Abstract
LRP130 (also known as LRPPRC) is an RNA-binding protein that is a constituent of postsplicing nuclear RNP complexes associated with mature mRNA. It belongs to a growing family of pentatricopeptide repeat (PPR) motif-containing proteins, several of which have been implicated in organellar RNA metabolism. We show here that only a fraction of LRP130 proteins are in nuclei and are directly bound in vivo to at least some of the same RNA molecules as the nucleocytoplasmic shuttle protein hnRNP A1. The majority of LRP130 proteins are located within mitochondria, where they are directly bound to polyadenylated RNAs in vivo. In vitro, LRP130 binds preferentially to polypyrimidines. This RNA-binding activity maps to a domain in its C-terminal region that does not contain any previously described RNA-binding motifs and that contains only 2 of the 11 predicted PPR motifs. Therefore, LRP130 is a novel type of RNA-binding protein that associates with both nuclear and mitochondrial mRNAs and as such is a potential candidate for coordinating nuclear and mitochondrial gene expression. These findings provide the first identification of a mammalian protein directly bound to mitochondrial RNA in vivo and provide a possible molecular explanation for the recently described association of mutations in LRP130 with cytochrome c oxidase deficiency in humans.
Transcription of protein-coding mRNAs in eukaryotic cells takes place in two distinct subcellular compartments, nuclei and mitochondria; in those cells that have them, it takes place also in chloroplasts. The vast majority of cellular mRNAs are synthesized in the nucleus, whereas mitochondria contain the genetic information for the transcription of only a few mRNAs (13 in mammalian cells), which are translated within the organelle into protein subunits of the respiratory chain (6, 45). The remaining mitochondrial proteins are encoded by nucleus-derived mRNAs and are imported posttranslationally into the organelle. Thus, mitochondrial proteins are products of translation from mRNAs encoded by both nuclear and mitochondrial genomes.
Nucleus-encoded mRNAs are transcribed as large precursors that undergo several processing steps before their export to the cytoplasm as mature mRNAs. These steps include addition of a 7-methylguanosine cap at the 5′ end, cleavage and polyadenylation of the 3′ end, and removal of introns through splicing. Throughout their maturation pathway, RNAs are associated with RNA-binding proteins as ribonucleoprotein (RNP) complexes. The proteins that are stably associated with nuclear RNAs have been extensively characterized and have been shown to participate in virtually all stages of mRNA maturation (16). In most cases, their RNA-binding activity resides in one or more distinct RNA-binding domains characterized by specific amino acid sequence motifs such as the RNP motif, KH domain, and RGG box (3). Through this binding they have an impact on the processing reactions occurring on the RNAs with which they associate (10, 16).
The specific set of proteins associated with an individual mRNA depends on the sequence characteristics of the RNA and changes in a processing-stage-dependent manner (10, 27). Analyses of RNP complexes associated with pre-mRNA and mRNA showed that there is extensive remodeling of their protein composition as the RNA matures. This remodeling includes recruitment of specific proteins at exon-exon junctions (18). Further remodeling leads to formation of nuclear mRNPs (nmRNPs), which are associated with mature mRNA and with shuttling RNA-binding proteins but from which nonshuttling hnRNP proteins are absent. In addition, these mRNPs contain specific proteins not found in pre-mRNA-associated RNPs, including alternatively spliced isoforms of hnRNP proteins and novel RNA-binding protein LRP130, which is discussed below (27).
Mitochondrial transcripts follow a maturation pathway quite distinct from that of nuclear RNAs. The circular mitochondrial DNA (mtDNA) is transcribed by a phage-like mitochondrial RNA polymerase into two large polycistronic primary transcripts complementary to each mtDNA strand (6, 45). Individual rRNAs and mRNAs are released from these transcripts by endonucleolytic cleavage and removal of intervening tRNA sequences (31). The cleavage reactions that yield mature tRNA 5′ ends are catalyzed by a mitochondrial RNase P, whose composition is controversial (38, 40). tRNA 3′ ends are generated by a distinct 3′ processing activity (21). Further processing of mitochondrial mRNAs in mammalian cells involves only polyadenylation of the 3′ end. rRNA maturation involves base modification and oligoadenylation at the 3′ end, while tRNAs undergo base modifications and addition of a CCA tail at the 3′ end (6, 45).
While the processing steps that lead to functional RNAs in mitochondria are quite well understood, little is known, especially for mammalian cells, about the proteins that associate with mitochondrial RNAs. By analogy to nuclear RNP complexes, and based on the paradigm set mainly through studies of the yeast Saccharomyces cerevisiae (9, 36), RNA-binding proteins would also be expected to participate and/or regulate every aspect of mitochondrial mRNA metabolism. A small number of mammalian mitochondrial proteins with well-characterized enzymatic activities, such as cis-aconitase, glutamate dehydrogenase, and enoyl-coenzyme A hydratase (1, 29, 37), have been shown to exhibit RNA-binding activity in vitro. However, it is not known whether they bind mitochondrial RNA directly in vivo. Interestingly, other candidate participants in mitochondrial RNA metabolism belong to a growing family of proteins that are not confined to this organelle but rather have dual localization in nuclei and in mitochondria. These include p32 (2), which is thought to function in regulating nuclear pre-mRNA splicing through associating with the splicing factor ASF/SF2 (33), while an involvement in mitochondrial RNA maturation has been reported for its trypanosome homologue (14). The putative human RNA helicase MDDX28 is also present in both organelles, but its function is unknown (47). Several yeast proteins with dual localization in mitochondria and nuclei have also been reported. They include Rna14p, a subunit of nuclear cleavage and polyadenylation factor I, which is mainly located in mitochondria (41), and Cca1p, Mod5p, and Trm1p, which catalyze various steps in the maturation of both nuclear and mitochondrial tRNAs (26).
LRP130, as shown in this work, belongs also to this family of proteins, with possible roles in both nuclear and mitochondrial RNA metabolism. LRP130, also known as LRPPRC (20), was originally identified as a lectin-binding protein overexpressed in hepatoblastoma HepG2 cells (15). The finding that it is an RNA-binding protein associated with nuclear mRNP complexes suggested that it has a function in mRNA-specific metabolic events (27). In agreement with this, the putative Drosophila melanogaster LRP130 homologue, BSF, has been implicated in stabilizing the nucleus-encoded bicoid mRNA by binding to a specific 3′ untranslated region element (22). More recently, on the other hand, mutations in the LRP130 gene were found to cause Leigh syndrome, French-Canadian type, which is a mitochondrial cytochrome c oxidase deficiency in humans, leading to the suggestion that LRP130 could also participate in mitochondrial RNA processing (28). Neither LRP130 nor Drosophila BSF contains any recognizable RNA-binding motifs in its primary amino acid sequence. However, both LRP130 and BSF are members of a family of proteins that contain multiple copies of the recently described pentatricopeptide repeat (PPR) motif (42). This ca. 35-amino-acid (aa) motif is present in more than 400 proteins in the databases. Importantly, the few other members of this family that have been functionally characterized are all implicated in RNA-processing events in mitochondria or chloroplasts (7, 12, 17, 23). Whether the PPR motif itself constitutes an RNA-binding domain, however, is not known.
Here, we show that, while a fraction of human LRP130 proteins are in the nucleus and associate with the shuttling hnRNP protein A1 in vivo through direct binding of both proteins to the same RNAs, the majority of LRP130 proteins are located within mitochondria. UV light-induced cross-linking shows that mitochondrial LRP130 is directly bound in vivo to mitochondrial polyadenylated RNA, implicating LRP130 in the metabolism of mitochondrial RNA. LRP130 binds preferentially to polypyrimidines in vitro. This RNA-binding activity maps to a C-terminal region of LRP130 that does not contain any of the previously described RNA-binding motifs and that likely constitutes a novel type of RNA-binding domain. These findings indicate that LRP130 belongs to a growing group of proteins with roles in RNA metabolism in both nuclei and mitochondria and provide a possible molecular explanation for the cytochrome c oxidase defects that result from mutations in LRP130.
MATERIALS AND METHODS
Cell culture.
HeLa cells were grown in monolayer culture in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum and 1% penicillin-streptomycin. Hybridoma cells were grown in serum-free medium (HyClone).
Plasmids.
