Abstract
A key transition in meiosis is the exit from prophase and entry into the nuclear divisions, which in the yeast Saccharomyces cerevisiae depends upon induction of the middle sporulation genes. Ndt80 is the primary transcriptional activator of the middle sporulation genes and binds to a DNA sequence element termed the middle sporulation element (MSE). Sum1 is a transcriptional repressor that binds to MSEs and represses middle sporulation genes during mitosis and early sporulation. We demonstrate that Sum1 and Ndt80 have overlapping yet distinct sequence requirements for binding to and acting at variant MSEs. Whole-genome expression analysis identified a subset of middle sporulation genes that was derepressed in a sum1 mutant. A comparison of the MSEs in the Sum1-repressible promoters and MSEs from other middle sporulation genes revealed that there are distinct classes of MSEs. We show that Sum1 and Ndt80 compete for binding to MSEs and that small changes in the sequence of an MSE can yield large differences in which protein is bound. Our results provide a mechanism for differentially regulating the expression of middle sporulation genes through the competition between the Sum1 repressor and the Ndt80 activator.
Meiosis and sporulation in Saccharomyces cerevisiae require the sequential expression of several classes of genes, analogous to the regulation of developmental pathways in higher eukaryotes (5, 13, 29). Many of the genes required for sporulation are expressed at specific times and levels in the pathway. Sporulation-specific genes can broadly be divided into early, middle, or late categories based on the timing of their expression (20). The expression of middle sporulation genes is important for controlling the meiotic G2-to-MI transition (6, 37). This transition is coupled to a key regulatory point after which cells become irreversibly committed to completing meiotic development (11). Genome-wide analyses of gene expression during sporulation identified more than 150 middle sporulation genes that are induced around the time that cells exit the pachytene stage of prophase and begin meiotic nuclear divisions (5, 29). Some middle sporulation genes encode B-type cyclins, components of the anaphase-promoting complex, and other cell cycle regulators that are required for the nuclear divisions. Other middle sporulation genes encode proteins required for spore morphogenesis, such as the Smk1 mitogen-activated protein kinase (12). Most of the middle sporulation genes contain a conserved sequence (gNCRCAAAW [lowercase indicates weak conservation]) in their promoters, termed the middle sporulation element (MSE), that is required to activate their transcription (9, 23).
The Ndt80 transcription factor binds to MSE DNA and is required to activate transcription of middle genes in vivo (5, 6). Cells lacking Ndt80 specifically arrest in meiosis at the pachytene stage before the nuclear divisions (37). NDT80 is itself a middle sporulation gene that is induced during prophase. Ndt80 is negatively regulated by the pachytene checkpoint, which monitors the completion of key events in meiosis, such as genetic recombination (6, 10, 25, 31, 35). Thus, Ndt80 plays a central role in controlling the transcriptional program and in processing signals that regulate the progression through meiosis and sporulation.
Members of a subset of the MSEs not only serve as Ndt80-dependent activator sites but also function as repressor elements during vegetative growth and at early times in the meiotic pathway (27, 36). For example, the SMK1 MSE is needed to repress transcription during vegetative growth and activate transcription during the middle stages of meiosis, functioning as a molecular switch that sets the timing of SMK1 expression.
MSE-mediated repression requires Sum1, a novel DNA-binding protein that binds some MSEs in the promoters of middle sporulation genes (36). Full repression of a subset of Sum1 regulated middle sporulation genes also requires Hst1 and Rfm1 (19). Hst1, like its close homolog Sir2, is an NAD+-dependent protein deacetylase (33). Evidence suggests that binding of Sum1 to MSE DNA can recruit the Hst1-Rfm1 complex and thereby promote targeted histone deacetylation and establishment of an inactive chromatin state (19, 28, 30).
During meiotic prophase the level of Sum1 protein in the cell decreases, presumably relieving repression of the promoters containing MSEs (18). SUM1 is required for full activity of the pachytene checkpoint, which blocks meiotic progression in the presence of recombination intermediates. Sum1 levels remain high when recombination is blocked. These data raise the possibility that the Sum1 and Ndt80 proteins are inversely regulated and play antagonistic roles in controlling meiotic progression.
To understand the regulation of the meiotic transcriptional program and the G2-to-MI transition, it is critical to determine how Sum1 and Ndt80 bind DNA and how they interact to carry out their functions at the MSE. Although both Ndt80 and Sum1 bind specifically to MSEs, neither protein shares significant sequence similarity with other characterized proteins. Our biochemical and physiological experiments reveal that Sum1 and Ndt80 have overlapping yet distinct sequence requirements for binding to and acting at MSEs. Genome-wide expression analysis shows that Sum1 is required for repression of more than 53 genes in mitotic cells. Most of the Sum1 targets are also middle sporulation genes and share a specific variant of the MSE. A competition between Sum1 and Ndt80 for binding to an MSE dictates whether the site acts as a repressor or an activator of transcription. Hence, the molecular switch that induces genes required for the meiotic G2-to-M transition depends on the relative affinities of each MSE site for Ndt80 and Sum1.
MATERIALS AND METHODS
Plasmids.
The MSE-lacZ reporter constructs were made as described previously and verified by sequence analysis (27). The C-terminally tagged His6-Sum1 bacterial expression plasmid was constructed by cloning a PCR-generated fragment of the SUM1 open reading frame (ORF) coding for residues 523 to 1062 into the NdeI-XhoI sites of pET21c (Novagen). The C-terminally tagged His6-Ndt80 bacterial expression plasmid was constructed by cloning a PCR-generated fragment of the NDT80 ORF coding for residues 1 to 409 into the NdeI-XhoI sites of pET21c (22). The C-terminally tagged SUM1-myc yeast expression plasmid was constructed by making an in-frame fusion of the full-length SUM1 ORF with 13 repeats of the myc epitope (EQKLISEEDL). A BglII restriction fragment containing a portion of the SUM1 ORF and the myc tag was then cloned into the BglII sites of a SUM1 genomic subclone, pJX62 (36). Positive clones were assayed for complementation of the sum1Δ null mutant phenotype for repression of transcription of the SMK1 MSE-lacZ reporter, pJX43 (27).
Liquid β-galactosidase assay.
All β-galactosidase activity assays in liquid were performed as described previously (8). The various MSE-lacZ plasmids were used to transform W303-1A (MATa ade2-1 trp-1-1 his3-11,15 can1-100 ura3-1 leu2-3,112) to measure Sum1-dependent repression. To determine Ndt80-dependent activation, these same reporter plasmids were used to transform MPY4 (MATa ade2-1 trp-1-1 his3-11,15 can1-100 ura3-1 leu2-3,112 sum1Δ::kanMX4 HIS3::pGAL1-10-NDT80). MPY4 is a derivative of strain JXY3 (36), which contains an integrated copy of the pGAL1-10-NDT80 expression plasmid (5). W303-1A cells were grown in SD medium lacking Ura (SD-Ura) medium to mid-log phase before being harvested for the β-galactosidase assay. MPY4 cells were grown in SRaf-Ura (2% raffinose) to saturation, diluted 1:100 in SRaf-Ura containing 2% galactose, grown overnight, and then assayed for β-galactosidase activity. Assays were performed in triplicate with independent transformants, and the standard deviations were less than 15%.
EMSA.
Proteins used for electrophoretic mobility shift assays (EMSAs) were prepared from BL21 Codon Plus cells transformed with plasmids expressing His6-tagged Sum1(523-1062) or Ndt80(1-409). Cells were grown to mid-log phase, protein expression was induced by addition of isopropyl-β-d-thiogalactopyranoside to a 1 mM concentration, and cells were allowed to grow at 37°C for 3 h. Cell lysis was carried out by French press, and the protein was purified using a nickel column (Novagen). Proteins were eluted from the column with 250 mM imidazole and were at least 90% pure as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels stained with Coomassie blue. Protein concentrations were determined by the Bradford assay using bovine serum albumin for the standard curve.
Probe preparation and EMSAs were carried out as previously described (36). Probes were made using the same oligonucleotides containing wild-type or mutant SMK1 MSEs that were cloned into the MSE-lacZ reporter vector. The percent binding activity of each mutant MSE was determined by calculating how much more (or less) protein was required to produce the same amount of DNA-protein complex with the mutant sites as was observed with the wild-type SMK1 MSE.
The Sum1 and Ndt80 competition experiments were performed using purified truncated forms of Sum1523-1062 and Ndt801-409. Prior to the addition of Ndt80, 18 μl of probe was mixed with 3 μl of dilution buffer or 3 μl of a dilution of Sum1 sufficient to bind approximately 50% of the SMK1 MSE DNA (final concentration, 0.12 μM). After a 30-min incubation, increasing amounts of Ndt80 protein (3 μl of twofold serial dilutions) were added to the reactions followed by incubation at room temperature for 30 min. Reactions were processed as described above.
Microarray and site analysis.
