Abstract
The stability of the p53 protein is regulated by Mdm2. By acting as an E3 ubiquitin ligase, Mdm2 directs the ubiquitylation of p53 and its subsequent degradation by the 26S proteasome. In contrast, the Mdmx protein, although structurally similar to Mdm2, cannot ubiquitylate or degrade p53 in vivo. To ascertain which domains determine this functional difference between Mdm2 and Mdmx and consequently are essential for p53 ubiquitylation and degradation, we generated Mdm2-Mdmx chimeric constructs. Here we show that, in addition to a fully functional Mdm2 RING finger, an internal domain of Mdm2 (residues 202 to 302) is essential for p53 ubiquitylation. Strikingly, the function of this domain can be fulfilled in trans, indicating that the RING domain and this internal region perform distinct activities in the ubiquitylation of p53.
The tumor suppressor protein p53 is a transcription factor that functions to inhibit cell proliferation and is therefore usually maintained at low levels in cells to allow normal growth. p53 is stabilized and activated in response to several forms of cellular stress, leading to the induction of a set of target genes involved in cell cycle arrest, DNA repair, or apoptosis, dependent on the type and strength of the signal (32). An important regulator of p53 activity is Mdm2, which inhibits the function of p53 both by abolishing its transcription-regulatory activity (4, 24) and by targeting p53 for degradation (10, 17). Mdm2 is an E3 ubiquitin ligase that directs the ubiquitylation of both p53 and Mdm2 (6, 11, 12), resulting in the degradation of both proteins by the 26S proteasome. Mdm2 is also a transcriptional target of p53 (21, 27), and so p53 and Mdm2 form a tight autoregulatory feedback loop, the importance of which has been genetically demonstrated by the ability of p53-null mice to rescue the embryonic lethality of mdm2-null mice (16, 25).
The Mdmx protein is structurally homologous to Mdm2 (29). The highest conservation between Mdmx and Mdm2 is found in three regions named CR1, CR2, and CR3. CR1 (residues 42 to 94) contains the p53-binding domain, CR2 (residues 301 to 329) spans a putative Zn finger domain, and CR3 (residues 444 to 483) harbors the RING finger domain required for ubiquitin ligase activity. In contrast to Mdm2, Mdmx contains no nuclear localization signal (NLS) or nuclear export signal (NES), and the acidic region (residues 237 to 274) is less well conserved at the primary amino acid level between the two proteins, although Mdmx also contains many acidic amino acids in the same region. Due to the lack of an NLS or NES, the subcellular localization of the Mdmx protein is determined by its association with other cellular proteins, providing a level of regulation of Mdmx activity. It has been shown by us and others that in several cell types Mdmx is mainly cytoplasmic but is efficiently translocated into the nucleus by coexpression of Mdm2 and/or p53 (8, 18, 22). In addition, a p53- and Mdm2-independent nuclear translocation after induction of DNA damage has been shown (18). However, in some cell types we find a constitutive predominantly nuclear localization of Mdmx (e.g., in C33A cells) (31). In spite of its inability to ubiquitylate or degrade p53 in vivo, Mdmx can inhibit transcription activation by p53 (15, 20, 30). Importantly, mdmx-null mice show embryonic lethality, a phenotype that can be rescued by p53 deficiency, showing that—like Mdm2—Mdmx is a critical regulator of p53 in vivo (7, 23, 26).
Considering the functional differences between Mdm2 and Mdmx, we generated a set of Mdm2-Mdmx hybrids to investigate the roles of different domains needed for p53 ubiquitylation and degradation. We show that, in addition to the RING finger, a central domain in Mdm2 is essential for p53 ubiquitylation.
MATERIALS AND METHODS
Plasmids.
Expression vectors for p53, a p53 NLS mutant, Mdm2, an Mdm2 NLS mutant, His-tagged ubiquitin, and pcDNA3.1-LacZ have been described previously (30). Expression vectors for Myc-Mdm2 and Myc-Mdm2Δp53 were made by PCR amplification on wild-type mouse Mdm2 cDNA with 5′ primers containing an EcoRI endonuclease recognition sequence, a translation initiation site, and the 9E10 Myc epitope coding sequence and with 3′ primers containing a stop codon and an EcoRI endonuclease recognition sequence. PCR fragments were cloned into pcDNA3.1 (Invitrogen).
To generate the mouse Mdmx-Mdm2 chimeric constructs, several endonuclease recognition sites were generated through a two-step PCR-based mutagenesis or the QuikChange mutagenesis method (Stratagene). The positions of the alterations and eventual amino acid sequence alterations are shown in Fig. 1A. All cDNAs were cloned into pcDNA3.1 (Invitrogen). All plasmids produced by PCR were checked by sequencing and restriction fragment analysis. The sequences of all primers used in the cloning or mutagenesis are available upon request.
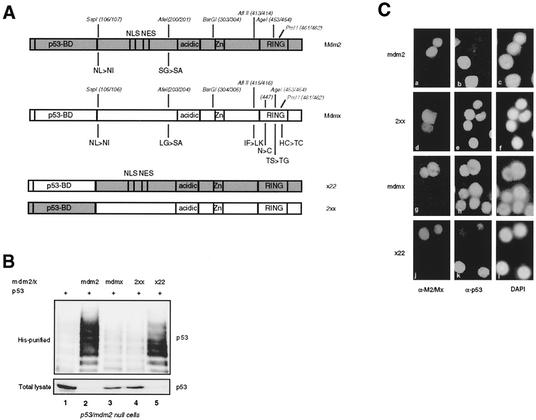
FIG. 1.
The p53-binding domains of Mdm2 and Mdmx are functionally similar. (A) Schematic representation of the Mdmx and Mdm2 proteins. Functional domains are indicated with the positions of the endonuclease recognition sites used to generate the chimeric constructs. Amino acid changes caused by the introduction of these sites are indicated. (B) p53- and mdm2-null cells (in 5-cm-diameter dishes) were transfected in duplicate with 200 ng of p53 without or with 1 μg of the indicated constructs in the presence of 1 μg of a His-tagged ubiquitin expression construct. Cells were either treated with 20 μM MG132 or mock treated 4 h before harvesting. The lysates from the MG132-treated cells were used to detect ubiquitylated p53 (His purified), while the lysates from the mock-treated cells were analyzed for total levels of p53. (C) C33A cells were transfected with the indicated constructs and analyzed by immunofluorescence 40 h after transfection. Mdm2 and 2xx were detected with the anti-Mdm2 antibody 4B2 (a and d), Mdmx was detected with the anti-Mdmx antibody 6B1A (g), and x22 was detected with the anti-Mdm2 antibody 2A10 (j). p53 was detected with the anti-p53 antibody FL393 (b, e, h, and k). Nuclei were stained with 4′,6′-diamidino-2-phenylindole (DAPI) (c, f, i, and l).