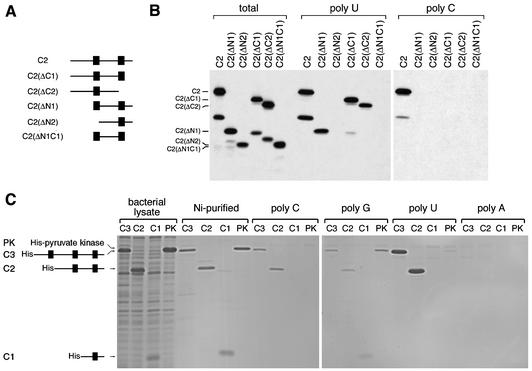
LRP130 cDNA in the pBluescript SK(+) vector was provided by W. L. McKeehan (Texas A&M University). PCR was used to introduce a NotI site at the 5′ end and a KpnI site in place of the stop codon, and the cDNA was subcloned into NotI and KpnI sites of the pcDNA3.1(-)/Myc-His vector (Invitrogen) in frame with the coding sequence for the Myc His tag. To generate N-terminal truncations, the LRP130 cDNA was digested with AvaI and BglII, SalI and BglII, StyI and BglII, EcoNI and BglII, or BsaBI and BglII. The 5′ end of each fragment was blunt ended and cloned into NdeI or XhoI (blunt ended) and BamHI sites of the pET15b vector (Novagen) to yield constructs C8, C7, C3, C2, and C1, respectively. Plasmids encoding further truncated forms of the C2 construct (see Fig. 7) were generated by PCR amplification of the corresponding cDNA fragments followed by ligation into the NdeI and BamHI sites of the pET15b vector (Novagen). The exact portions of LRP130 represented by each construct are as follows: C2ΔN1, aa 1015 to 1273; C2ΔN2, aa 1050 to 1273; C2ΔC1, aa 879 to 1228; C2ΔC2, aa 879 to 1193; C2ΔN1C1, aa 1015 to 1228. The numbers correspond to the positions of the amino acids at the end of each construct, based on the predicted sequence of the protein deduced from the deposited cDNA (GenBank accession no. M92439).
FIG. 7.
The C-terminal fragment (C2) of LRP130 is the minimal region of the protein that exhibits intrinsic RNA-binding activity. (A) Schematic diagram depicting the truncated LRP130 constructs used in panel B. (B) Truncated LRP130 constructs were produced by in vitro transcription-translation and incubated with immobilized poly(U) or poly(C). Bound proteins were then resolved by SDS-PAGE and visualized by autoradiography. Lanes “total” contain 20% of the input material used in the binding reactions shown in the poly(U) and poly(C) lanes. (C) The indicated LRP130 constructs, as well as pyruvate kinase (as a control), were expressed in bacteria and purified as described in Materials and Methods. Equal amounts of each purified protein were incubated with the indicated immobilized RNA homopolymers. Fifteen percent of the input material and the total amount of protein bound to each homopolymer were resolved by SDS-PAGE and visualized by staining with Coomassie blue.
Expression of LRP130 in Escherichia coli and production of MAbs.
BL21(DE3) E. coli cells were transformed with C8, C3, C2, or C1 construct plasmids encoding the C-terminal 90-, 59-, 43-, and 17-kDa regions of LRP130, respectively, fused to a His tag. Production of the proteins was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 2.5 to 3 h at 30°C. The recombinant proteins were purified with Ni-nitrilotriacetic acid-agarose beads (Qiagen) under denaturing (C8 construct) or nondenaturing (constructs C3, C2, and C1) conditions, according to the manufacturer's instructions. The C8 and C3 recombinant proteins were used as antigens for production of monoclonal antibodies (MAbs). Hybridoma production and screening were done as previously described (34). The epitopes for 9C9 and 4C12 were mapped by immunoprecipitation of C-terminally truncated forms of LRP130 produced by in vitro translation and by Western blot analysis of N-terminally truncated forms of LRP130 produced in bacteria (data not shown). The epitope for 4C12 maps between aa 453 and 600 of the full-length sequence (based on accession no. M92439), and the 9C9 epitope is located at the C-terminal end encompassing aa 1115 to 1273 (data not shown).
Immunofluorescence microscopy.
HeLa cells were fixed and stained as previously described (34) by using the purified 9C9 MAb at ∼2.5 μg/ml or a similar concentration of nonimmune SP2/O myeloma immunoglobulins. Staining was observed with a Zeiss Axiophot microscope and was recorded by using the same exposure conditions for both antibodies. For colocalization studies, HeLa cells were grown for 30 min in the presence of 200 nM Mitotracker Red CMXRos (Molecular Probes), followed by 5 min in regular medium prior to fixation and staining.
Subcellular fractionation.
HeLa cells were homogenized in hypotonic RSB-10 buffer (10 mM Tris-Cl [pH 7.4], 10 mM NaCl, 2.5 mM MgCl2) with a Dounce homogenizer. All steps were carried out at 4°C. Sucrose and EDTA were added to final concentrations of 250 and 1 mM, respectively, and nuclei were pelleted at 600 × g for 5 min. The supernatant was separated into a mitochondrial pellet and soluble cytoplasm by centrifugation at 17,000 × g for 15 min. The mitochondrial pellet was resuspended in isotonic sucrose buffer (250 mM sucrose, 10 mM Tris-Cl [pH 7.4], 1 mM EDTA). The mitochondrial fraction was layered onto a sucrose 1.0 to 1.5 M step density gradient and centrifuged at 20,000 rpm for 20 min in a Beckman L-70 ultracentrifuge. Fractions (0.5 ml) were collected from the top, and equal volumes from each fraction were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting. For protease digestion, proteinase K was added to aliquots of mitochondrial fractions at 1 μg of proteinase/μg of protein, with or without Triton X-100 at a final concentration of 0.5%. The samples were incubated on ice for 30 min, and the reaction was stopped by addition of phenylmethylsulfonyl fluoride to 5 mM. The samples were boiled in SDS-PAGE sample buffer and analyzed by SDS-PAGE and immunoblotting.
UV light-induced cross-linking of proteins to RNA in living cells.
Cross-linking of proteins to RNA in vivo by UV light irradiation of cells was performed as previously described (27) except that UV-irradiated cells were fractionated into nuclear and mitochondrial fractions as described above prior to oligo(dT) selection. For nuclease digestion, the mitochondrial fraction was treated for 15 min at 30°C with 200 U of micrococcal nuclease/ml-1 mM CaCl2 in the presence or absence of Triton X-100 (0.5%). Cross-linked complexes from each fraction were selected by oligo(dT) chromatography, and cross-linked proteins were analyzed by SDS-PAGE and immunoblotting, as previously described (27). For immunoprecipitation of cross-linked complexes, UV-irradiated cells were fractionated into soluble cytoplasmic and Triton X-100-extracted fractions as described previously (27) and cross-linked complexes were selected from the Triton X-100-extracted fraction. Prior to elution, the oligo(dT) columns were washed with a low-SDS-concentration binding buffer (10 mM Tris-Cl [pH 7.4], 1 mM EDTA, 0.05% SDS, 0.5 M LiCl). The salt concentration of the material eluted from the oligo(dT) column was adjusted to 100 mM NaCl, and the eluted material was subjected to immunoprecipitation with MAbs for 1 h at 4°C. The immunoprecipitated complexes were digested for 1 h at 30°C with a mixture of RNase A at 25 μg/ml and micrococcal nuclease at 400 U/ml for subsequent analysis of the released proteins by SDS-PAGE and immunoblotting.
Immunopurification of RNP complexes.
HeLa cells were lysed in RSB-100 (10 mM Tris-Cl [pH 7.4], 100 mM NaCl, 2.5 mM MgCl2) containing 0.5% Triton X-100 and were sonicated twice for 5 s on ice with a microtip sonicator (model XL2015; Heat Systems, Farmingdale, N.Y.). The sonicated material was layered onto a 30% sucrose cushion in RSB-100 and centrifuged at 4,000 × g for 15 min. The supernatant was collected and clarified further by centrifugation for 5 min at 8,000 × g in a microcentrifuge. RNP complexes were immunopurified from the supernatant with antibodies as previously described (34).
Gel electrophoresis and immunoblot analysis.
