Abstract
Mitogen-activated protein kinases (MAPKs) play key roles in differentiation, growth, proliferation, and apoptosis. Although MAPKs have been extensively studied, the precise function, specific substrates, and target genes of each MAPK are not known. These issues could be addressed by sole activation of a given MAPK, e.g., through the use of constitutively active MAPK enzymes. We have recently reported the isolation of eight hyperactive mutants of the Saccharomyces cerevisiae MAPK Hog1, each of which bears a distinct single point mutation. These mutants acquired high intrinsic catalytic activity but did not impose the full biological potential of the Hog1 pathway. Here we describe our attempt to obtain a MAPK that is more active than the previous mutants both catalytically and biologically. We combined two different activating point mutations in the same gene and found that two of the resulting double mutants acquired unusual properties. These alleles, HOG1D170A,F318L and HOG1D170A,F318S, induced a severe growth inhibition and had to be studied through an inducible expression system. This growth inhibition correlated with very high spontaneous (in the absence of any stimulation) catalytic activity and strong induction of Hog1 target genes. Furthermore, analysis of the phosphorylation status of these active alleles shows that their acquired intrinsic activity is independent of either phospho-Thr174 or phospho-Tyr176. Through fluorescence-activated cell sorting analysis, we show that the effect on cell growth inhibition is not a result of cell death. This study provides the first example of a MAPK that is intrinsically activated by mutations and induces a strong biological effect.
Mitogen-activated protein kinase (MAPK) signaling pathways play important roles in differentiation, growth, proliferation, and apoptosis. The MAPK module has been highly conserved throughout evolution and is present in all eukaryotic cells (20, 26, 38, 48, 54). It consists of three kinase proteins: a MAPK kinase kinase (MAPKKK) that activates MAPK kinase (MAPKK), which in turn activates the relevant MAPK by phosphorylating it on both Thr and Tyr residues in an appropriate Thr-X-Tyr motif (7, 17, 27). Phosphorylated activated MAPKs translocate from the cytoplasm into the nucleus, where many of their substrates reside. However, they phosphorylate cytoplasmatic and membrane proteins as well (26, 27).
In mammalian cells, three major families of MAPKs have been identified, the ERK, JNK, and p38 families (24, 31, 45). Each family, as well as each member within the family, responds differently to various activating conditions (14, 15, 24, 30, 35). As many MAPKs are activated in response to various external signals, albeit to different degrees, it is rather difficult to study the precise role of a given MAPK.
A direct way to reveal the biochemical and biological activities of a given MAPK would be selective activation of that molecule in vivo. This approach could be applied through expression of constitutively active forms of the MAPK of interest. Strategies to produce such molecules included construction of MAPKK-MAPK fusion genes (46, 56) and a variety of genetic approaches (3, 5, 6, 21). The MAPKK-MAPK fusion proteins were indeed found to be active, but the genetic screens usually provided MAPK molecules carrying point mutations that manifested only residual activities. A recent study showed that combining some of these mutations into one MAPK protein (ERK2 was used) resulted in a synergism that increased the catalytic activity (16).
Recently, the isolation of eight different hyperactive mutants of the Saccharomyces cerevisiae MAPK Hog1 was reported (1). Hog1 is a yeast homolog of the mammalian MAPKs p38 and JNK (18, 22). It is phosphorylated and activated by the MAPKK Pbs2 (4). The Pbs2/Hog1 cascade is activated under osmotic stress and is essential for survival under these conditions (4). The active Hog1 mutants were isolated in a screen planned to identify HOG1 alleles that allow pbs2Δ cells to survive under osmotic stress. Each of these mutants harbors a distinct single point mutation and was shown to possess intrinsic catalytic activity that is independent of dual phosphorylation. Most importantly, a human p38 protein carrying equivalent mutations also manifested an increased catalytic activity that was independent of upstream regulation (1).
Biochemical analysis of the eight active Hog1 mutants (e.g., level of activity, responsiveness to salt, and phosphorylation level) revealed distinct properties suggesting that the different alleles may be activated through different mechanisms (1).
Although very active, the alleles carrying the single activating mutations do not seem to possess the maximal activity possible. This notion is based on previous observations that hyperactivation of the HOG1 pathway is lethal to yeast cells (28, 29, 40). As the active Hog1 mutants did not impose lethality, it may be that they do not fully activate Hog1 downstream targets and are therefore not an optimal research tool. In order to develop Hog1 enzymes that fully potentiate the HOG1 cascade, we combined different activating mutations in the same gene, thereby potentially combining two activating mechanisms. We hoped for an additive if not a synergistic effect. A battery of dually mutated Hog1 molecules was prepared. Two of these double-mutant Hog1 enzymes acquired extremely high catalytic activity, induced elevated expression levels of Hog1 target genes, and imposed a severe growth arrest on yeast cells.
MATERIALS AND METHODS
Construction of six double mutants containing the N-terminal Y68H mutation.
A HOG1 5′ primer, bearing the Y68H mutation and the most 5′ PstI site of the HOG1 gene, and a 3′ primer termed BamHI3′ HOG1 (see primer sequences below), were used to amplify HOG1 genes in a series of PCRs. The templates used were HOG1 clones each bearing a different activating mutation of the six C-terminal mutations (1). The resulting PCR products were fragments containing both Y68H and one of the C-terminal mutations. These were digested with PstI, and the fragments obtained (still retaining both mutations) were ligated into a Bluescript SK+-HA-HOG1 plasmid, which was also digested with PstI. In turn, each of the resulting HA-HOG1 double mutants were digested from the SK+ plasmid by using NotI and HindIII and ligated into the PES86+ yeast expression vector, which was digested with the same enzymes. PES86+ is a derivative of pADNS (13), in which the BamHI fragment of pADNS was ligated (following a fill-in Klenow reaction) to the large PvuII fragment of pRS426 (11).
Primer sequences used in the PCRs.
The following primer sequences were used in the PCRs: 5′ HA-HOG1: 5′-TCC ACT GCA GTG CTG GCC AAA AGG ACA CAT CGT GAA C-3′; BamHI 3′ HOG1: 5′-CTA GCC GGA TCC TTA CTG TTG GAA CTC ATT AGC-3′.
Construction of six double mutants containing the N-terminal D170A mutation.
The plasmid PES86+-HA-HOG1D170A (1) was digested with NotI and SalI, resulting in a backbone bearing the vector fused to the N-terminal half of the HOG1 gene containing the D170A mutation. The same double digestion was applied to each of the six C-terminal mutants providing six fragments, each harboring a different C-terminal mutation. Each of these fragments was ligated to the PES86+-HA-HOG1D170A digested with NotI and SalI.
All twelve dually mutated clones were sequenced to verify the existence of both point mutations and the absence of any other mutation.
Construction of MET3-inducible double mutants.
MET3-inducible vectors were constructed by inserting each of the 12 dually mutated HOG1 genes, as HindIII+NotI fragments removed from the PES86+-based plasmids, into the Met 425 plasmid, bearing the MET3 promoter digested with the same enzymes. The plasmid used is a derivative of the p425 MET25 plasmid in which the MET25 promoter was replaced with the MET3 promoter (34). The resulting expression cassettes MET3-HOG1 were transferred as KpnI+SacI fragments, to pRS426 plasmids digested with the same enzymes.
To construct plasmids bearing the various HOG1 alleles mutated in one of the phosphoacceptors, we used Bluescript plasmids carrying HOG1wt or HOG1D170A into which either the T174A or Y176F mutation was previously inserted (information available upon request). These vectors were digested by HindIII and SalI, resulting in an insert comprising the N terminus of the HOG1 gene mutated in one of the phosphoacceptors alone or jointly with the proximal D170A mutation. This insert was then ligated into the relevant pRS426-MET3-HOG1 backbones (described above) identically digested.
Construction of integrative HOG1 alleles.
For construction of single-copy integrative plasmids harboring HOG1 alleles under the control of the endogenous HOG1 promoter (490 bp), the various HA-HOG1 fragments were digested out of pBs15 vectors by using BamHI and ClaI and ligated into the pRS306 vector digested identically. The construction of pBs15 was performed as follows. In order to introduce a HindIII restriction site to the 3′ end of the HOG1 promoter, the HOG1 gene was amplified in a PCR with the following primers: forward primer (Fwd), 5′-CCA TCG ATT GAA GGA AAT AAG AGG-3′; and reverse primer (Rev), 5′-GGC CCA AGC TTT ATT ATA TAC GAT AGT TGT AGT TTT-3′. This PCR fragment was digested with ClaI and HindIII and ligated into a Bluescript vector that harbors the coding sequence of HA-HOG1. This vector was termed pBs14. Next, pBs14 was cut with ClaI and AatII and ligated into pRS426-HOG1, which was digested with the same restriction enzymes. This new vector, which harbors a hemagglutinin (HA)-tagged full-length HOG1 gene (including promoter and terminator), was termed pRS1. pBs15 was subsequently constructed by digesting pRS1 with BamHI and ClaI and ligating the HOG1 fragment into Bluescript.
Yeast strains and media.