Cell lines, cell culture, and transfection.
H1299 cells were cultured in RPMI medium with 10% fetal bovine serum (FBS). Approximately 8 h prior to transfection, the medium was changed to Dulbecco's modified Eagle's medium (DMEM) with 10% FBS. C33A cells were maintained in DMEM with 10% FBS. H1299 and C33A cells were transfected by the calcium phosphate coprecipitation method as described previously (31). p53- and mdm2-null cells were cultured in DMEM with 10% FBS and transfected with Fugene6 transfection reagent (Roche Molecular Diagnostics) according to the manufacturer's protocol.
In vivo ubiquitylation assay.
Forty hours after transfection, cells were washed twice and scraped in ice-cold phosphate-buffered saline (PBS). Ten percent (10%) of the cell suspension was lysed in IPB 0.7 (20 mM triethanolamine [pH 7.4], 0.7 M NaCl, 0.5% NP-40, 0.2% sodium deoxycholic acid) or in Giordano buffer (50 mM Tris [pH 7.4], 250 mM NaCl, 0.1% Triton X-100, 5 mM EDTA). Lysis of the remaining 90% of the cells, subsequent isolation of His-ubiquitin-conjugated protein, and analysis of eluates and total lysates by Western blotting were performed as described previously (30).
Antibodies.
The following antibodies were used: the anti-p53 mouse monoclonal antibody DO-1, the anti-p53 rabbit polyclonal antibody FL393, the anti-Myc mouse monoclonal antibody 9E10 (all from Santa Cruz Biotechnology), the anti-Mdm2 mouse monoclonal antibodies 4B2 and 2A10, the anti-Mdmx mouse monoclonal antibody 6B1A (31), and the anti-Mdmx rabbit polyclonal antibody p57. p57 is a mouse Mdmx-specific rabbit serum raised against an internal peptide of Mdmx (amino acids [aa] 179 to 195). LacZ was detected with the mouse monoclonal antibody D19-2F3-2 (Roche Molecular Diagnostics).
Immunofluorescence.
Forty hours after transfection, C33A cells were washed twice with PBS, fixed for 15 min in 4% paraformaldehyde in PBS, and permeabilized for 10 min with 0.2% Triton X-100 in PBS at room temperature. Incubation of the cells with primary and secondary antibodies was performed as described previously (31).
RESULTS
In previous studies it was found that Hdmx appeared to be incapable of enhancing the ubiquitylation of p53 in vivo (30, 31). To further investigate whether Hdmx would have any detectable ubiquitin ligase activity, two different in vitro ubiquitylation assays were performed by use of UbcH5 as the E2 enzyme. In both an autoubiquitylation assay and a p53 ubiquitylation assay, Hdmx failed to display any significant activity, while all known E3 ligases tested (e.g., Mdm2 and Praja) showed self-ubiquitylation activity and Mdm2 could ubiquitylate p53. These results strongly suggest that the RING domain of Hdmx does not act as an E3 ubiquitin ligase, although the possibility that a different E2 enzyme is essential for Hdmx to function as such cannot be completely excluded. To study which domains determine this functional difference between Mdmx and Mdm2, we decided to generate Mdm2-Mdmx chimeric constructs.
Generation and nomenclature of the Mdm2-Mdmx chimeric constructs.
To investigate in more detail the domains in Mdm2 necessary for in vivo ubiquitylation and degradation of p53, we made mouse Mdm2-Mdmx chimeras. Initially we divided the Mdm2-Mdmx proteins into three parts: the p53-binding domain (p53-BD; aa 1 to 106), the middle domain (aa 107 to 303), and the zinc finger/RING finger domain (ZF/RF; aa 304 to 489). These three regions are also the basis of our nomenclature for the different chimeras; e.g., x22 contains the p53-BD of Mdmx and the middle domain and ZF/RF of Mdm2 (Fig. 1A). Subsequently, the middle and ZF/RF domains were each divided into two parts, and the order of these exchanges is indicated in parentheses; e.g., 2(2-x)2 contains the p53-BD, the NLS/NES region, and the ZF/RF region of Mdm2 but the acidic domain-containing region (AD) of Mdmx. To construct these chimeras, endonuclease recognition sites were created in Mdmx and Mdm2 cDNAs on analogous sites, with minimal changes on the amino acid level (Fig. 1A). Amino acid changes in Mdmx were chosen such that the sequence would be more like Mdm2. The few amino acid changes that were introduced into Mdm2 did not affect the function of full-length Mdm2 (data not shown).
The p53-binding domains of Mdmx and Mdm2 are functionally similar.
We and others have shown that the p53-binding domains of Hdm2 and Hdmx bind p53 with similar requirements (3, 8). To investigate whether these domains can functionally replace each other regarding ubiquitylation and degradation of p53, they were exchanged between Mdmx and Mdm2 (Fig. 1A).