Yeast cultures were grown in yeast extract-peptone-dextrose to mid-log phase, cells were harvested by centrifugation, and total and poly(A) RNA was isolated as described previously (7) (http://www.microarrays.org/protocols.html). The wild-type strains used were W303-1A and LNY435, a diploid derivative of W303-1A. Strains JXY3 (MATa) and JXY13 (MATa/MATα) are derivatives of W303-1A and contain the sum1Δ::kanMX4 deletion (36). The poly(A) RNA was used as a template for reverse transcription to make cDNA containing amino-allyl dUTP (Sigma), and then the modified dUTP was covalently linked to Cy3 or Cy5 fluorophores (Amersham Pharmacia) as described previously (3, 7) (http://www.microarrays.org/protocols.html). Poly(A) RNA samples from wild-type and sum1 mutant strains were processed in parallel and labeled with different fluorophores. The two differentially labeled cDNA samples were mixed together before hybridization to the same whole-genome microarray as described previously (3, 7, 16). Microarrays were scanned with a GenePix 4000A microarray scanner (Axon Instruments, Union City, Calif.), and quantitative analysis was done using GenePix software.
Three experiments were performed to compare the mRNA levels in wild-type and sum1 mutant strains. In experiment 1, poly(A) RNA samples from haploid wild-type (W303-1A) and haploid sum1 (JXY3) were labeled with Cy5 and Cy3, respectively. Experiment 2 was similar to experiment 1 except that the fluorophores were reversed. The microarray in experiment 3 was hybridized to Cy3-labeled cDNA made from a diploid wild-type strain (LNY435) and Cy5-labeled cDNA from a diploid sum1 strain (JXY13).
Data analysis was aided by several software programs, including GenePix (Axon Instruments), Microsoft Excel, and FileMaker Pro. The genes that were induced in the sum1Δ strain were compared with a set of 161 genes that are induced during the middle stages of sporulation. We have generated this set of middle genes by analyzing both published data sets of gene expression during sporulation (5, 29). Roughly, this set was defined by the overlap between the Chu et al. clusters, called early middle, middle, and mid-late, and the Primig et al. clusters, called 5 and 6. This list is available upon request. The identification of potential regulatory sites for each ORF in the genome was aided by a software tool called Promoter, written by J. DeRisi (available at http://derisilab.ucsf.edu). The regulatory region of a gene was provisionally defined as the 600 bp upstream of the ATG start site. Given a set of MSE sequences, the frequency of each base at each position in the site was calculated using the DNA Consensus software program written by T. Ognibene.
For analysis of sequence bias in the MSE sites found in coregulated gene sets, it was important to maximize the number of sequences used in the analysis in order to allow better statistics. Yet it was also critical to exclude noise where possible; one potential source of noise was genes with multiple MSE sites, since in some cases only one site need be functional to account for the observed transcriptional behavior. To address these opposing challenges, the following criteria were applied in choosing MSE sites to include in the sequence analysis for Tables 3, 4, 5, and 6. (A) First, each member of a set of genes with similar functional behavior was examined to identify all the MSES sites (DSYGWCAYWDW) and MSEN sites (VNDNCRCAAW) in the 600 bp upstream of the start codon. (B) At most, one site per gene was included in the sequence analysis. (C) If a gene had only one site, that site was included regardless of whether it was an MSES site or an MSEN site. (D) If a gene had more than one site, a unique site that satisfied the criteria for both an MSES and an MSEN site was chosen. (E) If a gene had more than one site and did not satisfy condition D, the locations of the sites were considered. Functional regulatory elements are usually within 250 to 300 bases of the start codon of yeast genes. If there was only one site located less than 300 bp from the ORF, it was chosen for inclusion in the analysis. (F) If a gene had more than one site and did not satisfy the conditions above, the sites were not included in the analysis.
TABLE 3.
| Base | 2 | 3 | G 4 | T 5 | G 6 | W 7 | C 8 | A 9 | C 10 | A 11 | A 12 | A 13 | N 14 | K 15 | 16 | 17 | 18 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 31 | 47 | 19 | 16 | 12 | 56 | 0 | 88 | 0 | 100 | 97 | 75 | 38 | 19 | 34 | 28 | 28 |
| G | 12 | 16 | 53 | 3 | 81 | 3 | 0 | 12 | 0 | 0 | 0 | 0 | 22 | 28 | 9 | 31 | 25 |
| C | 22 | 12 | 28 | 28 | 0 | 9 | 100 | 0 | 100 | 0 | 0 | 0 | 16 | 12 | 34 | 16 | 12 |
| T | 34 | 25 | 0 | 53 | 6 | 31 | 0 | 0 | 0 | 0 | 3 | 25 | 25 | 41 | 22 | 25 | 34 |
Values are the percentages of each base among 32 MSEs found upstream of genes that are derepressed in the sum1 mutant (see Materials and Methods for details).
Consensus base and position number are shown above each column. This more narrow consensus was chosen using the preferences noted in this table (see Materials and Methods for details). Bases that exhibit statistically significant differences from the control set of sequences are shown in bold and underlined. W: A or T; N: A, G, C, or T; K: G or T. Statistical comparison of these base preferences to the control set (see Table 6) yielded the following chi-square test P values: position 5, 0.016; 6, 7.8 × 10−6; 7, 0.029; 9, 0.014; and 15, 0.037.
TABLE 4.
| Base | 2 | A 3 | G 4 | Y 5 | G 6 | W 7 | C 8 | A 9 | C 10 | A 11 | A 12 | A 13 | A 14 | D 15 | 16 | 17 | 18 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 31 | 58 | 21 | 10 | 10 | 46 | 0 | 88 | 0 | 92 | 88 | 81 | 52 | 25 | 40 | 38 | 35 |
| G | 13 | 15 | 54 | 4 | 85 | 6 | 0 | 12 | 0 | 0 | 8 | 0 | 12 | 27 | 12 | 13 | 19 |
| C | 19 | 8 | 25 | 38 | 0 | 6 | 100 | 0 | 96 | 0 | 0 | 0 | 17 | 4 | 29 | 13 | 15 |
| T | 38 | 19 | 0 | 48 | 6 | 42 | 0 | 0 | 4 | 8 | 4 | 19 | 19 | 44 | 19 | 35 | 31 |
Values are the percentages of each base among 52 MSEs found upstream of genes that were observed to be bound by Sum1 protein (17) (see Materials and Methods for details).
The consensus (shown at top with position numbers) was chosen using the preferences noted in this table (see Materials and Methods for details). Bases that exhibit statistically significant differences from the control set of sequences are shown in bold and underlined. W: A or T; N: A, G, C, or T; K: G or T; D: A, G, or T. Statistical comparison of these base preferences to the control set (Table 6) yielded the following chi square test P values: Position 3, 0.031; 4, 0.0061; 5, 1.8 × 10−4; 6, 1.7 × 10−9; 7, 0.018; 9, 7.5 × 10−5; 14, 0.044; and 15, 0.0012.
TABLE 5.
| Base | 2 | 3 | 4 | Y 5 | G 6 | N 7 | C 8 | A 9 | C 10 | A 11 | A 12 | A 13 | A 14 | 15 | 16 | 17 | 18 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 30 | 37 | 34 | 18 | 25 | 36 | 0 | 82 | 0 | 97 | 95 | 76 | 58 | 36 | 25 | 43 | 38 |
| G | 22 | 17 | 36 | 4 | 50 | 17 | 0 | 18 | 0 | 0 | 5 | 0 | 16 | 18 | 14 | 18 | 21 |
| C | 17 | 21 | 30 | 30 | 0 | 13 | 100 | 0 | 95 | 0 | 0 | 0 | 9 | 16 | 25 | 13 | 9 |
| T | 30 | 25 | 0 | 47 | 25 | 34 | 0 | 0 | 5 | 3 | 0 | 24 | 17 | 30 | 36 | 25 | 32 |
Values are the percentages of each base among 76 MSEs from middle sporulation genes that are not derepressed in a sum1 mutant (see Materials and Methods for details).
This more narrow consensus was chosen using the preferences noted in this table (see Materials and Methods for details). Bases that exhibit statistically significant differences from the control set of sequences are shown in bold and underlined. Y: C or T; N: A, G, C, or T. Statistical comparison of these base preferences to those in the control set (Table 6) yielded the following chi square test P values: Position 5, 0.0011; 6, 0.0058; 9, 2.5 × 10−4; and 14, 0.0013.
TABLE 6.
Control: base preferences of MSEs from all other genesa
| Base | % at position:
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | |
| A | 30 | 36 | 32 | 33 | 40 | 34 | 0 | 59 | 0 | 91 | 91 | 62 | 34 | 39 | 33 | 33 | 34 |
| G | 19 | 25 | 34 | 14 | 34 | 21 | 0 | 41 | 0 | 0 | 4 | 0 | 17 | 16 | 22 | 22 | 19 |
| C | 24 | 14 | 34 | 22 | 0 | 13 | 100 | 0 | 89 | 0 | 0 | 0 | 14 | 19 | 19 | 21 | 19 |
| T | 27 | 25 | 0 | 30 | 25 | 32 | 0 | 0 | 11 | 9 | 6 | 38 | 35 | 26 | 26 | 24 | 29 |
To generate the narrow consensus sequences shown in Tables 3, 4, and 5, the following criteria were applied to the data shown in the table for each position. The chi-square test was used to compare the frequency of bases observed in the set of control sites (Table 6) to that observed in the set of sites from a coregulated class of genes. Base preferences with a P value of 0.11 or less were included in the consensus. In addition, if a single base was observed in 50% or more of the sites and the next-most-prevalent base was less than 30%, then the base was included in the consensus.
Western blotting.
Yeast lysates for Western blot analysis were prepared as previously described (18). Proteins were boiled, separated on a sodium dodecyl sulfate-8% polyacrylamide gel, transferred to nitrocellulose membranes, probed using the 9E10 anti-myc antibody, and detected using ECL Western blotting (Amersham Pharmacia Biotech).