The S. cerevisiae strains used were the pbs2Δ strain MAY1 (MATa ura3-52 lys2-801amber ade2-101ochre leu2-Δ1 his3-Δ200 pbs2-Δ2::LEU2) and the hog1Δ strain JBY13 (MATa leu2 ura3 his3 trp1 ade2 lys2 hog1::TRP1). Both strains were obtained from M. C. Gustin, Rice University. SP1hog1Δ, SP1pbs2Δ, and SP1hog1Δpbs2Δ strains were produced by disrupting the relevant genes in the SP1 strain (13) (MATa his3 leu2 ura3 trp1 ade8 canr). A MAY1 strain expressing the kinase dead form of pbs2 (PBS2KD) was created by integrating a pRS303 plasmid bearing the PBS2K389M allele into the HIS3 locus of the genome. The same plasmid was also integrated into the SP1Δpbs2 strains already carrying the various integrated HOG1 alleles (as explained above).
Cultures were maintained on YPD (1% yeast extract, 2% Bacto Peptone, 2% glucose), or on the synthetic medium YNB-URA or YNB-LEU [0.17% yeast nitrogen base without amino acids and NH4(SO4)2, 0.5% ammonium sulfate, 2% glucose, and 40 mg of adenine, histidine, tryptophan, lysine, and uracil or leucine per liter, and 160 mg of methionine/liter for noninducing conditions (or without methionine for induction)]. To induce Hog1 expression in liquid media, cultures were grown to logarithmic phase (optical density at 600 nm [OD600] of 0.5 to 1.0) at 30°C. Then, cells were split in half and collected by centrifugation followed by resuspension of one half in the same media and the other half in media lacking methionine. Cultures were further grown in media lacking methionine for 2 h.
Transformation and dot plating.
DNA was introduced into yeast cells according to the method of Schiestel and Gietz (50). Cells were plated on YNB-URA or YNB-LEU supplemented with 160 mg of methionine per liter. pRS306-based plasmids (single-copy vectors) were linearized with StuI prior to transformation to allow integration into the URA3 locus.
To test the ability of the transformants to grow when Hog1 expression was induced, yeast cultures were grown in noninducing media, washed with sterile double-distilled water (DDW), and resuspended in methionine-free media. The culture was then sequentially diluted and plated onto plates containing or lacking methionine. Cells in which the HOG1 genes were integrated into the genome were grown in liquid YPD, serially diluted, and plated on YPD plates supplemented with high NaCl concentrations (0.7, 0.8, and 0.9 M).
Preparation of native and denatured cell lysates and Western blotting.
Cell cultures were grown to an OD600 of 0.5 to 1.0 in media supplemented with 160 mg of methionine per liter. Cultures were collected, split, and resuspended in media containing or lacking methionine. In assays for which salt induction is also indicated, cells were exposed to 1 M NaCl for 10 min 1 h after methionine removal. For lysates, preparation cells were centrifuged at 4°C and the pellet was washed with 30 ml of ice-cold DDW. The pellet was resuspended in 1 to 2 volumes of ice-cold lysis buffer (50 mM Tris-HCl [pH 7.4], 0.25 M NaCl, 0.1% NP-40, 5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin per ml, 10 μg of trypsin inhibitor per ml, 10 μg of pepstatin A per ml, 250 μg of benzamidine per ml, 1 mM sodium vanadate, 10 mM NaF, 1 mM p-nitro phenyl phosphate [Sigma], 10 mM β-glycerol-P). Six hundred milligrams of glass beads was added and 8 × 1′ vortexing was applied. Samples were centrifuged at 800 × g for 5 min, and the supernatant was centrifuged again at 15,000 × g for 15 min at 4°C. Supernatants were aliquoted in small volumes (∼200 μl) and frozen immediately in liquid nitrogen. Denatured cell lysates were prepared by using trichloroacetic acid precipitation as described previously (1).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), Western blotting, and enhanced chemiluminescence reaction for identification of the HA-Hog1 protein and the phosphorylated Hog1 were performed as described by Sambrook et al. (47) and by Bell et al. (1). The antibody for the HA tag, 12CA5, is a monoclonal antibody derived from mouse. A dilution of 1/1,000 was used. For identification of the dually phosphorylated Hog1, commercial rabbit anti-P-p38 antibody (New England Biolabs) was used (diluted 1/2,500). For identification of threonine or tyrosine phosphorylation, rabbit antiphosphothreonine (Zymed) diluted 1/1,000 or mouse antiphosphotyrosine (4G10) diluted 1/100 was used. All secondary antibodies were diluted 1/10,000.
Kinase assay and quantification.
In vitro kinase assays were performed as previously described (1), except that following SDS-PAGE proteins were transferred to nitrocellulose by use of a semidry blotter. The kinase assay blots were quantified by using a FUJIFILM FLA-3000 phosphoimager through Image Gauge V3.45 program. Normalization to protein levels was done by NIH Image 1.62 software.
Northern blot analysis.
Total RNA was isolated at the time points indicated in the figure and separated by electrophoresis as described previously (47). Blots were hybridized in a buffer containing 7% SDS, 0.5 M sodium phosphate buffer [pH 7.2], and 1 mM EDTA. The signal was quantified using a phosphoimager (Bio-Rad Molecular Imager FX). The primers used for generating the PCR fragments, which were used as probes, were as follows: STL1 Fwd, 5′-TAA GCA GAA CCA GTC ACT GG-3′; STL1 Rev, 5′-GTA GAT TGT TGC GAA GAC CC-3′; IPP1 Fwd, 5′-CCA GAC AAA TTG GTG CCA AG-3′; IPP1 Rev, 5′-GAA CCG GAG ATG AAG AAC CA-3′; GPD1 Fwd, 5′-AAC TTC CGG CCA CTT GAA TG-3′; GPD1 Rev, 5′-ATC ATG TCC GGC AGG TTC TT-3′.
Probe fragments were labeled with Megaprime DNA labeling system (Amersham).
Membranes were stripped by incubation in boiling 0.1% SDS. This was performed a few times until radioactivity was no longer detected.
Dye exclusion analysis.
For propidium iodide (PI) staining, cultures were grown as described above for protein extraction. One-milliliter samples of 107 cells/ml were collected at the indicated time points and pelleted by centrifugation for 1 min at 17,900 × g. Pellets were resuspended in 1 ml of 50 mM Tris (pH 7.5). Prior to measurements performed with a FACSCalibur flow cytometer (Becton Dickinson, San Jose, Calif.), 55 μl of 0.5-mg/ml PI was mixed in and kept on ice. An FL2 filter was used for reading PI fluorescence. By utilizing the CellQuest software, the percentages of PI− (living) and PI+ (dead) cells were determined. PI-stained cells showed fluorescence 1,000 to 10,000 times the magnitude of the unstained cells.
RESULTS
HOG1 alleles, carrying two activating mutations expressed from a constitutive promoter, do not allow growth of colonies.
Each of the active Hog1 mutants studied so far harbored a single point mutation (1). To assess the possibility that the combination of two activating point mutations in a single Hog1 protein may enhance its intrinsic activity, we constructed 12 Hog1 molecules, each harboring two activating mutations (Fig. 1). Some of the point mutations present in these combinations (e.g., D170A, F318L, and F318S) were shown to possess the highest catalytic activity among the eight mutants isolated (1).
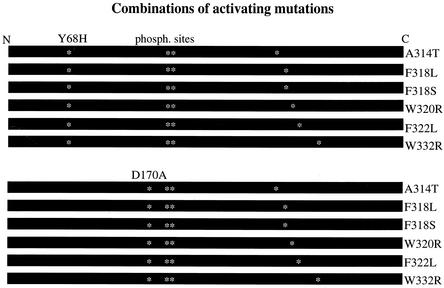
FIG. 1.
Scheme of the 12 double mutants constructed. The N-terminal mutation is noted above each group of plasmids, and each C-terminal mutation is noted on the right side of the construct. Asterisks mark the positions of the mutations and the phosphoacceptors (Thr 174 and Tyr 176).
Each of the 12 double mutants (Fig. 1) was cloned downstream from the strong constitutive ADH1 promoter on a 2μm-based plasmid (see Materials and Methods). We tried to introduce the resulting plasmids to hogΔ and pbs2Δ cells. Some plasmids gave rise to normal colonies, while others were unable to provide transformants. Yet others gave rise to colonies that failed to grow on medium supplemented with salt (Table 1). HOG1D170A,F318L and HOG1D170A,F318S alleles manifested the most severe effect (Table 1) and were selected for further studies.
TABLE 1.
Numbers of transformants obtained with active HOG1 alleles (under the control of a strong constitutive promoter) and their ability to support growth under osmotic pressure
| HOG1 allele |
hog1Δ cells
|
pbs2Δ cells
|
||
|---|---|---|---|---|
| Approx no. of colonies/μg of DNA | Growth on 1.3 M NaClb | Approx no. of colonies/μg of DNA | Growth on 1.3 M NaClb | |
| Y68H,A314T | A few thousand | + | A few thousand | + |
| Y68H,F318L | A few thousand | + | A few thousand | + |
| Y68H,F318S | A few thousand | + | A few thousand | + |
| Y68H,W320R | A few thousand | + | A few thousand | + |
| Y68H,F322L | A few thousand | + | A few thousand | + |
| Y68H,W332L | A few thousand | + | A few thousand | − |
| D170A,A314T | Hundreds of tiny colonies | + | Hundreds of tiny colonies | + |
| D170A,F318L | No coloniesa | Hundreds of tiny colonies | + | |
| D170A,F318S | No coloniesa | No coloniesa | ||
| D170A,W320R | Hundreds of tiny colonies | − | Hundreds of tiny colonies | + |
| D170A,F322L | Hundreds of tiny colonies | − | Hundreds of tiny colonies | + |
| D170A,W332L | Hundreds of tiny colonies | − | Hundreds of tiny colonies | + |
In these transformations, 30 to 70 big colonies appeared and were considered a consequence of rearrangement events. Some of these colonies were tested on salt and were found negative.
