Abstract
The inhibins are gonadal transforming growth factor β superfamily protein hormones that suppress pituitary follicle-stimulating hormone (FSH) synthesis. Recently, betaglycan and inhibin binding protein (InhBP/p120, also known as the product of immunoglobulin superfamily gene 1 [IGSF1]) were identified as candidate inhibin coreceptors, shedding light on the molecular basis of how inhibins may affect target cells. Activins, which are structurally related to the inhibins, act within the pituitary to stimulate FSH production. Betaglycan increases the affinity of inhibins for the activin type IIA (ACVR2) receptor, thereby blocking activin binding and signaling through this receptor. InhBP/p120 may not directly bind inhibins but may interact with the activin type IB receptor, ALK4, and participate in inhibin B's antagonism of activin signaling. To better understand the in vivo functions of InhBP/p120, we characterized the InhBP/p120 mRNAs and gene in mice and generated InhBP/p120 mutant mice by gene targeting in embryonic stem cells. InhBP/p120 mutant male and female mice were viable and fertile. Moreover, they showed no alterations in FSH synthesis or secretion or in ovarian or testicular function. These data contribute to a growing body of evidence indicating that InhBP/p120 does not play an essential role in inhibin biology.
The inhibins and activins were identified as gonadal factors which regulate the synthesis and secretion of pituitary follicle-stimulating hormone (FSH) (6, 10, 18, 22, 29, 34, 40, 47). As their names indicate, activins stimulate and inhibins suppress FSH production. Activins and inhibins are structurally related dimeric members of the transforming growth factor β (TGF-β) superfamily (11). Inhibins are heterodimers of an α subunit and one of two β (βA or βB) subunits. Activins are dimers of two inhibin-activin β subunits: activin A (βA-βA), activin B (βB-βB), or activin AB (βA-βB) (34). Since their discovery, activins have been shown to play diverse roles in various tissues, both in adult and developing animals (1, 7, 10, 30, 35, 43). In addition, much has been learned regarding their mechanisms of signal transduction. Like other members of the TGF-β superfamily, activins signal through a hetero-oligomeric serine/threonine receptor complex and SMAD proteins. Activins bind to one of two type II receptors, ACVR2 or ACVR2B (3, 31). Upon ligand binding, the type II receptor phosphorylates the activin type IB receptor, activin receptor-like kinase 4 (ALK4) (4, 39, 41). Activated ALK4 phosphorylates the intracellular signaling proteins, SMAD2 and SMAD3, which then dissociate from the receptor complex and bind to the co-SMAD, SMAD4 (25). The activated SMAD2-SMAD4 or SMAD3-SMAD4 complex translocates to the nucleus, where it interacts with cofactor proteins to stimulate or repress target gene transcription (14, 15, 46). TGF-β superfamily ligands have also been shown to signal through SMAD-independent pathways, but this is not as well characterized (2).
The mechanisms of inhibin action are not well understood, although two models have emerged in the literature (6, 26, 27, 37, 42). First, inhibins may act principally by preventing activin signaling. Specifically, inhibins compete with activins for binding to activin type II receptors. In the case of pituitary FSH regulation, locally produced activin B constitutively stimulates FSH production (17, 36). Therefore, blockade of this action by inhibins would reduce FSH production. In the second model, it is thought that inhibins bind distinct membrane-anchored receptor proteins that propagate inhibin-specific intracellular signals. The available data are more consistent with the first model than with the second. Both activins and inhibins bind the activin type II receptors via their β subunits; however, unlike the activins, inhibins do not recruit the type I signaling receptor into the complex (26). Thus, inhibins can antagonize the actions of the activins by blocking activin binding and downstream signaling. Because inhibins bind to the type II receptors with about a 10-fold-lower affinity than do the activins (3, 12, 28, 31, 45), it has been argued that inhibins may bind to coreceptor proteins that increase their affinity for the type II receptors (26). Indeed, betaglycan, the TGF-β type III receptor, was recently shown to form a high affinity ternary complex with inhibin A and ACVR2 which is resistant to disruption by activin A (27).
A second protein has also been identified as an inhibin coreceptor candidate. Inhibin binding protein or p120 (InhBP/p120), encoded by a gene on the X chromosome originally designated immunoglobulin superfamily gene 1 (IGSF1), was shown by affinity chromatography to bind inhibin A (16, 33). Subsequently, InhBP/p120 was shown to interface with the activin receptor complex via a direct interaction with ALK4 (13). Interestingly, conventional receptor binding assays have been unsuccessful in demonstrating direct binding of inhibin A or inhibin B to full-length InhBP/p120 expressed either alone or in combination with the various activin type I and type II receptors (12). Still, InhBP/p120 is highly expressed in the anterior pituitary gland, and its mRNA levels are regulated across the rat estrous cycle in a manner consistent with its putative role in inhibin's regulation of FSH (8).
Two isoforms of InhBP/p120, InhBP-L and InhBP-S, have been characterized in rats and humans (8, 20, 33, 38). InhBP-L is a large membrane-bound protein consisting of 12 C-type immunoglobulin (Ig)-like domains, a transmembrane domain, and a short cytoplasmic tail. The 12 Ig-like domains are organized in groups of five and seven, separated by a short hydrophobic linker region. InhBP-S is predicted to be a truncated, soluble form of the protein. It contains the signal peptide and first two Ig-like domains observed in InhBP-L but is truncated at its C terminus because of retention of an intronic sequence in the corresponding mRNA that introduces a premature stop codon (8). The functions of these two different forms of InhBP/p120 are not yet known.
To characterize more definitively the role of InhBP/p120 in inhibin biology and FSH regulation, we generated an InhBP/p120 mutant mouse model by gene targeting in embryonic stem (ES) cells. We hypothesized that if InhBP/p120 functions as an inhibin coreceptor, FSH levels would be increased in these mutant mice because of a reduced sensitivity of pituitary gonadotropes to the negative feedback effects of gonadally derived inhibins. Moreover, male and female mice deficient in the inhibin α subunit, which are devoid of both inhibin A and B, have elevated FSH levels and develop gonadal sex cord stromal tumors (32). Therefore, we predicted that mice deficient in InhBP/p120 may similarly develop gonadal tumors. The results show that in the absence of the two major pituitary forms of InhBP/p120, InhBP-S and InhBP-L, these mutant mice are fertile, have no obvious alterations in pituitary FSH synthesis or release, and exhibit normal gonadal physiology and histology.
MATERIALS AND METHODS
Cloning of mouse InhBP/p120 gene and characterization of InhBP/p120 cDNA.
A rat cDNA probe corresponding to exons 12 to 14 was used to screen 500,000 plaques from a 129S6/SvEv mouse genomic library. A single clone (mIR-10-1) was isolated, and its 15.8-kb insert was subcloned into the NotI site of pGEM11Zf (Promega, Madison, Wis.). A restriction map was generated by using standard methods. The InhBP/p120 gene maps to the X chromosome in mice and humans. Recently, mouse X chromosome genomic sequences containing the entire InhBP/p120 gene were deposited in GenBank (accession number AL731789). Our restriction map was confirmed by comparison to this sequence. The exon-intron structure of the InhBP/p120 gene was determined by aligning the mouse cDNA sequence (see below) with the AL731789 genomic sequence and subcloned fragments of mIR-10-1. mIR-10-1 extended 15.8 kb from approximately 3.3 kb 5′ of exon 1 to the end of intron 14 (of the 20-exon gene), and fragments from this clone were used to generate the gene targeting vector (see below; Fig. 1).
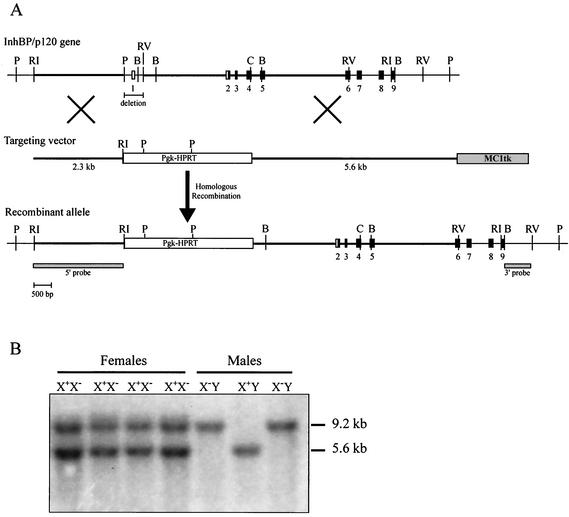
FIG. 1.
