Abstract
For nearly a century, cancer has been blamed on somatic mutation. But it is still unclear whether this mutation is aneuploidy, an abnormal balance of chromosomes, or gene mutation. Despite enormous efforts, the currently popular gene mutation hypothesis has failed to identify cancer-specific mutations with transforming function and cannot explain why cancer occurs only many months to decades after mutation by carcinogens and why solid cancers are aneuploid, although conventional mutation does not depend on karyotype alteration. A recent high-profile publication now claims to have solved these discrepancies with a set of three synthetic mutant genes that “suffices to convert normal human cells into tumorigenic cells.” However, we show here that even this study failed to explain why it took more than “60 population doublings” from the introduction of the first of these genes, a derivative of the tumor antigen of simian virus 40 tumor virus, to generate tumor cells, why the tumor cells were clonal although gene transfer was polyclonal, and above all, why the tumor cells were aneuploid. If aneuploidy is assumed to be the somatic mutation that causes cancer, all these results can be explained. The aneuploidy hypothesis predicts the long latent periods and the clonality on the basis of the following two-stage mechanism: stage one, a carcinogen (or mutant gene) generates aneuploidy; stage two, aneuploidy destabilizes the karyotype and thus initiates an autocatalytic karyotype evolution generating preneoplastic and eventually neoplastic karyotypes. Because the odds are very low that an abnormal karyotype will surpass the viability of a normal diploid cell, the evolution of a neoplastic cell species is slow and thus clonal, which is comparable to conventional evolution of new species.
For a century, cancer has been blamed on some kind of “somatic mutation” (1, 2). But it is still unclear whether this mutation is aneuploidy, an abnormal balance or number of chromosomes, or gene mutation, although the two hypotheses make very different testable predictions. According to their very different mutagenic ranges, nature uses gene mutation for minor adjustments within a species (3, 4) but reserves alteration of chromosome numbers for major discontinuous alterations such as the generation of new species (5, 6). Indeed, the complexity of cancer-specific phenotypes, such as abnormal cellular and nuclear morphology, metabolism, growth, DNA indices ranging from 0.5 to >2, invasiveness, metastasis, and neoantigens (7–9), is more compatible with aneuploidy altering the dosage of thousands of regulatory and structural genes than with gene mutations. Moreover, the exceedingly slow kinetics from carcinogen treatment to carcinogenesis (8, 9) are more compatible with the evolution of a new species than with gene mutation, which is instantaneous.
Nevertheless, currently most researchers assume that cancer is caused by certain gene mutations and that these mutations are caused by carcinogens (8, 10–12). This hypothesis makes six testable predictions: (i) carcinogens function as mutagens; (ii) mutations are cancer specific; (iii) cancer-specific genes are able to transform normal human or animal cells into cancer cells; (iv) transformation is coincident with mutation, i.e., carcinogen treatment; (v) cancer phenotypes are as stable as conventional mutations; (vi) cancers are diploid, because gene mutations do not depend on karyotype alterations for expression.
However, these predictions have been difficult to meet. (i) There is a growing list of nongenotoxic carcinogens, including asbestos, Ni2+, hormones, butter yellow, arsenic, acrylamide, urethan, etc. (13–17). (ii) No cancer-specific gene mutations have been found yet (18–23). According to a recent commentary (“How many mutations does it take to make a tumor?”), “There are no oncogenes or tumor suppressor genes that are activated or deleted from all cancers. Even tumors of a single organ rarely have uniform genetic alterations, although tumor types from one specific organ have a tendency to share mutations.” (24). Moreover, mutations that are relatively specific, e.g., ras and p53, are not shared by all cells of the same tumor (25–31). (iii) No genes have been isolated from cancers that transform normal human or animal cells into cancer cells (17, 22, 32–37), and the spontaneous loss of mutant ras, a presumed oncogene, does not revert the phenotype of a cancer cell back to normal (38, 39). (iv) The latent periods between carcinogen treatment and cancer are exceedingly long, ranging from many months to decades (8, 13), although carcinogen-mediated mutation is instantaneous (40, 41). (v) The phenotypes of cancer cells are notoriously unstable (30, 42–44), and (vi) virtually all solid cancers are aneuploid (45–49).
Recognizing these difficulties with the mutation hypothesis, Weinberg and coworkers have tried to save the troubled hypothesis with a high-profile publication, which claims that mutation with a set of three “defined” genes “suffices to convert normal human cells into tumorigenic cells” (50). Unable to isolate a sufficient set of genes from natural cancers “after more than 15 years of trying” (37), the authors “defined” these genes on the basis of established tumor virus models. Assuming that human cancer is caused by the cooperation of two cellular “oncogenes” and one “immortalization” gene, the authors have synthesized two hypothetical cancer genes from the oncogenic viruses, simian virus (SV)40 and Harvey sarcoma virus, and a hypothetical immortalization gene from the human telomerase. About “60 population doublings” after the introduction of the first of these genes, the tumor (T) antigen of SV40, tumorigenicity was observed. In view of this and assuming that the tumor cells were “polyclonal” for the added genes, the authors concluded that no “additional genetic alterations were required” for tumorigenesis and “that identical rules will be found to apply to autochthonously arising human tumor cells” (50).
As an alternative solution of the troubled mutation hypothesis, we and others have recently proposed that aneuploidy is the somatic mutation that causes cancer (43, 51–54). Aneuploidy was originally proposed over a century ago as a cause of cancer (55, 56) but was abandoned as a cause when the karyotypes of clonal cancers were found to be nonclonal (7, 42, 57–61). However, in light of our new two-stage mechanism, the aneuploidy hypothesis can now resolve all of the contradictions generated by the gene-mutation hypothesis listed above (51, 53). In stage one, carcinogens (or mutant genes) cause aneuploidy, and in stage two, the aneuploid karyotype evolves autocatalytically, because aneuploidy destabilizes the karyotype, generating ever new and eventually tumorigenic karyotypes (see below).
