Abstract
The mitotic exit network (MEN), a Ras-like signaling cascade, promotes the release of the protein phosphatase Cdc14 from the nucleolus and is essential for cells to exit from mitosis in Saccharomyces cerevisiae. We have characterized the functional domains of one of the MEN components, the protein kinase Cdc15, and investigated the role of these domains in mitotic exit. We show that a region adjacent to Cdc15's kinase domain is required for self-association and for binding to spindle pole bodies and that this domain is essential for CDC15 function. Furthermore, we find that overexpression of CDC15 lacking the C-terminal 224 amino acids results in hyperactivation of MEN and premature release of Cdc14 from the nucleolus, suggesting that this domain within Cdc15 functions to inhibit MEN signaling. Our findings indicate that multiple modes of MEN regulation occur through the protein kinase Cdc15.
Exit from mitosis is the final cell cycle transition that leads to the production of two genetically identical daughter cells. Proper execution of this transition is essential for each of the two daughter cells to receive a complete complement of the genome and, thus, to maintain ploidy. In all eukaryotes examined to date, exit from mitosis is brought about by the inactivation of mitotic cyclin-dependent kinase (mitotic CDK) activity (reviewed in reference 26). In budding yeast (Saccharomyces cerevisiae), the protein phosphatase Cdc14 induces the inactivation of mitotic CDKs by reversing CDK-dependent phosphorylation on proteins important for promoting mitotic CDK inactivation (14, 15, 41; reviewed in reference 2).
Cdc14 is regulated by an inhibitor, Cfi1/Net1. Cfi1/Net1 inhibits and sequesters Cdc14 in the nucleolus throughout most of the cell cycle (34, 42). Only during anaphase and telophase is Cdc14 released from its inhibitor (34, 36, 42). To date, two pathways have been identified that control Cdc14 localization: the FEAR network (Cdc 14 early anaphase release) (30, 35, 37, 44) and the mitotic exit network (MEN) (reviewed in references 2, 17, and 24). The FEAR network appears to be activated at the metaphase-anaphase transition and promotes Cdc14 release from the nucleolus during early anaphase. MEN is thought to be activated when the nucleus migrates into the daughter cell during anaphase (3, 29) and is essential for promoting and sustaining release of Cdc14 from the nucleolus during late stages of anaphase. Temperature-sensitive mutations in MEN components cause cells to arrest in telophase with Cdc14 sequestered in the nucleolus and high levels of mitotic CDK activity (15, 32, 34, 38, 41).
MEN is a signal transduction pathway comprised of a GTPase, Tem1; the putative guanine nucleotide exchange factor Lte1; the two-component GTPase-activating protein complex Bub2-Bfa1; the protein kinases Cdc15, Dbf2, and Cdc5; a Dbf2-associated factor, Mob1; and a scaffold protein, Nud1 (reviewed in reference 2). Localization of MEN components to spindle pole bodies (SPBs) and in vitro assays for Dbf2 kinase activation have been used to order MEN components into a pathway (3, 4, 10, 19, 22, 23, 40, 43, 45). These analyses revealed that Tem1 binds to and functions upstream of Cdc15, which activates Dbf2 in a Mob1-dependent manner. The polo kinase Cdc5 regulates the activity of several MEN components (13, 19, 40).
Most MEN components have been shown to localize to SPBs (reviewed in reference 2). Tem1, Bub2, and Bfa1 predominantly localize to the SPB that migrates into the daughter cell during anaphase (3, 6, 9, 29). Cdc5 localizes to both SPBs during metaphase and anaphase (33), whereas Cdc15 and Dbf2 localize to both SPBs only during anaphase (4, 10, 25, 40, 43). Localization of Dbf2 to SPBs coincides with activation of Dbf2 kinase activity, suggesting that SPB association of Dbf2 is a prerequisite for Dbf2 activation (8, 40). Localization of most MEN components to SPBs requires the SPB component Nud1. Cells carrying a temperature-sensitive allele in NUD1 arrest in telophase with Tem1, Cdc15, and Dbf2 mislocalized, suggesting that SPB localization of MEN components is important for promoting mitotic exit (3, 11, 40).
Three cellular events have been identified that control MEN activation: spindle position, mitotic spindle damage, and DNA damage. All are thought to regulate Tem1 through Lte1 and Bub2-Bfa1 (reviewed in references 2, 12, and 24). However, although Bub2-Bfa1 and Lte1 are important for controlling MEN activity, several lines of evidence suggest that MEN is regulated at additional levels. Neither LTE1 nor BUB2 nor BFA1 is essential for viability. Furthermore, in an unperturbed cell cycle, neither deletion of BUB2 or BFA1 nor overexpression of LTE1 causes a cell cycle defect or a premature release of Cdc14 from the nucleolus (3). Thus, other components of MEN are likely subject to regulation.
One MEN component known to be a target of regulation is the protein kinase Cdc15. Cdc14 released from the nucleolus by the FEAR network during early anaphase causes dephosphorylation of Cdc15, allowing it to become a more potent activator of mitotic exit (16, 25, 35, 43). In order to identify if additional modes of Cdc15 regulation exist, we have determined how different domains of Cdc15 function to regulate mitotic exit. We mapped the region of Cdc15 required for association with SPBs to a domain adjacent to the kinase domain. Our analyses also show that Cdc15 self-associates and that this self-association is mediated through a region that overlaps with the SPB localization domain of Cdc15. This domain is essential for viability, suggesting that SPB association and/or self-association is required for CDC15 function. Finally, we find that overexpression of CDC15 lacking the C-terminal 224 amino acids (aa), but not overexpression of full-length CDC15, causes hyperactivation of MEN as judged by elevated levels of Dbf2 kinase activity and premature release of Cdc14 from the nucleolus. Our results suggest the presence of at least three functional domains within Cdc15 aside from the kinase domain: an SPB association domain, a self-association domain, and a domain that inhibits MEN signaling.
MATERIALS AND METHODS
Growth conditions and yeast strains.
All strains are derivatives of strain W303 (A2587) and are listed in Table 1. To construct CDC15 truncation strains, the starting diploid strain A4267 (W303 diploid, TEM1-3MYC) was used. The GAL1 promoter and the DNA sequence encoding green fluorescent protein (GFP) were inserted either upstream of, or at positions internal to, one genomic copy of CDC15 by a PCR-based method (21). C-terminal truncations were then created using the diploid strains with GAL1-driven CDC15 constructs and insertion of a stop codon at positions internal to CDC15. To obtain haploid derivatives used in synchronous α-factor release experiments, spores from diploid strains were sonicated and mated to A3932 (cdc15::HIS3 leu2::CDC15-HA3-LEU2, SLJ511; a gift of D. Morgan), and appropriate diploids were selected and sporulated to give strains with the GAL1 CDC15 constructs at the CDC15 locus and CDC15-3HA at the LEU2 locus. Further details of strains and strain construction can be obtained upon request. Growth conditions for individual experiments are described in the figure legends. Where growth medium is unspecified, cells were grown in yeast extract-peptone (YEP) plus 2% glucose.
TABLE 1.
