Abstract
C/EBP family members contribute to the induction of the interleukin-12 p40 gene and the genes encoding several other mediators of inflammation. Here, we show by chromatin immunoprecipitation that C/EBPβ binds the p40 promoter following lipopolysaccharide stimulation of peritoneal macrophages. However, three modes of C/EBPβ regulation reported in other cell types were not detected, including alternative translation initiation, nuclear translocation, and increased DNA binding following posttranslational modification. In contrast, C/EBPβ concentrations greatly increased following stimulation via MAP kinase-dependent induction of C/EBPβ gene transcription. Increased C/EBPβ concentrations were unimportant for p40 induction, however, as transcription of the p40 gene initiated before C/EBPβ concentrations increased. Furthermore, disruption of C/EBPβ upregulation by a MAP kinase inhibitor only slightly diminished p40 induction. Phosphopeptide mapping revealed that endogenous C/EBPβ in macrophages is phosphorylated on only a single tryptic peptide containing 14 potential phosphoacceptors. This peptide was constitutively phosphorylated in primary and transformed macrophages, in contrast to its inducible phosphorylation in other cell types in response to Ras and growth hormone signaling. Altered-specificity experiments supported the hypothesis that C/EBPβ activity in macrophages does not require an inducible posttranslational modification. These findings suggest that, although C/EBPβ contributes to the induction of numerous proinflammatory genes, it is fully active in unstimulated macrophages and poised to stimulate transcription in conjunction with other factors whose activities are induced.
The important role of interleukin-12 (IL-12) in the development of T helper type 1 (Th1) cells is well established (23, 62, 63). Th1 cells that develop in an IL-12-containing milieu produce cytokines that promote macrophage activation and cytotoxic-T-cell maturation, which contribute to the generation of an effective cell-mediated response against intracellular pathogens (62). Mice that lack genes encoding IL-12 subunits and mice that have been treated with neutralizing IL-12 antibodies succumb to unusually low doses of intracellular pathogens (23, 64).
Transcription of the gene encoding the p40 subunit of the heterodimeric IL-12 cytokine is induced in macrophages, dendritic cells, and B cells following activation with bacterial products and intracellular parasites. Prior to induction, the murine IL-12 p40 promoter is contained within a positioned nucleosome, which is rapidly and selectively remodeled upon cell activation (66). Efficient activity of the p40 promoter requires multiple control elements (36, 41, 46, 75). In the murine promoter, the most important control elements identified in transient-transfection assays bind the Rel and C/EBP families of transcription factors. A p50/c-Rel heterodimer is likely to stimulate p40 transcription through the Rel site (51), and a C/EBPβ-containing complex appears to activate transcription through the C/EBP site. The latter hypothesis is supported by the presence of an abundant gel shift complex containing C/EBPβ in assays performed with a DNA probe containing the p40 C/EBP site and extracts from lipopolysaccharide (LPS)-stimulated macrophages (46). Nevertheless, peritoneal macrophages and an immortalized macrophage line from C/EBPβ−/− mice (24, 58) continue to produce IL-12 in response to LPS. Most likely, a closely related C/EBP protein, C/EBPδ, compensates for the C/EBPβ deficiency.
C/EBPβ is a member of the CCAAT/enhancer-binding protein (C/EBP) family of bZIP transcription factors that binds DNA as dimers. C/EBPβ was independently discovered by several groups and has also been named LAP, NF-IL-6, IL-6-DBP, AGP/EBP, NF-M, and CRP2 (2, 11, 14, 18, 28, 47, 68). Three C/EBPβ isoforms (called Full, LAP, and LIP in this study) (19) are translated from a single mRNA species through the alternative use of three translation initiation codons within the same open reading frame. The three proteins share 145 C-terminal residues that contain the basic DNA-binding domain and leucine zipper dimerization helix. The full-length protein and LAP contain 151 and 130 additional amino acids at the N terminus, respectively, and function as transcriptional activators. The LIP protein, which contains only the 145 C-terminal residues and lacks a transactivation domain, is believed to function as a transcriptional repressor in a dominant-negative fashion (19).
In macrophages, C/EBPβ is involved in the inducible expression of several genes that are important for inflammation and immunity, including the CD14, IL-12 p40, IL-1β, IL-6, tumor necrosis factor alpha, IL-8, lactoferrin, MCP-1, macrophage colony-stimulating factor, granulocyte colony-stimulating factor, mim1, myeloperoxidase, lysozyme, and nitric oxide synthase genes (48, 49). However, despite the well-documented importance of C/EBPβ in macrophages, most studies of its regulation have been performed in hepatocytes and adipocytes, where it is an important regulator of differentiation, proliferation, and the acute-phase response. Whether C/EBPβ activity is regulated in a similar manner in macrophages has not been fully investigated.
From studies performed with hepatocytes, adipocytes, and other cell types, a number of different transcriptional, posttranscriptional, translational, and posttranslational mechanisms have been proposed to regulate C/EBPβ activity. The potential mode of regulation that has been reported most frequently is increased C/EBPβ protein concentrations, usually due to an induction of C/EBPβ gene transcription (2, 3, 11, 12, 13, 16, 29, 34, 37, 38, 54, 70, 72, 74). Regulation by nuclear translocation (15, 28, 40, 73) and alternative translation initiation (4, 19) have also been reported. Furthermore, phosphorylation has been proposed to regulate C/EBPβ activity. Cyclic AMP-dependent kinase (40), retrovirus-derived oncogenic kinases (28, 57), mitogen-activated protein (MAP) kinase (42, 76), protein kinase C (60), protein kinase A (61), Ca2+/calmodulin-dependent kinase (65), and p90 ribosomal S kinase (RSK) (8) are among the kinases that have been shown to mediate phosphorylation of recombinant or overexpressed C/EBPβ. However, the only kinase that has been shown to phosphorylate endogenous murine C/EBPβ is p90 RSK (8). Finally, it has been suggested that C/EBPβ activation involves derepression of its transcriptional activation and DNA-binding domains, possibly in response to phosphorylation (31, 69).
In striking contrast to these numerous modes of regulation, Hu et al. (26) showed that constitutive expression of the bZIP domain of C/EBPβ is sufficient for LPS induction of the endogenous IL-6 and MCP-1 genes in a B-lymphoblastic line that fails to express endogenous C/EBP proteins. This led to the hypothesis that a specific C/EBPβ induction mechanism may not be required for the rapid activation of some endogenous target genes. It is noteworthy that, in addition to its functions as a transcription factor, C/EBPβ plays an important antiapoptotic role in hepatic stellate cells. Buck et al. (9) showed that phosphorylation of endogenous murine C/EBPβ by RSK on threonine 217 (T217) allows it to associate with procaspases 1 and 8, thereby inhibiting their processing and blocking the apoptotic cascade. Consistent with these results, C/EBPβ-deficient stellate cells undergo enhanced programmed cell death.
In this study, we investigated the mechanisms responsible for regulating C/EBPβ activity in LPS-stimulated macrophages, with an emphasis on the induction of IL-12 p40 transcription. Because almost all of the studies cited above relied on overexpression, we attempted, whenever possible, to analyze properties of endogenous C/EBPβ. The results reveal that endogenous C/EBPβ is phosphorylated and that transcription of the C/EBPβ gene is strongly upregulated in LPS-stimulated macrophages. However, the phosphorylation is constitutive and the transcriptional upregulation occurs too slowly to contribute to the initial induction of IL-12 p40 transcription. Together, these and other results suggest that the signaling pathways that allow C/EBPβ to contribute to IL-12 p40 induction may not target C/EBPβ itself. Rather, C/EBPβ appears to be fully functional in unstimulated macrophages, allowing it to contribute to inducible transcription via physical or functional interactions with coactivators and other DNA-binding proteins that are available and/or active only following LPS stimulation.
MATERIALS AND METHODS
Plasmids.
To generate the C/EBPβ promoter-reporter, a C/EBPβ promoter fragment (−121 to +15) was amplified by PCR from mouse genomic DNA using upstream and downstream primers containing KpnI and BglII sites, respectively. The product was inserted into a pCAT basic vector (Promega) that had been modified to contain KpnI and BglII sites in the polylinker. The reporter plasmid containing the C/EBP site multimer was a gift from T. Ronni (University of California, Los Angeles [UCLA]). The altered-specificity p40 promoter was generated by cleaving the −350 p40 promoter-chloramphenicol acetyltransferase (CAT) plasmid (46) with SpeI, which excised a 25-bp fragment containing the C/EBP site, followed by insertion of an oligonucleotide containing a recognition site for a bZIP protein found in Arabidopsis thaliana, GBF (44, 55). The wild-type and altered-specificity promoters were also placed into the pCAT enhancer vector (Promega) using PstI and XbaI sites. The expression plasmid for the altered-specificity protein was constructed by ligating the region encoding the N-terminal portion of LAP (residues 22 to 223) with that encoding the GBF bZIP region (residues 222 to 315; the GBF expression plasmid was a generous gift from Karambir Singh). Specifically, the LAP region was amplified using upstream and downstream primers containing XhoI and SacI sites, respectively. The GBF region was amplified using upstream and downstream primers containing SacI and ApaI sites, respectively. The products were ligated into the pcDNA3 expression vector (Invitrogen) cleaved with XhoI and ApaI.
Reagents and antibodies.
SB203580 was purchased from Calbiochem, the Phospho-CREB antibody was purchased from Upstate Biotechnology (no. 06-519), the CREB antibody was purchased from New England BioLabs (no. 9192), and the C/EBPβ antibody was purchased from Santa Cruz Biotechnology (no. sc-150x). Polyclonal antisera against a purified glutathione-S-transferase (GST)-C/EBPβ fusion protein (residues 22 to 195) were raised in rabbits by Animal Pharm (Healdsburg, Calif.).
Cell lines and macrophages.
The RAW264.7 and J774 murine macrophage cell lines (American Type Culture Collection) were maintained in Dulbecco modified Eagle medium (Gibco) supplemented with 10% low-endotoxin fetal bovine serum (Omega Scientific). The cells were activated as required with 10 μg of LPS (Sigma)/ml. C57BL/6 or C57BL/6-C3H mice were injected intraperitoneally with 1.5 ml of thioglycolate (Becton Dickinson). After 3 to 5 days, peritoneal exudate cells were isolated. Red blood cells were lysed by incubation with ACK lysis buffer (0.15 M NH4Cl, 1 mM KHCO3, 0.1 mM EDTA, pH 7.3) on ice for 10 min. The cells were washed twice with phosphate-buffered saline (PBS), resuspended in medium, counted, and plated. The macrophages were allowed to adhere to tissue culture plates, and nonadherent cells were removed by washing the plates with PBS. The cells were activated with 10 U of recombinant murine gamma interferon (IFN-γ) (PharMingen)/ml and 10 μg of LPS/ml. C/EBPβ−/− mice were a generous gift of Valeria Poli (52), Xiao-Hong Sun, and Gretchen Darlington.
ELISA.
IL-12 p40 concentrations were measured by sandwich enzyme-linked immunosorbent assay (ELISA) using 3.5 μg of purified rat anti-mouse IL-12 p40/p70 (PharMingen)/ml, 200 ng of biotin anti-mouse IL-12 p40/p70 (PharMingen)/ml, and a 1:5,000 dilution of poly-HRP80-streptavidin (Research Diagnostics). Recombinant murine IL-12 was used as the standard (R&D Systems). ELISAs were developed using the ABTS microwell peroxidase substrate system (Kirkegaard and Perry Laboratories).
Transient transfections and CAT assays.
RAW264.7 cells were transfected using the SuperFect transfection reagent (Qiagen) as previously described (7). Cell extracts were prepared using cell lysis buffer (Promega), and total protein was measured using the Coomassie protein assay reagent (Pierce). Fifty micrograms of total protein was used for each CAT assay, in accordance with the Promega protocol.
Cell extracts and gel shift assays.
