Abstract
The hypoxia-inducible factors 1α (HIF-1α) and 2α (HIF-2α) have extensive structural homology and have been identified as key transcription factors responsible for gene expression in response to hypoxia. They play critical roles not only in normal development, but also in tumor progression. Here we report on the differential regulation of protein expression and transcriptional activity of HIF-1α and -2α by hypoxia in immortalized mouse embryo fibroblasts (MEFs). We show that oxygen-dependent protein degradation is restricted to HIF-1α, as HIF-2α protein is detected in MEFs regardless of oxygenation and is localized primarily to the cytoplasm. Endogenous HIF-2α remained transcriptionally inactive under hypoxic conditions; however, ectopically overexpressed HIF-2α translocated into the nucleus and could stimulate expression of hypoxia-inducible genes. We show that the factor inhibiting HIF-1 can selectively inhibit the transcriptional activity of HIF-1α but has no effect on HIF-2α-mediated transcription in MEFs. We propose that HIF-2α is not a redundant transcription factor of HIF-1α for hypoxia-induced gene expression and show evidence that there is a cell type-specific modulator(s) that enables selective activation of HIF-1α but not HIF-2α in response to low-oxygen stress.
Animals respond to low oxygen tension by increasing the transcription of a number of genes, including many of those involved in glycolysis, oxygen delivery, and vasculogenesis (3). A master intracellular regulator responsible for induction of these genes is the hypoxia-inducible factor (HIF), a heterodimeric transcription factor consisting of HIF-α and -β (40). The first isoform of HIF-α, HIF-1α, was originally discovered as a high-affinity DNA binding protein localized to the 3′ hypoxia-responsive element (HRE) of the erythropoietin gene (45). Two additional HIF-α subunits have subsequently been cloned and named HIF-2α (also EPAS-2, HLF, HRF, and MOP2) (5, 7, 15, 43) and HIF-3α (10). HIF-1α and -2α have high sequence identity and similar organization of their functional domains; both contain basic helix-loop-helix and proline active site (PAS) domains in their N termini as well as two transcription activation domains and an inhibitory domain in their C termini (5, 43).
Biochemical analyses with ectopic overexpression and in vitro DNA binding analysis have demonstrated that HIF-1α and HIF-2α can function as indistinguishable transcriptional factors that enhance the expression of the same sets of target genes (43, 47). One prominent difference between HIF-1α and -2α is observed in their spatial expression patterns. HIF-1α is believed to be a universal master regulator for hypoxia-inducible gene expression along with its partner, HIF-1β, as they are expressed in a wide range of cell types (46). In contrast, HIF-2α transcription is highest in certain tissues; the highest expression of HIF-2α mRNA is observed in alveolar epithelial cells in lung, but the mRNA is also seen in endothelial cells of various tissues, such as brain, heart, kidney, and liver (5, 7). HIF-1α−/− embryos die before embryonic day 11.5 (E11.5) and display defects in neural fold formation, cephalic vascularization, and the cardiovascular system, whereas HIF-2α−/− embryos die between E13.5 and E16.5 and sometimes survive postnatally (19, 34, 36, 44). These results suggest that the two HIF-α isoforms play separate but essential roles during embryonic development. Moreover, a number of studies have shown that inactivation of HIF-1α completely abolishes induction of HIF target genes (2, 36). One possible explanation is a recent suggestion that HIF-2α is primarily responsible for hypoglycemia-induced gene expression (2).
It is well documented that HIF-1 activity is determined primarily by the protein stability of the α subunits. HIF-α subunits are constitutively expressed at the mRNA level, but their proteins are usually present at low levels in normoxia due to oxygen-dependent ubiquitination, leading to protein degradation by the 26S proteasome (16, 22). Hydroxylation on specific proline residues (Pro-402 and Pro-564) within the oxygen-dependent degradation domain (ODD) of HIF-α proteins is catalyzed by several isoforms of proline hydroxylases in normoxia and is the rate-limiting step for interaction with the von Hippel-Lindau protein (VHL)-elongin B and C-Cullin 2 complex (VBC); this interaction leads to ubiquitination and protein degradation (6, 18, 20, 21). HIF-α E3 ligase activity can be reconstituted with purified VHL, elongin B and C, and Cullin-2 proteins (24). However, it is still unknown exactly how many proteins compose the actual HIF-α E3 ligase complex within the cell, as an increasing number of proteins are being identified as constituents of VBC complexes (4, 12). For instance, recent work has shown that interaction of HIF-α subunits with VHL is mediated by two separate domains within the ODD, each containing a conserved proline residue (Pro-402 in the N-terminal ODD and Pro-564 in the C-terminal ODD) for enzyme-mediated hydroxylation (32). The two domains require different components for efficient interaction with VHL and subsequent ubiquitination. Evidence for this comes from the demonstration that additional factors are needed for interaction of the N-terminal ODD with VHL; these are present in RCC4 cell extracts but not in reticulocyte lysates (32).
HIF-α subunits have a conserved asparagine residue (Asn-803) within the C-terminal activation domain. This residue is hydroxylated by an asparaginyl hydroxylase, termed factor inhibiting HIF-1 (FIH-1), under normoxic conditions (27, 28). Dehydroxylation of this site enhances recruitment of the p300/CBP coactivator, a critical component for HIF-dependent transcriptional activation (27, 31). Decreasing cellular oxygen levels lead to a retardation of hydroxylation on the proline residues in the ODD and on the asparagine residue in the C-terminal activation domain. By this mechanism, intracellular oxygen concentrations can control both the expression level and transcriptional activity of HIF-α proteins.
HIF-1α and -2α have been used interchangeably for in vitro hydroxylation analysis, and there appears to be no difference between them in terms of oxygen-dependent regulation in published studies (6, 18, 20, 27). Here, we report on cell type-specific differential regulation of protein stabilization and transcriptional activation of endogenous HIF-1α and -2α in response to hypoxia in genetically manipulated mouse embryo fibroblasts (MEFs). We show that hypoxia exclusively regulates the expression and transcriptional activity of HIF-1α in these cells. We demonstrate that endogenous HIF-2α is expressed at constant levels regardless of oxygenation and is primarily localized to the cytoplasm. In MEFs, endogenous HIF-2α is not capable of stimulating transcription of the known HIF-1 target genes phosphoglycerate kinase (PGK), Glut-1, and vascular endothelial growth factor (VEGF) in response to hypoxia or hypoglycemia; however, transiently overexpressed HIF-2α has potent transcriptional activity on these targets. We show that the inhibitory effect of FIH-1 overexpression is restricted to the transcriptional activity of HIF-1α and does not affect HIF-2α. These data suggest that activation of HIF-2α signaling pathways is determined by control elements other than intracellular oxygen concentration and that these differ from the factors controlling HIF-1α function.
MATERIALS AND METHODS
Materials.
Cobalt chloride, deferoxamine mesylate, sodium azide, sodium orthovanadate, insulin, actinomycin D, and MG-132 were purchased from Sigma-Aldrich (St. Louis, Mo.). Geldanamycin was from Calbiochem (San Diego, Calif.).
Cell cultures.
The immortalized wild-type and HIF-1α null MEFs were prepared as previously described (37). To generate HIF-2α null MEFs, embryos were collected at E12.5 from HIF-2α+/− heterozygous crosses (44). Each embryo was genotyped separately for the experiment. VHL null MEFs were generated from conditionally targeted mouse embryos homozygous for the VHL conditional allele (11) and infected with adenovirus expressing Cre recombinase as previously described (37). Unless otherwise noted, cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, Calif.) with high glucose, supplemented with 10% fetal bovine serum, 100 U of penicillin, and 100 μg of streptomycin per ml of medium. For hypoxic treatment, cells were exposed to 10% CO2 and 0.5% O2 balanced with N2 in a Sanyo 3 gas incubator.
Plasmids.
Murine HIF-2α cDNA was amplified from C57BL/6 mouse brain cDNAs with the forward primer 5′-TCGGAGGGCCACGGCGACAATGACAG-3′,and the reverse primer 5′-AAGTGAAGCTGGCAGGTCAAGACGGC-3′ and cloned into the pGEM-T vector. A mammalian expression vector for Myc-tagged HIF-2α was generated by subcloning of HIF-2α cDNA into the pcDNA3.1/Myc-His plasmid (Invitrogen). The correct HIF-2α cDNA was confirmed by sequencing the entire coding sequence.