In view of this, we have analyzed the human tumor cells generated by Weinberg and coworkers for direct and indirect evidence of “additional genetic alterations,” above all aneuploidy. Indeed, the cells proved to be clonal and highly aneuploid, as predicted by the aneuploidy hypothesis. According to this hypothesis, we suggest that the T antigen and perhaps the other added genes generated aneuploidy and that aneuploidy initiated karyotype evolution, which “after 60 population doublings” would eventually generate clones of tumorigenic cells.
Materials and Methods
Cells.
A tumorigenic human kidney-derived cell line, termed HA1 ER, alias HEK (50), and a tumorigenic human fibroblast-derived cell line, termed BJ ELR, were kindly provided by Robert Weinberg (Massachusetts Institute of Technology, Cambridge, MA). The origin of these cells was recently described (50). The cells were propagated in DMEM supplemented with 10% FCS and antibiotics, following published procedures (51).
Chromosome Analysis.
The preparation of metaphase chromosomes after treatment of the two human tumor cell lines for 2–3 h with 0.6 μg/ml colcemid (Karymax, GIBCO/BRL) has been described by us recently (51, 62).
Results and Discussion
Analysis of the Generation and Genetic Structure of Human Tumor Cells Generated with “Three Defined Genetic Elements.”
To determine whether mutation by three defined genes does indeed “suffice to convert normal human cells into tumorigenic cells,” we first investigated the generation and then the genetic structure of these cells. According to Weinberg and coworkers, these tumor cells were generated from normal human embryonic kidney cells and fibroblasts by sequential infections with the three genes inserted into murine retroviral vectors. The first of the three artificial genes to be introduced was the coding region of the large T-antigen gene of the simian DNA tumor virus, SV40, inserted together with a selectable drug-resistance gene into a murine retrovirus. After selection for drug resistance, the surviving cells were infected with an artificial, presumably immortalizing, human telomerase gene, again together with a drug-resistance gene in the above retrovirus vector. After another round of drug selection, the surviving cells were infected with the second hypothetical human oncogene, i.e., the coding region of a mutant human ras gene (which is isogenic with that of the murine Harvey sarcoma virus) together with a drug-resistance gene in an analogous retroviral vector.
After a lengthy period of more than 60 cell generations since the introduction of the first of these genes, the T antigen of SV40, two tumorigenic cell lines, termed BJ ELR and HEK, alias HA1 ER (see Materials and Methods), were obtained. Assuming that these cells were “polyclonal,” because large percentages of cells were rendered drug resistant with the synthetic genes, the authors suggested that the resulting “tumorigenic growth . . . is not a consequence of additional, rare stochastic events . . .”, and that “additional genetic alterations” were not required (50).
However, there are three reasons to suggest that “rare stochastic events” and “additional genetic alterations” were necessary for tumorigenesis in this system. (i) The first is that, contrary to the authors' assumption, the tumorigenic cells were clonal. This is evident from the discrete retroviral integration sites shown in Fig. 4b of their paper (50). The figure shows about six bands, which is exactly what one would expect from three consecutive clonal viral infections, assuming one restriction site inside the retroviral DNA vectors (the paper did not describe restriction enzyme maps of the viral constructs used). Instead, even one, and certainly three, consecutive polyclonal infections would have generated a smear of viral-cell DNA hybrid molecules. Thus the tumor cells were clonal, signaling a “rare stochastic event” or an “additional genetic alteration.” This conclusion was confirmed by preliminary evidence for marker chromosomes in each of the two tumor cell lines generated by the study. Some marker chromosomes of the BJ line included elements from three or more different chromosomes.
(ii) The second argument for additional genetic alterations is derived from the long latent period required for tumorigenicity by these genes, i.e., “approximately 60 population doublings” from the introduction of the first of these genes, the T antigen of SV40. Because 60 cell doublings generate 1018 cells out of one, the equivalent of 10,000 human bodies, this time period does not preclude additional genetic events. By contrast, the expression of coinfected drug-resistance genes was instantaneous.
Even if one allows three cell generations for each round of retrovirus infection and three generations for each subsequent drug selection, the cells could have been rendered tumorigenic after only 18 generations. If vectors with both of the presumed transforming genes, T antigen and ras, had been used, transformed cells could have been selected in one step on the basis of the reported “morphological transformation.” Those cells could have then been treated with the presumably immortalizing telomerase gene in a second round of infection. Alternatively, vectors could have been possibly constructed carrying all three of these genes, which would have permitted the generation of tumorigenic cells in one step, obviating the need for introducing drug resistance altogether. Surprisingly, Weinberg and coworkers have just acknowledged in another Nature publication that the HA1, alias HEK, line “immortalized spontaneously” after transfection with T antigen (63). Thus, this line could have been rendered tumorigenic without the telomerase and thus presumably faster than described recently (50).
Moreover, previous studies by others also call into question Weinberg and coworkers' hypothesis that the T antigen of SV40 depends on two other gene products to render human cells tumorigenic. Three of these studies report that SV40 alone is sufficient (64–66), and a fourth reports that a combination of SV40 and the ras gene of Kirsten sarcoma virus is sufficient to initiate tumorigenicity, although again only after lag periods of multiple cell generations (67).
It would appear that the lengthy procedure of sequential infection with three hypothetically cooperating cancer genes is not a sufficient explanation for the long lag periods between the introduction of hypothetical cancer genes and tumorigenesis, particularly because the T antigen seems to be sufficient to initiate tumorigenesis on its own. It is, however, possible that the additional genes used by Weinberg and coworkers to generate tumor cells may have accelerated tumorigenicity by the SV40 T antigen. Whatever the correct explanation (see below), the process allows plenty of time for “additional genetic alterations,” for example, aneuploidy.
