Abstract
Stage-specific transcriptional programs are an integral feature of cell cycle regulation. In the budding yeast Saccharomyces cerevisiae, over 120 genes are coordinately induced in late G1 phase by two heterodimeric transcription factors called SBF and MBF. Activation of SBF and MBF is an upstream initiator of key cell cycle events, including budding and DNA replication. SBF and MBF regulation is complex and genetically redundant, and the precise mechanism of G1 transcriptional activation is unclear. Assays using SBF- and MBF-specific reporter genes revealed that the STB1 gene specifically affected MBF-dependent transcription. STB1 encodes a known Swi6-binding protein, but an MBF-specific function had not been previously suspected. Consistent with a specific role in regulating MBF, a STB1 deletion strain requires SBF for viability and microarray studies show a decrease in MBF-regulated transcripts in a swi4Δ mutant following depletion of Stb1. Chromatin immunoprecipitation experiments confirm that Stb1 localizes to promoters of MBF-regulated genes. Our data indicate that, contrary to previous models, MBF and SBF have unique components and might be distinctly regulated.
In the budding yeast Saccharomyces cerevisiae, commitment to enter the mitotic cell cycle occurs during late G1 phase at a point called Start which is analogous to the restriction point in mammalian cells (42). Start is marked by the coordinate induction of a large subset of genes that promote entry into the mitotic cell cycle (49, 51). Approximately 12% of yeast genes are cell cycle regulated, and expression of almost half of these genes peaks at the G1/S phase transition (27, 29). Transcriptional activation of genes in late G1 phase is largely dependent on two heterodimeric transcription factors called SBF and MBF (reviewed in reference 54). These two complexes share a common regulatory subunit, Swi6, which is tethered to DNA via its binding partners, Swi4 and Mbp1. Activation of SBF and MBF transcription initiates key cell cycle events, including budding, DNA synthesis, and spindle pole body duplication. The SBF (SCB-binding factor) complex activates transcription mainly through a cis-acting sequence element called SCB (for Swi4/6 cell cycle box). Genes activated by SBF include those encoding G1 cyclins (CLN1, CLN2, PCL1, and PCL2), the HO endonuclease gene, SWE1, which encodes a protein kinase, and a number of genes required for cell wall biosynthesis (29, 54). The MBF (MCB-binding factor) complex recognizes the MCB (for Mlu1 cell cycle box) element and activates G1-specific transcription of the S-phase cyclin genes, CLB5 and CLB6, and many genes required for DNA synthesis, including CDC9, POL1, RNR1, and CDC21 (29, 54). SBF and MBF have been shown (via genome-wide chromatin immunoprecipitation [ChIP] experiments) to bind to 235 gene promoters (29). Interestingly, this group of genes includes only 21% of the genes known to be induced at the G1/S transition (27, 29). However, among those SBF and/or MBF targets are 16 transcription factors which in turn may potentially regulate or influence the expression of thousands of genes, including those expressed in late G1 phase (27, 29).
Passage through Start and activation of SBF and MBF both require the cyclin-dependent kinase (Cdk), Cdc28, and one of three G1 cyclins, Cln1, Cln2, or Cln3. Although any one of the three G1 cyclins is sufficient to drive Start, genetic studies indicate a key role for Cln3 in activating SBF and MBF (53, 59). ChIP analyses have shown that promoter binding of SBF is crucial for the subsequent recruitment of the Srb mediator, TFIIB, as well as Kin28 (9). The Srb/mediator complex is recruited to promoters in the absence of Cdc28 kinase activity, whereas PolII, TFIIB, and Kin28 are only recruited in its presence (9). There is no evidence, however, that Cln3-Cdc28 acts to directly phosphorylate or interact with components of SBF or MBF (59). Moreover, a cln3Δ mutant strain is viable and still undergoes SBF- and MBF-dependent transcription, albeit at a larger cell size, indicating that alternative mechanisms must function to activate SBF/MBF (53). Our laboratory and others have uncovered a number of alternative activators of G1 transcription (11, 16, 25, 37, 58), but the precise mechanisms of activation and regulation of SBF and MBF remain unclear.
SBF and MBF transcriptional regulation is complex, and the distinction between SBF-controlled genes and MBF-controlled genes is not absolute. For example, CLN2 expression is not entirely abolished in mutants lacking SWI4 or SCB elements within the CLN2 promoter (10, 52) and CLN1 expression is regulated via SBF binding to MCB promoter elements (44). Consistent with redundancy in their roles, SWI4 and MBP1 are not essential genes; however, a strain lacking both genes arrests in G1 phase (32). Several observations suggest that the genetic redundancy of SWI4 and MBP1 reflects a functional redundancy at the level of DNA binding. First, Swi4 and Mbp1 share 50% identity in their DNA binding domains (32, 49). Second, isolated DNA binding domains from Swi4 and Mbp1 bind to both SCB and MCB sequences in vitro (32, 44) and Swi4 and Mbp1 bind overlapping sets of gene promoters in wild-type cells (29, 49). Finally, genome-wide ChIP studies have shown that Mbp1 and Swi4 share 34% of their target genes, suggesting that these proteins are at least partially redundant in wild-type populations (49). Despite their similarities, several differences exist between SWI4 and MBP1. For example, cells possessing null mutations in swi4 are viable but are slow growing and enlarged and exhibit defects in cell integrity and bud emergence (20). swi4Δ deletion mutants are also temperature sensitive and arrest primarily as unbudded cells with one nucleus and 2 N DNA content (20). Unlike swi4Δ mutants, mbp1Δ mutants appear to be similar to wild-type cells (32). Furthermore, swi4Δ swi6Δ double mutants are inviable and arrest prior to DNA synthesis while mbp1Δ swi6Δ mutants proliferate and phenotypically resemble swi6Δ mutants (32). Given these differences, it is possible that the regulatory mechanisms required for activation of SBF and MBF also differ. However, the genetically and functionally redundant nature of G1 control mechanisms has made it difficult to unravel the mechanisms of SBF and MBF regulation.
In this paper, we implicate Stb1 as a specific regulator of MBF-dependent transcription. STB1 encodes a Swi6-interacting protein which was previously identified as a regulator of Start transcription (25). However, the precise function of STB1 in G1 transcription is not known and an MBF-specific function was not previously appreciated. Consistent with a specific role in regulating MBF, a STB1 deletion strain requires the SBF subunit, Swi4, for viability and microarray studies showed a decrease in MBF-regulated transcripts in a swi4Δ mutant following depletion of Stb1. ChIP experiments confirmed that Stb1 localizes to promoters of MBF-regulated genes. Our data suggest that contrary to previous models, MBF and SBF might have unique components and might be distinctly regulated.
MATERIALS AND METHODS
Strains and media.
S. cerevisiae strains used in this study are listed in Table 1. Standard methods and media were used for yeast growth and transformation and strain construction (23). Minimal medium (synthetic dextrose [SD]) with appropriate amino acid supplements was used for maintaining plasmids in yeast transformants and for genetic selection (23). All gene disruptions were achieved by homologous recombination at their chromosomal loci using standard PCR-based methods (35).
TABLE 1.
S. cerevisiae strains used in this studya
| Strain | Genotype | Reference or source |
|---|---|---|
| BY263 | MATatrp1 leu2 his3 ura3 lys2 ade2 | 38 |
| BY184 | MATα swi4ΔHIS3 SCB::lacZ | 3 |
| BY185 | MATα swi6ΔHIS3 SCB::lacZ | 3 |
| BY551 | MATambp1ΔTRP1 | This study |
| BY805 | MATα stb1ΔURA3 | 25 |
| BY806 | MATα stb1ΔTRP1 | 25 |
| BY822 | MATastb1ΔTRP1 cln3ΔURA3 | 25 |
| BY1106 | MATα stb1ΔURA3 bck2ΔKAN | This study |
| BY1110 | MATabck2ΔKAN | This study |
| BY1298 | MATastb1ΔTRP1 | This study |
| BY1693 | MATα stb1ΔTRP1 swi4ΔHIS3 + pBA314 | This study |
| BY1694 | MATα stb1ΔTRP1 swi4ΔHIS3 + pBA417 | This study |
| BY1695 | MATα stb1ΔTRP1 swi4ΔHIS3 + pBA1044 | This study |
| BY1830 | MATα stb1ΔTRP1 mec1 + pBA1220 | This study |
Strains listed are isogenic to the parent BY263 unless otherwise indicated. BY263 is of S288C origin.
Plasmids.
The plasmids used in this study are listed in Table 2. Plasmid pBA314 was constructed by digesting pBA313 (36) with BglII. The resulting SWI4 fragment was then cloned into a BamHI-digested YEp13 vector. The STB1 gene was amplified by PCR from plasmid PBA1010 (25) using the primers 5′STB1BglII (5′-CCGGAGATCTATCACGCGAAAATGCAAG-3′) and 3′STB1BglII (5′-CCGGAGATCTGCCGTCAACGATCAATCA-3′) to generate a plasmid expressing full-length STB1 from the MET25-repressible promoter. The PCR product was digested with BglII and cloned into a BamHI-digested p415 MET25 vector (40) to create plasmid pBA1044. Plasmid pBA1220 was constructed through digestion of pBA1010 (25) with EcoRI and NotI. The resulting STB1 fragment was then cloned into an EcoRI/NotI-digested pRS426 vector (48). A plasmid expressing an N-terminal fusion of glutathione S-transferase (GST) to full-length Stb1 was constructed by amplification of the STB1 gene by PCR as described above. The PCR product was digested with BglII and cloned into a BamHI-digested pGEX-3X vector (Pharmacia), resulting in plasmid pBA1598.