A luciferase reporter with the p35srj promoter (pGL-SRJ) was generated by ligation of double-stranded oligonucleotides (sense sequence, 5′-CTAGCTAGCTAGGTGTGCGCGTGGTGCCATACGGGACGTGCAGCTACGTGCCCACCCCGCTCGAGCGG-3′; the hypoxia response element is underlined) into the pGL3 promoter after digestion with NheI and XhoI (1). The DNA encoding the C terminus of HIF-2α was amplified with the forward primer 5′-CCCCTTAAGGCCCCCACCCCAGGAGAT-3′ and the reverse primer 5′-TGCTCTAGAGCATCAGGTGGCCTGGTCCAGAGC-3′, digested with AflII and XbaI, and then ligated into the similarly digested p(HA)HIF-1α to generate p(HA)HIF1N2C.
To generate the expression vector for the fusion protein of the N terminus of HIF-2α and the C terminus of HIF-1α, p(Flag)HIF2N1C, the HIF-2α N-terminal coding sequence was amplified with the forward primer 5′-CGGGGTACCCCGATGGACTACAAGGACGACGATGACAAGATGACAGCTGACAAGGAG-3′ and the reverse primer 5′-CCCCTTAAGC-AACTGGGCCAGTTCCTCG, digested with KpnI and AflII, and ligated into the similarly digested p(HA)HIF-1α. The mammalian expression vector for FIH-1 was generated by cloning PCR products with the primer set 5′-CCGCTCGAGATGGCGGCGACGGCAGCC-3′ and 5′-CGGGGTACCGTTGTAACGGCCTTTAATCATG-3′ from mouse brain cDNA into the pcDNA3.1/Myc-His vector.
Western blot analysis.
Cells were harvested in ice-cold phosphate-buffered saline and centrifuged down to the pellet, which was frozen in liquid nitrogen and transferred into a −80°C freezer until needed. The cytoplasmic fraction was obtained from the supernatant of cell lysates in hypotonic lysis buffer (20 mM HEPES [pH 7.9], 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol, 1 mM dithiothreitol, 1 mM Na3VO4, 10 mM NaF, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktails). The nuclear pellet was resuspended in high-salt buffer (hypotonic lysis buffer with 0.42 M NaCl) and extracted for 20 min on ice with occasional vortexing. Nuclear extracts were obtained from the supernatant after centrifugation. A total of 50 μg (for whole-cell extract) or 25 μg (for either the cytoplasmic or nuclear extract) of protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by Western blotting with a mouse anti-HIF-1α or rabbit anti-HIF-2α antibody (Novus Biologicals, Littleton, Colo.). Typically, the transferred membrane was probed with anti-HIF-2α antibody and then the same membrane was reprobed with anti-HIF-1α. The specificity of polyclonal anti-HIF-2α antibody was confirmed with mouse HIF-2α expressed by with the T7-coupled TNT in vitro transcription and translation system (Promega, Madison, Wis.).
Reverse transcription and quantitative PCR.
First-strand cDNA was synthesized from 1 μg of total RNA isolated with Trizol reagent (Invitrogen) by the SuperScript first-strand synthesis system (Invitrogen) with the oligo(dT) primer. For real-time PCR analysis, the diluted cDNAs were amplified in TaqMan Universal PCR Master Mix by with the ABI Prism 7700 sequence detector (PE Applied Biosystems, Branchburg, N.J.) with the following primer and probe sets: for PGK, the primer sequences were CAAATTTGATGAGAATGCCAAGACT and TTCTTGCTGCTCTCAGTACCACA and the probe sequence was 6-carboxyfluorescein (FAM) TATACCTGCTGGCTGGATGGGCTTGGACT-black hole quencher (BHQ); for Glut-1, the primer sequences were GGGCATGTGCTTCCAGTATGT and ACGAGGAGCACCGTGAAGAT and the probe sequence was 6-FAM-CAACTGTGCGGCCCCTACGTCTTC-BHQ; for VEGF, the primer sequences were AGTCCCATGAAGTGATCAAGTTCA and ATCCGCATGATCTGCATGG and the probe sequence was 6-FAM-TGCCCACGTCAGAGAGCAACATCAC-BHQ; and for hypoxanthine phosphoribosyltransferase (HPRT), the primer sequences were TTATCAGACTGAAGAGCTACT and TTACCAGTGTCAATTATATCTTCAACAATC and the probe sequence was 6-FAM-TGAGAGATCATCTCCACCAATAACTTTTATGTCC-BHQ.
Transient transfection for reporter gene assay.
The hypoxia response element-luciferase (HRE-Luc) reporter gene (41) was transfected into MEFs with FuGene6 (Roche, Indianapolis, Ind.) according to the manufacturer's protocols along with pRL-CMV for normalization (Promega). After recovery from transfection, the cells were treated with either hypoxia or no-glucose medium for 22 h. Firefly and Renilla luciferase activities were measured with the dual luciferase assay system (Promega).
Immunofluorescence analysis.
Cells were grown on Lab-Tek II chamber slides (Nalge Nunc International, Naperville, Ill.) overnight and then treated with either normoxia or 0.5% O2 for 4 h. At the end of the treatment, the cells were washed three times with ice-cold phosphate-buffered saline, fixed with methanol, and probed with a 1:250 dilution of polyclonal anti-HIF-2α (Novus). The primary antibody was located with fluorescein isothiocyanate-labeled goat anti-rabbit immunoglobulin G (Vector Laboratories, Inc., Burlingame, Calif.).
RESULTS
Constitutive expression of HIF-2α in mouse embryo fibroblasts.
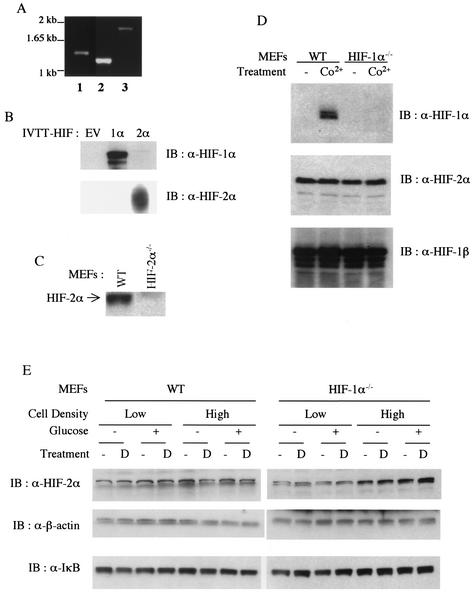
Northern blot analyses have shown that induction of most hypoxia-responsive genes is abolished in HIF-1α null MEFs (37). This might result from a lack of expression of HIF-2α in these fibroblasts. In order to confirm expression of HIF-2α, expression of its mRNA was verified by reverse transcription-PCR, and the resulting product was sequenced to ensure the presence of wild-type transcript (Fig. 1A). Expression of HIF-2α protein was examined by Western blot analysis with polyclonal anti-HIF-2α antibody. The specificity of this antibody was confirmed by immunoblot analysis with in vitro-transcribed and -translated HIF-2α (Fig. 1B) and whole-cell extracts from wild-type and HIF-2α null MEFs (44) (Fig. 1C). Similar levels of HIF-2α protein were then detected in whole-cell extracts of wild-type and HIF-1α null MEFs (Fig. 1D). Surprisingly, expression of HIF-2α protein was evident under normoxic conditions regardless of the presence of HIF-1α, and cobalt chloride treatment did not further increase its level, although HIF-1α was upregulated in cobalt-treated wild-type MEFs (Fig. 1D). HIF-1β (also called ARNT) is also expressed in these cells, and its expression was not affected by cobalt chloride or the presence of HIF-1α, in agreement with published data (Fig. 1D, bottom panel).
FIG. 1.
Constitutive expression of HIF-2α in T-antigen-transformed mouse embryonic fibroblasts. (A) Reverse transcription-PCR analysis of HIF-2α mRNA in MEFs. Reverse-transcribed first-strand cDNAs from wild-type (WT) MEFs were amplified with primer sets covering the 5′ half (lanes 1 and 2) and the 3′ half (lane 3) of the HIF-2α open reading frame. (B and C) Specificity of mouse monoclonal anti-HIF-1α and rabbit polyclonal anti-HIF-2α antibodies. Polyclonal anti-HIF-2α shows specificity to in vitro-transcribed and -translated (IVTT) mouse HIF-2α (B) and endogenous HIF-2α which is present in wild-type but absent from HIF-2α null cells (C). EV, empty vector. (D) Western blot (IB) analysis of whole-cell extracts of untreated (lanes 1 and 3) and CoCl2-treated (100 μM, 4 h; lanes 2 and 4) wild-type (lanes 1 and 2) and HIF-1α null MEFs (lanes 3 and 4) with anti-HIF-1α (upper), anti-HIF-2α (middle), and anti-ARNT (bottom) antibodies. (E) Effect of glucose deprivation on HIF-2α expression in MEFs at two different seeding densities. Both wild-type and HIF-1α null MEFs were incubated in either high-glucose (4.5 g/liter) or glucose-free Dulbecco's modified Eagle's medium with or without deferoxamine (100 μM; lanes D) for 4 h before being harvested for whole-cell extraction.