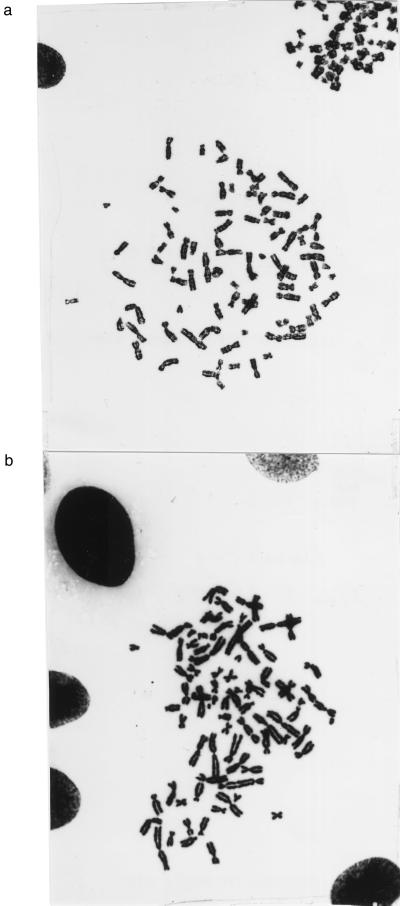
(iii) In an effort to obtain direct evidence for “additional genetic alterations,” we have analyzed the chromosomal structures of the two cell lines (Materials and Methods). It was found that the BJ ELR line was about 70% aneuploid. Among 32 cells, two were hypodiploid, each with 45 chromosomes, 10 were diploid or pseudodiploid with 46 chromosomes, and 20 were hyperdiploid with chromosome numbers between 74 and 91. The HA1 ER line was also about 70% aneuploid. Among 28 cells, two were hypodiploid, with 41 and 44 chromosomes, 8 were diploid or pseudodiploid, and 18 were hyperdiploid with chromosome numbers ranging from 48 to 89. A representative metaphase showing a HA1 cell with 78 chromosomes and a BJ cell with 82 chromosomes is shown in Fig. 1.
Figure 1.
The chromosomes of a representative aneuploid cell of the tumorigenic human HA1 ER (a) and BJ ELR (b) cell lines. Metaphase chromosomes of these cells were prepared as described previously (51) and photographed ×630 after Giemsa staining. The HA1 ER metaphase shown contains 78 and the BJ ELR metaphase contains 82 chromosomes.
Thus, (i) the clonality of the tumor cells, (ii) the long unexplained latent periods between the introduction of the transforming genes and tumorigenesis, and (iii) the aneuploidy of the tumor cells are all incompatible with the gene mutation hypothesis, which predicts the instant appearance of diploid polyclonal tumor cells after the addition of a sufficient set of transforming genes (whatever the correct number of a sufficient set). In the following, we first introduce the practically forgotten hypothesis that aneuploidy is a somatic mutation that causes cancer, then we demonstrate that this hypothesis provides a coherent explanation for how the methods used by Weinberg and coworkers would transform normal human cells to aneuploid tumor cells.
Tumorigenesis by Aneuploidy.
(i) The aneuploidy–cancer hypothesis.
Aneuploidy was first proposed to be the cause of cancer over a century ago. The hypothesis was based on Hansemann's observations of asymmetric mitoses in epithelial cancers and on Boveri's evidence that aneuploidy generates abnormal phenotypes in developing sea urchin embryos (55, 56, 68).
However, subsequent research has called the aneuploidy hypothesis into question because: (i) The quest for a cancer-specific aneuploidy has failed, revealing instead a “confusing plethora” of karyotypes even in cancers that are clonal for parental or somatic gene mutations (60). (ii) Aneuploidy has been found in noncancerous cells, e.g., Down's syndrome, preneoplastic cells, and rare cells, particularly of aging people or animals (59, 62, 69, 70), although this is relatively minor compared with that found in cancer. (iii) Rare studies from before chromosome banding and particularly from before comparative genomic hybridization claimed diploid cancers (a claim that may be correct only for retroviral cancers; see below). In view of this, most cancer researchers have abandoned the aneuploidy hypothesis in favor of gene mutation. Their reasoning is that aneuploidy must be a consequence of transformation, although this view is incompatible with aneuploidy in noncancerous cells (7, 42, 57–61).
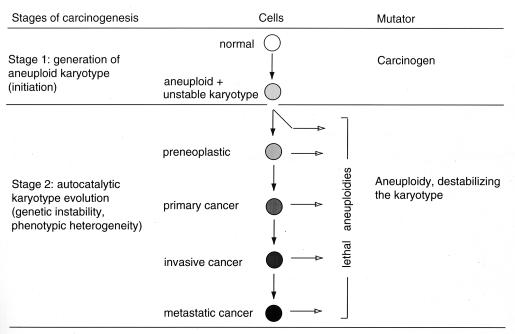
At variance with these views, we have recently proposed a two-stage mechanism that reconciles the nonclonal karyotypes with the clonal gene mutations of cancers and that also accounts for the long latent periods from carcinogens to cancers and for the existence of aneuploidy in noncancerous cells (43, 51, 53, 62, 71) (Fig. 2). The proposal runs as follows: (i) Stage one. Carcinogens induce aneuploidy by chemically or physically altering one or more of the many proteins of the spindle apparatus or the chromosomes, as we and others have already demonstrated (15, 51, 62, 72, 73). It is also possible that genotoxic carcinogens induce aneuploidy by mutating specific mitosis genes (44). However, this proposal predicts mutation-specific karyotype instabilities. Instead, the instabilities of the karyotypes of cancer cells, even those from a given clonal cancer, are heterogeneous but directly proportional to their degree of aneuploidy (43). (ii) Stage two. Aneuploidy destabilizes the karyotype and thus initiates an autocatalytic karyotype evolution. This process would generate lethal, preneoplastic and eventually neoplastic karyotypes (43, 51, 53, 54). The source of the karyotype instability is the imbalance that aneuploidy imparts on the spindle apparatus, e.g., abnormal ratios of spindle proteins and chromosomal proteins (74, 75) and abnormal structures and numbers of centrosomes (52, 54, 76, 77). The imbalanced spindle will cause chromosome nondisjunction and thus autocatalytically regroup the karyotype, a process that has been termed “chromosome error propagation” (78). The resulting “genetic instability” explains the heterogeneous karyotypes that are a hallmark of cancer (9, 43, 60). Thus, cancers are clonal for aneuploidy (and certain gene mutations) but not for a particular karyotype.