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Reference or source |
|---|---|---|
| pLGΔSS | CYC1::lacZ (lacking UAS) | 21 |
| pBA251 | 4× SCB::lacZ | 2 |
| pBA487 | 4× MCB::lacZ | 56 |
| pBA314 | 2μm SWI4-URA3 | This study |
| pRS425 | 2μm LEU2 vector | 48 |
| pBA417 | 2μm SWI4-LEU2 | 36 |
| pBA1010 | 2μm STB1-LEU2 | 25 |
| pBA1044 | p415 MET25 + STB1 | This study |
| pBA1220 | 2μ STB1-URA3 | This study |
| pBA1282 | pRSET-B + STB1ΔN | 25 |
| pBA1598 | pGEX-3X + STB1 | This study |
| pBA1622 | pRSET-B + STB1 | This study |
| pBA1623 | pRSET-B + STB1ΔC | This study |
| pBA1624 | CEN MEC1-LEU2 | This study |
| pBA1625 | CEN RNR1-LEU2 | This study |
| pBA1626 | p415 MET25 + STB1ΔN | This study |
| pBA1627 | p415 MET25 + STB1ΔC | This study |
To generate a plasmid expressing an N-terminal His tag fused to full-length Stb1, the STB1 gene was amplified by PCR as described above and the PCR product was digested with BglII and cloned into a BamHI-digested pRSET-B vector (Invitrogen), resulting in plasmid pBA1622. Using the primers 5′STB1BglII (see above) and 3′STB1ΔCBglII (5′-GGGGGAAGATCTTGTTCCATGATGTGATGAC-3′), a plasmid expressing an N-terminal His-tagged Stb1ΔC fusion protein was created by amplification of a fragment of the STB1 gene (lacking the final 630 nucleotides) from plasmid pBA1010 (25). The PCR product was digested with BglII and cloned into a BamHI-digested pRSET-B vector (Invitrogen), resulting in plasmid pBA1623. A plasmid expressing STB1ΔC from the MET25-repressible promoter was constructed through amplification of STB1ΔC, digestion with BglII, and cloning into a BamHI-digested p415 MET25 vector, thereby creating plasmid pBA1627.
A portion of the STB1 gene (lacking the first 210 amino acids) was amplified by PCR from plasmid pBA1010 (25) using the primers 5′STB1ΔNBglII (5′-GGGGGAAGATCTATGCTTGGTTTAAGTAATGTCC-3′) and 3′STB1ΔNBglII (5′-GGGGGAAGATCTTCAATCAGTGAGTTTGTCAT-3′) to generate a plasmid expressing a truncated form of STB1 (STB1ΔN) from the MET25-repressible promoter. The PCR product was digested with BglII and cloned into a BamHI-digested p415 MET25 vector, resulting in plasmid pBA1626. Plasmids pBA1624 and pBA1625, containing the MEC1 and RNR1 genes, respectively, were isolated from a LEU2-YCp50-derived yeast genomic DNA library (8) (American Type Culture Collection, Manassas, Va.) (ATCC no. 77162).
β-Galactosidase assays.
Liquid β-galactosidase assays were performed on frozen pellets as described previously (38). Data are presented as the mean values from triplicate experiments.
ChIP assays.
Cultures (10 ml) of cells were grown to an optical density at 600 nm (OD600) of 0.6. Strains containing MCB::lacZ or SCB::lacZ reporter plasmids (Fig. 1B and C) were grown in SD-URA medium, while strains which did not harbor a plasmid (see Fig. 6) were grown in yeast extract-peptone-dextrose (YEPD) medium. Formaldehyde cross-linking and preparation of whole-cell extracts were performed as previously described (4). Using approximately 2 mg of extract and 5 μl of affinity-purified Stb1 polyclonal antibodies, immunoprecipitations were performed as described elsewhere (4, 25). Precipitates derived from strains containing the MCB::lacZ reporter, SCB::lacZ reporter, and vector plasmids (Fig. 1B and C) were washed four times for 10 min each time in 1 ml of lysis buffer and four times for 10 min each time in 1 ml of Tris-buffered saline (4). Precipitates derived from strains that did not harbor a plasmid (see Fig. 6) were washed twice for 10 min each time in 1 ml of lysis buffer and twice for 10 min each time in 1 ml of Tris-buffered saline. Finally, the samples were processed for DNA purification and PCR amplification of immunoprecipitated DNA was carried out as previously described (4). The PCR primers used for the amplification of promoter regions in PHO5, RNR1, and plasmid pLGΔSS were as follows: PHO5-F (5′-CCTGGCGACTATGGTATTTC-3′) (4), PHO5-R (5′-TTCACTGACAGTCTGCAAGG-3′) (4), RNR1-F (5′-TCAATGCTGAACTTTCTATGG-3′), RNR1-R (5′-TATTCTAAAACGTGAGCTGCA-3′), ΔSS-F (5′-GATGCGGCCAGCAAAACTAA-3′), and ΔSS-R (5′-ATATGATCATGTGTCGTCGC-3′). PCR products were separated on 2% agarose gels.
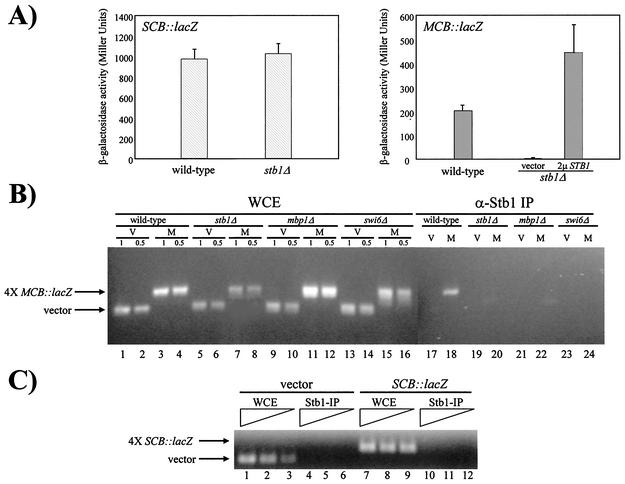
FIG. 1.
A stb1Δ mutant is defective in MCB::lacZ but not SCB::lacZ expression. (A) Wild-type (BY263) and stb1Δ (BY806) cells were transformed with the SCB::lacZ plasmid (pBA251, striped bars) or the MCB::lacZ plasmid (pBA487, solid bars). In MCB::lacZ assays, stb1Δ cells were also transformed with a 2μm STB1 (2μ STB1) plasmid (pBA1010) or a vector plasmid (pRS425). Strains were grown at 30°C to log phase, cell lysates were prepared, and β-galactosidase activity was measured. Depicted activity values represent the means of three experiments, and error bars indicate standard deviations for three experiments. (B) Stb1 localizes to MCB promoter elements. Wild-type (BY263), stb1Δ (BY806), mbp1Δ (BY551), and swi6Δ (BY185) strains containing a vector plasmid (pLGΔSS) (V) or an MCB::lacZ construct (pBA487) (M) were grown to mid-log phase and cross-linked with formaldehyde. Whole-cell extracts (WCE) were prepared (lanes 1 to 16) and subjected to immunoprecipitation (IP) using α-Stb1 affinity-purified antibodies (α-Stb1 IP) (lanes 17 to 24). PCR was performed on immunoprecipitated samples and on twofold serial dilutions (1× and 0.5×) of the WCE samples to amplify associated DNA. Primers used in this assay hybridized to the 3′ end of the URA3 gene and to sequences upstream of lacZ in both vector (pLGΔSS) and MCB::lacZ (pBA487) plasmids. (C) Stb1 does not localize to SCB promoter elements. Wild-type (BY263) strains containing a vector plasmid (pLGΔSS) or an SCB::lacZ construct (pBA251) were grown to mid-log phase and cross-linked with formaldehyde. Whole-cell extracts (WCE) were prepared (lanes 1 to 3 and 7 to 9) and subjected to immunoprecipitation (IP) using α-Stb1 affinity purified antibodies (α-Stb1-IP) (lanes 4 to 6 and 10 to 12). PCR was performed on immunoprecipitated samples and on twofold serial dilutions of the WCE samples to amplify associated DNA. The primers used to amplify associated DNA were the same as those described for panel B.
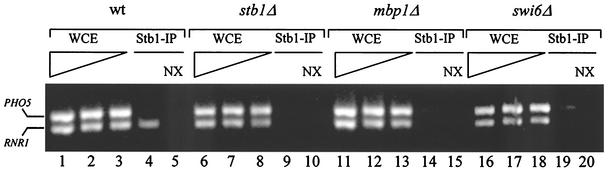
FIG. 6.
Stb1 localizes to the RNR1 promoter. Wild-type (wt) (BY263), stb1Δ (BY806), mbp1Δ (BY551), and swi6Δ (BY185) strains were grown to mid-log phase and cross-linked with formaldehyde. Cross-linked and non-cross-linked (NX) whole-cell extracts (WCE) were prepared and subjected to immunoprecipitation (IP) with α-Stb1 antibodies (Stb1-IP). PCR was performed on serial dilutions of the WCE samples (lanes 1 to 3, 6 to 8, 11 to 13, and 16 to 18) and on immunoprecipitated samples (lanes 4 to 5, 9 to 10, 14 to 15, and 19 to 20) to amplify associated DNA. Primers used for multiplex PCR were designed to flank the PHO5 promoter or the MCB elements of the RNR1 promoter.
Stb1 Western blotting analysis.
A stb1Δ (BY806) strain harboring STB1 (pBA1044), STB1ΔN (pBA1626), STB1ΔC (pBA1627), or a vector (p415 MET25) and a stb1Δswi4Δ mutant (BY1695) harboring a STB1 plasmid (pBA1044) were grown in SD-[LEU, MET] medium at 30°C to an OD600 of 0.5. Log-phase samples (10 ml) were taken for analysis. BY1695 cells were then diluted to an OD600 of 0.1 into SD-LEU medium with 5 mM methionine and grown at 30°C. Samples (10 ml) were taken at 2, 4, and 8 h time points. Samples (10 ml) of wild-type (BY263) and stb1Δ (BY806) strains (grown in the presence of 5 mM methionine for 8 h) were also harvested to determine endogenous Stb1 protein levels. Protein extracts and Stb1 Western blotting was performed as previously described (25).
DNA microarray analysis.
Yeast strains were grown in SD-[LEU, MET] medium at 30°C to an OD600 of 0.5. The cells were then diluted to an OD600 of 0.1 in SD-[LEU, MET] and SD-LEU medium supplemented with 5 mM methionine and grown for 8 h at 30°C (OD600, approximately 0.6). Cells were harvested by centrifugation and quickly frozen in liquid nitrogen. Total RNA and poly(A+) RNA were isolated as previously described (28). DNA microarrays consisting of approximately 97% genome coverage were probed with differentially labeled cDNA pools from a stb1Δ swi4Δ MET25pr-STB1 strain (BY1695) grown in the absence or presence of 5 mM methionine as previously described (28). Arrays were obtained from the Ontario Cancer Institute Microarray facility (www.microarrays.ca). Hybridized arrays were scanned using a Gene Pix 4000B scanner (Axon Instruments).
Northern blot analysis.
Yeast strains were grown as described above. RNA was isolated, and Northern blotting was performed as described previously (39). Probes used for Northern blot analysis included a 600-bp EcoRI-HindIII fragment of the ACT1 gene (38) and a 1.7-kb BglII-EcoRI fragment of the RNR1 gene (14). For RNA quantitation, Northern blots were exposed on a Molecular Dynamics screen, scanned using a Molecular Dynamics PhosphorImager, and analyzed using ImageQuant software, version 3.3 (Molecular Dynamics).
stb1Δ synthetic lethal screen.