It has been reported that normoxic HIF-2α expression levels in HeLa cells at or near confluence is much higher than that in cells seeded at low density (47). Thus, we investigated whether inducible expression of HIF-2α depends on cell density in MEFs. Figure 1E shows that HIF-2α induction was similar regardless of cell density, although cells at higher densities expressed more HIF-2α. Taken together, these data suggest that MEFs express significant amounts of HIF-2α message and protein and that expression is not regulated by hypoxia mimetics in these cells.
Lack of induction of HIF target genes in response to hypoxia or hypoglycemia in HIF-1α null cells.
Recently, Brusselmans et al. suggested that HIF-1α and -2α have different roles in stimulating expression of hypoxia-inducible genes in response to hypoxia or hypoglycemia (2). They found that in murine embryonic stem (ES) cells, glucose deprivation increased the expression of a number of hypoxia-inducible genes and determined that HIF-2α plays a role in hypoglycemia-induced expression of Glut-1 and VEGF (2). To determine if these findings also held in MEF cells, we examined the effect of glucose deprivation on HIF-2α expression in these cells. Interestingly, HIF-2α protein expression was not changed by glucose deprivation in MEFs (Fig. 1C).
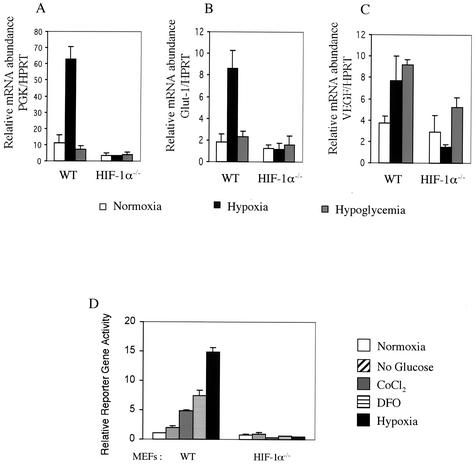
Steady-state mRNA levels from each cell type were analyzed quantitatively by real-time PCR of reverse-transcribed cDNAs. Hypoxia did increase the mRNA expression of three genes examined in wild-type MEFs, PGK, Glut-1, and VEGF (Fig. 2A to C). This induction pattern was totally abolished in HIF-1α null cells, confirming that HIF-1α is the only functional transcription factor for these hypoxia-inducible genes in this cell type. In contrast to ES cells, hypoglycemia had no effect on the expression of PGK and Glut-1 in either wild-type or HIF-1α null MEFs (Fig. 2A and B). The only gene that we found to be induced by hypoglycemia was that for VEGF, which was increased by incubation with glucose-free medium twofold in wild-type MEFs but not in HIF-1α null cells (Fig. 2C). These data indicate that neither hypoxia nor hypoglycemia stimulates HIF-2α signaling pathways to accomplish HIF target gene induction in MEFs.
FIG. 2.
Lack of transcriptional activity of constitutively expressed endogenous HIF-2α in MEFs in response to hypoxia or hypoglycemia. (A to C) Hypoxia-inducible gene expression profile in wild-type (WT) and HIF-1α null MEFs. Total RNAs were prepared from cells incubated in normoxia, hypoxia (0.5% O2), or hypoglycemia conditions for 24 h for reverse transcription. The first-strand cDNAs were then amplified with primers and TaqMan-labeled probes for PGK (A), Glut-1 (B), and VEGF (C) in real-time PCR analysis. Each signal was normalized against that for HPRT and is shown on the y axis (bars represent the standard deviation of triplicate analyses). (D) HRE reporter gene assay in transiently transfected MEFs. MEFs were transfected with the HRE-luciferase reporter and pRL-CMV and treated with normoxia, glucose-free medium, 100 μM cobalt chloride, 100 μM deferoxamine, or hypoxia (0.5% O2) for 22 h before being harvested. The y axis shows normalized firefly luciferase over Renilla luciferase activity relative to the wild-type normoxic response (bars represent the standard deviation of triplicate analyses).
The role of endogenous HIF-2α in transcriptional activation was further investigated by a transient-transfection assay with an HRE reporter gene (pHRE-Luc), which has six tandem repeats of HIF-1 binding sequences from the VEGF promoter (41). HRE-mediated transcription was activated by cobalt chloride, iron chelators, and hypoxia in wild-type cells; none of these induced HRE reporter gene activity when HIF-1α was inactivated (Fig. 2D). Consistent with our mRNA analyses of VEGF, glucose deprivation resulted in only a twofold induction of HRE reporter gene activity; this was also abolished in HIF-1α null cells. Taken together, these data suggest that HIF-1α is regulated by the intracellular oxygen concentration, whereas endogenous HIF-2α does not participate in hypoxic responses through gene induction despite its significant expression in MEFs. Furthermore, hypoglycemia itself does not initiate intracellular signaling pathways involving HIF-2α in MEFs.
Endogenous HIF-2α is compartmentalized in the cytoplasm of MEFs regardless of oxygen level.
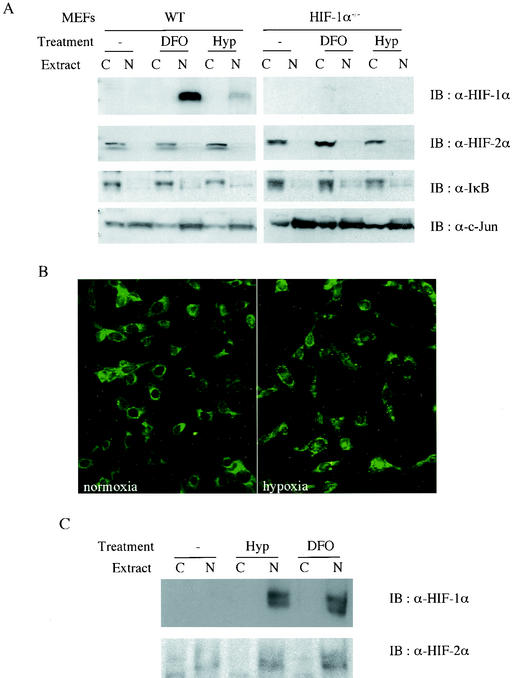
In order to elucidate the mechanism by which these immortalized MEFs keep endogenous HIF-2α inactive, we decided to examine each step of the intracellular signaling pathway for HIF-α activation. First, the intracellular location of HIF-2α was determined by subcellular fractionation of MEFs. HIF-1α was localized exclusively to nuclear extracts of wild-type MEFs after stabilization by hypoxia or an iron chelator (Fig. 3A, upper panel). Surprisingly, however, endogenous HIF-2α was detected only in cytoplasmic extracts of these cells, and exposure to hypoxia or an iron chelator was unable to translocate HIF-2α into the nucleus (Fig. 3A, lower panel).
FIG. 3.
Cytoplasmic localization of endogenous HIF-2α in MEFs even after hypoxia treatment. Subcellular fractionation of MEFs (A) and 293 cells (C). Cells were treated or not with either 0.5% O2 hypoxia or 100 μM deferoxamine for 4 h before being harvested. (B) Immunofluorescence analysis of endogenous HIF-2α in MEFs. Wild-type MEFs were grown on the glass slide chamber and treated or not with 0.5% O2 for 4 h before fixation. HIF-2α was probed with polyclonal anti-HIF-2α antibody and detected with fluorescein isothiocyanate-labeled anti-rabbit immunoglobulin antibody.
The intracellular distribution of endogenous HIF-2α was also investigated by immunofluorescence analysis with anti-HIF-2α antibody, which demonstrated that HIF-2α immunoreactivity was detected mainly in the cytoplasm of MEFs regardless of oxygen concentration (Fig. 3B). This was in sharp contrast to the subcellular location of HIF-1α and -2α in other well-characterized cell lines. For example, in the human embryonic kidney cell line 293, HIF-1α and -2α had identical patterns of protein expression and subcellular localization; both proteins showed increased expression after treatment with hypoxia or iron chelators and were localized primarily to the nucleus following these treatments (Fig. 3C). These data indicate that restriction of endogenous HIF-2α to the cytoplasm is a cell type-specific event that apparently has the capacity to discriminate between HIF-2α and HIF-1α.