+, growth observed; −, no growth observed.
HOG1D170A,F318L and HOG1D170A,F318S alleles induce growth arrest in hog1Δ cells and rescue pbs2Δ cells when expressed from the native HOG1 promoter as single integrated copies.
The inability of HOG1D170A,F318L and HOG1D170A,F318S mutants to give rise to colonies may stem from their increased catalytic activity, in combination with overexpression induced by the ADH1 promoter and the 2μm element. To test this possibility, we constructed a series of vectors in which transcription of the different HOG1 genes was driven by the native HOG1 promoter. In addition, each of these vectors was integrated as a single copy into the genome of either hog1Δ, pbs2Δ, or hog1Δpbs2Δ cells. The integrative constructs gave rise to a similar number of transformants in all strains. Cells carrying these integrated constructs were tested for their ability to grow under high osmotic pressure (Fig. 2).
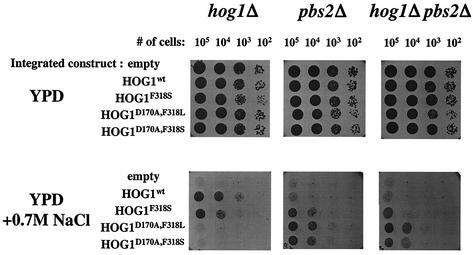
FIG. 2.
HOG1D170A,F318L and HOG1D170A,F318S mutated alleles rescue pbs2Δ cells when expressed from the native HOG1 promoter as single integrated copies. Plasmids bearing wild-type or mutant HOG1 expressed under the control of the endogenous HOG1 promoter were integrated as single copies into the genome of SP1hog1Δ, SP1pbs2Δ, or SP1hog1Δpbs2Δ strains. Cells carrying the integrated genes were grown to logarithmic phase on YPD media, serially diluted to the specified numbers, and plated on either YPD plates (upper panel) or YPD plates supplemented with 0.7 M salt (lower panel).
When tested on high salt concentrations (0.7, 0.8, and 0.9 M NaCl), hog1Δ cells harboring the HOG1wt gene as well as the HOG1F318S allele were able to grow. However, hog1Δ cells expressing the double mutants HOG1D170A,F318L and HOG1D170A,F318S did not grow under osmotic pressure (Fig. 2). When expressed in pbs2Δ or hog1Δ pbs2Δ cells, the dually mutated alleles did support growth on salt (Fig. 2). In fact, they rescued pbs2Δ and hog1Δ pbs2Δ cells much better than did the HOG1F318S allele (Fig. 2).
The ability of the dually mutated Hog1 proteins to rescue pbs2Δ and hog1Δpbs2Δ cells as single copies expressed from their native promoter suggests that they acquired some additional properties over the mutants harboring just one mutation. At these expression levels, the singly mutated HOG1F318S allele only partially rescued hog1Δpbs2Δ cells (Fig. 2). The inability of the integrated versions of HOG1D170A,F318L and HOG1D170A,F318S alleles to rescue hog1Δ cells is probably a result of their extreme activity that imposes growth arrest under high osmotic stress (see results of catalytic activity measurements below). The fact that these double mutants did not give rise to transformants when expressed at high levels (Table 1) supports this notion.
Inducible expression of Hog1 double mutants severely impedes cell growth.
To directly test the link between expression of the double mutants and retardation of colony propagation, we ligated the dually mutated HOG1 alleles downstream from the MET3 inducible promoter. This promoter is shut off in the presence of methionine and induced by its removal (9, 10). These constructs were introduced into the hog1Δ and pbs2Δ yeast strains, and transformants were plated onto YNB (-URA) plates supplemented with 160 mg of methionine per liter to suppress expression of the Hog1 molecules. Under these conditions, thousands of colonies with normal appearance were obtained with all constructs introduced into both strains.
To test whether expression of the double mutants indeed affects cell growth, cultures harboring the various MET3-driven constructs were plated onto YNB plates lacking methionine. On these plates, the growth of hog1Δ cells harboring alleles HOG1D170A,F318L or HOG1D170A,F318S was severely affected (Fig. 3A, plates on the left). In contrast, normal growth was observed for hog1Δ cells expressing wild-type Hog1 or the hyperactive mutants Hog1D170A, Hog1F318L, or Hog1F318S under the same conditions (Fig. 3A). pbs2Δ cells expressing Hog1D170A,F318L or Hog1D170A,F318S also showed growth retardation on plates lacking methionine, although the effect was less pronounced (Fig. 3A, plates on the right). Notably, all cultures grew normally on plates supplemented with methionine (Fig. 3A, top plates). In both hog1Δ and pbs2Δ cells harboring Hog1D170A,F318L or Hog1D170A,F318S, some degree of growth recovery was observed after a prolonged incubation period. Within 4 days, the growth of pbs2Δ cells harboring the dually mutated alleles was nearly fully recovered. In hog1Δ cells, however, only tiny undeveloped colonies were observed following several days of incubation (data not shown).
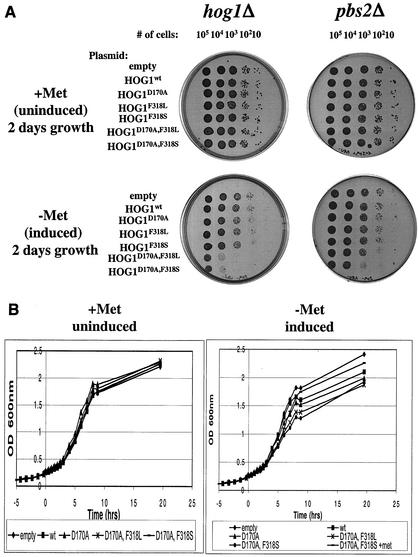
FIG.3.
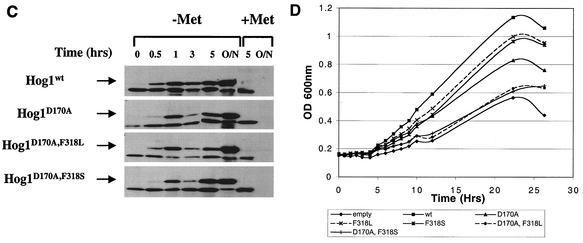
Expression of dually mutated Hog1 proteins severely impedes yeast growth. (A) Cells used were of the hog1Δ (plates on left) and the pbs2Δ (plates on right) strains harboring plasmids expressing the indicated HA-tagged Hog1 variants, under the control of the MET3 promoter. Cultures were grown in the presence of methionine to logarithmic phase, counted, washed with water, resuspended in inducing (methionine-free) media, serially diluted to the specified numbers, and plated onto the indicated plates (+Met or −Met). Plates were scanned 48 h after plating. (B) hog1Δ cells harboring the same constructs as above were grown overnight in the presence of methionine and diluted to an OD600 of 0.1 in the same media. At an OD600 of 0.25 (time zero in the graph), each culture was split in two, washed with DDW, and resuspended in media supplemented (+Met) or not (−Met) with methionine. Similar results were obtained in several independent experiments with a standard of deviation of <10% for all strains. (C) Protein extracts were precipitated by trichloroacetic acid from the cultures shown in Fig. 2B at the indicated time points, separated on SDS-PAGE, blotted, and probed with anti-HA antibodies. (O/N, overnight) (D) hog1Δ cells harboring the indicated plasmids were grown overnight in media containing methionine, diluted to an OD600 of 0.15 into media lacking methionine, and incubated for 1 h. Then (time zero in the graph), cultures were collected and resuspended in methionine-free media supplemented with 1 M NaCl. Similar results were obtained in three independent experiments.
The effect of methionine removal (and induction of the different Hog1 molecules) was also tested in liquid media with hog1Δ cells (Fig. 3B). When methionine was present in the media, all clones showed very similar growth rates and reached similar cell densities (Fig. 3B, left panel). Upon methionine removal, cells expressing active Hog1 mutants showed a reduction in growth rate (Fig. 3B, right panel). Most significantly affected were hog1Δ cells expressing the dually mutated Hog1 proteins. Western blot analysis verified that the MET3 system provided rapid induction of Hog1 protein expression following methionine removal (Fig. 3C). The effect of active Hog1 on cell growth in liquid medium was transient and was significant between 3 and 9 h after removal of methionine. Ten hours after methionine removal, all cultures manifested similar growth rates. Cells expressing the Hog1D170A,F318L or the Hog1D170A,F318S alleles, however, reached a lower cell density (Fig. 3B). Although similar in principle, the growth inhibition observed in the liquid media (Fig. 3B) is somewhat less severe than that seen on plates (Fig. 3A). Given the known physiological and anatomical differences between cultures grown in liquid and solid media (33, 36, 49), the different effects of the active HOG1 alleles under different culture conditions is not surprising (see Discussion).