Cytoplasmic and nuclear extracts were prepared by a modification of the method of Dignam et al. (21). Briefly, cells were washed two times with cold buffer A (10 mM HEPES, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 1 μM pepstatin A, 1 μg of aprotinin/ml, 4 μg of leupeptin/ml, 1 mM NaF, 10 mM sodium vanadate, 50 mM β-glycerol) and were then resuspended in buffer A plus 0.1% NP-40 (two to three pellet volumes) and lysed on ice for 2.5 min. The nuclei were pelleted by centrifugation, and the supernatant was saved (cytoplasmic extract). The nuclei were resuspended in cold buffer C (20 mM HEPES, pH 7.9, 420 mM NaCl, 1.5 mM MgCl2, 25% glycerol, and the protease and phosphatase inhibitors mentioned above [one to two pellet volumes]) and were rocked at 4°C for 20 min. After centrifugation, the supernatant was saved (nuclear extract). Whole-cell extracts were prepared by boiling the cells for 5 min in lysis buffer (2.86% sodium dodecyl sulfate [SDS], 285 mM Tris, pH 6.8). Protein concentrations were determined using the Micro BCA protein assay reagent kit (Pierce). C/EBPβ was prepared by in vitro transcription-translation using the TNT T7 coupled reticulocyte lysate system (Promega).
The gel shift probe was prepared by 5′-end labeling a double-stranded oligonucleotide containing the p40 C/EBP site (5′-GACACTAGTTTTCAGTGTTGCAATTGAGACTAGTCAGTTTCT-3′). Gel shift assays were conducted as described previously (46). C/EBPα, -β, -δ, and -ɛ antibodies for supershift experiments were from Santa Cruz Biotechnology (no. sc-61, sc-150, sc-151, and sc-158).
Primer extension.
J774 cells (107) or primary peritoneal macrophages (1.5 × 107) were lysed on ice for 5 min in 350 μl of NP-40 lysis buffer (150 mM NaCl, 10 mM Tris [pH 7.8], 0.65% NP-40). The cytoplasmic extract was combined with 400 μl of Bonner RNA extraction buffer (7 M urea, 0.35 M NaCl, 10 mM Tris [pH 7.6], 10 mM EDTA, 1% SDS), followed by two phenol-chloroform (pH 7.0) extractions, one chloroform extraction, and ethanol precipitation. Total RNA (10 to 30 μg) was hybridized for 90 min with a 32P-labeled IL-12 p40-specific primer (5′-TTACCTTGCTTTGCTGCGAGCTGC-3′) at 60°C or a C/EBPβ-specific primer (5′-CAGCAGGCGGTGCATGAACGCG-3′) at 45°C. Following reverse transcription using SuperScript II reverse transcriptase (Gibco BRL), the DNA products were separated on an 8% denaturing polyacrylamide gel.
Phosphopeptide mapping.
J774 or RAW264.7 cells (107) or thioglycolate-elicited peritoneal macrophages (5 × 107) were incubated in phosphate-free Dulbecco modified Eagle medium (Gibco) supplemented with 10% dialyzed fetal bovine serum (Omega Scientific) and 1.0 to 2.5 mCi of [32P]orthophosphate/ml for 4 h. During the last 5 to 60 min of incubation, 10 U of IFN-γ/ml and/or 10 μg of LPS/ml was added as appropriate. The cells were resuspended in SDS buffer (50 mM Tris [pH 6.8], 1% SDS, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 1 μM pepstatin A, 1 μg of aprotinin/ml, 4 μg of leupeptin/ml, 1 mM NaF, 10 mM sodium vanadate, 50 mM β-glycerol) and were lysed by being boiled for 5 min. The extract was diluted 10-fold with SDS-free RIPA buffer (150 mM NaCl, 50 mM NaH2PO4 [pH 7.5], 0.025% deoxycholate, 1% NP-40, and the protease and phosphatase inhibitors mentioned above). C/EBPβ was then immunoprecipitated and separated by SDS-polyacrylamide gel electrophoresis (PAGE). After the immunoprecipitate was transferred to nitrocellulose, in vivo-labeled LAP and/or LIP was excised from the membrane and digested with trypsin (Worthington). The digested peptides were washed, spotted on a thin-layer chromatography plate (EM Science), and separated by thin-layer electrophoresis in the first dimension followed by chromatography in the second dimension (6).
ChIP assay.
Unactivated and activated cells were fixed at room temperature for 10 min by adding formaldehyde directly to the culture medium to a final concentration of 1%. The reaction was stopped by adding glycine at a final concentration of 0.125 M for 5 min at room temperature. After three ice-cold PBS washes, the cells were collected and lysed for 10 min on ice in cell lysis buffer {5 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid) [pH 8.0], 85 mM KCl, 0.5% NP-40, protease inhibitors}. The nuclei were resuspended in nucleus lysis buffer (50 mM Tris-HCl [pH 8.1], 10 mM EDTA, 1% SDS, protease inhibitors) and incubated on ice for 10 min. Chromatin was sheared into 500- to 1,000-bp fragments by sonication and was then precleared with protein A-Sepharose beads. The purified chromatin was diluted with chromatin immunoprecipitation (ChIP) dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl [pH 8.1], 167 mM NaCl, protease inhibitors) and immunoprecipitated overnight at 4°C using 10 μg of anti-C/EBPβ antibody (no. sc-150x; Santa Cruz), or 5 μl of anti-GST rabbit immune serum. Immune complexes were collected with protein A-Sepharose beads and were then washed and eluted. After protein-DNA cross-linking was reversed and the DNA was purified, the presence of selected DNA sequences was assessed by PCR. The primer pairs were as follows: IL-12 primers 2 (5′-GAGTTAGCGACAGGGAAGAGGAGAG-3′) and 53 (5′-CCTGGGATTTCGACGTCTATATTCCCTCTGT-3′) and 5TdT intron1 (5′-GAGGACATCCCCAAGGGACCCAAG-3′) and 3TdT intron2 (5′-CACCCTCCCTGCTCCAAAGTAGCA-3′). The PCR conditions were as follows: 94°C for 180 s; 29 cycles at 94°C for 30 s, 64°C for 30 s, and 72°C for 60 s; and final elongation at 72°C for 10 min. The PCR products were resolved on a 2% agarose gel.
Reverse transcription-PCR of primary transcripts and mRNAs.
RNA was extracted from unactivated and activated peritoneal macrophages using TRI-reagent (MRC). RNA was then treated with RNase-free DNase I and purified by phenol-chloroform extraction, followed by ethanol precipitation. For reverse transcription, the following primers were used: IL-12 p40, 5′-CTTGATGTTGAACTTCAAGTCC-3′ (complementary to exon 4), and GAPDH (glyceraldehyde-3-phosphate dehydrogenase), 5′-ATTTCTCGTGGTTCACAC-3′. cDNAs were subjected to PCR using the following primers: IL-12 p40, 5′-CGTGCACTGAGGCTCAGAAATGTTTC-3′ (intron 3), and 5′-TTTCTTTGCACCAGCCATGAGC-3′ (exon 4); and GAPDH, 5′-TCCAGTATGACTCCACTC-3′ and the reverse transcription primer. The PCR conditions were as follows: IL-12 p40, 94°C for 180 s; 34 cycles of 94°C for 60 s, 64°C for 60 s, and 72°C for 60 s; final elongation at 72°C for 10 min; and GAPDH, 94°C for 180 s; 19 cycles of 94°C for 60 s, 55°C for 60 s, and 72°C for 60 s; final elongation at 72°C for 10 min. The PCR products were resolved on a 1% agarose gel (50).
RESULTS
C/EBPβ associates with the endogenous IL-12 p40 promoter in LPS-stimulated macrophages.
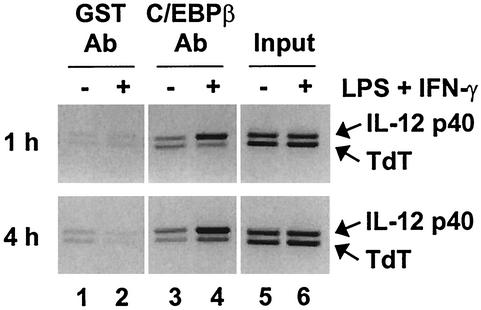
Despite previous evidence that C/EBPβ may contribute to the induction of the IL-12 p40 promoter (46), C/EBPβ−/− mice and cell lines have been reported to secrete normal levels of IL-12 (24, 58). Normal IL-12 p40 expression was also observed in peritoneal macrophages from C/EBPβ−/− mice using our LPS-plus-IFN-γ stimulation conditions (data not shown). A likely explanation for these findings is that other members of the C/EBP family, including C/EBPδ, may be redundant with or compensate for the loss of C/EBPβ. To determine whether C/EBPβ directly binds the endogenous IL-12 p40 promoter, ChIP assays were performed with peritoneal macrophages. When C/EBPβ antibodies were used, the p40 PCR product was greatly enriched in stimulated cells relative to the control product (derived from the T-cell-specific tdt gene) (Fig. 1). The p40 product was not enriched when a GST control antiserum was used. These results demonstrate that C/EBPβ associates with the endogenous p40 promoter after stimulation with LPS plus IFN-γ. The inducible binding of C/EBPβ is consistent with genomic-footprint results, which demonstrated that the C/EBP site in the endogenous p40 promoter is efficiently occupied only in stimulated cells (66).
FIG. 1.
C/EBPβ associates with the IL-12 p40 promoter in LPS-stimulated macrophages. ChIP assays were performed with cross-linked chromatin from wild-type peritoneal macrophages that were unstimulated (−) (lanes 1, 3, and 5) or stimulated (+) with LPS plus IFN-γ for 1 or 4 h (lanes 2, 4, and 6). Immunoprecipitations were performed with GST control antibodies (Ab) (lanes 1 and 2) or C/EBPβ antibodies (lanes 3 and 4). Precipitation of IL-12 p40 and TdT promoter fragments was monitored by PCR. Input samples are in lanes 5 and 6.
C/EBPβ activity may be regulated by transcriptional upregulation but not by alternative translation initiation, nuclear translocation, or inducible DNA binding.
As described above, numerous studies have suggested that C/EBPβ activity in multiple cell types, including LPS-stimulated macrophages, is regulated by transcriptional induction of the C/EBPβ gene (2, 3, 11, 12, 13, 16, 29, 34, 37, 38, 54, 70, 72, 74). Other studies suggested that C/EBPβ activity may be induced by alternative translation initiation, which could lead to increased ratios of functional and nonfunctional isoforms (4, 19), by rapid nuclear translocation (15, 28, 40, 73), or by posttranslational modifications that induce DNA-binding activity (8, 15, 28, 31, 40, 42, 57, 60, 65, 69, 76).
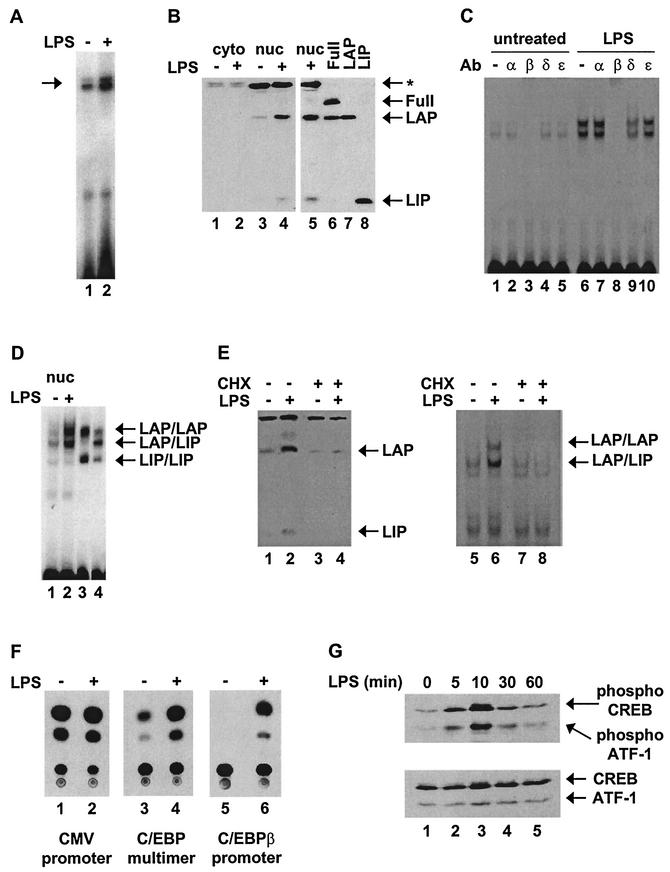
The results shown in Fig. 2 demonstrate that C/EBPβ gene transcription is induced by LPS in the J774 macrophage cell line, but regulation by the other three mechanisms was not observed. J774 cells were suitable for these initial studies because they produce substantial quantities of IL-12 p40 mRNA and protein following LPS stimulation (66). First, C/EBPβ mRNA levels were monitored by primer extension using a radiolabeled primer complementary to a sequence near the 5′ end of the gene. The results revealed an increase in C/EBPβ mRNA 4 h after activation with LPS (Fig. 2A). Western blot analyses revealed two C/EBPβ isoforms in J774 nuclear extracts (Fig. 2B, lanes 3 and 4). By comparison with in vitro-translated C/EBPβ isoforms (Fig. 2B, lanes 6 to 8), the isoforms in J774 cells were found to correspond to LAP and LIP, with the Full isoform undetectable. Titrations of multiple unactivated and activated extracts revealed that the concentrations of LAP increased two- to fivefold by 4 h postactivation (data not shown).