Nuclear translocation of HIF-2α is dependent on a bipartite nuclear localization signal in the C terminus (13, 30). We examined the sequence of HIF-2α cDNA in these cells to determine whether any mutation had occurred in this conserved nuclear localization sequence. The bipartite nuclear localization signal 710-KLKLKR-X27-KRMKS-747 (underlining shows the conserved bipartite nuclear localization signal) was not altered in these MEFs, however (data not shown) (13, 30). In addition, other crucial amino acid residues, including the two proline residues in the ODD and the asparagine residue required for FIH-1 hydroxylation, were unmutated in these MEFs (data not shown).
HIF-2α does not undergo 26S proteasome-dependent protein degradation.
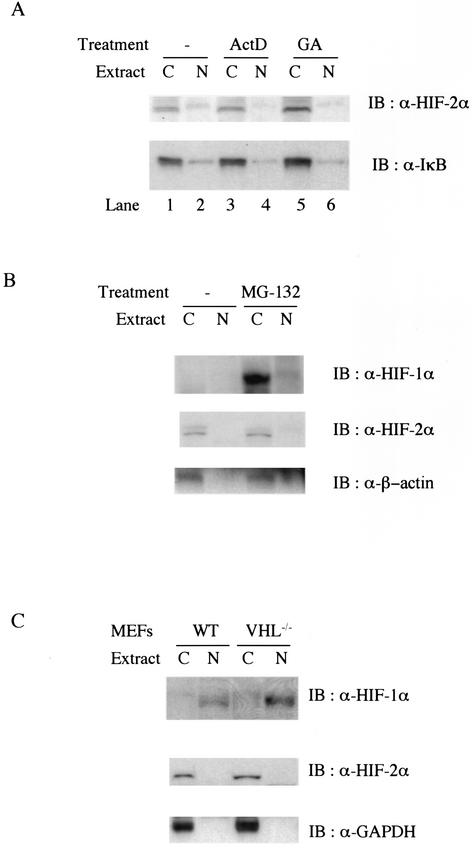
It has been shown that VHL undergoes constitutive nuclear-cytoplasmic trafficking in a transcription-dependent but oxygen-independent manner and that this process is required for ubiquitination, nuclear export, and subsequent protein degradation of HIF-1α in reoxygenated cells (9). This implies that accelerated VHL-dependent nuclear export might lead to steady-state cytoplasmic accumulation of endogenous HIF-2α in MEFs. In order to test this possibility, subcellular fractionation was performed following treatment with actinomycin D, which is known to block nuclear export of VHL and HIF-1α upon reoxygenation (9). However, inhibition of transcription did not change the subcellular location of HIF-2α, indicating that nucleocytoplasmic shuffling of VHL is not responsible for HIF-2α accumulation within the cytoplasm of MEFs (Fig. 4A, lanes 3 and 4).
FIG. 4.
(A) Protein expression level and subcellular location of endogenous HIF-2α are not affected by inhibition of transcription or heat shock protein 90 activity. Wild-type MEFs were untreated (lanes 1 and 2) or treated with 10 μg of actinomycin D per ml (ActD, lanes 3 and 4) or 10 μM geldanamycin (GA, lanes 5 and 6). Then 25 μg of either cytoplasmic (lanes C) or nuclear (lanes N) protein was analyzed by Western blot (IB) analysis with anti-HIF-2α (upper panel) or anti-IκB (lower panel) antibodies. (B) Endogenous HIF-2α escapes 26S proteasome-dependent protein turnover in normoxic MEFs. Wild-type (WT) MEFs were treated with 50 μM MG-132 for 4 h before being harvested. Then 25 μg of either cytoplasmic or nuclear protein was analyzed for Western blot analysis with anti-β-actin (lower panel) or anti-HIF-2α (middle panel), and the same membrane was reprobed with anti-HIF-1α (upper panel). (C) Expression and subcellular location of HIF-1α and -2α in wild-type and VHL null primary MEFs. Cell extracts from wild-type and VHL null primary MEFs were analyzed by Western blot analysis with anti-HIF-1α (upper panel), anti-HIF-2α (middle panel), and anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (lower panel).
The molecular chaperone HSP90 has been implicated as a key component in protecting HIF-1α from VHL-independent ubiquitination and protein degradation in VHL-deficient renal cell carcinoma (RCC) lines (17). We thus wished to determine whether endogenous HIF-2α is able to escape VHL-dependent protein degradation and accumulate in the cytosol with the assistance of HSP90s. Inhibition of HSP90 activity by geldanamycin has been shown to decrease the level of stabilized HIF-1α in response to hypoxia (17, 26, 33). However, geldanamycin treatment failed to change the level of HIF-2α expression or its intracellular location, excluding the involvement of HSP90 in stabilizing and anchoring endogenous HIF-2α to the cytoplasm in these MEFs (Fig. 4A, compare lanes 5 and 6 with lanes 1 and 2).
Next, we decided to investigate whether there is a separate pool of endogenous HIF-2α that still undergoes oxygen-dependent degradation. In order to test this possibility, 26S proteasome-dependent protein degradation in wild-type MEFs was blocked with a specific inhibitor, MG-132, before harvest. Western blot analysis with cell extracts showed that 26S proteasome inhibition resulted in upregulation of HIF-1α protein in the cytoplasm in normoxic cells (Fig. 4B, upper panel). Blocking proteasome activity failed to change the levels of HIF-2α significantly, arguing that the majority of endogenous HIF-2α did not undergo 26S proteasome-dependent protein degradation (Fig. 4B, lower panel); this may account for the constitutive accumulation of HIF-2α seen under normoxia.
Loss of VHL does not promote further accumulation and nuclear localization of HIF-2α but does allow HIF-1α stabilization and nuclear localization under normoxia.
Recognition of HIF-α isoforms by VHL has been regarded as a rate-limiting step for their polyubiquitination and efficient degradation by the 26S proteasome and is primarily regulated by proline hydroxylation (21). The role of VHL in HIF-α stability has been shown clearly in VHL-null RCCs, which exhibit deregulated expression of HIF-α but regain the ability to regulate protein stability of both endogenous HIF-2α and ectopically expressed HIF-1α by expression of exogenous VHL (20, 21).
To study further whether VHL function differentially affects the relative protein stability and localization of HIF-1α and HIF-2α, we generated primary MEFs with a conditional deletion of VHL (11, 37) and examined the expression pattern of HIF-1α and -2α in these cells (Fig. 4C). As in RCCs, ablation of VHL expression in fibroblasts led to the stabilization of HIF-1α in normoxic cells (Fig. 4C, upper panel). However, VHL deletion altered neither the expression level nor the subcellular location of HIF-2α (Fig. 4C, middle panel), indicating that oxygen-independent expression of HIF-2α results from an inability of VHL to recruit HIF-2α for ubiquitination in MEFs. These data, together with those in Fig. 4B, support the notion that HIF-2α does not conform to the well-characterized VHL-mediated signaling pathway that HIF-1α adopts. Moreover, in MG-132-treated cells, HIF-1α became stabilized but remained in the cytoplasm under normoxic conditions, whereas, in contrast, HIF-1α in VHL null MEFs accumulated within the nucleus regardless of the oxygenation status. This indicates that VHL and, by extension, ubiquitination can negatively influence the nuclear localization of HIF-1α, probably via a constitutive nucleocytoplasmic shuffling (11); it further implies that hypoxic signaling is not required for nuclear localization of HIF-1α. Taken together, these data suggest that VHL does indeed play a key role in regulating oxygen-dependent protein stability and subcellular location of HIF-1α but affects HIF-2α differently.
Hypoxia-independent transcriptional activity of overexpressed HIF-2α in immortalized MEFs.