Strain list
| Strain | Relevant genotype |
|---|---|
| A1411 | MATaCDC14-3HA |
| A1828 | MATaTEM1-3MYC |
| A2587 | (K699) MATaade2-1 leu2-3 ura3 trp1-1 his3-11,15 can1-100 GAL |
| A3932 | MATacdc15::HIS3 CDC15-3HA::LEU2 |
| A4376 | MATa/α TEM1/TEM1-3MYC cdc15::GAL-GFP-CDC15 [267-974]/CDC15 |
| A4403 | MATa/α TEM1/TEM1-3MYC cdc15::GAL-GFP-CDC15 [267-750]/CDC15 |
| A4441 | MATa/α TEM1/TEM1-3MYC cdc15::GAL-GFP-CDC15/CDC15 |
| A4677 | MATaTEM1-3MYC CDC15-3HA::LEU2 cdc15::GAL-GFP-CDC15 [1-376] |
| A4678 | MATα TEM1-3MYC CDC15-3HA::LEU2 cdc15::GAL-GFP-CDC15 [1-376] |
| A4680 | MATaTEM1-3MYC CDC15-3HA::LEU2 cdc15::GAL-GFP-CDC15 [267-429] |
| A4683 | MATα CDC15-3HA::LEU2 cdc15::GAL-GFP-CDC15 |
| A4685 | MATaTEM1-3MYC CDC15-3HA::LEU2 cdc15::GAL-GFP-CDC15 [267-974] |
| A4686 | MATαTEM1-3MYC CDC15-3HA::LEU2 cdc15::GAL-GFP-CDC15 [267-974] |
| A4687 | MATaTEM1-3MYC CDC15-3HA::LEU2 cdc15::GAL-GFP-CDC15 [267-550] |
| A4690 | MATaCDC15-3HA::LEU2 cdc15::GAL-GFP-CDC15 [1-273] |
| A4691 | MATaTEM1-3MYC CDC15-3HA::LEU2 cdc15::GAL-GFP-CDC15 [1-550] |
| A4692 | MATα TEM1-3MYC CDC15-3HA::LEU2 cdc15::GAL-GFP-CDC15 [1-550] |
| A4695 | MATaCDC15-3HA::LEU2 cdc15::GAL-GFP-CDC15 [267-750] |
| A5051 | MATaTEM1-3MYC CDC15-3HA::LEU2 cdc15::GAL-GFP-CDC15 [267-702] |
| A5254 | MATa/α cdc15::GAL-GFP-CDC15 [361-974]/CDC15 |
| A5401 | MATaCDC15-13MYC::HIS3 |
| A5464 | MATaCDC15-3HA::LEU2 cdc15::GAL-GFP-CDC15 [361-974] |
| A5466 | MATaCDC15-3HA::LEU2 cdc15::GAL-GFP-CDC15 [395-974] |
| A5568 | MATaDBF2-3MYC CDC15-3HA::LEU2 cdc15::GAL-GFP-CDC15 |
| A5571 | MATaDBF2-3MYC CDC15-3HA::LEU2 cdc15::GAL-GFP-CDC15 [267-750] |
| A5574 | MATaTEM1-3MYC CDC15-3HA::LEU2 cdc15::GAL-GFP-CDC15 [1-750] |
| A5575 | MATα CDC15-3HA::LEU2 cdc15::GAL-GFP-CDC15 [1-550] |
| A5576 | MATaCDC15-3HA::LEU2 cdc15::GAL-GFP-CDC15 [1-702] |
| A5578 | MATa TEM1-3MYC CDC15-3HA3::LEU2 cdc15::GAL-GFP-CDC15 [1-464] |
| A5580 | MATaCDC15-3HA::LEU2 cdc15::GAL-GFP-CDC15 [1-429] |
| A5909 | MATaCDC15-3HA::LEU2 cdc15::GAL-GFP-CDC15 [361-702] |
| A5914 | MATaCDC15-3HA::LEU2 cdc15::GAL-GFP-CDC15 [395-702] |
| A5965 | MATaCDC14-3HA CDC15-3HA::LEU2 cdc15::GAL-GFP-CDC15 |
| A5966 | MATaCDC14-3HA CDC15-3HA::LEU2 cdc15::GAL-GFP-CDC15 [1-750] |
| A5968 | MATaCDC14-3HA CDC15-3HA::LEU2 cdc15::GAL-GFP-CDC15 [267-750] |
| A6130 | MATaDBF2-3MYC CDC15-3HA::LEU2 cdc15::GAL-GFP-CDC15 [1-750] |
| A6501 | MATaCDC15-3HA::LEU2 CDC15-13MYC::HIS3 |
| A6502 | MATaCDC15 [Δ362-464]-3HA::LEU2 CDC15-13MYC::HIS3 |
| A6503 | MATaCDC15 [Δ465-550]-3HA::LEU2 CDC15-13MYC::HIS3 |
| A6542 | MATaTEM1-3MYC CDC15-3HA::LEU2 |
| A6543 | MATaTEM1-3MYC CDC15 [Δ362-464]-3HA::LEU2 CDC15-13MYC::HIS3 |
| A6544 | MATaTEM1-3MYC CDC15 [Δ465-550]-3HA::LEU2 CDC15-13MYC::HIS3 |
| A6546 | MATacdc15-2 CDC15-3HA::LEU2 |
| A6547 | MATacdc15-2 CDC15 [Δ362-464]-3HA::LEU2 |
| A6548 | MATacdc15-2 CDC15 [Δ465-550]-3HA::LEU2 |
| A8371 | MATacdc15::GAL-GFP-CDC15 |
| A8372 | MATacdc15::GAL-GFP-CDC15 [1-750] |
| A8374 | MATα cdc15::GAL-GFP-CDC15 [1-702] |
| A8375 | MATacdc15::GAL-GFP-CDC15 [1-550] |
| A8778 | MATα GAL-SPO12::URA3 cdc15::GAL-GFP-CDC15 [267-750] |
| A8853 | MATaGAL-SPO12::URA3 cdc15::GAL-GFP-CDC15 [267-702] |
Strains containing CDC15 with internal deletions were constructed by inserting plasmids containing the deletion construct into the LEU2 locus. Plasmids lacking aa 362 to 464 (pCDC15 [Δ362-464]-3HA) or aa 465 to 550 (pCDC15 [Δ465-550]-3HA) were constructed by standard cloning techniques with pRS305-CDC15-3HA plasmid (a gift of D. Morgan). This plasmid contains CDC15 expressed from the endogenous promoter C-terminally tagged with the 3-hemagglutinin (3HA) epitope. GAL1-driven versions of these constructs were created by insertion of a PCR-derived GAL1-GFP upstream of the start codon of the integrated plasmids (21).
Immunofluorescence and fluorescence imaging.
Cells expressing GFP fusion proteins were fixed for 10 min in 3.7% formaldehyde-0.1 M potassium phosphate, pH 6.4, followed by 1 min of fixation in 70% ethyl alcohol. The cell pellet was resuspended in 1 ng of DAPI (4′,6′-diamidino-2-phenylindole)/ml in 0.1 M potassium phosphate, pH 7.5. Cells were stored at 4°C until analysis. Fixation and immunofluorescence assays to analyze mitotic spindles were performed as previously described (42).
Coimmunoprecipitation techniques, immunoblot analysis, and Dbf2 and Cdc15 kinase assays.
For coimmunoprecipitation, 50 ml of mid-log-phase cells was harvested and washed with 10 mM Tris, pH 7.5. Pellets were resuspended in 200 μl of NP-40 lysis buffer (150 mM NaCl, 50 mM Tris [pH 7.5], 1% NP-40, 1 mM dithiothreitol, 1 mM Pefaflock [Roche], 10 μg of TLCK [Nα-p-tosyl-l-lysine chloromethyl ketone; Sigma]/ml, 10 μg of TPCK [tosylsulfonyl phenylalanyl chloromethyl ketone; Sigma]/ml, 10 μg of pepstatin A [Sigma]/ml, 10 μg of leupeptin [Sigma]/ml, 40 μg of aprotinin [Sigma]/ml). Glass beads (Sigma) were added, and samples were vortexed for 10 min on a Vibrax vortexer. Extracts were then centrifuged at 10,000 × g three times for 8 min each. One milligram of precleared extract in 200 μl of NP-40 lysis buffer was used for immunoprecipitation. and 4 μl of antibody was added and incubated for 1 h at 4°C (either mouse monoclonal anti-GFP [Chemicon], mouse monoclonal anti-HA.11 [Covance], or mouse monoclonal anti-MYC 9E10 [Covance] was used). Twenty-five microliters of protein G-Sepharose beads (Pierce) (for GFP and HA immunoprecipitations) or 25 ml of protein A-Sepharose (Amersham) (for MYC immunoprecipitations) was used and incubated at 4°C for 2 h. Supernatant was removed, and beads were then washed five times with NP-40 lysis buffer. Sample buffer was added, and samples were boiled and run on sodium dodecyl sulfate-polyacrylamide gels followed by transfer to nitrocellulose membrane (VWR).
Western blot analysis was conducted using the following antibodies and dilutions in phosphate-buffered saline-Tween (PBST): primary 1/1,000 polyclonal rabbit anti-GFP (AbCam), primary 1/1,000 polyclonal rabbit anti-MYC (Gramsch), primary 1/1,000 mouse monoclonal anti-HA.11 (Covance), primary 1/1,000 rabbit polyclonal anti-Clb2, primary 1/1,000 polyclonal anti-Cdc28, secondary 1/2,000 horseradish peroxidase (HRP)-conjugated anti-rabbit immunoglobulin (Amersham), and secondary 1/2,000 HRP-conjugated anti-mouse immunoglobulin (Amersham). Membranes were blocked in PBST plus 5% milk for 1 h and then incubated with primary antibodies diluted in PBST plus 2% bovine serum albumin and incubated overnight at 4°C. Washes in PBST were followed by incubation with secondary antibodies diluted in PBST plus 1% bovine serum albumin and 1% milk for 5 h at room temperature. Supersignal West Pico chemiluminescence substrate from Pierce was used to detect HRP.