These chimeras, x22 and 2xx, were coexpressed with p53 in p53- and mdm2-null cells, with wild-type Mdm2 and Mdmx as controls. Duplicate transfected dishes were either incubated with the proteasome inhibitor MG132 (to allow detection of p53 ubiquitylation) or mock treated (to allow detection of p53 degradation) 4 h prior to harvesting of the cells. The degradation and in vivo ubiquitylation of p53 were investigated as described previously (30, 33). The results show that the x22 hybrid can ubiquitylate and degrade p53 as efficiently as Mdm2 (Fig. 1B, lanes 2 and 5), while the 2xx hybrid, like Mdmx, lacks both activities (Fig. 1B, lanes 3 and 4). The Mdmx and 2xx proteins are both cytoplasmic and nuclear in the p53- and mdm2-null cells. However, directing the expression of these proteins exclusively to the nucleus by fusing the simian virus 40 NLS at the N terminus does not change the inability to ubiquitylate or degrade p53 (data not shown). To confirm these activities in an independent assay, the same constructs were expressed in C33A cells in order to investigate their abilities to degrade endogenous mutant p53. Again, only Mdm2 (Fig. 1Ca, b, and c) and the x22 chimera (Fig. 1Cj, k, and l) degraded p53. A protein is scored as being able to degrade p53 when at least 80% of strongly positive cells show a significant reduction in the p53 signal. We found with the in vivo ubiquitylation assay that the ubiquitylation of endogenous mutant p53 in C33A cells is increased upon transfection of Mdm2 or a degrading chimera, while a nondegrading Mdm2 mutant or chimera has no effect (data not shown). This result indicates that degradation of p53, as scored by immunofluorescence, and enhanced ubiquitylation are correlated. As mentioned above, in the C33A cells the Mdmx protein, as well as 2xx, is mainly nuclear. This localization is dependent on an intact Mdmx RING domain (see also below). Since these results show that the origin of the p53-binding domain does not affect the ubiquitylation and degradation of p53, most subsequent chimeras tested contained the p53-binding domain of Mdm2, allowing simultaneous detection of the Mdm2-Mdmx chimeras with the anti-Mdm2 antibody 4B2.
A complete Mdm2 RING is essential for efficient ubiquitylation and degradation of p53.
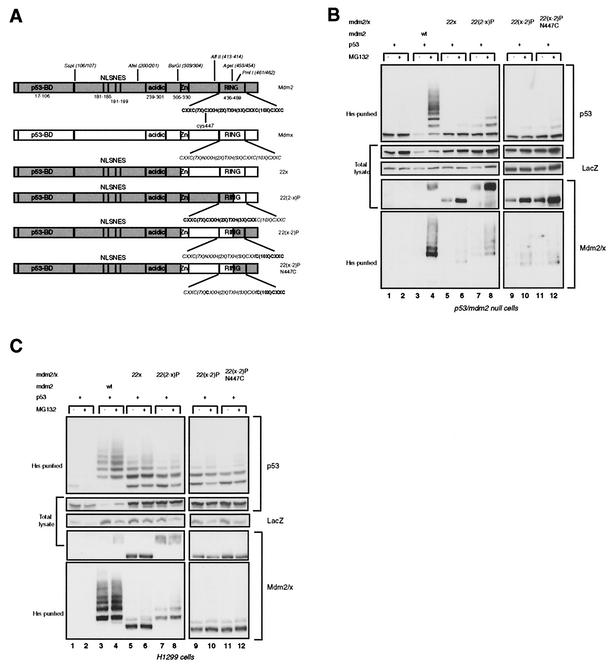
As a basis for further chimeras we exchanged the ZF/RF domain of Mdm2 for that of Mdmx, creating 22x (Fig. 2A). This chimeric protein, as well as the derived RING finger chimeras (see below), contains the NLS/NES region of Mdm2 and is predominantly located in the nucleus. As expected, the 22x chimeric protein could not ubiquitylate or degrade p53 in p53- and mdm2-null cells or in H1299 cells (Fig. 2B and C, lanes 5 and 6). In H1299 cells some lower-molecular-weight ubiquitylated p53 species were almost always detected; these are most likely the result of an interaction with the endogenous Hdm2 (30). However, this ubiquitylation is insufficient to target p53 for degradation, a finding supported by the observation that treatment with a proteasome inhibitor does not lead to stronger accumulation of ubiquitylated p53. The RING fingers of Mdmx and Mdm2 are rather well conserved on the amino acid level (51% identity), and so we investigated whether any part of the Mdmx RING could replace the function of the Mdm2 RING. First, we generated chimeras by use of the PmlI site (residues 461 and 462 in Mdm2), generating 22(2-x)P and 22(x-2)P (Fig. 2A). Fang et al. have shown that cysteine 449 of Hdm2 (C447 in Mdm2) is essential for the degradation of p53 by Hdm2 (6). The analogous position in Mdmx contains an asparagine (N447), and so we mutated N447 to C447 within the 22(x-2)P chimera. The activities of these chimeras were tested following cotransfection with p53 into p53- and mdm2-null cells and H1299 cells. All the chimeric proteins that contain part of the Mdmx RING lost the ability to ubiquitylate and degrade p53, although the 22(2-x)P protein showed some residual p53 ubiquitylation (Fig. 2B and C, top panels). Furthermore, no significant self-ubiquitylation activity was detected (Fig. 2B and C, bottom panels). 22(2-x)P showed some minor higher-molecular-weight forms in p53- and mdm2-null cells, possibly indicating some self-ubiquitylation (Fig. 2B, bottom panel, lanes 7 and 8), but the half-lives of all these chimeric proteins, as measured by a cycloheximide pulse-chase experiment with H1299 cells, were much prolonged compared to that of Mdm2 (data not shown). These results show that the C-terminal 27 aa of the Mdm2 protein (aa 463 to 489), only 14 of which are part of the RING domain, are essential for the ubiquitin ligase activity. The inability of these chimeric proteins to degrade p53 was confirmed by transfection into C33A cells and investigation of the level of endogenous p53 by immunofluorescence (data not shown).
FIG. 2.
Mdm2-Mdmx swaps at the C terminus of the RING finger inhibit ubiquitin ligase activity. (A) Schematic representation of Mdmx-Mdm2 chimeras with exchanges at the C terminus of the RING domain. (B) p53- and mdm2-null cells were transfected in duplicate with 200 ng of p53, with or without 1 μg of the indicated constructs, and with 1 μg of the His-ubiquitin expression vector and 1 μg of CMV-LacZ. Cells were either treated with 20 μM MG132 or mock treated 4 h before harvesting. Western blot analysis was performed on ubiquitylated proteins (His-purified) and total cell lysates. p53 was detected with DO-1, Mdm2 and chimeric proteins were detected with 4B2, and LacZ was detected with D19-2F3-2. (C) H1299 cells (in 9.4-cm-diameter dishes) were transfected with 500 ng of p53, with or without 2 μg of constructs, and with 2 μg of a His-ubiquitin expression vector and 2 μg of CMV-LacZ. Transfected cells were treated, and lysates were subsequently analyzed, as described for panel B.