Generation of InhBP/p120 mutant mice. (A) Schematic representation of the targeting strategy used to generate InhBP/p120-deficient mice. The 428-bp PstI-EcoRV fragment containing exon 1 (and the transcription start site) was replaced with the Pgk-HPRT positive selection cassette. The 5′ and 3′ homology arms are indicated by the thick horizontal lines. Exons appear as boxes and are numbered. Noncoding and coding sequences are indicated in white and black, respectively. The positions of the 5′ internal and 3′ external probes used to screen the ES cells are indicated. B, BamHI; C, ClaI; P, PstI; RI, EcoRI; and RV, EcoRV. (B) Genotyping of offspring was performed by Southern blot analysis of tail genomic DNA digested with EcoRI and EcoRV and hybridized with a cDNA probe corresponding to sequences in exons 2 through 6. The wild-type and recombinant alleles detected in this manner are 5.6 and 9.2 kb, respectively. Note the presence of both alleles in heterozygous females (X+X−). Because InhBP/p120 is X-linked, males possess either a wild-type (X+Y) or recombinant (X−Y) allele.
The mouse cDNA was characterized by reverse transcription-PCR (RT-PCR) with C57BL/6JC57BL/6JJ mouse pituitary RNA by using primers derived from the cloned rat and human cDNA sequences. The 5′ and 3′ ends were determined by using rapid amplification of cDNA ends (RACE) RT-PCR with gene-specific primers and by following the instructions of the manufacturers (Ambion, Austin, Tex.; Invitrogen, Carlsbad, Calif.). PCR products were ligated into a TA cloning vector (pCR3.1; Invitrogen) and sequenced with ABI dye terminator cycle sequencing (Foster City, Calif.). Sequence contigs were assembled by using Sequencher for the Macintosh and Windows (GeneCodes, Ann Arbor, Mich.).
Construction of targeting vectors and generation of mutant mice.
The targeting vector (Fig. 1A) for the InhBP/p120 gene was designed to replace a 428-bp region including exon 1 (PstI-to-EcoRV fragment) with a Pgk-HPRT selection cassette. The 5′ and 3′ arms of homology were 2.3 and 5.6 kb, respectively, and an MC1tk selection cassette was incorporated adjacent to the 3′ arm. Twenty-five micrograms of the KpnI-linearized targeting vector was electroporated into AB2.2 ES cells derived from 129S6/SvEv mice. Cells were cultured in selective media containing hypoxanthine-aminopterin-thymidine and 1-(2′-deoxy-2′-fluoro-β-d-arabinofuranosyl)-5′-iodouracil (FIAU). Ninety-four of the surviving clones were screened by genomic Southern blot analysis. PstI-digested DNA was electrophoresed, transferred onto a nylon membrane, and hybridized with 5′ internal and 3′ external [32P]dCTP-labeled probes (Rediprime II kit; Amersham Pharmacia Biotech, Piscataway, N.J.). Eleven correctly targeted clones (11.7%) were identified and expanded. ES cells containing the Igsf1tm1Zuk allele (mice derived from these cells are herein called InhBP/p120 mutant or knockout mice) were injected into C57BL/6JC57BL/6JJ blastocysts to generate chimeric mice identifiable by their coat colors. ES cell culture conditions and blastocyst injection have been described previously (9, 32).
Chimeras were bred to C57BL/6J or 129S6/SvEv mice to give rise to (C57BL/6J × 129S6/SvEv)F1 hybrid mice or to 129S6/SvEv inbred mice, respectively. Chimeras derived from two independent ES clones transmitted the targeted allele to offspring and were used to establish two separate mouse colonies. Mice were genotyped by Southern blot analysis of EcoRI/EcoRV-digested tail DNA by using a labeled InhBP/p120 cDNA probe (Fig. 1B). The probe template was amplified from mouse pituitary cDNA with direct (5′-GCA AAG CCT TCA AGA CTT GG-3′) and reverse (5′-CCC CTG TAT GAC CCA TCA TA-3′) primers located in exons 2 and 6 of the mouse InhBP/p120 gene, respectively.
RNA isolation.
For Northern blot analysis of InhBP/p120 expression in mutant and wild-type mice, six 8-week-old mice of each sex and genotype were sacrificed and the pituitary glands were removed. Tissues were pooled by genotype and sex and immediately preserved in RNAlater reagent (Ambion) before RNA extraction. Total RNA was subsequently extracted with TRI reagent (Sigma, St. Louis, Mo.) by following the manufacturer's instructions. For RT-PCR analysis, three 8-week-old mice of each sex and genotype were sacrificed and pituitaries were pooled as described above. RNA was isolated with TRI reagent. All mice in these studies were handled in accordance with local and federal guidelines.
Northern blot and RT-PCR analyses of InhBP/p120 knockout mice.
For Northern blot analyses, 15 μg of total RNA from male and female wild-type and InhBP/p120 knockout mouse pituitaries was electrophoresed on a MOPS (morpholinepropanesulfonic acid)-formaldehyde agarose gel by using standard techniques. RNA was then transferred overnight onto a positively charged nylon filter (Nytran Supercharge; Schleicher & Schuell, Keene, N.H.) by capillary action with 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). The filter was hybridized sequentially with [32P]dCTP random prime-labeled (Ready-To-Go labeling beads; Amersham Pharmacia) cDNA probes corresponding to the open reading frame of the rat InhBP-S cDNA (GenBank accession number AF322217), the 1,069-bp EcoRI fragment between bp 1662 and 2731 of the rat InhBP-L cDNA (GenBank accession number AF322216), 381 bp of the mouse FSHβ cDNA (GenBank accession number NM_008045), and bp 401 to 595 of the cDNA for the rat ribosomal protein L19 (RPL19; GenBank accession number NM_031103). The latter probe was used to confirm equal loading of the RNA samples on the gel. The filter was hybridized by using conditions described previously (44) and stripped with boiling 0.5% sodium dodecyl sulfate between hybridizations.
For RT-PCR analyses, 2 μg of total pituitary RNA from male and female wild-type and mutant mice was reverse transcribed into cDNA by using 100 U of Moloney murine leukemia virus reverse transcriptase (MMLV-RT) in the presence of 20 U of RNasin, 500 μM deoxynucleoside triphosphates, and 100 ng of random hexamer primers (all reagents from Promega). Negative control (RT−) samples were run identically except that the RT enzyme was omitted. In the subsequent PCR, these controls demonstrated that amplified products were derived from RNA and not contaminating genomic DNA in the samples. Several PCRs were run on each RT or RT− sample with the primers indicated in Table 1. Four microliters of the 40-μl RT or RT− sample was combined with 46 μl of a master mix including 2.5 U of Taq polymerase (Promega), PCR primers (400 nM each), 200 μM deoxynucleoside triphosphates, 1× PCR buffer, and 1.5 mM MgCl2 and subjected to 28 or 35 cycles of amplification with an iCycler (Bio-Rad, Hercules, Calif.). Ten microliters of each product was run on 1.2% agarose gels containing ethidium bromide. Gels were photodocumented with a digital camera interfaced with an IBM Think Pad computer with Kodak digital science one-dimensional image analysis software (version 2.0.2).
TABLE 1.
PCR primers used to analyze InhBP/p120 isoform expression in wild-type and mutant micea
| Primer(s) corresponding to: | Sequence | Amplicon size (bp) | PCR cycle no. |
|---|---|---|---|
| InhBP-L-mInhBP-3 | 5′-CTAACATCTGGTCACATCGC | 397 | 35 |
| 3′-AGCACAAACTCCTTAGTTGA | |||
| InhBP-S | 5′-ATGATGCTTCGGACCTTCAC | 672 | 35 |
| 3′-AATTGRCAGATGCCAGCAGCC | |||
| mInhBP-4 | 5′-TTTTTTTCTGGTCTCGCTAG | 222 | 35 |
| 3′-AGAAGGGCTTCTGCACCAGA | |||
| RPL19 | 5′-CTGAAGGTCAAAGGGAATGTG | 195 | 28 |
| 3′-GGACAGAGTCTTGATGATCTC |
Some of these primers were designed for use in rats or humans and are not perfect matches for the mouse. Bases listed in boldface do not match those from the mouse sequence.
Characterization of novel InhBP/p120 transcripts in mouse pituitary.