FIG.2.
C/EBPβ protein and DNA-binding activity are upregulated in LPS-stimulated J774 cells. (A) C/EBPβ mRNA was detected by primer extension using 30 μg of total RNA from unstimulated (−) or LPS-stimulated (4 h) (+) J774 cells. (B) C/EBPβ isoforms Full, LAP, and LIP were detected by Western blotting using 60 μg of cytoplasmic (cyto) and nuclear (nuc) extracts from unstimulated or LPS-stimulated (4 h) J774 cells (lanes 1 to 5). Also shown are in vitro-translated C/EBPβ isoforms (lanes 6 to 8). The arrows indicate the locations of LAP, LIP, and a nonspecific protein recognized by the antibody (*). (C) Gel shift and antibody (Ab) supershift assays were performed with a radiolabeled probe containing the C/EBP site from the IL-12 p40 promoter and nuclear extracts from unstimulated or LPS-stimulated (4 h) J774 cells. Binding reactions were performed with no antibody or 2 μg of antibody against C/EBPα, -β, -δ, or -ɛ. (D) A gel shift assay was performed with nuclear extracts from unstimulated or LPS-stimulated (4 h) J774 cells (lanes 1 and 2). As markers, the LAP and LIP isoforms were translated in vitro separately to allow formation of LAP/LAP and LIP/LIP homodimers and were then mixed (lane 3). LAP and LIP were also cotranslated to allow the formation of LAP/LIP heterodimers, as indicated (lane 4). (E) Western blot and gel shift assays were performed with nuclear extracts from unstimulated or LPS-stimulated (4 h) J774 cells. CHX was added (+) to the cells where indicated. (F) RAW264.7 cells were transfected with a CAT reporter plasmid driven by either a constitutive cytomegalovirus (CMV) promoter (lanes 1 and 2), a trimer of the IL-12 p40 C/EBP site upstream of a TATA-Inr core promoter (lanes 3 and 4), or a C/EBPβ promoter (lanes 5 and 6). Transfected cells were left unstimulated or were stimulated with LPS for 24 h. The cells were then harvested, and CAT assays were performed. (G) Whole-cell extracts (100 μg) from unstimulated or LPS-stimulated J774 cells were analyzed by Western blotting using antibodies that recognize phosphorylated CREB (top) or total CREB (bottom).
The Western blot data suggest that C/EBPβ activity is not regulated by alternative translation initiation in J774 cells, because the concentration of LAP substantially exceeded that of LIP both before and after stimulation (Fig. 2B, lanes 3 and 4). The Western blot data also suggest that C/EBPβ activity is not regulated by nuclear translocation, as the C/EBPβ protein was undetectable in cytoplasmic fractions both before and after activation (Fig. 2B, lanes 1 and 2). Confocal immunofluorescence showed that C/EBPβ is restricted to the nucleus before and after stimulation (data not shown). The conclusion that nuclear translocation does not occur is supported by evidence that nuclear LAP concentrations increased with much slower kinetics than reported in the nuclear translocation studies performed with other cell types (see below) (40, 28, 73, 15). Furthermore, LAP concentrations increased by similar magnitudes and with similar kinetics in nuclear extracts and in cells boiled directly in SDS sample buffer (data not shown), suggesting that LAP was not sequestered in membrane fractions in unactivated cells.
Consistent with the Western blot data, two gel shift complexes observed with a probe containing the C/EBP site from the IL-12 p40 promoter were significantly enhanced (two- to fivefold in multiple experiments) in extracts from LPS-activated J774 cells (Fig. 2C, lanes 1 and 6). Because the magnitudes of the increases in the gel shift and Western blot experiments were comparable, the results strongly suggest that the increased DNA-binding activity does not require an inducible posttranslational modification. Preincubation of the extracts from activated cells with C/EBPβ antibodies abolished both gel shift complexes (Fig. 2C, lane 8), suggesting that both contain C/EBPβ isoforms. C/EBPα and C/EBPɛ antibodies had no effect on complex formation (lanes 7 and 10), whereas C/EBPδ antibodies diminished both bands to a moderate extent (lane 9). Because the C/EBPδ antibody did not cross-react with recombinant C/EBPβ (data not shown), both bands may contain some C/EBPβ-C/EBPδ heterodimers.
To identify the C/EBPβ isoform(s) within each gel shift band, the complexes obtained with J774 nuclear extracts were analyzed adjacent to complexes obtained with in vitro-translated LAP and LIP (Fig. 2D). Lane 3 shows the LAP and LIP homodimeric complexes obtained when LAP and LIP were translated separately and then mixed. Lane 4 shows the predominant LAP-LIP heterodimer obtained when LAP and LIP were cotranslated. By comparison with the complexes obtained when using J774 nuclear extracts, one can conclude that the J774 upper band (Fig. 2D, lanes 1 and 2) corresponds primarily to LAP homodimers. Although the migration of the lower band corresponds to that of a LAP/LIP heterodimer, the high abundance of this complex relative to the low abundance of LIP in nuclear extracts is difficult to explain, leaving open the possibility that the lower complex in both unstimulated and stimulated cells contains a heterodimer between LAP and an unidentified protein.
To determine whether the increase in protein levels observed in stimulated macrophages (Fig. 2B) was truly responsible for the enhancement of C/EBPβ DNA-binding activity (Fig. 2C), as opposed to a posttranslational modification, experiments were conducted in the presence of cycloheximide (CHX). CHX prevented the increase in C/EBPβ protein concentration normally seen in activated macrophages (Fig. 2E, lanes 1 to 4). In the absence of this increase, C/EBPβ binding activity did not increase (Fig. 2E, lanes 5 to 8).
Previous studies revealed that p40 promoter induction requires an intact C/EBP element (46). To study the activity of this element in isolation, three copies were inserted into a CAT reporter upstream of a minimal promoter containing consensus TATA and Inr elements. Transient-transfection experiments were performed in RAW264.7 macrophages because J774 cells could not be transfected at an efficiency that allowed detectable reporter activity. RAW264.7 cells produce less IL-12 p40 protein than J774 cells (66) but can be transfected at much higher efficiency and were used successfully for a previous analysis of the murine IL-12 p40 promoter (46). The validity of using RAW264.7 cells is supported by the fact that repetition of the experiments shown in Fig. 2B, C, D, and G with RAW264.7 cells yielded results that were very similar to those obtained with J774 cells (data not shown). In transiently transfected RAW264.7 cells, the activity of the reporter plasmid containing three C/EBP recognition sites was strongly enhanced following LPS stimulation (Fig. 2F, lanes 3 and 4), consistent with the upregulation of C/EBPβ mRNA, protein, and DNA-binding activity.
The increase in C/EBPβ mRNA upon LPS activation of macrophages suggests that transcription from the C/EBPβ gene may be induced. To examine this possibility further, the murine C/EBPβ promoter was isolated and inserted upstream of a CAT reporter gene. In transfected RAW264.7 cells, the activity of the resulting plasmid was strongly enhanced by LPS stimulation (Fig. 2F, lanes 5 and 6).
Previous studies demonstrated that C/EBPβ promoter activity is induced during monocyte and hepatocyte activation (5, 43). These studies revealed that the induction requires the binding of CREB to a critical element in the C/EBPβ promoter. The Western blot results in Fig. 2G, performed with phospho-CREB antibodies (which also interact with phospho-ATF-1 [top]), demonstrate that CREB is rapidly phosphorylated in LPS-stimulated J774 cells, consistent with a previous report (10). Taken together, the results in Fig. 2 are consistent with the hypothesis that C/EBPβ activity is enhanced in LPS-stimulated macrophages by the induction of C/EBPβ gene transcription, resulting in increased C/EBPβ mRNA, protein, and DNA-binding activity. The induction of C/EBPβ gene transcription in macrophages is likely to require a signal transduction pathway that results in CREB phosphorylation. Interestingly, the results suggest that, in macrophages, C/EBPβ is not regulated by alternative translation initiation, nuclear translocation, or a posttranslational modification that influences DNA-binding activity.
C/EBPβ upregulation does not contribute to the initial induction of p40 transcription.
Consistent with the results of several previous studies, the above data suggest that C/EBPβ activity is regulated by transcriptional induction of the C/EBPβ gene. However, the relevance of this regulatory mechanism for IL-12 p40 induction (or for the rapid induction of most other C/EBP target genes) has not been established. To address this issue, the time course of IL-12 p40 gene induction and C/EBPβ association with the endogenous locus were compared to the time course of C/EBPβ upregulation. As shown in Fig. 1, C/EBPβ association with the endogenous p40 locus was readily apparent 1 h after LPS stimulation of peritoneal macrophages. This result is consistent with genomic footprinting results, which showed that the C/EBP site is occupied 1 h after stimulation (66).
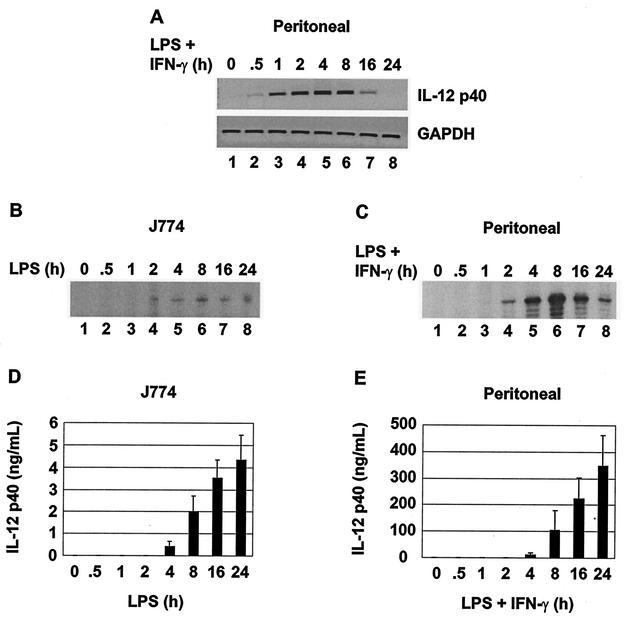
To determine the time course of IL-12 p40 transcription, p40 primary transcripts were first monitored by semiquantitative PCR, using primers complementary to intronic sequences. Because introns are usually rapidly excised and degraded, this method is thought to provide an approximate measure of transcription initiation frequency, similar to the information provided by nuclear run-on assays (33, 50). p40 primary transcripts were readily detected in peritoneal macrophages 0.5 h after LPS stimulation and reached a peak 2 to 4 h poststimulation (Fig. 3A). Mature p40 mRNA was detected by primer extension 2 h poststimulation in both peritoneal macrophages and J774 cells (Fig. 3B and C). IL-12 p40 protein was detected by ELISA 4 h after stimulation of both peritoneal macrophages and J774 cells (Fig. 3D and E). The results of these four assays support the conclusion that the induction of p40 transcription occurs rapidly and is accompanied by C/EBPβ binding to the p40 promoter.
FIG. 3.
Time course of C/EBPβ association and IL-12 p40 expression. (A) Primary p40 transcripts and GAPDH mRNA were monitored by intron PCR in unstimulated or LPS-plus-IFN-γ-stimulated peritoneal macrophages. (B) p40 mRNA was monitored by primer extension in LPS-stimulated J774 cells. (C) p40 mRNA was monitored by primer extension in LPS-plus-IFN-γ-stimulated peritoneal macrophages. (D) p40 protein was monitored by ELISA in supernatants from LPS-stimulated J774 cells. (E) p40 protein was monitored by ELISA in supernatants from LPS-plus-IFN-γ-stimulated peritoneal macrophages.