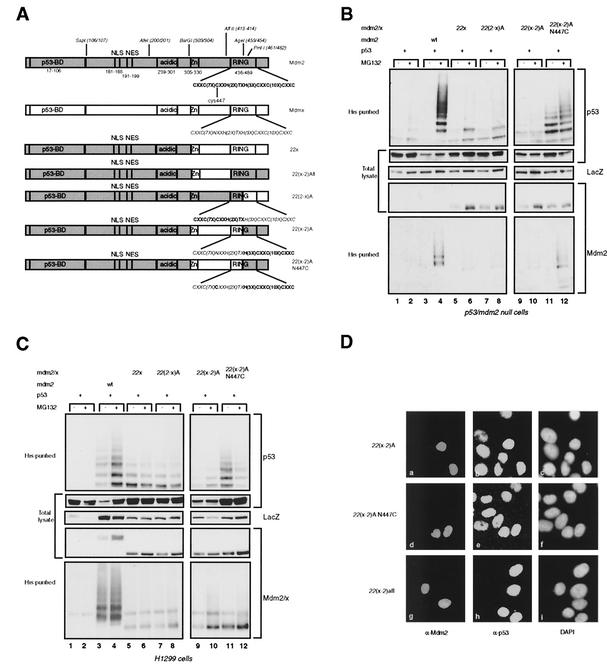
To investigate whether a small N-terminal part of the Mdm2 RING could be functionally replaced by the analogous part of the Mdmx RING, we created chimeras by use of the AgeI site at aa 453 and 454, generating 22(2-x)A and 22(x-2)A. The 22(x-2)A construct was also made with the N447C mutation (Fig. 3A). These chimeras were coexpressed with p53 in p53- and mdm2-null cells and H1299 cells, and it was found that all these chimeras are impaired in the ability to ubiquitylate and degrade p53 (Fig. 3B and C, top four panels). Transfection of the 22(x-2)A N447C construct resulted in the detection of ubiquitylated p53 species (Fig. 3B and C, top panels, lanes 11 and 12). However, the amount of ubiquitylated p53 did not increase upon treatment with the proteasome inhibitor and p53 was not degraded, suggesting that the ubiquitylation observed was not sufficient for degradation. The lack of degradation can be explained by the fact that the high-molecular-weight ubiquitylated p53 proteins were underrepresented relative to the low-molecular-weight ubiquitylated p53 species, compared to the ubiquitylation pattern of p53 observed after coexpression with Mdm2. In addition, relative to the total level of p53, the level of ubiquitylated species is much lower than that seen after coexpression of Mdm2. Taken together, these results indicate that a complete Mdm2 RING is necessary for efficient polyubiquitylation of p53 such that it can be degraded by the proteasome. The failure of these chimeric proteins to degrade p53 was confirmed by expression in C33A cells (Fig. 3D). Furthermore, these chimeric proteins were also virtually unable to self-ubiquitylate in both cell types (Fig. 3B and C, bottom panels), thereby enhancing their half-lives compared to that of Mdm2 (data not shown).
FIG. 3.
Mdm2-Mdmx exchanges at the N terminus of the RING finger prevent complete p53 ubiquitylation. (A) Schematic representation of the Mdmx-Mdm2 chimeras with exchanges at the N terminus of the RING or containing the complete Mdm2 RING. (B and C) p53- and mdm2-null cells (B) and H1299 cells (C) were transfected in duplicate with the indicated constructs; ubiquitylated proteins and total lysates were analyzed as described for Fig. 2B. (D) Immunofluorescence analysis of C33A cells transfected with 22(x-2)A, 22(x-2)A N447C, or 22(x-2)Afl. Cells were stained for expression of the chimeric proteins with 4B2 (a, d, and g) and for expression of p53 with FL393 (b, e, and h). Nuclei were stained with 4′,6′-diamidino-2-phenylindole (DAPI) (c, f, and i).
The results obtained with the 22(x-2)A and 22(x-2)A N447C chimeras do not formally prove that the defect in p53 ubiquitylation and degradation is due to modification of the function of the RING domain, since a contribution of the region containing the Zn finger and the linker sequences to the RING finger cannot be excluded. To investigate this possibility, the complete Mdm2 RING was substituted for the Mdmx RING in 22x, generating 22(x-2)Afl (Fig. 3A). Expression of this chimeric protein in C33A cells resulted in degradation of the endogenous p53 (Fig. 3D), indicating either that the region between aa 304 and 413 of Mdm2 is not important for p53 degradation and ubiquitylation or that the analogous region of Mdmx can fulfill the same function.
A central part of Mdm2 is essential for ubiquitylation of p53.