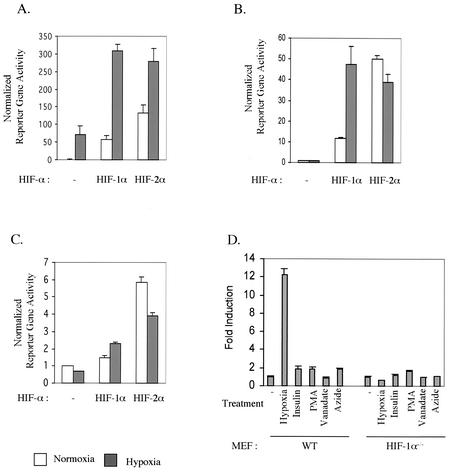
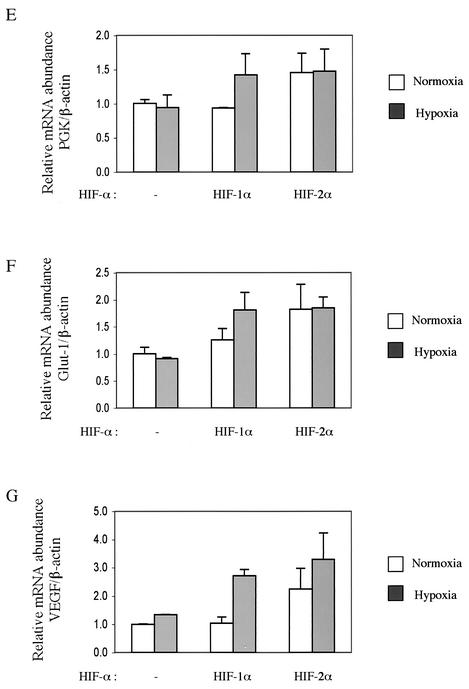
We next asked whether murine HIF-2α has hypoxia-inducible transcriptional activity in immortalized MEFs when it is overexpressed. Cells were transfected with an HRE reporter gene together with either an empty, HIF-1α, or HIF-2α expression vector. Consistent with published findings, transfection of 293 cells with HIF-2α led to a hypoxia-induced increase in HRE reporter gene activity at a level similar to that induced by HIF-1α (Fig. 5A). Unexpectedly, in HIF-1α null MEFs, overexpressed HIF-2α exhibited very strong constitutive transcriptional activity under normoxic conditions; this activity was comparable to hypoxic HIF-1α transcriptional activity (Fig. 5B and C). Hypoxic treatment was unable to further induce transcriptional activity of overexpressed HIF-2α from two different HRE reporter genes (Fig. 5B and C), verifying that murine HIF-2α is a functional but hypoxia-independent transcriptional factor for HRE-containing promoters in these cells. Overexpression of HIF-1α and -2α also gave rise to similar patterns of expression in endogenous HIF-1 target genes (Fig. 5E to G).
FIG. 5.
Potent transcriptional activity of ectopically overexpressed HIF-2α. (A and B) HIF-1α and -2α were overexpressed along with the HRE-luciferase reporter in 293 cells (A) and HIF-1α null MEFs (B). Then, the cells were treated with either normoxia or 0.5% O2 hypoxia for 22 h before being harvested for the reporter gene assay. (C) HIF-1α null MEFs were transfected with HIF-1α or HIF-2α along with the pGL-SRJ reporter. (D) Endogenous HIF-2α remains inactive after treatment of MEFs with various stimulators. HIF-1α null MEFs were transfected with a HRE-luciferase reporter and treated with various intracellular signaling pathway modulators to stimulate HIF-2α activity Relative HRE reporter gene activity is shown as induction over that of untreated cells. WT, wild type; PMA, phorbol myristate acetate. (E to G) Overexpressed HIF-2α can induce the expression of some endogenous HIF target genes. HIF-1α null MEFs were transfected with either an empty, HIF-1α, or HIF-2α expression vector and treated with either normoxia or 0.5% O2 hypoxia for 22 h before being harvested for total RNA extraction. Expression of PGK, Glut-1, and VEGF was determined as described for Fig. 2 and normalized against that of β-actin.
As overexpressed HIF-2α has transcriptional activity comparable to that of HIF-1α in fibroblasts, it is tempting to hypothesize that endogenous HIF-2α is also a functional transcription factor that is activated by stimuli other than hypoxia and utilizes a separate signaling pathway. In fact, there have been many reports of nonhypoxic activators of HIF-1. These include insulin, vanadate, nitric oxide, and reactive oxygen species generated by mitochondrial inhibition (8, 39, 48). In addition, some kinase-mediated signal transduction pathways have been suggested to be involved in HIF signaling; mitogen-activated protein kinase and p38-dependent phosphorylation on HIF-1α enhances transcriptional activity, and activation of phosphatidylinositol 3-kinase/AKT signaling is also known to activate HIF signaling (35, 42, 49). Therefore, we decided to investigate whether constitutively expressed HIF-2α is activated by any one of these stimuli in MEFs.
HIF-1α null MEFs were transfected with an HRE reporter and then treated with various reagents known to activate HIF-1. As shown in Fig. 5D, most of the reagents tested failed to stimulate HRE reporter gene activity except phorbol myristate acetate (PMA), which induced activity approximately twofold over that in untreated cells. However, this induction level was very low compared to that of wild-type cells treated with hypoxia. Likewise, cotransfection of HIF-1α null MEFs with various kinase expression vectors (mitogen-activated protein kinase kinases 1 and 3, p38, and AKT) did not stimulate HRE reporter gene activity (data not shown).
Nuclear localization of ectopically overexpressed HIF-2α.
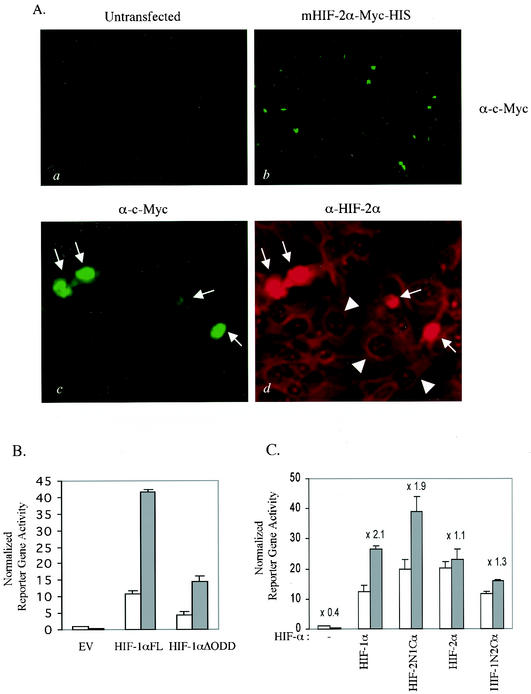
We then determined the intracellular location of overexpressed HIF-2α. Full-length murine HIF-2α was tagged with the c-Myc epitope at the C terminus and transfected into wild-type MEFs for immunofluorescence analysis (Fig. 6A). Anti-c-Myc antibody detection recognized exogenous HIF-2α only in transfected MEFs (Fig. 6A, panels a and b). Examination under higher magnification revealed that overexpressed Myc-tagged HIF-2α was localized to the nucleus (marked with arrows in Fig. 6A, panel c). Double staining with anti-HIF-2α and anti-Myc antibodies demonstrated that endogenous HIF-2α remained in the cytoplasm in untransfected cells (marked with arrowheads in Fig. 6A, panel d), while much more intense signals for both HIF-2α and the c-Myc tag were observed in the nucleus of cells transfected with exogenous HIF-2α expression vectors (arrows in Fig. 6A, panel d). These data suggest that nuclear localization may be a rate-limiting step that determines activation of HIF-2α as a functional transcriptional factor for HIF-1 target genes. It also suggests that overexpressed HIF-2α can bypass this step and facilitate expression of HIF target genes regardless of oxygen status.
FIG. 6.
(A) Nuclear accumulation of overexpressed HIF-2α. Wild-type MEFs were grown on a glass slide chamber and transfected with pcDNA3.1/mHIF-2α-Myc-His. After fixation, the cells were double stained with monoclonal anti-c-Myc and polyclonal anti-HIF-2α antibodies, which were located by fluorescein isothiocyanate-labeled anti-mouse immunoglobulin G and Texas Red-labeled anti-rabbit immunoglobulin G, respectively. Low magnification of anti-Myc staining of untransfected (a) and Myc-tagged, HIF-2α-transfected (b) MEFs. Higher magnification of MEFs transfected with Myc-tagged HIF-2α shows the nuclear location of overexpressed Myc-tagged HIF-2α (indicated by arrows) only in transfected cells (c) and cytoplasmic compartmentalization of endogenous HIF-2α (marked with arrowheads, d). (B) Constitutively expressed HIF-1α still requires the hypoxic signal to further induce an HRE reporter gene. HIF-1α null MEFs were transiently transfected with either the empty vector (EV), full-length HIF-1α (HIF-1αFL), or ODD-deleted HIF-1α (HIF-1αΔODD) together with an HRE-luciferase reporter. (C) The C-terminal half of HIF-2α is responsible for the lack of hypoxia-inducible gene expression. HIF-1α null MEFs were transiently transfected with a vector expressing either full-length HIF-1α (HIF-1αFL), full-length HIF-2α (HIF-2αFL), a fusion protein of the N-terminal half of HIF-2α and the C-terminal half of HIF-1α (HIF-2N1C), or a fusion protein of the N-terminal half of HIF-1α and the C-terminal half of HIF-2α (HIF-1N2C) together with an HRE-luciferase reporter. Values above bars are factors of difference between hypoxic treatment and normoxia.