A Northern blot of 15 μg of wild-type C57BL/6J male mouse pituitary RNA was generated as described above. The blot was hybridized sequentially with [32P]dCTP-labeled cDNA probes corresponding to the rat InhBP-S cDNA open reading frame (as described above), a 204-bp fragment corresponding to exon 13b (see Results) of the mouse InhBP/p120 gene, and a RT-PCR-generated fragment corresponding to sequences in exons 18 through 20 of the mouse InhBP/p120 gene (primer sequences, 5′-GGAAGGGAAAGGCATTGGAAAC-3′ and 5′-TGGTTACACTCTTCAAGGACTACGG-3′). The blot was stripped with boiling 0.5% sodium dodecyl sulfate between hybridizations. The results of these analyses revealed four distinct transcripts in mouse pituitary. Two transcripts corresponded to the previously characterized long and short forms of InhBP (InhBP-L and InhBP-S). Two novel transcripts (both about 2.7 kb) were further characterized by 3′ RACE and long-range RT-PCR (Invitrogen).
Morphological and histological analyses.
Three mice of each sex and genotype were sacrificed and tissues were collected immediately into 10% buffered formalin (pituitaries and ovaries) or Bouin's fixative (testes). After overnight fixation, tissues were embedded in paraffin blocks, sectioned, and stained with hematoxylin and eosin. Embedding and staining were performed by using standard procedures in the Baylor College of Medicine Pathology Core Services laboratory.
Serum analyses.
Fifteen mice of each sex and genotype were anesthetized by inhalation of isoflurane (Abbott Laboratories, North Chicago, Ill.), and blood was recovered by closed cardiac puncture. Serum samples were separated in Microtainer tubes (Becton Dickinson, Franklin Lakes, N.J.) and stored at −80°C prior to analysis. FSH and luteinizing hormone (LH) measurements were made at the University of Virginia Ligand Core Facility (Specialized Cooperative Centers Program in Reproduction Research). Differences in hormone levels between genotypes were tested by using nonpaired t tests, and statistical significance was assessed relative to P of <0.05.
Nucleotide sequence accession numbers.
cDNA sequences for the mouse InhBP-L (accession number AY227771), InhBP-S (accession number AY227772), mInhBP-3 (accession number AY227773), and mInhBP-4 (accession number AY227774) genes were submitted to GenBank and assigned the indicated accession numbers.
RESULTS
Mouse InhBP cDNA and genomic organization.
The mouse InhBP-L cDNA is 4,403 bp, with an open reading frame of 3,951 bp predicted to encode a protein of 1,317 amino acids. The mouse InhBP-L amino acid sequence shares 95 and 84% sequence identity with the orthologs in rat and human. The mouse InhBP-L mRNA is derived from 20 exons spanning 15.2 kb of genomic sequence (Fig. 2A). The sizes of the exons and introns as well as the splice junctions are summarized in Table 2. In all cases, with one exception, the canonical splice donor (gt) and splice acceptor (ag) dinucleotide sequences are observed. (Lowercase letters are used for intron nucleotides.) The splice donor in intron 7 is gc rather than gt. Interestingly, the same dinucleotide is observed at this splice donor site in the human InhBP/p120 gene (20). Overall, the exon-intron structure as well as the length of each segment is well conserved in mice and humans. A previous report indicated that the human InhBP-L gene (referred to as Ig domain-containing gene 1 [IGDC1]) consists of 19 exons spanning less than 20 kb (20). Our analysis of the human InhBP cDNA (GenBank accession numbers Y10523 and AF034198) in comparison with the relevant genomic sequence of human Xq25-26.2 (GenBank accession number AL590806) indicates that the human gene spans 15.8 kb and comprises 20 exons, similar to the mouse InhBP/p120 gene. The previous study observed that exon 19 contained about 550 bp and encoded both the transmembrane and intracellular domains. Our analysis indicates that human exon 19 is 124 bp and encodes the transmembrane domain and a small portion of the intracellular domain. This exon is followed by a 151-bp intron and the 423-bp exon 20, which encodes the majority of the intracellular domain. This is the same organization that we observe for the mouse InhBP/p120 gene. In addition, in both the mouse and human genes, the open reading frame begins in exon 2 and terminates in exon 20. In mice, exon 1 contains 87 bp of the 179-nucleotide 5′ untranslated region (UTR). The 276-nucleotide 3′ UTR is contained entirely within exon 20. A consensus polyadenylation sequence (AAUAAA) is located 32 nucleotides 5′ of the poly(A) tail.
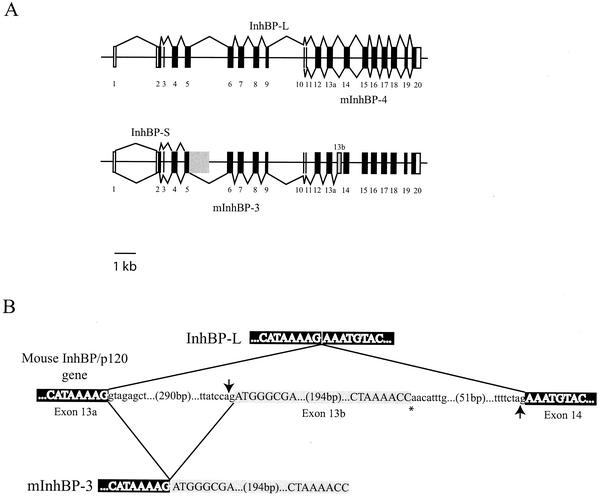
FIG. 2.
Schematic representation of the mouse InhBP/p120 gene and mRNA organization. (A) The mouse InhBP/p120 gene consists of 20 exons and spans 15.2 kb. Exons are numbered and depicted as boxes or vertical lines. Unfilled boxes and filled boxes represent noncoding and coding sequences, respectively. The top of the panel shows the splicing of InhBP-L and mInhBP-4 genes. The former comprises exons 1 through 20, while the latter consists of exons 10 through 20 and is believed to derive from an alternative promoter in intron 9. Both InhBP-L and mInhBP-4 genes use exon 13a. The bottom of the panel shows the splicing of InhBP-S and mInhBP-3 genes. The former comprises exons 1 through 5 as well as part of intron 5 (shaded box). The mInhBP-3 gene comprises exons 1 through 13a plus exon 13b (grey box) at its 3′ end. (B) Schematic representation of the splicing events that give rise to either InhBP-L (top) or mInhBP-3 (bottom). The 3′ end of exon 13a and 5′ end of exon 14 are indicated in white type with black shading. Exon 13b is highlighted with grey shading. In the InhBP-L transcript, the 3′ splice acceptor (ag) at the end of intron 13 (indicated by the upward-pointing arrow) is used. In the mInhBP-3 transcript, an alternative 3′ splice acceptor within intron 13 (indicated by the downward-pointing arrow) is used. The asterisk marks the 3′ end of the mInhBP-3 sequence, where the poly(A) tail begins. Intronic sequences are indicated in lowercase.
TABLE 2.
Mouse InhBP-L cDNA splice junctionsa
| Exon | Exon length (bp) | 5′ splice donor | Intron length (bp) | 3′ splice acceptor |
|---|---|---|---|---|
| 1 | 87 | TTCTACTACCgtaagtacct | 2,245 | cttcttttagATTCTACTAC |
| 2 | 129 | CTTTGCATTTgtaagttcaa | 129 | ctcttctcagGGCTCAATCC |
| 3 | 27 | ACTTCACTGGgtaagtgtct | 349 | ggaatggcagCAGTGGAGTC |
| 4 | 282 | GAAGCACCAGgtgagatggc | 300 | ttttttccagGCCAACTCCC |
| 5 | 288 | GTGGTAGCAGgtgggtgtgg | 1,804 | ccctttctagGTCTCTACCC |
| 6 | 285 | TGGGTGACTGgtaagatatg | 234 | ctcatcctagACACTTTCCC |
| 7 | 294 | ATGGTTGTAGgcaagtgatc | 520 | tccctttcagCTTGGCCCAG |
| 8 | 252 | GGACCAGCAGgtgagaagat | 312 | ctggtcacagGCTTCCTCAC |
| 9 | 121 | TGAGGCTCAGgtaatttgct | 1,863 | gtctcgctagAGAAGCCTGG |
| 10 | 71 | CTTTGCTGTGgtgagtacaa | 82 | tcttttgcagGACTGTGCAA |
| 11 | 33 | GAGGAGATTGgtgagttttg | 384 | ctgtttctagAAATAGTCAT |
| 12 | 291 | GTGGGGACAGgtaagagaaa | 220 | tgttttctagACATCCTTCC |
| 13a | 279 | GTCATAAAAGgtagagctgg | 583 | tcttttctagAAATGTACCC |
| 14 | 288 | GTTGTGACAGgttagggagg | 728 | ttcattttagAATTCTACCC |
| 15 | 288 | TGGGTGACTGgtaagaacag | 178 | ctctgtctagATACATTCCC |
| 16 | 288 | ATAGTCACTGgtaaggggat | 196 | tttcttatagGTTTACTCCC |
| 17 | 288 | TGGGTGACTGgtaaggttct | 131 | ctggttacagATAAGCCCCC |
| 18 | 279 | GGAGCTGCAGgtgagtggat | 388 | tataccacagGGCCTGTTGC |
| 19 | 124 | TTCGGACTAGgtaaatactc | 147 | ctttatacagGGGCTCTGAG |
| 20 | 412 |
The table shows the DNA sequences at the exon-intron boundaries for mouse InhBP-L cDNA. Lowercase letters represent introns. Sequences of exons 1 to 20 were determined by aligning the mouse InhBP-L cDNA (GenBank accession number AY227771) with the genomic sequence from the mouse X chromosome (GenBank accession number AL731789 and our unpublished data). Note that the splice donor site in intron 7 (underlined) does not conform to the consensus sequence. The same gc dinucleotide is observed in the human intron 7 splice donor site.