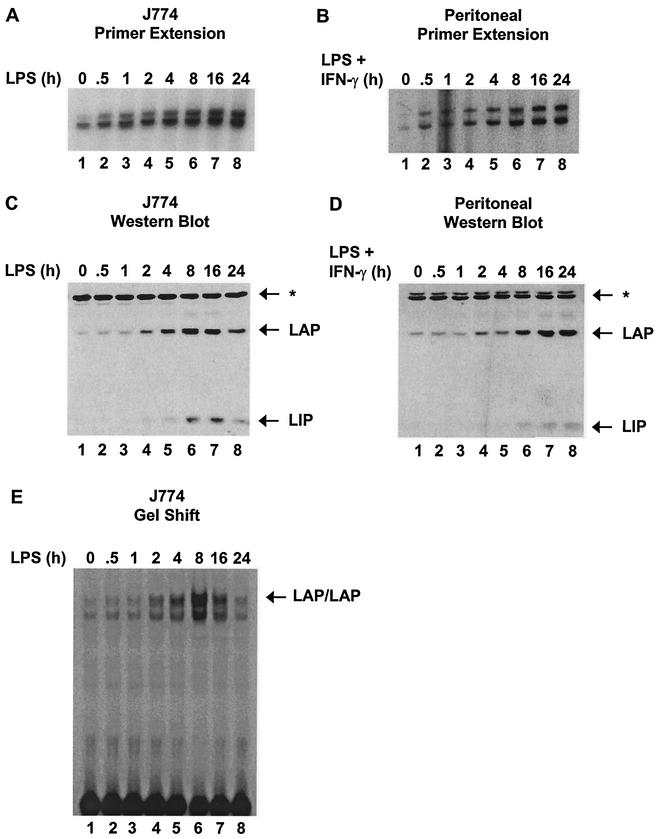
In contrast to the above-mentioned results, C/EBPβ mRNA and protein were readily detected in unstimulated J774 cells and peritoneal macrophages (Fig. 4A to D). C/EBPβ mRNA concentrations increased gradually beginning 0.5 h after stimulation, but protein concentrations did not begin to increase until 2 h poststimulation. C/EBPβ DNA-binding activity, monitored by gel shift in nuclear extracts from J774 cells, also failed to increase until 2 h poststimulation (Fig. 4E). The fact that C/EBPβ protein and DNA-binding activity have not increased 1 h after stimulation, when C/EBPβ is efficiently associated with the endogenous promoter and when active IL-12 p40 transcription is observed, strongly suggests that C/EBPβ upregulation is not involved in the initial induction of p40 transcription.
FIG. 4.
Time courses of C/EBPβ upregulation. (A) C/EBPβ mRNA from LPS-stimulated J774 cells was monitored by primer extension. (B) C/EBPβ mRNA from LPS-plus-IFN-γ-stimulated peritoneal macrophages was monitored by primer extension. (C) C/EBPβ isoforms (LAP and LIP) in nuclear extracts (60 μg) from LPS-stimulated J774 cells were monitored by Western blotting. A protein that cross-reacts with the antibody is indicated (*). (D) C/EBPβ isoforms (LAP and LIP) in nuclear extracts (60 μg) from LPS-plus-IFN-γ-stimulated peritoneal macrophages were monitored by Western blotting. (E) C/EBPβ DNA-binding activity was monitored by gel shift, using nuclear extracts (6 μg) from LPS-stimulated J774 cells.
To explore the significance of these results in greater depth, the number of C/EBPβ molecules per cell was determined by quantitative Western analysis of whole-cell extracts, using purified recombinant C/EBPβ as a standard (data not shown). The concentration of recombinant C/EBPβ was determined by comparing a dilution series with a titration of known amounts of bovine serum albumin on Coomassie-stained and silver-stained SDS gels. The results revealed that each unstimulated macrophage contains ∼250,000 C/EBPβ molecules, similar to the concentration of NF-κB p65 in murine macrophages and the concentration of Ikaros in murine thymocytes (data not shown).
The most reasonable hypothesis to explain these results is that the concentration of C/EBPβ in unstimulated cells is sufficient for the induction of IL-12 p40 transcription and for the inducible binding of C/EBPβ to the endogenous p40 promoter. It remains possible that the increased C/EBPβ protein concentrations observed 2 to 16 h after stimulation contribute to the frequency of p40 transcription initiation at these later times, although the initiation frequency appears to increase only slightly after the 1-h time point (Fig. 3A).
Role of p38 MAP kinase in C/EBPβ upregulation and IL-12 p40 induction.
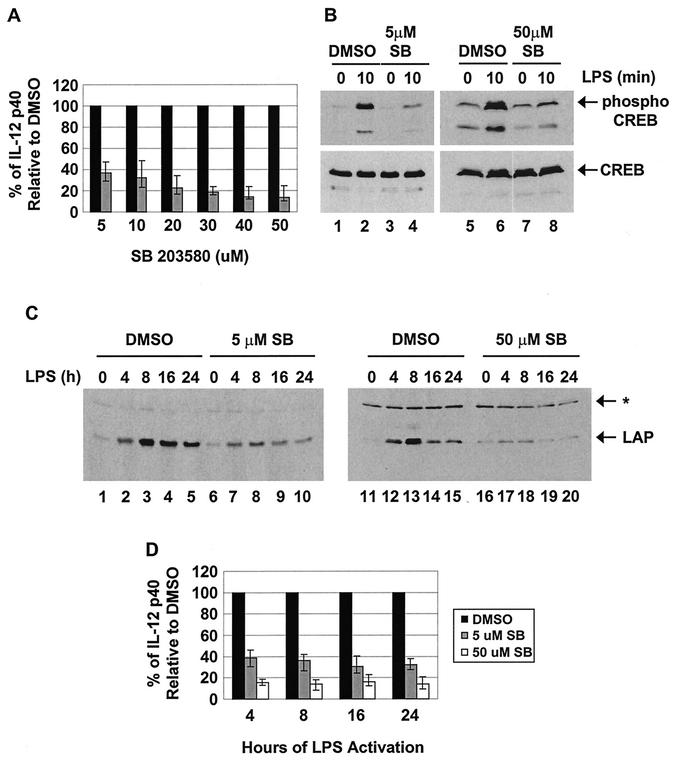
Previous studies demonstrated that the p38 MAP kinase pathway is essential for optimal expression of the IL-12 p40 gene. Specifically, p40 transcription was reduced in LPS-stimulated macrophages from Mkk3−/− mice (35) and in cell lines and primary macrophages treated with a specific inhibitor of the p38 pathway, SB203580 (22). Because CREB has been shown to activate the C/EBPβ promoter and has been suggested to be a downstream target of p38 (17, 27, 71), we considered the possibility that p38 might enhance p40 transcription by upregulating C/EBPβ transcription. The following experiments demonstrated that CREB phosphorylation and C/EBPβ upregulation are greatly reduced by the p38 inhibitor, SB203580. We make use of this observation to examine in greater depth the relationship between p38 activity, C/EBPβ upregulation, and IL-12 p40 induction.
As demonstrated previously (22), SB203580 partially inhibited IL-12 p40 expression in LPS-stimulated J774 cells, as monitored by ELISA (Fig. 5A). Consistent with the previous suggestion that CREB phosphorylation is a downstream target of the p38 pathway (17, 27, 71), low and high concentrations of SB203580 greatly reduced phospho-CREB accumulation in LPS-stimulated J774 cells, as determined by Western blotting (Fig. 5B). Importantly, the induction of LAP protein expression was also greatly reduced in the presence of low and high concentrations of SB203580 (Fig. 5C).
FIG. 5.
C/EBPβ is unlikely to be a relevant target of p38 MAP kinase during IL-12 p40 induction. (A) J774 cells were treated for 1 h with different concentrations of SB203580 (shaded bars) or an equivalent volume of the solvent dimethyl sulfoxide (DMSO) (solid bars). The cells were then stimulated with LPS (24 h) in the presence of inhibitor, and p40 protein secretion was monitored by ELISA. The results represent the means (± standard deviations) from three separate experiments. (B) J774 cells were treated for 1 h with different concentrations of SB203580 (SB) or an equivalent volume of DMSO. Phosphorylated CREB and total CREB were then monitored by Western blotting in whole-cell lysates (100 μg) from unstimulated or LPS-stimulated (10 min) cells. (C) J774 cells were treated for 1 h with different concentrations of SB203580 or an equivalent volume of DMSO. LAP was then monitored by Western blotting in nuclear extracts (60 μg) from unstimulated or LPS-stimulated C/EBPβ cells. (D) J774 cells were treated for 1 h with SB203580 or an equivalent volume of DMSO. IL-12 p40 protein was monitored in culture supernatants from LPS-stimulated cells. The data are expressed as percentages of p40 production in DMSO-treated cells.
The above-mentioned results suggest that the p38 pathway may enhance IL-12 p40 expression by stimulating CREB phosphorylation, which then leads to the upregulation of C/EBPβ expression. Our ability to block C/EBPβ upregulation using the SB203580 inhibitor and our knowledge that the C/EBPβ protein is not upregulated until 2 to 4 h poststimulation (Fig. 3 and 4) suggest a strategy for testing this hypothesis, based on the following prediction: if the C/EBPβ gene is indeed the p38 target that is responsible for the stimulation of p40 transcription by the p38 pathway, then the SB203580 inhibitor should inhibit IL-12 p40 production much more strongly at late time points following LPS stimulation. To test this prediction, the magnitude of the SB203580 effect was monitored at different times following LPS stimulation of J774 cells (Fig. 5D). An ELISA was used for this analysis because of its enhanced sensitivity. The results revealed that the magnitudes of inhibition by SB203580 were similar at the earliest time points at which p40 protein is detectable (4 h) and at late time points (16 and 24 h). Since the upregulation of C/EBPβ is unlikely to be important for IL-12 p40 production at the 4-h time point, p38 is likely to act on a different, as-yet-unidentified protein that contributes to p40 induction. The reduced expression of the p40 gene at late time points in the presence of SB203580 may be due, in part, to the effect of SB203580 on C/EBPβ upregulation. However, a simpler model is that the important p38 target protein is the same at both the early and late time points and that C/EBPβ upregulation is not important for p40 induction even at the late time points.
Analysis of C/EBPβ regulation using an altered-specificity approach.
The above-mentioned results provide strong evidence that C/EBPβ's contribution to IL-12 p40 induction does not involve alternative translation initiation, nuclear translocation, a posttranslational modification that enhances DNA-binding activity, or the upregulation of C/EBPβ expression. Another possible regulatory mechanism is that a posttranslational modification may enhance the transcriptional activation properties of C/EBPβ. This is a particularly attractive mechanism because numerous kinases have been reported to modify recombinant or overexpressed C/EBPβ (see the introduction).
To examine the possibility that a posttranslational modification is required for the induction of IL-12 p40 transcription by C/EBPβ, altered-specificity experiments were performed. These experiments separate the potential effect of an inducible posttranslational modification from the effect of C/EBPβ upregulation, as the altered-specificity protein is expressed at similar concentrations in both unactivated and activated cells. This approach also has the potential to reveal contributions from any type of inducible posttranslational modification, not just phosphorylation.
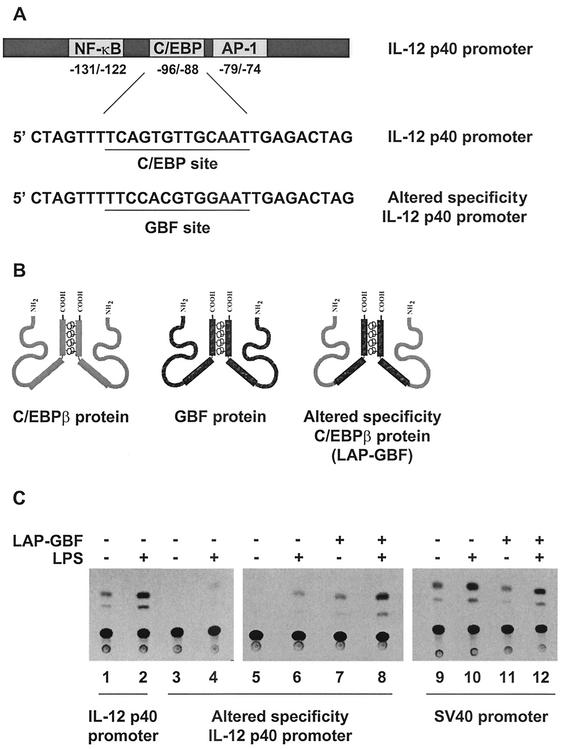
To perform the altered-specificity experiments, a recognition site for the A. thaliana GBF protein was inserted into a p40-CAT reporter plasmid (−350 to +55) in place of the native C/EBP site (Fig. 6A) (25, 44, 53). Murine C/EBP proteins apparently cannot bind the introduced GBF site, as demonstrated by the reduced activity of the altered promoter relative to the wild-type p40 promoter in a transient-transfection assay performed in RAW264.7 cells (Fig. 6C, lanes 1 to 4). To generate a C/EBPβ variant capable of binding the GBF site, the bZIP region of LAP was substituted for that of the GBF protein (Fig. 6B). Cotransfection of RAW264.7 cells with the altered-specificity promoter-reporter plasmid and the expression plasmid for the LAP-GBF fusion protein restored promoter activity in unstimulated and LPS-stimulated RAW264.7 cells to levels comparable to those observed with the wild-type promoter (Fig. 6C, lanes 1 to 8). In contrast, coexpression of the altered protein had no effect on the unrelated simian virus 40 promoter, which is known to be moderately inducible in RAW264.7 cells (Fig. 6C, lanes 9 to 12) (7).