A stb1Δ strain (BY806) harboring a high-copy STB1-URA3 plasmid (pBA1220) was subjected to ethyl methanesulfonate mutagenesis as described elsewhere (60). Surviving cells were screened for sensitivity to 5-fluoroortic acid (5-FOA) at 25°C. 5-FOA sensitivity indicates dependence on STB1 for viability and reflects the lethality generated by deletion of STB1 in combination with extragenic mutations (i.e., synthetic lethality). STB1 deletion strains harboring multiple extragenic mutations were identified by mating ethyl methanesulfonate-generated mutants to a wild-type strain (BY263). Diploids were grown in the presence of 5-FOA to promote loss of the STB1-URA3 plasmid and were subsequently sporulated. Two mutants (sls1 and sls2) were identified by tetrad analysis that segregated 2:2 for viability, indicating that these stb1Δ strains contained a single extragenic mutation and required STB1 for viability (data not shown). Synthetic lethal mutants were characterized genetically as previously described (60). Mutants were cloned by complementation using a yeast genomic DNA library in a LEU2-YCp50 derivative (8). Using a tagged MEC1 strain (data not shown), allelism of sls1 with MEC1 was confirmed by tetrad analysis.
Protein expression and purification and far-Western binding analysis.
Full-length Stb1, Stb1ΔN, and Stb1ΔC proteins were expressed from Escherichia coli harboring the expression plasmids pBA1622, pBA1282, and pBA1623, respectively. Protein expression was induced for 2 h at 30°C following addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Whole-cell extracts were prepared by harvesting 5-ml cultures and resuspending the pellets in 0.5 ml of 1× sodium dodecyl sulfate (SDS) sample buffer (50 mM Tris-HCl [pH 6.8], 10% glycerol, 2% SDS, 1% 2-mercaptoethanol). Volumes (10 μl) of whole-cell extracts were subjected to polyacrylamide gel electrophoresis (PAGE) (7.5% polyacrylamide) and transferred to nitrocellulose by means of a semidry transfer apparatus (Bio-Rad). Immunoblots were probed as described previously (25). Swi6 labeling and far-Western assays were performed as described below. GST-Stb1 was purified as previously described (39) from E. coli harboring the expression plasmid pBA1598. Recombinant kinase complexes were expressed and purified as described previously (41, 50). Using 1 μl of kinase/μg of substrate, approximately 1 μg of GST-Stb1 was phosphorylated in vitro. The reaction mixtures contained 20 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 2 mM NaF, 0.1 mg of bovine serum albumin/ml, 1 mM ATP, and 1 mM dithiothreitol in a volume of 20 μl. Following incubation at 30°C for 3 h, an additional 1 μl of kinase was added and the reaction mixtures were incubated at 30°C for a further 19 h. Unphosphorylated and phosphorylated GST-Stb1 was subjected to PAGE (7.5% polyacrylamide) and transferred to nitrocellulose by means of a semidry transfer apparatus (Bio-Rad). Immunoblots were probed as described previously (25). To produce full-length Swi6, the plasmid template pBA513 (3) was used as recommended in the instructions for the T7 TnT coupled reticulocyte lysate system (Promega). Far-Western assays using [35S]methionine (Mandel)-labeled Swi6 were carried out as previously described (22).
Microscopy.
Cells were grown in YEPD medium to log phase and observed at a magnification of ×630 with Normarski optics and a Micromax 1300y high-speed digital camera (Princeton Instruments, Trenton, N.J.) mounted on a Leica DM-LB microscope. Images from the camera were analyzed with Metaview software (Universal Imaging, Media, Pa.). Where indicated, the percentage of budded cells in each sample was determined by counting at least 300 cells per sample.
FACS.
The DNA content of strains BY263, BY805, BY1106, and BY1110 was analyzed by fluorescence-activated cell sorting (FACS) as described elsewhere (55).
RESULTS
MCB-driven UAS activity is dependent on STB1.
Owing to the complexity and genetic redundancy of SBF and MBF regulation, it is difficult to assign Start-specific transcription to SBF or MBF on the basis of consensus promoter elements in an upstream region. However, the observed redundancy in SBF and MBF function can be overcome by studying the ability of synthetic SCB and MCB elements to activate transcription of a reporter gene. CYC1::lacZ reporter genes lacking an upstream activating sequence (UAS) and driven by multiple synthetic SCB or MCB oligonucleotides are specifically dependent on SBF or MBF, respectively (2, 56). For example, deletion of MBP1 does not affect SCB::lacZ expression and, conversely, deletion of SWI4 causes only a modest defect in MCB::lacZ expression (56).
To determine the requirement for Stb1 in G1-specific transcription, MCB::lacZ and SCB::lacZ reporter gene plasmids were transformed into wild-type and stb1Δ mutant strains and β-galactosidase activities were measured in vitro (Fig. 1A). The SCB sequence was able to function efficiently as a UAS in both wild-type and stb1Δ cells (Fig. 1A). Conversely, the MCB sequence was unable to function as a UAS in the stb1Δ mutant strain. A 200-fold decrease in β-galactosidase activity was observed in a stb1Δ mutant compared to that seen in wild-type cells (Fig. 1A). β-Galactosidase activity was restored in stb1Δ mutants upon overexpression of STB1 (Fig. 1A). We interpret the large difference in STB1 requirement for the activation of transcription from the MCB sequences as an indication of the specific requirement for STB1 in this process.
Stb1 localizes to MCB promoter elements.
Since STB1 appeared to be required for MCB-dependent reporter gene expression, we directly examined the association of Stb1 with MCB synthetic promoter elements. ChIP assays were performed using affinity-purified Stb1 polyclonal antibodies (Fig. 1B). Wild-type, stb1Δ, mbp1Δ, and swi6Δ cells harboring the MCB::lacZ construct or a control vector were harvested during exponential growth phase. The cells were fixed with formaldehyde, and chromatin was immunoprecipitated using Stb1 antibodies (25). The abundance of specific DNA sequences within the immunoprecipitated material was measured using PCR and a vector-specific primer pair flanking the synthetic MCB elements. The CYC1::lacZ vector lacking a UAS was used as a negative control, since it does not contain any MCB elements. MCB elements and the control vector were detected in the input whole-cell extracts (Fig. 1B, lanes 1 to 16). Using cross-linked extracts derived from a wild-type strain (Fig. 1B, lane 18), specific PCR enrichment of a 380-bp fragment containing MCB promoter elements was detected in ChIPs, and this enrichment was dependent on the presence of STB1, MBP1, and SWI6 (Fig. 1B, lanes 20, 22, and 24). STB1 is transcribed in mbp1Δ and swi6Δ as well as swi4Δ mutants (reference 25 and data not shown), indicating that the inability of Stb1 to localize to MCB elements in the absence of MBF is not due to inadequate expression of STB1. Vector DNA was not enriched in the ChIP assay performed using affinity-purified Stb1 antibodies (Fig. 1B, lanes 17, 19, 21, and 23). Furthermore, DNA enrichment was not detected in ChIP assays performed using non-cross-linked lysates (data not shown). Similar ChIP assays were performed using a wild-type strain harboring an SCB::lacZ construct (Fig. 1C). In contrast to our findings with MCB promoter elements, no PCR enrichment of vector or SCB synthetic elements was observed in ChIP assays performed using affinity-purified Stb1 antibodies (Fig. 1C, lanes 4 to 6 and lanes 10 to 12). We therefore conclude that Stb1 specifically localizes to MCB elements in an MBF-dependent manner.
An STB1 deletion strain requires SWI4 for viability.
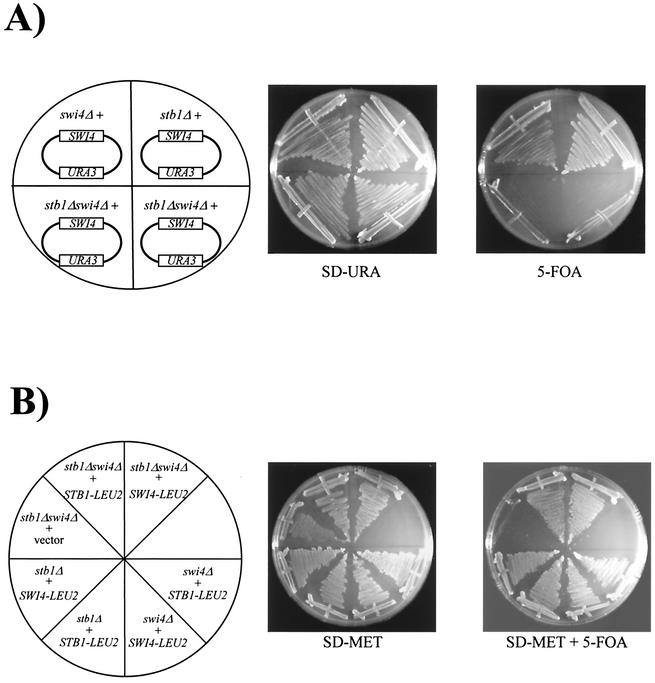
The results of β-galactosidase and ChIP assays suggest that STB1 functions to activate MBF-dependent transcription specifically. However, previous efforts examining SBF- and MBF-dependent expression of chromosomally encoded genes failed to reveal any defects in a stb1Δ mutant (25). The lack of transcriptional defects might be explained by functional redundancy between MBF (which requires Stb1) and SBF. We therefore reexamined STB1 genetic interactions. First, we constructed a stb1Δ swi4Δ double mutant (Fig. 2). This strain lacks functional SBF, and G1 transcription is entirely dependent on MBF; thus, a stb1Δ swi4Δ mutant may be more sensitive to defects in MBF regulation. A stb1ΔTRP1 mutant strain was mated to a swi4ΔHIS3 strain harboring a high-copy-number SWI4-URA3 plasmid. All haploid double mutants isolated by tetrad dissection contained the high-copy SWI4 plasmid, and double mutants were subsequently tested for sensitivity to 5-FOA (Fig. 2A). Expression of the URA3 gene causes lethality when cells are grown in the presence of 5-FOA (23); cells that are dependent on SWI4 for viability are sensitive to 5-FOA, since they cannot lose the SWI4-URA3 plasmid. Unlike stb1Δ and swi4Δ single mutants, stb1Δ swi4Δ double mutants were sensitive to 5-FOA, indicating that SWI4 was required for viability of the double mutant (Fig. 2A). We then rescued the 5-FOA sensitivity by expressing SWI4 or STB1 from LEU2-based plasmids (Fig. 2B). Complementation of stb1Δ swi4Δ 5-FOA sensitivity confirmed that viability of the double-mutant strain is dependent on SWI4 and STB1. Thus, consistent with an MBF-dependent function, STB1 is required for viability in the absence of SWI4. Furthermore, expression of CLN1 partially rescued the 5-FOA sensitivity of the stb1Δ swi4Δ double mutant (data not shown), suggesting that the lethality of stb1Δ swi4Δ mutants stems from inadequate G1 cyclin levels. However, similar to swi4Δ mbp1Δ mutants (32), the rescued double-mutant cells grew more slowly than either stb1Δ or swi4Δ single mutants and had abnormal morphology, suggesting that deletion of STB1 and SWI4 affects additional genes (data not shown). These results provide genetic evidence that STB1 is required for MBF-dependent transcription.