The mouse InhBP-S cDNA was amplified by RT-PCR from adult pituitary RNA. The open reading frame encodes a 232-amino-acid predicted protein that is 95 and 87% identical to the InhBP-S (or pituitary gland-specific factor-2 [PGSF2]) sequence described in rats and humans, respectively (8, 38).
Novel InhBP/p120 isoforms in mouse pituitary.
In addition to the InhBP-L and InhBP-S isoforms described previously in humans and rats (8, 20, 33, 38), we identified two novel InhBP/p120 mRNA variants in mouse pituitary glands (Fig. 2 and 3). With the use of 3′ RACE, a product of approximately 1.1 kb was amplified with a sense primer (5′-CTAACATCTGGTCACATCGC-3′) directed against sequences in exon 8 and with the abridged universal anchor primer (Invitrogen). Nested PCR, with a sense primer in exon 9 (5′-AAGCCTGGTTGTTGGGAACA-3′) and the abridged universal anchor primer, was used to confirm that the amplified product corresponded to the InhBP/p120 gene. A product of approximately 0.9 kb was amplified. Upon sequencing, the product was observed to contain InhBP-L cDNA sequences from exon 9 through the end of exon 13 plus 210 bp of intron 13 at its 3′ end (Fig. 2B). Interestingly, this intron 13 sequence was not continuous with exon 13 within the genomic sequence but began 307 bp into intron 13 (Fig. 2B). An alternative splice acceptor site appears to be used within the intron, resulting in a novel exon being added to the 3′ end of this transcript (referred to as exon 13b in Fig. 2). A consensus polyadenylation signal occurs 29 bases 5′ of the poly(A) tail in this variant. PCR was used to amplify a 204-bp region within exon 13b corresponding to the 3′ end of this variant. The product was used as a probe for Northern blotting of total RNA from mouse pituitary. A single minor transcript of 2.7 kb was detected (Fig. 3B). This transcript corresponds in size to one detected with a 5′ probe that also detected both InhBP-L and InhBP-S transcripts (Fig. 3A). These data suggested that the transcript containing exon 13b might extend 5′ through exon 1.
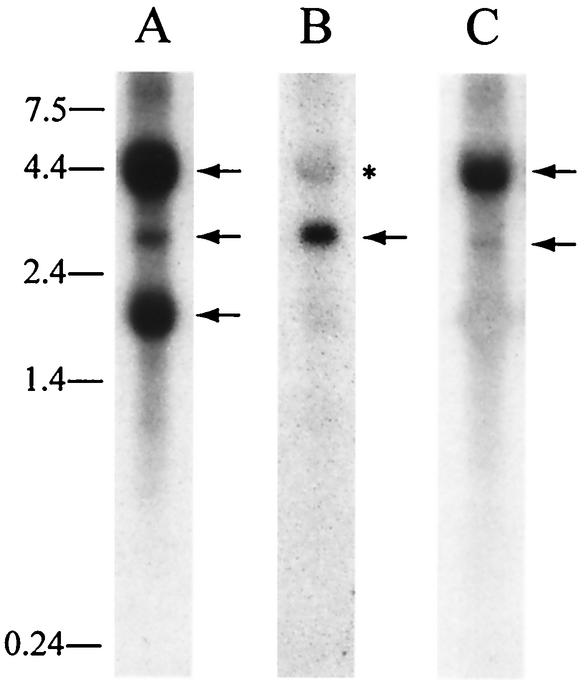
FIG. 3.
InhBP/p120 transcripts in mouse pituitary gland. Northern blot analyses indicate the presence of four major InhBP/p120 RNA species in adult mouse pituitary. (A) A probe directed against the open reading frame of the rat InhBP-S gene detected transcripts of 4.4 (InhBP-L, top arrow), 2.7 (mInhBP-3, middle arrow), and 1.8 (InhBP-S, bottom arrow) kb. (B) A 204-bp probe corresponding to exon 13b detected a transcript of 2.7 kb (mInhBP-3, arrow). The asterisk indicates the InhBP-L transcript that was incompletely stripped from a previous hybridization. (C) A probe directed against 3′ sequences in exons 18 through 20 detected transcripts of 4.4 (InhBP-L, top arrow) and 2.7 (mInhBP-4, bottom arrow) kb. RNA molecular size standards (in kilobases) are indicated at the left of the figure.
Using long-range RT-PCR with a forward primer directed against the 5′ UTRs of the InhBP-L and InhBP-S genes (5′-ACTACACCAGAGGGCCTCCA-3′) and a reverse primer within exon 13b (5′-AGGAAGAAGTCCGCTATATGC-3′), we amplified a fragment of 2,513 bp. Upon sequencing, the PCR product was determined to contain exons 2 through 13a, as described for the InhBP-L cDNA, plus the novel exon 13b. The open reading frame of this variant encodes a protein of 762 amino acids, predicted to correspond to the first seven Ig-like loops and the hydrophobic linker region between loops 5 and 6 of InhBP-L, followed by eight novel amino acids encoded by exon 13b. The predicted protein lacks a carboxy-terminal hydrophobic domain, and therefore it is unlikely to be linked to the membrane by a glycosylphosphatidylinositol moiety or a carboxy-terminal transmembrane domain. A web-based informatics package (http://www.cbs.dtu.dk/services/TMHMM/) predicts that the hydrophobic linker region may contain two transmembrane helices, but we do not yet know if this form of the protein is membrane bound or soluble. Hereafter we refer to this variant as mInhBP-3 (Fig. 2).
A fourth variant of InhBP/p120 was observed in mouse pituitary RNA hybridized with a probe directed against 3′ sequences in the cDNA. As shown in Fig. 3C, this probe detected the InhBP-L transcript and a minor transcript of approximately 2.7 kb. While the size of this minor transcript was indistinguishable from that of the mInhBP-3 transcript when analyzed by Northern blotting, the probe was directed against sequences in exons 18 to 20, which are absent from the mInhBP-3 transcript (Fig. 2A). We have observed similar transcripts in Northern blots of rat pituitary, testis, and adrenal RNAs hybridized with a homologous probe (data not shown). Since this transcript, but not that of the InhBP-L or InhBP-S gene, is expressed in rat adrenal gland (8), we hypothesized that it might derive from an alternative (intronic) promoter that is active in the rat adrenal gland. In mice, exons 10 to 20 of the InhBP-L gene contain approximately 2.7 kb of sequence. Therefore, we further hypothesized that if this putative intronic promoter is also used in mice, it may reside within the large intron 9 (1,863 and 1,876 bp in mouse and human, respectively) and drive transcription of a 2.7-kb mRNA that would comprise predominantly exons 10 to 20 of the InhBP-L gene.
To test this hypothesis, we screened the mouse expressed sequence tag (EST) database by BLASTN by using the mouse sequence at the end of intron 9 and beginning of exon 10. Three cDNA clones from mouse embryo or neonatal head (GenBank accession numbers AI787545, BB575799, and BB613891) contained the continuous intron 9-exon 10 sequence. Based on sequences derived from these EST clones, we designed PCR primers against the 3′ end of mouse intron 9 and in exon 20 (within the 3′ UTR of the InhBP-L gene) to amplify this variant from adult mouse pituitary. Upon sequencing of the amplified product, we determined that exons 10 to 20 were included in this variant and were spliced as in the InhBP-L gene. The open reading frame was 2,325 bp. The predicted protein contains the final seven Ig-like loops, transmembrane domain, and cytoplasmic domain observed in InhBP-L. Interestingly, the second hydrophobic domain (encoded by exons 10 and 11) in the linker region of InhBP-L represents the N terminus of this variant and is predicted to function as a signal peptide in this form of the protein. We refer to this variant as mInhBP-4 and predict that it encodes a membrane-bound protein.