When expressed from single-copy integrated genes, Hog1D170A,F318L and Hog1D170A,F318S induced growth arrest only in the presence of salt (Fig. 2). In the MET3 system it seems that significant, but not complete growth retardation was caused by their expression alone in the absence of any osmotic stress (Fig. 3A and B). Western blot analysis showed that the HOG1 alleles are indeed expressed at levels in the MET3 system that were significantly higher than those of the integrated alleles (data not shown).
To test whether this growth inhibition could be further enhanced by osmotic stress, hog1Δ cells carrying the various MET3-driven HOG1 alleles were grown in liquid cultures that were supplemented with 1 M NaCl 2 h after removal of methionine (Fig. 3D). Under these conditions, the growth differential between the various clones was more pronounced than that observed without salt (compare panels B and D of Fig. 3). Cells expressing Hog1wt overcame the osmotic stress and entered a steep growth phase after approximately 5 h. Similar kinetics were observed with cells expressing the three single mutants tested, although their growth rate was slower. In contrast, hog1Δ cells carrying the dually mutated alleles (HOG1D170A,F318L or HOG1D170A,F318S) grew very poorly, at a rate similar to that of the osmosensitive hog1Δ cells carrying a control plasmid (Fig. 3D).
Combining the results obtained with the MET3-based inducible constructs (Fig. 3) with those obtained with the constitutive expression vectors (Table 1) and the integrative constructs (Fig. 2), it seems that HOG1D170A,F318L and HOG1D170A,F318S alleles impose a severe growth retardation. This effect probably stems from intrinsic activity of these molecules, since their expression was sufficient to induce it. The effect could be further enhanced, however, by high salt concentrations.
The catalytic activity of the double mutants is significantly higher than that of the single mutants.
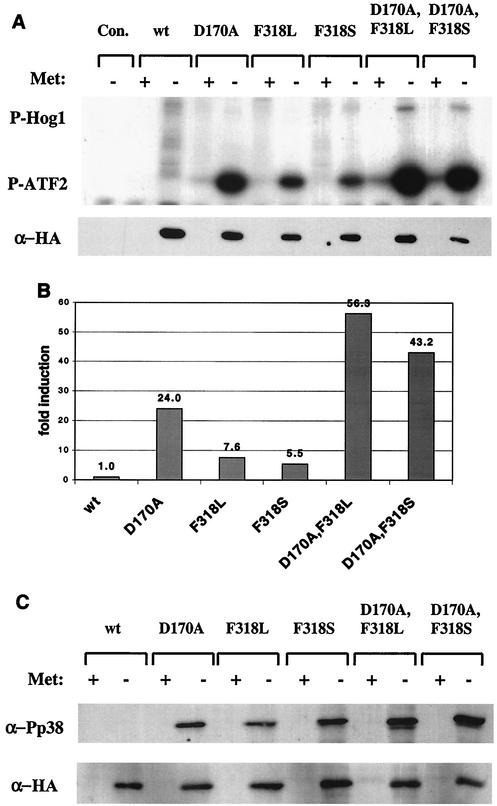
The results above demonstrated that the double mutants induced a severe growth inhibition. This effect may stem from strong catalytic activity or from another property of these molecules. To test the catalytic activity of the double mutants, we immunoprecipitated Hog1 proteins from yeast cells before and after methionine removal and monitored their ability to phosphorylate GST-ATF2 in vitro (Fig. 4 and 5).
FIG. 4.
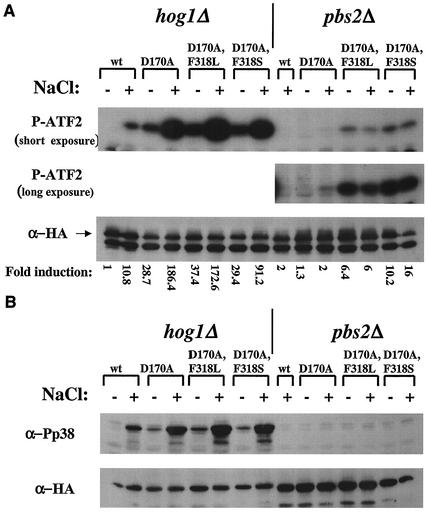
Hog1 double mutants immunoprecipitated from hog1Δ cells are catalytically active in vitro and are dually phosphorylated in the absence of salt. (A) The various Hog1 proteins expressed in hog1Δ cells under the MET3 promoter were immunoprecipitated and assayed in vitro using glutathione S-transferase (GST)-ATF2 as a substrate. Immunoprecipitation was performed on lysates prepared from cells grown in the presence (+) or absence (−) of methionine. The upper panel is an autoradiogram of the in vitro kinase assay. The top bands show autophosphorylated Hog1 proteins, and the bottom bands show the phosphorylated GST-ATF2 substrate. The lower panel is a Western blot performed on samples of the immunoprecipitated proteins. (B) Quantification (using a phosphorimager) of the above kinase assay blot. The activity of each sample was normalized to its protein level and is shown as induction compared to Hog1 wild-type activity, which was set as 1 (in the absence of methionine). (C) Phosphorylation state of the various proteins expressed in hog1Δ cells under the control of the MET3 promoter. The upper panel portrays the level of dually phosphorylated Hog1 as measured by a Western blot of whole-cell lysates using anti-phospho-p38 antibodies. The lower panel shows the same bolt after stripping and reprobing with anti-HA antibodies.
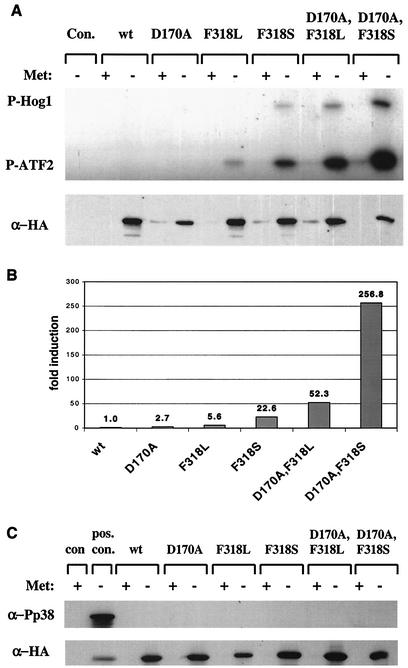
FIG. 5.
Hog1 double mutants immunoprecipitated from pbs2Δ cells are catalytically active in vitro but are not phosphorylated. The experimental procedures for this figure were identical to those described in the legend of Fig. 4, with the exception that they were done with pbs2Δ cells. (A) In vitro kinase assay (upper panel) and Western blot analysis (lower panel) of immunoprecipitated Hog1 proteins. (B) Quantification of above kinase assay. (C) Anti-phospho-p38 and anti-HA Western blot analysis of whole-cell lysates. The positive control (pos. con.) is a lysate extracted from hog1Δ cells carrying Hog1 single mutant D170A 10 min after 1 M salt induction.
As shown in Fig. 4A, Hog1wt proteins immunoprecipitated from hogΔ cells showed no activity. Single Hog1 mutants Hog1D170A, Hog1F318L, and Hog1F318S showed high catalytic activity, as expected (1). Hog1 double mutants immunoprecipitated from hog1Δ cells portrayed very high catalytic activity compared to those of the single mutants (see quantification in Fig. 4B). Also, the double mutants exhibited some degree of autophosphorylation in this assay (Fig. 4A). These very high activities of Hog1D170A,F318L and Hog1D170A,F318S proteins may account for their biological effect.
Having observed the autophosphorylation activity of the double mutants, we measured the steady state levels of phosphorylation of the mutants through Western blot analysis (Fig. 4C). As expected, we were unable to detect any phosphorylation of the Hog1wt protein. Strikingly, however, all mutants (single and double) were found to be highly dually phosphorylated even though the cells were not exposed to osmotic stress (Fig. 4C). To determine whether this phosphorylation is a result of autophosphorylation or rather Pbs2 mediated, we tested phosphorylation levels of the same Hog1 molecules in pbs2Δ cells. As the dual phosphorylation is completely abolished in pbs2Δ cells (Fig. 5C), the phosphorylation observed in hog1Δ cells must be Pbs2 dependent.
The strong and spontaneous Pbs2-dependent phosphorylation of the double mutants is intriguing because the mutants were originally isolated as Pbs2-independent active molecules. All the mutants are indeed biologically active in the absence of Pbs2 (1) (Fig. 2). The question remains whether the catalytic activity of the mutants is dependent on Pbs2 phosphorylation. To address this matter, we measured the activity of the mutants in pbs2Δ cells (Fig. 5).
The activity of the single mutants was low in pbs2Δ cells compared to their activity in hogΔ cells. Yet, their activity was clearly measurable whereas activity of Hog1wt was not. Most importantly, the activity of the dually mutated proteins expressed in pbs2Δ cells was very high (Fig. 5A and B). Given that none of the mutants were dually phosphorylated in this strain (Fig. 5C), it seems to us that spontaneous phosphorylation of the mutants, observed in hog1Δ cells (Fig. 4C), plays no role in establishing the high basal activity of the mutants. This activity is an intrinsic independent property of the active alleles. Pbs2-mediated phosphorylation further enhances this activity, contributing to the differences in activity levels observed when the same alleles are expressed in hog1Δ or pbs2Δ cells. This notion also coincides with the observations that all effects on growth were more severe in hog1Δ cells than in pbs2Δ cells.