SDS-PAGE and immunoblotting were carried out as previously described (35). The following antibodies were used for immunoblot analysis: 4C12 and 9C9 (anti-LRP130; described here), 4B10 (anti-hnRNP A1) (35), 4F4 (anti-hnRNP C1/C2) (5), 1A5 (anti-hnRNP K/J) (25), 7G2 (antinucleolin) (34), anti-PABP1 (13), and anti-hsp60 (StressGen).
In vitro transcription-translation.
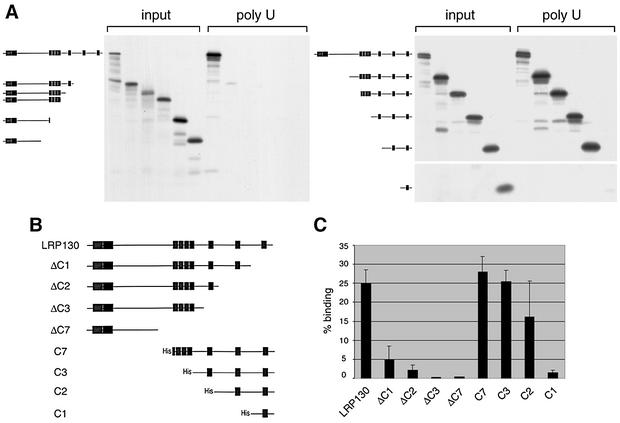
Full-length LRP130 and N-terminally truncated forms were synthesized by coupled in vitro transcription-translation using the TNT reticulocyte lysate system (Promega), with T7 RNA polymerase, in accordance with the manufacturer's instructions. The C-terminally truncated forms LRP130 ΔC1, ΔC2, ΔC3, and ΔC7 were produced by runoff transcription of the full-length LRP130 cDNA plasmid (see above) after digestion with BsaBI, EcoRI, SacI, or PvuII, respectively. Additional C-terminally truncated constructs that were used for the experiment shown in Fig. 6A were produced similarly after digestion with BspHI or SalI.
FIG. 6.
The C-terminal region of LRP130 is necessary and sufficient for RNA binding. (A) Full-length LRP130 and truncated constructs (depicted schematically at the left [filled boxes, predicted PPR motifs]) were produced by in vitro transcription-translation and incubated with immobilized poly(U). Twenty percent of the input material and the material bound to poly(U) were resolved by SDS-PAGE and visualized by autoradiography. (C) Selected constructs (schematically depicted in panel B.) were used in RNA-binding assays as described for panel A. The binding of each construct to the RNA was quantified and plotted as the percentage of input protein that bound RNA. Error bars, standard deviations of three to six independent experiments.
RNA-binding assays.
Total HeLa extract or 5 μl of each in vitro translation reaction mixture was added to agarose-bound RNA homopolymers (Sigma) in 300 μl of RSB-100 buffer containing 0.5% Triton X-100. Binding reaction mixtures were incubated for 20 min at 4°C, and reaction products were washed four times with the above buffer. Bound proteins were eluted by boiling in SDS-PAGE sample buffer and analyzed by SDS-PAGE. Proteins were detected by immunoblotting. When in vitro-translated proteins were used, the gel was fixed and dried and bound proteins were visualized by autoradiography. Quantification of the signals was performed with a phosphorimager.
RESULTS
Production of MAbs against LRP130.
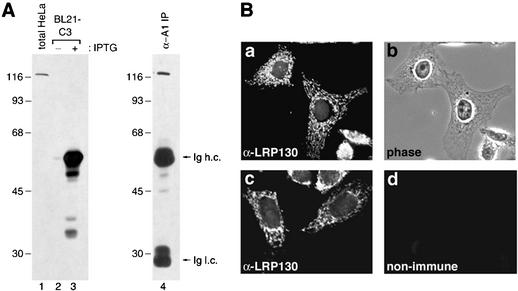
We recently identified LRP130 as an RNA-binding protein in nuclear mRNP complexes that likely represent late stages of nuclear mRNA maturation (27). To characterize in detail the properties and functional significance of LRP130, we generated MAbs against it. The antigens used were portions of LRP130 expressed in bacteria and subsequently purified (see Materials and Methods). Two hybridoma clones, 9C9 and 4C12, produced antibodies that specifically bound human LRP130. Western blot analysis using the 9C9 MAb shows that it binds specifically a single protein band of ca. 130 kDa in a total HeLa cell lysate (Fig. 1A, lane 1), as well as in RNP complexes associated with hnRNP A1 (Fig. 1A, lane 4), in agreement with our previous identification of LRP130 in these complexes by mass spectrometry (27). Furthermore, 9C9 binds specifically the bacterially expressed C3 fragment of LRP130, which was used as the antigen, only in lysates from bacteria induced to produce the protein (Fig. 1A, lanes 2 and 3). Identical results were obtained with the 4C12 MAb, which binds a different epitope on LRP130 than 9C9 (see Materials and Methods), demonstrating that both antibodies react specifically with human LRP130.
FIG. 1.
MAbs against human LRP130. (A) Lysates from HeLa cells (lane 1) and from bacteria transformed with the C3 construct plasmid, encoding the 59-kDa C-terminal portion of LRP130, prior to or after induction with IPTG (lanes 2 and 3), and proteins from nmRNP complexes immunoprecipitated with a MAb against hnRNP A1 (lane 4) were resolved by SDS-PAGE and immunoblotted with the 9C9 MAb. Molecular mass markers, in kilodaltons, are indicated on the left. Ig h.c. and Ig l.c., antibody heavy and light chains, respectively. (B) Immunofluorescence localization of the LRP130 protein. HeLa cells grown on glass coverslips were fixed, permeabilized, and stained with MAb 9C9 (a and c) or with nonimmune myeloma immunoglobulins (d). (b) Phase microscopy image of the cells shown in panel a. All immunostaining images were obtained by using the same exposures.
Immunofluorescence microscopy of HeLa cells with 9C9 revealed a punctate cytoplasmic staining, as well as nuclear staining of relatively lower intensity (Fig. 1Ba and c). Both cytoplasmic and nuclear signals were specific, since no staining was observed with nonimmune antibodies under the same conditions (Fig. 1Bc and d), and identical staining patterns were observed with the MAb 4C12 (data not shown).
LRP130 is predominantly localized in mitochondria.
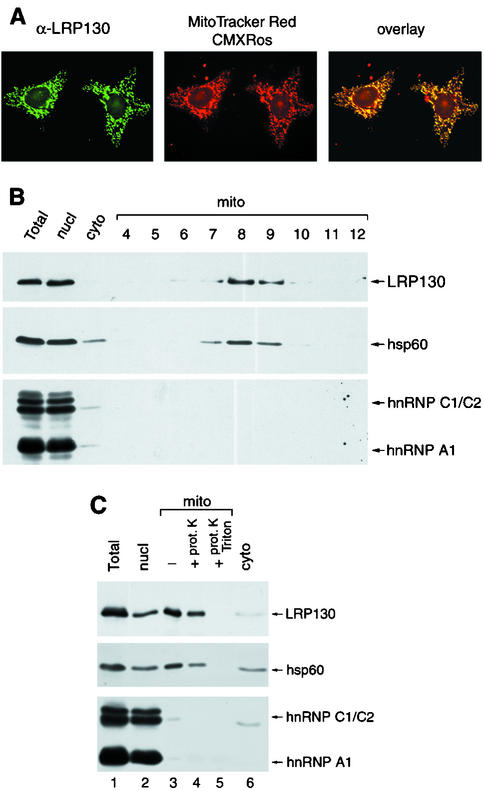
Because the cytoplasmic distribution of LRP130 resembles that of mitochondrial components, we tested whether cytoplasmic LRP130 colocalizes with mitochondria. HeLa cells were grown in the presence of Mitotracker Red CMXRos to visualize mitochondria and were subsequently stained with the 9C9 MAb. The distribution of LRP130 overlapped virtually entirely with the mitochondrion-specific dye (Fig. 2A), indicating that LRP130 is associated with mitochondria.
FIG. 2.