FIG. 2.
SWI4 is required for viability of a stb1Δ deletion strain. (A) stb1Δ (BY1298), swi4Δ (BY184), and stb1Δ swi4Δ (BY1693) mutants harboring a high-copy SWI4-URA3 plasmid (pBA314) were grown on SD-URA medium or on SD medium containing 5-FOA. (B) stb1Δ (BY1298), swi4Δ (BY184) and stb1Δ swi4Δ (BY1694 and BY1695) mutants containing a SWI4-URA3 plasmid and harboring either a SWI4-LEU2 (pBA417), MET25pr-STB1-LEU2 (pBA1044), or vector (pRS425) plasmid were grown in the absence (SD-MET) or presence (SD-MET + 5-FOA) of 5-FOA.
An stb1Δ bck2Δ double mutant accumulates in the G1 phase of the cell cycle.
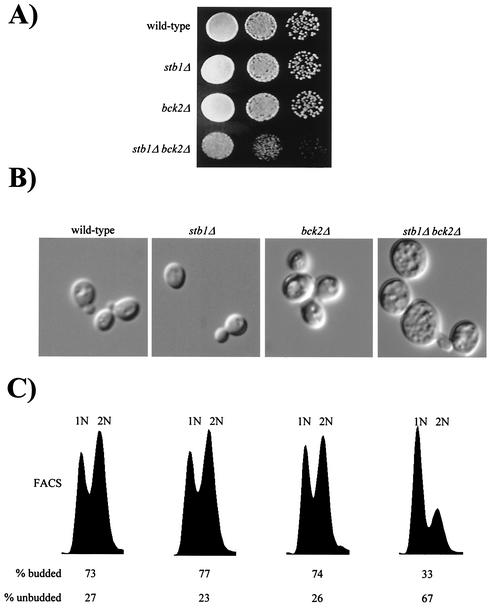
In an effort to reveal the genetic pathway defined by STB1, we expanded our genetic tests to include other known regulators of Start transcription. The Cln3-Cdc28 kinase is required for efficient activation of transcription at Start (12, 53, 59). However, CLN3 is not absolutely required for activation of SBF- and MBF-dependent gene expression. In the absence of CLN3, activation of Start transcription is delayed and other regulators, including STB1 and BCK2, are required for proper cell cycle progression and G1-specific transcription. Indeed, both stb1Δ cln3Δ and bck2Δ cln3Δ double mutants show pronounced G1 accumulation and a severe growth defect (11, 16, 25, 58). We extended this analysis by assessing the phenotype of a stb1Δ bck2Δ double-mutant strain (Fig. 3). When cultured in rich or minimal medium, the growth rates of both stb1Δ and bck2Δ mutants were comparable to that of the wild-type strain, whereas the stb1Δ bck2Δ double mutant grew much more slowly (Fig. 3A and data not shown). Analysis of cell morphology and DNA content in log phase cultures revealed that stb1Δ bck2Δ double-mutant cells accumulated in G1 phase as predominantly large unbudded cells (Fig. 3B and C). These observations support the notion that in addition to acting in a parallel pathway to CLN3 (25), STB1 might also function in parallel to BCK2 to regulate Start transcription.
FIG. 3.
Growth characteristics of the stb1Δ bck2Δ double mutant. (A) Slow-growth phenotype of a stb1Δ bck2Δ mutant strain. Tenfold serial dilutions were prepared from wild-type (BY263), stb1Δ (BY805), bck2Δ (BY1110), and stb1Δ bck2Δ (BY1106) cultures, plated onto YEPD medium, and incubated at 30°C. (B) Morphology of wild-type, stb1Δ, bck2Δ, and stb1Δ bck2Δ strains. Wild-type (BY263), stb1Δ (BY805), bck2Δ (BY1110), and stb1Δ bck2Δ (BY1106) strains were grown to mid-log phase in rich medium. The cells were viewed with Nomarski optics and photographed. (C) DNA content as measured by FACS analysis of samples shown in panel B. The positions of cells with G1 or G2 DNA contents are indicated by 1N or 2N, respectively. The percentages of budded and unbudded cells are indicated below the FACS profile.
MBF-dependent transcription is defective in a stb1Δ swi4Δ strain.
If STB1 is required for MBF-specific transcription, defects in MCB-driven gene expression may be revealed in the absence of functional SBF. Thus, to obtain a view of gene expression patterns in a stb1Δ swi4Δ mutant, we examined the genome-wide transcriptional consequences of depleting STB1 expression in a swi4Δ mutant. To accomplish this, viability of the stb1Δ swi4Δ double-mutant strain was maintained through ectopic expression of STB1 from the repressible MET25 promoter (Fig. 2B). Western blot analysis revealed that STB1 levels expressed from the MET25 promoter were drastically reduced (compared to those of endogenously expressed STB1) but not eliminated in the presence of 5 mM methionine (Fig. 4A, lanes 5 and 6). Limited expression of STB1 was sufficient for proliferation of the double-mutant strain (data not shown). The ability to limit rather than eliminate STB1 expression was useful, since it allowed us to isolate total RNA from living cells. Under these conditions, transcriptional defects are attributable to decreased Stb1 levels rather than cell death.
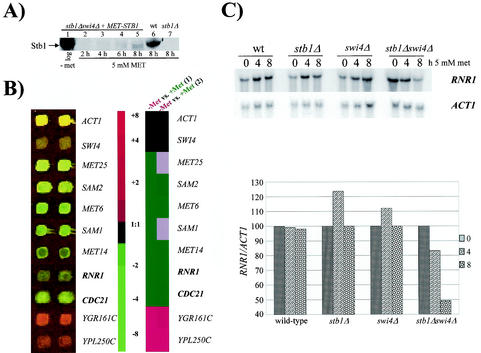
FIG. 4.
MBF-regulated genes are repressed in a stb1Δ swi4Δ double mutant. (A) Anti-Stb1 Western blot analysis of extracts prepared from a stb1Δ swi4Δ strain harboring a MET25pr-STB1 plasmid (BY1695; lanes 1 to 5), a wild-type (wt) strain (BY263; lane 6), and a stb1Δ strain (BY806; lane 7). Log-phase stb1Δ swi4Δ strain plus MET25pr-STB1 cultures were grown in the absence of methionine (lane 1) and subsequently grown in medium supplemented with 5 mM methionine (lanes 2 to 5). Wild-type and stb1Δ cultures were also grown in medium supplemented with 5 mM methionine (lanes 6 to 7). (B) A stb1Δ swi4Δ strain containing a MET25pr-STB1 construct (BY1695) was grown to mid-log phase in the absence of methionine. Cultures were then diluted and grown in the absence or presence of 5 mM methionine. cDNAs derived from cultures grown in the absence of methionine were labeled with Cy3 fluor, while cDNAs from cultures grown in 5 mM methionine were labeled with a Cy5 fluor. The results for a subset of genes whose expression levels changed at least threefold are shown. Green indicates methionine-dependent repression, while red indicates genes induced in the presence of 5 mM methionine. The reciprocal experiment was also performed (data not shown). (C) RNR1 is repressed in a stb1Δ swi4Δ double mutant. Wild-type (wt) (BY263), stb1Δ (BY806), swi4Δ (BY184), and stb1Δ swi4Δ (BY1695) mutants (containing a MET25pr-STB1 plasmid) were grown to mid-log phase in the absence of methionine (t = 0). Strains were subsequently grown in minimal medium containing 5 mM methionine. Aliquots were taken at 4- and 8-h time points. Total RNA was isolated from cells and probed with radiolabeled RNR1 and ACT1. The histogram depicts quantitation of the Northern blot. The RNR1 signal was quantified by phosphorimager analysis, and the values were normalized to the ACT1 loading control before plotting.
Cultures were grown to mid-log phase in the absence or presence of 5 mM methionine, and total RNA derived from these cultures was subjected to DNA microarray analysis (Fig. 4B). Expression levels of genes such as ACT1, which is neither cell cycle periodic nor regulated by methionine, remained unchanged (Fig. 4B). A 1:1 hybridization ratio was also observed for SWI4, since it was deleted from both samples (Fig. 4B). As expected, a large number of genes exhibited differential expression patterns. The majority of these genes function either directly or indirectly in methionine biosynthesis or metabolic pathways (Fig. 4B). However, differential expression of genes, other than those involved in methionine-regulated pathways, was also observed. In particular, expression of RNR1 and CDC21 was repressed three- and fivefold, respectively, in the presence of 5 mM methionine (Fig. 4B). RNR1 and CDC21 both function in DNA replication and metabolism; RNR1 encodes the large subunit of the ribonucleotide reductase complex and CDC21 encodes thymidylate synthase. Cell cycle expression of both RNR1 and CDC21 is dependent on MBF, since the late-G1-specific periodicity of these genes is abolished in swi6Δ and mbp1Δ mutants but is unaffected in swi4Δ mutants (32, 56). We interpret modest transcriptional defects as significant, since STB1 expression was reduced but not eliminated in the stb1Δ swi4Δ mutant. Furthermore, although periodic transcription is abolished, strains lacking MBP1 produce constitutive levels of mRNA from MCB-driven genes such as CDC21 (32). Given these results, we did not expect to observe large differences in the abundance of MBF-dependent transcripts in stb1Δ swi4Δ mutants. Thus, depletion of STB1 in the absence of SWI4 was sufficient to cause transcriptional defects in at least two previously characterized MBF-regulated genes as determined by microarray analysis.