FIG. 6.
Use of an altered-specificity assay to monitor the potential role of C/EBPβ posttranslational modifications. (A) Sequence of the p40 promoter encompassing the C/EBP site. Also shown is the same sequence with the C/EBP site replaced by a GBF binding site. (B) Schematic representations of the C/EBPβ protein, GBF protein, and LAP-GBF fusion protein. (C) RAW264.7 cells were transfected with a CAT reporter plasmid containing the wild-type p40 promoter (−350 to +55; lanes 1 and 2), the altered-specificity p40 promoter (lanes 3 to 8), or a simian virus 40 (SV40) promoter (lanes 9 to 12) either alone (lanes 1 to 6, 9, and 10) or with (+) the LAP-GBF fusion protein (lanes 7, 8, 11, and 12). CAT assays were performed with extracts from unstimulated (−) or LPS-stimulated (24 h) (+) cells.
The ability of LAP-GBF to enhance the activity of the altered p40 promoter suggests that this altered-specificity protein can effectively substitute for wild-type C/EBPβ. However, the results failed to provide evidence of an essential inducible posttranslational modification, because the LAP-GBF protein yielded comparable increases in promoter activity in both unstimulated and stimulated cells. That is, LAP-GBF enhanced promoter activity eightfold in unstimulated cells (Fig. 6C, compare lanes 5 and 7) and sevenfold in stimulated cells (compare lanes 6 and 8). If the protein acquired a posttranslational modification during stimulation that substantially enhanced its ability to activate transcription from the IL-12 p40 promoter, a much greater effect of the constitutively expressed LAP-GBF protein should be observed in stimulated cells. It is noteworthy that the induction of promoter activity observed in both the absence and presence of LAP-GBF (compare lanes 5 and 6 and lanes 7 and 8) is presumably due to the induction of Rel proteins and other proteins that contribute to p40 promoter induction (46).
Although these results suggest that C/EBPβ activity does not require an inducible posttranslational modification, two important caveats must be noted. First, these experiments involved transient-transfection assays and protein overexpression, leaving open the possibility that an inducible posttranslational modification is needed for activation of the endogenous p40 gene by physiological concentrations of C/EBPβ. Second, the replacement of the C/EBPβ bZIP domain with the comparable domain from GBF leaves open the possibility that a posttranslational modification targets this domain of C/EBPβ.
Constitutive phosphorylation of endogenous C/EBPβ in peritoneal macrophages and J774 cells.
To examine in greater detail the possibility that C/EBPβ is phosphorylated upon LPS stimulation, phosphopeptide-mapping experiments were performed with endogenous C/EBPβ. Despite numerous studies demonstrating phosphorylation of C/EBPβ in vitro or following overexpression, the first phosphopeptide map of endogenous murine C/EBPβ was reported only recently in conjunction with an analysis of C/EBPβ regulation in hepatocytes (8). That analysis revealed two phosphopeptides that were strongly inducible in response to transforming growth factor alpha, one of which corresponded to T217.
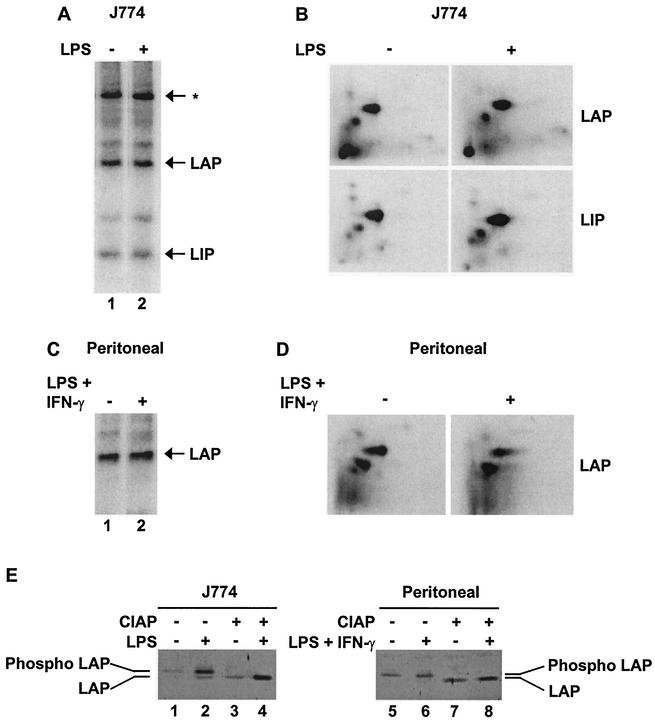
To analyze phosphorylation of endogenous C/EBPβ, J774 cells, RAW264.7 cells, and peritoneal macrophages were labeled in vivo with [32P]orthophosphate. Extracts were then prepared from unstimulated cells and cells that were stimulated with LPS or LPS plus IFN-γ for 20 min. Following immunoprecipitation with C/EBPβ antibodies, LAP was found to be phosphorylated equivalently before and after stimulation (Fig. 7A [J774] and C [peritoneal]; RAW264.7, data not shown). In J774 and RAW264.7 cells (Fig. 7A and data not shown), LIP was also phosphorylated equivalently before and after stimulation. In primary cells, LIP was not monitored because the experiments were performed with a different antibody that does not recognize LIP but results in more efficient immunoprecipitation of LAP.
FIG. 7.
Murine C/EBPβ is constitutively phosphorylated in macrophages. (A) J774 cells were incubated with [32P]orthophosphate for 4 h. Phosphorylation of LAP and LIP from unstimulated (−) and LPS-stimulated (20 min) (+) cells was monitored by SDS-PAGE, followed by transfer to nitrocellulose and exposure to film. (B) LAP and LIP were excised from the membrane, digested with trypsin, spotted on a thin-layer chromatography plate, and separated by electrophoresis in the first dimension and chromatography in the second dimension. (C) Phosphorylation of LAP in unstimulated and LPS-plus-IFN-γ-stimulated (20 min) peritoneal macrophages was monitored as described for panel A. The C/EBPβ antibody used for this experiment does not bind LIP. (D) Phosphopeptide mapping of LAP from peritoneal macrophages was performed as decribed for panel B. (E) Untreated and calf intestine alkaline phosphatase (CIAP)-treated LAP were monitored by Western blotting in extracts from unstimulated or LPS-stimulated (4 h) J774 cells or in extracts from unstimulated or LPS-plus-IFN-γ-stimulated (4 h) peritoneal macrophages.
Subsequent analysis by two-dimensional tryptic phosphopeptide mapping revealed the same two phosphopeptide spots in unstimulated and stimulated cells (Fig. 7B [J774] and D [peritoneal]; RAW264.7, data not shown). Similar results were obtained when the stimulation time was varied from 5 to 60 min (data not shown). The same two constitutive spots were observed in eight independent experiments with J774 cells, three experiments with RAW264.7 cells, and two experiments with peritoneal macrophages.
As controls, ELISAs were performed in most experiments with parallel samples of cells to confirm that p40 protein was not expressed before stimulation but was efficiently expressed after stimulation (data not shown). In one experiment, ELISAs were performed directly with supernatants from radiolabeled unstimulated and stimulated cells to confirm that the radioactivity was incapable of activating p40 expression. Furthermore, intracellular flow cytometry experiments revealed that LPS-plus-IFN-γ stimulation of peritoneal macrophages induced IL-12 p40 expression in almost 100% of the cells (reference 66 and data not shown), demonstrating that inefficient activation is not responsible for our inability to detect inducible phosphopeptides.
To examine in greater depth the phosphorylation state of C/EBPβ in unstimulated and stimulated cells, we took advantage of the observation that phosphorylated and unphosphorylated forms can be distinguished by their differential migration rates on SDS protein gels (45). By Western blotting, no significant difference in the migration of LAP was detected in nuclear extracts from unstimulated and stimulated J774 cells (Fig. 7E, lanes 1 and 2) or peritoneal macrophages (lanes 5 and 6). To confirm that phosphorylation affects LAP migration under our gel conditions, the extracts were pretreated with alkaline phosphatase, which increased migration to similar extents in all four extracts (Fig. 7E, lanes 3, 4, 7, and 8). Consistent with the altered-specificity and phosphopeptide-mapping results, these results suggest that C/EBPβ is not inducibly phosphorylated upon macrophage stimulation.
Multiple residues within one tryptic peptide are constitutively phosphorylated.
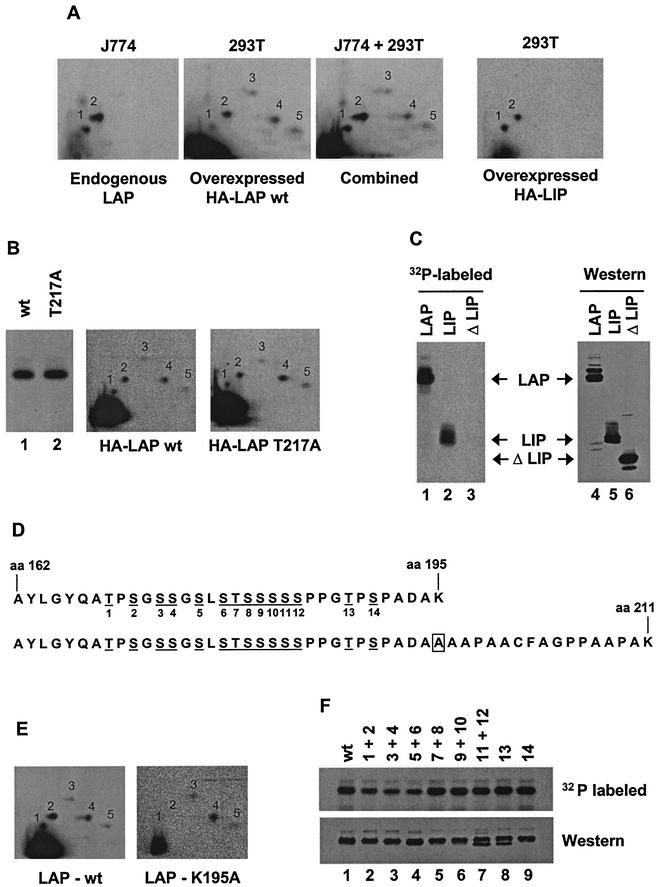
Although we could find no evidence that endogenous C/EBPβ is inducibly phosphorylated in murine macrophages, it is important to identify the sites of constitutive phosphorylation to determine the functional relevance of phosphorylation and, eventually, to identify the relevant kinases. The HEK 293T line was used for this purpose because phosphopeptide mapping of transiently expressed murine LAP revealed phosphopeptides that comigrated with the two endogenous phosphopeptides from murine macrophages (Fig. 8A, two left-hand maps). The comigration was confirmed by spotting the phosphopeptides from the endogenous protein together with the phosphopeptides from the transiently expressed protein on the same plate (Fig. 8A, map J774 + 293T, spots 1 and 2). Phosphopeptide mapping of the truncated form of C/EBPβ (LIP) confirmed that the phosphorylated residues are located within the C-terminal half of LAP (Fig. 8A, right-hand map).
FIG.8.