Figure 2.
A two-stage model for how carcinogens may cause cancer via aneuploidy. Stage one: a carcinogen “initiates” carcinogenesis by generating an aneuploid cell. Stage two: aneuploidy destabilizes symmetric chromosome segregation, because it unbalances spindle and chromosomal proteins and centrosome numbers by unbalancing their chromosomal templates (see text). As a result, aneuploidy initiates autocatalytic karyotype variation and evolution, which generates new lethal, preneoplastic and eventually neoplastic karyotypes. The autocatalytic karyotype evolution would explain the previously unresolved carcinogen-independent transformation of an “initiated” preneoplastic into a neoplastic cell (8, 9, 62). The notorious long latent periods from initiation to carcinogenesis would be a consequence of the low probability of generating by chance a karyotype that can outperform normal cells. Autocatalytic karyotype variation would also explain the notorious genetic instability and phenotypic heterogeneity of cancer cells (9, 43).
According to our proposal, the long latent periods from the initial aneuploidization to cancer reflect the low probability of evolving by chance a neoplastic karyotype, which surpasses the viability of a normal diploid cell. Boveri estimated the odds for a neoplastic karyotype to be as low as winning the “lottery” (56). The low probability of this evolution also explains why most cancers are clonal for either germinal or somatic mutations, including “marker” (rearranged) chromosomes (8, 9, 47, 60). This view also explains the existence of nonneoplastic aneuploidy and predicts that there is a threshold of aneuploidy for cancer (Fig. 2) (53, 62).
The low probability of a neoplastic among randomly generated aneuploid karyotypes also explains why fusion of cancer cells with normal cells often, but not always, generates nononcogenic cell hybrids (9). Such fusions would destroy the rare neoplastic chromosome combination. But typically such hybrids regain neoplastic properties by differential loss of chromosomes (9), driven by the karyotypic instability of aneuploid cells.
(ii) Correlative evidence.
Since Hansemann's observations of asymmetric mitoses in cancers in 1890 (55), aneuploidy has been observed in virtually all of the over 5,000 solid human cancers that have been analyzed (45–48). The correlations between solid cancers and aneuploidy are so tight that neither of the two textbooks of cancer cytogenetics, i.e., Heim and Mitelman's Cancer Cytogenetics (60) and Sandberg's The Chromosomes in Human Cancer and Leukemia (47), lists confirmed examples of solid cancers that are diploid or euploid. According to a recent survey, all “of over 2,400 human solid tumors” analyzed by comparative genomic hybridization were aneuploid with regard to either segments or complete complements of chromosomes (49). In view of this, Oshimura and Barrett commented that “a better correlation with cell transformation is observed with induction of aneuploidy than of point mutations” (15). And the cytogeneticist Atkin asked in 1990, “Are human cancers ever diploid?” (79). Thus, aneuploidy meets the equivalent of the first of Koch's postulates, i.e., a perfect correlation, as a cause of cancer.
However, the claim that aneuploidy is cancer specific must be balanced by the nonneoplastic and preneoplastic aneuploidies that are in fact part of our hypothesis (see above). According to our hypothesis, neoplastic aneuploidy differs from nonneoplastic aneuploidy quantitatively and qualitatively, i.e., we postulate an as yet poorly defined threshold for neoplastic aneuploidy (53, 62) (Fig. 2). Aneuploidy below this threshold would involve few and predominantly small chromosomes, e.g., Down's syndrome with a trisomy or monosomy of chromosome no. 21, whereas aneuploidies above this threshold, i.e., cancer, would involve many or all chromosomes (53).
(iii) Functional evidence.
Because cellular behavior is determined by the balance of thousands of regulatory and structural genes, which is maintained by a species-specific set of chromosomes, aneuploidy inevitably generates abnormal phenotypes, independent of gene mutation, because it unbalances large fractions of these genes. The effects of aneuploidy on the phenotypes of cells would be analogous to those of randomizing assembly lines of an automobile factory on cars, i.e., cars with abnormal ratios of normal (rather than mutated) wheels, bodies, and engines.
Proof of this principle was first experimentally obtained by Boveri, who found that aneuploidy generates “pathological,” including “lethal,” phenotypes in developing sea urchins (68). Some of these phenotypes had actually been so abnormal that they were termed “tumors” by Boveri (68). More recently, aneuploidy has been confirmed experimentally as a dominant mutator of eukaryotes that is independent of gene mutation in plants (80), yeast (81), and Drosophila (82). Likewise, spontaneous congenital aneuploidies have been identified as the single cause of serious abnormalities, e.g., Down's syndrome, which is caused by monosomy or trisomy of chromosome no. 21 (83–86).
Indeed, one special form of aneuploidy appears to be nature's most definitive and far-ranging mutation, i.e., speciation. Because a species is defined by a specific number of chromosomes (5, 6), speciation falls within the definition of aneuploidy. This applies to aneuploid cancer as well as to the minor noncancerous aneuploidies described above. It follows that cancer is, by definition, a species of its own. It differs from authentic species in that it is parasitic, i.e., it is unable to function independently. Thus there is ample proof of the principle that aneuploidy is a gene mutation-independent and far-ranging mutator of eukaryotic cells and therefore a plausible cause of cancer.
By contrast, the range of gene mutation is highly restricted in vivo. Because virtually all enzymes and functions of cells are integrated into kinetically linked biochemical assembly lines (3, 87) or signal pathways (12, 50), and because all enzymes work far below saturating concentration, i.e., at only a small fraction of their capacity (3, 87), rare positive or activating mutations of enzymes or of hypothetical oncogenes are very effectively buffered in vivo via supplies and demands of unmutated upstream and downstream enzymes (3, 4, 87). For example, transfecting 10 to 50 copies of singular enzymes of the tryptophan pathway into yeast increases the yield of tryptophan no more than 2–30% (4). Likewise, negative mutations of enzymes or of the hypothetical functions of tumor suppressor genes (12) are buffered in vivo by increased substrate concentrations from kinetically linked upstream enzymes and by decreased product concentrations from downstream enzymes of the respective assembly lines (4, 87). Even null mutations are buffered in diploid cells by a second unmutated allele. Drawing on the car-factory analogy, the mutation of individual genes would be equivalent to mutating individual workers in an assembly line who work at only a small fraction of their capacity. Both activated and inactivated workers would be buffered by unmutated workers working upstream and downstream and by redundant capacity.