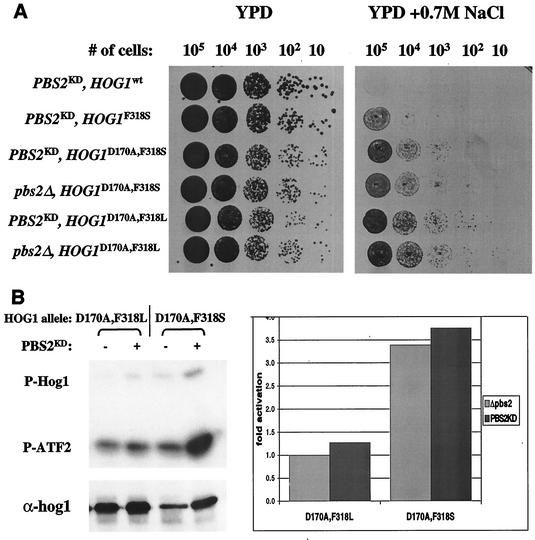
Since the hyperactive HOG1 alleles were not phosphorylated and still showed significant intrinsic catalytic activity in pbs2Δ cells, the possibility remains that their increased activity in hog1Δ cells is also phosphorylation independent. Namely, Pbs2 contributes to their activity (in hog1Δ cells) as a scaffold protein and not as a catalyst (39). To test this possibility, we expressed a “kinase dead” allele of PBS2 (PBS2KD) in pbs2Δ cells that also express the integrated HOG1D170A,F318L or HOG1D170A,F318S genes. If PBS2KD supports an increase in catalytic activity of the double mutants, the relevant clones would not grow under osmotic stress, as was the case for hog1ΔPBS2 cells expressing these double mutants (Fig. 2). Yet, as shown in Fig. 6A, strains expressing PBS2KD and the various HOG1 alleles grew under osmotic stress as well as their parental pbs2Δ cells. Also, catalytic activity of the two doubly mutated hyperactive Hog1 enzymes was not significantly affected by the presence of kinase-dead PBS2 allele (Fig. 6B).
FIG. 6.
Pbs2 scaffold function is not required for the double mutants' biological or catalytic activity. (A) Plasmids bearing HOG1 wild type or mutants expressed under the control of the endogenous HOG1 promoter were integrated as single copies into the genome of SP1pbs2Δ or SP1pbs2KD strains. Cells carrying the integrated genes were grown to logarithmic phase on YPD media, serially diluted to the specified numbers and plated onto either YPD plates (left panel) or YPD plates supplemented with 0.7 M salt (right panel). (B) pbs2Δ cells of the MAY1 strain, containing or lacking an integrated PBS2KD allele, were transformed with MET3-based plasmids harboring either HOG1D170A,F318L or HOG1D170A,F318S. Hog1 proteins were immunoprecipitated and assayed in vitro by using glutathione S-transferase (GST)-ATF2 as a substrate. Immunoprecipitation was performed on lysates prepared from cells grown in the absence of methionine. The upper left panel is an autoradiogram of the in vitro kinase assay. The top bands are autophosphorylated Hog1 proteins, and the bottom bands are phosphorylated GST-ATF2 substrate. The lower left panel shows a Western blot performed with the immunoprecipitated proteins. The right hand panel is the quantification (using a phosphorimager) of the kinase assay blot. The activity of each sample was normalized to its protein level and is shown as increase in activation compared to the HOG1D170A,F318S activity in pbs2Δ cells, which was set as 1.
In summary, when expressed in either pbs2Δ or hog1Δ cells, the double mutants (in particular HOG1D170A,F318S) are significantly more active than the single mutants. This high catalytic activity is well correlated with the growth retardation induced by the double mutants and not by the single mutants (Fig. 3). Catalytic and biological activities are more pronounced in hog1Δ cells, due to further activation by Pbs2, which functions as a catalyst and not as a scaffold protein.
Intrinsic activity of the double mutants is independent of either Thr174 or Tyr176 phosphorylation.
The results presented above show that the double mutants are biologically and catalytically active in pbs2Δ cells in which they are not dually phosphorylated (Fig. 5). Current understanding of MAPK activation suggests that dual phosphorylation at the phosphorylation lip is an absolute requirement for stabilizing an active confomer and increasing catalysis (41, 44). We wished to test whether the double mutants may require phosphorylation of at least one of the phosphoacceptors (Thr174 or Tyr176). We therefore produced HOG1 alleles that carry a mutation in either Thr174 (substituted to Ala) or Tyr176 (changed to Phe) in addition to the two activating mutations.
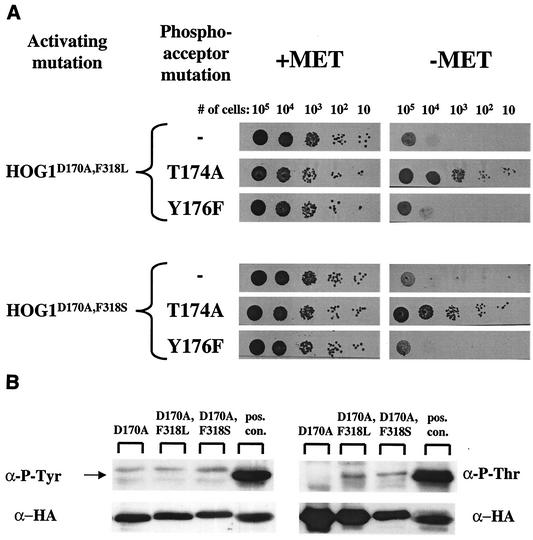
When expressed in hog1Δ cells under the MET3 promoter, HOG1Y176F,D170A,F318L and HOG1Y176F,D170A,F318S induced growth inhibition to the same degree as their parental double mutants (Fig. 7A). It therefore appears that the Tyr176 phosphoacceptor does not play a role in the double mutants' biological effect. To further test this idea, tyrosine phosphorylation was directly tested in HOG1 alleles expressed in pbs2Δ cells. All Hog1 proteins tested were not phosphorylated on tyrosine (Fig. 7B, left panel).
FIG. 7.
The double mutants' intrinsic activity is independent of either Thr174 or Tyr176 phosphorylation. (A) Cells of the hog1Δ strain harboring MET3-based plasmids encoding Hog1 double mutants (activating mutation) with a mutated phosphoacceptor (phosphoacceptor mutation) were used. Cultures were grown in the presence of methionine to logarithmic phase, counted, washed with water, resuspended in inducing (methionine-free) media, serially diluted to the specified numbers, and plated on the indicated plates (+Met or −Met). Plates were scanned 48 h after plating. (B) Western blot analysis of whole-cell lysates using anti-phospho-Tyr (left panel) or of immunoprecipitated Hog1 proteins using anti-phospho-Thr (right panel). The positive control (pos. con.) is a lysate extracted from hog1Δ cells carrying Hog1 single mutant D170A 10 min after 1 M salt induction.
Unlike what was observed with the Tyr176 mutants, HOG1T174A,D170A,F318L and HOG1T174A,D170A,F318S grew normally in the presence or absence of methionine (Fig. 7A), suggesting that the T174A mutation abolished the capability of the double mutants to impair growth. These results strongly suggested that an intact Thr174 is essential for the activity of the mutants and raised the question as to whether this residue is somehow phosphorylated and thereby contributes to the catalytic properties of the mutants. Direct measurement of Thr phosphorylation of the double mutants revealed very low phosphorylation levels (Fig. 7B, right panel). These phosphorylation levels are particularly low when compared to those measured in Hog1 proteins extracted from osmotically challenged cells (Fig. 7B, pos. con. lanes). It could be that the intrinsic specific activity of the few Thr monophosphorylated molecules is so high that this small fraction could account for the properties portrayed by the double mutants. However, we believe that it is more likely that Thr174 serves as an essential structural motif and not as a phosphoacceptor in the active mutants (see also Discussion).
To verify that the detected phospho-Thr is indeed the Thr174 residue, Hog1 proteins carrying activating mutations in addition to the T174A mutation were probed with α-phospho-Thr. No signal was detected (data not shown).
The catalytic activity of the double mutants is further enhanced by osmotic shock.
The observation that addition of salt further enhances the growth retardation effect induced by the double mutants (Fig. 2 and 3D) suggested that salt may increase the catalytic activity of these molecules. To test this idea, various Hog1 proteins were expressed for 2 h (by removal of methionine) in hog1Δ or pbs2Δ cells that were subsequently exposed for 10 min to 1 M NaCl. Hog1 proteins were then immunoprecipitated from cell lysates, and their activity was monitored in vitro (Fig. 8).
FIG. 8.
Activity and phosphorylation of Hog1 double mutants is further enhanced by osmotic stress in hog1Δ cells but not in pbs2Δ cells. hog1Δ or pbs2Δ cells expressing the indicated Hog1 molecules were grown in the absence of methionine for 2 h. Half of each culture was subjected for 10 min to 1 M NaCl. (A) The upper panels show two exposures of the same autoradiogram of a kinase assay performed with the various Hog1 proteins immunoprecipitated from cells treated (+) or untreated (−) with 1 M NaCl. The lower panel shows a Western blot of the immunoprecipitated lysates. (B) Western blot analysis of whole-cell lysates prepared from the indicated strains. The upper panel shows the level of dually phosphorylated Hog1 when anti-phospho-p38 antibodies are used. The lower panel shows the total Hog1 protein levels when anti-HA antibodies are used.