Immunoprecipitates for kinase assays were obtained as described above except that 60 mM β-glycerophosphate, 0.1 mM NaVO3, and 15 mM para-nitrophenylphosphate were also added to NP-40 lysis buffer and protease inhibitors. Cdc15 kinase assays were performed as described in reference 16. Dbf2 kinase assays were performed as described in reference 40.
RESULTS
The first 550 aa of Cdc15 are sufficient to support viability when overexpressed.
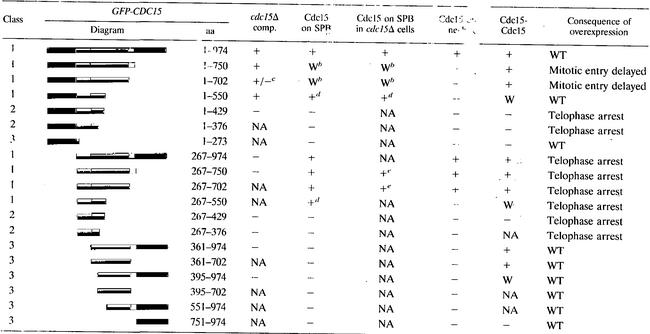
To identify the functional domains within CDC15, we first determined which region of the protein was essential for viability. We constructed N-terminal as well as C-terminal truncations of CDC15 and expressed these truncations from the galactose-inducible GAL1-10 promoter and, to facilitate detection of the protein, fused the GFP to the N terminus of the truncated proteins. The ability of various Cdc15 truncations to support growth in cells carrying a deletion of the endogenous copy of CDC15 was then assayed in the presence of galactose. We found that cells carrying truncations GFP-Cdc15 [1-750], GFP-Cdc15 [1-702], and GFP-Cdc15 [1-550] as the sole source of CDC15 were able to grow in the presence of galactose (Table 2). Further truncation of the protein as well as deletion of the kinase domain, which is located within the first 267 aa of the protein, led to loss of Cdc15 function (Table 2). We conclude that the first amino acids, 1 to 550, are sufficient to support cell division, at least when the protein is overproduced.
TABLE 2.
Cdc15 localization and self-association in cells overexpressing various CDC15 truncationsa
Abbreviations: comp., complementation; W, weak association; WT, wild type; NA, not analyzed.
Localizes to SPBs but also forms large aggregates in the cytoplasm.
Allows approximately 50% of spores to germinate and form colonies in the absence of full-length CDC15.
Localizes to SPBs but also along the mitotic spindle.
Localization was analyzed for cells overexpressing SPO12 from the GAL1-10 promoter to ensure survival.
The kinase and C-terminal domains of Cdc15 are dispensable for its localization to the SPB.
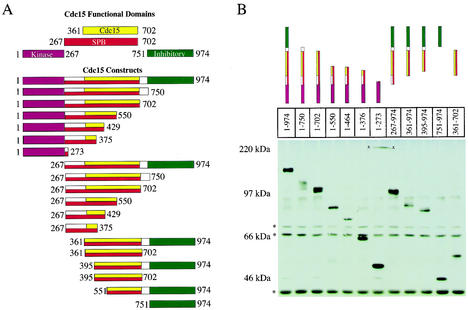
The association of Cdc15 with SPBs is cell cycle regulated. SPB localization occurs during anaphase and depends on TEM1 (4, 25, 40, 43). To identify regions within Cdc15 that are required for its association with SPBs, we examined the localization of various GFP-Cdc15 truncation mutants (Fig. 1A). As endogenous Cdc15 is exceedingly difficult to detect in cells, we employed the GAL1 promoter to drive expression of the CDC15 truncations (Fig. 1B).
FIG. 1.
Functional domains of Cdc15 and expression levels of various truncation constructs. (A) The N-terminal kinase domain (aa 1 to 267) is shown in purple; the self-association domain (aa 361 to 702) is shown in yellow; the SPB localization domain (aa 267 to 702) is shown in red, and the inhibitory domain (aa 751 to 974) is shown in green. (B) Anti-GFP Western blot analysis from strains grown in YEP plus 2% raffinose and induced with 2% galactose for 2.5 h. (*, cross-reacting bands; x, nonspecific mark on membrane). The following strains were used: A4683, A5575, A5576, A4692, A5578, A4678, A4690, A4685, A5464, A5466, A6456, and A5909.
The association of the overexpressed full-length GFP-Cdc15 with SPBs during the cell cycle was indistinguishable from that of the endogenous Cdc15 protein tagged with either 3HA or 13MYC epitopes (40; data not shown). GFP-Cdc15 localized to SPBs during anaphase and telophase but not significantly during other cell cycle stages (Fig. 2A). Previous data and our analysis of live cells showed that GFP-Cdc15 also localized to the bud neck, which is not detected by indirect in situ immunofluorescence with Cdc15-3HA or Cdc15-13MYC fusions, presumably due to fixation conditions (Table 2) (4, 25, 40, 43).
FIG. 2.
The aa 267 to 702 of Cdc15 are sufficient for SPB localization. (A) Cells were grown to mid-log phase in YEP plus 2% raffinose and induced for 2.5 h with 2% galactose, and Cdc15-GFP was examined in cells producing GFP-Cdc15 [1-974, full-length] (A4441), GFP-Cdc15 [267-974] (A4376), GFP-Cdc15 [267-750] (A4403), or GFP-Cdc15 [361-974] (A5254). (B) Cells were grown to mid-log phase in YEP plus 2% raffinose. Galactose was then added, and the accumulation of GFP-Cdc15 [1-974, full-length] (A4683) and GFP-Cdc15 [361-974] (A5464) was determined at the indicated times. (C) Cells of genotypes described for panel B were induced with galactose for 4 h, and the localization of GFP-Cdc15 full-length and GFP-Cdc15 [361-974] was determined.
Having established that overexpressed GFP-Cdc15 associates with SPBs in a cell cycle-dependent manner, we made a series of truncations to determine which region in Cdc15 was required for its association with this organelle. Because CDC15 is an essential gene, the Cdc15 truncations employed in this analysis were expressed in cells also containing full-length CDC15 under its endogenous promoter (summarized in Fig. 1A). GFP-Cdc15 lacking the N-terminal kinase domain (GFP-Cdc15 [267-974]) localized to both SPBs (Fig. 2; Table 1). A C-terminal deletion of this construct demonstrated that aa 751 to 974 were also dispensable for SPB localization, as GFP-Cdc15 [267-750] localized to both SPBs (Fig. 2A; Table 2). The localization of GFP-Cdc15 [267-750] to SPBs appeared even stronger than that of the full-length protein. Truncation GFP-Cdc15 [267-702] also localized to SPBs but was somewhat reduced compared to truncation GFP-Cdc15 [267-750] but more pronounced than full-length Cdc15 (Table 2). Truncations GFP-Cdc15 [1-750], GFP-Cdc15 [1-702], GFP-Cdc15 [1-550], GFP-Cdc15 [267-750], and GFP-Cdc15 [267-702] also localized to SPBs in the absence of endogenous full-length Cdc15 (Table 2), indicating that association of these truncated proteins is not mediated through binding to endogenous full-length Cdc15. (Note that cells carrying CDC15 truncations lacking the kinase domain as the sole source of CDC15 were kept alive by overexpression of the FEAR network component SPO12 from the GAL1-10 promoter.) Further removal of the N terminus or C terminus resulted in defects or loss of SPB localization (Table 2). GFP-Cdc15 [267-550] localized to SPBs but was also found in a punctate pattern along the mitotic spindle (data not shown). Further truncation N-terminally or C-terminally resulted in loss of SPB localization (Table 2). An example of such a truncation, GFP-Cdc15 [361-974], is shown in Fig. 2B and C. Whereas full-length Cdc15 was detected on SPBs of anaphase cells 3 h after galactose induction, GFP-Cdc15 [361-974] was not detected even after 23 h of induction. Notably, a Cdc15 truncation that has previously been shown to include amino acids sufficient for Tem1 binding (GFP-Cdc15 [267-376] [1]) did not localize to SPBs (Table 2; data not shown). Our results demonstrate that aa 267 to 702 of GFP-Cdc15 are necessary and sufficient for a wild-type pattern of SPB association. Thus, the region within Cdc15 required for SPB association is much larger than that reported to be sufficient for Tem1 binding, suggesting that mechanisms other than Tem1 binding are necessary for a stable association of Cdc15 with SPBs.