Thus, only homozygous null mutations are likely to alter the phenotype in vivo, but those are unlikely starting material of cancer cells. According to the cancer researcher Cairns, “one of the problems is that most mutations lead to loss of function, rather than creation of new function” (8). It would appear that aneuploidy, by altering at once the dosage of thousands of structural and regulatory genes, offers the only simple solution to the generation of the many complex abnormal phenotypes of cancer cells, and to the exceedingly slow kinetics of carcinogenesis mentioned above (53, 88).
Aneuploidy, the “Additional Genetic Alteration” Required by the Three Genes and by SV40 to Transform Normal Human Cells.
In light of the aneuploidy hypothesis, we suggest that one or more of the synthetic genes used by Weinberg and coworkers to generate human tumor cells functioned as aneuploidizing agents and that the resulting aneuploidy initiated the karyotype evolution that subsequently generated clonal neoplastic karyotypes. As pointed out above, such cancers are clonal for preexisting or carcinogen-induced gene mutations and for aneuploidy but not for a particular karyotype, because of the inherent karyotype instability of aneuploid cells. Likewise, the tumor cells prepared by Weinberg and coworkers were clonal for the integration sites of the three added genes but nonclonal for the resulting aneuploidy.
Indeed, SV40 or its cloned T antigen alone (89) has been known since 1962 to be a particularly efficient aneuploidogen in human cells (90–93). The mechanism of aneuploidization is thought to be induction of chromosome nondisjunction, possibly because T antigen displaces histone proteins from chromosomal DNA and unwinds nucleosomally organized DNA (94) and thus blocks the normal chromosomal binding sites for tubulin fibers. This preneoplastic aneuploidy, then, evolves into neoplastic aneuploidy not only autocatalytically (see Fig. 2) but also by the continued aneuploidizing action of the viral T antigen. It would follow that the T antigen does not directly cause and does not maintain transformation but initiates transformation via aneuploidy after a latent period of many cell generations, just like a chemical carcinogen. But in contrast to a chemical, the T antigen is a replicating carcinogen.
It is consistent with this proposal that, despite extensive efforts, a mutant of SV40 or of the related Polyoma virus that is temperature sensitive for the maintenance of tumorigenic transformation was never found (95). [There is evidence for the loss of SV40 from transformed lines but no evidence for the simultaneous loss of tumorigenicity (96).] The aneuploidy hypothesis also offers an explanation for the “60 population doublings” that it took from the introduction of the SV40 T-antigen gene to tumorigenicity. Because the probability is low of generating by chance a karyotype that is more viable than a normal diploid one, even in the presence of the T antigen, the evolution of a tumorigenic karyotype is slow, compatible with a “rare stochastic event,” and the resulting tumor is typically clonal. This confirms and extends the view that neither the T antigen alone nor the three genes used to generate the tumor cells were “sufficient” for tumorigenesis, but they were sufficient to initiate aneuploidy, which eventually generated neoplastic karyotypes.
The same explanation probably applies to the previously known but unexplained long latent period of 2–4 mo between infection and subsequent transformation of diploid primary human cells by SV40 (90, 91). Aneuploid karyotype evolution also explains the kinetics of tumorigenesis and pathogenesis of mice carrying T antigen with a weak promoter in the germ line. Such mice develop aneuploid tumors within 3–9 mo (97, 98).
There is, however, a class of genes, the oncogenes of retroviruses, that meet all predictions of the gene mutation hypothesis: (i) transformation within one or a few cell generations; (ii) transformation depending on continued function of viral protein; (iii) transforming function of chromosomal DNA; (iv) polyclonal tumors; and (v) diploid tumors (at least initially) (71, 95, 99–101). But despite enormous efforts, including those by Weinberg and coworkers, there is as yet no evidence for such genes outside of rare oncogenic retroviruses (22, 35, 36, 100, 102).
Conclusions
Because two tumorigenic cell lines generated by Weinberg and coworkers from normal human cells with a combination of three artificial genes appeared to be clonal and were highly aneuploid, and because both had apparently acquired tumorigenicity only many generations after the introduction of these genes, we call into question the claim that these genes are sufficient for tumorigenicity. Instead, we propose that one or more of these genes induced preneoplastic aneuploidy and that autocatalytic and SV40 T-antigen-driven karyotype evolution eventually generated neoplastic aneuploidy. It would follow that aneuploidy was necessary for transformation in this system. Our proposal also predicts that the normal human cells became aneuploid before tumorigenicity, as we have recently demonstrated for chemical carcinogenesis (62). Our proposal can be refuted readily by generating either diploid tumor cells within one generation after the introduction of the three artificial genes of Weinberg and coworkers (50) or, more relevant for natural carcinogenesis, after the introduction of an appropriate set of unaltered mutant genes from natural cancers.
Acknowledgments
We thank Harvey Bialy (Science Editor, Nature Biotechnology, New York), Michael Botchan, Dan Koshland, and Richard Strohman (University of California, Berkeley), and Robert Weinberg (Massachusetts Institute of Technology, Cambridge, MA) for critical comments on the manuscript; George McNamara (Applied Spectral Imaging, Carlsbad, CA) for help with spectral karyotype analysis; and Robert Leppo (philanthropist, San Francisco), the Abraham and Phyllis Katz Foundation (New York), a foundation that prefers to remain anonymous, and several private donors for support.