The activity of all Hog1 proteins extracted from hog1Δ cells was further increased in response to salt (Fig. 8A). In addition, a significant increase in their dual phosphorylation level was observed following the osmotic stress (Fig. 8B). When Hog1 proteins were isolated from pbs2Δ cells, neither enhancement of catalytic activity nor increase in dual phosphorylation was observed (Fig. 8A and B). Thus, Pbs2-dependent phosphorylation is responsible for the enhanced, but not the basal, catalytic activity of the mutants. These results confirm the notion that Pbs2 functions on the mutants as a catalyst.
Hog1 double mutants spontaneously induce high expression of target genes.
One of the goals of developing a hyperactive MAPK is the study of its specific downstream targets and biological functions. Therefore, we wished to test whether the active mutants isolated in our studies spontaneously and accurately activate known Hog1 target genes (23, 42, 43). We also wanted to test whether the double mutants induce abnormal levels of these genes in an attempt to explain the growth arrest. To this end, we performed Northern blot analysis, examining the expression of known Hog1 target genes, STL1 (mnemonic for sugar transporter-like protein) and GPD1 (mnemonic for glycerol-3-phosphate dehydrogenase). RNA was prepared from hog1Δ cells containing the different HOG1 alleles at different time points after methionine removal.
The kinetics of STL1 mRNA accumulation following methionine removal showed that expression of the double mutant Hog1D170A,F318S induced very high expression of this gene compared with that of wild-type Hog1 or the single mutant F318S (Fig. 9). Induction of STL1 in cells expressing Hog1wt, Hog1F318S, and Hog1D170A,F318S peaked at 75 min after methionine removal. STL1 mRNA levels in cells expressing the single mutant were about twofold higher than the levels measured in cells expressing wild-type Hog1. In cells expressing the double Hog1 mutant STL1 levels were 3.5-fold higher.
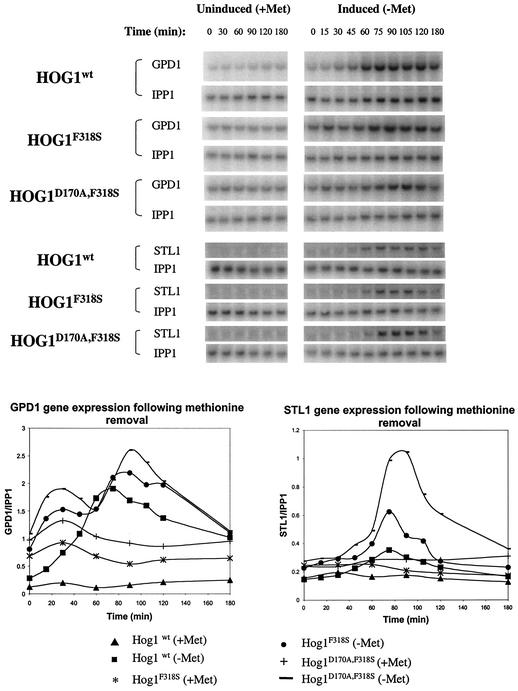
FIG. 9.
The Hog1 double mutant induces high expression of target genes in the absence of osmotic stress. hog1Δ cells harboring plasmids with the indicated HOG1 alleles were grown to logarithmic phase in the presence of methionine. Then, cultures were split and methionine was removed from one half (time zero was after the split and methionine removal). Samples for RNA extraction were taken at the indicated time points. Northern blots (upper panels) were hybridized with 32P-labeled probes corresponding to the Hog1 target genes GPD1, STL1, or the IPP1 gene that was used as a loading control. IPP1 was hybridized to the blots after stripping. Blots were quantified (by phosphorimager) and were normalized to IPP1 levels and plotted against time (lower panel).
GPD1 mRNA showed somewhat different kinetics, but as in the case of STL1 mRNA, it was induced to very high levels when the Hog1 double mutant was expressed (Fig. 9). Interestingly, the hyperactive Hog1 molecules induced high expression of GPD1 in the presence of methionine. This may reflect some leakiness of the methionine-inducible system, the consequence of which can be detected only with the expression of hyperactive Hog1 molecules. The fact that low expression levels of the active Hog1 mutants are sufficient to induce target genes (time zero in Fig. 9) underscores the power of these molecules.
In summary, HOG1 alleles carrying the two mutations are much more effective than HOG1wt or alleles carrying a single mutation in spontaneous induction of Hog1 target genes. These results demonstrate that the intrinsic catalytic activity acquired by the mutants (single and double) is sufficient to activate the entire pathway up to its target genes. Since these events occur in the absence of any osmotic stress, the double mutants could be used to selectively study the biochemistry and biology of Hog1.
The observation that the expression pattern of STL1 peaked and then declined to basal levels in all strains tested may be related to the ability of cells expressing the double mutants to recover after initial growth arrest (Fig. 3B and data not shown). This observation also suggests that the hyperactive Hog1 mutants are permissive, just like Hog1wt, to regulation by the feedback machinery responsible for compromising the HOG1 pathway 1 to 2 h after stimulation with salt.
We also examined transcription of GPD1 in hog1Δ cells that constitutively overexpressed wild-type Hog1 or Hog1F318S from the ADH1 promoter in a 2μm-based plasmid. Double mutants could not be studied using a constitutive promoter, since no colonies were obtained (Table 1). These experiments revealed an unexpected expression pattern. Following exposure to salt, cells expressing wild-type Hog1 manifested the expected fourfold-greater induction of GPD1 mRNA. Cells expressing Hog1F318S, however, failed to induce a significant increase in GPD1 expression (data not shown). These results indicate that when constitutively expressed, the effects of the active Hog1 molecules on Hog1 target genes are negative, probably as part of a long-term adaptation response. It seems, therefore, that for accurately studying the short-term functions of Hog1, inducible expression systems (such as the MET3) should be used in combination with the active mutants.
The growth inhibition effect of the double mutants is not a result of cell death.
The inability of cells expressing the HOG1D170A,F318L and HOG1D170A,F318S alleles to grow normally (Table 1 and Fig. 2 and 3) could result from deleterious effects on essential cellular components that lead to lethality. The observation that growth inhibition is partially reversible (Fig. 3B and data not shown) challenges this possibility. We wished to test this point in more detail and measured the effect of expression of the active mutants on cell viability.
hog1Δ cells harboring a control plasmid or MET3-based plasmids coding for either HOG1WT or HOG1D170A,F318S were grown in the presence of methionine, and samples were taken at different time points from the cultures and stained with PI. Cells were not fixed or permeabilized prior to staining so that only dead cells would be stained (dye exclusion method [19]). Under these experimental conditions, we observed very low levels of PI staining in all cultures (Table 2). Transferring the cultures to media lacking methionine did not change these levels (Table 2), indicating that the very strong activity of the Hog1 variants did not induce lethality. This was also the case for more-prolonged (up to 18 h) expression of the HOG1 alleles (data not shown).
TABLE 2.
Percentage of dead cells (PI positive) following induced expression of HOG1WT or HOG1D170A,F318S
| Plasmid | % of dead cellsa
|
|||
|---|---|---|---|---|
| In presence of methionine | After methionine has been removed for indicated time (h)
|
|||
| 1.5 | 3 | 5 | ||
| Control | 13.3 | 6.4 | 2.6 | 6.1 |
| Coding for | ||||
| HOG1WT | 8.5 | 3.2 | 2.1 | 3.2 |
| HOG1D170A,F318S | 13.2 | 5.5 | 4.2 | 4.5 |
Results are the averages from two independent experiments.
DISCUSSION
This paper described the development of hyperactive variants of the MAPK Hog1. These novel molecules were shown to induce high levels of Hog1 target genes and severe growth inhibition when expressed in yeast cells. These effects, which were obtained in the absence of any external stimulation, show that the alleles studied here, HOG1D170A,F318L and HOG1D170A,F318S, could serve as strong tools for the study of the specific downstream effects of Hog1. Hyperactive variants that induce biological effects have not been described so far for members of the MAPK family. We believe that the knowledge obtained here regarding Hog1 could be applied for producing active forms of other MAPKs (see below).