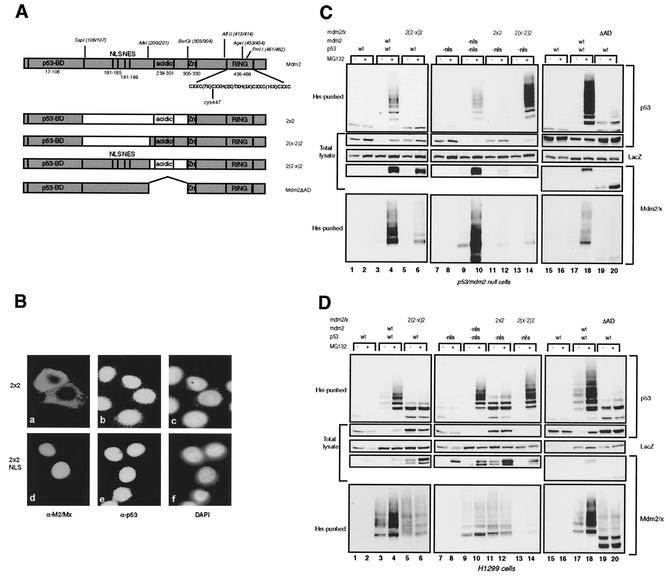
Since the RING finger of Mdm2 is responsible for ubiquitin ligase activity, we investigated if substituting the ZF/RF domain of Mdmx for that of Mdm2 in 2xx, generating 2x2 (Fig. 4A), would result in p53 ubiquitylation and degradation. Since this chimeric protein lacks the NLS of Mdm2 and contains the RING of Mdm2, it is mainly localized in the cytoplasm. Transfection into C33A cells showed the cytoplasmic localization and lack of degradation of endogenous p53 (Fig. 4Ba, b, and c). Fusion of the simian virus 40 NLS signal at the N terminus resulted in nuclear localization but not in degradation (Fig. 4Bd, e, and f). The 2x2 construct was also cotransfected with p53 into p53- and mdm2-null cells (Fig. 4C) and H1299 cells (Fig. 4D) in order to further examine the ubiquitylation pattern and degradation of p53. Since the 2x2 protein is cytoplasmic, in this transfection assay a p53 mutant lacking the main NLS was used, and an Mdm2 mutant lacking the NLS was used as positive control. We and others have shown previously that these mutant proteins are indeed predominantly cytoplasmic but that ubiquitylation and degradation of p53 still occurs very efficiently in the cytoplasm (1, 30, 34, 35). The 2x2 protein is virtually incapable of ubiquitylating p53, and no degradation was observed (Fig. 4C and D, top two panels, lanes 11 and 12). Importantly, the ubiquitin ligase activity per se of the 2x2 chimera remained, since self-ubiquitylation could still be detected (Fig. 4C and D). The amount of ubiquitylated 2x2 species appears to be small in the p53- and mdm2-null cells, but this is probably caused by the fact that the 2x2 protein is very unstable in these cells, and even after MG132 treatment, accumulation of the protein is much less than that of Mdm2. In H1299 cells the 2x2 protein appears to be more stable, an observation for which we have no explanation at the moment. To further define which part within the middle domain is important for p53 ubiquitylation and degradation, the NLS/NES region or the acidic domain-containing region of Mdm2 was substituted for that of Mdmx in this chimera, creating 2(2-x)2 and 2(x-2)2, which accumulate in the nucleus and cytoplasm, respectively (Fig. 4A). It was found with both cell types that the 2(x-2)2 chimera very efficiently ubiquitylates and degrades p53 (Fig. 4C and D, top two panels, lanes 13 and 14) while the 2(2-x)2 chimera is unable to do so (lanes 5 and 6), showing a critical importance of the Mdm2 acidic domain-containing region.
FIG. 4.
A central part in Mdm2 is essential for p53 ubiquitylation. (A) Schematic representation of the Mdmx-Mdm2 chimeras containing swaps in the central part and of the Mdm2ΔAD deletion mutant. (B) Immunofluorescence analysis of C33A cells transfected with 2x2 or NLS-2x2. Cells were stained for 2x2 with 4B2 (a and d) and for p53 with FL393 (b and e); nuclei were stained with 4′,6′-diamidino-2-phenylindole (DAPI) (c and f). (C and D) p53- and mdm2-null cells (C) and H1299 cells (D) were transfected in duplicate with the indicated constructs. Ubiquitylated proteins and total lysates were analyzed as described for Fig. 2B.
Apparently, the Mdm2 NLS/NES-containing region either is dispensable for this function or has a function that can be fulfilled by the analogous region of Mdmx. These results were confirmed by transfection into C33A cells. Interestingly, although the 2(x-2)2 protein is mainly cytoplasmic, in a proportion (10 to 20%) of the transfected C33A cells the endogenous p53 was still degraded (data not shown). The same observation was made with the Mdm2 NLS mutant (data not shown). Both chimeras still showed self-ubiquitylation, apparently low compared to that of Mdm2 (Fig. 4C and D, bottom panels, lanes 5, 6, 13, and 14). However, the levels of ubiquitylated species relative to total levels are similar after expression of Mdm2 or the different chimeras. Like 2x2, these chimeric proteins are expressed at low levels because of a short half-life in p53- and mdm2-null cells.
It has been reported that certain deletion mutants of Mdm2 lacking part of the acidic domain were impaired in the degradation, but not the ubiquitylation, of p53 (1, 36). The fact that the 2(2-x)2 chimera, in contrast, is unable to ubiquitylate p53 could mean that the acidic domain-containing region of Mdmx has a dominant-negative effect on the ubiquitylation of p53 by Mdm2. To investigate this possibility, we deleted this region (aa 202 to 302) from Mdm2, creating Mdm2ΔAD (Fig. 4A). Coexpression of Mdm2ΔAD with p53 resulted in the same pattern of ubiquitylation as that of 2(2-x)2, with only the lower-weight bands of ubiquitylated p53 detected (Fig. 4C and D, upper panels, lanes 19 and 20). This result led us to conclude that at least part of the acidic domain-containing region is necessary for the ubiquitylation of p53, in addition to the function essential for the degradation of ubiquitylated p53. In contrast to the chimeric proteins with exchanges in the middle domain, Mdm2ΔAD also showed impaired self-ubiquitylation (Fig. 4C and D, bottom panel, lanes 19 and 20), suggesting that the acidic domain-containing region of Mdmx can fulfill a function necessary for this activity.
The acidic domain and the RING finger can cooperate in trans.
In the transfection studies in C33A cells we encountered an interesting result. It had been shown previously that, in contrast to its location in C33A cells, Hdmx is mainly cytoplasmic in most other cell types, e.g., U2OS cells, but Mdm2 could efficiently translocate Hdmx into the nucleus (22). We wondered whether the cytoplasmic chimeric proteins could also be translocated in this way. To that end we expressed xx2 and 22x, either alone or together, in C33A cells. Figure 5B shows that xx2 is cytoplasmic (Fig. 5Ba, b, and c) and 22x is nuclear (Fig. 5Bj, k, and l). When these two chimeric proteins were coexpressed, xx2 was translocated into the nucleus (Fig. 5Bd, e, and f). In these cotransfections more than 80% of the transfected cells expressed both proteins. Double staining of the cells from the same transfections with anti-Mdmx and anti-p53 or with anti-Mdm2 and anti-p53 antibodies showed that, as expected, neither xx2 (Fig. 5Bg, h, and i) nor 22x (Fig. 5Bj, k, and l) was able to degrade the endogenous p53. However, after cotransfection of xx2 and 22x, endogenous p53 was either absent or present at very low levels in the cells expressing xx2 in the nucleus (Fig. 5Bm, n, and o). The same observation was made for the majority of the 22x-positive cells from the same cotransfection (data not shown). Taken together with the results obtained with the 2(x-2)2 and 2(2-x)2 hybrids, these findings allowed us to hypothesize that the two important domains for p53 ubiquitylation, the acidic domain-containing region and the RING finger, would be able to function independently in a trans fashion. To investigate this possibility, further transfections into p53- and mdm2-null cells were performed. It was found that, when either Mdm2ΔAD or 2(2-x)2 was coexpressed with 22x, p53 was efficiently ubiquitylated and degraded, while these proteins individually did not have these capacities (Fig. 5C). Transfection of the same chimeras into C33A cells confirmed these results. In C33A cells additional transfections were performed with an Mdm2 mutant from which the p53-binding domain (aa 1 to 111) was deleted (M2Δp53) but which contained an N-terminal Myc tag. Since M2Δp53 cannot bind p53, it failed to degrade p53 (Fig. 5Bs, t, and u). As a control to ensure that the Myc tag did not interfere with Mdm2 function, a Myc-tagged Mdm2 was also transfected and was found to degrade p53 (data not shown). Coexpression of M2Δp53 with 22x in C33A cells resulted in degradation of p53 (Fig. 5Bp, q, and r), indicating that the complementing proteins do not necessarily both require the ability to bind p53. These results strongly indicate that the RING domain and a central part in Mdm2 comprise two domains that perform distinct functions in the ubiquitylation of p53.