FIH-1 exerts its inhibitory effect on HIF-1α-dependent but not HIF-2α-dependent transactivation.
It has been shown that deletion of the ODD or mutation of the conserved proline residues leads to constitutive expression of HIF-α isoforms, although transcriptional activation of these mutants is still regulated by oxygen (16, 32). Recent work has shown that FIH-1-mediated asparagine hydroxylation acts independently of ubiquitination to regulate HIF-1α transcriptional activity in an oxygen-dependent manner (14, 28). As in other cells, HIF-1α containing a deletion of the ODD (HIF-1α/ΔODD) still showed hypoxia-inducible gene expression in fibroblasts (Fig. 6B) (16). This was in contrast to the transcriptional activity of HIF-2α, which exhibited no hypoxia inducibility (Fig. 5B and C).
This result prompted us to ask whether HIF-2α escapes the asparagine hydroxylation-dependent regulation of transcriptional activity. Two different HIF-1α and -2α fusion proteins were generated: HIF-2N1C, a fusion protein of the N-terminal half of HIF-2α and the C-terminal half of HIF-1α, and HIF-1N2C, generated by fusing the N-terminal half of HIF-1α to the C-terminal half of HIF-2α. Reporter gene assays of transiently transfected HIF-1α null MEFs with constructs expressing these fusion proteins demonstrated that the lack of hypoxia-inducible transcriptional activity seen in endogenous HIF-2α stemmed from its C terminus. This region harbors both its ODD and FIH-1 interaction domains (Fig. 6C).
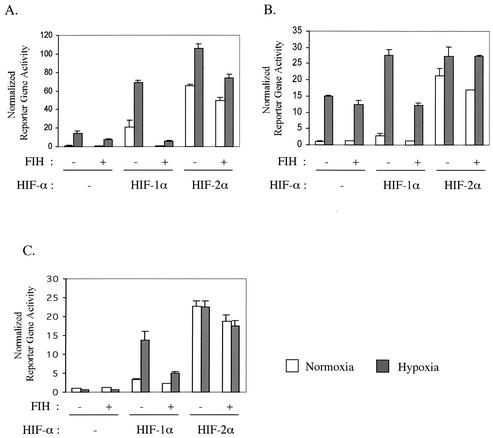
We then tested HIF-2α response to the overexpression of FIH-1. In agreement with published data, overexpression of FIH-1 decreased the transcriptional activity of the endogenous HIF-αs in the human embryonic kidney 293 cell line (31) (Fig. 7A). Overexpression of FIH-1 also inhibited the transcriptional activity of overexpressed HIF-1α, down to the level of endogenous HIF-αs, but it was much less effective in suppressing exogenous HIF-2α-mediated transcription (Fig. 7A). Interestingly, this selective inhibition pattern by FIH-1 was even more pronounced in MEFs (Fig. 7B and C). The normoxic and hypoxic transcriptional activity of overexpressed HIF-2α was only marginally affected by overexpression of FIH-1, whereas HIF-1α transcriptional activity was reduced to 50% by FIH-1 overexpression (Fig. 7B and C). These data demonstrate that FIH-1 is able to selectively inhibit the transcriptional activity of HIF-1α in MEFs with little effect on HIF-2α-driven transcription.
FIG. 7.
Isoform-specific inhibitory effect of FIH-1 overexpression on transcriptional activity of HIF-1α and -2α. 293 cells (A), wild-type MEFs (B), and HIF-1α null MEFs (C) were transfected with the HRE-luciferase reporter and pRL-CMV along with either empty vector or the HIF-1α or -2α and FIH-1 expression vector. After recovery from transfection, the cells were incubated in 20% O2 (normoxia) or 0.5% O2 (hypoxia) for 22 h before being harvested. The y axis shows normalized firefly luciferase over Renilla luciferase activity relative to the wild-type normoxic response (bars represent the standard deviations of triplicate analyses).
DISCUSSION
We show here that HIF-2α escapes from oxygen-dependent protein degradation and accumulates in significant amounts in immortalized MEFs under normoxia, whereas HIF-1α is subject to tight regulation by oxygen levels. We further demonstrate that oxygen-independent expression of HIF-2α results from a lack of VHL-dependent ubiquitination and proteasome-dependent protein degradation (Fig. 4B and C).
The relationships between subcellular location and ubiquitination/degradation of HIF-αs have been investigated extensively but are still unclear. Initially, it was thought that hypoxia ushers HIF-1α into the nucleus, where it is protected from protein degradation by dimerization with ARNT (23). A minute-scale kinetic analysis of the subcellular location of HIF-α with VHL-positive and -negative RCCs showed that a newly translated HIF-1α was imported very quickly into the nucleus and that the primary location for HIF-α hydroxylation and ubiquitination was indeed the nucleus (9). During deactivation of HIF-1α signaling upon reoxygenation, HIF-1α forms a complex with VHL and becomes polyubiquitinated in the nucleus. It is then quickly exported into the cytoplasm for degradation. According to this model, cytoplasmic trapping of HIF-2α may be sufficient to avoid ubiquitination and subsequent protein degradation in normoxia.
Cytoplasmic compartmentalization of endogenous HIF-2α under both normoxic and hypoxic conditions also appears to be responsible for silenced transcriptional activity, as overexpressed HIF-2α can be translocated into the nucleus and stimulate HRE-dependent gene induction (Fig. 4B and 5A). This argues that the rate-determining step for activation of endogenous HIF-2α is nuclear localization. It should be noted that the nuclear localization signal of HIF-2α overlaps the transcription-inhibitory domain, and overexpression of the peptides spanning this region increase the transcriptional activity of HIF-1α, probably by titrating out the transcription-inhibitory factor (38). This transcription-inhibitory factor might be able to inhibit the transcriptional activity of HIF-αs by prohibiting nuclear translocation, as it has the potential to mask the nuclear localization signal. One model for the function of this factor is that it possesses a higher affinity for the transcription-inhibitory domain of HIF-2α than for that of HIF-1α; this could provide the MEFs with specificity for HIF-2α.
Thus, we propose that in certain cell types, HIF-2α activity is not regulated by a the classical hypoxia sensing pathway, i.e., the newly identified proline hydroxylases and VBC complex, but rather by a novel regulatory mechanism that exploits the intracellular spatial distribution of HIF-2α as an on-off switch. Indeed, modulation of nuclear localization is a very common strategy for mammalian cells to control gene expression. One of the clearest examples is the signaling pathway for activation of NF-κB, which is anchored in the cytosol as an inactive component of an IκB-containing complex in unstimulated cells (25). Activation of cells with proinflammatory cytokines, viruses, or bacteria initiates the intracellular signaling pathways leading to the activation of IκB kinase. Phosphorylation of IκB by IκB kinase enables E3RSIκB ligase to recognize and polyubiquitinate IκB for degradation. This leads to liberation of NF-κB for nuclear localization and subsequent activation of its target genes (25). It would be interesting to see if an HIF-2α nuclear localization blocker is also regulated by ubiquitin-dependent protein degradation in a similar manner.
All of the nonhypoxic HIF activators that we tested failed to stimulate HRE reporter gene activity in HIF-1α null MEFs. It has been suggested that HIF-2α functions as a transcription factor responding to the availability of glucose rather than oxygen, which can cause apoptosis under hypoglycemic conditions (2). However, since there was no evidence of accumulation of HIF-2α protein after glucose deprivation in ES cells in the study of Brusselmans et al. (2) that first proposed this role for the transcription factor, it is not clear how this type of HIF-2α-dependent regulation occurs. We show that glucose deprivation influences neither HIF-2α protein levels nor gene expression in fibroblasts, arguing against the role of HIF-2α-mediated gene induction during glucose deprivation in this cell type.
A surprising finding is that modulation of HIF-α transcriptional activity by FIH-1 has specificity. Our data indicate that the FIH-1-mediated asparagine hydroxylation signaling pathway controls HIF-1α but not HIF-2α in immortalized MEFs. This may result from a lower affinity of HIF-2α for FIH-1, although this needs to be determined. Unexpectedly, we found that FIH-1 overexpression can repress HIF-1α-mediated gene induction in hypoxia and normoxia; this suggests that there might be a mechanism other than cofactor recruitment by which FIH-1 modulates HIF-1α transactivation.