Northern blot analysis confirmed results obtained from DNA microarray experiments (Fig. 4C). Wild-type, stb1Δ, swi4Δ, and stb1Δ swi4Δ strains (harboring a MET25pr-STB1 plasmid) were grown to exponential phase in medium lacking methionine. Cultures were subsequently grown in medium containing 5 mM methionine, and RNR1 expression was analyzed in samples harvested at time points 4 and 8 h after addition of methionine. RNR1 transcript levels remained constant in wild-type, stb1Δ, and swi4Δ strains (Fig. 4C). However, consistent with our microarray experiments, the stb1Δ swi4Δ double-mutant strain exhibited a gradual decrease in RNR1 expression. Following 8 h of growth in 5 mM methionine, a twofold decrease in RNR1 transcription was detected in the stb1Δ swi4Δ strain relative to that seen with wild-type or single mutant strains (Fig. 4C).
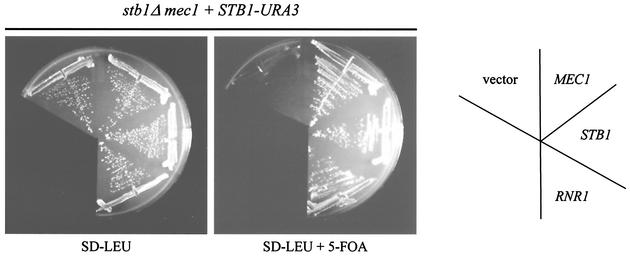
Taken together, results from DNA microarray and Northern analyses indicate that STB1 affects expression of at least a subset of MBF target genes, including RNR1. We made a genetic observation that further supports this view. In a separate series of experiments, we performed a stb1Δ synthetic lethal screening and isolated a mec1 mutant that requires STB1 for viability (Fig. 5; see Materials and Methods). MEC1 encodes an essential kinase which is involved in the G1, S, and G2 cell cycle checkpoint pathways in budding yeast (reviewed in reference 13). The 5-FOA sensitivity of a stb1Δ mec1 double mutant (harboring a high-copy STB1-URA3 plasmid) was rescued by expression of MEC1 or STB1 from a LEU2-based plasmid (Fig. 5). In addition to that of STB1 and MEC1, expression of RNR1 also rescued the stb1Δ mec1 phenotype (Fig. 5). Consistent with our microarray and Northern blot analyses, this result suggests that the lethality of the stb1Δ mec1 mutant is due, at least in part, to inadequate expression of RNR1.
FIG. 5.
RNR1 rescues the lethality of a stb1Δ mec1 mutant. A stb1Δ mec1 strain (BY1830) containing a STB1-URA3 plasmid (pBA1220) and harboring either a STB1-LEU2 (pBA1010), MEC1-LEU2 (pBA1624), RNR1-LEU2 (pBA1625), or vector (pRS425) plasmid was grown in the absence (SD-LEU) or presence (SD-LEU + 5-FOA) of 5-FOA.
Microarray analyses were also employed to examine the genome-wide effects of STB1 overexpression. Ectopic expression of both STB1 and MBP1 caused only modest effects on gene expression, as might be expected due to the tight cell cycle regulation of MBF target genes (data not shown). Nonetheless, statistical analysis of these expression experiments revealed a significant correlation between STB1 and MBP1 overexpression profiles compared to that of relevant control experiments, consistent with a role for STB1 in regulating MBF-dependent transcription (data not shown).
Stb1 localizes to the RNR1 promoter.
Since microarray and Northern analyses suggested that STB1 is required for RNR1 transcription, we directly examined the association of Stb1 with the RNR1 promoter. ChIP assays were performed in which wild-type, stb1Δ, mbp1Δ, and swi6Δ cells were harvested during the exponential-growth phase. Cells were fixed with formaldehyde, and chromatin was isolated using affinity-purified Stb1 polyclonal antibodies. The abundance of specific DNA sequences within the immunoprecipitates was measured using PCR and the appropriate primer pairs. Reaction mixtures contained two sets of primers, enabling us to simultaneously measure the relative abundances of Stb1 at RNR1 and PHO5 promoters. For RNR1 measurements, the primer pairs were designed to straddle previously identified MCB elements (15). Specific PCR enrichment of RNR1 promoter DNA was detected in ChIPs from a wild-type strain (Fig. 6, lane 4). PHO5 was used as a negative control, since there are no detectable SCB or MCB elements in its promoter and the time of maximal PHO5 expression does not coincide with SBF and MBF activity (4, 51). DNA from RNR1 and PHO5 promoters was detected in formaldehyde cross-linked whole-cell extracts (Fig. 6, lanes 1 to 3, 6 to 8, 11 to 13, and 16 to 18). As expected, control assays using non-cross-linked lysates were not enriched for promoter-specific DNA (Fig. 6, lanes 5, 10, 15, and 20) and Stb1 antibodies did not cause enrichment of PHO5 promoter-specific DNA (Fig. 6, lanes 4, 9, 14, and 19). Moreover, ChIP assays using stb1Δ, mbp1Δ, and swi6Δ strains and affinity-purified Stb1 antibodies were not enriched for RNR1 promoter DNA (Fig. 6, lanes 9, 14, and 19). These experiments suggest that Stb1 specifically localizes to MCB elements located within the RNR1 promoter. Consistent with our hypothesis, Stb1 requires Swi6 and Mbp1 to associate with MCB promoter elements, suggesting that Stb1 functions in the context of MBF.
Interaction with Swi6 is required for Stb1 function.
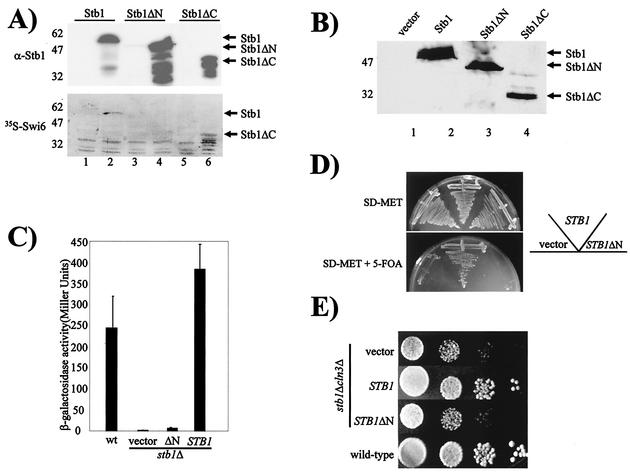
Stb1 was previously characterized as a Swi6-binding protein (25), but the biological relevance of the Stb1-Swi6 interaction was not understood. We used far-Western analysis to further examine the Stb1-Swi6 physical interaction (Fig. 7A). Whole-cell extracts were prepared from bacterial cells expressing either full-length STB1 (Stb1), truncated Stb1 protein lacking 70 amino acids at the N terminus (Stb1ΔN) (25), or a truncated form of Stb1 lacking the C-terminal 210 amino acids (Stb1ΔC). Uninduced (Fig. 7A, lanes 1, 3, and 5) and IPTG-induced (Fig. 7A, lanes 2, 4, and 6) extracts were blotted to nitrocellulose and probed with affinity-purified Stb1 antibodies or [35S]methionine-labeled Swi6 (Fig. 7A). The anti-Stb1 immunoblot revealed that all three forms of Stb1 protein were expressed following IPTG induction (Fig. 7A, upper panel). Following incubation with 35S-labeled Swi6, a difference in binding between full-length and truncated forms of Stb1 was observed. Unlike full-length Stb1 and Stb1ΔC, which bound Swi6 directly (Fig. 7A, lanes 2 and 6), a Stb1ΔN-Swi6 interaction was not observed (Fig. 7A, lane 4). This suggests that the first 70 N-terminal amino acids of the Stb1 protein are required for Swi6 binding. We next expressed full-length STB1, STB1ΔN, and STB1ΔC from the repressible MET25 promoter in yeast transformants (Fig. 7B). Since all three forms of the Stb1 protein were expressed in both a stb1Δ mutant (Fig. 7B) and a wild-type strain (data not shown), we asked whether the Stb1ΔN protein was able to complement STB1-dependent phenotypes (Fig. 7C, D, and E). As described above, a stb1Δ mutant is defective in MCB-driven UAS activity (Fig. 1A). Unlike expression of full-length STB1, which restores MCB::lacZ activity to wild-type levels, STB1ΔN does not complement the defect in MBF-dependent reporter gene expression (Fig. 7C). Furthermore, expression of STB1ΔN does not complement the 5-FOA sensitivity of a stb1Δ swi4Δ double mutant, demonstrating that full-length Stb1 is required for viability in the absence of Swi4 (Fig. 7D). The inability to complement these phenotypes suggests that the MBF-specific function of STB1 is dependent on its interaction with Swi6. Consistent with these observations, Stb1ΔN also fails to complement the slow growth phenotype of a stb1Δ cln3Δ double mutant (Fig. 7E), indicating that Swi6 binding is also required for Stb1 function in G1 cell cycle progression.
FIG. 7.
Interaction with Swi6 is required for Stb1 function. (A) Uninduced (lanes 1, 3, and 5) and IPTG-induced (lanes 2, 4, and 6) bacterial whole-cell extracts were prepared from cells expressing full-length STB1 (pBA1622; lanes 1 and 2), STB1ΔN (pBA1282; lanes 3 and 4) and STB1ΔC (pBA1623; lanes 5 and 6). Extracts were subjected to SDS-PAGE, and duplicate gels were transferred to nitrocellulose. One blot was probed with α-Stb1 antibodies (α-Stb1), while a second blot was probed with in vitro-translated [35S]methionine Swi6 (35S-Swi6), as indicated to the left of the photograph. (B) Anti-Stb1 Western blot analysis of extracts prepared from a stb1Δ mutant (BY806) harboring a p415 MET25 vector (lane 1) or a STB1 (pBA1044, lane 2), STB1ΔN (pBA1626, lane 3), or STB1ΔC (pBA1627, lane 4) plasmid. Cultures were grown to mid-log phase in medium lacking methionine. (C) Wild-type (wt) (BY263) and stb1Δ (BY806) cells were transformed with the MCB::lacZ plasmid (pBA487). The stb1Δ strain was also transformed with a vector (p415 MET25) or a STB1 (pBA1044) or STB1ΔN (pBA1626) plasmid. Strains were grown at 30°C to log phase in the absence of methionine, cell lysates were prepared, and β-galactosidase activity was measured. Depicted activity values represent the means of three experiments, and error bars indicate standard deviations for three experiments. (D) A stb1Δ swi4Δ (BY1693) mutant containing a SWI4-URA3 plasmid and harboring either a vector (p415 MET25) or a STB1-LEU2 (pBA1044) or STB1ΔN-LEU2 (pBA1626) plasmid was grown in the absence (SD-MET) or presence (SD-MET + 5-FOA) of 5-FOA. (E) Tenfold serial dilutions were prepared from a wild-type (BY263) and a stb1Δ cln3Δ (BY822) strain harboring a vector (p415 MET25) or a STB1 (pBA1044) or STB1ΔN (pBA1626) plasmid, plated onto medium lacking methionine, and incubated at 30°C.