Abbreviations
- SV40
simian virus 40
- T antigen
tumor antigen
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.040529797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.040529797
References
- 1.Tyzzer E E. J Cancer Res. 1916;1:125–155. [Google Scholar]
- 2.Strong L C. Br J Cancer. 1949;3:97–108. doi: 10.1038/bjc.1949.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kacser H, Burns J A. Genetics. 1981;97:639–666. doi: 10.1093/genetics/97.3-4.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornish-Bowden A. In: Biotechnology. Rehm H-J, Reed G, editors. New York: VCH; 1995. pp. 121–136. [Google Scholar]
- 5.Yosida T H. Cancer Genet Cytogenet. 1983;8:153–179. doi: 10.1016/0165-4608(83)90047-x. [DOI] [PubMed] [Google Scholar]
- 6.O'Brien S, Menotti-Raymond M, Murphy W, Nash W, Wirnberg J, Stanyon R, Copeland N, Jenkins N, Womack J, Marshall Graves J. Science. 1999;286:458–481. doi: 10.1126/science.286.5439.458. [DOI] [PubMed] [Google Scholar]
- 7.Braun A C. The Cancer Problem. A Critical Analysis and Modern Synthesis. New York: Columbia Univ. Press; 1969. [Google Scholar]
- 8.Cairns J. Cancer: Science and Society. San Francisco: Freeman; 1978. [Google Scholar]
- 9.Pitot H C. Fundamentals of Oncology. New York: Dekker; 1986. [Google Scholar]
- 10.Lewin B. Genes V. Oxford, U.K.: Oxford Univ. Press; 1994. [Google Scholar]
- 11.Lodish H, Baltimore D, Berk A, Zipursky S L, Matsudaira P, Darnell J. Molecular Cell Biology. New York: Freeman; 1995. [Google Scholar]
- 12.Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson J D. Molecular Biology of the Cell. New York: Garland; 1994. [Google Scholar]
- 13.Berenblum I, Shubik P. Br J Cancer. 1949;3:109–118. doi: 10.1038/bjc.1949.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burdette W J. Cancer Res. 1955;15:201–226. [PubMed] [Google Scholar]
- 15.Oshimura M, Barrett J C. Environ Mutagen. 1986;8:129–159. doi: 10.1002/em.2860080112. [DOI] [PubMed] [Google Scholar]
- 16.Ashby J. In: Tumor Promotors: Biological Approaches for Mechanistic Studies and Assay Systems. Langenbach R, et al., editors. New York: Raven; 1988. pp. 417–430. [Google Scholar]
- 17.Lijinsky W. Environ Mol Mutagen. 1989;14:78–84. doi: 10.1002/em.2850140615. [DOI] [PubMed] [Google Scholar]
- 18.Hollstein M, Rice K, Greenblatt M S, Soussi R, Fuchs R, Sorlie T, Hovig E, Smith-Sorensen B, Montesano R, Harris C C. Nucleic Acids Res. 1994;22:3551–3555. [PMC free article] [PubMed] [Google Scholar]
- 19.Vogelstein B, Fearon E R, Stanley B A, Hamilton R, Kern S E, Preisinger A C, Leppert M, Nakamura Y, White R, Smits A M M, et al. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 20.Cooper G M. Oncogenes. Boston: Jones and Bartlett; 1990. [Google Scholar]
- 21.Strauss B S. Cancer Res. 1992;52:249–253. [PubMed] [Google Scholar]
- 22.Duesberg P H, Schwartz J R. Prog Nucleic Acid Res Mol Biol. 1992;43:135–204. doi: 10.1016/s0079-6603(08)61047-8. [DOI] [PubMed] [Google Scholar]
- 23.Haber D A, Fearon E R. Lancet. 1998;351:SII1–SII8. doi: 10.1016/s0140-6736(98)90326-9. [DOI] [PubMed] [Google Scholar]
- 24.Boland C R, Ricciardello L. Proc Natl Acad Sci USA. 1999;96:14675–14677. doi: 10.1073/pnas.96.26.14675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konishi N, Hiasa Y, Matsuda H, Tao M, Tsuzuki T, Hayashi I, Kitahori Y, Shiraishi T, Yatani R, Shimazaki J, et al. Am J Pathol. 1995;147:1112–1122. [PMC free article] [PubMed] [Google Scholar]
- 26.Shibata D, Schaeffer J, Li Z H, Capella G, Perucho M X. J Natl Cancer Inst. 1993;85:1058–1063. doi: 10.1093/jnci/85.13.1058. [DOI] [PubMed] [Google Scholar]
- 27.Al-Mulla F, Going J J, Sowden E T, Winter A, Pickford I R, Birnie G D. J Pathol. 1998;185:130–138. doi: 10.1002/(SICI)1096-9896(199806)185:2<130::AID-PATH85>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 28.Roy-Burman P, Zheng J, Miller G J. Mol Med Today. 1997;3(11):476–482. doi: 10.1016/S1357-4310(97)01126-X. [DOI] [PubMed] [Google Scholar]
- 29.Kuwabara S, Ajioka Y, Watanabe H, Hitomi J, Nishikura K, Hatakeyama K. Jap J Cancer Res. 1998;89:405–410. doi: 10.1111/j.1349-7006.1998.tb00578.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heppner G, Miller F R. Int Rev Cytol. 1998;177:1–56. doi: 10.1016/s0074-7696(08)62230-5. [DOI] [PubMed] [Google Scholar]
- 31.Giaretti W, Monaco R, Pujic N, Rapallo A, Nigro S, Geido E. Am J Pathol. 1996;149:237–245. [PMC free article] [PubMed] [Google Scholar]
- 32.Augenlicht L H, Wahrman M Z, Halsey H, Anderson L, Taylor J, Lipkin M. Cancer Res. 1987;47:6017–6021. [PubMed] [Google Scholar]