LRP130 is predominantly an inner mitochondrial protein. (A) HeLa cells grown in the presence of Mitotracker Red dye were fixed, permeabilized, and stained with the 9C9 MAb. Distribution of LRP130 was visualized with a fluorescein isothiocyanate (FITC)-conjugated secondary antibody (left) and the Mitotracker dye by direct fluorescence (middle). Overlay of the two images shows colocalization of the two signals in yellow (right). (B) HeLa cells were fractionated into nuclear (nucl), mitochondrial (mito), and cytosolic (cyto) fractions. The mitochondrial fraction was further fractionated on a sucrose density step gradient (fractions 4 to 12). Proteins from each fraction, corresponding to an equal number of cells, or equal volumes of fractions 4 to 12, were analyzed by immunoblotting with antibodies against LRP130 (4C12), hsp60, hnRNP C1/C2 (4F4), and hnRNP A1 (4B10). (C) HeLa cells were fractionated as in panel B. The mitochondrial fraction was left untreated (lane 3) or was digested with proteinase K in the absence (lane 4) or presence (lane 5) of Triton X-100. Proteins from each fraction, corresponding to an equal number of cells, were resolved by SDS-PAGE and analyzed as described for panel B.
The mitochondrial association of LRP130 was confirmed by subcellular fractionation. Mitochondria were separated from nuclei and soluble cytoplasm by differential centrifugation (see Materials and Methods), and the mitochondrial fraction was enriched further on a sucrose density step gradient. Western blot analysis of the resulting fractions showed that LRP130 cofractionates with the mitochondrial matrix protein hsp60 (43) (Fig. 2B). The lack of detectable hnRNP C1/C2 and A1 in these fractions indicates the absence of significant nuclear contamination. LRP130 associated with mitochondria was resistant to digestion by proteinase K (Fig. 2C, lane 4) unless membrane integrity was disrupted with Triton X-100 (Fig. 2C, lane 5), a property consistent with that of an inner mitochondrial protein. The small reduction in the amount of LRP130 upon proteinase digestion in the absence of detergent (Fig. 2C, lanes 3 and 4) can be attributed to disruption of a small proportion of the mitochondria during the experimental manipulations, as a similar decrease is observed for hsp60. Thus, these results indicate that LRP130 is not simply adsorbed onto the cytoplasmic side of the outer mitochondrial membrane and therefore show that LRP130 is an inner mitochondrial protein.
LRP130 is bound to polyadenylated mitochondrial RNAs in vivo.
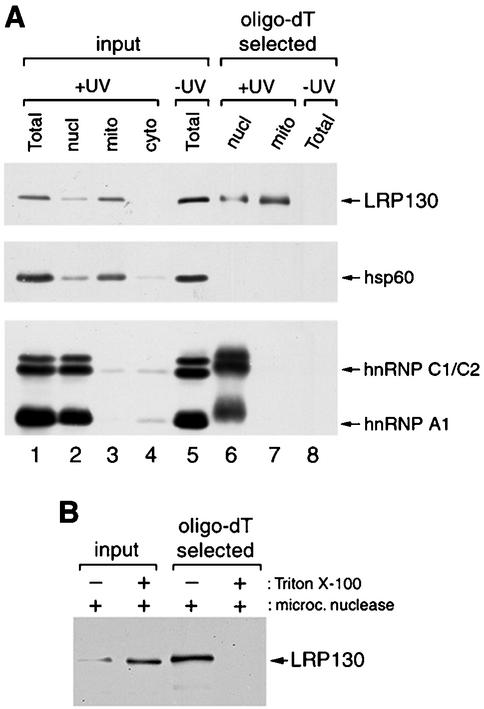
We showed previously that LRP130 is bound in vivo to polyadenylated RNAs (27). The predominantly mitochondrial location of LRP130 prompted us to test whether these RNAs included also mitochondrial transcripts. To do so, live HeLa cells were irradiated with UV light to induce covalent cross-links between RNAs and their bound proteins. The cells were subsequently fractionated into nuclear, mitochondrial, and soluble cytoplasmic fractions as described for Fig. 2C. Nuclear and mitochondrial fractions were then subjected to oligo(dT) chromatography under protein-denaturing conditions, in order to select poly(A) RNAs from each fraction together with proteins covalently cross-linked to them. As shown in Fig. 3A, LRP130 is cross-linked to poly(A) RNA in a mitochondrial fraction that is largely devoid of detectable nuclear contamination, as indicated by the absence of cross-linked hnRNP proteins (Fig. 3A, lane 7). Its retention on the oligo(dT) column reflects specific cross-linking of the protein to poly(A)-containing RNAs, as underscored by the fact that no LRP130 is detected if UV irradiation is omitted (Fig. 3A, lane 8), as well as by the absence of hsp60, a mitochondrial protein with no reported RNA-binding activity, from the cross-linked complexes (Fig. 3A, lane 7).
FIG. 3.
LRP130 is bound to mitochondrial polyadenylated RNAs in vivo. (A) Nuclear and mitochondrial fractions from UV-irradiated HeLa cells and total extract from control cells were fractionated by oligo(dT) chromatography under protein-denaturing conditions to select polyadenylated RNAs with cross-linked proteins (lanes 6 to 8). Proteins in the selected RNPs were released by digestion with RNase and analyzed by immunoblotting with antibodies against LRP130, hsp60, hnRNP C1/C2, and hnRNP A1. Abbreviations are as for Fig. 2. (B) The mitochondrial fraction from UV-irradiated cells was treated with micrococcal nuclease in the presence or absence of Triton X-100. Cross-linked RNP complexes were selected by oligo(dT) chromatography, and bound proteins were released and analyzed as described for panel A by using the anti-LRP130 MAb 4C12. Input lanes contain approximately 1% of the material loaded on the oligo(dT) columns. The smaller amount of LRP130 in the lane corresponding to mitochondria treated in the absence of Triton X-100 is most likely due to the fact that, under these conditions, the nuclease cannot gain access to the LRP130-cross-linked RNAs. The cross-linked LRP130-RNA complexes that resist nuclease digestion would, in turn, be too large to enter the gel.
To distinguish whether the RNA to which LRP130 was cross-linked in the mitochondrial fraction is within mitochondria or on their cytoplasmic sides, mitochondria from UV-irradiated cells were treated with micrococcal nuclease in the presence or absence of detergent, prior to selection of cross-linked RNPs. As shown in Fig. 3B, LRP130 was still selected by oligo(dT) chromatography after nuclease treatment of intact mitochondria, while it was no longer selected if the integrity of the mitochondrial membranes was disrupted with Triton X-100 during nuclease digestion. Thus, the RNAs to which LRP130 is cross-linked in this fraction are membrane enclosed, and, taken with the above data, this shows that LRP130 is bound in vivo to mitochondrial RNA.
Binding of LRP130 to hnRNP A1-associated RNAs is direct and occurs in vivo.
LRP130 cross-linked to polyadenylated RNA was readily detected in the nuclear fraction in the experiments described above (Fig. 3A, lane 6). However, we could not determine from these results to what extent this reflected the binding of LRP130 to nuclear mRNA, since we could not recover intact nuclei without contaminating mitochondria (Fig. 3A, lane 2). An association of LRP130 with nucleus-encoded RNAs was previously deduced from its RNA-dependent association with nuclear mRNP complexes and from the fact that its Drosophila homologue, BSF, binds to the nucleus-encoded bicoid mRNA. In agreement with such an association with nuclear RNAs, our immunofluorescence results (Fig. 1) show that LRP130 is present in nuclei in addition to mitochondria. The availability of anti-LRP130 antibodies allowed us to address directly its association with nmRNPs in vivo.