The use of GFP-Cdc15 fusions also enabled us to determine which region within Cdc15 was necessary for its association with the bud neck. We found that GFP-Cdc15 [267-702] was the smallest fragment able to localize to the bud neck (Table 2). We conclude that SPB and bud neck localization are mediated through a similar region within Cdc15.
Cdc15 self-associates and is mediated by aa 360 to 702.
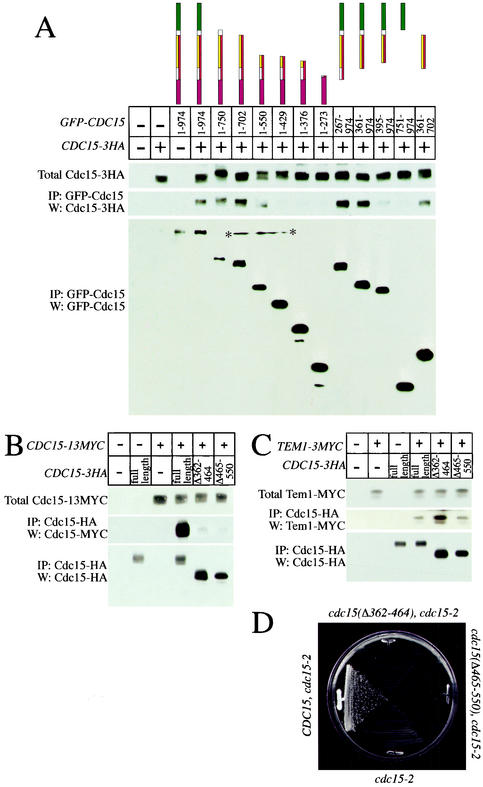
The importance of dimerization in the regulation of kinases has been highlighted in recent years with the discovery that Raf is positively regulated through dimerization and that PAK is negatively regulated through a transautoinhibitory dimerization mechanism (7, 28). To determine whether Cdc15 could associate with itself, we performed coimmunoprecipitation analyses with GFP-Cdc15 expressed from the GAL1 promoter and endogenous Cdc15 tagged with 3HA epitopes. The GFP-Cdc15 fusion immunoprecipitated Cdc15-3HA and vice versa (Fig. 3A; data not shown). Endogenous Cdc15-3HA also immunoprecipitated Cdc15-13MYC produced at endogenous levels (Fig. 3B), indicating that the association between overproduced GFP-Cdc15 and Cdc15-HA was not an artifact of overexpression. We conclude that Cdc15 self-associates. Whether this self-association is direct or is mediated by another factor is unknown.
FIG. 3.
Cdc15 contains a domain that mediates self-association. (A) Cells were grown in YEP plus 2% raffinose and induced with 2% galactose for 2.5 h, and GFP-Cdc15 constructs were immunoprecipitated to determine the amount of associated Cdc15-3HA. (Top) Amount of Cdc15-3HA in 50 μg of whole-cell extracts; (middle and bottom) amount of Cdc15-3HA immunoprecipitated with GFP-Cdc15 and amount of immunoprecipitated GFP-Cdc15 construct, respectively, each from 500 μg of cell extract (*, nonspecific mark on membrane). The following strains were used: A2587, A3932, A4441, A4683, A5575, A5576, A4692, A5578, A4678, A4690, A4685, A5464, A5466, A6456, and A5909. (B) Loss of aa 362 to 464 or 465 to 550 disrupts the ability of Cdc15-3HA to immunoprecipitate Cdc15-13MYC. Cells were grown in yeast extract-peptone-dextrose to mid-log phase, and Cdc15-3HA was immunoprecipitated. (Top) Amount of Cdc15-13MYC in extracts; (middle) amount of Cdc15-13MYC immunoprecipitated with Cdc15-3HA; (bottom) amount of immunoprecipitated Cdc15-3HA. The following strains were used: A2587, A3932, A5401, A6501, A6502, and A6503. (C) Loss of self-association domains does not affect the ability of Cdc15 to interact with Tem1. Cells were grown in yeast extract-peptone-dextrose to mid-log phase, and Cdc15-3HA was immunoprecipitated. (Top) Amount of Tem1-3MYC in extracts; (middle) amount of Tem1-3MYC immunoprecipitated with Cdc15-3HA; (bottom) amount of immunoprecipitated Cdc15-3HA. The following strains were used: A2587, A1828, A6542, A6543, and A6544. (D) The self-association domain is essential for viability. Strains expressing either wild-type CDC15 (A6546 [cdc15-2 CDC15-3HA]) or CDC15 lacking amino acids required for self-association (A6547 [cdc15-2 CDC15(Δ362-464-3HA)] and A6548 [cdc15-2 CDC15(Δ465-550-3HA)]) were streaked to single colonies on yeast extract-peptone-dextrose plates and grown at 37°C for 36 h.
To determine the region of Cdc15 that mediates interaction with itself, various GFP-Cdc15 truncations produced from the GAL1 promoter were tested for their ability to immunoprecipitate Cdc15-3HA. Removal of the first 360 aa of the GFP-Cdc15 product did not significantly affect the interaction of GFP-Cdc15 with Cdc15-3HA (Fig. 3A; Table 2), indicating that the kinase domain and the Tem1 binding domain (as defined by reference 1) were dispensable for this interaction. The removal of the C terminus (aa 703 to 974) also did not significantly affect self-association, as GFP-Cdc15 [361-702] efficiently immunoprecipitated Cdc15-3HA (Fig. 3A; Table 2). Further deletion of C-terminal and N-terminal regions reduced the ability of Cdc15 to self-associate. GFP-Cdc15 [1-550] retained some ability to bind to Cdc15-3HA; however, GFP-Cdc15 [1-429] did not (Fig. 3A; Table 2). Our results show that region aa 361 to 702 is sufficient for Cdc15 to self-associate.
To further define the self-association domain within Cdc15, we removed two regions within Cdc15, aa 362 to 464 (Cdc15-3HA [Δ362-464]) and 465 to 550 (Cdc15-3HA [Δ465-550]), and assayed the ability of these constructs expressed from the endogenous promoter to bind to Cdc15-13MYC (also expressed from the endogenous promoter). Removal of aa 362 to 464 or 465 to 550 dramatically reduced the interaction of Cdc15-3HA with Cdc15-13MYC (Fig. 3B). Deletion of these regions also affected CDC15 function, as the cdc15-3HA [Δ362-464] and cdc15-3HA [Δ465-550] constructs did not complement the temperature-sensitive lethality of a cdc15-2 allele (Fig. 3D). Loss of function was not due to degradation of an unstable protein at 37°C, since protein levels of Cdc15-3HA [Δ362-464] and Cdc15-3HA [Δ465-550] were comparable to that of wild-type protein at 37°C (data not shown). Truncations Cdc15-3HA [Δ362-464] and Cdc15-3HA [Δ465-550] retained the ability to immunoprecipitate Tem1 (Fig. 3C) and retained in vitro kinase activity (data not shown), and overexpression of CDC15 lacking either region caused a dominant-negative cell cycle arrest in late anaphase/telophase, indicating that the proteins were not grossly misfolded. We conclude that aa 361 to 702 are required and sufficient for Cdc15 to self-associate and that this region is essential for Cdc15 function.
Cdc15-3HA is active as a kinase when GFP-Cdc15 is overexpressed.
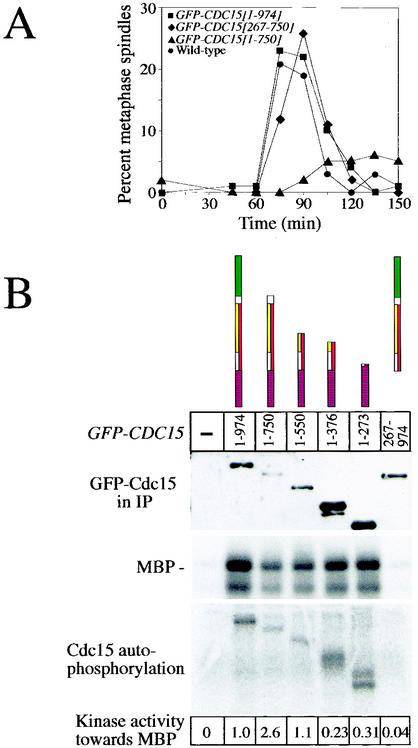
To investigate whether self-association of Cdc15 negatively regulates its kinase activity, as previously described for Pak1 kinase (20), we determined the consequences of overexpressing GFP-CDC15 on the kinase activity associated with immunoprecipitated Cdc15-3HA. We reasoned that, if self-association were inhibitory, overproduced GFP-Cdc15 should cause a higher fraction of Cdc15-3HA to be complexed with other Cdc15 molecules, thus leading to a decrease in Cdc15-3HA-associated kinase activity. When full-length GFP-CDC15 was overexpressed, the kinase activity associated with Cdc15-3HA immunoprecipitates was not decreased but instead slightly increased (2.0-fold; Fig. 4). The increase in Cdc15-3HA-associated kinase activity was consistently seen in multiple experiments and was dependent on the kinase domain of GFP-CDC15 since overexpression of GFP-CDC15 [267-974] did not lead to an increase in kinase activity. Additionally, the elevation in kinase activity was dependent on the self-association domain. GFP-Cdc15 [1-376], a construct which has wild-type kinase activity (Fig. 6B) but lacks the self-association domain (Table 2; Fig. 3A), also did not lead to an increase in kinase activity associated with Cdc15-3HA (Fig. 4). Our results suggest that Cdc15 is not inhibited by self-association.