RESULTS
MSE sequence requirements for Ndt80 and Sum1 functions are distinct.
To understand meiotic transcriptional regulation, it is critical to determine whether Ndt80 and Sum1 bind to identical or overlapping DNA sequences. The Ndt80 activator specifically binds to MSE DNA. Mutations that change multiple bases in the MSE consensus sequence (DNCRCAAAW) cause a decrease in Ndt80-dependent transcriptional activation in vivo and DNA binding in vitro (6). We have previously shown that the Sum1 repressor binds to a subset of MSEs and a mutation that alters multiple bases in the MSE consensus disrupts Sum1 binding in vitro (36). Interestingly, while some MSEs are targets for both Ndt80 and Sum1, other MSEs appear to function only as Ndt80-dependent activator sites or to function mainly as Sum1-dependent repressor sites. These results suggest that the two proteins may have different sequence specificities for binding DNA.
To examine the differential specificity of these proteins, we made a series of nucleotide substitutions within the SMK1 MSE in the context of a heterologous promoter driving the expression of lacZ. The effects of these mutations on Sum1-dependent repression were determined by measuring β-galactosidase activity in mitotic cells. The mutant MSEs were also assayed for Ndt80-dependent activation in mitotic cells in which Ndt80 was ectopically expressed using the GAL1 UAS (5). The binding affinities of Ndt80 and Sum1 to the mutant MSEs were assayed directly using an EMSA.
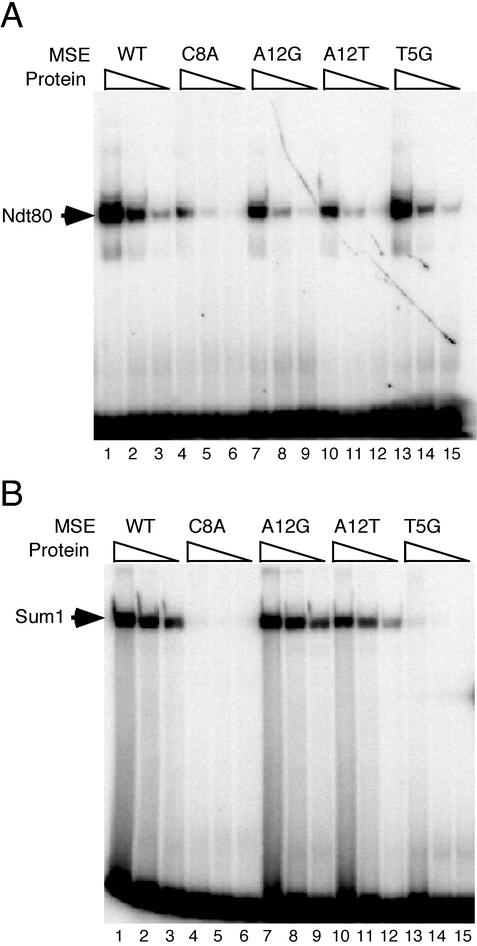
Many of the single-base-pair substitutions within the previously proposed MSE consensus sequence (nucleotides 6 to 14 in Table 1) exhibited significant decreases in Ndt80-dependent activation. For example, the G6C, C8A, A9C/T, C10A/G/T, A11T/C, and A12T substitutions each decreased Ndt80-dependent activation by fivefold or more, indicating specific sequence requirements at these positions. Our data also confirm the suggestion that position 7 can be any base (N) because each of the substitutions at this position had essentially wild-type activity. In fact, the T7C and T7A mutants showed slightly higher levels of activation, suggesting that the native SMK1 MSE is not optimal for Ndt80 function. In general, base-pair substitutions outside of the MSE, both 5′ and 3′ to the consensus, had little or no effect on activation. These data support the model that a sequence similar to the previously proposed MSE consensus sequence is the target site for Ndt80-dependent activation.
TABLE 1.
Repression, activation, and DNA binding of variant SMK1 MSE sitesa
| Substitution | 2468 1012 1416 18 C A A G T G T C A C A A A T T A G T G V N D N C R C A A W D S Y G W C A Y W D W | % Activity of:
|
|||
|---|---|---|---|---|---|
| Ndt80
|
Sum1
|
||||
| Act.b | Boundc | Rep.d | Boundc | ||
| WT | 100 | 100 | 100 | 100 | |
| A2C | C | 87 | 83 | 54 | 25 |
| A3C | C | 31 | 83 | 3 | 10 |
| A3T | T | 55 | 111 | 75 | 63 |
| A3G | G | 76 | 56 | 188 | 100 |
| G4T | T | 38 | 50 | 3 | 1.4 |
| G4C | C | 38 | 59 | 58 | 8 |
| G4A | A | 18 | 71 | 34 | 50 |
| T5A | A | 57 | 83 | 1 | 0.4 |
| T5G | G | 73 | 40 | 2 | 0.4 |
| T5C | C | 62 | 63 | 80 | 67 |
| G6A | A | 38 | 111 | 3 | 0.1 |
| G6C | C | 20 | 28 | 4 | 1 |
| G6T | T | 58 | 56 | 2 | 0.4 |
| T7C | C | 269 | 143 | 8 | 7 |
| T7A | A | 118 | 200 | 188 | 100 |
| T7G | G | 77 | 100 | 19 | 1 |
| C8T | T | 50 | 24 | 4 | 0.05 |
| C8G | G | 28 | 4 | ||
| C8A | A | 16 | 8 | 2 | 0.6 |
| A9G | G | 57 | 29 | 48 | 25 |
| A9T | T | 19 | 13 | ||
| A9C | C | 20 | 17 | 3 | 1 |
| C10T | T | 14 | 2 | 92 | 3 |
| C10G | G | 16 | 50 | ||
| C10A | A | 20 | 2 | 3 | 3 |
| A11T | T | 18 | 12 | 105 | 20 |
| A11C | C | 16 | 11 | ||
| A11G | G | 31 | 13 | 6 | 24 |
| A12T | T | 10 | 18 | 188 | 40 |
| A12C | C | 28 | 1 | ||
| A12G | G | 37 | 14 | 4 | 111 |
| A13T | T | 77 | 31 | 188 | 125 |
| A13C | C | 23 | 2 | ||
| A13G | G | 40 | 42 | 3 | 40 |
| T14G | G | 124 | 166 | 21 | 83 |
| T14A | A | 148 | 87 | ||
| T15G | G | 58 | 100 | 188 | 100 |
| A16C | C | 187 | 125 | ||
| G17T | T | 97 | 94 | ||
The consensus sequences for Ndt80 (MSEN) (middle sequence at top of table) and Sum1 (MSES) (bottom sequence at top of table) activity and binding based upon the in vivo and in vitro analyses of mutations in the SMK1 MSE (top sequence) (WT) shown in this table. A base was included in the consensus if it gave a value of 50% or more in the in vitro or the in vivo assay or both. D: A, G or T; V: A, G, or C; N: A, G, C or T W: A or T; R: A or G; S: G or C; Y: C or T.
The percent activity for transcriptional activation (Act.) by Ndt80 was calculated by comparing the activation of the mutant sites to activation of the wild-type site (defined as 100%) when Ndt80 is ectopically expressed. Ndt80 activated the wild-type SMK1 MSE reporter 6.2 fold (18.7 β-galactosidase units) over the reporter lacking the site (3 β-galactosidase units) when NDT80 expression was induced in a sum1 mutant strain.
The percent binding (Bound) was calculated by measuring the ability of Sum1 or Ndt80 to bind to each mutant sequence compared to binding to the wild-type sequence. Several concentrations of protein were tested with each mutant site.
The percent activity for repression (Rep.) by Sum1 was calculated by measuring reporter gene expression and comparing the repression conferred by each mutant site to the repression of the wild-type site (defined as 100%). The wild-type site represses the reporter 46-fold (0.4 β-galactosidase units) compared to a reporter lacking the site (18.3 β-galactosidase units) in a wild type SUM1 strain.
As observed with Ndt80-dependent activation, Sum1-dependent repression was impaired by many of the base-pair substitutions within the MSE consensus, as measured by increases in the expression of the lacZ reporter (Table 1). For example, the C8A/G/T, A9C, C10A, and A11G mutations derepressed the MSE reporter 17- to 50-fold. Substitutions 3′ to the consensus site, such as T15G, A16C, and G17T, had no effect on repression. In contrast, substitutions made 5′ to the MSE, such as A3C, G4T, and T5A/G, caused significant losses in repression, illustrating that positions outside of the MSE consensus sequence are required for repression but not for activation. While any of the four bases were able to function at position 7 for Ndt80-dependent activation, Sum1-mediated repression strongly favored A or T at this position. Both Ndt80 and Sum1 activity were optimal with a G at position 6, but mutating this site decreased Sum1 activity 25- to 50-fold, while these substitutions decreased Ndt80 activity only twofold to fivefold. Taken together, these data indicate that the sequence requirements for repression overlap with the requirements for activation. However, there are also distinct sequence requirements for activation and repression.