The extreme hyperactivity of the mutants was obtained through the use of a simple strategy, i.e., combining two mutations previously shown to render Hog1 catalytically active (1) in the same gene. Several of the double mutants produced imposed various degrees of growth defects (Table 1). Some of the double mutants induced growth inhibition only when plated on salt. We believe that these alleles were not intrinsically active at sufficient levels and that further salt induced activation was necessary to impose growth inhibition. However, the HOG1D170A,F318L and HOG1D170A,F318S alleles manifested the most severe effects. The protein products of these two alleles portrayed intrinsic catalytic activity, sufficient to impose growth inhibition on the cells (Fig. 4 to 6). Both alleles carried mutations in D170 and F318. D170 is located at the phosphorylation lip (7, 53, 55), while F318 is located at the L16 domain (53, 55). Current understanding of the mode of activation of MAPKs suggests that MAPKK phosphorylation induces a significant movement of the phosphorylation lip so that it interacts with the L16 domain (7, 12). This hypothesis is based on the three-dimensional structure of ERK2 (active and nonactive) (7, 25) and p38 (8, 53). This interaction is stabilized mainly by hydrogen and ionic bonds between the phosphothreonine at the phosphorylation lip and various residues in L16. It could be that the activating point mutations identified in Hog1 (1) stabilized to a certain degree some interactions between the phosphorylation lip and L16, thereby mimicking the active conformation. Combining such stabilizing mutations may result in even stronger interactions between those domains, increasing the stability of the active conformation. The catalytic activity of Hog1D170A,F318L and Hog1D170A,F318S is indeed significantly higher than that of Hog1D170A, Hog1F318S, or Hog1F318L. The fact that intact Thr174 (but not Tyr176) is essential for activity of the mutants and probably serves as a structural anchor and not a phosphoacceptor (Fig. 6B) strongly supports the idea that the activating mutations stabilize an active conformer.
The significant difference between the activities of the double mutants and the single mutants is best observed in pbs2Δ cells (Fig. 4 and 5). As these cells cannot activate Hog1 alleles, this difference can only stem from an increase in the intrinsic activities of the double mutants. In hog1Δ cells, Pbs2-dependent phosphorylation (Fig. 4 and 8) enhances activity of all hyperactive Hog1 alleles, and therefore the difference between the activities of the double and single mutants is less appreciated.
The observation that in hog1Δ cells all active mutants (but not Hog1wt) are dually phosphorylated in the absence of external stimulation is unexpected. Under these conditions the mutated Hog1 molecules are active, since they possess intrinsic activity, while Pbs2 is not expected to be active. Hence, it could be that the active Hog1 variants induce a positive feedback loop that culminates in activation of Pbs2.
D170 is a residue conserved in many MAPKs including mammalian p38s and ERKs. It would be interesting to test the effect of mutating this residue in other MAPKs. Previous attempts to use the information obtained from the activating Hog1 mutations to produce active human p38α were successful and provided p38α variants that are 70- to 100-fold more active than native p38 in the absence of any upstream phosphorylation (1). The specific activity of the mutated p38α molecules, tested as recombinant proteins produced in Escherichia coli, is, however, about 25% of that of the dually phosphorylated p38α (data not shown). The experience with Hog1 described here suggests that combining several activating mutations into a single p38α cDNA would create a molecule with stronger specific activity. The combinations used here in HOG1 could not be directly applied to p38, as the Phe residue at position 318 in HOG1 is not conserved in p38. For production of hyperactive p38α mutants, other mutations should be tested in combination with the equivalent of D170. Preliminary catalytic results with doubly mutated p38α are promising (data not shown). Phe318 is conserved in members of the ERK family of MAPKs, including ERK1, ERK2, and ERK5. In these molecules it should be possible to combine mutations equivalent to D170A and F318L or F318S used here.
A combination of other mutations was shown to increase specific activity of ERK2 (16). In this report various mutations identified in several genetic screens were combined, and some of the combinations gave rise to ERK2 molecules, which were active in vitro and in vivo. The most significant combination was that of L73P and S151D. These residues are not part of the domains that we found to be important for MAPK activation. The specific activity of those mutated ERK2 molecules was rather low (16), but the data could support the notion that combinations of more than one mutation may be required to produce hyperactive forms of MAPKs.
The catalytic activity of the singly mutated Hog1 is severalfold higher than that of the phosphorylated Hog1WT (1) (Fig. 6). Yet, this activity, which is sufficient to rescue pbs2Δ cells, had only minor effects on cell growth. Only the double mutants that possess even higher activity had a severe effect on growth. It will be interesting to find out whether there are particular substrates or target genes of Hog1 that are responsible for the growth inhibition and whether they are significantly more affected by the double mutants. As Hog1 is activated to different levels and shows different kinetics of activation when exposed to low versus high salt concentrations (51), it could be that the effects of the single and double mutants reflect the two degrees of response.
Growth inhibition following expression of the double mutants is most dramatic on plates and less severe on liquid media (Fig. 2). We believe that the effect on plates is more relevant, because in nature microorganisms usually grow on solid surfaces on which they form multicellular colonies. Several aspects of the physiology, metabolism, and gene expression are different in cells grown on solid surfaces compared to cells grown in liquid (32, 33, 36, 37). On solid media, diffusion of chemicals and movement of cells are limited. As a result cells interact and form colonies with organized anatomy (2, 49, 52). It could be that yeast cells expressing doubly mutated Hog1 produce a growth inhibitory agent (glycerol?) that is less effective if dispersed by shaking and diffusion in liquid media.
Acknowledgments
This study was supported by grants from the Israel Science Foundation, the Israel Cancer Research Fund, and the chief scientist of the ministry of health of the state of Israel. Part of the work was performed at a Marie Curie training site, at the Göteborg Yeast Centre, contract no. QLK3-CT2000-60036.
We thank Irit Marbach for constructing yeast strains, Marcus Krantz for important advice and support, and Yael Friedman, Melanie Grably, Alexander Levitzki, and Ariel Stanhill for comments on the manuscript.
G.Y. and M.B. contributed equally to the study.
REFERENCES
- 1.Bell, M., R. Capone, I. Pashtan, A. Levitzki, and D. Engelberg. 2001. Isolation of hyperactive mutants of the MAPK p38/Hog1 that are independent of MAPK kinase activation. J. Biol. Chem. 276:25351-25358. [DOI] [PubMed] [Google Scholar]
- 2.Ben-Jacob, E., I. Cohen, and D. L. Gutnick. 1998. Cooperative organization of bacterial colonies: from genotype to morphotype. Annu. Rev. Microbiol. 52:779-806. [DOI] [PubMed] [Google Scholar]
- 3.Bott, C. M., S. G. Thorneycroft, and C. J. Marshall. 1994. The sevenmaker gain-of-function mutation in p42 MAP kinase leads to enhanced signalling and reduced sensitivity to dual specificity phosphatase action. FEBS Lett. 352:201-205. [DOI] [PubMed] [Google Scholar]
- 4.Brewster, J., T. De Valoir, N. Dwyer, E. Winter, and M. Gustin. 1993. An osmosensing signal transduction pathway in yeast. Science 259:1760-1763. [DOI] [PubMed] [Google Scholar]
- 5.Brill, J. A., E. A. Elion, and G. R. Fink. 1994. A role for autophosphorylation revealed by activated alleles of FUS3, the yeast MAP kinase homolog. Mol. Biol. Cell. 5:297-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunner, D., N. Oellers, J. Szabad, W. H. Biggs III, S. L. Zipursky, and E. Hafen. 1994. A gain-of-function mutation in Drosophila MAP kinase activates multiple receptor tyrosine kinase signaling pathways. Cell 76:875-888. [DOI] [PubMed] [Google Scholar]
- 7.Canagarajah, B. J., A. Khokhlatchev, M. H. Cobb, and E. J. Goldsmith. 1997. Activation mechanism of the MAP kinase ERK2 by dual phosphorylation. Cell 90:859-869. [DOI] [PubMed] [Google Scholar]
- 8.Chang, C. I., B. E. Xu, R. Akella, M. H. Cobb, and E. J. Goldsmith. 2002. Crystal structures of MAP kinase p38 complexed to the docking sites on its nuclear substrate MEF2A and activator MKK3b. Mol. Cell 9:1241-1249. [DOI] [PubMed] [Google Scholar]
- 9.Cherest, H., P. Kerjan, and Y. Surdin-Kerjan. 1987. The Saccharomyces cerevisiae MET3 gene: nucleotide sequence and relationship of the 5′ non-coding region to that of MET25. Mol. Gen. Genet. 210:307-313. [DOI] [PubMed] [Google Scholar]
- 10.Cherest, H., N. T. Nguyen, and Y. Surdin-Kerjan. 1985. Transcriptional regulation of the MET3 gene of Saccharomyces cerevisiae. Gene 34:269-281. [DOI] [PubMed] [Google Scholar]