FIG. 4.
Overexpression of full-length GFP-CDC15 does not affect associated kinase activity of Cdc15-3HA. Cells were grown in YEP plus 2% raffinose overnight to mid-log phase. Cultures were diluted to an optical density at 600 nm of 0.2 and arrested in metaphase with 15 μg of nocodazole/ml for 1.5 h. An additional 15 μg of nocodazole/ml was added to prevent cells from escaping the metaphase arrest, and cells were induced for 2.5 h with 2% galactose (except lane 3, which is uninduced). The amount of kinase activity associated with Cdc15-3HA was assayed by immunoprecipitation followed by a kinase assay with MBP as an artificial substrate. The following strains were used: A2587, A3932, A4683, A4686, and A4678.
FIG. 6.
Expression of Cdc15 lacking the C terminus causes a delay in entry into mitosis and an increase in specific kinase activity. (A) Strains were arrested for 3 h in G1 with the α-factor pheromone (5 μg/ml) and induced with 2% galactose for the last hour. Cells were then synchronously released into YEP plus 2% raffinose and 2% galactose, and the percentage of cells with metaphase spindles was determined at the indicated time. Circles, strain A3932; squares, strain A4441; triangles, strain A5574; and diamonds, strain A4695. (B) Cultures were prepared as described in the legend to Fig. 4, GFP-Cdc15 proteins were immunoprecipitated, and associated kinase activity was measured with MBP as an artificial substrate. (Top) Western blot analysis of the amount of GFP-Cdc15 proteins present in immunoprecipitates used for kinase assays; (middle) amount of 32P-labeled MBP from in vitro kinase assays; (bottom) autophosphorylation occurring on immunoprecipitated GFP-Cdc15. The amount of 32P-labeled MBP was determined relative to the amount of GFP-Cdc15 protein present in the immunoprecipitates. The following strains were used: A3932, A4683, A5575, A4692, A4678, A4690, and A4686.
Overexpression of Cdc15 lacking the kinase domain causes cell cycle arrest in telophase.
During our localization studies we noticed that overexpression of CDC15 lacking the kinase domain (GFP-CDC15 [267-974]) upon release from a pheromone-induced G1 block caused cells to arrest in late anaphase/telophase with DNA masses segregated and long mitotic spindles (Fig. 5A; Table 2). The fact that these cells also expressed full-length Cdc15 from its endogenous promoter suggested that this construct functioned in a dominant-negative manner. Overproduction of truncations GFP-Cdc15 [267-750], GFP-Cdc15 [267-702], and GFP-Cdc15 [267-550] also caused an anaphase/telophase arrest (Fig. 5B; Table 2). In contrast, overproduction of full-length GFP-Cdc15 or the same constructs with the kinase domain present did not cause cell cycle arrest (Fig. 5; Table 2). Our data suggest that truncations GFP-Cdc15 [267-750], GFP-Cdc15 [267-702], and GFP-Cdc15 [267-550], henceforth called class 1 truncations, while retaining the ability to bind to at least some important binding partners must outcompete endogenous full-length Cdc15 and sequester the binding partners into a kinase-inactive complex, thereby preventing exit from mitosis.
FIG. 5.
Several GFP-CDC15 truncation constructs cause a telophase arrest upon overexpression. Strains were arrested for 3 h in G1 with the α-factor pheromone (5 μg/ml) and induced with 2% galactose for the last hour. Cells were synchronously released into YEP plus 2% raffinose and 2% galactose, and samples were taken at indicated time points. (A) The key at right indicates strains A3932, A4441, and A4376 (top to bottom, respectively). Dashed lines represent metaphase spindles; solid lines represent anaphase/telophase spindles. (B) Growth conditions were as described for panel A, and the percentages of anaphase and telophase spindles are analyzed. The leftmost panel depicts class 1 Cdc15 constructs. The key shows strains A3932, A5574, A4695, A5576, A5051, A4691, and A4687 (top to bottom, respectively). Class 2 Cdc15 constructs are shown in the middle panel. The key shows strains A3932, A5580, A4680, A4677, and A6373 (top to bottom, respectively). The rightmost panel depicts class 3 Cdc15 constructs. The key shows strains A3932, A5464, A5909, A5466, A5914, A6456, and A4690 (top to bottom, respectively).
Our analysis of the consequences of overexpressing various GFP-Cdc15 truncations for cell cycle progression revealed two more classes of Cdc15 truncations. Class 2 truncations (GFP-Cdc15 [1-429], GFP-Cdc15 [267-429], GFP-Cdc15 [1-376], GFP-Cdc15 [267-376], GFP-Cdc15-3HA [Δ362-464], and GFP-Cdc15-3HA [Δ465-550]) also caused a late anaphase/telophase arrest when overproduced (Fig. 5B; data not shown). In contrast to class 1 truncations, however, cell cycle arrest (or delay in the case of GFP-CDC15 [267-376]) was also observed when the kinase domain was present in the truncation (Fig. 5B; Table 2; data not shown). Class 2 truncations that contained the N-terminal kinase domain possessed kinase activity as judged by their ability to phosphorylate myelin basic protein (MBP) in vitro (Fig. 6B; data not shown). Thus, loss of kinase activity is not likely to be responsible for the dominant-negative phenotype caused by overexpressing these truncations. Perhaps, class 2 truncations retain the ability to bind one or more binding partners and therefore outcompete the endogenously expressed Cdc15-3HA. However, they seem to lose the ability to interact with other binding partners important for mitotic exit, as the presence of the kinase domain is not sufficient to alleviate the telophase arrest.
Class 3 truncations, in contrast to class 1 and 2 truncations, did not interfere with cell cycle progression. This class included GFP-CDC15 [1-273], GFP-CDC15 [361-974], GFP-CDC15 [361-702], GFP-CDC15 [395-974], GFP-CDC15 [395-702], GFP-CDC15 [551-974], and GFP-CDC15 [751-974] (Fig. 5B; Table 2). We have found two modes of dominant-negative inhibition by overexpression of Cdc15 truncated proteins: one that results simply from a lack of the kinase domain (class 1) and a second which is independent of the presence or absence of the kinase domain (class 2). The class 2 arrest seems to result from the loss of aa 430 to 550, since GFP-CDC15 [1-550] does not cause a telophase arrest but GFP-CDC15 [1-429] does cause a telophase arrest (Fig. 5B; Table 2).
Overexpression of Cdc15 leads to loss of Tem1 from SPBs.
The telophase arrest phenotype of class 1 truncations lacking the kinase domain and class 2 truncations could be explained if they were able to bind Tem1 and sequester the protein into a nonfunctional complex. To test this possibility, we made use of the observation that overexpression of GFP-CDC15 [1-974] leads to loss of Tem1 from SPBs, likely through binding to Tem1 and titrating the protein away from SPBs. We examined the localization of Tem1 in cells overexpressing various class 1, 2, and 3 truncations. In cells expressing class 1 and 2 CDC15 truncations, Tem1 was no longer found on SPBs (Table 3). It is unlikely that class 1 and 2 truncations compete with Tem1 for SPB binding, as some of these constructs (GFP-CDC15 [267-429] and GFP-CDC15 [267-376]) did not themselves localize to SPBs (Table 2). Class 3 mutants did not interfere with Tem1 localization and did not cause cells to arrest in telophase (Tables 1 and 2). We interpret these findings to mean that one reason for the telophase arrest caused by overexpression of class 1 truncations lacking the kinase domain and class 2 truncations is that they sequester Tem1 into a nonfunctional complex. However, it is important that at least some of the truncations are likely to also cause a telophase arrest by interfering with other aspects of Cdc15 function (see Discussion).