Next, we conducted DNA-binding assays with the mutant MSEs and purified Ndt80 and Sum1 proteins. Representative binding assays are shown in Fig. 1, and the relative DNA-binding affinities for many of the mutant sites are listed in Table 1. There is a strong correlation between the Ndt80 DNA-binding affinity in vitro and transcriptional activation in vivo. Mutations in the MSE that caused large decreases in transcriptional activation also showed significant decreases in Ndt80-binding affinity. For example, the C8A, A9C, C10A/T, A11T, and A12T mutations caused greater-than-fivefold decreases in both activation and Ndt80 DNA-binding affinity. In contrast, substitutions on either side of the consensus site, such as A2C, A3C/T/G, T14G, and T15G, had less effect on Ndt80-dependent activation and did not substantially affect Ndt80 binding. Our results agree well with a less comprehensive analysis of Ndt80-binding affinities with mutants of the SPS4 MSE (14).
FIG. 1.
Mutations in SMK1 MSE alter Sum1 and Ndt80 binding affinity. EMSA was performed with wild-type and mutant SMK1 MSEs using fivefold serial dilutions of purified His6-tagged Ndt801-409 (A) and Sum1523-1062 (B). Results from four representative mutants are shown. The SMK1 wild-type site is in lanes 1 to 3; C8A mutant, lanes 4 to 6; A12G, lanes 7 to 9; A12T, lanes 10 to 12; and T5G, lanes 13 to 15. The percent binding of each mutant site was determined in comparison to the wild type and is shown in Table 1.
There is also a strong correlation between the effects of mutations on transcriptional repression in vivo and Sum1-binding affinity in vitro. For example, the A12T and A13T mutations did not affect repression, nor do they exhibit any significant decrease in binding activity compared to the wild-type site. Mutations within the MSE consensus that cause a decrease in repression, such as T7C/G, C8A/T, A9C, and C10A, also cause large decreases in Sum1-binding affinity. Mutations outside of the MSE consensus that caused a decrease in repression, such as A3C, G4T, and T5A/G, also displayed decreases in Sum1-binding affinity.
Although most of the mutants showed similar effects in vitro and in vivo, there were a few interesting exceptions to the correlation between Sum1 DNA binding and repression. The A12G, A13G, and to some extent A11G mutant sites showed large reductions in Sum1-dependent repression but had little or no effect on Sum1 DNA-binding affinity. This suggests that although Sum1 binding is required for repression it is not sufficient. One possibility is that an additional factor(s) is required to bind to this region to mediate repression.
The mutational analysis shows that there is a clear overlap between Sum1 and Ndt80 in the sequence requirements for transcriptional regulation and for protein binding. However, the mutagenesis also demonstrates that there are sequence requirements that are specific for MSE-mediated activation and Ndt80 binding and others that are specific for repression and Sum1 binding. We conclude that Ndt80-dependent activation requires a sequence that loosely correlates with the previously proposed consensus of DNCRCAAAW (5, 23). Based upon the in vivo analyses of mutations in the SMK1 MSE, we propose an overlapping but distinct consensus sequence for Sum1-mediated repression and binding. To facilitate further sequence analysis (see below), we initially defined the Ndt80 and Sum1 sites rather broadly (Table 1). For each position, a specific base was included in the consensus if the variant site containing that base gave a value above 50% in the in vivo assay or the in vitro assay or both. Application of these criteria yielded an Ndt80 site, termed MSEN, with the sequence VNDNCRCAAW and a Sum1 site, MSES, with the sequence DSYGWCAYWDW (N, A/G/C/T; D, A/G/T; V, A/G/C; W, A/T; R, A/G; S, G/C; Y, C/T).
A validation of the MSEN and MSES consensus definitions is provided by examining the upstream regulatory region of NDT80 itself. The MSEs located at −78 and −221 upstream of the NDT80 gene are known to be required for Ndt80-dependent transcription, but only the −78 MSE is capable of Sum1-mediated repression (24, 36). In accordance with these observations, both MSEs satisfy the definition of MSEN, but only the one that serves as a Sum1 target, located at −78, satisfies the definition of MSES. Of the 161 middle sporulation genes identified by DNA microarray analysis, 117 of their upstream regulatory regions contain a match to the previously defined MSE consensus DNCRCAAAW (5, 29), 132 contain at least one MSEN, and 63 contain at least one MSES. The MSEN and MSES sites are found upstream of 50% and 15% of the genes in yeast, respectively. Thus, these sites are too broad to have any predictive value for identifying additional targets of Sum1 regulation.
Genes that are derepressed in a sum1 mutant contain matches to the MSE.
To identify genes that are repressed by Sum1, we compared gene expression in wild-type and sum1Δ strains. We examined the mitotic mRNA levels of nearly all yeast genes simultaneously by hybridization to DNA microarrays. Three experiments gave similar results. The 53 genes that were derepressed in the sum1 mutant relative to the wild-type control strain are shown in Table 2. A gene was counted as derepressed if its transcript level was at least twofold higher for the sum1 mutant than for the wild type in all three experiments. (A less stringent definition of Sum1-regulated genes yields 80 genes that were derepressed more than 1.5-fold in each experiment and more than 2-fold on average.) Among the 53 genes that were strongly derepressed in the sum1 mutant, 31 are middle sporulation genes. Many of these genes code for proteins involved in the synthesis of the spore wall, for example, SMK1, SPS1, GIP1, CDA1, and DTR1.
TABLE 2.
Genes that are derepressed in a sum1Δ strain
| ORF | Gene |
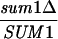 a a
|
SD | Expb | Expc | Ndt80d
|
Sum1e
|
||
|---|---|---|---|---|---|---|---|---|---|
| No. | Location(s) | No. | Location | ||||||
| YHR091C | MSR1 | 29.5 | 7.3 | ||||||
| YAL018C | YAL018C | 26.6 | 10.2 | M | 5a | 1 | 434 | 1 | 434 |
| YGR059W | SPR3 | 22.9 | 4.8 | M | 5a | 2 | 27,294 | 2 | 21,294 |
| YCL026C-A | FRM2 | 20.6 | 9.2 | 1 | 233 | 1 | 185 | ||
| YDR522C | SPS2 | 13.8 | 2.2 | M | 5a | 1 | 582 | ||
| YNL318C | HXT14 | 11.8 | 6.1 | M | 5a | 2 | 135, 486 | 3 | 235, 241, 462 |
| YLR307W | CDA1 | 10.6 | 3.0 | M | 5a | 3 | 119, 163, 370 | ||
| YBL098W | BNA4 | 10.5 | 3.6 | 1 | 562 | ||||
| YLR343W | YLR343W | 9.2 | 1.9 | M | 5a | 1 | 150 | 2 | 150, 560 |
| YGL138C | YGL138C | 9.1 | 1.0 | M | 1 | 463 | |||
| YFL012W | YFL012W | 9.1 | 1.7 | M | 5a | 3 | 52, 170, 438 | 1 | 170 |
| YHR090C | YNG2 | 8.7 | 1.3 | 1 | 350 | ||||
| YOR365C | YOR365C | 7.7 | 6.6 | M | 5b | 1 | 479 | 1 | 497 |
| YBR045C | GIP1 | 7.4 | 2.4 | M/L | 5a | 3 | 189, 347, 569 | 2 | 145, 368 |
| YLR232W | YLR232W | 7.4 | 2.8 | ||||||
| YJL037W | YJL037W | 7.3 | 1.1 | M | 5a | 2 | 281, 562 | 1 | 432 |
| YDR523C | SPS1 | 7.1 | 2.4 | M | 5a | 1 | 242 | 1 | 217 |
| YNL319W | YNL319W | 6.9 | 1.2 | M | N | 1 | 547 | ||
| YLR231C | BNA5 | 6.9 | 4.5 | 1 | 60 | ||||
| YER106W | MAM1 | 6.9 | 1.5 | M | 5b | 3 | 160, 292, 318 | 2 | 154, 160 |
| YJL038C | YJL038C | 6.8 | 2.3 | M | 5a | 2 | 275, 560 | 1 | 145 |
| YJR025C | BNA1 | 6.5 | 2.7 | 2 | 279, 465 | 2 | 353, 465 | ||
| YGL230C | YGL230C | 6.5 | 3.1 | M | 5a | 2 | 170, 406 | ||
| YHR147C | MRPL6 | 6.5 | 2.1 | 1 | 578 | ||||
| YFR023W | PES4 | 6.3 | 2.8 | M | 5a | 3 | 38, 235, 249 | 1 | 249 |
| YDL114W | YDL114W | 5.9 | 1.2 | M | 5a | 3 | 125, 360, 384 | 3 | 47, 125, 360 |
| YGR273C | YGR273C | 5.8 | 3.3 | M | 2 | 202, 496 | 1 | 496 | |
| YOL047C | YOL047C | 5.8 | 1.2 | M | 5a | 2 | 255, 448 | ||
| YJL043W | YJL043W | 5.6 | 3.2 | 1 | 114 | ||||
| YJR078W | BNA2 | 5.3 | 2.2 | 2 | 222, 299 | 1 | 130 | ||
| YBR180W | DTR1 | 5.0 | 2.9 | M/L | 5a | 1 | 145 | ||
| YGL015C | YGL015C | 4.7 | 1.0 | M | 5a | 4 | 475, 495,··· | 2 | 130, 475 |
| YKL178C | STE3 | 4.7 | 3.7 | 3 | 276, 519, 576 | 3 | 274, 276, 282 | ||
| YBR040W | FIG1 | 4.5 | 2.1 | 1 | 214 | ||||
| YPR054W | SMK1 | 4.4 | 2.0 | M | 5a | 2 | 84, 312 | 1 | 84 |
| YOL024W | YOL024W | 4.1 | 2.5 | M | 5a | 2 | 70, 104 | ||
| YIR027C | DAL1 | 4.1 | 1.0 | 3 | 405, 469, 562 | ||||
| YMR096W | SNZ1 | 3.9 | 0.6 | ||||||
| YOL091W | SPO21 | 3.9 | 0.4 | M | 5b | 1 | 122 | 2 | 128, 142 |
| YGR259C | YGR259C | 3.8 | 1.1 | ||||||
| YGR260W | TNA1 | 3.6 | 0.2 | ||||||
| YLL005C | SP075 | 3.5 | 0.6 | M | n | 1 | 505 | 1 | 499 |
| YLR040C | YLR040C | 3.4 | 1.0 | ||||||
| YGL170C | SPO74 | 3.4 | 1.2 | M | 5a | 1 | 492 | 1 | 492 |
| YAL067C | SEO1 | 3.4 | 1.7 | 3 | 99, 349,425 | 1 | 349 | ||
| YOR214C | YOR214C | 3.2 | 1.0 | E/M | 5a | 1 | 367 | ||
| YCR105W | ADH7 | 3.1 | 0.7 | ||||||
| YJR079W | YJR079W | 3.0 | 0.7 | 1 | 492 | 1 | 573 | ||
| YOR255W | YOR255W | 2.9 | 0.4 | M | 5a | 3 | 56, 78, 254 | 2 | 50,78 |
| YLR041W | YLR041W | 2.8 | 0.7 | ||||||
| YLR136C | TIS11 | 2.7 | 1.0 | 5 | 278, 355, 419, 445, 549 | ||||
| YBR250W | YBR250W | 2.3 | 0.4 | M | 5b | 2 | 178, 184 | 2 | 178, 184 |
| YJR107W | YJR107W | 2.2 | 0.1 | M | 5a | 1 | 298 | 1 | 292 |
Derepression in a sum1Δ strain compared to that for a wild-type strain. The average value from three experiments is shown.
