amino Acid Transport, Mechanism and Control. Kilberg M S, Haussinger D, editors. New York: Plenum; 1992. pp. 3–30. [Google Scholar]
- 33.Tatusova T A, Madden T L. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]