The constitutive C/EBPβ phosphorylation sites are within one tryptic peptide. (A) Phosphopeptide maps generated with endogenous LAP (J774 cells) and overexpressed LAP (293T cells) are shown individually (left-hand two maps) and when the two samples were run together (J774 + 293T). A map of overexpressed LIP (293T cells) is also shown (right). HA, hemagglutinin; wt, wild type. (B) Phosphorylation of wild-type LAP and the T217A mutant LAP in 293T cells was monitored by SDS-PAGE (left) and by two-dimensional phosphopeptide mapping (middle and right). (C) Phosphorylation of LAP, LIP, and ΔLIP in 293T cells was monitored by SDS-PAGE. A Western blot shows the overall abundances of the three proteins. (D) Sequence of tryptic peptide phosphorylated in vivo (top), along with sequence of tryptic peptide created by K195A mutation (bottom). aa, amino acid. Potential phosphoacceptors are underlined and numbered. The box corresponds to the residue altered in LAP-K195A. (E) Phosphopeptide maps of wild-type LAP and the K195A mutant LAP in 293T cells. (F) Phosphorylation of LAP mutants that alter potential phosphoacceptors between residues 162 and 195 was monitored by SDS-PAGE of radiolabeled protein (top) and by Western blotting (bottom). Potential phosphoacceptors altered in each mutant protein are indicated above each lane.
To address the possibility that the endogenous protein may be phosphorylated on T217, as was recently observed in mouse hepatocytes (8), a protein carrying a single substitution at T217 (T217A) was analyzed alongside the wild-type protein. Both proteins were phosphorylated to similar extents (Fig. 8B, left), and the phosphopeptide maps were very similar (Fig. 8B, middle and right). Examination of the C/EBPβ sequence revealed a cluster of potential phosphoacceptor sites in the N-terminal region of LIP (Fig. 8D). Deletion of these amino acids resulted in a loss of C/EBPβ phosphorylation in 293T cells (Fig. 8C, lanes 1 to 3), although the expression level was similar to those of LAP and LIP (Fig. 8C, lanes 4 to 6).
To verify that this deleted region was the target of phosphorylation in the context of full-length LAP, we took advantage of the fact that all 14 serines and threonines in this cluster were located within one tryptic fragment (Fig. 8D, top sequence). The trypsin cleavage site located at amino acid 195 of the wild-type protein was mutated from a lysine to an alanine, preventing trypsin cleavage at the site but allowing cleavage at the next lysine, located at amino acid 211 (Fig. 8D, bottom sequence). This mutation resulted in the production of a much larger tryptic fragment. If the cluster of 14 phosphoacceptors was truly the target of constitutive phosphorylation, forcing the production of a larger tryptic fragment containing these sites should greatly alter the migration of the phosphopeptides. As shown in Fig. 8E, the K195A mutation eliminated the two phosphopeptides that correspond to the endogenous phosphopeptides (spots 1 and 2) but did not affect three other phosphopeptides that are observed only with the overexpressed protein (spots 3 to 5). (The larger tryptic fragments created by this mutation were probably not detected because they remained at the origin or migrated off the plate.) These results provide strong evidence that spots 1 and 2 were both derived from the same tryptic fragment (residues 162 to 195).
To determine which of the 14 serines or threonines within the relevant fragment are phosphorylated, mutations were made that spanned the region. The first 12 potential phosphoacceptor sites were altered in pairs, and the last 2 were mutated individually. Transient expression and labeling revealed that none of the mutations completely abrogated phosphorylation (Fig. 8F). Mutation of potential phosphoacceptors 7 to 10 and 14 had no effect on phosphorylation (lanes 5, 6, and 9), whereas mutations in phosphoacceptors 1 to 6 reduced overall phosphorylation by approximately twofold (lanes 2 to 4). Phosphopeptide maps of these mutants showed that spots 1 and 2 were both retained, however (data not shown). Interestingly, mutation of potential phosphoacceptors 11 and 12 or 13 eliminated phosphorylation of a fraction of the mutant protein, as demonstrated by the detection of a faster-migrating species on the Western blot that was not labeled with [32P]orthophosphate (lanes 7 and 8). This suggested that these three residues may be the major phosphorylation sites. However, simultaneous disruption of all three residues (11 to 13) did not decrease overall phosphorylation and did not further increase the fraction of protein that escaped phosphorylation (data not shown).
Further attempts to determine precisely which residues within this fragment are phosphorylated were unsuccessful. Simultaneous mutation of phosphoacceptors 1 to 4 retained spots 1 and 2, whereas both spots were lost when phosphoacceptors 1 to 6 were altered simultaneously (data not shown). However, we suspect that the structure of the latter mutant protein was severely compromised because it was unable to bind DNA in gel shift assays (data not shown). In contrast, DNA-binding activity was unaltered by phosphatase treatment of the wild-type protein, which alters the migration of the protein (Fig. 7E) and eliminates labeling with [32P]orthophosphate (data not shown). Finally, it is worth noting that residue T188 (phosphoacceptor 13 [Fig. 8D]) corresponds to a consensus recognition site for ERK kinases, which previously were found to phosphorylate overexpressed C/EBPβ in response to Ras signaling (42). In our experiments, efficient phosphorylation was retained when this phosphoacceptor was altered, but it remains possible, if not likely, that ERK phosphorylation is important for the constitutive phosphorylation observed in macrophages (see Discussion).
Taken together, our findings suggest that multiple residues between amino acids 162 and 195 can be phosphorylated upon overexpression in 293T cells. It is important to note that the residues phosphorylated on the endogenous C/EBPβ protein in macrophages may or may not be as complex as those observed in 293T cells. Nevertheless, the present results have succeeded in localizing the sites of constitutive C/EBPβ phosphorylation in macrophages to a single tryptic fragment extending from residues 162 to 195.
DISCUSSION
C/EBPβ was identified as the major protein species capable of binding in vitro to functionally important C/EBP sites in the promoters for a large number of genes that are rapidly induced during macrophage activation, including the IL-12 p40 gene (reviewed in references 48 and 49). The frequent occurrence of binding sites led to the expectation that the C/EBPβ protein or gene would be a direct target of signal transduction pathways involved in inducible transcription during macrophage activation. This hypothesis received support from a large number of studies showing that C/EBPβ can be modified by a variety of inducible kinases in vitro and following overexpression in vivo and that C/EBPβ concentrations greatly increased following macrophage activation (see the introduction). Furthermore, studies performed with other cell types provided evidence that C/EBPβ can be regulated by inducible nuclear translocation or alternative translation initiation. In striking contrast, a study by Hu and colleagues found, using a transformed B-cell line that lacks most or all C/EBP family members, that the minimal bZIP domain of C/EBPβ was sufficient for restoring inducible transcription of the endogenous IL-6 and MCP-1 genes (26). This suggests that the contribution of C/EBPβ proteins to inducible transcription in at least one cell type may not require direct regulation.
In this study, we focused on an examination of endogenous C/EBPβ in primary macrophages and the J774 cell line. Despite the inducible binding of C/EBPβ to the IL-12 p40 promoter and previous evidence that it contributes to the inducible transcription of many other mediators of inflammation and immunity in macrophages, the results provide strong support for the hypothesis that C/EBPβ is fully active in unstimulated macrophages and is poised to activate transcription after other relevant factors are induced (26). Specifically, C/EBPβ is present at a relatively high abundance in the nuclei of unstimulated macrophages, the upregulation of C/EBPβ protein occurs well after the inducible binding of C/EBPβ to the IL-12 p40 promoter and well after transcription of the IL-12 p40 gene and several other putative target genes is induced, and endogenous C/EBPβ appeared to be modified only by constitutive phosphorylation in primary macrophages and macrophage cell lines. Furthermore, disruption of C/EBPβ gene induction by a p38 MAP kinase inhibitor had little effect on IL-12 p40 induction, and altered-specificity experiments failed to reveal the involvement of a posttranslational modification.
If C/EBPβ is indeed fully active in unstimulated cells, it is important to consider how its association with the endogenous IL-12 p40 promoter might be rapidly induced, as shown by ChIP and genomic footprinting. One possibility is that C/EBPβ binding must be preceded by remodeling of the positioned nucleosome encompassing the p40 promoter (66). Although indirect evidence that remodeling must precede C/EBP binding was previously provided (67), genomic footprinting also showed that occupancy of the C/EBP site remains inducible in a stably transfected promoter-reporter plasmid that appears to be associated with constitutively remodeled nucleosomes (67). Thus, inducible binding of C/EBPβ may not require inducible remodeling. An alternative possibility is that binding of C/EBPβ to the p40 promoter depends on cooperative interactions with a DNA-binding protein whose activity is induced, such as the NF-κB or AP-1 proteins that bind the p40 promoter (36, 41, 46, 75). The substantial defects in promoter activity observed when any of these sites are disrupted demonstrate that the proteins synergize with each other functionally. Physical interactions and cooperative binding by the C/EBP, NF-κB, and AP-1 proteins have been described previously (20, 30, 32, 39, 56), but we have been unable to demonstrate that these proteins bind cooperatively to the p40 promoter (data not shown). In addition, it was previously shown that inducible occupancy of the C/EBP site in the endogenous p40 promoter was retained in c-Rel−/− macrophages that do not express the IL-12 p40 gene, suggesting the cooperative binding with the p50/c-Rel complex that is believed to interact with the NF-κB site is not required for C/EBP binding (67). Nevertheless, the possibility that C/EBPβ binds cooperatively with another protein factor in vivo, such as NF-κB, has not been formally excluded. A third possibility is that C/EBPβ interacts with an inducible cofactor that stabilizes binding to the p40 promoter. A fourth possibility is that C/EBPβ may itself be modified by a phosphorylation event or by another posttranslational modification (e.g., acetylation or glycosylation) that was not detected in our phosphopeptide-mapping experiments and that was not required for activity in our altered-specificity experiments.
It is important to note that the mechanism of C/EBPβ induction that is relevant to the activation of endogenous target genes cannot be studied by examining the events involved in upregulation of a transiently transfected promoter-reporter plasmid containing C/EBP binding sites. The activity of such a plasmid is indeed upregulated following LPS stimulation. However, because reporter activity is typically measured 24 to 48 h after stimulation, the upregulation is likely to be strongly dependent on the upregulation of the C/EBPβ gene, which does not appear to be involved in the rapid induction of endogenous genes.
The biological relevance of the slow but substantial increase in C/EBPβ protein concentrations following LPS stimulation is difficult to predict. The high abundance of the protein in unstimulated cells (∼250,000 copies per cell) is likely to be sufficient for the activation of endogenous target genes, as the abundance is comparable to or higher than those of other transcription factors, such as NF-κB p65, Ikaros, and Sp1. It is not clear why the high copy number observed 8 h after stimulation (∼2,500,000 copies per cell) would be required. These high concentrations may be required for the transcription of genes that are induced at later time points. Alternatively, a high concentration may be needed for a completely different function of C/EBPβ, such as its role in preventing apoptosis by interacting with procaspases 1 and 8 (9). A large fraction of C/EBPβ is also localized to foci of pericentromeric heterochromatin in interphase cells, for reasons that remain unknown (reference 59 and data not shown). Perhaps a high concentration is needed for its function at these foci.
The significance of the constitutive phosphorylation is also difficult to predict. The most attractive possibility is that these phosphorylation events are required for transcriptional activation and that an autoinhibitory mechanism prevents the unphosphorylated protein from activating transcription. This possibility is supported by the fact that the phosphorylation sites are in close proximity to a region identified by Williams and colleagues that regulates an autoinhibitory activity (69). One prediction was that mutation of the constitutive phosphorylation sites would inhibit transactivation. Indeed, we found that disruption of 6 of the 14 phosphoacceptors within the targeted phosphopeptide eliminated transactivation. However, this mutation also inhibited DNA binding, whereas removal of the phosphates by phosphatase treatment had no effect on DNA binding. These results suggest that the six-residue mutation disrupted the structure of the protein in a manner that may be unrelated to the true physiological functions of the phosphorylation sites. Further investigation of the relevance of the constitutive phosphates will require the identification of the specific combination of phosphorylation sites.