Timing of expression during sporulation based on microarray analysis from Chu et al. (5).
Timing of expression during sporulation based on microarray analysis from Primig et al. (29).
Number and locations of Ndt80 sites in the promoter. The site was defined loosely as VNDNCRCAAW; location is given as number of bases upstream of the start codon.
Number and location(s) of Sum1 sites in the promoter. The site was defined loosely as DSYGWCAYWDW; location is given as number of bases upstream of the start codon. Note that sequences at some locations satisfy the definitions of both an Ndt80 site and a Sum1 site.
Twenty-nine of the fifty-three strongly Sum1-dependent genes have at least one MSES site, and 44 have at least one MSEN site. Eight genes have no apparent MSE sequences (Table 2). Although an MSEN site is found in 50% of the upstream regulatory regions in the genome, it is enriched in the middle sporulation gene promoters (81%) and the Sum1-dependent promoters (83%). Similarly, the MSES sequence is found in 15% of the promoters in the genome, 55% of the Sum1-dependent promoters, and 39% of the middle sporulation gene promoters. Recently, genome-wide DNA binding of Sum1 in yeast cells was examined by chromatin immunoprecipitation (17). Sum1 was found upstream of about 95 genes, of which 75% contain an MSEN site and 55% contain an MSES site.
We sought a better understanding of the genomic sequences where Sum1 and Ndt80 may act. Specifically, we wanted to determine if the sequences of the MSEs in the promoters of the different classes of genes identified in the microarray analysis correlate with whether these genes are regulated by Sum1 and Ndt80. Also, we hoped to arrive at more narrowly defined consensus sequences for the MSE(s). Thus, we compared the MSEs in the promoters of Sum1-dependent genes, the MSEs in Sum1-independent middle sporulation genes, and the MSEs in Sum1-bound genes (17) to look for differences in sequence. For these analyses, we considered all MSEN and MSES sites in each class of genes. A complication is that many promoters contain more than one MSE. Our previous studies showed that Sum1-dependent and Sum1-independent MSEs can exist in a single promoter. To avoid bias and limit noise, we chose to restrict our analysis to genes that have a single MSE or to genes in which one MSE (among several) appears to be the best candidate. Application of the criteria for selecting MSEs to include in the analysis (see Materials and Methods) yielded 32 Sum1-dependent candidate sites, 52 Sum1-bound candidate sites, and 76 middle gene sites that are independent of Sum1.
We aligned the MSE and flanking sequences (positions 1 to 18, as shown in Table 1) from the 32 Sum1-regulated genes and determined the frequency of each base at each position (Table 3). We performed the same analysis on the 52 MSEs from genes bound by Sum1 (Table 4) and the 76 MSEs from middle sporulation genes that are not derepressed in the sum1Δ strain (Table 5). For comparison, a random subset of genes that do not fall into one of the three functional classes was subjected to the same procedure for identification and analysis of MSEs in order to generate base preferences found in 234 control MSEs (Table 6). The base preferences in each functional set (Tables 3, 4, and 5) were compared to those in the control set of genes not regulated by Sum1 or Ndt80 (Table 6). Statistically significant differences in base preferences between the functional sets and the control set were noted at several positions (see tables).
The base preferences found in MSE sequences upstream of Sum1-regulated genes were similar to those in Sum1-bound genes, even though the overlap of these two classes is incomplete. Of 95 genes bound by Sum1, only 22 were found to be Sum1 dependent (see Discussion for possible explanations). Thus, 31 of the 53 Sum1-repressible genes were not identified as bound to Sum1 (at least not within the P-value limit of 0.005). Both classes of genes contained MSE sites with statistically significant nucleotide biases at positions 5, 6, 7, 9, and 15. For example, at position 6, the MSEs from both functional classes were far less likely to have A or C and more likely to have G than is expected from a random sampling of MSEs (P value of 7.8 × 10−6 and 1.7 × 10−9 for Sum1-dependent and Sum1-bound genes, respectively). This bias against A and C agrees with the observation that changing the G found at this position in the SMK1 MSE to A or C causes a 30- to 50-fold decrease in repression and a 250- to 1,000-fold decrease in binding of Sum1 (Table 1). This analysis yields the consensus GTGWCACAAANK for the MSE sites found upstream of Sum1-repressible genes. The Sum1-bound genes give a more specific but similar consensus of AGYGWCACAAAAD. Thus, we propose a revised MSES consensus of AGYGWCACAAAAD.
The MSE sequences upstream of middle genes that are not derepressed in the sum1 mutant exhibited strong nucleotide biases relative to the control sample at positions 5, 9, and 14. For example, at position 9 there was a bias against G and a corresponding enrichment of A (P = 2.5 × 10−4). Accordingly, the earlier results demonstrated that changing the A at position 9 in the SMK1 MSE to G decreased reporter gene activation nearly twofold and impaired Ndt80 binding more than threefold (Table 1). However, the base preferences at positions 5 and 14 could not be predicted from the earlier results measuring Ndt80 activity in vitro and in vivo. The middle gene MSE base preferences perhaps reflect not only the constraints of Ndt80 binding and action but also additional constraints we do not yet understand. We propose a new middle gene MSEM consensus of YGNCACAAAA, which extends farther 5′ than the Ndt80 MSEN consensus, proposed to be DNCACAAAA.
The middle gene MSEM consensus is similar to but broader than the revised MSES consensus. The MSES consensus is more specific at positions 3, 4, 7, and 15, where the MSEM sequence can be any base, just as Sum1 activity is more strongly impaired by changes in the SMK1 MSE at positions 3, 4, and 7 than Ndt80 activity (Table 1). Also, the preference for G at position 6 is stronger in the Sum1-associated MSEs than in the middle gene MSEs, in accordance with the more severe consequences for Sum1 binding and activity of mutating the base at position 6. Thus, it appears that actual functional MSE sites in the genome may fall along a gradient between the MSES and MSEN consensus sequences that can be defined from Table 1. It is likely that sequences in the genome that show stronger similarity to the MSES function as strong repressor sites while sites that match MSEN are better activators.
Sum1 and Ndt80 compete for binding to the SMK1 MSE.