TABLE 3.
Tem1 localization in cells overexpressing various CDC15 truncations
An inhibitory domain within the C terminus of Cdc15.
Two truncations caused defects in progression through early stages of the cell cycle. Expression of Cdc15 lacking the C-terminal 224 aa (GFP-CDC15 [1-750]) led to defects in mitotic entry as judged by the reduction in formation of metaphase and anaphase spindles in these cells (Fig. 5B and 6A). Cells expressing this truncation resembled that of cells lacking CFI1/NET1, which have an elongated bud morphology (40) (data not shown), suggesting that Cdc14 may be hyperactive in these cells. Truncation GFP-CDC15 [1-702] also impaired progression through mitosis, albeit to a lesser extent (Fig. 5B). This finding raised the possibility that the MEN was hyperactive in these cells. Since the MEN antagonizes CDK activity, cells in which the MEN is overactive have delayed entry into and progression through mitosis (reviewed in reference 2). To test this idea, we first determined whether GFP-Cdc15 [1-750] was more active as a kinase than was full-length GFP-Cdc15. Cdc15 kinase activity does not fluctuate during the cell cycle as judged by in vitro kinase assays (16). However, since expression of some constructs causes arrest in telophase, cells were arrested in metaphase by using nocodazole to ensure that cells were in the same cell cycle stage when kinase activity was assayed. The specific activity of GFP-Cdc15 [1-750] was 2.6-fold higher than that of full-length GFP-Cdc15 (Fig. 6B), suggesting that the protein was indeed hyperactive as a protein kinase. We also noted that GFP-Cdc15 [1-750] caused hyperphosphorylation of endogenous full-length Cdc15 (Fig. 3A). Whether this phosphorylation affects Cdc15 kinase activity or other aspects of Cdc15's function is, at present, unclear.
To determine whether GFP-CDC15 [1-750] caused hyperactivation of the MEN, we compared Dbf2-associated kinase activity in GFP-CDC15 [1-750]-expressing cells with that of cells expressing full-length Cdc15 (GFP-CDC15 [1-974]). Cultures of wild-type, GFP-CDC15 [1-974], GFP-CDC15 [1-750], and GFP-CDC15 [267-750] (lacking the kinase domain) cells were released from a pheromone-induced G1 arrest in the presence of galactose to induce production of the various Cdc15 constructs. In wild-type cells, Dbf2-associated kinase activity was, as previously reported (39), low during G1 and S phase and early mitosis but high during anaphase and exit from mitosis (105 to 120 min after release; Fig. 7A). Full-length CDC15 did not affect Dbf2-associated kinase activity in vitro (Fig. 7A), indicating that overexpression of CDC15 does not further activate Dbf2 kinase. In contrast, expression of GFP-CDC15 [1-750] caused a dramatic increase in Dbf2-associated kinase activity. Dbf2 was activated as early as 45 min after release from the pheromone block and stayed high for the duration of the experiment (Fig. 7A). The increase in Dbf2 kinase activity was not due to higher expression of GFP-CDC15 [1-750], as the amount of GFP-Cdc15 [1-750] protein produced was actually lower than that of full-length Cdc15 (Fig. 1B and 6B). Furthermore, the increase in Dbf2 kinase activity depended on the presence of the kinase domain of GFP-CDC15 [1-750]. GFP-CDC15 [267-750] lacking the kinase domain in fact prevented activation of Dbf2, which is consistent with the observation that this construct causes a dominant-negative telophase arrest (Fig. 5B). Expression of GFP-CDC15 [1-750] also caused a decrease in Clb2-associated kinase activity (data not shown), a finding that further indicates that GFP-CDC15 [1-750] causes hyperactivation of the MEN.
FIG. 7.
Removal of a C-terminal domain of Cdc15 causes hyperactivation of MEN. (A) Strains were arrested for 3 h in G1 with the α-factor pheromone (5 μg/ml) and induced with 2% galactose for the last hour. Cells were synchronously released into YEP plus 2% raffinose and 2% galactose, and Dbf2 kinase activity and the amount of Dbf2 and Cdc28 (loading control) protein were determined at the indicated times. The following strains were used: A6131, A6130, A5568, and A5571. (B and C) Strains were grown to mid-log phase in YEP plus 2% raffinose and arrested in metaphase with 15 μg of nocodazole/ml for 1.5 h. An additional 7.5 μg of nocodazole/ml was added to maintain the metaphase arrest, and expression of GFP-CDC15 constructs was induced with 2% galactose. Samples were taken at indicated time points to determine the percentage of cells with Cdc14 released from the nucleolus. The image shown in panel C shows Cdc14 localization 90 min after galactose addition. Cdc14 staining is shown in red, and DNA is shown in blue. The following strains were used: A1411, A5965, A5966, and A5968.
Cdc14 is released prematurely from the nucleolus in cells expressing GFP-Cdc15 [1-750].
Activation of the MEN ultimately triggers Cdc14 release from its inhibitor Cfi1/Net1 in the nucleolus. We therefore tested whether overexpression of GFP-CDC15 [1-750] promoted release of Cdc14 from the nucleolus at times when Cdc14 is normally sequestered. Wild-type cells and cells overexpressing full-length GFP-CDC15, GFP-CDC15 [1-750], and GFP-CDC15 [267-750] lacking the kinase domain were arrested in metaphase with the microtubule-depolymerizing drug nocodazole (Fig. 7B and C). Overexpression of GFP-CDC15 [1-750] in the arrest caused release of Cdc14 from the nucleolus, in a manner dependent on its kinase activity, while overexpression of GFP-Cdc15 [1-974] did not (Fig. 7B and C). Our results show that removal of the C-terminal 224 aa leads to CDC15 being hyperactive when overexpressed, suggesting that the C terminus of Cdc15 functions in the inhibition of MEN signaling.
We also examined whether GFP-CDC15 [1-750] expressed from the endogenous promoter caused hyperactivation of the MEN. When expressed at endogenous levels GFP-CDC15 [1-750] failed to complement the temperature-sensitive lethality of a cdc15-2 mutation (data not shown). The truncation GFP-Cdc15 [1-750] appeared to be less stable than full-length GFP-Cdc15 as judged by the ability of the protein to accumulate in cells when produced from the GAL1 promoter (Fig. 1B and 6B). This suggests that a less stable truncated protein, and thus the presence of lower levels of this truncation in the cell, could account for the failure of the Cdc15 [1-750] expressed at endogenous levels to sustain cell division. Consistent with this idea is the finding that GFP-CDC15 [1-750] expressed from the GAL1 promoter not only suppressed the temperature-sensitive lethality of a cdc15-2 mutant but also supported cell division as the sole source of CDC15 in the cell (Table 2).
DISCUSSION
Cdc15 is similar to members of the Ste20/PAK family of protein kinases, which are regulated by modular domains present in their noncatalytic region (reviewed in reference 5). In this study, we defined the various functional domains within Cdc15 and determined their role in exit from mitosis. We show that a region of Cdc15 adjacent to the kinase domain promotes SPB binding and self-association. Our deletion analysis also identified a region within the C terminus of Cdc15 which when deleted causes an increase in specific activity of Cdc15, hyperactivation of the MEN protein kinase Dbf2, and premature release of Cdc14 from the nucleolus, at least when the protein is overproduced. Our findings suggest the presence of a MEN-inhibitory function in the C terminus of Cdc15.
SPB localization of Cdc15 is mediated through a large region of Cdc15 adjacent to the kinase domain.
When expressed at endogenous levels, Cdc15 is difficult to detect within the cell by indirect in situ immunofluorescence (4, 25, 40, 43). To facilitate Cdc15's detection and the construction of N-terminal truncations, we expressed GFP-CDC15 fusions from the GAL1 promoter. We carefully compared the localization of endogenous Cdc15 with that of overexpressed Cdc15 and found that the cell cycle-regulated association of Cdc15 with SPBs and the bud neck was not affected when CDC15 was overexpressed, indicating that at least this aspect of Cdc15 regulation is preserved under conditions of overexpression.