Cln-Cdc28-dependent phosphorylation inhibits the Stb1-Swi6 interaction.
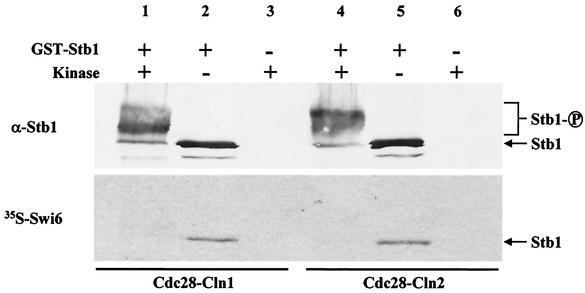
Previous studies have suggested that the Cln3-Cdc28 complex is a major activator of SBF- and MBF-dependent transcription (53, 59). However, Cln3 does not appear to phosphorylate or interact with components of either SBF or MBF, suggesting that Cln3-dependent activation is indirect (59). Stb1 was previously identified as a Swi6-interacting protein by affinity chromatography and coimmunoprecipitation experiments (25), and Swi6 binding is likely required for Stb1 function in G1 phase (Fig. 7). Stb1 is also a phosphoprotein in yeast, and Ho et al. have shown that Stb1 is an excellent substrate for Cln-Cdc28 kinase complexes in vitro and in vivo (25). Thus, Cln-Cdc28 complexes might regulate MBF through phosphorylation of Stb1. To explore this possibility, we employed far-Western analysis to examine the effects of Cln-dependent phosphorylation on the Stb1-Swi6 interaction. Using Cln1-Cdc28 or Cln2-Cdc28 kinase purified from insect cells, purified recombinant GST-Stb1 was phosphorylated in vitro (Fig. 8). Unphosphorylated Stb1 and Stb1 phosphorylated by Cln-Cdc28 were blotted to nitrocellulose and probed with affinity-purified Stb1 antibodies or [35S]methionine-labeled Swi6 (Fig. 8). Cln-dependent phosphorylation of Stb1 was confirmed by Western blot analysis, since phosphorylated Stb1 migrated more slowly than the unphosphorylated protein (Fig. 8, upper panel, lanes 1 and 4). Following incubation with 35S-labeled Swi6, a clear difference in binding between phosphorylated and unphosphorylated Stb1 was observed. Unphosphorylated Stb1 was able to bind Swi6 (Fig. 8, lanes 2 and 5), whereas Stb1 which had been phosphorylated by Cdc28-Cln1 or Cdc28-Cln2 failed to interact with Swi6 (Fig. 8, lanes 1 and 4). Therefore, Cln-dependent phosphorylation inhibits the Stb1-Swi6 interaction in vitro and, hence, may play an inhibitory role in regulating MBF-dependent transcription (see Discussion).
FIG. 8.
Cln1- and Cln2-dependent phosphorylation of Stb1 inhibits the Swi6-Stb1 interaction in vitro. Bacterially expressed GST-Stb1 was phosphorylated in vitro with Cdc28-Cln1 (lane 1) or Cdc28-Cln2 (lane 4) purified from insect cells. Phosphorylated Stb1 (lanes 1 and 4) and unphosphorylated Stb1 (lanes 2 and 5) were subjected to SDS-PAGE. Duplicate gels were transferred to nitrocellulose. One blot was probed with α-Stb1 antibodies (α-Stb1). A second blot was probed with in vitro-translated [35S]methionine Swi6 (35S-Swi6).
DISCUSSION
In vivo footprinting experiments have revealed that SBF and MBF are bound to promoter elements throughout the G1 phase (24, 33). Since transcription only occurs late in G1 phase, DNA binding is not sufficient and specific activation of SBF and MBF must occur. So far, intense genetic scrutiny has failed to reveal activation mechanisms or regulatory differences between SBF and MBF complexes. In this paper, we describe a series of experiments whose results suggest a role for Stb1 in the regulation of MBF-specific transcription at Start.
A role for Stb1 as a component of MBF.
With the exception of the HO gene, whose expression is completely dependent on the presence of SBF (6, 7), only synthetic SCB- and MCB-driven reporter genes exhibit a specific dependence on SBF and MBF, respectively (2, 56). We found that a strain lacking STB1 was specifically defective in MCB::lacZ expression. STB1 had been previously implicated as a cell cycle regulator (25); a stb1Δ cln3Δ mutant is slow growing and is severely delayed in G1 phase, accumulating a high fraction of large unbudded cells with 1 N DNA content (25). Based on these results, it was concluded that STB1 is required for G1/S-phase progression and functions in a pathway parallel to that of CLN3 to activate G1-specific transcription. However, the precise role for STB1 in transcriptional activation remained unclear, since conventional Northern analyses failed to reveal defects in transcript levels or cell cycle periodicity of G1-regulated genes in a stb1Δ mutant (25). The MCB::lacZ defect described here represents direct evidence that Stb1 functions as a transcriptional activator.
In the absence of SWI4, G1-specific transcription and cell cycle progression is dependent on MBF (32). We found that STB1, like MBP1, has an overlapping essential function with SWI4. Unlike the case of a stb1Δ swi4Δ mutant, no obvious additional phenotypes were found in stb1Δ mbp1Δ or stb1Δ swi6Δ strains. Although these genetic results do not preclude a role for STB1 in regulating SBF-dependent genes, they strongly support our other experiments showing that the primary role for STB1 in G1 transcription involves MBF. Consistent with an MBF-specific function, we also observed a clear reduction in transcription of two MBF-regulated genes, RNR1 and CDC21, when Stb1 levels were limiting. This result may indicate that STB1 is required for transcription of only a subset of MBF targets. Alternatively, our experimental conditions may create a bias for identifying the MBF-regulated genes which are most sensitive to Stb1 levels.
Our observation that a mec1 mutant requires STB1 for viability is also consistent with an MBF-specific role for STB1. Since MEC1 is required for viability in our strain background (S288C), we infer that the mec1 mutation we identified is a hypomorphic allele. Although we have not yet pursued a detailed molecular characterization of the sls allele of MEC1, our RNR1 suppression data allow us to formulate a model for the stb1Δ mec1 genetic interaction. The RNR1 gene product is limiting for ribonucleotide reductase activity in yeast extracts and is therefore tightly regulated (57, 61). One form of RNR1 regulation occurs at the transcriptional level. Ho et al. previously showed that SBF is involved in up-regulating RNR gene expression in response to DNA damage (26). However, periodic expression of RNR1 (in the absence of DNA damage) is dependent on MBF rather than SBF (32, 56). In addition to transcriptional regulation, Rnr1 is also regulated at the posttranslational level (61). Rnr1 binds to a protein known as Sml1 which inhibits its activity when DNA synthesis is not required (61). During S phase, a signal is transmitted to relieve Sml1 inhibition of Rnr1; Mec1 appears to remove the inhibitory effect of Sml1 on Rnr1 during S phase, thus facilitating DNA replication (61). We suggest that the Sml1-mediated negative control of Rnr1 is not relieved in our stb1Δ mec1 mutant. Hence, inhibition of Rnr1 activity coupled with reduced RNR1 gene expression (in the absence of STB1 [Fig. 4 and 5]) may result in cellular dNTP levels which are inadequate to support DNA synthesis.
Stb1 localizes to the RNR1 promoter, and association with MCB element-containing promoters is indirect and dependent on the presence of Mbp1 and Swi6. Consistent with this finding, efforts to purify MBF from whole-cell extracts revealed that an unidentified protein (with a molecular weight similar to that of Stb1) copurified with Mbp1 and Swi6 (32) and Stb1 copurifies with Swi6 and Mbp1 when Swi6 is isolated from yeast extracts by means of an affinity-tagging protocol (N. Krogan and J. Greenblatt, personal communication). Although our ChIP experiments further implicate Stb1 as a component of MBF, biochemical studies have shown that Mbp1-Swi6 and Swi4-Swi6 complexes produced by in vitro translation are sufficient for DNA binding (32, 45). Thus, we propose that Stb1 localization to MCB-driven promoters functions to regulate MBF-dependent transcription rather than mediate MBF-DNA binding.
Consistent with our findings, analyses of genome-wide ChIP and microarray studies have provided additional evidence suggesting that Stb1 acts as a direct regulator of G1-specific transcription (34). However, this strictly computational approach failed to identify the RNR1 promoter as a significant Stb1 target, while our results clearly show Mbp1-dependent localization of Stb1 to the RNR1 promoter. Statistical analysis of genomic binding data also suggested that Stb1 does not share common promoter targets with Swi6 (34). In contrast to this interpretation, Stb1 is a Swi6-binding protein (25) and observations from this study indicate that Stb1 association with chromatin is dependent on the presence of Swi6 and that an interaction with Swi6 is important for the cell cycle-dependent function of Stb1. It is difficult to address Stb1 specificity by means of analysis of genome-wide ChIP experiments, since both MBF and SBF were capable of binding most of the identified Stb1 target promoters in these experiments (34). Statistical analyses of genome-wide expression and binding data can provide many testable hypotheses with regard to the transcriptional circuitry of budding yeast (34). However, given the nature of genomic experiments, more-focused investigations are required to confirm specific details proposed by these models.
Swi6 is a common subunit of both SBF and MBF. Ho et al. previously showed that the Swi6 ankyrin repeat domain is required for the Stb1-Swi6 interaction (25). Mutational analyses of Swi6 ankyrin repeats identified several point mutants that retained the ability to bind DNA in the context of SBF and MBF (17). However, these mutants failed to induce SBF- and MBF-dependent gene expression, indicating that the Swi6 ankyrin repeat domain is important for transcriptional activation (17, 19, 46). In similarity to stb1Δ mutants, most Swi6 ankyrin repeat domain mutants showed severe defects in MCB reporter gene expression while the SCB reporter gene was less affected (17). These results imply that the Swi6-Mbp1 interaction or MBF activity might have a stronger dependence on the ankyrin domain, and this dependence may account for the observed MBF-specific function of STB1. Alternatively, Stb1, in addition to interacting with the Swi6 ankyrin repeat domain, may associate directly with Mbp1. Homologous ankyrin repeat domains are also found in the central region of Swi4, and Mbp1 and far-Western analysis revealed that Stb1 can bind Mbp1 directly in vitro (M. Costanzo, unpublished data). Whether nonconserved residues in the ankyrin domain of Mbp1 mediate specific interactions with Stb1 in vivo requires further investigation.