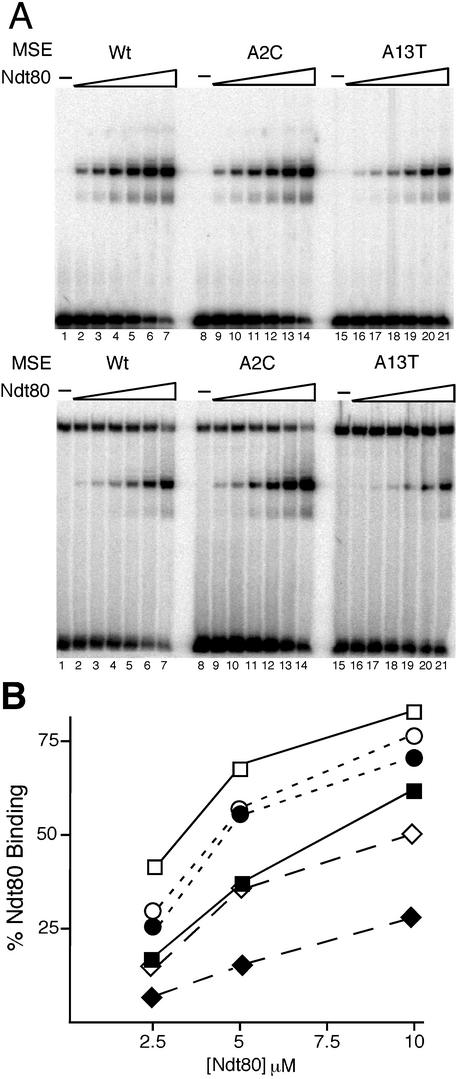
Sum1 is required to repress expression of 20 to 25% of the middle sporulation genes during vegetative growth, while Ndt80 is required for activation of most of the middle sporulation genes during meiosis (6, 36). Since the Ndt80 binding site in the SMK1 MSE overlaps the Sum1 binding site, it was critical to determine whether these proteins can bind simultaneously or instead compete for MSE occupancy. To answer this question we measured DNA-protein interactions by EMSAs in which increasing amounts of Ndt80 protein were added to a DNA-binding reaction with a constant concentration of Sum1 (Fig. 2). Simultaneous binding of both proteins is expected to generate a band above the Sum1-DNA complex that corresponds to a ternary Sum1-Ndt80-DNA complex. On the other hand, if there is competition for this site, then we would expect to see a decrease in the amount of Sum1-bound DNA as the concentration of Ndt80 increases. At low concentrations of Ndt80 (Fig. 2A, bottom panel, lanes 2 and 3), there was little change in the intensity of the Sum1-DNA complex. However, as Ndt80 levels increased, the amount of DNA in complex with Sum1 decreased significantly, concomitant with the appearance of an Ndt80-DNA complex (lanes 4 to 7). These data demonstrate that Ndt80 directly competes with Sum1 for binding to the MSE in vitro. When Ndt80 was added to roughly the same concentration as Sum1, we observed equal amounts of DNA bound to Sum1 and Ndt80. This is in accordance with the minimal differences in the apparent disassociation constants that we have determined from EMSAs using the SMK1 MSE and Sum1 alone or Ndt80 alone (1.3 × 10−7 M and 1.9 × 10−7 M, respectively) (Fig. 1 and 2A and data not shown).
FIG. 2.
Ndt80 competes with Sum1 for binding to the SMK1 MSE in vitro. (A) EMSAs of Ndt80 binding are shown in the absence of Sum1 (top) or in the presence (bottom) of a constant amount of purified Sum1523-1062. Ndt801-409 protein was titrated into the reactions in twofold increments ranging from 0.011 to 0.35 μM final concentrations per reaction (lanes 2 to 7, 9 to 14, and 16 to 21). No Ndt80 was added to lanes 1, 8, and 15 in either panel. The reactions in the bottom panel contain a constant amount of Sum1 at a 0.12 μM final concentration per reaction. Lanes 1 to 7 show competition for binding to the wild-type SMK1 MSE, lanes 8 to 14 show the SMK1 MSE A2C mutant site, and lanes 15 to 21 show the SMK1 MSE A13T mutant site. (B) The percent of probe bound by Ndt80 in each of the competition experiments was quantitated using a phosphorimager. Data are presented for the three highest amounts of Ndt80 in each titration (lanes 5 to 7, 12 to 14, and 19 to 21). Open symbols indicate levels of Ndt80binding in the absence of Sum1 (data from EMSA in panel A, top). Solid symbols indicate binding in the presence of Sum1 (data from EMSA in panel A, bottom). Squares are data for binding to the wild-type SMK1 MSE; circles, A2C mutant site; and diamonds, A13T site.
We have previously shown that the strength of the Sum1-mediated repression and Ndt80-mediated activation varies for different MSEs (36). We therefore predict that the ability of Ndt80 to inhibit Sum1 binding depends on the relative Sum1 and Ndt80 binding affinities for a particular site. To test this model, we assayed competition between Sum1 and Ndt80 for binding to SMK1 MSEs with mutations that alter the binding affinity of each protein (Fig. 2). As expected, Ndt80 is better able to compete with Sum1 in binding to a site with the A2C mutation, which decreases Sum1 DNA-binding affinity. In contrast, a higher concentration of Ndt80 is required to inhibit Sum1 binding to the A13T site, which slightly increases Sum1 binding affinity and weakens Ndt80 binding. These results show how differences in the MSE sequences that alter the relative binding affinities of Sum1 and Ndt80 can have large effects on whether the site is occupied by Ndt80 or Sum1.
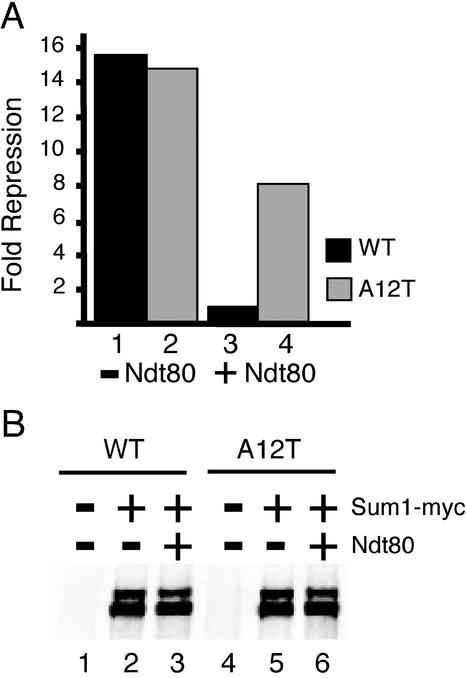
It is possible that the relative binding affinities of Ndt80 and Sum1 for variant MSEs in vitro play a regulatory role in the conversion of a repressed middle sporulation gene to a transcriptionally active state. To test this model, we assayed the level of repression of a reporter gene controlled by a wild-type or a mutant (A12T) SMK1 MSE. The A12T mutant binds Sum1 and represses transcription at or near wild-type levels but exhibits severe defects in both Ndt80-dependent activation and Ndt80 binding (Table 1). These reporters were introduced into a sum1Δ GAL1-10-NDT80 strain that also harbored a pSUM1-myc yeast expression plasmid that fully complements a sum1Δ mutation. In the absence of Ndt80 (growth in glucose), both the wild-type and A12T mutant MSE repressed transcription of the lacZ reporter 15-fold (Fig. 3A, lanes 1 and 2). When Ndt80 was induced with galactose, there was a nearly 18-fold decrease in repression mediated by the wild-type MSE (lane 3). In contrast, there was only a 1.8-fold decrease in repression of the mutant A12T reporter (lane 4). These results are consistent with a model in which Ndt80 is able to displace Sum1 from the wild-type SMK1 MSE, allowing for derepression of the wild-type reporter, but is unable to derepress the reporter controlled by the A12T site due to decreased Ndt80-binding affinity.
FIG. 3.
Sum1 and Ndt80 compete for binding to the SMK1 MSE in vivo. (A) Sum1-dependent repression of lacZ reporter expression in the presence and absence of Ndt80. Reporter plasmids containing wild-type (lanes 1 and 3) or A12T mutant (lanes 2 and 4) SMK1 MSE were cotransformed with a SUM1-myc plasmid into sum1Δ strains. One host strain had an integrated copy of a GAL-NDT80 expression construct (MPY4, lanes 3 and 4), and the other did not (JXY3, lanes 1 and 2). Cells were grown in galactose, and the expression levels of the reporter were measured using β-galactosidase liquid assays. The repression of each reporter construct was calculated by comparison with a reporter lacking an MSE in the same strain background. (B) Western blot of Sum1-myc from lysates containing wild-type (lanes 1 to 3) and A12T mutant (lanes 4 to 6) MSE reporters. Lysates in lanes 2, 3, 5, and 6 were from strains that contained the SUM1-myc-tagged gene expressed from the SUM1 promoter. Lanes 1 and 4 are lysates from strains containing an empty vector, pRS415. Lanes 2 and 5 were lysates from strain JXY3, lacking the GAL-NDT80 construct, and lanes 3 and 6 are lysates from strain MPY4, which contains the GAL-NDT80 construct.
It has been shown that Sum1 levels decrease during prophase (18). This corresponds to the time when Ndt80 is induced. One possible explanation for our observation that Ndt80 reduces Sum1-mediated repression is that Ndt80 may cause a decrease in Sum1 protein levels. We monitored Sum1 protein levels by Western blot analysis in the mitotic cells expressing NDT80 from the GAL1 UAS (Fig. 3B). There was no decrease in Sum1-myc protein levels in the presence of Ndt80 from those in the absence of Ndt80 (lanes 2 versus 3 and lanes 5 versus 6). These results demonstrate that derepression of the wild-type MSE reporter in the presence of Ndt80 occurs when the Sum1-myc levels are high, suggesting that Ndt80 can bind the MSE in vivo and prevent Sum1 binding.
DISCUSSION
Most of the genes that are coordinately expressed during the middle stages of the sporulation pathway contain MSEs in their promoters (5). However, there are significant differences in the regulatory functions of specific MSEs (6, 10, 36). We have previously shown that some but not all MSEs function as Sum1-dependent repressor sites. There are also variations in the level of Ndt80-dependent transcriptional activation from the different sites (36). In this paper we have investigated the DNA sequence specificity of Sum1 and Ndt80 binding and the interactions between these two proteins. Our observations suggest a model for how Ndt80 and Sum1 function to regulate target genes during the middle stages of sporulation.