Recently, it was proposed that binding of VHL to the proline-hydroxylated HIF-1α ODD facilitates the recruitment of FIH-1 to the HIF-1α C-terminal activation domain and assembly of an HIF-α-VHL-FIH-1 ternary complex, which allows asparagine hydroxylation (29). In this model, the oxygen-sensing capability of FIH-1 is not built into the FIH-1 active site but rather depends on an interaction of VHL with the HIF-1α ODD (29). This model is in good agreement with the results that we describe. We have shown that HIF-2α expression is not dependent on the presence of VHL or proteasome activity; this presumably reflects the selectively decreased interaction of VHL with the HIF-2α ODD. Currently, we are investigating whether normoxic HIF-2α is modified by hydroxylation at its conserved proline and/or arginine residues to determine how the differential activation of this molecule occurs.
We have shown here that HIF-1α and -2α undergo differential regulation of their expression and activation in fibroblasts, although both are capable of induction of a similar set of downstream target genes. HIF-1α functions as a hypoxia-sensing transcription factor in mammalian cells; HIF-2α regulates hypoxia-inducible genes in a more complicated manner. There is clearly evidence for multiple signaling pathways regulating the protein expression and activation of HIF-2α. HIF-2α functions as a hypoxia-inducible transcription factor in the RCC and 293 cell lines, but in other cells, such as the MEFs described here, HIF-2α is no longer a hypoxia-inducible factor and escapes oxygen-dependent protein degradation and FIH-1-dependent transcription inhibition. This appears to be modulated by an unidentified factor which constrains HIF-2α subcellular location. Thus, it is essential to evaluate the differential programs of expression and activation of HIF-1α and -2α in order to understand the full range of hypoxia-induced gene expression.
Acknowledgments
We thank H. F. Bunn for the generous gifts of HIF-1α expression vectors.
This work was supported by NIH grant CA82515.
REFERENCES
- 1.Bhattacharya, S., C. L. Michels, M. K. Leung, Z. P. Arany, A. L. Kung, and D. M. Livingston. 1999. Functional role of p35srj, a novel p300/CBP binding protein, during transactivation by HIF-1. Genes Dev. 13:64-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brusselmans, K., F. Bono, P. Maxwell, Y. Dor, M. Dewerchin, D. Collen, J. M. Herbert, and P. Carmeliet. 2001. Hypoxia-inducible factor-2α (HIF-2α) is involved in the apoptotic response to hypoglycemia but not to hypoxia. J. Biol. Chem. 276:39192-39196. [DOI] [PubMed] [Google Scholar]
- 3.Bunn, H. F., and R. O. Poyton. 1996. Oxygen sensing and molecular adaptation to hypoxia. Physiol. Rev. 76:839-885. [DOI] [PubMed] [Google Scholar]
- 4.Clifford, S. C., D. Astuti, L. Hooper, P. H. Maxwell, P. J. Ratcliffe, and E. R. Maher. 2001. The pVHL-associated SCF ubiquitin ligase complex: molecular genetic analysis of elongin B and C, Rbx1 and HIF-1α in renal cell carcinoma. Oncogene 20:5067-5074. [DOI] [PubMed] [Google Scholar]
- 5.Ema, M., S. Taya, N. Yokotani, K. Sogawa, Y. Matsuda, and Y. Fujii-Kuriyama. 1997. A novel basic helix-loop-helix-PAS factor with close sequence similarity to hypoxia-inducible factor 1α regulates the VEGF expression and is potentially involved in lung and vascular development. Proc. Natl. Acad. Sci. USA 94:4273-4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Epstein, A. C., J. M. Gleadle, L. A. McNeill, K. S. Hewitson, J. O'Rourke, D. R. Mole, M. Mukherji, E. Metzen, M. I. Wilson, A. Dhanda, Y. M. Tian, N. Masson, D. L. Hamilton, P. Jaakkola, R. Barstead, J. Hodgkin, P. H. Maxwell, C. W. Pugh, C. J. Schofield, and P. J. Ratcliffe. 2001. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107:43-54. [DOI] [PubMed] [Google Scholar]
- 7.Flamme, I., T. Frohlich, M. von Reutern, A. Kappel, A. Damert, and W. Risau. 1997. HRF, a putative basic helix-loop-helix-PAS-domain transcription factor is closely related to hypoxia-inducible factor-1 α and developmentally expressed in blood vessels. Mech. Dev. 63:51-60. [DOI] [PubMed] [Google Scholar]
- 8.Gao, N., M. Ding, J. Z. Zheng, Z. Zhang, S. S. Leonard, K. J. Liu, X. Shi, and B. H. Jiang. 2002. Vanadate Induced expression of hypoxia-inducible factor 1a and vascular endothelial growth factor through phosphatidylinositol 3-kinase/Akt pathway and reactive oxygen species. J. Biol. Chem. 277:31963-31971. [DOI] [PubMed]
- 9.Groulx, I., and S. Lee. 2002. Oxygen-dependent ubiquitination and degradation of hypoxia-inducible factor requires nuclear-cytoplasmic trafficking of the von Hippel-Lindau tumor suppressor protein. Mol. Cell. Biol. 22:5319-5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gu, Y. Z., S. M. Moran, J. B. Hogenesch, L. Wartman, and C. A. Bradfield. 1998. Molecular characterization and chromosomal localization of a third α-class hypoxia-inducible factor subunit, HIF3α. Gene Expr. 7:205-213. [PMC free article] [PubMed] [Google Scholar]
- 11.Haase, V. H., J. N. Glickman, M. Socolovsky, R. Jaenisch. 2001. Vascular tumors in livers with targeted inactivation of the von Hippel-Lindau tumor suppressor. Proc. Natl. Acad. Sci. USA 98:1583-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen, W. J., M. Ohh, J. Moslehi, K. Kondo, W. G. Kaelin, and W. J. Welch. 2002. Diverse effects of mutations in exon II of the von Hippel-Lindau (VHL) tumor suppressor gene on the interaction of pVHL with the cytosolic chaperonin and pVHL-dependent ubiquitin ligase activity. Mol. Cell. Biol. 22:1947-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hara, S., C. Kobayashi, and N. Imura. 1999. Nuclear localization of hypoxia-inducible factor-2α in bovine arterial endothelial cells. Mol. Cell. Biol. Res. Commun. 2:119-123. [DOI] [PubMed] [Google Scholar]
- 14.Hewitson, K. S., L. A. McNeill, M. V. Riordan, Y. M. Tian, A. N. Bullock, R. W. Welford, J. M. Elkins, N. J. Oldham, S. Bhattacharya, J. M. Gleadle, P. J. Ratcliffe, C. W. Pugh, and C. J. Schofield. 2002. Hypoxia-inducible factor (HIF) asparagine hydroxylase is identical to factor inhibiting HIF (FIH) and is related to the cupin structural family. J. Biol. Chem. 277:26351-26355. [DOI] [PubMed]
- 15.Hogenesch, J. B., W. K. Chan, V. H. Jackiw, R. C. Brown, Y. Z. Gu, M. Pray-Grant, G. H. Perdew, and C. A. Bradfield. 1997. Characterization of a subset of the basic-helix-loop-helix-PAS superfamily that interacts with components of the dioxin signaling pathway. J. Biol. Chem. 272:8581-8593. [DOI] [PubMed] [Google Scholar]
- 16.Huang, L. E., J. Gu, M. Schau, and H. F. Bunn. 1998. Regulation of hypoxia-inducible factor 1α is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc. Natl. Acad. Sci. USA 95:7987-7992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Isaacs, J. S., Y.-J. Jung, E. G. Mimnaugh, A. Martinez, F. Cuttitta, and L. M. Neckers. 2002. Hsp90 regulates a von Hippel Lindau-independent hypoxia-inducible factor-1α-degradative pathway. J. Biol. Chem. 277:29936-29944. [DOI] [PubMed] [Google Scholar]
- 18.Ivan, M., K. Kondo, H. Yang, W. Kim, J. Valiando, M. Ohh, A. Salic, J. M. Asara, W. S. Lane, and W. G. Kaelin, Jr. 2001. HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292:464-468. [DOI] [PubMed] [Google Scholar]