A role for STB1 in regulating G1-specific transcription.
Based on our results, we present a model for G1-specific transcription whereby SBF and MBF transcription factors are differentially regulated (Fig. 9). CLN3 is the major activator of Start transcription and likely activates both SBF and MBF through Swi6 (53, 59). However, a cln3Δ mutant is viable and still undergoes SBF- and MBF-dependent transcription, indicating the existence of alternate mechanisms to activate G1-specific transcription and promote entry into S phase (43, 53). Consistent with this model, Ho et al. previously showed that STB1 functions in a pathway parallel to that of CLN3 to regulate G1-specific transcription (25).
FIG. 9.
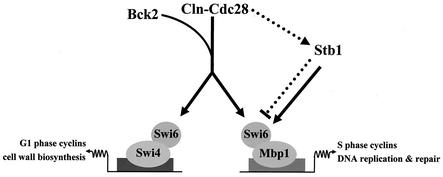
A model for Stb1-dependent regulation of MBF transcription. Our model predicts that Stb1 functions in an alternative pathway to activate G1-specific transcription. Cln3-Cdc28 is the major activator of SBF- and MBF-dependent transcription, whereas Stb1 and Bck2 function as alternate activators of Start transcription. Unlike Cln3 and Bck2, Stb1 specifically regulates MBF. Prior to Start, Stb1 is predominantly unphosphorylated and interaction with Swi6 serves to activate MBF-dependent transcription (solid lines). Immediately following the time of maximal Start-specific transcription, Cln-Cdc28-dependent phosphorylation of Stb1 mediates dissociation of the Stb1-MBF complex, resulting in termination of MBF-specific transcription (dotted lines).
In addition to STB1, the BCK2 (bypass of C-kinase mutation) gene also appears to be involved in an alternative pathway(s). In similarity to a stb1Δ cln3Δ strain, bck2Δ cln3Δ double mutants grow slowly, accumulate in G1 phase, and show reduced levels of Start transcription (11, 16, 58). Unlike that of stb1Δ mutants, which do not have obvious transcriptional defects (25), expression of SBF- and MBF-regulated genes is modestly delayed, but not abolished, in bck2Δ mutants (11). Furthermore, overexpression of BCK2 results in transcriptional induction of several SBF and MBF target genes (11, 16, 58). These observations suggest that BCK2 likely activates both SBF- and MBF-dependent transcription.
We found that in similarity to bck2Δ cln3Δ and stb1Δ cln3Δ strains, stb1Δ bck2Δ double mutants are also slow growing and accumulate in the G1 phase of the cell cycle. The G1 delay phenotype observed in these mutants suggests that there are at least three independent pathways for activation of Start transcription. Our present work suggests that STB1, unlike CLN3 and BCK2, functions specifically to regulate MBF-dependent transcription. Therefore, in the absence of STB1 and CLN3 or BCK2, at least two SBF/MBF activation pathways are compromised, resulting in reduced levels and/or timing of Start transcription and a delay in G1 phase. Consistent with this prediction, activation of RNR1, CLN1, and CLN2 transcription was significantly delayed in a stb1Δ cln3Δ double mutant (25).
Our work also suggests that phosphorylation of Stb1 might regulate its ability to associate with MBF. Stb1 is both a Cln-Cdc28 substrate and a Swi6-interacting protein (25), and our far-Western analysis revealed that Cln-dependent phosphorylation of Stb1 inhibits the Swi6-Stb1 interaction in vitro. Since Swi6 binding is likely required for Stb1 function, we propose that phosphorylation of Stb1 might play a role in down-regulating MBF-dependent transcription (Fig. 9). Previous studies demonstrated that Stb1 phosphorylation is cell cycle periodic, with maximal phosphorylation occurring after Start transcription but prior to DNA replication (25). Hence, it is possible that Cln-dependent phosphorylation mediates the dissociation of Stb1 from MBF, resulting in down-regulation of MCB-driven gene expression. SBF repression is likely mediated by Clb2-Cdc28 kinase activity (1, 47), and DNA microarray analysis has also shown that Clb2 overexpression results in repression of a large number of G1 phase-regulated genes (51). Previous studies have also shown that Clb2 interacts with the ankyrin repeat domain of Swi4 (47). Thus, in similarity to that by Clb2, Stb1-dependent regulation of MBF transcription may also be mediated through its interaction with ankyrin repeats.
Analogy between budding yeast MBF and E2F in higher eukaryotes.
Despite MBP1 having conserved functional and sequence homologues in distantly related yeasts (5), no homologues have yet been identified in higher eukaryotes. However, members of the E2F/DP1 family of transcription factors may be considered functionally analogous to MBF (30). E2F is under the control of the retinoblastoma protein, Rb, which binds and inhibits E2F (18). The Rb-E2F interaction is regulated by phosphorylation; in noncycling cells or in early G1 phase, Rb is hypophosphorylated and inhibits E2F activity. Conversely, in late G1 phase, Rb is progressively phosphorylated by cyclin-Cdk complexes and, consequently, its affinity for E2F diminishes. The release of Rb triggers the activation of E2F target genes, which allows cells to progress through the G1/S transition (18). In this report, we propose that Stb1 plays a role as an MBF-specific regulator and that analogous to Cdk-dependent phosphorylation of Rb, Cdc28-dependent phosphorylation might inhibit the interaction between Stb1 and MBF. Therefore, cyclin-Cdk complexes appear to affect E2F- and MBF-dependent transcription, at least in part, through regulation of protein-protein interactions between transcription factors and regulatory proteins such as Rb and Stb1. Rb inhibits E2F transcriptional activation, to some extent, by recruiting chromatin remodeling factors such as histone deacetylases and members of the SWI/SNF complex (18). Similarly, Stb1 was found to interact with the Sin3-Rpd3 histone deacetylase complex (31). However, our data suggest that, unlike Rb, Stb1 functions as an activator of MBF-dependent transcription. A direct role for Stb1 in transcriptional repression remains unclear.
Acknowledgments
We are grateful to Helena Friesen, Bri Lavoie, and Grant Brown for comments on the manuscript. We thank Jason Moffat, Timothy Hughes, Mark Robinson, and Jeff Pootoolal for technical assistance and statistical analysis of microarray experiments.
M.C. held a Doctoral Award from the Canadian Institutes of Health Research (CIHR). This work was supported by an operating grant to B.A. from the CIHR.
REFERENCES
- 1.Amon, A., M. Tyers, B. Futcher, and K. Nasmyth. 1993. Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell 74:993-1007. [DOI] [PubMed] [Google Scholar]
- 2.Andrews, B. J., and I. Herskowitz. 1989. Identification of a DNA binding factor involved in cell cycle-control of the yeast HO gene. Cell 57:21-29. [DOI] [PubMed] [Google Scholar]
- 3.Baetz, K., and B. Andrews. 1999. Regulation of cell cycle transcription factor Swi4 through auto-inhibition of DNA binding. Mol. Cell. Biol. 19:6729-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baetz, K., J. Moffat, J. Haynes, M. Chang, and B. Andrews. 2001. Transcriptional coregulation by the cell integrity mitogen-activated protein kinase Slt2 and the cell cycle regulator Swi4. Mol. Cell. Biol. 21:6515-6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breeden, L. 1996. Start-specific transcription in yeast. Curr. Top. Microbiol. Immunol. 208:95-127. [DOI] [PubMed] [Google Scholar]
- 6.Breeden, L., and G. E. Mikesell. 1991. Cell cycle-specific expression of the SWI4 transcription factor is required for cell cycle regulation of HO transcription. Genes Dev. 5:1183-1190. [DOI] [PubMed] [Google Scholar]
- 7.Breeden, L., and K. Nasmyth. 1987. Cell cycle control of the yeast HO gene: cis- and trans-acting regulators. Cell 48:389-397. [DOI] [PubMed] [Google Scholar]
- 8.Christianson, T. W., R. S. Sikorski, J. H. Dante, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110:119-122. [DOI] [PubMed] [Google Scholar]
- 9.Cosma, M. P., S. Panizza, and K. Nasmyth. 2001. Cdk1 triggers association of RNA polymerase to cell cycle promoters only after recruitment of the mediator by SBF. Mol. Cell 7:1213-1220. [DOI] [PubMed] [Google Scholar]
- 10.Cross, F., M. Hoek, J. D. McKinney, and A. H. Tinkelenberg. 1994. Role of Swi4 in cell cycle regulation of CLN2 expression. Mol. Cell. Biol. 14:4779-4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DiComo, C. J., H. Chang, and K. T. Arndt. 1995. Activation of CLN1 and CLN2 G1 cyclin expression by BCK2. Mol. Cell. Biol. 15:1835-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dirick, L., T. Bohm, and K. Nasmyth. 1995. Roles and regulation of the Cln-Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. EMBO J. 14:4803-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elledge, S. 1996. Cell cycle checkpoints: preventing an identity crisis. Science 274:1664-1672. [DOI] [PubMed] [Google Scholar]
- 14.Elledge, S. J., and R. W. Davis. 1990. Two genes differentially regulated in the cell cycle and by DNA-damaging agents encode alternative regulatory subunits of ribonucleotide reductase. Genes Dev. 4:740-751. [DOI] [PubMed] [Google Scholar]
- 15.Elledge, S. J., Z. Zhou, J. B. Allen, and T. A. Navas. 1993. DNA damage and cell cycle regulation of ribonucleotide reductase. Bioessays 15:333-339. [DOI] [PubMed] [Google Scholar]
- 16.Epstein, C., and F. Cross. 1994. Genes that can bypass the CLN requirement for Saccharomyces cerevisiae cell cycle START. Mol. Cell. Biol. 14:2041-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ewaskow, S. P., J. M. Sidorova, J. Hendle, J. C. Emery, D. E. Lycan, K. Y. J. Zhang, and L. L. Breeden. 1998. Mutation and modeling analysis of the Saccharomyces cerevisiae Swi6 ankyrin repeats. Biochemistry 37:4437-4450. [DOI] [PubMed] [Google Scholar]