- 33.Thraves P, Reynolds S, Salehi Z, Kim W K, Yang J H, Rhim J S, Dritschilo A. In: Neoplastic Transformation in Human Cell Culture. Rhim J S, Dritschilo A, editors. Totowa, NJ: Humana; 1991. pp. 93–101. [Google Scholar]
- 34.Stanbridge E J. Annu Rev Genet. 1990;24:615–657. doi: 10.1146/annurev.ge.24.120190.003151. [DOI] [PubMed] [Google Scholar]
- 35.Duesberg P. Science. 1995;267:1407–1408. doi: 10.1126/science.7794335. [DOI] [PubMed] [Google Scholar]
- 36.Hua V Y, Wang W, K, Duesberg P H. Proc Natl Acad Sci USA. 1997;94:9614–9619. doi: 10.1073/pnas.94.18.9614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weitzman J B, Yaniv M. Nature (London) 1999;400:401–402. doi: 10.1038/22637. [DOI] [PubMed] [Google Scholar]
- 38.Albino A P, Le Strange R, Oliff A I, Furth M E, Old L J. Nature (London) 1984;308:69–72. doi: 10.1038/308069a0. [DOI] [PubMed] [Google Scholar]
- 39.Plattner R, Anderson M J, Sato K Y, Fasching C L, Der C J, Stanbridge E J. Proc Natl Sci Acad USA. 1996;93:6665–6670. doi: 10.1073/pnas.93.13.6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muller H J. Science. 1927;66:84–87. doi: 10.1126/science.66.1699.84. [DOI] [PubMed] [Google Scholar]
- 41.Brookes P, Lawley P D. Nature (London) 1964;202:781–784. doi: 10.1038/202781a0. [DOI] [PubMed] [Google Scholar]
- 42.Nowell P C. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 43.Duesberg P, Rausch C, Rasnick D, Hehlmann R. Proc Natl Acad Sci USA. 1998;95:13692–13697. doi: 10.1073/pnas.95.23.13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cahill D P, Kinzler K W, Vogelstein B, Lengauer C. Trends Biol Sci. 1999;24:M57–M60. [PubMed] [Google Scholar]
- 45.Mitelman F. Catalogue of Chromosome Aberrations in Cancer. New York: Wiley; 1994. [DOI] [PubMed] [Google Scholar]
- 46.Mitelman F, Mertens F, Johansson B. Nat Genet. 1997;15:S417–S474. doi: 10.1038/ng0497supp-417. [DOI] [PubMed] [Google Scholar]
- 47.Sandberg A A. The Chromosomes in Human Cancer and Leukemia. New York: Elsevier; 1990. [Google Scholar]
- 48.Mertens F, Johansson B, Hoeglund M, Mitelman F. Cancer Res. 1997;57:2765–2780. [PubMed] [Google Scholar]
- 49.Gebhart E, Liehr T. Int J Oncol. 2000;16:383–399. doi: 10.3892/ijo.16.2.383. [DOI] [PubMed] [Google Scholar]
- 50.Hahn W C, Counter C M, Lundberg A S, Beijersbergen R L, Brooks M W, Weinberg R A. Nature (London) 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 51.Li R, Yerganian G, Duesberg P, Kraemer A, Willer A, Rausch C, Hehlmann R. Proc Natl Acad Sci USA. 1997;94:14506–14511. doi: 10.1073/pnas.94.26.14506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brinkley B R, Goepfert T M. Cell Motil Cytoskel. 1998;41:281–288. doi: 10.1002/(SICI)1097-0169(1998)41:4<281::AID-CM1>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 53.Rasnick D, Duesberg P. Biochem J. 1999;340:621–630. [PMC free article] [PubMed] [Google Scholar]
- 54.Duesberg P. Science. 1999;284:2091–2092. doi: 10.1126/science.284.5423.2089f. [DOI] [PubMed] [Google Scholar]
- 55.Hansemann D. Virchows Arch A Pathol Anat Histol. 1890;119:299–326. [Google Scholar]
- 56.Boveri T. Zur Frage der Entstehung maligner Tumoren. Jena, Germany: Fischer; 1914. [Google Scholar]
- 57.Fardon J C. Science. 1953;117:441–445. doi: 10.1126/science.117.3043.441. [DOI] [PubMed] [Google Scholar]
- 58.DiPaolo J A. In Vitro. 1975;11:89–96. doi: 10.1007/BF02624081. [DOI] [PubMed] [Google Scholar]
- 59.Harnden D G, Taylor A M R. In: Advances in Human Genetics. Harris H, Hirschhorn K, editors. New York: Plenum; 1979. pp. 1–70. [Google Scholar]
- 60.Heim S, Mitelman F. Cancer Cytogenet. New York: Wiley; 1995. [Google Scholar]
- 61.Bauer K H. Das Krebsproblem. Berlin: Springer; 1963. [Google Scholar]
- 62.Duesberg, P., Li, R., Rasnick, D., Rausch, C., Willer, A., Kraemer, A., Yerganian, G. & Hehlmann, R. (2000) Cancer Genet. Cytogenet., in press. [DOI] [PubMed]
- 63.Hahn W C, Stewart S A, Brooks M W, York S G, Eaton E E, Kurachi A, Beijersbergen R L, Knoll J H M, Meyerson M, Weinberg R A. Nat Med. 1999;5:1164–1170. doi: 10.1038/13495. [DOI] [PubMed] [Google Scholar]
- 64.Koprowski H, Croce C. Proc Natl Acad Sci USA. 1977;74:1142–1146. doi: 10.1073/pnas.74.3.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stanbridge E J, Der C J, Doersen C-J, Nishimi R Y, Peehl D M, Weissman B E, Wilkinson J. Science. 1982;215:252–259. doi: 10.1126/science.7053574. [DOI] [PubMed] [Google Scholar]
- 66.Choi K H, Tevethia S S, Shin S. Cytogenet Cell Genet. 1983;36:633–640. doi: 10.1159/000131987. [DOI] [PubMed] [Google Scholar]
- 67.Walen K, Arnstein P. In Vitro Cell Dev Biol. 1986;22:57–65. doi: 10.1007/BF02623534. [DOI] [PubMed] [Google Scholar]