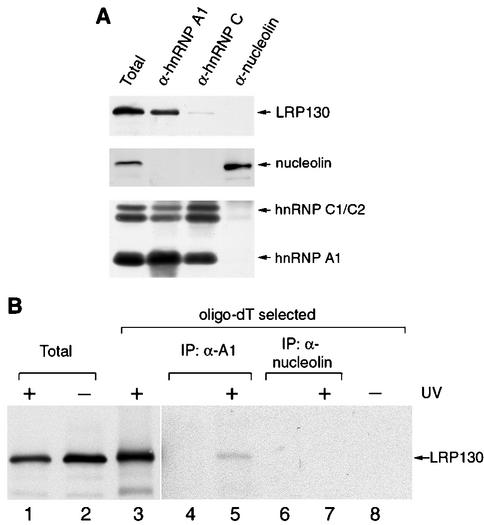
To test the specificity of the association of LRP130 with hnRNP A1-associated nmRNP complexes, we isolated different RNP complexes associated with a variety of other RNA-binding proteins and their bound RNAs from whole HeLa cell lysates and tested them for the presence of LRP130 (Fig. 4A). If LRP130 nonspecifically interacted with RNAs during the isolation or exchanged with other types of RNPs, it would be expected to be present in all RNP complexes regardless of the type of RNA that they contain. Pre-mRNA- and mRNA-containing RNPs associated with hnRNP A1 or with hnRNP C1/C2 and rRNA-containing RNPs associated with nucleolin were isolated with MAbs directed against the corresponding proteins. Western blot analysis confirmed that LRP130 is present in RNP complexes associated with hnRNP A1. Only a small amount was detected in hnRNP C-associated complexes upon long exposures of the blots, and virtually none was detected in association with nucleolin (Fig. 4A). Quantification of the recovered RNA labeled with [3H]uridine in parallel experiments showed that all of the complexes contained similar amounts of RNA (data not shown). Therefore, the presence of LRP130 in hnRNP A1-associated RNPs most likely represents its specific association with nmRNPs but not pre-mRNPs or rRNPs.
FIG. 4.
Binding of LRP130 to hnRNP A1-associated RNAs is specific and occurs in vivo. (A) RNP complexes were immunopurified from HeLa cell lysates with antibodies against hnRNP A1 (4B10), hnRNP C1/C2 (4F4), and nucleolin (7G2). Proteins in the isolated complexes were resolved by SDS-PAGE and analyzed by immunoblotting with the indicated antibodies. Five percent of the material used for immunoprecipitation was loaded in lane Total. (B) Cross-linked RNP complexes from HeLa cells exposed to UV light were selected from a Triton X-100-extracted subcellular fraction as described in the text and subjected to immunoprecipitation with antibodies against hnRNP A1 or nucleolin (lanes 5 and 7). Proteins in the immunopurified complexes were released by digestion with RNase, resolved by SDS-PAGE, and analyzed by immunoblotting with an antibody against LRP130. Lanes 2 and 8, lysates from cells that were not exposed to UV light; lanes Total, 1% of the material loaded on the oligo(dT) columns; lanes 4 and 6, mock immunopurifications (IP) in which HeLa cell material was omitted; lanes 3 and 8, total oligo(dT)-eluted material from cross-linked or control cells equal to the amount used for the immunopurifications shown in lanes 5 and 7.
As an even more stringent test in vivo of whether LRP130 and hnRNP A1 associate simultaneously with the same RNAs, we again used UV cross-linking as described above. In this case, however, cross-linked complexes that eluted from the oligo(dT) column were first subjected to immunoprecipitation with MAbs against hnRNP A1 or against nucleolin as a negative control. Because the proteins in the eluted complexes are denatured and covalently cross-linked to RNA, there is no possibility of A1 or LRP130 associating with RNA during the extraction process unless they were bound to those RNAs during irradiation in vivo. The presence of LRP130 in immunoprecipitated, cross-linked complexes was then tested by immunoblotting. Coimmunoprecipitation of two proteins would demonstrate that the two were directly bound to the same RNA(s) in vivo. LRP130 coimmunoprecipitated with hnRNP A1 (Fig. 4B, lane 5), but not with nucleolin (Fig. 4B, lane 7). Coimmunoprecipitation of LRP130 with hnRNP A1 depended on prior UV irradiation of the cells (Fig. 4B, lanes 5 and 8) as well as on the integrity of the RNA, since RNase digestion prior to immunoprecipitation significantly reduced the amount of LRP130 that remained associated with hnRNP A1 (data not shown). The relatively small amount of LRP130 associated with hnRNP A1, compared to the total amount of LRP130 cross-linked to RNA (Fig. 4B, lanes 3 and 5), can be attributed at least in part to the likelihood that most of the cross-linked LRP130 is associated with mitochondrial transcripts, as well as to the low efficiency of the cross-linking of hnRNP A1 to RNA. These data demonstrate that in vivo LRP130 and hnRNP A1 are directly bound to the same RNA(s), and this in turn indicates that LRP130, in addition to binding to mitochondrial transcripts, is also directly bound to nucleus-encoded mRNAs (see Discussion).
Binding of LRP130 to RNA is mediated by a discrete C-terminal region that excludes most of the PPR motifs.
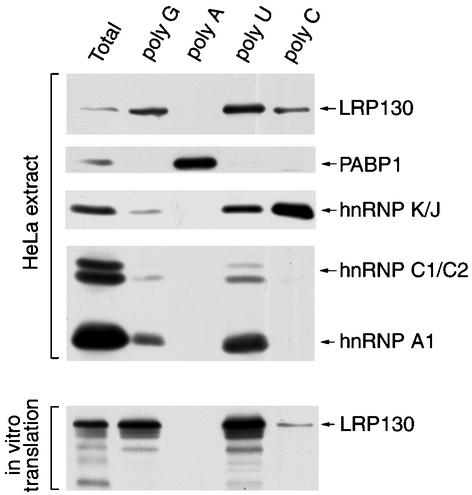
Although LRP130 binds RNA directly, its primary sequence did not reveal any previously characterized RNA-binding motifs (3). To characterize the RNA-binding properties of LRP130 in more detail and to set up a suitable assay to delineate its RNA-binding domain, we tested its binding in vitro to immobilized RNA homopolymers, as done previously for other RNA-binding proteins (44). This revealed that LRP130 binds with distinct specificity to different RNA homopolymers. LRP130 from HeLa cell extracts bound efficiently to poly(U) and less efficiently to poly(G) and poly(C), whereas it didn't associate detectably with poly(A) (Fig. 5). LRP130 produced by in vitro transcription and translation exhibited the same RNA-binding specificity as the endogenous HeLa cell protein (Fig. 5), indicating that, at least as far as binding to RNA homopolymers is concerned, the in vitro-translated protein has properties similar to those of the endogenous protein.
FIG. 5.
Specificity of LRP130 toward RNA homopolymers. (Top [HeLa extract]) HeLa cell lysate was incubated with the indicated agarose-bound RNA homopolymers. Bound proteins were resolved by SDS-PAGE and analyzed by immunoblotting with antibodies against LRP130, PABP1, hnRNP K/J, hnRNP C1/C2, and hnRNP A1. (Bottom) LRP130 produced by in vitro transcription-translation was incubated with the indicated agarose-bound RNA homopolymers. Bound protein was resolved by SDS-PAGE and visualized by autoradiography. Lane Total represents approximately 10% of the input material used in each binding reaction.
To delineate the RNA-binding domain of LRP130, constructs with progressive deletions from the C or N terminus of LRP130 were generated by in vitro transcription-translation of the corresponding cDNAs and their poly(U)-binding activity was tested (Fig. 6A). The PPR motifs were used as a reference, and deletions were designed to serially remove clusters or individual PPR motifs from each end of the protein. The binding of each truncated protein to poly(U) was quantified as the percentage of bound protein compared to that present in the input material. As shown in Fig. 6C, deletion of one or more PPR motifs from the C-terminal end of the protein (constructs ΔC1, ΔC2, ΔC3, and ΔC7) drastically reduced the binding to poly(U). By contrast, removing the two N-terminal PPR clusters (constructs C7 and C3), as well as removing the third PPR motif from the end (construct C2), did not have any detectable effect on binding to poly(U). A protein containing only the last PPR motif (construct C1) did not show significant RNA-binding activity. Similar results were obtained when using poly(G) or poly(C) as the ligand (data not shown), whereas none of the truncated proteins showed binding to poly(A) under these conditions, in agreement with the results obtained with full-length LRP130 (Fig. 5).
To delineate in more detail the boundaries of the LRP130 RNA-binding domain within the C2 fragment, N- and C-terminally truncated forms of this fragment were produced and tested for binding to immobilized RNA homopolymers (Fig. 7A and B). Deletions from either end of the C2 fragment abolished any detectable binding to poly(C). By contrast, constructs where either the N-terminal (C2ΔN1) or the C-terminal (C2ΔC1) regions flanking the remaining PPR motifs were deleted continued to bind to poly(U). Furthermore, the most C-terminal PPR motif could be removed (C2ΔC2) without abolishing the binding to poly(U), provided that the entire remaining portion of the C2 fragment was retained. We interpret these differences in binding to poly(C) and poly(U) as reflecting the weaker binding of LRP130 to the former, as suggested by the fact that binding to poly(C) is more salt sensitive than binding to poly(U) (S. Mili and S. Piñol-Roma, unpublished observations). However, even for poly(U), deletions from either end of the C2 fragment initially decrease the efficiency of binding [constructs C2(ΔN1) and C2 (ΔC2)] and eventually abolish it when they are combined [construct C2(ΔN1C1)] or when they exceed a certain length [C2(ΔN2)]. Thus, of all the constructs tested here, the C2 fragment represents a minimal RNA-binding domain of LRP130. Furthermore, these results indicate that regions of this fragment including (but not restricted to) the two predicted PPR motifs act synergistically to achieve high-affinity binding to RNA.