- 18.Ferreira, R., I. Naguibneva, L. L. Pritchard, S. Ait-Si-Ali, and A. Harel-Bellan. 2001. The Rb/chromatin connection and epigenetic control: opinion. Oncogene 20:3128-3133. [DOI] [PubMed] [Google Scholar]
- 19.Foord, R. I., I. Taylor, S. Sedgwick, and S. Smerdon. 1999. X-ray structural analysis of the yeast cell cycle regulator Swi6 reveals variations of the ankyrin fold and has implications for Swi6 function. Nat. Struct. Biol. 6:157-165. [DOI] [PubMed] [Google Scholar]
- 20.Gray, J. V., J. P. Ogas, Y. Kamada, M. Stone, D. E. Levin, and I. Herskowitz. 1997. A role for the Pkc1 MAP kinase pathway of Saccharomyces cerevisiae in bud emergence and identification of a putative upstream regulator. EMBO J. 16:4924-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guarente, L. 1983. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 101:181-191. [DOI] [PubMed] [Google Scholar]
- 22.Guichet, A., J. W. R. Copeland, M. Erdelyi, D. Hlousek, P. Zavorsky, J. Ho, S. Brown, A. Percival-Smith, H. M. Krause, and A. Ephrussi. 1997. The nuclear receptor homologue Ftz-F1 and the homeodomain protein Ftz are mutually dependent cofactors. Nature 385:548-552. [DOI] [PubMed] [Google Scholar]
- 23.Guthrie, C., and G. R. Fink (ed.). 1991. Methods in enzymology, vol. 194. Guide to yeast genetics and molecular biology. Academic Press, Inc., San Diego, Calif. [PubMed]
- 24.Harrington, L. A., and B. J. Andrews. 1996. Binding to the yeast Swi4,6-dependent cell cycle box, CACGAAA, is cell cycle regulated in vivo. Nucleic Acids Res. 24:558-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ho, Y., M. Costanzo, L. Moore, R. Kobayashi, and B. Andrews. 1999. Regulation of transcription at the Saccharomyces cerevisiae Start transition by Stb1, a Swi6-binding protein. Mol. Cell. Biol. 19:5267-5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho, Y., S. Mason, R. Kobayashi, M. Hoekstra, and B. Andrews. 1997. Role of the casein kinase I isoform, HRR25, and the cell cycle regulatory transcription factor, SBF, in the transcriptional response to DNA damage in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94:581-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horak, C. E., N. M. Luscombe, J. Qian, P. Bertone, S. Piccirillo, M. Gerstein, and M. Snyder. 2002. Complex transcriptional circuitry at the G1/S transition in Saccharomyces cerevisiae. Genes Dev. 16:3017-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang, D., J. Moffat, and B. Andrews. 2002. Dissection of a complex phenotype by functional genomics reveals roles for the yeast cyclin-dependent protein kinase Pho85 in stress adaptation and cell integrity. Mol. Cell. Biol. 22:5076-5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iyer, V. R., C. E. Horak, C. S. Scafe, D. Botstein, M. Snyder, and P. O. Brown. 2001. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409:533-538. [DOI] [PubMed] [Google Scholar]
- 30.Johnson, D., and R. Schneider-Broussard. 1998. Role of E2F in cell cycle control and cancer. Front. Biosci. 3:447-448. [DOI] [PubMed] [Google Scholar]
- 31.Kasten, M. M., and D. J. Stillman. 1997. Identification of the Saccharomyces cerevisiae genes STB1-STB5 encoding Sin3p binding proteins. Mol. Gen. Genet. 256:376-386. [DOI] [PubMed] [Google Scholar]
- 32.Koch, C., T. Moll, M. Neuberg, H. Ahorn, and K. Nasmyth. 1993. A role for the transcription factors Mbp1 and Swi4 in progression from G1 to S phase. Science 261:1551-1557. [DOI] [PubMed] [Google Scholar]
- 33.Koch, C., A. Schleiffer, G. Ammerer, and K. Nasmyth. 1996. Switching transcription on and off during the yeast cell cycle: Cln/Cdc28 kinases activate bound transcription factor SBF (Swi4/Swi6) at Start, whereas Clb/Cdc28 kinases displace it from the promoter in G2. Genes Dev. 10:129-141. [DOI] [PubMed] [Google Scholar]
- 34.Lee, T. I., N. J. Rinaldi, F. Robert, D. T. Odom, Z. Bar-Joseph, G. K. Gerber, N. M. Hannett, C. T. Harbison, C. M. Thompson, I. Simon, J. Zeitlinger, E. G. Jennings, H. L. Murray, B. Gordon, B. Ren, J. J. Wyrick, J. Tagne, T. L. Volkert, E. Fraenkel, D. K. Gifford, and R. A. Young. 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298:799-804. [DOI] [PubMed] [Google Scholar]
- 35.Longtine, M. S., A. McKenzie, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 36.Macpherson, N., V. Measday, L. Moore, and B. Andrews. 2000. A taf17 mutant requires the Swi6 transcriptional activator for viability and shows defects in cell cycle-regulated transcription. Genetics 154:1561-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Madden, K., Y.-J. Sheu, K. Baetz, B. Andrews, and M. Snyder. 1997. SBF cell cycle regulator as a target of the yeast PKC-MAP kinase pathway. Science 275:1781-1784. [DOI] [PubMed] [Google Scholar]
- 38.Measday, V., L. Moore, J. Ogas, M. Tyers, and B. Andrews. 1994. The PCL2 (ORFD)-PHO85 cyclin-dependent kinase complex: a cell cycle regulator in yeast. Science 266:1391-1395. [DOI] [PubMed] [Google Scholar]
- 39.Measday, V., L. Moore, R. Retnakaran, J. Lee, M. Donoviel, A. Neiman, and B. Andrews. 1997. A family of cyclin-like proteins that interact with the Pho85 cyclin-dependent kinase. Mol. Cell. Biol. 17:1212-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mumberg, D., R. Muller, and M. Funk. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:119-122. [DOI] [PubMed] [Google Scholar]
- 41.Nash, P., X. Tang, S. Orlicky, Q. Chen, F. B. Gertler, M. D. Mendenhall, F. Sicheri, T. Pawson, and M. Tyers. 2001. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nat. Cell Biol. 414:514-521. [DOI] [PubMed] [Google Scholar]
- 42.Nasmyth, K. 1996. At the heart of the budding yeast cell cycle. Trends Genet. 12:405-412. [DOI] [PubMed] [Google Scholar]
- 43.Nasmyth, K., and L. Dirick. 1991. The role of SWI4 and SWI6 in the activity of G1 cyclins in yeast. Cell 66:995-1013. [DOI] [PubMed] [Google Scholar]
- 44.Partridge, J. F., G. E. Mikesell, and L. L. Breeden. 1997. Cell cycle-dependent transcription of CLN1 involves Swi4 binding to MCB-like elements. J. Biol. Chem. 272:9071-9077. [DOI] [PubMed] [Google Scholar]
- 45.Primig, M., S. Sockanathan, H. Auer, and K. Nasmyth. 1992. Anatomy of a transcription factor important for the start of the cell cycle in S. cerevisiae. Nature 358:593-597. [DOI] [PubMed] [Google Scholar]
- 46.Sedgwick, S. G., I. A. Taylor, A. C. Adam, A. Spanos, S. Howell, B. A. Morgan, M. K. Treiber, N. Kanuga, G. R. Banks, R. Foord, and S. J. Smerdon. 1998. Structural and functional architecture of the yeast cell-cycle transcription factor Swi6. J. Mol. Biol. 281:763-775. [DOI] [PubMed] [Google Scholar]
- 47.Siegmund, R. F., and K. Nasmyth. 1996. The Saccharomyces cerevisiae Start-specific transcription factor Swi4 interacts through the ankyrin repeats with the mitotic Clb2/Cdc28 kinase and through its conserved carboxy terminus with Swi6. Mol. Cell. Biol. 16:2647-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 12:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simon, I., J. Barnett, N. Hannett, C. T. Harbison, N. J. Rinaldi, T. L. Volkert, J. J. Wyrick, J. Zeitlinger, D. K. Gifford, T. S. Jaakkola, and R. A. Young. 2001. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell 106:697-708. [DOI] [PubMed] [Google Scholar]
- 50.Skowyra, D., K. L. Craig, M. Tyers, S. J. Elledge, and J. W. Harper. 1997. F-box proteins function as receptors to recruit phosphorylated substrates to E3 ubiquitin-ligase complexes. Cell 91:209-219. [DOI] [PubMed] [Google Scholar]
- 51.Spellman, P. T., G. Sherlock, M. Q. Zhang, R. I. Vishwanath, K. Anders, M. B. Eisen, P. O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9:3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stuart, D., and C. Wittenberg. 1994. Cell cycle-dependent transcription of CLN2 is conferred by multiple distinct cis-acting regulatory elements. Mol. Cell. Biol. 14:4788-4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stuart, D., and C. Wittenberg. 1995. CLN3, not positive feedback, determines the timing of CLN2 transcription in cycling cells. Genes Dev. 9:2780-2794. [DOI] [PubMed] [Google Scholar]
- 54.Tyers, M., and P. Jorgensen. 2000. The cell cycle, p. 58-105. In P. Fantes and J. Beggs (ed.), The yeast nucleus: frontiers in molecular biology. Oxford University Press, Oxford, United Kingdom.
- 55.Tyers, M., G. Tokiwa, and B. Futcher. 1993. Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J. 12:1955-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Verma, R., J. Smiley, B. Andrews, and J. Campbell. 1992. Regulation of the yeast DNA replication genes through the MluI cell cycle box is dependent on SWI6. Proc. Natl. Acad. Sci. USA 89:9479-9483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, P. J., A. Chabes, R. Casagrande, X. C. Tian, L. Thelander, and T. C. Huffaker. 1997. Rnr4p, a novel ribonucleotide reductase small-subunit protein. Mol. Cell. Biol. 17:6114-6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wijnen, H., and B. Futcher. 1999. Genetic analysis of the shared role of CLN3 and BCK2 at the G1-S transition in Saccharomyces cerevisiae. Genetics 153:1131-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wijnen, H., A. Landman, and B. Futcher. 2002. The G1 cyclin Cln3 promotes cell cycle entry via the transcription factor Swi6. Mol. Cell. Biol. 22:4402-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu, D., D. J. Fields, S.-J. Tang, A. Moris, B. P. Bobechko, and J. D. Friesen. 1998. Synthetic lethality of yeast slt mutations with U2 small nuclear RNA mutations suggests functional interactions between U2 and U5 snRNPs that are important for both steps of pre-mRNA splicing. Mol. Cell. Biol. 18:2055-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao, X., E. G. D. Muller, and R. Rothstein. 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2:329-340. [DOI] [PubMed] [Google Scholar]