FIG. 5.
The central part of Mdm2 can function in a trans manner. (A) Schematic representation of the constructs used in the transfections. The Mdm2Δp53 construct carries the 9E10 Myc epitope at the N terminus. (B) C33A cells were transfected with the indicated constructs, and expression of the constructs and of p53 was evaluated by immunofluorescence. The xx2 protein was detected with the anti-Mdmx antibody p57 (a and d) or 6B1A (g and m). The 22x protein was stained with the anti-Mdm2 antibody 4B2 (e and j), and Mdm2Δp53 was stained with the anti-Myc antibody 9E10 (p and s). p53 was detected with antibody FL393 (h, k, n, q, and t), and nuclei were stained with 4′,6′-diamidino-2-phenylindole (DAPI). (C) p53- and mdm2-null cells were transfected in duplicate with the indicated constructs; ubiquitylated proteins and total lysates were analyzed as described for Fig. 2B.
DISCUSSION
In this study we investigated the roles of different domains in Mdm2 in the ubiquitylation and degradation of p53. In contrast to earlier studies, in which parts of Mdm2 were deleted (1, 33), we swapped domains between the structurally homologous Mdm2 and Mdmx proteins. It has been shown previously that Mdmx has no detectable in vivo ubiquitin ligase activity toward p53 (27). In addition, in vitro ubiquitylation studies revealed no detectable ubiquitin ligase activity of Mdmx, either toward p53 or toward itself (data not shown). Exchanges within and around the RING finger of Mdm2 and Mdmx indicated that a complete Mdm2-derived RING is necessary for efficient polyubiquitylation and degradation of p53. To our surprise, a chimeric Mdm2-Mdmx protein containing only aa 202 to 302 of Mdmx could not significantly ubiquitylate p53, although it still showed self-ubiquitylation. Deletion of the same region from Mdm2 affected the ability to ubiquitylate p53 similarly. However, this deletion mutant also showed an impaired ability to self-ubiquitylate, and it accumulated to higher levels than wild-type Mdm2. The latter observations correlate well with earlier reports showing that Mdm2 mutants lacking small parts within this region have greater stability than wild-type Mdm2 (1, 36). These data suggest that the analogous region in Mdmx can fulfill a function that is necessary for self-ubiquitylation but cannot provide an activity essential for efficient p53 ubiquitylation. The mechanism by which the central region of Mdm2 affects p53 ubiquitylation is still elusive. It has been reported that the ARF protein inhibits p53 ubiquitylation by binding to Mdm2 (33). However, Mdm2 deletion mutants (Δ222-272 and Δ217-247) (1, 36) that should not bind ARF can still ubiquitylate p53. This finding indicates that ARF binding and deletion of sequences that are needed for ARF binding have distinct functional effects. However, it is possible that the inhibition of p53 ubiquitylation by ARF has a stronger effect on the structure of the Mdm2 protein than deletion of a number of amino acids. In addition, it has been reported that pRB can inhibit the degradation of p53 by Mdm2 (13). Since the pRB-binding domain on Mdm2 partly overlaps with the region deleted in our Mdm2ΔAD mutant, it could be hypothesized that pRB binding to Mdm2 has the same effect as deletion of the middle region and thus represses the ubiquitylation of p53. Our preliminary results indicate that strong overexpression of pRB can partially inhibit p53 ubiquitylation, but the effect is much less than that observed after deletion of the central domain. Both ARF and pRB could, through their interaction with Mdm2, prevent the binding of other proteins essential in ubiquitylation. Binding of p300 in the central region of Mdm2 has also been shown to stimulate the ability of Mdm2 to degrade p53. However, the loss of p300 binding apparently leads not to decreased ubiquitylation but to a degradation defect of the ubiquitylated p53 (36). Similarly, mutations of several phosphorylatable serines within the acidic domain of Mdm2 have the same effect in that only degradation, not ubiquitylation, of p53 is impaired (2).