As the proteins used in these experiments were produced in reticulocyte lysates, it was possible that their observed binding to RNA did not reflect direct interaction but was instead mediated by their association with another RNA-binding protein from the lysate. To clarify this issue, several of these constructs were expressed in bacteria, purified, and tested for binding to the RNA homopolymers. The RNA-binding activity of the purified proteins was indistinguishable from that of the corresponding proteins produced by in vitro translation (Fig. 7C), confirming that the observed binding reflects direct interaction of LRP130 and of its RNA-binding domain with RNA.
Taken together, these results show that the C-terminal region of LRP130 is both necessary and sufficient to confer RNA-binding activity to levels similar to those for the full-length protein and therefore likely represents the RNA-binding domain of the protein. This region of LRP130 does not show any similarity to previously described RNA-binding motifs and therefore likely constitutes a novel type of RNA-binding domain.
DISCUSSION
We previously identified LRP130 as an RNA-binding protein associated with postsplicing nuclear mRNP complexes, which suggested that it participates in aspects of RNA metabolism specific to processed nuclear mRNA (27). In agreement with this, the D. melanogaster protein BSF, a likely homologue of LRP130, was found to participate in regulating the stability of the nucleus-encoded bicoid mRNA (22). We show here that, surprisingly, most of the cellular LRP130 proteins are localized inside mitochondria, where they are directly bound to polyadenylated RNAs in vivo. These findings indicate strongly that LRP130 associates with both nucleus-encoded and mitochondrial mRNAs. This provides, to our knowledge, the first identification in mammalian cells of a protein that is bound directly to mitochondrial RNAs in vivo. Furthermore, in vitro experiments indicate that this association is likely mediated by a novel type of RNA-binding domain that does not contain any of the recognizable RNA-binding motifs described to date.
The mitochondrial location of LRP130 is supported by immunolocalization and subcellular fractionation and by its inaccessibility to exogenously added proteases unless membranes are disrupted (Fig. 2). Both the Drosophila BSF protein and mouse LRP130 (22, 46) were reported to localize in part to cytoplasmic structures which, given our findings, are likely to correspond to mitochondria. In vivo cross-linking and nuclease protection experiments show that LRP130 is bound to RNA within mitochondria (Fig. 3), supporting the conclusion that LRP130 binds mitochondrially encoded RNAs. Other mitochondrial proteins with well-characterized enzymatic activities such as glutamate dehydrogenase, aconitase, and enoyl-coenzyme A hydratase have been shown to have RNA-binding activity in vitro (1, 29, 37). It will be of interest to determine whether they also bind mitochondrial RNA in vivo. An important next step in determining the function of LRP130 in mitochondria will be to identify the RNAs to which it is bound. The in vivo cross-linking and oligo(dT) selection results show that at least a subset of the RNAs bound by LRP130 are polyadenylated RNAs. The most obvious candidates for these RNAs are mitochondrial mRNAs, which contain about 60 to 80 adenylate residues (30). Additional mitochondrial RNAs that are oligoadenylated include the 12S and 16S rRNAs (11). Our results do not discern, however, whether all or only a subset of these RNAs are bound by LRP130, nor do they address whether nonpolyadenylated mitochondrial RNAs are also bound by it. We are currently investigating the identity of the RNAs to which LRP130 is bound.
While LRP130 is predominantly located in mitochondria, a detectable fraction is also found in nuclei (Fig. 1). Similarly, a recent report on mouse LRP130 also showed that a fraction of it localizes to the nucleus (46). This nuclear location is consistent with the association of LRP130 with nucleus-associated mRNPs (27). Furthermore, we have demonstrated here that LRP130 is associated with hnRNP A1 in vivo by direct cross-linking of the two proteins to the same mRNAs (Fig. 4). We consider it most likely that the RNAs bound by both LRP130 and hnRNP A1 are nucleus encoded, given the association of hnRNP A1 with nucleus-encoded RNAs. We note, however, that a different shuttling protein, hnRNP K, was recently reported to associate with mitochondrial RNA, although a direct binding to mitochondrial RNA in vivo was not demonstrated (32). This raises the question of whether hnRNP A1 associates similarly with mitochondrial RNA to which LRP130 is also bound. While we cannot formally exclude this possibility, we consider it highly unlikely given that we cannot detect any hnRNP A1 in mitochondria by either immunofluorescence microscopy or subcellular fractionation (Fig. 2). In addition, other proteins associated with LRP130 in mitochondrial RNPs do not also associate with hnRNP A1 (S. Mili and S. Piñol-Roma, unpublished data).
Our findings support a role for LRP130 in both nuclear and mitochondrial mRNA metabolism, and perhaps in coordinating gene expression between the two organelles. Studies with its Drosophila homologue suggest that LRP130 is involved in regulating mRNA stability (22). More recently, a number of interaction studies using primarily yeast two-hybrid approaches have implicated it in a broader range of cellular processes that include cytoplasmic events involving signal transduction and chromosome stability (20, 46). A dual localization and function in nuclei and mitochondria have been described for several other proteins and RNP complexes involved in RNA or DNA metabolism, including p32, the DEAD box protein MDDX28, and the yeast proteins Rna14p, Trm1, Cca1, and Mod5 (see the introduction). In addition, the mammalian RNase P RNP participates in the processing or modification of both nuclear and mitochondrial tRNAs (38). Similarly, RNase MRP is mostly concentrated in nucleoli, where it is required for 5.8S rRNA processing, and is also found in mitochondria, where it functions in cleavage of RNA primers during replication of mtDNA (19).
The dual localization of LRP130 implies the presence of targeting signals within the protein that direct it to nuclei or to mitochondria. A mitochondrial targeting signal (MTS) was not readily identifiable in the amino acid sequence of LRP130 initially deduced from the deposited cDNA (accession no. M92439). However, subsequent searches of expressed sequence tag databases revealed that the sequence of the deposited cDNA is incomplete, in that it is truncated at the 5′ end and lacks an additional ca. 380 nucleotides that encode the actual first 125 aa of LRP130. This additional sequence is also predicted by the National Center for Biotechnology Information Annotation Project (note that LRP130 is also referred to as LRPPRC; accession no. XM_031527). This additional amino-terminal sequence is highly conserved between human, mouse, and fruit fly LRP130 homologues, and we recently found that it contains a bona fide MTS that is both necessary and sufficient for mitochondrial location of LRP130 (S. Mili and S. Piñol-Roma, unpublished data). Sequences that could function as nuclear localization signals or as nuclear export sequences for the CRM1 export receptor have also been observed by us and others (20) in LRP130, and work to determine whether they indeed function in the nucleocytoplasmic traffic of LRP130 is in progress. Several mechanisms to account for the targeting of proteins to both nuclei and mitochondria, including use of alternative transcription and/or translation initiation, have been described (8, 26). Work to determine how the subcellular targeting sequences of LRP130 operate in its dual localization and whether they are subjected to regulation is in progress.