- 19.Iyer, N. V., L. E. Kotch, F. Agani, S. W. Leung, E. Laughner, R. H. Wenger, M. Gassmann, J. D. Gearhart, A. M. Lawler, A. Y. Yu, and G. L. Semenza. 1998. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 α. Genes Dev. 12:149-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaakkola, P., D. R. Mole, Y. M. Tian, M. I. Wilson, J. Gielbert, S. J. Gaskell, A. Kriegsheim, H. F. Hebestreit, M. Mukherji, C. J. Schofield, P. H. Maxwell, C. W. Pugh, and P. J. Ratcliffe. 2001. Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292:468-472. [DOI] [PubMed] [Google Scholar]
- 21.Kaelin, W. G., Jr. 2002. How oxygen makes its presence felt. Genes Dev. 16:1441-1445. [DOI] [PubMed] [Google Scholar]
- 22.Kallio, P. J., W. J. Wilson, S. O'Brien, Y. Makino, and L. Poellinger. 1999. Regulation of the hypoxia-inducible transcription factor 1α by the ubiquitin-proteasome pathway. J. Biol. Chem. 274:6519-6525. [DOI] [PubMed] [Google Scholar]
- 23.Kallio, P. J., K. Okamoto, S. O'Brien, P. Carrero, Y. Makino, H. Tanaka, and L. Poellinger. 1998. Signal transduction in hypoxic cells: inducible nuclear translocation and recruitment of the CBP/p300 coactivator by the hypoxia-inducible factor-1α. EMBO J. 17:6573-6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamura, T., S. Sato, K. Iwai, M. Czyzyk-Krzeska, R. C. Conaway, and J. W. Conaway. 2000. Activation of HIF1α ubiquitination by a reconstituted von Hippel-Lindau (VHL) tumor suppressor complex. Proc. Natl. Acad. Sci. USA 97:10430-10435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karin, M., and M. Delhase. 2000. The IκB kinase (IκΚ) and NF-κB: key elements of proinflammatory signalling. Semin. Immunol. 12:85-98. [DOI] [PubMed] [Google Scholar]
- 26.Katschinski, D. M., L. Le, D. Heinrich, K. F. Wagner, T. Hofer, S. G. Schindler, and R. H. Wenger. 2002. Heat induction of the unphosphorylated form of hypoxia-inducible factor-1α is dependent on heat shock protein-90 activity. J. Biol. Chem. 277:9262-9267. [DOI] [PubMed] [Google Scholar]
- 27.Lando, D., D. J. Peet, D. A. Whelan, J. J. Gorman, and M. L. Whitelaw. 2002. Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science 295:858-861. [DOI] [PubMed] [Google Scholar]
- 28.Lando, D., D. J. Peet, J. J. Gorman, D. A. Whelan, M. L. Whitelaw, and R. K. Bruick. 2002. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 16:1466-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, C., S. J. Kim, D. G. Jeong, S. M. Lee, and S. E. Ryu. 2003. Structure of human FIH-1 reveals a unique active site pocket and interaction sites for HIF-1 and von Hippel-Lindau. J. Biol. Chem. 27:7558-7563. [DOI] [PubMed] [Google Scholar]
- 30.Luo, J. C., and M. Shibuya. 2001. A variant of nuclear localization signal of bipartite-type is required for the nuclear translocation of hypoxia-inducible factors (1α, 2α and 3α). Oncogene 20:1435-1444. [DOI] [PubMed] [Google Scholar]
- 31.Mahon, P. C., K. Hirota, and G. L. Semenza. 2001. FIH-1: a novel protein that interacts with HIF-1α and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 15:2675-2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masson, N., C. Willam, P. H. Maxwell, C. W. Pugh, and P. J. Ratcliffe. 2001. Independent function of two destruction domains in hypoxia-inducible factor-α chains activated by prolyl hydroxylation. EMBO J. 20:5197-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minet, E., D. Mottet, G. Michel, I. Roland, M. Raes, J. Remacle, and C. Michiels. 1999. Hypoxia-induced activation of HIF-1: role of HIF-1α-Hsp90 interaction. FEBS Lett. 460:251-256. [DOI] [PubMed] [Google Scholar]
- 34.Peng, J., L. Zhang, L. Drysdale, and G. H. Fong. 2000. The transcription factor EPAS-1/hypoxia-inducible factor 2α plays an important role in vascular remodeling. Proc. Natl. Acad. Sci. USA 97:8386-8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richard, D. E., E. Berra, E. Gothie, D. Roux, and J. Pouyssegur. 1999. p42/p44 mitogen-activated protein kinases phosphorylate hypoxia-inducible factor 1α (HIF-1α) and enhance the transcriptional activity of HIF-1. J. Biol. Chem. 274:32631-32637. [DOI] [PubMed] [Google Scholar]
- 36.Ryan, H. E., J. Lo, and R. S. Johnson. 1998. HIF-1 α is required for solid tumor formation and embryonic vascularization. EMBO J. 17:3005-3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ryan, H. E., M. Poloni, W. McNulty, D. Elson, M. Gassmann, J. M. Arbeit, and R. S. Johnson. 2000. Hypoxia-inducible factor-1α is a positive factor in solid tumor growth. Cancer Res. 60:4010-4015. [PubMed] [Google Scholar]
- 38.Sang, N., J. Fang, V. Srinivas, I. Leshchinsky, and J. Caro. 2002. Carboxyl-terminal transactivation activity of hypoxia-inducible factor 1α is governed by a von Hippel-Lindau protein-independent, hydroxylation-regulated association with p300/CBP. Mol. Cell. Biol. 22:2984-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Semenza, G. L. 2001. Hif-1, O2, and the 3 phds: how animal cells signal hypoxia to the nucleus. Cell 107:1-3. [DOI] [PubMed] [Google Scholar]
- 40.Semenza, G. L. 1999. Regulation of mammalian O2 homeostasis by hypoxia-inducible factor 1. Annu. Rev. Cell Dev. Biol. 15:551-578. [DOI] [PubMed] [Google Scholar]
- 41.Shibata, T., A. J. Giaccia, and J. M. Brown. 2000. Development of a hypoxia-responsive vector for tumor-specific gene therapy. Gene Ther. 7:493-498. [DOI] [PubMed] [Google Scholar]
- 42.Sodhi, A., S. Montaner, V. Patel, M. Zohar, C. Bais, E. A. Mesri, and J. S. Gutkind. 2000. The Kaposi's sarcoma-associated herpes virus G protein-coupled receptor up-regulates vascular endothelial growth factor expression and secretion through mitogen-activated protein kinase and p38 pathways acting on hypoxia-inducible factor 1α. Cancer Res. 60:4873-4880. [PubMed] [Google Scholar]
- 43.Tian, H., S. L. McKnight, and D. W. Russell. 1997. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 11:72-82. [DOI] [PubMed] [Google Scholar]
- 44.Tian, H., R. E. Hammer, A. M. Matsumoto, D. W. Russell, and S. L. McKnight. 1998. The hypoxia-responsive transcription factor EPAS1 is essential for catecholamine homeostasis and protection against heart failure during embryonic development. Genes Dev. 12:3320-3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, G. L., B. H. Jiang, E. A. Rue, and G. L. Semenza. 1995. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. USA 92:5510-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wiener, C. M., G. Booth, and G. L. Semenza. 1996. In vivo expression of mRNAs encoding hypoxia-inducible factor 1. Biochem. Biophys. Res. Commun. 225:485-488. [DOI] [PubMed] [Google Scholar]
- 47.Wiesener, M. S., H. Turley, W. E. Allen, C. Willam, K. U. Eckardt, K. L. Talks, S. M. Wood, K. C. Gatter, A. L. Harris, C. W. Pugh, P. J. Ratcliffe, and P. H. Maxwell. 1998. Induction of endothelial PAS domain protein-1 by hypoxia: characterization and comparison with hypoxia-inducible factor-1α. Blood 92:2260-2268. [PubMed] [Google Scholar]
- 48.Zelzer, E., Y. Levy, C. Kahana, B. Z. Shilo, M. Rubinstein, and B. Cohen. 1998. Insulin induces transcription of target genes through the hypoxia-inducible factor HIF-1α/ARNT. EMBO J. 17:5085-5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zundel, W., C. Schindler, D. Haas-Kogan, A. Koong, F. Kaper, E. Chen, A. R. Gottschalk, H. E. Ryan, R. S. Johnson, A. B. Jefferson, D. Stokoe, and A. J. Giaccia. 2000. Loss of PTEN facilitates HIF-1-mediated gene expression. Genes Dev. 14:391-396. [PMC free article] [PubMed] [Google Scholar]