One intriguing question is the relationship between the constitutive phosphorylation of C/EBPβ observed in macrophages in this study and the inducible phosphorylation of the same peptide observed previously in fibroblasts (42, 45). The initial study (42) showed, by phosphopeptide mapping, that overexpressed human C/EBPβ was inducibly phosphorylated by ERK1/2 at T235 (corresponding to murine T188) in response to Ras signaling and was constitutively phosphorylated at S231 (murine S184). Human T235 and murine T188 are both contained within consensus ERK1/2 recognition sequences. The constitutive phosphorylation at S231 by an unknown kinase was disrupted by mutation of T235 (42). A recent study of growth hormone signaling, using antibodies specific for phosphorylated murine T188, revealed that this residue is rapidly and transiently phosphorylated in response to growth hormone signaling (45). The results shown in Fig. 8F suggest that a small percentage of the overexpressed C/EBPβ in 293T cells may be phosphorylated on T188 and S184 but that most of the phosphates are on other residues. Two-dimensional maps prepared with C/EBPβ phosphorylated in vitro by ERK1 have confirmed that this kinase can phosphorylate C/EBPβ at T188 and that the peptide phosphorylated at this position comigrates with spot 1 on the two-dimensional maps (data not shown) (Fig. 8A). However, spots 1 and 2 were both retained at levels comparable to those observed with the wild-type protein when a T188 mutant protein was analyzed in vivo (data not shown). This result provides further evidence that most of the phosphorylation in vivo occurs at other sites within the same tryptic peptide, at least with overexpressed C/EBPβ.
Two additional observations suggest that ERK phosphorylation of T188 is not responsible for the constitutive phosphorylation of endogenous C/EBPβ in macrophages. First, ERK activity is rapidly and transiently induced in macrophages (data not shown). Second, pretreatment of macrophages with the ERK inhibitor U0126 or PD98059 had no effect on phosphopeptide spots 1 and 2 or on the induction of IL-12 p40 expression (data not shown). One hypothesis that may account for these results is that, during macrophage development, T188 may be transiently phosphorylated by ERK1/2, which may promote phosphorylation of other nearby sites by other kinases, with these secondary phosphorylations stably retained in unstimulated mature macrophages. The stable, constitutive phosphorylation of C/EBPβ in mature macrophages may help keep it active and poised to stimulate gene transcription in combination with other inducible factors. Further advances in mass spectrometry technology will be required before it will be possible to determine which sites within this dense cluster of potential phosphoacceptors are phosphorylated on endogenous C/EBPβ.
A final unresolved issue is the relevance of our results to the many other C/EBPβ phosphorylation events that have been reported. It perhaps is most relevant that only one other phosphorylation event, at T217 in mouse and the corresponding residue S105 in rat, has been demonstrated by two-dimensional phosphopeptide mapping of endogenous C/EBPβ (8). Although T188 phosphorylation was originally observed by two-dimensional mapping only with the overexpressed protein (42), phosphorylation at this site on the endogenous protein was recently confirmed by using antibodies that are specific for phosphorylated T188 (45). Other phosphorylation events that have been proposed on the basis of experiments performed in vitro or with overexpressed kinases and C/EBPβ await confirmation from studies of the endogenous protein.
Acknowledgments
We thank Martina Buck, Philip Cohen, Roger Davis, Tony Hunter, Peter Johnson, and Valeria Poli for helpful suggestions and Valeria Poli, Xiao-Hong Sun, and Gretchen Darlington for providing C/EBPβ-deficient mice. We are also grateful to Valerie Schuman and Dana Russell for performing the thioglycolate injections and to Peter Johnson, Shomyseh Sanjabi, and Tianyi Wang for critical reading of the manuscript.
This work was supported by the USPHS National Research Service Award GM07185, a Warsaw Fellowship, and a UCLA Dissertation Year Fellowship to M.N.B. S.T.S. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Agre, P., P. F. Johnson, and S. L. McKnight. 1989. Cognate DNA binding specificity retained after leucine zipper exchange between GCN4 and C/EBP. Science 246:922-926. [DOI] [PubMed] [Google Scholar]
- 2.Akira, S., H. Isshiki, T. Sugita, O. Tanabe, S. Kinoshita, Y. Nishio, T. Nakajima, T. Hirano, and T. Kishimoto. 1990. A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. EMBO J. 9:1897-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alam, T., M. R. An, and J. Papaconstantinou. 1992. Differential expression of three C/EBP isoforms in multiple tissues during the acute phase response. J. Biol. Chem. 267:5021-5024. [PubMed] [Google Scholar]
- 4.An, M. R., C.-C. Hsieh, P. D. Reisner, J. P. Rabek, S. G. Scott, D. T. Kuninger, and J. Papaconstantinou. 1996. Evidence for posttranslational regulation of C/EBPα and C/EBPβ isoform expression during the lipopolysaccharide-mediated acute-phase response. Mol. Cell. Biol. 16:2295-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berrier, A., G. Siu, and K. Calame. 1998. Transcription of a minimal promoter from the NF-IL6 gene is regulated by CREB/ATF and SP1 proteins in U937 promonocytic cells. J. Immunol. 161:2267-2275. [PubMed] [Google Scholar]
- 6.Boyle, W. J., P. van der Geer, and T. Hunter. 1991. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 201:110-149. [DOI] [PubMed] [Google Scholar]
- 7.Brightbill, H. D., S. E. Plevy, R. L. Modlin, and S. T. Smale. 2000. A prominent role for Sp1 during lipopolysaccharide-mediated induction of the IL-10 promoter in macrophages. J. Immunol. 164:1940-1951. [DOI] [PubMed] [Google Scholar]
- 8.Buck, M., V. Poli, P. van der Geer, M. Chojkier, and T. Hunter. 1999. Phosphorylation of rat serine 105 or mouse threonine 217 in C/EBPβ is required for hepatocyte proliferation induced by TGFα. Mol. Cell 4:1087-1092. [DOI] [PubMed] [Google Scholar]
- 9.Buck, M., V. Poli, T. Hunter, and M. Chojkier. 2001. C/EBP beta phosphorylation by RSK creates a functional XEXD caspase inhibitory box critical for cell survival. Mol. Cell 8:807-816. [DOI] [PubMed] [Google Scholar]
- 10.Caivano, M., B. Gorgoni, P. Cohen, and V. Poli. 2001. The induction of cyclooxygenase-2 mRNA in macrophages is biphasic and requires both CCAAT enhancer-binding protein β (C/EBPβ) and C/EBPδ transcription factors. J. Biol. Chem. 276:48693-48701. [DOI] [PubMed] [Google Scholar]
- 11.Cao, Z., R. M. Umek, and S. L. McKnight. 1991. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 5:1538-1552. [DOI] [PubMed] [Google Scholar]
- 12.Cardinaux, J. R., and P. J. Magistretti. 1996. Vasoactive intestinal peptide, pituitary adenylate cyclase-activating peptide, and noradrenaline induce the transcription factors CCAAT/enhancer binding protein (C/EBP)-beta and C/EBP delta in mouse cortical astrocytes: involvement in cAMP-regulated glycogen metabolism. J. Neurosci. 16:919-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cardinaux, J. R., I. Allaman, and P. J. Magistretti. 2000. Pro-inflammatory cytokines induce the transcription factors C/EBPβ and C/EBPδ in astrocytes. Glia 29:91-97. [PubMed] [Google Scholar]
- 14.Chang, C.-J., T.-T. Chen, H.-Y. Lei, D.-S. Chen, and S.-C. Lee. 1990. Molecular cloning of a transcription factor, AGP/EBP, that belongs to members of the C/EBP family. Mol. Cell. Biol. 10:6642-6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chinery, R., J. A. Brockman, D. T. Dransfield, and R. J. Coffey. 1997. Antioxidant-induced nuclear translocation of CCAAT/Enhancer-binding Protein β. J. Biol. Chem. 272:30356-30361. [DOI] [PubMed] [Google Scholar]
- 16.Clarkson, R. W. E., C. M. Chen, S. Harrison, C. Wells, G. E. O. Muscat, and M. J. Waters. 1995. Early responses of trans-activating factors to growth hormone in preadipocytes: differential regulation of CCAAT Enhancer-Binding Protein-β (C/EBPβ) and C/EBPδ. Mol. Endocrinol. 9:108-120. [DOI] [PubMed] [Google Scholar]
- 17.Deak, M., A. D. Clifton, L. M. Lucocq, and D. R. Alessi. 1998. Mitogen-and stress-activated protein kinase-1 (MSK1) is directly activated by MAPK and SAPK2/p38, and may mediate activation of CREB. EMBO J. 17:4426-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Descombes, P., M. Chojkier, S. Lichtsteiner, E. Falvey, and U. Schibler. 1990. LAP, a novel member of the C/EBP gene family, encodes a liver-enriched transcriptional activator protein. Genes Dev. 4:1541-1551. [DOI] [PubMed] [Google Scholar]
- 19.Descombes, P., and U. Schibler. 1991. A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell 67:569-579. [DOI] [PubMed] [Google Scholar]
- 20.Diehl, J. A., and M. Hannink. 1994. Identification of a C/EBP-Rel complex in avian lymphoid cells. Mol. Cell. Biol. 14:6635-6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng, G.-J., H. S. Goodridge, M. M. Harnett, X.-Q. Wei, A. V. Nikolaev, A. P. Higson, and F.-Y. Liew. 1999. Extracellular signal-related kinase (ERK) and p38 mitogen-activated protein (MAP) kinases differentially regulate the lipopolysaccharide-mediated induction of inducible nitric oxide synthase and IL-12 in macrophages: Leishmania phosphoglycans subvert macrophage IL-12 production by targeting ERK MAP kinase. J. Immunol. 163:6403-6412. [PubMed] [Google Scholar]
- 23.Gately, M. K., L. M. Renzetti, J. Magram, A. S. Stern, L. Adorini, V. Gubler, and D. H. Presky. 1998. The interleukin-12/interleukin-12-receptor system: role in normal and pathological immune responses. Annu. Rev. Immunol. 16:495-521. [DOI] [PubMed] [Google Scholar]
- 24.Gorgoni, B., D. Maritano, P. Marthyn, M. Righi, and V. Poli. 2002. C/EBPβ gene inactivation causes both impaired and enhanced gene expression and inverse regulation of IL-12 p40 and p35 mRNAs in macrophages. J. Immunol. 168:4055-4062. [DOI] [PubMed] [Google Scholar]
- 25.Hill, C. S., R. Marais, S. John, J. Wynne, S. Dalton, and R. Treisman. 1993. Functional analysis of a growth factor-responsive transcription factor complex. Cell 73:395-406. [DOI] [PubMed] [Google Scholar]
- 26.Hu, H.-M., Q. Tian, M. Baer, C. J. Spooner, S. C. Williams, P. F. Johnson, and R. C. Schwartz. 2000. The C/EBP bZIP domain can mediate lipopolysaccharide induction of the proinflammatory cytokines interleukin-6 and monocyte chemoattractant protein-1. J. Biol. Chem. 275:16373-16381. [DOI] [PubMed] [Google Scholar]
- 27.Iordanov, M., K. Bender, T. Ade, W. Schmid, C. Sachsenmaier, E. Engel, M. Gaestel, H. J. Rahmsdorf, and P. Herrlich. 1997. CREB is activated by UVC through a p38/HOG-1-dependent protein kinase. EMBO J. 16:1009-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katz, S., L. E. Kowenz, C. Muller, K. Meese, S. A. Ness, and A. Leutz. 1993. The NF-M transcription factor is related to C/EBPβ and plays a role in signal transduction, differentiation and leukemogenesis of avian myelomonocytic cells. EMBO J. 12:1321-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kinoshita, S., S. Akira, and T. Kishimoto. 1992. A member of the C/EBP family, NF-IL6 beta, forms a heterodimer and transcriptionally synergizes with NF-IL6. Proc. Natl. Acad. Sci. USA 4:1473-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klampfer, L., T. H. Lee, W. Hsu, J. Vilcek, and S. Chen-Kiang. 1994. NF-IL6 and AP-1 cooperatively modulate the activation of the TSG-6 gene by tumor necrosis factor alpha and interleukin-1. Mol. Cell. Biol. 14:6561-6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kowenz-Leutz, E., G. Twamley, S. Ansieau, and A. Leutz. 1994. Novel mechanism of C/EBPβ (NF-M) transcriptional control; activation through derepression. Genes Dev. 8:2781-2791. [DOI] [PubMed] [Google Scholar]