By Northern blot analysis, we have been unable to confirm that the 2.7-kb transcript detected with the 3′ probe (Fig. 3C) corresponds to mInhBP-4 because the only unique sequence in this variant is the 21-bp sequence at the end of intron 9. Unfortunately, its short length and the presence of a poly(T) segment (Table 3) make this a poor probe candidate. However, the pattern of InhBP/p120 expression in the knockout mouse pituitaries is consistent with the proposition that mInhBP-3 and mInhBP-4 are distinct variants and that the latter is derived from a unique intronic promoter (see below; Fig. 4).
TABLE 3.
Intron 9-exon 10 junctions in rats, mice, and humansa
| Species | Sequences of intron 9-exon 10 junction |
|---|---|
| Rat | ttttttttctggtctcactagAGAAGCCTGG |
| Mouse | ttttttttctggtctcgctagAGAAGCCTGG |
| Human | tctcttttctggtctcactagAGAAGCCTGG |
The 3′ 21 bp of intron 9 (lowercase) and 5′ 10 bp of exon 10 (uppercase) in rat, mouse, and human InhBP genes are shown. Nucleotides in bold are different from those in the other two species. The intron 9 sequence, which is absent from InhBP-L, InhBP-S, and mInhBP-3 cDNAs, is located at the 5′ end of the mInh-BP-4 cDNA. mInhBP-4 gene transcription initiation appears to occur within intron 9.
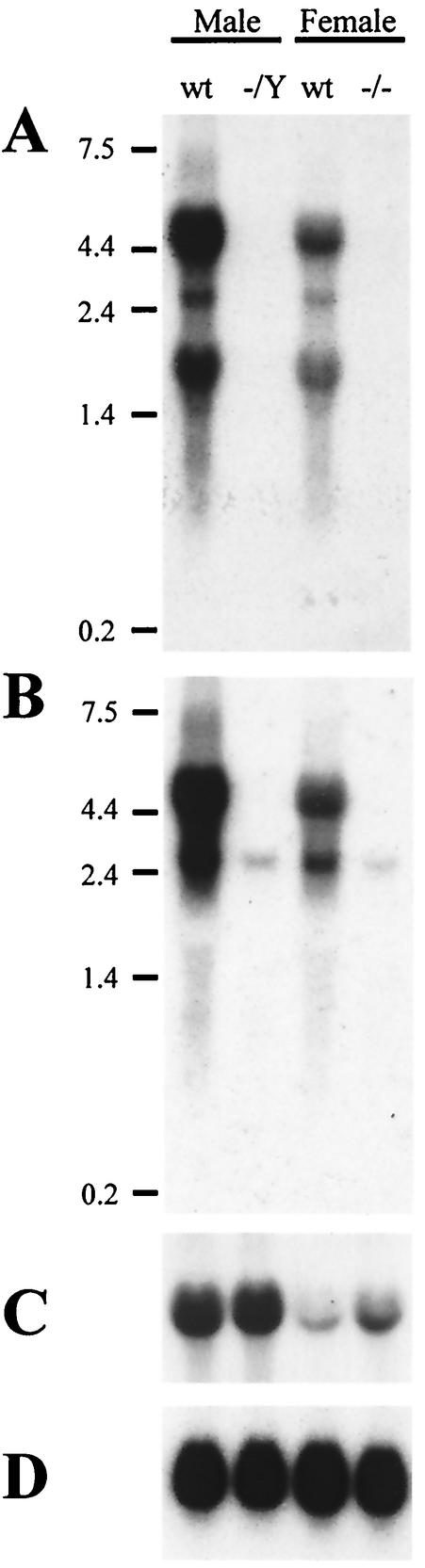
FIG. 4.
InhBP/p120 expression in wild-type (wt) and knockout pituitaries. (A) A Northern blot shows expression of InhBP-L (4.4 kb), mInhBP-3 (2.7 kb), and InhBP-S (1.8 kb) in wild-type, but not mutant, mouse pituitaries of males (−/Y) and females (−/−). (B) The same blot hybridized with a 3′ probe shows expression of InhBP-L (4.4 kb) in wild-type, but not knockout, mice. This probe also detected a minor mInhBP-4 band (2.7 kb) in both genotypes. (C) FSHβ mRNA levels were higher in males than in females of both genotypes. The mutation did not significantly affect FSHβ expression in knockout mice relative to that in wild-type animals. (D) Probing of the blot with a rat RPL19 cDNA indicated equal loading of RNA in all lanes.
Generation of InhBP/p120 mutant mice.
As described in Materials and Methods, a targeting vector with 7.9 kb of total homology to the wild-type InhBP/p120 gene was designed to replace a 428-bp segment containing the noncoding exon 1 with a Pgk-HPRT selection cassette (Fig. 1A). Because this exon contains the transcription start site (as determined by 5′ RNA ligase-mediated [RLM]-RACE), we reasoned that this strategy would produce a null mutation for the InhBP-L, InhBP-S, and mInhBP-3 transcripts (and corresponding proteins). The mInhBP-4 transcript (described above) was identified only subsequent to production of the knockout mice; therefore, no attempt had been made to produce a deletion that would affect intron 9 and exon 10, where mInhBP-4 gene transcription and translation are initiated. Nonetheless, because the InhBP-S mRNA terminates in intron 5 and the mInhBP-4 mRNA starts in intron 9, a deletion of 4.7 kb would have been required to directly affect all four transcripts. In retrospect, the strategy employed was optimal in terms of affecting the most numerous and abundant transcripts (see below). As described, ES cells surviving the positive-negative selection were targeted at an efficiency of 11.7%. Because InhBP/p120 is X-linked and ES cells with the targeted allele are hemizygous for the mutation, the gene does not appear to be necessary for ES cell viability. Targeted ES cells were expanded and injected into C57BL/6J blastocysts to generate chimeras identifiable by coat colors.
Genotype distributions and breeding data.
Offspring from chimeras with the agouti coat color were genotyped by Southern blot analysis to confirm inheritance of the targeted X chromosome InhBP/p120 allele. Twelve cages with heterozygous females breeding to wild-type males were then established to generate mutant males, and the sexes and genotypes of offspring were recorded (Fig. 1B). From these mating cages, a total of 135 females and 144 males were born, approximating the expected 1:1 ratio. Of these offspring, 109 males were genotyped to identify 54 wild-type (X+Y) and 55 mutant (knockout; X−Y) males. Similarly, 10 cages with heterozygote females breeding to knockout males were established to yield 134 females and 119 males. Of the female offspring from matings of X+X− and X−Y mice, 107 were genotyped to identify 50 heterozygous (X+X−) and 57 homozygous mutant (knockout; X−X−) females. Thus, expected sex ratios and Mendelian genotype distributions of offspring were found in mice carrying this X chromosome mutation. These data indicate that the mutant allele does not affect sexual differentiation or viability.
InhBP/p120 knockout males and females of the C57BL/6J/129S6/SvEv genetic background were bred with control mice to assess their fertility. In these studies, 10 hemizygous (X−Y) males were bred for up to 6 months. These males sired an average of 0.86 ± 0.19 litter per month with an average of 7.47 ± 1.46 pups per litter. No males were infertile. Similarly, 10 female homozygous (X−X−) knockout mice were monitored for up to 6 months of breeding. All fostered litters, producing an average of 0.95 ± 0.24 litter per month and an average of 8.23 ± 1.69 pups per litter. Heterozygous (X+X−) females produced an average of 0.84 ± 0.10 litter per month with an average of 7.71 ± 1.53 pups in each litter. None of these values deviated significantly from normal (23, 24).
InhBP mRNA expression in mutant mice.
Since the InhBP/p120 mutant males and females were fertile, we tested whether the mutation had disrupted the production of the InhBP/p120 mRNA forms. InhBP/p120 mRNA was detected in wild-type male and female pituitaries by using a probe that detects InhBP-L, InhBP-S, and mInhBP-3 (Fig. 4A). All three transcripts were more abundantly expressed in male than in female mouse pituitaries. Importantly, none of these transcripts were detected by Northern blotting in the pituitary glands of mutant males (X−Y) or females (X−X−). These data demonstrated that the targeting strategy produced a null mutation for these three forms of InhBP/p120.