- 68.Boveri T. In: Foundations of Experimental Embryology. Willier J M, Oppenheimer B H, editors. Englewood Cliffs, NJ: Prentice-Hall; 1964. pp. 74–97. [Google Scholar]
- 69.Galloway S M, Buckton K E. Cytogenet Cell Genet. 1978;20:78–96. doi: 10.1159/000130842. [DOI] [PubMed] [Google Scholar]
- 70.Hagmar L, Bonassi S, Stromberg U, Brogger A, Knudsen L, Norppa H, Reuterwall C. Cancer Res. 1998;58:4117–4121. [PubMed] [Google Scholar]
- 71.Duesberg P, Rasnick D, Li R, Winters L, Rausch C, Hehlmann R. Anticancer Res. 1999;19:4887–4906. [PubMed] [Google Scholar]
- 72.Jensen K G, Oenfelt A, Poulsen H E, Doehmer J, Loft S. Carcinogenesis. 1993;14:2115–2118. doi: 10.1093/carcin/14.10.2115. [DOI] [PubMed] [Google Scholar]
- 73.Matsuoka A, Ozaki M, Takeshita K, Sakamoto H, Glatt H, Hayashi M, Sofuni T. Mutagenesis. 1997;12:365–372. doi: 10.1093/mutage/12.5.365. [DOI] [PubMed] [Google Scholar]
- 74.Burke D, Gasdaska P, Hartwell L. Mol Cell Biol. 1989;9:1049–1059. doi: 10.1128/mcb.9.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mayer V W, Aguilera A. Mutat Res. 1990;231:177–186. doi: 10.1016/0027-5107(90)90024-x. [DOI] [PubMed] [Google Scholar]
- 76.Lingle W L, Lutz W H, Ingle J N, Maihle N J, Salisbury J L. Proc Natl Acad Sci USA. 1998;95:2950–2955. doi: 10.1073/pnas.95.6.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pihan G A, Purohit A, Wallace J, Knecht H, Woda B, Quesenberry P, Doxsey S J. Cancer Res. 1998;58:3974–3985. [PubMed] [Google Scholar]
- 78.Holliday R. Trends Genet. 1989;5:42–45. doi: 10.1016/0168-9525(89)90020-6. [DOI] [PubMed] [Google Scholar]
- 79.Atkin N B, Baker M C. Cytogenet Cell Genet. 1990;53:58–60. doi: 10.1159/000132895. [DOI] [PubMed] [Google Scholar]
- 80.Matzke M A, Mittelsten-Scheid O, Matzke A J M. BioEssays. 1999;21:761–767. doi: 10.1002/(SICI)1521-1878(199909)21:9<761::AID-BIES7>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 81.Hartwell L. Cell. 1992;71:543–546. doi: 10.1016/0092-8674(92)90586-2. [DOI] [PubMed] [Google Scholar]
- 82.Lindsley D L, Sandler L, Baker B S, Carpenter A T C, Denell R E, Hall J C, Jacobs P A, Gabor Miklos G L, Davis B K, Gethmann R C, et al. Genetics. 1972;71:157–184. doi: 10.1093/genetics/71.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lejeune J, Turpin R, Gautier M. Ann Genet. 1959;2:41–49. [Google Scholar]
- 84.Sandler L, Hecht F. Am J Hum Genet. 1973;25:332–339. [PMC free article] [PubMed] [Google Scholar]
- 85.Shapiro B L. Am J Med Genet. 1983;14:241–269. doi: 10.1002/ajmg.1320140206. [DOI] [PubMed] [Google Scholar]
- 86.Epstein C. The Consequences of Chromosome Imbalance: Principles, Mechanisms, and Models. Cambridge, U.K.: Cambridge Univ. Press; 1986. [Google Scholar]
- 87.Fell D. Understanding the Control of Metabolism. London: Portland; 1997. [Google Scholar]
- 88.Cornish-Bowden A. Nat Biotechnol. 1999;17:641–643. doi: 10.1038/10854. [DOI] [PubMed] [Google Scholar]
- 89.Ray F A, Peabody D S, Cooper J L, Cram L S, Kraemer P M. J Cell Biochem. 1990;42:13–31. doi: 10.1002/jcb.240420103. [DOI] [PubMed] [Google Scholar]
- 90.Shein H M, Enders J F. Proc Natl Acad Sci USA. 1962;48:1164–1172. doi: 10.1073/pnas.48.7.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Koprowski H, Ponten J A, Jensen F, Ravdin R D, Moorehead P S, Saksela E. J Cell Comp Physiol. 1962;59:281–292. [Google Scholar]
- 92.Todaro G J, Wolman S R, Green H. J Cell Comp Physiol. 1963;62:257–265. doi: 10.1002/jcp.1030620305. [DOI] [PubMed] [Google Scholar]
- 93.Wolman S R, Steinberg M L, Defendi V. Cancer Genet Cytogenet. 1980;2:39–46. [Google Scholar]
- 94.Ramsperger U, Stahl H. EMBO J. 1995;14:3215–3225. doi: 10.1002/j.1460-2075.1995.tb07324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tooze J. The Molecular Biology of Tumour Viruses. Plainview, NY: Cold Spring Harbor Lab. Press; 1973. [Google Scholar]
- 96.Steinberg B, Pollack R, Topp W, Botchan M. Cell. 1978;13:19–32. doi: 10.1016/0092-8674(78)90134-4. [DOI] [PubMed] [Google Scholar]
- 97.Keough R, Powell B, Rogers G. J Cell Sci. 1995;108:957–966. doi: 10.1242/jcs.108.3.957. [DOI] [PubMed] [Google Scholar]
- 98.Hanahan D. Annu Rev Genet. 1988;22:479–519. doi: 10.1146/annurev.ge.22.120188.002403. [DOI] [PubMed] [Google Scholar]
- 99.Mitelman F. In: Chromosomes and Cancer. German J, editor. New York: Wiley; 1974. pp. 675–693. [Google Scholar]
- 100.Duesberg P H. Cancer Res. 1987;47:1199–1220. [PubMed] [Google Scholar]
- 101.Martin G S. Nature (London) 1970;227:1021–1023. doi: 10.1038/2271021a0. [DOI] [PubMed] [Google Scholar]
- 102.Duesberg P H. Nature (London) 1983;304:219–226. doi: 10.1038/304219a0. [DOI] [PubMed] [Google Scholar]