While LRP130 binds RNA directly, exhaustive analyses of its amino acid sequence have not revealed any previously described RNA-binding motifs. The most readily recognizable amino acid sequence motif in LRP130 is the PPR motif (42), of which there are 11 predicted copies in LRP130. Notably, the few other proteins with PPR motifs that have been characterized to date have been implicated in RNA metabolism in mitochondria or chloroplasts. The Zea mays Crp1 protein is involved in mRNA processing and in regulation of translation in chloroplasts (12), and a radish chloroplast protein highly similar to Crp1 was found to bind RNA (17). Similarly, S. cerevisiae Pet309p is required for the translation and for the production and/or stability of mitochondrial COX1 mRNA (23). Neurospora crassa Cya5p, related in sequence to Pet309p, is also required for posttranscriptional steps in the production of Cox1 (7). In this context, the PPR motif had emerged initially as a candidate RNA-binding motif, as its predicted structure is such that a tandem array of PPR motifs could form a superhelix enclosing a groove whose width and charge could accommodate a single strand of RNA (42). Therefore, we were surprised that 9 out of the 11 PPR motifs in LRP130 could be deleted without impairing detectably its RNA-binding activity, as the carboxy-terminal region of the protein that includes only the last two PPR motifs exhibits RNA-binding activity similar to that of the entire protein (Fig. 6). However, we cannot exclude the possibility that the portion of LRP130 containing the first nine PPR motifs has no detectable RNA-binding activity because this particular construct is misfolded or because it binds to RNAs with sequence or secondary structure characteristics different from the ones tested here. In any case, the results presented here show that the C-terminal region of LRP130 forms an independently folded RNA-binding domain and that the PPR motifs present in this region, while they may still be involved in binding, are not the only requirements for this activity. We are currently exploring the specific amino acid sequence requirements for the RNA-binding activity of the domain that we have delineated and whether the PPR motifs per se contribute to this activity. Note also that, since the homopolymers used here are rather artificial substrates, definitive confirmation that the RNA-binding domain delineated here mediates the binding of LRP130 to mitochondrial and nuclear RNAs in vivo awaits identification of its specific target binding sites in mitochondrial RNA as well as in nuclear RNA.
The available information points to the PPR motif as a signature of a large family of proteins involved in organellar RNA metabolism (42). Our results indicate strongly that most of the PPR motifs of LRP130 are dispensable for its RNA-binding activity and that it is unlikely that the sole function of PPR motifs is to define an RNA-binding domain. This suggests that they may mediate another functional aspect(s) of this family of proteins. Such additional roles may include targeting proteins within organelles and/or providing protein-protein interaction surfaces. For example, both yeast Pet309p and a PPR-containing 50-kDa protein from Nicotiana tabacum associate with membranes (4, 24). It has been predicted that many of the PPR-containing proteins in Arabidopsis thaliana also associate with membranes (42). As all the PPR-containing proteins that have been functionally characterized participate in RNA stability or translation (which at least in yeast is tightly associated with the inner mitochondrial membrane [9]), this raises the possibility that PPR motifs may be involved in the association of proteins with organellar membranes. An exception would be provided by Crp1p, which does not show significant membrane association (12). PPR motifs could also function in protein-protein interactions, in a manner similar to the closely related tetratricopeptide motifs. An intriguing observation is the presence of a PPR motif at the N termini of many metazoan mitochondrial RNA polymerases (39). The analogous N-terminal region of the yeast mitochondrial RNA polymerase interacts with Nam1p, a protein involved in mitochondrial RNA processing and translation, thus possibly coupling mitochondrial transcription to RNA processing (39). It could be envisioned that the PPR motifs of metazoan mitochondrial RNA polymerases could fulfill a similar role by recruiting mitochondrial RNA-processing factors. We note in addition that, to our knowledge, a possible role for these other PPR-containing proteins in nuclear RNA metabolism was not excluded.
Our findings that human PPR-containing protein LRP130 is an RNA-binding protein will enable us to gain a better understanding of this family of proteins and also open the way for a thorough exploration of the role of RNA-binding proteins in human mitochondrial gene expression. Furthermore, the dual localization of LRP130 in nuclei and in mitochondria raises the possibility that it participates in coordinating gene expression in these two organelles. Finally, these findings provide a framework for a detailed understanding of the molecular processes affected by the LRP130 mutations that underlie the French-Canadian type of Leigh syndrome, a cytochrome c oxidase deficiency in humans (28).
Acknowledgments
We are grateful to Konstadinos Moissoglu for valuable discussions and for suggesting the experiment in Fig. 4B, to Wolfgang Hübner for assistance with the expressed sequence tag database searches and analyses, and Gillian Small for assistance with mitochondrial preparations. We thank Fabio Triolo for helpful discussions during the course of this work; Wallace McKeehan for the LRP130 cDNA; Scott Henderson for assistance with microscopy; and Jeanne Hirsch, Bettina Winckler, Charles Query, and members of our laboratory for critical reading of the manuscript.
Microscopy was performed at the MSSM-Microscopy Shared Resource Facility, supported, in part, with funding from NIH-NCI shared resources grant (1 R24 CA095823-01). This work was supported by a grant (GM53468) from the NIH to S.P.-R.
REFERENCES
- 1.Brennan, L. E., J. Nakagawa, D. Egger, K. Bienz, and C. Moroni. 1999. Characterisation and mitochondrial localisation of AUH, an AU-specific RNA-binding enoyl-CoA hydratase. Gene 228:85-91. [DOI] [PubMed] [Google Scholar]
- 2.Brokstad, K. A., K. H. Kalland, W. C. Russell, and D. A. Matthews. 2001. Mitochondrial protein p32 can accumulate in the nucleus. Biochem. Biophys. Res. Commun. 281:1161-1169. [DOI] [PubMed] [Google Scholar]
- 3.Burd, C. G., and G. Dreyfuss. 1994. Conserved structures and diversity of functions of RNA-binding proteins. Science 265:615-621. [DOI] [PubMed] [Google Scholar]
- 4.Chang P.-F. L., B. Damsz, A. K. Kononowicz, M. Reuveni, Z. Chen, Y. Xu, K. Hedges, C. C. Tseng, N. K. Singh, M. L. Binzel, M. L. Narasimhan, P. M. Hasegawa, and R. A. Bressan. 1996. Alterations in cell membrane structure and expression of a membrane-associated protein after adaptation to osmotic stress. Physiol. Plant. 98:505-516.D. B. [Google Scholar]
- 5.Choi, Y. D., and G. Dreyfuss. 1984. Monoclonal antibody characterization of the C proteins of heterogeneous nuclear ribonucleoprotein complexes in vertebrate cells. J. Cell Biol. 99:1997-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clayton, D. A. 1991. Replication and transcription of vertebrate mitochondrial DNA. Annu. Rev. Cell Biol. 7:453-478. [DOI] [PubMed] [Google Scholar]
- 7.Coffin, J. W., R. Dhillon, R. G. Ritzel, and F. E. Nargang. 1997. The Neurospora crassa cya-5 nuclear gene encodes a protein with a region of homology to the Saccharomyces cerevisiae PET309 protein and is required in a post-transcriptional step for the expression of the mitochondrially encoded COXI protein. Curr. Genet. 32:273-280. [DOI] [PubMed] [Google Scholar]
- 8.Danpure, C. J. 1995. How can the products of a single gene be localized to more than one intracellular compartment? Trends Cell Biol. 5:230-238. [DOI] [PubMed] [Google Scholar]
- 9.Dieckmann, C. L., and R. R. Staples. 1994. Regulation of mitochondrial gene expression in Saccharomyces cerevisiae. Int. Rev. Cytol. 152:145-181. [DOI] [PubMed] [Google Scholar]
- 10.Dreyfuss, G., V. N. Kim, and N. Kataoka. 2002. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 3:195-205. [DOI] [PubMed] [Google Scholar]