A 3′ probe was hybridized to the same blot. InhBP-L and mInhBP-4 were detected in wild-type pituitaries (Fig. 4B). Interestingly, while InhBP-L was absent, the minor mInhBP-4 band was detected in both male and female knockout pituitaries. The probe used here detected both mInhBP-3 and mInhBP-4 transcripts, so the 2.7-kb band detected in wild-type pituitaries reflected a combination of the two transcripts, while the band in knockout animals represented the mInhBP-4 transcript alone (see also RT-PCR analysis below). Based on these results, it was not possible to conclude whether or not the mutation affected mInhBP-4 mRNA levels. Subsequent analyses (see below; Fig. 5) indicated comparable mInhBP-4 expression in the two genotypes (at least for females).
FIG. 5.
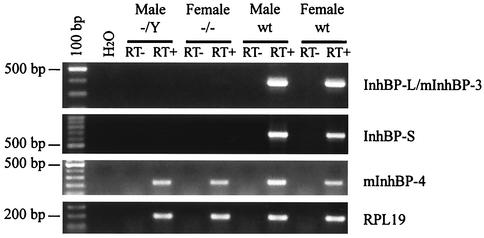
RT-PCR analysis of InhBP/p120 isoform expression in mouse pituitaries. RT-PCR analysis confirms the lack of InhBP-L, mInhBP-3, and InhBP-S expression in mutant male and female pituitary glands (top two panels). mInhBP-4 is expressed in males and females of both genotypes (third panel). RPL19 mRNA levels are similar in wild-type and mutant mice (bottom panel). No bands were detected in the H2O only or RT− samples. wt, wild-type.
To confirm that the mutation was not hypomorphic for the InhBP-L, InhBP-S, and mInhBP-3 transcripts, we employed more sensitive RT-PCR analyses to confirm the absence of these three transcripts in mutant mice. Primers directed against sequences in exons 8 and 13a, which recognize both InhBP-L and mInhBP-3 transcripts, amplified the predicted product in wild-type but not mutant pituitaries (Fig. 5, top panel). Similarly, primers designed to recognize InhBP-S amplified the predicted product in wild-type but not mutant pituitaries (Fig. 5, second panel). Using primers directed against sequences in intron 9 and exon 12, we amplified a product of 222 bp in both wild-type and knockout pituitaries (Fig. 5, third panel) corresponding to the 5′ end of the mInhBP-4 transcript. mInhBP-4 mRNA levels were slightly higher in wild-type males than in the other three groups. This result was confirmed by Northern blotting (data not shown). Thus, the mutation does not appear to affect mInhBP-4 expression in females, although it may minimally suppress expression in males.
FSHβ mRNA and serum gonadotropin levels.
If InhBP/p120 functions as an inhibin coreceptor, its absence in the pituitary would be predicted to increase FSH synthesis because of a decrease in pituitary sensitivity to inhibins. We therefore measured FSHβ subunit mRNA levels by Northern blotting in knockout and wild-type pituitaries to test this hypothesis. FSHβ mRNA levels were higher in males than in randomly cycling females (Fig. 4C). Whereas levels were comparable in wild-type and mutant males, there was a trend for increased FSHβ expression in knockout females relative to that in wild-type females (Fig. 4C). However, a similar analysis on a second set of pooled pituitaries failed to reproduce this pattern of expression (data not shown), suggesting that the physiological condition of the mice used (e.g., estrous cycle stage) and not the mutation per se may have affected FSHβ expression. RPL19 expression was comparable in all four samples, indicating equal loading of the RNA samples (Fig. 4D).
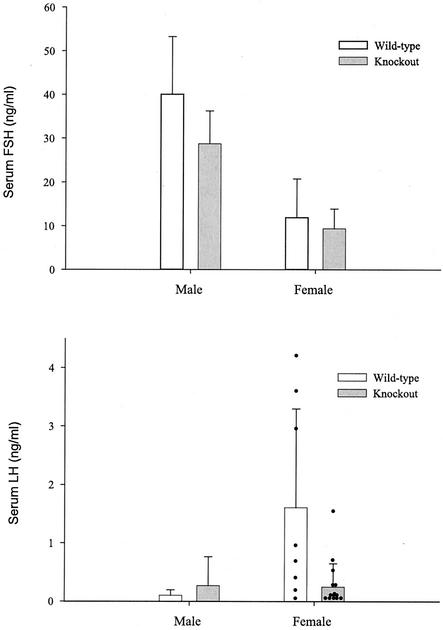
Blood samples were collected from male and randomly cycling female wild-type and knockout mice to assess circulating gonadotropin levels. Consistent with the sex-linked difference in pituitary FSHβ mRNA levels, serum FSH concentrations were higher in males than in females. Neither FSH levels nor LH levels in serum differed significantly between genotypes (Fig. 6).
FIG. 6.
Serum gonadotropin levels. Radioimmunoassays for FSH (top) and LH (bottom) indicated that serum gonadotropin levels were not significantly affected by the mutation (data reflect the means ± standard deviations). Note that in the LH graph, individual data points are presented for females (black dots). While the two groups do not differ significantly, the large variability observed in the female wild-type group appears to result from elevated LH levels in three animals. An LH surge or pulse was observed in only one of the knockout females. The animals used in these studies were randomly cycling, and these data reflect the normal variability observed when animals are sampled in this fashion.
Mutant mice were sacrificed at 8 weeks of age to assess pituitary and gonadal histology. No abnormalities were observed, and all pituitary lineages, stages of spermatogenesis, and stages of folliculogenesis were evident (data not shown). Northern blot analyses failed to show differences in levels of LHβ, thyroid-stimulating hormone β, growth hormone, prolactin, or pro-opiomelanocortin (POMC) expression in wild-type and knockout animals (data not shown).
DISCUSSION
Since its initial identification as a putative inhibin coreceptor (16), InhBP/p120's role in inhibin biology has been investigated at length (8, 12, 13, 21). Various forms of InhBP/p120 are expressed at high levels in the pituitary glands of all mammalian species examined to date (e.g., mice, rats, humans, rhesus monkeys, sheep, and cows) and to a much lower extent in other tissues (8, 20, 33). The high specificity of InhBP/p120 expression within the pituitary is consistent with its hypothesized role in inhibin action, given that inhibins appear to target the pituitary gland and few other tissues. Moreover, in female rats, InhBP-L and InhBP-S mRNA levels fluctuate across the estrous cycle such that levels are high during metestrus and diestrus (when serum FSH levels are low) and low during the secondary FSH surge on the morning of estrus (8). In vitro studies in heterologous cell lines indicate that InhBP-L interacts with the activin type IB receptor, ALK4, and appears to potentiate inhibin B's antagonism of activin A-stimulated gene transcription (13). More recent studies have investigated the inhibin binding properties of InhBP-L (12). Surprisingly, and in apparent contrast to the results of the original affinity chromatography analysis that was used to identify InhBP/p120 (16), InhBP-L does not bind inhibin A or inhibin B when expressed alone or in combination with various activin receptors in heterologous cell lines. These latter observations cast some doubt on the role of InhBP/p120 as an inhibin coreceptor (5).
To characterize more thoroughly the role of InhBP/p120 in inhibin biology and reproduction, we generated InhBP/p120 mutant mice by gene targeting in ES cells. Our targeted mutation strategy eliminated exon 1 of the 20-exon mouse InhBP/p120 gene, which includes the transcription start site. Northern blot analyses indicate a complete absence of InhBP-L and InhBP-S transcripts in both male and female mutant mice, while levels of both transcripts are abundant in wild-type littermates. These results are corroborated by the results of more sensitive RT-PCR analyses.
Northern blot analysis with a 5′ probe also indicates the presence of a 2.7-kb transcript not previously identified in rats (8). Because this represents the third form of InhBP/p120 characterized and because it may be unique to mice, we named the form mInhBP-3. The mInhBP-3 transcript comprises the first 13 exons observed in InhBP-L plus an additional exon (exon 13b) derived from the sequence in intron 13. Exon 13b appears to be incorporated into this transcript through the use of an alternative 3′ splice acceptor site upstream of the splice site used for exon 14 in the InhBP-L transcript. The transcript terminates in exon 13b, as a consensus polyadenylation sequence provides the necessary signal for the addition of a poly(A) tail prior to the start of exon 14. This variant is predicted to encode a protein comprising the first seven Ig-like loops observed in InhBP-L. It is not clear whether this predicted protein is membrane bound or soluble. mInhBP-3 mRNA is also undetectable in pituitaries of both male and female mutant mice.