- 32.LeClair, K. P., M. A. Blanar, and P. A. Sharp. 1992. The p50 subunit of NF-kappa B associates with the NF-IL6 transcription factor. Proc. Natl. Acad. Sci. USA 89:8145-8149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lipson, K. E., and R. Baserga. 1989. Transcriptional activity of the human thymidine kinase gene determined by a method using the polymerase chain reaction and an intron-specific probe. Proc. Natl. Acad. Sci. USA 86:9774-9777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu, P. C. C., D. Y. Dunlap, and F. Matsumura. 1998. Suppression of C/EBPα and induction of C/EBPβ by 2,3,7,8-tetrachlorodibenzo-p-dioxin in mouse adipose tissue and liver. Biochem. Pharmacol. 55:1647-1655. [DOI] [PubMed] [Google Scholar]
- 35.Lu, H.-T., D. D. Yang, M. Wysk, E. Gatti, I. Mellman, R. J. Davis, and R. A. Flavell. 1999. Defective IL-12 production in mitogen-activated protein (MAP) kinase kinase 3 (Mkk3)-deficient mice. EMBO J. 18:1845-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma, X., J. M. Chow, G. Gri, G. Carra, F. Gerosa, S. F. Wolf, R. Dzialo, and G. Trinchieri. 1996. The interleukin-12 p40 gene promoter is primed by interferon-γ in monocytic cells. J. Exp. Med. 183:147-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacDougald, O. A., P. Cornelius, R. Liu, and M. D. Lane. 1995. Insulin regulates transcription of the CCAAT/Enhancer binding protein (C/EBP) α, β, and δ genes in fully-differentiated 3T3-L1 adipocytes. J. Biol. Chem. 270:647-654. [DOI] [PubMed] [Google Scholar]
- 38.Matsuno, F., S. Chowdhury, T. Gotoh, K. Iwase, H. Matsuzaki, K. Takatsuki, M. Mori, and M. Takiguchi. 1996. Induction of the C/EBP beta gene by dexamethasone and glucagons in primary-cultured rat hepatocytes. J. Biochem. 119:524-532. [DOI] [PubMed] [Google Scholar]
- 39.Matsusaka, T., K. Fujikawa, Y. Nishio, N. Mukaida, K. Matsushima, T. Kishimoto, and S. Akira. 1993. Transcription factors NF-IL6 and NF-kappa B synergistically activate transcription of the inflammatory cytokines, interleukin 6 and interleukin 8. Proc. Natl. Acad. Sci. USA 90:10193-10197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Metz, R., and E. Ziff. 1991. cAMP stimulates the C/EBP-related transcription factor rNFIL-6 to translocate to the nucleus and induce c-fos transcription. Genes Dev. 5:1754-1766. [DOI] [PubMed] [Google Scholar]
- 41.Murphy, T., M. Cleveland, P. Kulesza, J. Magram, and K. Murphy. 1995. Regulation of interleukin 12 p40 expression through an NF-kB half-site. Mol. Cell. Biol. 15:5258-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakajima, T., S. Kinoshita, T. Sasagawa, K. Sasaki, M. Naruto, T. Kishimoto, and S. Akira. 1993. Phosphorylation at threonine 235 by ras-dependent mitogen activated protein cascade is essential for transcription factor NF-IL6. Proc. Natl. Acad. Sci. USA 90:2207-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niehof, M., M. P. Manns, and C. Trautwein. 1997. CREB controls LAP/CEBPβ transcription. Mol. Cell. Biol. 17:3600-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olive, M., S. C. Williams, C. Dezan, P. F. Johnson, and C. Vinson. 1996. Design of a C/EBP-specific, dominant-negative bZIP protein with both inhibitory and gain-of-function properties. J. Biol. Chem. 271:2040-2047. [DOI] [PubMed] [Google Scholar]
- 45.Piwien-Pilipuk, G., O. MacDougald, and J. Schwartz. 2002. Dual regulation of phosphorylation and dephosphorylation of C/EBPβ modulate its transcriptional activation and DNA binding in response to growth hormone. J. Biol. Chem. 277:44557-44565. [DOI] [PubMed] [Google Scholar]
- 46.Plevy, S., J. Gemberling, S. Hsu, A. Dorner, and S. T. Smale. 1997. Multiple control elements mediate activation of the murine and human interleukin 12 p40 promoters: evidence of functional synergy between C/EBP and Rel proteins. Mol. Cell. Biol. 17:4572-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poli, V., F. P. Mancini, and R. Cortese. 1990. IL-6DBP, a nuclear protein involved in interleukin-6 signal transduction, defines a new family of leucine zipper proteins related to C/EBP. Cell 63:643-653. [DOI] [PubMed] [Google Scholar]
- 48.Poli, V. 1998. The role of C/EBP isoforms in the control of inflammatory and native immunity functions. J. Biol. Chem. 273:29279-29282. [DOI] [PubMed] [Google Scholar]
- 49.Ramji, D. P., and P. Foka. 2002. CCAAT/Enhancer binding proteins: Structure, function and regulation. Biochem. J. 365:561-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reddy, S. T., R. S. Gilbert, W. Xie, S. Luner, and H. R. Herschman. 1994. TGF-beta inhibits both endotoxin-induced prostaglandin synthesis and expression of the TIS10/prostaglandin synthase 2 gene in murine macrophages. J. Leukoc. Biol. 55:192-200. [DOI] [PubMed] [Google Scholar]
- 51.Sanjabi, S., A. Hoffmann, H.-C. Liou, D. Baltimore, and S. T. Smale. 2000. Selective requirement for c-Rel during IL-12 p40 gene induction in macrophages. Proc. Natl. Acad. Sci. USA 97:12705-12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Screpanti, I., L. Romani, P. Musiani, A. Modesti, E. Fattori, D. Lazzaro, C. Sellitto, S. Scarpa, D. Bellavia, G. Lattanzio, F. Bistoni, L. Frati, R. Cortese, A. Gulino, G. Ciliberto, F. Costantini, and V. Poli. 1995. Lymphoproliferative disorder and imbalanced T-helper response in C/EBPβ-deficient mice. EMBO J. 14:1932-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shah, P. C., E. Bertolino, and H. Singh. 1997. Using altered specificity Oct-1 and Oct-2 mutants to analyze the regulation of immunoglobulin gene transcription. EMBO J. 16:7105-7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen, B.-J., C.-J. Chang, H.-S. Lee, W.-H. Tsai, L.-H. Miau, and S.-C. Lee. 1997. Transcriptional induction of the agp/ebp (c/ebpβ) gene by hepatocyte growth factor. DNA Cell Biol. 16:703-711. [DOI] [PubMed] [Google Scholar]
- 55.Siberil, Y., P. Doireau, and P. Gantet. 2001. Plant bZIP G-box binding factors. Modular structure and activation mechanisms. Eur. J. Biochem. 268:5655-5666. [DOI] [PubMed] [Google Scholar]
- 56.Stein, B., and A. S. Baldwin Jr. 1993. Distinct mechanisms for regulation of the interleukin-8 gene involves synergism and cooperativity between C/EBP and NF-kappa B. Mol. Cell. Biol. 13:7191-7198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sterneck, E., C. Muller, S. Katz, and A. Leutz. 1992. Autocrine growth induced by kinase type oncogenes in myeloid cells requires AP-1 and NF-M, a myeloid specific, C/EBP-like factor. EMBO J. 11:115-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tanaka, T., S. Akira, K. Yoshida, M. Umemoto, Y. Yoneda, N. Shirafuji, H. Fujiwara, S. Suematsu, N. Yoshida, and T. Kishimoto. 1995. Targeted disruption of the NF-IL6 gene discloses its essential role in bacterial killing and tumor cytotoxicity by macrophages. Cell 80:353-361. [DOI] [PubMed] [Google Scholar]
- 59.Tang, Q. Q., and M. D. Lane. 1999. Activation and centromeric localization of CCAAT/enhancer-binding proteins during the mitotic clonal expansion of adipocyte differentiation. Genes Dev. 13:2231-2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trautwein, C., C. Caelles, P. van der Geer, T. Hunter, M. Karin, and M. Chojkier. 1993. Transactivation by LAP/NF-IL6 is enhanced by phosphorylation of its activation domain. Nature 364:544-547. [DOI] [PubMed] [Google Scholar]
- 61.Trautwein, C., P. van der Geer, M. Karin, T. Hunter, and M. Chojkier. 1994. Protein kinase A and C site-specific phosphorylations of LAP (NF-IL6) modulate its binding affinity to DNA recognition elements. J. Clin. Investig. 93:2554-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trinchieri, G. 1995. Interleukin-12: a proinflammatory cytokine with immunoregulatory functions that bridge innate resistance and antigen-specific adaptive immunity. Annu. Rev. Immunol. 13:251-276. [DOI] [PubMed] [Google Scholar]
- 63.Trinchieri, G., and F. Gerosa. 1996. Immunoregulation by interleukin-12. J. Leukoc. Biol. 59:505-511. [DOI] [PubMed] [Google Scholar]
- 64.Tripp, C. S., M. K. Gately, J. Hakimi, P. Ling, and E. R. Unanue. 1994. Neutralization of IL-12 decreases resistance to Listeria in SCID and C.B-17 mice. Reversal by interferon-γ. J. Immunol. 152:1883-1887. [PubMed] [Google Scholar]
- 65.Wegner, M., Z. Cao, and M. G. Rosenfeld. 1992. Calcium-regulated phosphorylation within the leucine zipper of C/EBPβ. Science 256:370-373. [DOI] [PubMed] [Google Scholar]
- 66.Weinmann, A. S., S. E. Plevy, and S. T. Smale. 1999. Rapid and selective remodeling of a positioned nucleosome during the induction of IL-12 p40 transcription. Immunity 11:665-675. [DOI] [PubMed] [Google Scholar]
- 67.Weinmann, A. S., D. M. Mitchell, S. Sanjabi, M. N. Bradley, A. Hoffmann, H.-C. Liou, and S. T. Smale. 2001. Nucleosome remodeling at the IL-12 p40 promoter is a TLR-dependent, Rel-independent event. Nat. Immunol. 2:51-57. [DOI] [PubMed] [Google Scholar]
- 68.Williams, S. C., C. A. Cantwell, and P. F. Johnson. 1991. A family of C/EBP-related proteins capable of forming covalently linked leucine zipper dimers in vitro. Genes Dev. 5:1553-1567. [DOI] [PubMed] [Google Scholar]
- 69.Williams, S. C., M. Baer, A. J. Dillner, and P. F. Johnson. 1995. CRP2(C/EBPβ) contains a bipartite regulatory domain that controls transcriptional activation, DNA binding and cell specificity. EMBO J. 14:3170-3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu, Z., Y. Xie, N. L. R. Bucher, and S. R. Farmer. 1995. Conditional ectopic expression of C/EBPβ in NIH-3T3 cells induces PPARγ and stimulates adipogenesis. Genes Dev. 9:2350-2363. [DOI] [PubMed] [Google Scholar]
- 71.Xing, J., J. M. Kornhauser, Z. Xia, E. A. Thiele, and M. E. Greenberg. 1998. Nerve growth factor activates extracellular signal-regulated kinase and p38 mitogen-activated protein kinase pathways to stimulate CREB serine 133 phosphorylation. Mol. Cell. Biol. 18:1946-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yeh, W.-C., Z. Cao, M. Classon, and S. L. McKnight. 1995. Cascade regulation of terminal adipocyte differentiation by three members of the C/EBP family of leucine zipper proteins. Genes Dev. 9:168-181. [DOI] [PubMed] [Google Scholar]
- 73.Yin, M., S. Q. Yang, H. Z. Lin, M. D. Lane, S. Chatterjee, and A. M. Diehl. 1996. Tumor necrosis factor α promotes nuclear localization of cytokine-inducible CCAAT/enhancer binding protein isoforms in hepatocytes. J. Biol. Chem. 271:17974-17978. [DOI] [PubMed] [Google Scholar]
- 74.Yukawa, K., T. Tanaka, S. Tsuji, and S. Akira. 1998. Expressions of CCAAT/enhancer-binding proteins β and δ and their activities are intensified by cAMP signaling as well as Ca2+/calmodulin kinase activation in hippocampal neurons. J. Biol. Chem. 273:31345-31351. [DOI] [PubMed] [Google Scholar]
- 75.Zhu, C., K. Gagnidze, J. H. Gemberling, and S. E. Plevy. 2001. Characterization of an activating-protein-1 binding site in the murine interleukin-12 p40 promoter. Demonstration of novel functional elements by a reductionist approach. J. Biol. Chem. 276:18519-18528. [DOI] [PubMed] [Google Scholar]
- 76.Zhu, S., K. Yoon, E. Sterneck, P. F. Johnson, and R. C. Smart. 2002. CCAAT/enhancer binding protein-β is a mediator of keratinocyte survival and skin tumorigenesis involving oncogenic ras signaling. Proc. Natl. Acad. Sci. USA 99:207-212. [DOI] [PMC free article] [PubMed] [Google Scholar]