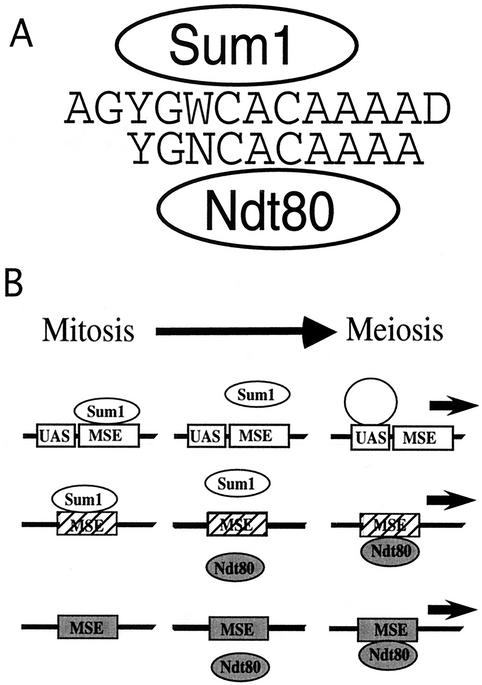
The results from the mutational analysis of the SMK1 MSE and genome-wide expression analysis support a model in which Ndt80 and Sum1 bind to overlapping but distinct sequences (Fig. 4A). Although many of the positions within the SMK1 MSE show sequence-specific requirements for binding by both Ndt80 and Sum1, the base pair preferences at several positions are different for the two proteins. These positions are likely to specify whether an MSE functions as a strong mitotic repressor site or a meiotic activator site or functions in both regulatory capacities (Fig. 4B). Subtle sequence differences among the MSEs from different genes, such as differences from the weakly conserved G at position 6 or at the less conserved position 7, may contribute to the varied levels of Ndt80-dependent activation of different middle sporulation genes. Likewise, genes with MSEs that contain an A at position 7 or a T at positions 12 or 13 may function as better Sum1-dependent repressor sites than the SMK1 MSE. These differences in binding affinity by Sum1 and Ndt80 are likely to specify the regulatory characteristics of a given MSE to achieve a spectrum of levels and timing of expression of genes regulated by these sites.
FIG. 4.
Models for Sum1 and Ndt80 regulation of the MSE. (A) Sum1 and Ndt80 bind to overlapping sequences within the MSE. The sequences are based on the mutational analysis and alignment of sites from the microarray analysis. Sequence letters include N (A, G, C, or T), Y (C or T), W (A or T), and D (A, G, or T). (B) Regulatory activity of different MSEs during the transition from mitotic growth to the middle stages of meiosis. MSEs that function as only a Sum1 repressor site (open boxes), only an Ndt80 activator site (shaded boxes), or as a site of both Sum1 repression and Ndt80 activation (hatched boxes) are shown. Around the time of meiotic prophase, Sum1 is degraded and Ndt80 is expressed. During this transition, Sum1 and Ndt80 may compete for binding to a subset of the MSEs.
The crystal structures of the Ndt80 DNA-binding domain alone and bound to DNA have recently been solved (14, 21). Mutations in the SMK1 MSE that have a large effect on the binding affinity in vitro and transcriptional activation in vivo are all at positions in which the Ndt80 protein makes base-specific contacts with the DNA. However, our results also show that substitutions of positions in which there are only contacts to the phosphate backbone also have a slight effect on transcriptional activation, suggesting that there is some indirect sequence specificity conferred by these contacts as well.
Of the 161 middle sporulation genes, only 31 are very strongly derepressed in the sum1 mutant (5, 29). Of the 130 remaining middle sporulation genes, 64 are expressed in a cell cycle-regulated manner or are required for normal mitotic growth (32). Many of these genes, such as the CLB genes, KAR1, CDC16, APC9, and ORC1, are expressed at the G2/M stage of the mitotic cell cycle and have essential cellular functions. The existence of MSEs that bind Ndt80 but not Sum1 provides a mechanism to allow this set of genes to be expressed during mitosis and also to be strongly induced during meiosis. Genes containing strong Sum1-binding MSEs are unlikely to have critical roles in mitotic growth under standard conditions.
It is likely that the list of genes that are derepressed in the sum1 strain (Table 2) is incomplete. There are several reasons for this. First, we only included in this list genes that were derepressed at least twofold in all three microarray experiments. However, if less-stringent criteria are applied (an average of twofold depression between all three experiments), then 14 of the additional 27 genes that would be included in the list are classified as middle sporulation genes. It is highly likely that Sum1 may have a role in regulating these genes.
A second explanation for why some middle sporulation genes are not derepressed in the sum1 mutant is that they may also be regulated by other mitotic repressors. For example, it has been previously shown that NDT80 expression is negatively regulated by both Sum1 and Ume6 (24, 36). The NDT80 promoter not only has two MSEs (one that is repressed by Sum1 and one that is not) but also contains two URS1 sites that recruit the Ume6-Sin3-Rpd3 repressor complex. We therefore did not expect, nor did we find, that NDT80 was expressed in the sum1 microarray expression experiments. Several of the middle sporulation genes that are not derepressed in the sum1 mutant strain contain matches to a URS1 consensus. It is likely that these genes are repressed by Ume6 during vegetative growth and therefore are not derepressed in the sum1 mutant. Combinatorial control of these promoters by Ume6- and Sum1-dependent repression may contribute to distinct regulatory characteristics of subsets of middle promoters. In accordance with this type of regulatory scheme, 73 genes that were identified to be bound by Sum1 (17), including NDT80 and 15 other middle genes, had the same transcript levels in the presence and absence of Sum1 under standard growth conditions.
A third explanation for why some middle sporulation genes are not derepressed in the sum1 mutant is that the transcription factors that activate these genes may not be present under the mitotic conditions tested, perhaps because the activators are meiosis specific. For example, Ndt80 may be the sole activator for some middle sporulation genes. Since NDT80 is not expressed in the sum1 mutant during vegetative growth, these target genes would not be expressed even when Sum1 is no longer repressing the promoter. The middle sporulation genes that are not derepressed in the sum1 mutant are more likely to be completely dependent on Ndt80 for their meiotic expression. In support of this model, deletion of NDT80 abolishes meiotic induction of 42.5% of the middle sporulation genes that are unaffected in vegetative sum1 cells (5). In contrast, the corresponding value is 13.3% for the middle sporulation genes that are derepressed in the sum1 mutant, suggesting that factors other than Ndt80 can activate these genes. It is likely that additional activators, like Ndt80, are functional only during meiosis (2, 9).
What is the physiological significance of Sum1 repression? Deletion of SUM1 leads to only a slight reduction in growth rate in some strain backgrounds (4, 36). This observation suggests that the Sum1-repressible gene set is not deleterious if improperly expressed during mitotic growth. In addition, diploid sum1 mutants complete meiosis and form spores, suggesting that under optimal conditions Sum1 repression is not essential for completion of this developmental program. Sum1 may, however, play a role in fine-tuning the transcriptional program during sporulation. Although deletion of SUM1 advances or increases expression of some middle sporulation genes during meiosis (24), the regulatory network appears to be sufficiently robust to support normal progression. Moreover, Sum1 is required for the meiotic pachytene checkpoint (18). Sum1-mediated repression may be required to delay chromosome segregation and spore formation when conditions are unfavorable.
Twenty-two of the fifty-three genes derepressed in the sum1 mutant are not classified as middle sporulation specific by DNA microarray analysis (5). This result suggests that Sum1 may also be involved in regulation of nonmeiotic processes. Interestingly, a number of the nonmeiotic genes that are derepressed in the sum1 mutant have been recently shown to be involved in NAD biosynthesis (26). These genes, BNA1, BNA2, BNA4, and BNA5, are also depressed in rfm1 and hst1 mutants, indicating that the Sum1-Rfm1-Hst1 complex is required for their repression (19). Hst1 is a member of the Sir2 class of NAD+-dependent histone deacetylases (15, 34). It has been shown that lower levels of NAD+ in a npt1 mutant cause derepression of Hst1-regulated promoters (18, 23). It is therefore possible that repression of the BNA genes by the Sum1-Rfm1-Hst1 repressor complex serves as a feedback loop to control the levels of NAD production in the cell.
Our data suggest that each MSE in the yeast genome consists of a central core sequence with flanking regions contributing to the specificity of the site (Fig. 4A). MSEs may fall in three broad functional categories, as shown in the model in Fig. 4B: vegetative repressor elements bound by Sum1, meiotic activator elements bound by Ndt80, and switch elements that have high-level affinity for both proteins. These results suggest that early in meiosis, as the concentration of Ndt80 increases relative to that of Sum1, the displacement of Sum1 by Ndt80 will happen earlier at some MSEs than at others, depending on small differences in sequence.
Another layer of complexity that must be superimposed on the Sum1/Ndt80 competition model involves changes in the amounts and activities of these proteins. Ndt80 is encoded by a meiosis-specific gene and can induce its own expression (6). Transcription of NDT80 and phosphorylation of Ndt80 are regulated by an assortment of protein kinases, including components of the pachytene checkpoint (1, 6, 10, 25, 31, 35, 37). In contrast, Sum1 is constitutively transcribed in mitotic and meiotic cells and is degraded specifically in meiotic cells (18). It has been suggested but not proven that its degradation is regulated by the pachytene checkpoint. Recent evidence suggests that the meiosis-specific kinase Ime2 is required both to stimulate Ndt80 activity and to antagonize Sum1 activity (1, 24, 31). Thus, the complex regulation of Ndt80 and Sum1 protein levels and activities, combined with their differential affinities for variant MSE, may account for the differences in timing and levels of transcription among the middle sporulation genes. The model proposed here may be a general mechanism for modulating expression of different members of a coregulated gene set in response to distinct developmental signals.