Despite the absence of InhBP-L, InhBP-S, and mInhBP-3 transcripts (all of which include exon 2, which encodes the N-terminal signal peptide), mutant male and female mice have no apparent deficits in reproductive function. Both male and female mutant mice produce normal litter sizes at frequencies indistinguishable from those of control mice of the same genetic background. Consistent with their normal fertility, neither male nor female mutant mice exhibit altered serum FSH or LH levels. An a priori prediction was that these mice would have elevated FSH levels because of a lowered sensitivity of the pituitaries to the inhibitory effects of gonadal inhibins. Not only were serum FSH levels unaffected by the mutation, we also did not detect any alteration of pituitary FSHβ mRNA levels. Histological examination of the pituitaries of the mutant mice did not reveal any obvious changes in cellular morphology. The mutation also does not appear to affect lactotrope, somatotrope, thyrotrope, or corticotrope function. In addition, the ovarian and testicular phenotypes of the mutant mice resemble those of their wild-type littermates and, unlike inhibin α-null mice (32), InhBP/p120 knockout mice do not develop gonadal tumors.
In the course of our investigation, we observed a fourth InhBP/p120 transcript in mouse pituitaries (encoding mInhBP-4). This variant is 2.7 kb, like the mInhBP-3 transcript, but is detected with a probe that lies 3′ from where the mInhBP-3 transcript terminates. We observed a similarly sized transcript in rat pituitaries and testes by using a comparable probe. Interestingly, this variant is expressed in rat adrenal gland, a tissue in which we have not detected other forms of InhBP/p120 by Northern blotting (8). This distinct pattern of expression of InhBP/p120 variants led us to hypothesize that mInhBP-4 is transcribed from an alternative promoter. In fact, using 5′ RLM-RACE analysis with rat testes, we mapped a transcription start site within intron 9 (data not shown). We screened the mouse EST database and located a clone in which the 5′ end also contains the analogous intron 9 sequence and the 3′ end continues through exon 20. Using primers corresponding to the ends of this EST clone, we amplified by RT-PCR the mInhBP-4 transcript from mouse pituitary RNA. The predicted protein encoded by this transcript corresponds to part of the hydrophobic linker region, the last seven Ig-like loops, the transmembrane domain, and the intracellular tail of InhBP-L. Although the normal NH2-terminal signal peptide is absent from a hypothetical InhBP-4 protein, the linker region, which constitutes the amino terminus of this variant, may function as a signal peptide, suggesting that this truncated form of the protein may be trafficked to the membrane. Although it is possible that we observe no alterations in reproductive phenotypes in the mutant mice because mInhBP-4 compensates for loss of the other forms of InhBP/p120, we consider this unlikely because mInhBP-4 mRNA levels are significantly lower than those of the dominant InhBP transcripts. In addition, the mInhBP-4 levels in the mutant mice were comparable to, if not lower than (males), those in wild-type controls. Nonetheless, in the future, it may be valuable to produce mutant mice that are deficient in all forms of InhBP/p120. Based on the results here, though, we predict that such mice would also be viable and fertile.
Although InhBP/p120 was initially identified by inhibin A affinity chromatography (16), there are now several pieces of data calling into question the role of this protein as an inhibin coreceptor. First, in standard receptor binding studies, InhBP-L fails to bind iodinated inhibin A or inhibin B when expressed alone or in combination with activin receptors ACVR2B and/or ALK4. Under the same experimental conditions, both forms of inhibin bind betaglycan with high affinity (12). Second, betaglycan, but not InhBP/p120, appears to be part of an inhibin A binding complex in mouse testis cell lines (TM3 and TM4) and rat pituitary cells (19, 21). Third, the data reported in the present study indicate that in the absence of three major pituitary forms of InhBP/p120 (InhBP-L, InhB-S, and mInhBP-3), there is no alteration in FSH synthesis or secretion or in fertility. In summary, the existing data fail to support the hypothesis that InhBP/p120 is an essential inhibin coreceptor. Future studies using the mutant mice described here may help determine the functions of the various forms of InhBP/p120.
Acknowledgments
Daniel J. Bernard and Kathleen H. Burns contributed equally to this work and share first authorship on this paper.
We acknowledge the assistance of the Ligand Core Facility, supported by NICHD/NIH through agreement U54 HD28934 as part of the Specialized Cooperative Centers Program in Reproduction Research, and T. Rajendra Kumar for his help in the evaluation of tissues from the knockout mice. Dan Philips also provided valuable technical assistance. Sally Camper (University of Michigan) and Dan Linzer (Northwestern University) supplied mouse POMC, prolactin, and growth hormone constructs.
The research was supported by grants CA60651 (M.M.M.) and HD037096 (T.K.W.) from the National Institutes of Health, which also provided research recovery funds after tropical storm Allison, and by a Lalor Foundation postdoctoral fellowship (D.J.B.). K.H.B. is a student in the Medical Scientist Training Program, supported in part by NIH grant T32GM-07330.
REFERENCES
- 1.Asashima, M., K. Kinoshita, T. Ariizumi, and G. M. Malacinski. 1999. Role of activin and other peptide growth factors in body patterning in the early amphibian embryo. Int. Rev. Cytol. 191:1-52. [DOI] [PubMed] [Google Scholar]
- 2.Attisano, L., and J. L. Wrana. 2002. Signal transduction by the TGF-beta superfamily. Science 296:1646-1647. [DOI] [PubMed] [Google Scholar]
- 3.Attisano, L., J. L. Wrana, S. Cheifetz, and J. Massague. 1992. Novel activin receptors: distinct genes and alternative mRNA splicing generate a repertoire of serine/threonine kinase receptors. Cell 68:97-108. [DOI] [PubMed] [Google Scholar]
- 4.Attisano, L., J. L. Wrana, E. Montalvo, and J. Massague. 1996. Activation of signalling by the activin receptor complex. Mol. Cell. Biol. 16:1066-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernard, D. J., S. C. Chapman, and T. K. Woodruff. 2002. Inhibin binding protein (InhBP/p120), betaglycan, and the continuing search for the inhibin receptor. Mol. Endocrinol. 16:207-212. [DOI] [PubMed] [Google Scholar]
- 6.Bernard, D. J., S. C. Chapman, and T. K. Woodruff. 2001. Mechanisms of inhibin signal transduction. Recent Prog. Horm. Res. 56:417-450. [DOI] [PubMed] [Google Scholar]
- 7.Bernard, D. J., and T. K. Woodruff. 2001. Genetic approaches to the study of pituitary follicle-stimulating hormone regulation, p. 297-317. In M. M. Matzuk, C. W. Brown, and T. R. Kumar (ed.), Transgenics in endocrinology. Humana Press, Totawa, N.J.
- 8.Bernard, D. J., and T. K. Woodruff. 2001. Inhibin binding protein in rats: alternative transcripts and regulation in the pituitary across the estrous cycle. Mol. Endocrinol. 15:654-667. [DOI] [PubMed] [Google Scholar]
- 9.Bradley, A. 1987. Production and analysis of chimeric mice—teratocarcinomas and embryonic stem cells: a practical approach, p. 113-151. In E. J. Robinson (ed.), Production and analysis of chimeric mice. Oxford, London, England.