It has recently been shown that acetylation of p53 prevents the ubiquitylation of these acetylated lysines and that deacetylation is required for Mdm2-mediated ubiquitylation and degradation (14, 19). It was found that Mdm2 interacts with an HDAC1-containing complex and recruits this complex to p53, leading to its deacetylation. It is tempting to speculate that the Mdm2ΔAD mutant can no longer recruit this HDAC1-containing complex, resulting in impaired deacetylation and, consequently, reduced ubiquitylation. This model could also explain the complementation that is observed when two proteins, each alone incapable of ubiquitylating and degrading p53, can perform these functions together. Complementation needs p53 binding by at least one of the proteins. An active RING domain and an intact acidic domain can be provided by separate proteins. It can be hypothesized that the enzymatic activities needed for full ubiquitylation of p53, deacetylation and ubiquitin ligation, are fulfilled by these two main domains. The RING directs the ubiquitin ligase activity, and the acidic domain is essential for recruitment of the HDAC1-containing complex. This would also imply that complementation can occur only when the complementing proteins are both in a complex with p53. The experiments for which results are shown in Fig. 5 were all performed with complementing proteins, of which one contained the RING of Mdm2 and the other contained the RING of Mdmx. These RING domains are known to form a strong interaction, and we have indeed found that these proteins can very efficiently be coimmunoprecipitated, in a complex together with p53 (data not shown). Although less efficiently, we can find some degradation of p53 in C33A cells with the combination of Mdm2Δp53 and Mdm2ΔAD (∼20% of the Mdm2ΔAD-expressing cells), correlating with a much less efficient coimmunoprecipitation. The interaction might still take place via Mdm2 RING finger homodimerization. In addition, we find a somewhat stronger effect (∼25% of Mdm2ΔAD-expressing cells) by the combination of Mdm2ΔAD with Mdm2Δp53ΔRING (the latter deletion mutant of Mdm2 lacks both the p53-binding domain and the RING finger). Also, these proteins can be coimmunoprecipitated, together with p53, although again with low efficiency. These data are in agreement with the results described in the accompanying report by Kawai et al. (16a), in which it is shown that an Mdm2ΔAD mutant, very similar to ours, can be complemented by a short Mdm2 protein essentially representing only the NLS/NES region and the acidic domain. We have no direct evidence for the mechanism that recruits the Mdm2Δp53ΔRING protein to the p53/Mdm2ΔAD complex. However, it has been found that the RING domain of Mdm2 can not only homodimerize but also interact with the Mdm2 central acidic region (5), and this interaction was reported not to inhibit the E3 ligase activity of Mdm2. Although perhaps less likely, the possibility that the central domain of Mdm2 is mediating a direct association with p53 is not excluded. Although the major binding sites within both Mdm2 and p53 have been localized to the N termini of the proteins, there is precedent for the contribution of other regions to the interaction. Shimizu et al. found that an Mdm2-RNA complex associates not only with the N-terminal part but also with the core domain of p53 (28). Mutations in the p53 core domain that affected this second association also affected the ubiquitylation of p53 by Mdm2, in a manner dependent on the presence of the N-terminal binding site in p53. The region in Mdm2 that binds to the p53 core was not determined, but the central domain of Mdm2 could be the second p53-binding site.
Apart from a recruitment function of proteins helping Mdm2 in the ubiquitylation of p53, it is possible that the central domain somehow induces a conformational shift in p53 in such a way that the lysines to be ubiquitylated are more available. It has been suggested previously that the ubiquitylation of p53 is a stepwise process accompanied by a conformational alteration (9). In conclusion, we have shown that a central domain in Mdm2 has a critical function in the ubiquitylation of p53. Elucidation of the mechanism of p53 regulation, including the Mdm2-mediated degradation, is important for understanding the tumor suppressor functions of p53 and may lead to additional targets for p53 reactivation in cancer therapy.
Acknowledgments
We thank A. Levine, D. Lane, and M. Oren for anti-Mdm2 antibodies and Mdm2 expression plasmids, and we thank G. Lozano for p53- and mdm2-null cells. We thank S. Grivell for practical assistance in some of the experiments.
This work was supported by grants from the Dutch Cancer Society (grant RUL 2001-2523 to E.M. and R.F.; grant RUL 96-1329 to R.S.) and from the AICR (grant 01-276 to P.D.G.).
REFERENCES
- 1.Argentini, M., N. Barboule, and B. Wasylyk. 2001. The contribution of the acidic domain of MDM2 to p53 and MDM2 stability. Oncogene 20:1267-1275. [DOI] [PubMed] [Google Scholar]
- 2.Blattner, C., T. Hay, D. W. Meek, and D. P. Lane. 2002. Hypophosphorylation of Mdm2 augments p53 stability. Mol. Cell. Biol. 22:6170-6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bottger, V., A. Bottger, C. Garcia-Echeverria, Y. F. Ramos, A. J. van der Eb, A. G. Jochemsen, and D. P. Lane. 1999. Comparative study of the p53-mdm2 and p53-MDMX interfaces. Oncogene 18:189-199. [DOI] [PubMed] [Google Scholar]
- 4.Chen, J., J. Lin, and A. J. Levine. 1995. Regulation of transcription functions of the p53 tumor suppressor by the mdm-2 oncogene. Mol. Med. 1:142-152. [PMC free article] [PubMed] [Google Scholar]
- 5.Dang, J., M. L. Kuo, C. M. Eischen, L. Stepanova, C. J. Sherr, and M. F. Roussel. 2002. The RING domain of Mdm2 can inhibit cell proliferation. Cancer Res. 62:1222-1230. [PubMed] [Google Scholar]
- 6.Fang, S., J. P. Jensen, R. L. Ludwig, K. H. Vousden, and A. M. Weissman. 2000. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem. 275:8945-8951. [DOI] [PubMed] [Google Scholar]
- 7.Finch, R. A., D. B. Donoviel, D. Potter, M. Shi, A. Fan, D. D. Freed, C. Y. Wang, B. P. Zambrowicz, R. Ramirez-Solis, A. T. Sands, and N. Zhang. 2002. mdmx is a negative regulator of p53 activity in vivo. Cancer Res. 62:3221-3225. [PubMed] [Google Scholar]