Acknowledgments
This work is supported by a Busch Predoctoral Fellowship to M.P.; the Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation Fellowship (DRG-1443) to K.R.B.; and grants from the National Institutes of Health (E.W. and A.K.V.). The microarray experiments were performed using the UCSF Core Facility for Genomics and Proteomics with funds from the Hughes Medical Institute and the Sandler Program in the Basic Sciences.
We are deeply indebted to Joe DeRisi for help with the microarray experiments and analysis and for sharing his Sporulation DataBase and Promoter software and to Edward Moler for advice on statistical analysis.
REFERENCES
- 1.Benjamin, K. R., C. Zhang, K. M. Shokat, and I. Herskowitz. Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes Dev. (in press). [DOI] [PMC free article] [PubMed]
- 2.Bogengruber, E., T. Eichberger, P. Briza, I. W. Dawes, M. Breitenbach, and R. Schricker. 1998. Sporulation-specific expression of the yeast DIT1/DIT2 promoter is controlled by a newly identified repressor element and the short form of Rim101p. Eur. J. Biochem. 258:430-436. [DOI] [PubMed] [Google Scholar]
- 3.Carroll, A. S., A. C. Bishop, J. L. DeRisi, K. M. Shokat, and E. K. O'Shea. 2001. Chemical inhibition of the Pho85 cyclin-dependent kinase reveals a role in the environmental stress response. Proc. Natl. Acad. Sci. USA 98:12578-12583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chi, M. H., and D. Shore. 1996. SUM1-1, a dominant suppressor of SIR mutations in Saccharomyces cerevisiae, increases transcriptional silencing at telomeres and HM mating-type loci and decreases chromosome stability. Mol. Cell. Biol. 16:4281-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu, S., J. DeRisi, M. Eisen, J. Mulholland, D. Botstein, P. O. Brown, and I. Herskowitz. 1998. The transcriptional program of sporulation in budding yeast. Science 282:699-705. [DOI] [PubMed] [Google Scholar]
- 6.Chu, S., and I. Herskowitz. 1998. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol. Cell 1:685-696. [DOI] [PubMed] [Google Scholar]
- 7.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 8.Gailus-Durner, V., C. Chintamaneni, R. Wilson, S. J. Brill, and A. K. Vershon. 1997. Analysis of a meiosis-specific URS1 site: sequence requirements and involvement of replication protein A. Mol. Cell. Biol. 17:3536-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hepworth, S. R., L. K. Ebisuzaki, and J. Segall. 1995. A 15-base-pair element activates the SPS4 gene midway through sporulation in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:3934-3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hepworth, S. R., H. Friesen, and J. Segall. 1998. NDT80 and the meiotic recombination checkpoint regulate expression of middle sporulation-specific genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:5750-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Honigberg, S. M., and R. E. Esposito. 1994. Reversal of cell determination in yeast meiosis: postcommitment arrest allows return to mitotic growth. Proc. Natl. Acad. Sci. USA 91:6559-6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krisak, L., R. Strich, R. S. Winters, J. P. Hall, M. J. Mallory, D. Kreitzer, R. S. Tuan, and E. Winter. 1994. SMK1, a developmentally regulated MAP kinase, is required for spore wall assembly in Saccharomyces cerevisiae. Genes Dev. 8:2151-2161. [DOI] [PubMed] [Google Scholar]
- 13.Kupiec, M., B. Byers, R. Esposito, and A. Mitchell. 1997. Meiosis and sporulation in Saccharomyces cerevisiae, p. 889-1036. In J. Pringle, J. Broach, and E. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces, vol. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 14.Lamoureux, J. S., D. Stuart, R. Tsang, C. Wu, and J. N. Glover. 2002. Structure of the sporulation-specific transcription factor Ndt80 bound to DNA. EMBO J. 21:5721-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Landry, J., A. Sutton, S. T. Tafrov, R. C. Heller, J. Stebbins, L. Pillus, and R. Sternglanz. 2000. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. USA 97:5807-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lashkari, D. A., J. L. DeRisi, J. H. McCusker, A. F. Namath, C. Gentile, S. Y. Hwang, P. O. Brown, and R. W. Davis. 1997. Yeast microarrays for genome wide parallel genetic and gene expression analysis. Proc. Natl. Acad. Sci. USA 94:13057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, T. I., N. J. Rinaldi, F. Robert, D. T. Odom, Z. Bar-Joseph, G. K. Gerber, N. M. Hannett, C. T. Harbison, C. M. Thompson, I. Simon, J. Zeitlinger, E. G. Jennings, H. L. Murray, D. B. Gordon, B. Ren, J. J. Wyrick, J. B. Tagne, T. L. Volkert, E. Fraenkel, D. K. Gifford, and R. A. Young. 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298:799-804. [DOI] [PubMed] [Google Scholar]
- 18.Lindgren, A., D. Bungard, M. Pierce, J. Xie, A. Vershon, and E. Winter. 2000. The pachytene checkpoint in Saccharomyces cerevisiae requires the Sum1 transcriptional repressor. EMBO J. 19:6489-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCord, R., M. Pierce, J. Xie, S. Wonkatal, C. Mickel, and A. K. Vershon. 2003. Rfm1, a novel tethering factor required to recruit the Hst1 histone deacetylase for repression of middle sporulation genes. Mol. Cell. Biol. 23:2009-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchell, A. P. 1994. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol. Rev. 58:56-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montano, S. P., M. L. Cote, I. Fingerman, M. Pierce, A. K. Vershon, and M. M. Georgiadis. 2002. Crystal structure of the DNA-binding domain from Ndt80, a transcriptional activator required for meiosis in yeast. Proc. Natl. Acad. Sci. USA 99:14041-14046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montano, S. P., M. Pierce, M. L. Cote, A. K. Vershon, and M. M. Georgiadis. 2002. Crystallographic studies of a novel DNA-binding domain from the yeast transcriptional activator Ndt80. Acta Crystallogr. D Biol. Crystallogr. 58:2127-2130. [DOI] [PubMed] [Google Scholar]
- 23.Ozsarac, N., M. J. Straffon, H. E. Dalton, and I. W. Dawes. 1997. Regulation of gene expression during meiosis in Saccharomyces cerevisiae: SPR3 is controlled by both ABFI and a new sporulation control element. Mol. Cell. Biol. 17:1152-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pak, J., and J. Segall. 2002. Regulation of the premiddle and middle phases of expression of the NDT80 gene during sporulation of Saccharomyces cerevisiae. Mol. Cell. Biol. 22:6417-6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pak, J., and J. Segall. 2002. Role of Ndt80, Sum1, and Swe1 as targets of the meiotic recombination checkpoint that control exit from pachytene and spore formation in Saccharomyces cerevisiae. Mol. Cell. Biol. 22:6430-6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Panozzo, C., M. Nawara, C. Suski, R. Kucharczyka, M. Skoneczny, A. M. Becam, J. Rytka, and C. J. Herbert. 2002. Aerobic and anaerobic NAD+ metabolism in Saccharomyces cerevisiae. FEBS Lett. 517:97-102. [DOI] [PubMed] [Google Scholar]
- 27.Pierce, M., M. Wagner, J. Xie, V. Gailus-Durner, J. Six, A. K. Vershon, and E. Winter. 1998. Transcriptional regulation of the SMK1 mitogen-activated protein kinase gene during meiotic development in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:5970-5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pijnappel, W. W., D. Schaft, A. Roguev, A. Shevchenko, H. Tekotte, M. Wilm, G. Rigaut, B. Seraphin, R. Aasland, and A. F. Stewart. 2001. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 15:2991-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Primig, M., R. M. Williams, E. A. Winzeler, G. G. Tevzadze, A. R. Conway, S. Y. Hwang, R. W. Davis, and R. E. Esposito. 2000. The core meiotic transcriptome in budding yeasts. Nat. Genet. 26:415-423. [DOI] [PubMed] [Google Scholar]
- 30.Rusche, L. N., and J. Rine. 2001. Conversion of a gene-specific repressor to a regional silencer. Genes Dev. 15:955-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sopko, R., S. Raithatha, and D. Stuart. 2002. Phosphorylation and maximal activity of Saccharomyces cerevisiae meiosis-specific transcription factor Ndt80 is dependent on Ime2. Mol. Cell. Biol. 22:7024-7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spellman, P. T., G. Sherlock, M. Q. Zhang, V. R. Iyer, K. Anders, M. B. Eisen, P. O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9:3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sutton, A., R. C. Heller, J. Landry, J. S. Choy, A. Sirko, and R. Sternglanz. 2001. A novel form of transcriptional silencing by Sum1-1 requires Hst1 and the origin recognition complex. Mol. Cell. Biol. 21:3514-3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanny, J. C., and D. Moazed. 2001. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: evidence for acetyl transfer from substrate to an NAD breakdown product. Proc. Natl. Acad. Sci. USA 98:415-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tung, K. S., E. J. Hong, and G. S. Roeder. 2000. The pachytene checkpoint prevents accumulation and phosphorylation of the meiosis-specific transcription factor ndt80. Proc. Natl. Acad. Sci. USA 97:12187-12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie, J., M. Pierce, V. Gailus-Durner, M. Wagner, E. Winter, and A. K. Vershon. 1999. Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J. 18:6448-6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu, L., M. Ajimura, R. Padmore, C. Klein, and N. Kleckner. 1995. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:6572-6581. [DOI] [PMC free article] [PubMed] [Google Scholar]