- 11.Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110:119-122. [DOI] [PubMed] [Google Scholar]
- 12.Cobb, M. H., and E. J. Goldsmith. 2000. Dimerization in MAP-kinase signaling. Trends Biochem. Sci. 25:7-9. [DOI] [PubMed] [Google Scholar]
- 13.Colicelli, J., B. C., T. Michaeli, K. O'Neill, M. Riggs, and M. Wigler. 1989. Isolation and characterization of a mammalian gene encoding a high-affinity cAMP phophodiesterase. Proc. Natl. Acad. Sci. USA 86:3599-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cowley, S., H. Paterson, P. Kemp, and C. J. Marshall. 1994. Activation of MAP kinase kinase is necessary and sufficient for PC12 differentiation and for transformation of NIH 3T3 cells. Cell 77:841-852. [DOI] [PubMed] [Google Scholar]
- 15.Davis, R. J. 2000. Signal transduction by the JNK group of MAP kinases. Cell 103:239-252. [DOI] [PubMed] [Google Scholar]
- 16.Emrick, M. A., A. N. Hoofnagle, A. S. Miller, L. F. Ten Eyck, and N. G. Ahn. 2001. Constitutive activation of extracellular signal-regulated kinase 2 by synergistic point mutations. J. Biol. Chem. 276:46469-46479. [DOI] [PubMed] [Google Scholar]
- 17.English, J., G. Pearson, J. Wilsbacher, J. Swantek, M. Karandikar, S. Xu, and M. H. Cobb. 1999. New insights into the control of MAP kinase pathways. Exp. Cell Res. 253:255-270. [DOI] [PubMed] [Google Scholar]
- 18.Galcheva-Gargova, Z., B. Derijard, I. H. Wu, and R. J. Davis. 1994. An osmosensing signal transduction pathway in mammalian cells. Science 265:806-808. [DOI] [PubMed] [Google Scholar]
- 19.Green, L. J., P. Marder, L. L. Mann, L. C. Chio, and W. L. Current. 1999. LY303366 exhibits rapid and potent fungicidal activity in flow cytometric assays of yeast viability. Antimicrob. Agents Chemother. 43:830-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gustin, M. C., J. Albertyn, M. Alexander, and K. Davenport. 1998. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62:1264-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall, J. P., V. Cherkasova, E. Elion, M. C. Gustin, and E. Winter. 1996. The osmoregulatory pathway represses mating pathway activity in Saccharomyces cerevisiae: isolation of a FUS3 mutant that is insensitive to the repression mechanism. Mol. Cell. Biol. 16:6715-6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han, J., J. D. Lee, L. Bibbs, and R. J. Ulevitch. 1994. A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science 265:808-811. [DOI] [PubMed] [Google Scholar]
- 23.Hohmann, S. 2002. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66:300-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ip, Y. T., and R. J. Davis. 1998. Signal transduction by the c-Jun N-terminal kinase (JNK)—from inflammation to development. Curr. Opin. Cell Biol. 10:205-219. [DOI] [PubMed] [Google Scholar]
- 25.Khokhlatchev, A. V., B. Canagarajah, J. Wilsbacher, M. Robinson, M. Atkinson, E. Goldsmith, and M. H. Cobb. 1998. Phosphorylation of the MAP kinase ERK2 promotes its homodimerization and nuclear translocation. Cell 93:605-615. [DOI] [PubMed] [Google Scholar]
- 26.Kyriakis, J. M., and J. Avruch. 2001. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol. Rev. 81:807-869. [DOI] [PubMed] [Google Scholar]
- 27.Lewis, T. S., P. S. Shapiro, and N. G. Ahn. 1998. Signal transduction through MAP kinase cascades. Adv. Cancer Res. 74:49-139. [DOI] [PubMed] [Google Scholar]
- 28.Maeda, T., M. Takekawa, and H. Saito. 1995. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science 269:554-558. [DOI] [PubMed] [Google Scholar]
- 29.Maeda, T., A. Y. Tsai, and H. Saito. 1993. Mutations in a protein tyrosine phosphatase gene (PTP2) and a protein serine/threonine phosphatase gene (PTC1) cause a synthetic growth defect in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:5408-5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mansour, S. J., W. T. Matten, A. S. Hermann, J. M. Candia, S. Rong, K. Fukasawa, G. F. Vande Woude, and N. G. Ahn. 1994. Transformation of mammalian cells by constitutively active MAP kinase kinase. Science 265:966-970. [DOI] [PubMed] [Google Scholar]
- 31.Marshall, C. J. 1994. MAP kinase kinase kinase, MAP kinase kinase and MAP kinase. Curr. Opin. Genet. Dev. 4:82-89. [DOI] [PubMed] [Google Scholar]
- 32.Mendelson, N. H., and B. Salhi. 1996. Patterns of reporter gene expression in the phase diagram of Bacillus subtilis colony forms. J. Bacteriol. 178:1980-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meunier, J. R., and M. Choder. 1999. Saccharomyces cerevisiae colony growth and ageing: biphasic growth accompanied by changes in gene expression. Yeast 15:1159-1169. [DOI] [PubMed] [Google Scholar]
- 34.Mumberg, D., R. Muller, and M. Funk. 1994. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22:5767-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ono, K., and J. Han. 2000. The p38 signal transduction pathway activation and function. Cell. Signal. 12:1-13. [DOI] [PubMed] [Google Scholar]
- 36.Palkova, Z., F. Devaux, M. Icicova, L. Minarikova, S. Le Crom, and C. Jacq. 2002. Ammonia pulses and metabolic oscillations guide yeast colony development. Mol. Biol. Cell 13:3901-3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palkova, Z., B. Janderova, J. Gabriel, B. Zikanova, M. Pospisek, and J. Forstova. 1997. Ammonia mediates communication between yeast colonies. Nature 390:532-536. [DOI] [PubMed] [Google Scholar]
- 38.Pearson, G., F. Robinson, T. Beers Gibson, B. E. Xu, M. Karandikar, K. Berman, and M. H. Cobb. 2001. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr. Rev. 22:153-183. [DOI] [PubMed] [Google Scholar]
- 39.Posas, F., and H. Saito. 1997. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science 276:1702-1705. [DOI] [PubMed] [Google Scholar]
- 40.Posas, F., S. M. Wurgler-Murphy, T. Maeda, E. A. Witten, T. C. Thai, and H. Saito. 1996. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component”osmosensor. Cell 86:865-875. [DOI] [PubMed] [Google Scholar]
- 41.Prowse, C. N., M. S. Deal, and J. Lew. 2001. The complete pathway for catalytic activation of the mitogen-activated protein kinase, ERK2. J. Biol. Chem. 276:40817-40823. [DOI] [PubMed] [Google Scholar]
- 42.Rep, M., M. Krantz, J. M. Thevelein, and S. Hohmann. 2000. The transcriptional response of Saccharomyces cerevisiae to osmotic shock. Hot1p and Msn2p/Msn4p are required for the induction of subsets of high osmolarity glycerol pathway-dependent genes. J. Biol. Chem. 275:8290-8300. [DOI] [PubMed] [Google Scholar]
- 43.Rep, M., V. Reiser, U. Gartner, J. M. Thevelein, S. Hohmann, G. Ammerer, and H. Ruis. 1999. Osmotic stress-induced gene expression in Saccharomyces cerevisiae requires Msn1p and the novel nuclear factor Hot1p. Mol. Cell. Biol. 19:5474-5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robbins, D. J., E. Zhen, H. Owaki, C. A. Vanderbilt, D. Ebert, T. D. Geppert, and M. H. Cobb. 1993. Regulation and properties of extracellular signal-regulated protein kinases 1 and 2 in vitro. J. Biol. Chem. 268:5097-5106. [PubMed] [Google Scholar]
- 45.Robinson, M. J., and M. H. Cobb. 1997. Mitogen-activated protein kinase pathways. Curr. Opin. Cell Biol. 9:180-186. [DOI] [PubMed] [Google Scholar]
- 46.Robinson, M. J., S. A. Stippec, E. Goldsmith, M. A. White, and M. H. Cobb. 1998. A constitutively active and nuclear form of the MAP kinase ERK2 is sufficient for neurite outgrowth and cell transformation. Curr. Biol. 8:1141-1150. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 48.Schaeffer, H. J., and M. J. Weber. 1999. Mitogen-activated protein kinases: specific messages from ubiquitous messengers. Mol. Cell. Biol. 19:2435-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scherz, R., V. Shinder, and D. Engelberg. 2001. Anatomical analysis of Saccharomyces cerevisiae stalk-like structures reveals spatial organization and cell specialization. J. Bacteriol. 183:5402-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schiestl, R. H., and R. D. Gietz. 1989. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16:339-346. [DOI] [PubMed] [Google Scholar]
- 51.Van Wuytswinkel, O., V. Reiser, M. Siderius, M. C. Kelders, G. Ammerer, H. Ruis, and W. H. Mager. 2000. Response of Saccharomyces cerevisiae to severe osmotic stress: evidence for a novel activation mechanism of the HOG MAP kinase pathway. Mol. Microbiol. 37:382-397. [DOI] [PubMed] [Google Scholar]
- 52.Varon, M., and M. Choder. 2000. Organization and cell-cell interaction in starved Saccharomyces cerevisiae colonies. J. Bacteriol. 182:3877-3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, Z., P. C. Harkins, R. J. Ulevitch, J. Han, M. H. Cobb, and E. J. Goldsmith. 1997. The structure of mitogen-activated protein kinase p38 at 2.1-A resolution. Proc. Natl. Acad. Sci. USA 94:2327-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Widmann, C., S. Gibson, M. B. Jarpe, and G. L. Johnson. 1999. Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol. Rev. 79:143-180. [DOI] [PubMed] [Google Scholar]
- 55.Wilson, K. P., M. J. Fitzgibbon, P. R. Caron, J. P. Griffith, W. Chen, P. G. McCaffrey, S. P. Chambers, and M. S. Su. 1996. Crystal structure of p38 mitogen-activated protein kinase. J. Biol. Chem. 271:27696-27700. [DOI] [PubMed] [Google Scholar]
- 56.Zheng, C., J. Xiang, T. Hunter, and A. Lin. 1999. The JNKK2-JNK1 fusion protein acts as a constitutively active c-Jun kinase that stimulates c-Jun transcription activity. J. Biol. Chem. 274:28966-28971. [DOI] [PubMed] [Google Scholar]