GFP-Cdc15 [267-550] was the smallest region of Cdc15 found to associate with SPBs. The protein, however, was also found in a punctate pattern along the mitotic spindle. Thus, while GFP-Cdc15 [267-550] retains some ability to associate with SPBs, only region 267 to 702 is sufficient for wild-type levels of SPB association. A previous study showed that removal of the C-terminal 135 aa (GFP-Cdc15 [1-839]) caused loss of SPB localization of overexpressed Cdc15-GFP (25). Our finding that GFP-Cdc15 [267-702] localizes to the SPB demonstrates that a domain sufficient for localization is present in this region. Perhaps aa 267 to 702 are masked in the GFP-Cdc15 [1-839] truncation employed by Menssen et al. (25).
Previous work has shown that Cdc15-3HA expressed from the endogenous promoter does not localize to SPBs in tem1-3 cells arrested in telophase at 37°C (40). While these data show that TEM1 is required for the association of Cdc15 with SPBs, several lines of evidence indicate that Tem1 is not the only anchor for Cdc15 at SPBs. First, Tem1 primarily localizes to the SPB entering the daughter cell while Cdc15 is localized symmetrically to both SPBs (3, 29, 40, 43). Second, aa 276 to 362 within Cdc15, which are sufficient for Tem1 interaction (1), are not sufficient for SPB association (Table 2). We speculate that TEM1 is required to induce binding of Cdc15 to the SPB and that this induction may require an initial binding of Tem1 to Cdc15.
Cdc15 self-associates.
Studies in recent years have shown that homodimerization plays an important role in the regulation of many protein kinases (5, 28, 31). Cdc15, like PAK kinases, also self-associates. However, in contrast to other PAK kinases, Cdc15 multimers possess kinase activity, indicating that multimerization is not inhibitory. In fact we observed an increase in Cdc15-3HA kinase activity when a GFP-Cdc15 fusion was overproduced. This may suggest that multimerization stimulates Cdc15 kinase activity. It is, however, also possible that more GFP-Cdc15 molecules associate with Cdc15-3HA when GFP-Cdc15 is overproduced.
It is not yet known whether self-association of Cdc15, which is mediated through aa 360 to 702, is essential for CDC15 function or whether deletion of this region causes loss of function by interfering with other aspects of Cdc15. As well as resulting in loss of self-association, GFP-Cdc15 [Δ362-464] and GFP-Cdc15 [Δ465-550] also led to loss of SPB association (A. J. Bardin, unpublished observations), raising the possibility that it is the inability of these constructs to bind to SPBs that causes this loss in CDC15 function. It is also possible that self-association is intricately related to SPB localization. Point mutants that disrupt self-association specifically will be necessary to determine this.
Another question that remains to be addressed is whether Cdc15 self-association is regulated. We observed that Cdc15 self-associates throughout the cell cycle (Bardin, unpublished). Furthermore, Cdc15 phosphorylation, which has previously been shown to be inhibitory and which is removed by the protein phosphatase Cdc14 during anaphase (16), also does not appear to affect self-association. Mutating the phosphorylation sites (Cdc15-7A [16]) in Cdc15 to amino acids that can no longer be phosphorylated does not affect self-association of Cdc15 (Bardin, unpublished).
Overexpression of Cdc15 lacking the kinase domain causes cell cycle arrest in telophase.
The overexpression of a large number of CDC15 truncations caused arrest in telophase despite endogenous full-length CDC15 being expressed. Further examination revealed two classes of truncations that caused such a phenotype. We believe that the telophase arrest caused by high levels of class 1 truncations is due to the ability of these truncations to sequester proteins important for mitotic exit into a complex which lacks kinase activity and is thus incapable of promoting mitotic exit. Consistent with this idea is the fact that all class 1 truncations no longer cause a telophase arrest when the kinase domain is present. In fact, these constructs with the kinase domain, when overexpressed, support cell viability even in the absence of full-length CDC15. Tem1, whose interaction with Cdc15 is mediated through aa 276 to 362 (1), could be one of the proteins sequestered by class 1 truncations. Indeed, the class 1 group of truncations titrates Tem1 away from SPBs. However, some truncations quite possibly also affect the function of other proteins important for MEN signaling, such as Dbf2 or Mob1.
In contrast to class 1 truncations, class 2 truncations cause telophase arrest irrespective of whether the kinase domain was present. GFP-CDC15 [1-550] does not disrupt progression through mitosis, whereas GFP-CDC15 [1-429] causes cell cycle arrest in telophase. It seems, therefore, that aa 430 to 550 cause the difference in phenotype. It is possible that aa 430 to 550 are important for Cdc15 kinase activity in vivo although they are not required for Cdc15 kinase activity in vitro. We favor the idea that class 2 truncations have kinase activity but are incapable of interacting with a subset of proteins whose association with Cdc15 is important for exit from mitosis and depends on aa 430 to 550. It seems that these truncations can, however, bind and titrate other important MEN components from the endogenous Cdc15, resulting in a dominant-negative telophase arrest. Class 2 truncations interact with Tem1 (Fig. 3C; Bardin, unpublished) and titrate it away from SPBs (Table 3), but the truncations are no longer able to bind to SPBs or self-associate, which could prevent MEN activation. Alternatively, Dbf2-Mob1 binding to Cdc15 could be affected. We have not been able to test whether Dbf2 or Mob1 binds to aa 430 to 550, as we were unable to reliably immunoprecipitate Dbf2 with Cdc15.
An inhibitory domain in the C terminus of Cdc15.
Removal of 224 aa from the C terminus of GFP-Cdc15 expressed from the GAL1 promoter results in hyperactivation of MEN as judged by the following four criteria. (i) GFP-Cdc15 [1-750] was found to have a higher specific activity towards MBP than full-length Cdc15; (ii) Dbf2 kinase activity was dramatically increased in cells overexpressing GFP-CDC15 [1-750]; (iii) GFP-CDC15 [1-750] causes release of Cdc14 from the nucleolus in nocodazole-arrested cells; and (iv) overexpression of GFP-CDC15 [1-750] led to a delay in entry into mitosis, which is a characteristic of cells with overactive Cdc14 (42). CDC15 [1-750] expressed from its endogenous promoter was not able to support growth. This is likely to be due to the protein being produced at lower levels or being more unstable than full-length Cdc15, as protein levels of GFP-Cdc15 [1-750] are lower than those of GFP-Cdc15 [1-974] (Fig. 1B).
Autoinhibitory regions of both Raf and PAK kinases function to prevent kinase activation until binding of GTP-bound GTPase occurs (18, 27, 28). How might the inhibitory region within Cdc15 function? Our data are consistent with the following two models: region 751 to 974 could make contacts either with a region of the same protein or a second molecule of Cdc15 and cause inhibition of the kinase activity, similar to that seen in Raf and PAK. Alternatively, an inhibitor of MEN signaling could bind to this region of Cdc15 and negatively regulate Cdc15 kinase activity or interaction of Cdc15 with other proteins. Determining how this region is involved in regulating MEN activity and whether it is itself a target of regulation will be of great interest. Finally we note that our data and those of others lend support to the idea that MEN signaling is regulated in part through Cdc15 (16, 25, 35, 43).
Acknowledgments
We are grateful to David Morgan for reagents. We thank Frank Solomon and members of the Amon lab for their critical reading of the manuscript. A.J.B. thanks members of the Amon lab and Pierre-Antoine Defossez for helpful discussions.