- 10.Burns, K. H., and M. M. Matzuk. 2002. Genetic models for the study of gonadotropin actions. Endocrinology 143:2823-2835. [DOI] [PubMed] [Google Scholar]
- 11.Chang, H., C. W. Brown, and M. M. Matzuk. 2002. Genetic analysis of the mammalian transforming growth factor-beta superfamily. Endocr. Rev. 23:787-823. [DOI] [PubMed] [Google Scholar]
- 12.Chapman, S. C., D. J. Bernard, J. Jelen, and T. K. Woodruff. 2002. Properties of inhibin binding to betaglycan, InhBP/p120 and the activin type II receptors. Mol. Cell. Endocrinol. 196:79-93. [DOI] [PubMed] [Google Scholar]
- 13.Chapman, S. C., and T. K. Woodruff. 2001. Modulation of activin signal transduction by inhibin B and inhibin-binding protein (InhBP). Mol. Endocrinol. 15:668-679. [DOI] [PubMed] [Google Scholar]
- 14.Chen, X., M. J. Rubock, and M. Whitman. 1996. A transcriptional partner for MAD proteins in TGF-beta signalling. Nature 383:691-696. [DOI] [PubMed] [Google Scholar]
- 15.Chen, X., E. Weisberg, V. Fridmacher, M. Watanabe, G. Naco, and M. Whitman. 1997. Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature 389:85-89. [DOI] [PubMed] [Google Scholar]
- 16.Chong, H., S. A. Pangas, D. J. Bernard, E. Wang, J. Gitch, W. Chen, L. B. Draper, E. T. Cox, and T. K. Woodruff. 2000. Structure and expression of a membrane component of the inhibin receptor system. Endocrinology 141:2600-2607. [DOI] [PubMed] [Google Scholar]
- 17.Corrigan, A. Z., L. M. Bilezikjian, R. S. Carroll, L. N. Bald, C. H. Schmelzer, B. M. Fendly, A. J. Mason, W. W. Chin, R. H. Schwall, and W. Vale. 1991. Evidence for an autocrine role of activin B within rat anterior pituitary cultures. Endocrinology 128:1682-1684. [DOI] [PubMed] [Google Scholar]
- 18.DePaolo, L. V. 1997. Inhibins, activins, and follistatins: the saga continues. Proc. Soc. Exp. Biol. Med. 214:328-339. [DOI] [PubMed] [Google Scholar]
- 19.Farnworth, P. G., C. A. Harrison, P. Leembruggen, K. L. Chan, P. G. Stanton, G. T. Ooi, N. A. Rahman, I. T. Huhtaniemi, J. K. Findlay, and D. M. Robertson. 2001. Inhibin binding sites and proteins in pituitary, gonadal, adrenal and bone cells. Mol. Cell. Endocrinol. 180:63-71. [DOI] [PubMed] [Google Scholar]
- 20.Frattini, A., S. Faranda, E. Redolfi, P. Allavena, and P. Vezzoni. 1998. Identification and genomic organization of a gene coding for a new member of the cell adhesion molecule family mapping to Xq25. Gene 214:1-6. [DOI] [PubMed] [Google Scholar]
- 21.Harrison, C. A., P. G. Farnworth, K. L. Chan, P. G. Stanton, G. T. Ooi, J. K. Findlay, and D. M. Robertson. 2001. Identification of specific inhibin A-binding proteins on mouse Leydig (TM3) and Sertoli (TM4) cell lines. Endocrinology 142:1393-1402. [DOI] [PubMed] [Google Scholar]
- 22.Knight, P. G. 1996. Roles of inhibins, activins, and follistatin in the female reproductive system. Front. Neuroendocrinol. 17:476-509. [DOI] [PubMed] [Google Scholar]
- 23.Kumar, T. R., Y. Wang, N. Lu, and M. M. Matzuk. 1997. Follicle stimulating hormone is required for ovarian follicle maturation but not male fertility. Nat. Genet. 15:201-204. [DOI] [PubMed] [Google Scholar]
- 24.Kumar, T. R., A. L. Wiseman, G. Kala, S. V. Kala, M. M. Matzuk, and M. W. Lieberman. 2000. Reproductive defects in gamma-glutamyl transpeptidase-deficient mice. Endocrinology 141:4270-4277. [DOI] [PubMed] [Google Scholar]
- 25.Lagna, G., A. Hata, A. Hemmati-Brivanlou, and J. Massague. 1996. Partnership between DPC4 and SMAD proteins in TGF-beta signalling pathways. Nature 383:832-836. [DOI] [PubMed] [Google Scholar]
- 26.Lebrun, J. J., and W. W. Vale. 1997. Activin and inhibin have antagonistic effects on ligand-dependent heteromerization of the type I and type II activin receptors and human erythroid differentiation. Mol. Cell. Biol. 17:1682-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis, K. A., P. C. Gray, A. L. Blount, L. A. MacConell, E. Wiater, L. M. Bilezikjian, and W. Vale. 2000. Betaglycan binds inhibin and can mediate functional antagonism of activin signalling. Nature 404:411-414. [DOI] [PubMed] [Google Scholar]
- 28.Martens, J. W., J. P. de Winter, M. A. Timmerman, A. McLuskey, R. H. van Schaik, A. P. Themmen, and F. H. de Jong. 1997. Inhibin interferes with activin signaling at the level of the activin receptor complex in Chinese hamster ovary cells. Endocrinology 138:2928-2936. [DOI] [PubMed] [Google Scholar]
- 29.Mather, J. P., A. Moore, and R. H. Li. 1997. Activins, inhibins, and follistatins: further thoughts on a growing family of regulators. Proc. Soc. Exp. Biol. Med. 215:209-222. [DOI] [PubMed] [Google Scholar]
- 30.Mathews, L. S. 1994. Activin receptors and cellular signaling by the receptor serine kinase family. Endocr. Rev. 15:310-325. [DOI] [PubMed] [Google Scholar]
- 31.Mathews, L. S., and W. W. Vale. 1991. Expression cloning of an activin receptor, a predicted transmembrane serine kinase. Cell 65:973-982. [DOI] [PubMed] [Google Scholar]
- 32.Matzuk, M. M., M. J. Finegold, J. G. Su, A. J. Hsueh, and A. Bradley. 1992. Alpha-inhibin is a tumour-suppressor gene with gonadal specificity in mice. Nature 360:313-319. [DOI] [PubMed] [Google Scholar]
- 33.Mazzarella, R., G. Pengue, J. Jones, C. Jones, and D. Schlessinger. 1998. Cloning and expression of an immunoglobulin superfamily gene (IGSF1) in Xq25. Genomics 48:157-162. [DOI] [PubMed] [Google Scholar]
- 34.Pangas, S. A., and T. K. Woodruff. 2000. Activin signal transduction pathways. Trends Endocrinol. Metab. 11:309-314. [DOI] [PubMed] [Google Scholar]
- 35.Phillips, D. J., K. L. Jones, J. Y. Scheerlinck, M. P. Hedger, and D. M. de Kretser. 2001. Evidence for activin A and follistatin involvement in the systemic inflammatory response. Mol. Cell. Endocrinol. 180:155-162. [DOI] [PubMed] [Google Scholar]
- 36.Roberts, V., H. Meunier, J. Vaughan, J. Rivier, C. Rivier, W. Vale, and P. Sawchenko. 1989. Production and regulation of inhibin subunits in pituitary gonadotropes. Endocrinology 124:552-554. [DOI] [PubMed] [Google Scholar]
- 37.Robertson, D. M., R. Hertan, and P. G. Farnworth. 2000. Is the action of inhibin mediated via a unique receptor? Rev. Reprod. 5:131-135. [DOI] [PubMed] [Google Scholar]
- 38.Tanaka, S., K. Tatsumi, K. Okubo, K. Itoh, S. Kawamoto, K. Matsubara, and N. Amino. 2002. Expression profile of active genes in the human pituitary gland. J. Mol. Endocrinol. 28:33-44. [DOI] [PubMed] [Google Scholar]
- 39.ten Dijke, P., H. Yamashita, H. Ichijo, P. Franzen, M. Laiho, K. Miyazono, and C. H. Heldin. 1994. Characterization of type I receptors for transforming growth factor-beta and activin. Science 264:101-104. [DOI] [PubMed] [Google Scholar]
- 40.Welt, C. K. 2002. The physiology and pathophysiology of inhibin, activin and follistatin in female reproduction. Curr. Opin. Obstet. Gynecol. 14:317-323. [DOI] [PubMed] [Google Scholar]
- 41.Willis, S. A., C. M. Zimmerman, L. I. Li, and L. S. Mathews. 1996. Formation and activation by phosphorylation of activin receptor complexes. Mol. Endocrinol. 10:367-379. [DOI] [PubMed] [Google Scholar]
- 42.Woodruff, T. K. 1999. Hope, hypothesis, and the inhibin receptor. Does specific inhibin binding suggest there is a specific inhibin receptor? Endocrinology 140:3-5. [DOI] [PubMed] [Google Scholar]
- 43.Woodruff, T. K. 1998. Regulation of cellular and system function by activin. Biochem. Pharmacol. 55:953-963. [DOI] [PubMed] [Google Scholar]
- 44.Wu, S., Q. Lu, and A. L. Kriz. 1995. Multiple-sandwich, one-step hybridization of Northern and Southern blots. BioTechniques 18:585-586. [PubMed] [Google Scholar]
- 45.Xu, J., K. McKeehan, K. Matsuzaki, and W. L. McKeehan. 1995. Inhibin antagonizes inhibition of liver cell growth by activin by a dominant-negative mechanism. J. Biol. Chem. 270:6308-6313. [DOI] [PubMed] [Google Scholar]
- 46.Yeo, C. Y., X. Chen, and M. Whitman. 1999. The role of FAST-1 and Smads in transcriptional regulation by activin during early Xenopus embryogenesis. J. Biol. Chem. 274:26584-26590. [DOI] [PubMed] [Google Scholar]
- 47.Ying, S. Y. 1988. Inhibins, activins, and follistatins: gonadal proteins modulating the secretion of follicle-stimulating hormone. Endocr. Rev. 9:267-293. [DOI] [PubMed] [Google Scholar]