- 11.Dubin, D. T., J. Montoya, K. D. Timko, and G. Attardi. 1982. Sequence analysis and precise mapping of the 3′ ends of HeLa cell mitochondrial ribosomal RNAs. J. Mol. Biol. 157:1-19. [DOI] [PubMed] [Google Scholar]
- 12.Fisk, D. G., M. B. Walker, and A. Barkan. 1999. Molecular cloning of the maize gene crp1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J. 18:2621-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorlach, M., C. G. Burd, and G. Dreyfuss. 1994. The mRNA poly(A)-binding protein: localization, abundance, and RNA-binding specificity. Exp. Cell Res. 211:400-407. [DOI] [PubMed] [Google Scholar]
- 14.Hayman, M. L., M. M. Miller, D. M. Chandler, C. C. Goulah, and L. K. Read. 2001. The trypanosome homolog of human p32 interacts with RBP16 and stimulates its gRNA binding activity. Nucleic Acids Res. 29:5216-5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou, J., F. Wang, and W. L. McKeehan. 1994. Molecular cloning and expression of the gene for a major leucine-rich protein from human hepatoblastoma cells (HepG2). In Vitro Cell. Dev. Biol. Anim. 30A:111-114. [DOI] [PubMed] [Google Scholar]
- 16.Krecic, A. M., and M. S. Swanson. 1999. hnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol. 11:363-371. [DOI] [PubMed] [Google Scholar]
- 17.Lahmy, S., F. Barneche, J. Derancourt, W. Filipowicz, M. Delseny, and M. Echeverria. 2000. A chloroplastic RNA-binding protein is a new member of the PPR family. FEBS Lett. 480:255-260. [DOI] [PubMed] [Google Scholar]
- 18.Le Hir, H., E. Izaurralde, L. E. Maquat, and M. J. Moore. 2000. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 19:6860-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, K., C. S. Smagula, W. J. Parsons, J. A. Richardson, M. Gonzalez, H. K. Hagler, and R. S. Williams. 1994. Subcellular partitioning of MRP RNA assessed by ultrastructural and biochemical analysis. J. Cell Biol. 124:871-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, L., and W. L. McKeehan. 2002. Sequence analysis of LRPPRC and its SEC1 domain interaction partners suggests roles in cytoskeletal organization, vesicular trafficking, nucleocytosolic shuttling, and chromosome activity. Genomics 79:124-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manam, S., and G. C. Van Tuyle. 1987. Separation and characterization of 5′- and 3′-tRNA processing nucleases from rat liver mitochondria. J. Biol. Chem. 262:10272-10279. [PubMed] [Google Scholar]
- 22.Mancebo, R., X. Zhou, W. Shillinglaw, W. Henzel, and P. M. Macdonald. 2001. BSF binds specifically to the bicoid mRNA 3′ untranslated region and contributes to stabilization of bicoid mRNA. Mol. Cell. Biol. 21:3462-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manthey, G. M., and J. E. McEwen. 1995. The product of the nuclear gene PET309 is required for translation of mature mRNA and stability or production of intron-containing RNAs derived from the mitochondrial COX1 locus of Saccharomyces cerevisiae. EMBO J. 14:4031-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manthey, G. M., B. D. Przybyla-Zawislak, and J. E. McEwen. 1998. The Saccharomyces cerevisiae Pet309 protein is embedded in the mitochondrial inner membrane. Eur. J. Biochem. 255:156-161. [DOI] [PubMed] [Google Scholar]
- 25.Marcu, A., B. Bassit, R. Perez, and S. Piñol-Roma. 2001. Heterogeneous nuclear ribonucleoprotein complexes from Xenopus laevis oocytes and somatic cells. Int. J. Dev. Biol. 45:743-752. [PubMed] [Google Scholar]
- 26.Martin, N. C., and A. K. Hopper. 1994. How single genes provide tRNA processing enzymes to mitochondria, nuclei and the cytosol. Biochimie 76:1161-1167. [DOI] [PubMed] [Google Scholar]
- 27.Mili, S., H. J. Shu, Y. Zhao, and S. Piñol-Roma. 2001. Distinct RNP complexes of shuttling hnRNP proteins with pre-mRNA and mRNA: candidate intermediates in formation and export of mRNA. Mol. Cell. Biol. 21:7307-7319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mootha, V. K., P. Lepage, K. Miller, J. Bunkenborg, M. Reich, M. Hjerrild, T. Delmonte, A. Villeneuve, R. Sladek, F. Xu, G. A. Mitchell, C. Morin, M. Mann, T. J. Hudson, B. Robinson, J. D. Rioux, and E. S. Lander. 2003. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc. Natl. Acad. Sci. USA 100:605-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nanda, S. K., and J. L. Leibowitz. 2001. Mitochondrial aconitase binds to the 3′ untranslated region of the mouse hepatitis virus genome. J. Virol. 75:3352-3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ojala, D., and G. Attardi. 1974. Expression of the mitochondrial genome in HeLa cells. XIX. Occurrence in mitochondria of polyadenylic acid sequences, “free” and covalently linked to mitochondrial DNA-coded RNA. J. Mol. Biol. 82:151-174. [DOI] [PubMed] [Google Scholar]
- 31.Ojala, D., J. Montoya, and G. Attardi. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature 290:470-474. [DOI] [PubMed] [Google Scholar]
- 32.Ostrowski, J., L. Wyrwicz, L. Rychlewski, and K. Bomsztyk. 2002. Heterogeneous nuclear ribonucleoprotein K protein associates with multiple mitochondrial transcripts within the organelle. J. Biol. Chem. 277:6303-6310. [DOI] [PubMed] [Google Scholar]
- 33.Petersen-Mahrt, S. K., C. Estmer, C. Ohrmalm, D. A. Matthews, W. C. Russell, and G. Akusjarvi. 1999. The splicing factor-associated protein, p32, regulates RNA splicing by inhibiting ASF/SF2 RNA binding and phosphorylation. EMBO J. 18:1014-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piñol-Roma, S. 1999. Association of nonribosomal nucleolar proteins in ribonucleoprotein complexes during interphase and mitosis. Mol. Biol. Cell 10:77-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piñol-Roma, S., Y. D. Choi, M. J. Matunis, and G. Dreyfuss. 1988. Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals an assortment of RNA-binding proteins. Genes Dev. 2:215-227. [DOI] [PubMed] [Google Scholar]
- 36.Poyton, R. O., and J. E. McEwen. 1996. Crosstalk between nuclear and mitochondrial genomes. Annu. Rev. Biochem. 65:563-607. [DOI] [PubMed] [Google Scholar]
- 37.Preiss, T., A. G. Hall, and R. N. Lightowlers. 1993. Identification of bovine glutamate dehydrogenase as an RNA-binding protein. J. Biol. Chem. 268:24523-24526. [PubMed] [Google Scholar]
- 38.Puranam, R. S., and G. Attardi. 2001. The RNase P associated with HeLa cell mitochondria contains an essential RNA component identical in sequence to that of the nuclear RNase P. Mol. Cell. Biol. 21:548-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodeheffer, M. S., B. E. Boone, A. C. Bryan, and G. S. Shadel. 2001. Nam1p, a protein involved in RNA processing and translation, is coupled to transcription through an interaction with yeast mitochondrial RNA polymerase. J. Biol. Chem. 276:8616-8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rossmanith, W., A. Tullo, T. Potuschak, R. Karwan, and E. Sbisa. 1995. Human mitochondrial tRNA processing. J. Biol. Chem. 270:12885-12891. [DOI] [PubMed] [Google Scholar]
- 41.Rouillard, J. M., C. Brendolise, and F. Lacroute. 2000. Rna14p, a component of the yeast nuclear cleavage/polyadenylation factor I, is also localised in mitochondria. Mol. Gen. Genet. 262:1103-1112. [DOI] [PubMed] [Google Scholar]
- 42.Small, I. D., and N. Peeters. 2000. The PPR motif—a TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 25:46-47. [DOI] [PubMed] [Google Scholar]
- 43.Soltys, B. J., and R. S. Gupta. 1996. Immunoelectron microscopic localization of the 60-kDa heat shock chaperonin protein (Hsp60) in mammalian cells. Exp. Cell Res. 222:16-27. [DOI] [PubMed] [Google Scholar]
- 44.Swanson, M. S., and G. Dreyfuss. 1988. Classification and purification of proteins of heterogeneous nuclear ribonucleoprotein particles by RNA-binding specificities. Mol. Cell. Biol. 8:2237-2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taanman, J. W. 1999. The mitochondrial genome: structure, transcription, translation and replication. Biochim. Biophys. Acta 1410:103-123. [DOI] [PubMed] [Google Scholar]
- 46.Tsuchiya, N., H. Fukuda, T. Sugimura, M. Nagao, and H. Nakagama. 2002. LRP130, a protein containing nine pentatricopeptide repeat motifs, interacts with a single-stranded cytosine-rich sequence of mouse hypervariable minisatellite Pc-1. Eur. J. Biochem. 269:2927-2933. [DOI] [PubMed] [Google Scholar]
- 47.Valgardsdottir, R., G. Brede, L. G. Eide, E. Frengen, and H. Prydz. 2001. Cloning and characterization of MDDX28, a putative dead-box helicase with mitochondrial and nuclear localization. J. Biol. Chem. 276:32056-32063. [DOI] [PubMed] [Google Scholar]