This research was supported by National Institutes of Health grant GM 56800 to A.A. A.A. is an Assistant Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Asakawa, K., S. Yoshida, F. Otake, and A. Toh-e. 2001. A novel functional domain of Cdc15 kinase is required for its interaction with Tem1 GTPase in Saccharomyces cerevisiae. Genetics 157:1437-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bardin, A. J., and A. Amon. 2001. Men and sin: what's the difference? Nat. Rev. Mol. Cell Biol. 2:815-826. [DOI] [PubMed] [Google Scholar]
- 3.Bardin, A. J., R. Visintin, and A. Amon. 2000. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell 102:21-31. [DOI] [PubMed] [Google Scholar]
- 4.Cenamor, R., J. Jimenez, V. J. Cid, C. Nombela, and M. Sanchez. 1999. The budding yeast Cdc15 localizes to the spindle pole body in a cell-cycle-dependent manner. Mol. Cell Biol. Res. Commun. 2:178-184. [DOI] [PubMed] [Google Scholar]
- 5.Dan, I., N. M. Watanabe, and A. Kusumi. 2001. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 11:220-230. [DOI] [PubMed] [Google Scholar]
- 6.Daum, J. R., N. Gomez-Ospina, M. Winey, and D. J. Burke. 2000. The spindle checkpoint of Saccharomyces cerevisiae responds to separable microtubule-dependent events. Curr. Biol. 10:1375-1378. [DOI] [PubMed] [Google Scholar]
- 7.Farrar, M. A., I. Alberol, and R. M. Perlmutter. 1996. Activation of the Raf-1 kinase cascade by coumermycin-induced dimerization. Nature 383:178-181. [DOI] [PubMed] [Google Scholar]
- 8.Fesquet, D., P. J. Fitzpatrick, A. L. Johnson, K. M. Kramer, J. H. Toyn, and L. H. Johnston. 1999. A Bub2p-dependent spindle checkpoint pathway regulates the Dbf2p kinase in budding yeast. EMBO J. 18:2424-2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fraschini, R., E. Formenti, G. Lucchini, and S. Piatti. 1999. Budding yeast Bub2 is localized at spindle pole bodies and activates the mitotic checkpoint via a different pathway from Mad2. J. Cell Biol. 145:979-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frenz, L. M., S. E. Lee, D. Fesquet, and L. H. Johnston. 2000. The budding yeast Dbf2 protein kinase localises to the centrosome and moves to the bud neck in late mitosis. J. Cell Sci. 113:3399-3408. [DOI] [PubMed] [Google Scholar]
- 11.Gruneberg, U., K. Campbell, C. Simpson, J. Grindlay, and E. Schiebel. 2000. Nud1p links astral microtubule organization and the control of exit from mitosis. EMBO J. 19:6475-6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoyt, M. A. 2000. Exit from mitosis: spindle pole power. Cell 102:267-270. [DOI] [PubMed] [Google Scholar]
- 13.Hu, F., Y. Wang, D. Liu, Y. Li, J. Qin, and S. J. Elledge. 2001. Regulation of the Bub2/Bfa1 GAP complex by Cdc5 and cell cycle checkpoints. Cell 107:655-665. [DOI] [PubMed] [Google Scholar]
- 14.Jaspersen, S. L., J. F. Charles, and D. O. Morgan. 1999. Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr. Biol. 9:227-236. [DOI] [PubMed] [Google Scholar]
- 15.Jaspersen, S. L., J. F. Charles, R. L. Tinker-Kulberg, and D. O. Morgan. 1998. A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. Mol. Biol. Cell 9:2803-2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaspersen, S. L., and D. O. Morgan. 2000. Cdc14 activates cdc15 to promote mitotic exit in budding yeast. Curr. Biol. 10:615-618. [DOI] [PubMed] [Google Scholar]
- 17.Jensen, S., M. Geymonat, and L. H. Johnston. 2002. Mitotic exit: delaying the end without FEAR. Curr. Biol. 12:R221-R223. [DOI] [PubMed] [Google Scholar]
- 18.Kerkhoff, E., and U. R. Rapp. 2001. The Ras-Raf relationship: an unfinished puzzle. Adv. Enzyme Regul. 41:261-267. [DOI] [PubMed] [Google Scholar]
- 19.Lee, S. E., L. M. Frenz, N. J. Wells, A. L. Johnson, and L. H. Johnston. 2001. Order of function of the budding-yeast mitotic exit-network proteins Tem1, Cdc15, Mob1, Dbf2, and Cdc5. Curr. Biol. 11:784-788. [DOI] [PubMed] [Google Scholar]
- 20.Lei, M., W. Lu, W. Meng, M. C. Parrini, M. J. Eck, B. J. Mayer, and S. C. Harrison. 2000. Structure of PAK1 in an autoinhibited conformation reveals a multistage activation switch. Cell 102:387-397. [DOI] [PubMed] [Google Scholar]
- 21.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 22.Luca, F. C., M. Mody, C. Kurischko, D. M. Roof, T. H. Giddings, and M. Winey. 2001. Saccharomyces cerevisiae Mob1p is required for cytokinesis and mitotic exit. Mol. Cell. Biol. 21:6972-6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mah, A. S., J. Jang, and R. J. Deshaies. 2001. Protein kinase Cdc15 activates the Dbf2-Mob1 kinase complex. Proc. Natl. Acad. Sci. USA 98:7325-7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCollum, D., and K. L. Gould. 2001. Timing is everything: regulation of mitotic exit and cytokinesis by the MEN and SIN. Trends Cell Biol. 11:89-95. [DOI] [PubMed] [Google Scholar]
- 25.Menssen, R., A. Neutzner, and W. Seufert. 2001. Asymmetric spindle pole localization of yeast Cdc15 kinase links mitotic exit and cytokinesis. Curr. Biol. 11:345-350. [DOI] [PubMed] [Google Scholar]
- 26.Morgan, D. O. 1999. Regulation of the APC and the exit from mitosis. Nat. Cell Biol. 1:E47-E53. [DOI] [PubMed] [Google Scholar]
- 27.Morrison, D. K., and R. E. Cutler. 1997. The complexity of Raf-1 regulation. Curr. Opin. Cell Biol. 9:174-179. [DOI] [PubMed] [Google Scholar]
- 28.Parrini, M. C., M. Lei, S. C. Harrison, and B. J. Mayer. 2002. Pak1 kinase homodimers are autoinhibited in trans and dissociated upon activation by Cdc42 and Rac1. Mol. Cell 9:73-83. [DOI] [PubMed] [Google Scholar]
- 29.Pereira, G., T. Hofken, J. Grindlay, C. Manson, and E. Schiebel. 2000. The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol. Cell 6:1-10. [PubMed] [Google Scholar]
- 30.Pereira, G., C. Manson, J. Grindlay, and E. Schiebel. 2002. Regulation of the Bfa1p-Bub2p complex at spindle pole bodies by the cell cycle phosphatase Cdc14p. J. Cell Biol. 157:367-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlessinger, J. 2000. Cell signaling by receptor tyrosine kinases. Cell 103:211-225. [DOI] [PubMed] [Google Scholar]
- 32.Shirayama, M., Y. Matsui, and A. Toh-e. 1994. The yeast TEM1 gene, which encodes a GTP-binding protein, is involved in termination of M phase. Mol. Cell. Biol. 14:7476-7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shirayama, M., W. Zachariae, R. Ciosk, and K. Nasmyth. 1998. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 17:1336-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shou, W., J. H. Seol, A. Shevchenko, C. Baskerville, D. Moazed, Z. W. Chen, J. Jang, H. Charbonneau, and R. J. Deshaies. 1999. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97:233-244. [DOI] [PubMed] [Google Scholar]
- 35.Stegmeier, F., R. Visintin, and A. Amon. 2002. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell 108:207-220. [DOI] [PubMed] [Google Scholar]
- 36.Straight, A. F., W. Shou, G. J. Dowd, C. W. Turck, R. J. Deshaies, A. D. Johnson, and D. Moazed. 1999. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell 97:245-256. [DOI] [PubMed] [Google Scholar]
- 37.Sullivan, M., and F. Uhlmann. 2003. A non-proteolytic function of separase links the onset of anaphase to mitotic exit. Nat. Cell Biol. 5:249-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Surana, U., A. Amon, C. Dowzer, J. McGrew, B. Byers, and K. Nasmyth. 1993. Destruction of the CDC28/CLB mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. EMBO J. 12:1969-1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toyn, J. H., and L. H. Johnston. 1994. The Dbf2 and Dbf20 protein kinases of budding yeast are activated after the metaphase to anaphase cell cycle transition. EMBO J. 13:1103-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Visintin, R., and A. Amon. 2001. Regulation of the mitotic exit protein kinases Cdc15 and Dbf2. Mol. Biol. Cell 12:2961-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Visintin, R., K. Craig, E. S. Hwang, S. Prinz, M. Tyers, and A. Amon. 1998. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell 2:709-718. [DOI] [PubMed] [Google Scholar]
- 42.Visintin, R., E. S. Hwang, and A. Amon. 1999. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature 398:818-823. [DOI] [PubMed] [Google Scholar]
- 43.Xu, S., H. K. Huang, P. Kaiser, M. Latterich, and T. Hunter. 2000. Phosphorylation and spindle pole body localization of the Cdc15p mitotic regulatory protein kinase in budding yeast. Curr. Biol. 10:329-332. [DOI] [PubMed] [Google Scholar]
- 44.Yoshida, S., K. Asakawa, and A. Toh-e. 2002. Mitotic exit network controls the localization of Cdc14 to the spindle pole body in Saccharomyces cerevisiae. Curr. Biol. 12:944-950. [DOI] [PubMed] [Google Scholar]
- 45.Yoshida, S., and A. Toh-e. 2001. Regulation of the localization of Dbf2 and Mob1 during cell division of Saccharomyces cerevisiae. Genes Genet. Syst. 76:141-147. [DOI] [PubMed] [Google Scholar]