- 8.Gu, J., H. Kawai, L. Nie, H. Kitao, D. Wiederschain, A. G. Jochemsen, J. Parant, G. Lozano, and Z. M. Yuan. 2002. Mutual dependence of MDM2 and MDMX in their functional inactivation of p53. J. Biol. Chem. 277:19251-19254. [DOI] [PubMed] [Google Scholar]
- 9.Gu, J., L. Nie, D. Wiederschain, and Z. M. Yuan. 2001. Identification of p53 sequence elements that are required for MDM2-mediated nuclear export. Mol. Cell. Biol. 21:8533-8546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haupt, Y., R. Maya, A. Kazaz, and M. Oren. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387:296-299. [DOI] [PubMed] [Google Scholar]
- 11.Honda, R., H. Tanaka, and H. Yasuda. 1997. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420:25-27. [DOI] [PubMed] [Google Scholar]
- 12.Honda, R., and H. Yasuda. 2000. Activity of MDM2, a ubiquitin ligase, toward p53 or itself is dependent on the RING finger domain of the ligase. Oncogene 19:1473-1476. [DOI] [PubMed] [Google Scholar]
- 13.Hsieh, J. K., F. S. Chan, D. J. O'Connor, S. Mittnacht, S. Zhong, and X. Lu. 1999. RB regulates the stability and the apoptotic function of p53 via MDM2. Mol. Cell 3:181-193. [DOI] [PubMed] [Google Scholar]
- 14.Ito, A., Y. Kawaguchi, C. H. Lai, J. J. Kovacs, Y. Higashimoto, E. Appella, and T. P. Yao. 2002. MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J. 21:6236-6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson, M. W., and S. J. Berberich. 2000. MdmX protects p53 from Mdm2-mediated degradation. Mol. Cell. Biol. 20:1001-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones, S. N., A. E. Roe, L. A. Donehower, and A. Bradley. 1995. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 378:206-208. [DOI] [PubMed] [Google Scholar]
- 16a.Kawai, H., D. Wiederschain, and Z.-M. Yuan. 2003. Critical contribution of the MDM2 acidic domain to p53 ubiquitination. Mol. Cell. Biol. 23:4939-4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kubbutat, M. H., S. N. Jones, and K. H. Vousden. 1997. Regulation of p53 stability by Mdm2. Nature 387:299-303. [DOI] [PubMed] [Google Scholar]
- 18.Li, C., L. Chen, and J. Chen. 2002. DNA damage induces MDMX nuclear translocation by p53-dependent and -independent mechanisms. Mol. Cell. Biol. 22:7562-7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, M., J. Luo, C. L. Brooks, and W. Gu. 2002. Acetylation of p53 inhibits its ubiquitination by Mdm2. J. Biol. Chem. 277:50607-50611. [DOI] [PubMed] [Google Scholar]
- 20.Little, N. A., and A. G. Jochemsen. 2001. Hdmx and Mdm2 can repress transcription activation by p53 but not by p63. Oncogene 20:4576-4580. [DOI] [PubMed] [Google Scholar]
- 21.Michael, D., and M. Oren. 2002. The p53 and Mdm2 families in cancer. Curr. Opin. Genet. Dev. 12:53-59. [DOI] [PubMed] [Google Scholar]
- 22.Migliorini, D., D. Danovi, E. Colombo, R. Carbone, P. G. Pelicci, and J. C. Marine. 2002. Hdmx recruitment into the nucleus by Hdm2 is essential for its ability to regulate p53 stability and transactivation. J. Biol. Chem. 277:7318-7323. [DOI] [PubMed] [Google Scholar]
- 23.Migliorini, D., E. Lazarini Denchi, D. Danovi, A. Jochemsen, M. Capillo, A. Gobbi, K. Helin, P. G. Pelicci, and J. C. Marine. 2002. Mdm4 (Mdmx) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Mol. Cell. Biol. 22:5527-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Momand, J., G. P. Zambetti, D. C. Olson, D. George, and A. J. Levine. 1992. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69:1237-1245. [DOI] [PubMed] [Google Scholar]
- 25.Montes de Oca Luna, R., D. S. Wagner, and G. Lozano. 1995. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378:203-206. [DOI] [PubMed] [Google Scholar]
- 26.Parant, J., A. Chavez-Reyes, N. A. Little, W. Yan, V. Reinke, A. G. Jochemsen, and G. Lozano. 2001. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat. Genet. 29:92-95. [DOI] [PubMed] [Google Scholar]
- 27.Prives, C. 1998. Signaling to p53: breaking the MDM2-p53 circuit. Cell 95:5-8. [DOI] [PubMed] [Google Scholar]
- 28.Shimizu, H., L. R. Burch, A. J. Smith, D. Dornan, M. Wallace, K. L. Ball, and T. R. Hupp. 2002. The conformationally flexible S9-S10 linker region in the core domain of p53 contains a novel MDM2 binding site whose mutation increases ubiquitination of p53 in vivo. J. Biol. Chem. 277:28446-28458. [DOI] [PubMed] [Google Scholar]
- 29.Shvarts, A., W. T. Steegenga, N. Riteco, T. van Laar, P. Dekker, M. Bazuine, R. C. van Ham, W. van der Houven van Oordt, G. Hateboer, A. J. van der Eb, and A. G. Jochemsen. 1996. MDMX: a novel p53-binding protein with some functional properties of MDM2. EMBO J. 15:5349-5357. [PMC free article] [PubMed] [Google Scholar]
- 30.Stad, R., N. A. Little, D. P. Xirodimas, R. Frenk, A. J. van der Eb, D. P. Lane, M. K. Saville, and A. G. Jochemsen. 2001. Mdmx stabilizes p53 and Mdm2 via two distinct mechanisms. EMBO Rep. 2:1029-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stad, R., Y. F. Ramos, N. Little, S. Grivell, J. Attema, A. J. van der Eb, and A. G. Jochemsen. 2000. Hdmx stabilizes Mdm2 and p53. J. Biol. Chem. 275:28039-28044. [DOI] [PubMed] [Google Scholar]
- 32.Vousden, K. H., and X. Lu. 2002. Live or let die: the cell's response to p53. Nat. Rev. Cancer 2:594-604. [DOI] [PubMed] [Google Scholar]
- 33.Xirodimas, D., M. K. Saville, C. Edling, D. P. Lane, and S. Lain. 2001. Different effects of p14ARF on the levels of ubiquitinated p53 and Mdm2 in vivo. Oncogene 20:4972-4983. [DOI] [PubMed] [Google Scholar]
- 34.Xirodimas, D. P., C. W. Stephen, and D. P. Lane. 2001. Cocompartmentalization of p53 and Mdm2 is a major determinant for Mdm2-mediated degradation of p53. Exp. Cell Res. 270:66-77. [DOI] [PubMed] [Google Scholar]
- 35.Yu, Z. K., R. K. Geyer, and C. G. Maki. 2000. MDM2-dependent ubiquitination of nuclear and cytoplasmic P53. Oncogene 19:5892-5897. [DOI] [PubMed] [Google Scholar]
- 36.Zhu, Q., J. Yao, G. Wani, M. A. Wani, and A. A. Wani. 2001. Mdm2 mutant defective in binding p300 promotes ubiquitination but not degradation of p53: evidence for the role of p300 in integrating ubiquitination and proteolysis. J. Biol. Chem. 276:29695-29701. [DOI] [PubMed] [Google Scholar]